Abstract
ATP-competitive mTOR kinase inhibitors (mTorKIs) are a new generation of mTOR-targeted agents with more potent anticancer activity than rapamycin in several tumor models. However, the sensitivity and resistance of cancer cells to mTorKIs remain poorly understood. In this study, we tested mTorKIs against a large panel of colorectal cancer (CRC) cell lines, and found that mTorKIs displayed broader anti-CRC activity than rapamycin, including CRC cells with K-Ras or B-Raf mutations, suggesting that these mTorKIs are particularly useful for CRCs resistant to EGFR inhibitors. Unexpectedly, we found that 40% CRC cell lines were intrinsically drug resistant. Moreover, we discovered an mTOR-independent 4E-BP1 phosphorylation that was correlated with mTorKI resistance. Altogether, our findings provide compelling preclinical support for testing mTorKIs in human CRC clinical trials. They further reveal the existence of significant intrinsic mTorKI drug resistance in cancer cells and suggest that 4E-BP1 phosphorylation is a predictive biomarker for mTorKI sensitivity and resistance.
Key words: mTOR, kinase, colorectal cancer, drug resistance, 4E-BP1, phosphorylation
Introduction
Colorectal cancer (CRC) is one of the most common human malignancies and is second in cancer-related death, responsible for 1.2 million new cases and over 600,000 deaths per year worldwide.1 It is even more prevalent in developed countries, accounting for 60% occurrence. Genetic heterogeneity of CRCs renders it a major therapeutic challenge. An exciting recent development is the finding that a subpopulation of CRC patients with amplification of epidermal growth factor receptor (EGFR) is responsive to EGFR-targeted therapy. Even these patients frequently encounter resistance to EGFR inhibitors due to genetic aberration in K-Ras.2 New therapies are much needed to improve the mortality of CRC patients.
mTOR is a central controller of cell growth and survival in response to growth factors, cytokines, hormones and nutrients.3,4 It is a PI3K-related kinase that forms two distinct protein complexes called mTOR complex 1 or mTORC1,5,6 and mTOR complex 2 or mTORC2.7 mTORC1 acts downstream of PI3K-Pten-Akt. In response to upstream stimuli, mTORC1 phosphorylates S6K1 and 4E-BP1 to stimulate protein synthesis,8 while mTORC2 phosphorylates AKT to promote cell survival.9 Genetic aberrations of the PI3K-mTOR pathway are among the most common events in human malignancies, resulting in hyperactivation of mTOR signaling and causing these cancer cells highly addictive to mTOR pathway.10 We reported that mTOR signaling is frequently hyper-activated in primary human CRC tumors, and RNAi-mediated knockdown of mTOR attenuated CRC tumor growth in vitro and in vivo.11 However, rapamycin was not effective against these CRC tumor models.12 These observations are consistent with our previous finding that rapamycin is only a partial inhibitor of TOR.13 Moreover, inhibition of mTORC1 triggers activation of feedback loops involving compensatory pathways such as AKT, which may enhance cancer cell survival in the presence of mTORC1 blockage.14–16 These results explain the low efficacy of rapamycin analogs (rapalogs) in clinical trials for several solid tumor types including CRC.17–19
We discovered that TOR kinase domain is required for both rapamycin-sensitive and rapamycin-insensitive functions, suggesting that the kinase domain is a more potent site for mTOR inhibition.13 Recently, several ATP-competitive mTOR kinase inhibitors (mTorKIs) were developed to block the activity of both mTOR complexes.19,20 In addition, some of these compounds originally developed as PI3K inhibitors but were later found to also inhibit mTOR kinase activity and are thus called mTOR-PI3K dual inhibitors. The latter is thought to have added advantage of negating the IRS1-PI3K-Akt negative feedback loop.19 Thus far, mTorKIs have been tested against a number of cancer models, including breast cancer, glioma, non-small cell lung carcinoma (NSCLC) and AML.19,21,22 However, they have not been explored in CRC models. Furthermore, initial research focused on validating them as useful anticancer agents. Sensitivity and resistance of cancer cells to this new class of targeted therapeutic agents is not understood. In the present study, we tested three representative mTorKIs against a large panel of 12 CRC cell lines with diverse origins, histological features and genetic backgrounds. Collectively, our results show that mTorKIs broad activity against CRC but also revealed significant intrinsic drug resistance. Importantly, we discovered an mTOR-independent 4E-BP1 phosphorylation that is strongly correlated with CRC resistance to mTorKIs.
Results
mTorKIs display broader anti-CRC activity than rapamycin.
To investigate anti-CRC effects of mTorKIs, we have assembled a large panel of 12 CRC cell lines that are representative of the heterogeneity of primary CRC tumors. They were derived from colorectal cancer with different histological features and origins (Table 1). In addition, they vary in the status of K-Ras, B-RAF, PIK3CA, PTEN, p53, APC and Smad4 that are oncogenes or tumor suppressors most commonly found with genetic aberrations in CRCs (Table 1). We compared BEZ235, PP242 and WYE354 with rapamycin for their ability to inhibit CRC cell growth. BEZ235 is a PI3K-mTOR dual inhibitor while PP242 and WYE354 are selective mTOR inhibitors. In agreement with a previous observation that CRC cells are poorly sensitive to rapamycin,12 10 CRC cell lines were completely resistant to rapamycin treatment, while only two (CACO2 and DLD1) were rapamycin-sensitive (Table 2). In contrast, the growth of 5 CRC cell lines was sensitive and 2 CRC cell lines partially sensitive to mTorKIs (Table 2), which represent 58% response rate, indicating that mTorKIs indeed have superior anti-CRC activity to rapamycin. Interestingly, most mTorKI-sensitive CRC cell lines contain K-Ras or B-Raf mutations that are known to confer resistance to EGFR inhibitors, suggesting that mTorKIs are useful in treatment of EGFR inhibitor-resistant patients. On the other hand, 5 CRC cell lines (SW620, COLO205, HCT116, HT29 and DLD1) or 42% CRC cell lines were mTorKI-resistant. This observation reveals that intrinsic drug resistance is potentially a major problem. PI3KCA and PTEN mutations have previously been implicated in drug sensitivity for rapamycin. However, there is no apparent correlation between these genetic aberrations and mTorKI-sensitivity (Tables 1 and 2).
Table 1.
Characteristics of various CRC cell lines
| Cell line | Pathological feature | Source | K-RAS | BRAF | PIK3CA | PTEN | p53 |
| CACO-247 | Poorly differentiated | Original tumor | wt | wt | N/A | wt | null |
| COLO-205* | Dukes' type D† | Ascites fluid | wt | mut | wt | wt | mut |
| DLD-1* | Dukes' type C† | Original tumor | mut | wt | mut | wt | mut |
| HCT116* | Poorly differentiated | Original tumor | mut | wt | mut | wt | wt |
| HT29* | Moderately well-differentiated | Original tumor | wt | mut | mut | wt | mut |
| KM-1248 | Dukes' type B, poorly differentiated | Original tumor | mut | wt | wt | mut | mut |
| LOVO49 | Dukes' type C, grade IV† | Metastatic site (left supraclavicular region) | mut | wt | wt | N/A | wt |
| RKO* | Poorly differentiated | Original tumor | wt | mut | mut | wt | wt |
| SW1116* | Dukes' type A, grade III† | Original tumor | mut | wt | wt | wt | mut |
| SW48* | Dukes' type C, grade IV† | Original tumor | wt | wt | wt | wt | wt |
| SW620* | Dukes' type C† | Lymph node metastasis | mut | wt | wt | wt | mut |
| SW480†50 | Dukes' type B | Original tumor | mut | wt | wt | wt | mut |
N/A, not available. Wt, wild type. Mut, mutant; *Information from www.sanger.ac.uk/genetics/CGP/cosmic.
Information from www.atcc.org.
Table 2.
Sensitivity and resistance of human CRC cell lines to mTOR inhibitors
| Rapamycin | BEZ235 | PP242 | WYE354 | |
| CACO2 | − | − | − | − |
| COLO205 | + | + | + | + |
| DLD1 | − | − | + | + |
| HCT116 | + | + | + | + |
| HT29 | + | +/− | + | + |
| KM12 | + | − | − | − |
| LOVO | + | − | − | − |
| RKO | + | − | +/− | − |
| SW1116 | + | + | +/− | +/− |
| SW48 | + | − | − | − |
| SW620 | + | + | + | + |
| SW480 | + | − | − | − |
Summary of the inhibitory effect of mTOR inhibitors on 12 human colorectal cancer cell lines as judged by growth inhibition using the optimized sulforhodamine B (SRB) assay. +, drug-resistant; −, drug-sensitive; +/−, partially drug-sensitive.
Differential response of 4E-BP1 phosphorylation to mTorKIs in drug-sensitive and -resistant CRC cells.
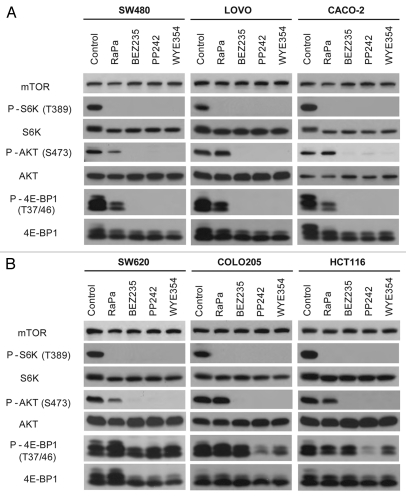
To gain an insight into the sensitivity and resistance of CRC cells to mTorKIs, we selected three most sensitive CRC cell lines (SW480, LOVO and CACO2) and three most resistant CRC cell lines (SW620, COLO205 and HCT116) to investigate how mTOR pathway responds to drug treatment. We found that BEZ235, PP242 and WYE354 blunted the phosphorylation of S6K1(T389) and AKT(S473), substrates of mTORC1 and mTORC2, respectively, in all six CRC cell lines (Fig. 1). In contrast, rapamycin only inhibited phosphorylation of S6K1(T389), but not AKT(S473). mTorKIs also completely abolished phosphorylation of 4E-BP1, another mTORC1 substrate in SW480, LOVO and CACO2 cells. In striking contrast, significant level of 4E-BP1 phosphorylation remains even after prolonged drug treatment in SW620, COLO205 and HCT116 cells. This observation demonstrates a strong correlation between 4E-BP1 phosphorylation and mTorKI resistance in CRC cells.
Figure 1.
Effect of BEZ235, PP242 and WYE354 on mTOR signaling in drug-sensitive and -resistant CRC cell lines. (A) Three most mTorKI-sensitive CRC cell lines (SW480, LOVO205 and CACO-2) were treated with rapamycin (Rapa), BEZ235, PP242 and WYE354 for 12 h. The effect of mTOR inhibitors on mTOR signaling was analyzed by substrate phosphorylation of mTORC1 and mTORC2 by western blot. (B) Three most mTorKI-resistant CRC cell lines (SW620, COLO205 and HCT116) were treated with rapamycin, BEZ235, PP242 and WYE354 for 12 h. The effect of mTOR inhibitors on mTOR signaling was analyzed by substrate phosphorylation of mTORC1 and mTORC2 by western blot.
Evaluation of mTorKIs using in vivo CRC models.
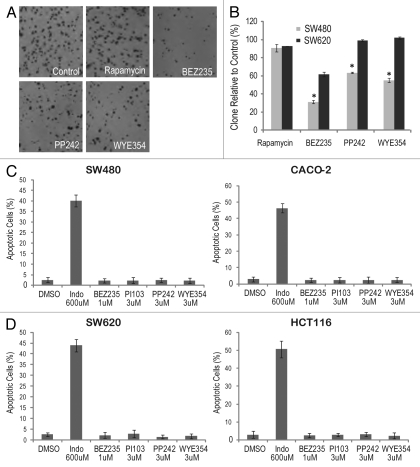
SW480 and SW620 are a pair of matched primary and metastatic CRC cell lines from the same patient, with SW480 derived from the initial tumor biopsy and SW620 from a subsequent metastatic lymph node cancer cells 6 mo after the disease recurrence.23 Furthermore, both cell lines were isolated prior to any chemotherapy.24 Because of the similar genetic background (both cell lines harbor mutations in K-RAS, p53, APC and SMAD4, but are wild type B-RAF, PIK3CA and PTEN), they are commonly used as isogenic pairs in CRC research.25,26 To further evaluate the anti-CRC effect of mTorKIs, we tested them in more physiologically relevant tumor models. They were first assayed in colony formation assay of SW480 and SW620 cells. BEZ235, PP242 and WYE354 significantly decreased the colony formation of SW480 cells (Fig. 2A and B). In contrast, PP242, WYE354 and rapamycin failed to attenuate colony formation in SW620 cells, and only BEZ235 showed moderate effect (Fig. 2A and B). It has been reported that mTorKIs induce apoptosis in certain tumor cell type such as leukemia and breast cancer.27,28 However, no significant cell death were observed in CRC cells treated with high drug doses (Fig. 2C and D), suggesting that mTorKIs are mainly cytostatic against CRCs.
Figure 2.
Differential anticancer effect of mTOR kinase inhibitors (mTorKIs) toward SW480 and SW620 cells. (A) SW480 and SW620 cells were cultured in soft agar in the absence or presence of different mTOR inhibitors. The anchorage-independent cell growth was analyzed by the ability of these cells to form colonies. A representative result is shown for SW480 cells. (B) Quantification of the soft-agar assay results are expressed as the ratio of colonies in treated vs. control cells. Data represent mean ± SD from three independent triplicate experiments. *p < 0.01, vs. control. (C) Two mTorKI-sensitive CRC cell lines (SW480 and CACO-2) were treated with high dose of mTorKIs for 72 h, and apoptotic cells were quantified. Indomethecin (Indo) was used as a positive control. Data represent means ± SD from three independent triplicate experiments. (D) Two mTorKI-resistant CRC cell lines (SW620 and HCT116) were treated with a high dose of mTorKIs for 72 h, and apoptotic cells were quantified. Indomethecin (Indo) was used as a positive control. Data represent means ± SD from three independent triplicate experiments.
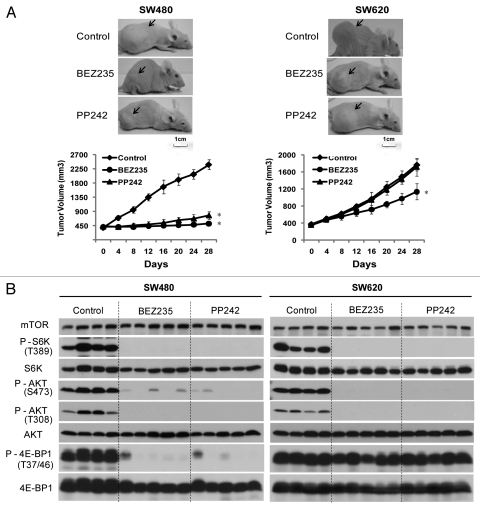
We further established SW480 and SW620 xenograft tumors in nude mice and investigated the therapeutic efficacy of BEZ235 and PP242. During the course of the experiment, animal weights were measured weekly, which showed minimal, non-statistically significant weight fluctuations in both drug-treated and control groups (data not shown), suggesting that chronic dosing with 45 mg/kg BEZ235 and 60 mg/kg PP242 was well tolerated by the tumor-bearing animals. Both BEZ235 and PP242 significantly attenuated SW480 tumor growth, with an average tumor volume of 517 ± 45 mm3 (p < 0.01) and 778 ± 114 mm3 (p < 0.01), respectively, by day 28 of treatment (Fig. 3A), while the vehicle-treated group had tumor volume of 2,389 ± 156 mm3. In agreement with lack of inducing apoptosis by mTorKIs in CRC cells, no tumor shrinkage was seen in treated animals. In contrast, SW620 tumors were essentially unresponsive to PP242 (1,715 ± 204 mm3 for PP242 vs. 1,768 ± 137 mm3 for control, p = 0.609), and only moderately inhibited by BEZ235 (1,136 ± 188 mm3 for BEZ235 vs. 1,768 ± 137 mm3 for control, p < 0.01) (Fig. 3A).
Figure 3.
mTorKIs inhibit 4E-BP1 phosphorylation in SW480 but not SW620 xenograft tumors. (A) Xenograft tumor mouse models were orally administered with BEZ235 at 45 mg/kg/day or PP242 at 60 mg/kg/day once daily for 28 d. Upper parts show representative tumors (as indicated by arrow-heads) in treated and control xenograft mice. Lower part shows tumor volume measurements (means ± SD; n = 8; *p < 0.01, vs. control). (B) Xenograft tumor mouse models derived from SW480 and SW620 were orally administered with BEZ235 at 45 mg/kg/day or PP242 at 60 mg/kg/day, once daily for 28 d. On day 28, CRC tumors were removed and analyzed for inhibition of PI3K-mTOR signaling by western blot. Four tumor samples from control group and 5 samples from each mTorKIs treatment group were shown here, with each lane representing an individual tumor sample.
The effect of BEZ235 and PP242 on mTOR signaling was analyzed after the last drug administration on day 28. In both tumors, BEZ235 and PP242 blunted the activity of mTORC1, mTORC2 and PI3K, as shown by the disappearance of P-S6K1(T389) and P-AKT(S473) signals, respectively (Fig. 3B), demonstrating that these agents achieved on-target inhibition of mTOR in vivo. 4E-BP1(T37/46) phosphorylation was also attenuated by both compounds in SW480 tumors. In contrast, BEZ235 and PP242 completely failed to inhibit 4E-BP1 phosphorylaiton in SW620 tumors (Fig. 3B). Together, these data show that SW480 and SW620 tumors are highly sensitive and resistant to mTorKIs, respectively, which is strongly correlated with the ability of mTorKIs to inhibit 4E-BP1 phosphorylation.
mTOR-independent 4E-BP1 phosphorylation in SW620 cells.
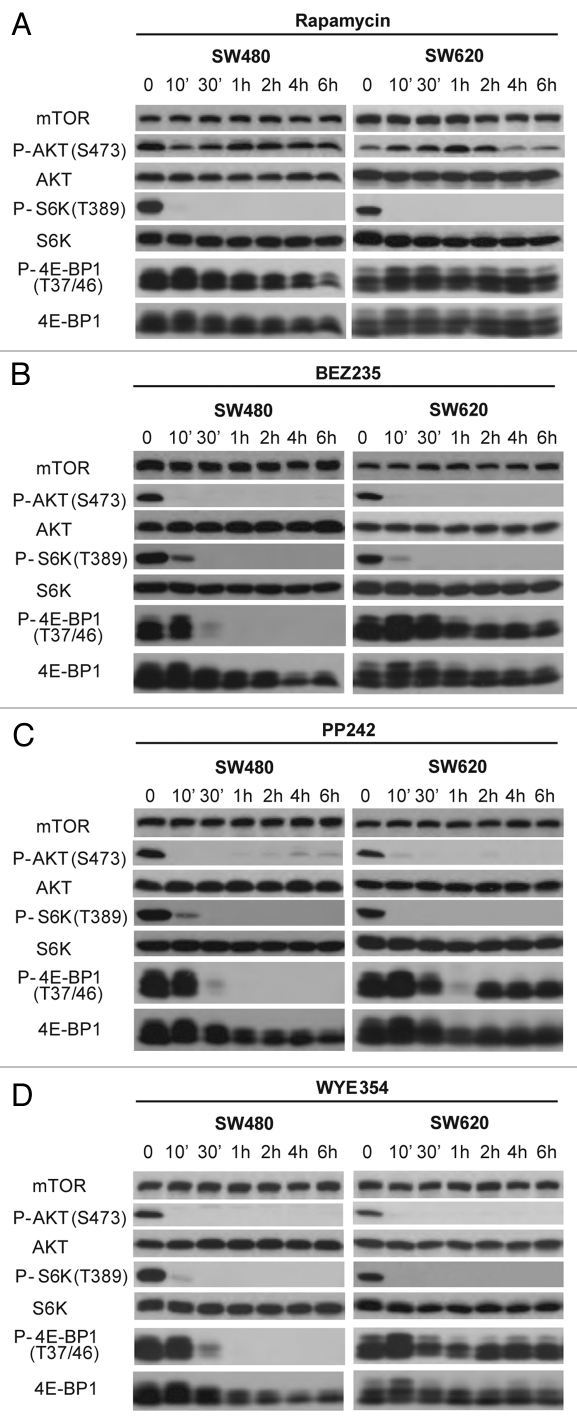
To understand the molecular basis of mTorKI action, we analyzed the kinetic changes of mTOR signaling in SW480 and SW620 cells in response to drug treatment. Upon addition of BEZ235, PP242 or WYE354, P-S6K1(T389) and P-AKT(S473) rapidly disappeared in both CRC cell lines and remained virtually undetectable throughout the time course, indicating that both mTOR complexes were rapidly and persistently inhibited (Fig. 4). P-4E-BP1(T37/46) signal also decreased to undetectable level in SW480 cells (Fig. 4). However, 4E-BP1 phosphorylation was only transiently inhibited in SW620 cells, and then quickly returned (Fig. 4). Because mTOR was catalytically inhibited throughout the course of the study as indicated by the blockage of S6K1(T389) and AKT(S473) phosphorylation, the re-appearance of 4E-BP1 phosphorylation is likely due to an mTOR-independent mechanism in SW620 cells.
Figure 4.
The kinetics of mTOR inhibition by mTorKIs in SW480 and SW620 cells. SW480 and SW620 cells were treated with mTOR inhibitors rapamycin (A), BEZ235 (B), PP242 (C) and WYE354 (D) for different times. The phosphorylation and total level of different mTOR substrates was determined by western blot.
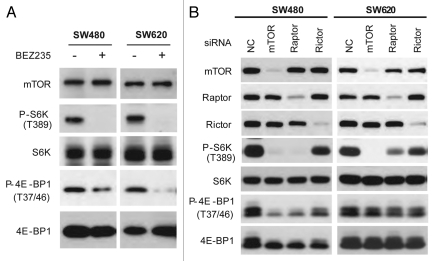
To verify whether 4E-BP1 re-phosphorylation is indeed mTOR-independent mechanism in SW620 cells, we performed in vitro kinase assay of mTOR isolated from SW480 and SW620 cells treated without or with BEZ235. BEZ235 treatment inhibited phosphorylation of recombinant 4E-BP1 as well as S6K1 by mTOR from both SW480 and SW620 cell lines (Fig. 5A). We further used siRNA to knock down mTOR complexes in SW480 and SW620 cells. siRNA-mediated suppression of mTOR or raptor, but not rictor inhibited 4E-BP1 and S6K1 phosphorylation in SW480 cells (Fig. 5B). In contrast, mTOR and raptor siRNAs did not affect 4E-BP1 phosphorylation in SW620 cells even though they effectively blocked S6K1 phosphorylation (Fig. 5B). This observation unequivocally demonstrates that mTOR kinase activity toward 4E-BP1 is inhibited by BEZ235 in both SW480 and SW620 cells, and 4E-BP1 re-phosphorylation in mTorKI-treated SW620 cells is mediated by an mTOR-independent mechanism.
Figure 5.
mTOR-independent 4E-BP1 phosphorylation in SW620 cells. (A) BEZ235 inhibits mTOR kinase activity toward 4E-BP1 in both SW480 and SW620 cells. SW480 and SW620 cells were treated with 100 nM BEZ235 or drug vehicle control (DMSO) for 6 h. mTOR was immunoprecipitation and assayed for in vitro kinase activity toward bacterial recombinant S6K1 and GST-4E-BP1 by western blot. (B) siRNA-mediated knockdown of mTORC1 inhibits 4E-BP1 phosphorylation in SW480 but not SW620 cells. SW480 and SW620 cells were transfected with siRNA targeting mTOR, raptor and rictor, respectively, for 48 h. Inhibition of mTOR signaling was analyzed by western blot.
Discussion
CRC is one of the most common human malignancies. Despite recent advances in EGFR-targeted therapy, it remains a leading cause of cancer-related death and urgently need new therapy. We have previously shown that siRNA-mediated knockdown of mTOR but not rapamycin potently inhibited CRC tumor models.11 Although these studies validated mTOR as a useful CRC drug target, they also showed the lack of anti-CRC efficacy by rapamycin.12 Therefore, more potent mTOR inhibitors are needed for effective mTOR-targeted CRC therapy. In this study, we tested several ATP-competitive mTOR kinase inhibitors against a large panel of 12 common CRC cell lines. They were effective in ∼60% CRC cell lines (7 out of 12), compared with 17% for rapamycin, clearly demonstrating that mTorKIs have much improved anti-CRC activity than rapamycin. Curiously, mTorKI sensitivity was not correlated with mutation of PI3KCA or PTEN that are known to cause mTOR activation, suggesting that they are not predictive biomarkers. Additionally, CRC cell lines carrying K-Ras mutation were largely sensitive to mTorKIs (Table 1). Because these mutations are known to cause resistance to EGFR-targeted therapy,29 mTorKIs are potentially useful to treat patients who have K-Ras or B-Raf mutations.
A surprising finding is that a large proportion of tested CRC cell lines (>40%) were mTorKI-resistant, which warrants considerable attention. Although mTorKIs achieved rapid and sustained on-target inhibition of mTOR in CRC cells, they only transiently attenuated 4E-BP1 phosphorylation in drug-resistant CRC cells. We further found that 4E-BP1 was re-phosphorylated in an mTOR-independent manner. 4E-BP1 is a major mTORC1 substrate that plays a pivotal role in mTORC1 signaling to control translation, cell proliferation and senescence.30,31 4E-BP1 phosphorylation has recently been implicated in rapamycin resistance in certain cancer cells.32–34 mTorKI was shown to abolish “rapamycin-resistant 4E-BP1 phosphorylation,” which was thought to be due to inhibition of a “rapamycin-insensitive mTORC1” by mTorKIs.35,36 Therefore, although P-4E-BP1 can be a useful predictor for both rapamycin- and mTorKI resistance, our observations indicate that the mechanism for drug-resistant 4E-BP1 phosphorylation is clearly distinct for the two classes of mTOR inhibitors. Identification of the alternative kinase responsible for 4E-BP1 re-phosphorylation will be an important future task.
Because of their anticancer potential, several mTorKIs are currently under early-stage clinical trials for lymphoma, advanced hepatocellular carcinoma, breast cancer, glioma and non-small cell lung carcinoma (NSCLC).19 However, their therapeutic activity toward CRC cells remains unclear. Our study with in vivo CRC models provides strong preclinical rationale for testing them in human CRC clinical trials. Our study revealed that the existence of significant intrinsic drug resistance in colorectal cancer cells, which warrants further study of intrinsic drug resistance in other cancer types, especially those in which mTorKIs are being tested in clinical trials. Because phosphorylation of S6K1, S6 and AKT was blunted by mTorKIs in all CRC cells, they can be useful pharmacodynamic (PD) biomarkers for ontarget inhibition. On the other hand, 4E-BP1 phosphorylation is strongly correlated with drug resistance, indicating that it is a potential biomarker for predicting drug resistance, which should provide valuable guidance for on-going and future human cancer clinical trials.
Materials and Methods
CRC cell lines and mTOR inhibitors.
Twelve human CRC cell lines (CACO-2, COLO-205, DLD-1, HCT116, HT29, KM-12, LOVO, RKO, SW1116, SW48, SW620 and SW480) were primarily obtained from ATCC. Table 1 summarizes the histological feature, origin and status of oncogene or tumor suppressors that are most commonly detected with genetic aberrations in CRCs (K-Ras, B-RAF, PIK3CA, PTEN and p53). The genetic information was queried from the literature, ATCC and the Catalogue of Somatic Mutations in Cancer (COSMIC, www.sanger.ac.uk/genetics/CGP/cosmic). The CRC cells were maintained in RPMI 1640 medium (GIBCO, #72400-120) supplemented with 10% fetal bovine serum and 5 mmol/L l-glutamine, at 37°C, 5% CO2. Rapamycin was purchased from LC laboratories (R-5000). BEZ235, PP242 and WYE354 were purchased from Chemdea (#CD0196, #CD0258, #CD0270). The compounds were dissolved in DMSO and diluted with cell culture medium. The final concentration of DMSO was less than 0.5%.
Growth, colony formation and apoptosis assays.
The growth of CRC cells and the inhibitory effect of mTOR inhibitors were determined by optimized sulforhodamine B (SRB) assay as described before in reference 37. All 12 cell lines were tested simultaneously and the experiment repeated twice. Each drug was tested at 6 drug concentrations with each concentration point representative of 10 replicate wells for each cell line. Briefly, cells were seeded in triplicate in 96-well plates at an initial density of 3 × 103 cells/well. After 12 h, cells were treated with various concentrations of mTOR inhibitors for 48 h. Cells were then fixed with 10% (wt/vol) trichloroacetic acid and stained with SRB solution (0.057% w/v in 1% acetic acid) for 30 min, after which the excess dye was removed by washing with 1% (vol/vol) acetic acid. The protein-bound dye was dissolved in 10 mM Tris solution for OD determination at 492 nm using a microplate reader. The relative growth was expressed as the percentage of the absorbance of treated vs. control cells and fitted to Pharmcology DoseResp using OriginPro 8.0 software to calculate IC50.
Soft agar colony formation assay was performed as described before in reference 38. Briefly, 1 × 103 cells were seeded in 0.35% Fisher low melt agar on a base of 0.7% Sigma agar in a 6-well plate. Culture dishes were then transferred sequentially to a refrigerator (4°C) for 15 min, to room temperature for 10 min, and then to the cell culture incubator. An upper layer of 0.5 ml culture medium containing drug (n = 3) or drug vehicle (n = 3) was applied to the cultures and changed every other day for two weeks. Cultures were stained with p-Iodonitroneotetrazolium violet (Sigma-Aldrich, #146-68-9) for two hours and then inspected and photographed using a MiniCount Colony Counter (Imaging Products International, #013100708). The colony number was expressed as the ratio of treated vs. control cells. Data represent mean ± SD from three independent triplicate experiments.
For apoptosis assay, CRC cells were treated with BEZ235 1 uM, PI103 3 uM, PP242 3 uM and WYE354 3 uM, for ∼72 h. Apoptosis was determined by acridine orange (AO) staining as described previously in reference 39. Calculated apoptotic rates after treatment are graphed and representative histograms of SW480, SW620, CACO-2 and HCT116 cells are shown. 0.1% DMSO was used as vehicle control. Indomethecin 600 uM (Indo) was used as a positive control, which can induce robust apoptosis in CRC cells based on our previous findings.39 Data represent means ± SD from three independent triplicate experiments.
Xenograft CRC tumor models.
Male BALB/c athymic nude mice (4–6 weeks old) were obtained from SIBS. They were injected subcutaneously into the right hind flank with 5 × 106 SW480 cells or SW620 cells to establish the CRC xenograft model. Seven days after injection, mice were randomized into 3 groups (8 animals per group). Group 1 was given 45 mg/kg BEZ235; group 2 was given 60 mg/kg PP242, and group 3 was given the vehicle used for administration (control). BEZ235 and PP242 in all animals was administered via oral gavage and freshly prepared daily just before administration. Prior to gavage, drugs were solubilized in 200 µl of NMP 10%/PEG300 90%. Treatment frequency was once daily for a total duration of 4 weeks. Bidimensional tumor measurements were taken every 3 d and mice were weighed once weekly. Tumor volume was calculated by the following formula: tumor volume (mm3) = (shorter diameter2 × longer diameter)/2 and are presented as means ± SD (n = 8).11 BEZ235 and PP242 were used according to previous studies, which were at much lower doses than the reported maximum tolerated doses.27,40,41 For analysis of signaling inhibition, tumor tissues were removed from the animals after administration of the last dose of drug, and immediately frozen in liquid nitrogen. Tissue extracts were prepared for analysis of PI3K-mTOR signaling by western blot. The animal studies were approved by the Institutional Animal Care and Use Committee and were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering.
Western blot, immunoprecipitation, in vitro kinase and RNA interference assays.
Western blotting was performed to examine PI3K-mTOR signaling as described previously in reference 42 and 43. mTOR antibody was described before in reference 44 and 45. Antibodies against Akt, S6K1, 4E-BP1, P-Akt(S473), P-Akt(T308), P-S6K(T389), P-4E-BP1(T37/46) were purchased from Cell Signaling Technology. The data were representative of several independent experiments. Cell lyses preparation and Immunoprecipitations were performed as previously described in reference 46. For mTOR in vitro kinase assay, CRC cells treated with BEZ235 100 nM or DMSO (vehicle control) for 6 h were lysed in ice-cold lysis buffer [40 mM HEPES, 2 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 0.3% CHAPS, and one tablet of EDTA-free protease inhibitors (Roche Diagnostics, #05056489001) per 25 ml]. mTOR was then immunoprecipitated and incubated with 150 ng bacterial recombinant S6K1 or GST-4E-BP1. For RNA interference assays, SW480 and SW620 cells cultured in 6-well plates were transfected with 100 nM short interfering RNA (siRNA) against mTOR, Raptor or Rictor using the DharmaFECTTM transfection agent (Dharmacon Inc., #T-2001) according to the manufacturer's instructions. At 48 h after siRNA transfection, cells were harvested and assessed by western blot analysis. The siRNA sequences are: mTOR siRNA: 5′-AUA GAA GCG AGU AGA CUC CUC-3′; Raptor siRNA: 5′-CUG UAA GAU CAG CCU CAU CUU-3′; Rictor siRNA: 5′-AAG AUA CGA UUC UUC ACA A-3′; Control siRNA 5′-ACG UGA CAC GUU CGG AGA ATT-3′.
Statistical analysis.
The data are representative of replicate experiments. Statistical analyses were performed using SPSS 11.0 software. Comparisons between groups were performed using the Student t-test. All statistical tests were conducted with a twosided significance level of 0.05.
Acknowledgements
We thank Janice Thomas, Tzung-Ju Wu and Jun-Hung Cho for technical assistance. The work was supported by an NIH grant (R01CA123391).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Dasari A, Messersmith WA. New strategies in colorectal cancer: biomarkers of response to epidermal growth factor receptor monoclonal antibodies and potential therapeutic targets in phosphoinositide-3-kinase and mitogen-activated protein kinase pathways. Clin Cancer Res. 2010;16:3811–3818. doi: 10.1158/1078-0432.CCR-09-2283. [DOI] [PubMed] [Google Scholar]
- 3.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Tsang CK, Qi H, Liu LF, Zheng XFS. Targeting mammalian target of rapamycin (mTOR) for health and diseases. Drug Discov Today. 2007;12:112–124. doi: 10.1016/j.drudis.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 6.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 7.Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10:2305–2316. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 10.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YJ, Dai Q, Sun DF, Xiong H, Tian XQ, Gao FH, et al. mTOR signaling pathway is a target for the treatment of colorectal cancer. Ann Surg Oncol. 2009;16:2617–2628. doi: 10.1245/s10434-009-0555-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YJ, Tian XQ, Sun DF, Zhao SL, Xiong H, Fang JY. Combined inhibition of MEK and mTOR signaling inhibits initiation and progression of colorectal cancer. Cancer Invest. 2009;27:273–285. doi: 10.1080/07357900802314893. [DOI] [PubMed] [Google Scholar]
- 13.Zheng XF, Florentino D, Chen J, Crabtree GR, Schreiber SL. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 14.Stoeltzing O. Dual-targeting of mTOR and HSP90 for cancer therapy: facing oncogenic feed-back-loops and acquired mTOR resistance. Cell Cycle. 2010;9:2051–2052. doi: 10.4161/cc.9.11.11924. [DOI] [PubMed] [Google Scholar]
- 15.Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 16.O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruppuso PA, Boylan JM, Sanders JA. The physiology and pathophysiology of rapamycin resistance: implications for cancer. Cell Cycle. 2011;10:1050–1058. doi: 10.4161/cc.10.7.15230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Don ASA, Zheng XFS. Recent clinical trials of mTOR-targeted cancer therapies. Rev Recent Clin Trials. 2011;6:24–35. doi: 10.2174/157488711793980147. [DOI] [PubMed] [Google Scholar]
- 19.Zhang YJ, Duan Y, Zheng XFS. Targeting the mTOR kinase domain: the second generation of mTOR inhibitors. Drug Discov Today. 2011;16:325–331. doi: 10.1016/j.drudis.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markman B, Dienstmann R, Tabernero J. Targeting the PI3K/Akt/mTOR pathway—beyond rapalogs. Oncotarget. 2010;1:530–543. doi: 10.18632/oncotarget.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altman JK, Sassano A, Platanias LC. Targeting mTOR for the treatment of AML. New agents and new directions. Oncotarget. 2011;2:510–517. doi: 10.18632/oncotarget.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martelli AM, Evangelisti C, Chiarini F, McCubrey JA. The phosphatidylinositol-3-kinase/Akt/mTOR signaling network as a therapeutic target in acute myelogenous leukemia patients. Oncotarget. 2010;1:89–103. doi: 10.18632/oncotarget.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leibovitz A, Stinson JC, McCombs WB, 3rd, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 24.Hewitt RE, McMarlin A, Kleiner D, Wersto R, Martin P, Tsokos M, et al. Validation of a model of colon cancer progression. J Pathol. 2000;192:446–454. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH775>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 25.Duranton B, Holl V, Schneider Y, Carnesecchi S, Gossé F, Raul F, et al. Polyamine metabolism in primary human colon adenocarcinoma cells (SW480) and their lymph node metastatic derivatives (SW620) Amino Acids. 2003;24:63–72. doi: 10.1007/s00726-002-0333-5. [DOI] [PubMed] [Google Scholar]
- 26.Liu K, McDuffie E, Abrams SI. Exposure of human primary colon carcinoma cells to anti-Fas interactions influences the emergence of pre-existing Fasresistant metastatic subpopulations. J Immunol. 2003;171:4164–4174. doi: 10.4049/jimmunol.171.8.4164. [DOI] [PubMed] [Google Scholar]
- 27.Janes MR, Limon JJ, So L, Chen J, Lim RJ, Chavez MA, et al. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 2010;16:205–213. doi: 10.1038/nm.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brachmann SM, Hofmann I, Schnell C, Fritsch C, Wee S, Lane H, et al. Specific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cells. Proc Natl Acad Sci USA. 2009;106:22299–22304. doi: 10.1073/pnas.0905152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 30.Chao SK, Horwitz SB, McDaid HM. Insights into 4E-BP1 and p53 mediated regulation of accelerated cell senescence. Oncotarget. 2011;2:89–98. doi: 10.18632/oncotarget.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yellen P, Saqcena M, Salloum D, Feng J, Preda A, Xu L, et al. High-dose rapamycin induces apoptosis in human cancer cells by dissociating mTOR complex 1 and suppressing phosphorylation of 4E-BP1. Cell Cycle. 2011;10:3948–3956. doi: 10.4161/cc.10.22.18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dowling RJO, Topisirovic I, Fonseca BD, Sonenberg N. Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim Biophys Acta. 2010;1804:433–439. doi: 10.1016/j.bbapap.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Choo AY, Blenis J. Not all substrates are treated equally: implications for mTOR, rapamycin resistance and cancer therapy. Cell Cycle. 2009;8:567–572. doi: 10.4161/cc.8.4.7659. [DOI] [PubMed] [Google Scholar]
- 35.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thoreen CC, Sabatini DM. Rapamycin inhibits mTORC1, but not completely. Autophagy. 2009;5:725–726. doi: 10.4161/auto.5.5.8504. [DOI] [PubMed] [Google Scholar]
- 37.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 38.Alley MC, Pacula-Cox CM, Hursey ML, Rubinstein LR, Boyd MR. Morphometric and colorimetric analyses of human tumor cell line growth and drug sensitivity in soft agar culture. Cancer Res. 1991;51:1247–1256. [PubMed] [Google Scholar]
- 39.Zhang YJ, Bao YJ, Dai Q, Yang WY, Cheng P, Zhu LM, et al. mTOR signaling is involved in indomethacin and nimesulide suppression of colorectal cancer cell growth via a COX-2 independent pathway. Ann Surg Oncol. 2011;18:580–588. doi: 10.1245/s10434-010-1268-9. [DOI] [PubMed] [Google Scholar]
- 40.Cao P, Maira SM, Garcia-Echeverria C, Hedley DW. Activity of a novel, dual PI3-kinase/mTor inhibitor NVP-BEZ235 against primary human pancreatic cancers grown as orthotopic xenografts. Br J Cancer. 2009;100:1267–1276. doi: 10.1038/sj.bjc.6604995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu TJ, Koul D, LaFortune T, Tiao N, Shen RJ, Maira SM, et al. NVP-BEZ235, a novel dual phosphatidylinositol-3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol Cancer Ther. 2009;8:2204–2210. doi: 10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drenan RM, Liu X, Bertram PG, Zheng XFS. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J Biol Chem. 2004;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Zheng XFS. Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol Biol Cell. 2007;18:1073–1082. doi: 10.1091/mbc.E06-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsang CK, Liu H, Zheng XF. mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle. 2010;9:953–957. doi: 10.4161/cc.9.5.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi JH, Bertram PG, Drenan R, Carvalho J, Zhou HH, Zheng XFS. The FKBP12-rapamycin-associated protein (FRAP) is a CLIP-170 kinase. EMBO Rep. 2002;3:988–994. doi: 10.1093/embo-reports/kvf197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 47.Trainer DL, Kline T, McCabe FL, Faucette LF, Feild J, Chaikin M, et al. Biological characterization and oncogene expression in human colorectal carcinoma cell lines. Int J Cancer. 1988;41:287–296. doi: 10.1002/ijc.2910410221. [DOI] [PubMed] [Google Scholar]
- 48.Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O'Meara S, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abubaker J, Bavi P, Al-Harbi S, Ibrahim M, Siraj AK, Al-Sanea N, et al. Clinicopathological analysis of colorectal cancers with PIK3CA mutations in Middle eastern population. Oncogene. 2008;27:3539–3545. doi: 10.1038/sj.onc.1211013. [DOI] [PubMed] [Google Scholar]
- 50.Wee S, Jagani Z, Xiang KX, Loo A, Dorsch M, Yao YM, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009;69:4286–4293. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]