Abstract
DNA methylation of tumor suppressor genes is a common feature of human cancer. The cyclin-dependent kinase inhibitor gene p16/Ink4A is hypermethylated in a wide range of malignant tissues and the p14/ARF gene located 20 kb upstream on chromosome 9p21 is also methylated in carcinomas. p14/ARF (ARF, alternative reading frame) does not inhibit the activities of cyclins or cyclin-dependent kinase complexes; however, the importance of the two gene products in the etiology of cancer resides in their involvement in two major cell cycle regulatory pathways: p53 and the retinoblastoma protein, Rb, respectively. Distinct first exons driven from separate promoters are spliced onto the common exons 2 and 3 and the resulting proteins are translated in different reading frames. Both genes are expressed in normal cells but can be alternatively or coordinately silenced when their CpG islands are hypermethylated. Herein, we examined the presence of methyl-CpG binding proteins associated with aberrantly methylated promoters, the distribution of acetylated histones H3 and H4 by chromatin immunoprecipitation assays, and the effect of chemical treatment with 5-aza-2′-deoxycytidine (5aza-dC) and trichostatin A on gene induction in colon cell lines by quantitative reverse transcriptase–PCR. We observed that the methyl-CpG binding protein MBD2 is targeted to methylated regulatory regions and excludes the acetylated histones H3 and H4, resulting in a localized inactive chromatin configuration. When methylated, the genes can be induced by 5aza-dC but the combined action of 5aza-dC and trichostatin A results in robust gene expression. Thus, methyl-CpG binding proteins and histone deacetylases appear to cooperate in vivo, with a dominant effect of DNA methylation toward histone acetylation, and repress expression of tumor suppressor genes hypermethylated in cancers.
Progression through the cell cycle is a complex process that is sequentially regulated by extracellular stimuli relayed by various intracellular signaling networks. For example, the G1/S-phase transition is regulated positively by cyclin-dependent kinases and negatively by the retinoblastoma protein Rb and inhibitors of cyclin-dependent kinases (1). Tumor-associated alterations of the retinoblastoma pathway include amplification of the cyclin D1 gene, CDK4 amplification, activation by point mutations, and inactivation of inhibitors of cyclin-dependent kinases, such as INK4a, INK4b, INK4c, and INK4d (2).
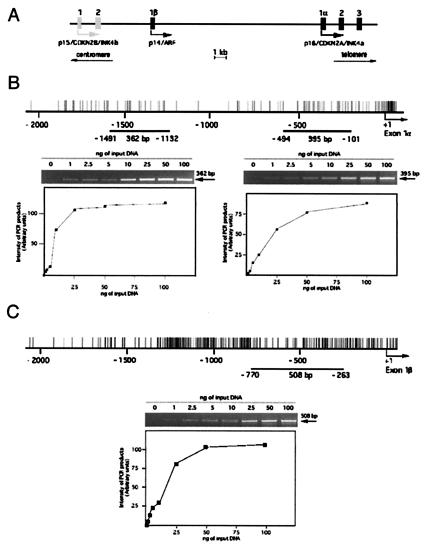
Two of the INK4 tumor suppressor proteins, p15/CDKN2B/Ink4b and p16/CDKN2A/Ink4a, have presumably arisen as a consequence of gene duplication during evolution (3). Indeed, the two human genes are located on 9p21 within 30 kb of one another in the same transcriptional orientation. In addition to gene duplication, the genomic structure of the region is complicated by the insertion of an additional exon (1β) between the first exon of the two genes (ref. 4 and Fig. 1A). Exon 1β is transcribed from an alternative promoter and is spliced to the second exon of the p16 gene. The cyclin-dependent kinase inhibitor p16 is encoded by exons 1α, 2, and 3, and p14/ARF is encoded by exons 1β, 2, and 3. Their amino acid sequences, however, are unrelated because the two proteins are translated in different reading frames (5). In human tumors, chromosome 9p21 is a major site of chromosomal abnormalities (3, 6). Alterations of p16 have been exhaustively described and homozygous deletions of the p16 locus can affect the integrity of the p14 gene. Conversely, specific deletions of exon 1β have been proposed as major hotspots for chromosomal aberrations in leukemias but have no effect on the expression of p16 (7, 8).
Figure 1.
Sensitivity of PCR amplification on sonicated DNA after DNA–protein cross-linking. (A) Schematic representation of the p15/p14/p16 locus on human chromosome 9p21. Shaded boxes, coding exons for p15/CDKN2B/INK4b (exons 1 and 2); solid boxes, coding exons for p14/ARF (exon 1β) and p16/CDKN2A/INK4a (exons 1α, 2, and 3); arrows, transcription initiation sites. The p16 and p14 promoters are separated by 20 kb and both are CpG sites. Distribution of CpG sites are represented by upward strokes for p16 (B) and p14 (C). Lines, regions amplified by PCR from the initiation of transcription of p16 (exon 1α) (B) or p14 (exon 1β) (C). Sensitivity of the PCR amplification was monitored for ChIP experiments in which the yield of PCR product depended on the amount of input DNA and 10% of the PCR mixture (total volume, 50 μl) from serial dilution of input DNA from HCT15 colon cells was analyzed on ethidium bromide-stained 2% agarose gels. The intensity of the bands of PCR products was measured by densitometry and plotted against the amount of input DNA (from 1 to 100 ng) for each set of primers. (B) The 362-bp band corresponds to the expected size of the PCR product for the Alu element (15 CpG sites, positions −1,491 to −1,132); the 395-bp band corresponds to the promoter of p16 (21 CpG sites, positions −494 to −101). (C) The 508-bp band corresponds to the expected size of the PCR product for the p14 promoter (43 CpG sites, positions −770 to −263).
Hypermethylation of the p16 CpG island is observed in many human tumors (5). Aberrant changes in methylation patterns have been detected in in situ lesions and invasive cancers, suggesting that hypermethylation of this tumor suppressor gene is an early event in cancer (9, 10). The human p14 promoter is embedded in a CpG island that is also aberrantly methylated in colon carcinomas (11, 12). In cancer cells, transcriptional silencing of tumor suppressor genes by methylation can be transiently reversed by treatment with the hypomethylating agent 5-aza-2′-deoxycytidine (5aza-dC), which restores minimal gene expression. However, the combination of 5aza-dC and trichostatin A (TSA), an inhibitor of deacetylase activity, results in a stronger stimulation, suggesting that DNA methylation and chromatin architecture act together to silence genes (13, 14).
Direct associations between DNA methylation and chromatin structure have been established after the identification of several complexes containing methyl-CpG binding proteins, chromatin remodeling factors, and histone deacetylases (15). CpG methylation and formation of nuclease-resistant chromatin structures are common characteristics of these complexes, but their target sequences and roles in gene silencing in vivo are unknown. Therefore, we have investigated whether methyl-CpG binding proteins are involved in silencing the p16/p14 locus in human colon carcinomas cell lines. We show that the methyl-CpG binding protein MBD2 binds to the 5′ regulatory ends of the p16 and p14 genes in human colon cancer cell lines and, thereby, contributes to gene repression.
Materials and Methods
Cell Culture.
Human colon cell lines HCT116, HCT15, and SW48 were grown in McCoy's 5a modified medium. The human cervix cell line HeLa was maintained in DMEM. All media (Life Technologies, Grand Island, NY) were supplemented with 10% FCS and grown at 37°C in a humidified atmosphere of 5% CO2/95% air.
Drug Treatments.
Cells were seeded at a low density 16 h before drug addition and then treated for 72 h with 1 μM 5aza-dC (Sigma). For TSA experiments, cells were treated with 100 nM TSA (Wako Biochemicals, Osaka) for 16 h. For the combination of drugs, cells were incubated with 5aza-dC for 16–24 h and then with TSA for an additional 16 h.
Semiquantitative Reverse Transcriptase–PCR (RT-PCR) Assays.
Total RNA was isolated in a single-step procedure by acid guanidium thiocyanate/phenol/chloroform extraction (16). Samples were amplified simultaneously for GAPDH and p16 or p14 as described (17) with the following primers: F-GAPDH, 5′-TCCCATCACCATCTTCCAG-3′; R-GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-ATGAGTCCTTCCACGATACC-3′; F-p16, 5′-CACGGCCGCGGCCCGGGGTC-3′; R-p16-p14, 5′-GGCCCGGTGCAGCACCACCA-3′; F-p14, 5′-GCCAGGGGCGCCCGCCGCTG-3′. Ten percent of the PCR products was analyzed on 2% agarose gels containing ethidium bromide (0.1 μg/ml), and signals were quantitated with CHEMIIMAGER 4400 LOW LIGHT IMAGING SYSTEM, VERSION 5.1 software (Alpha Innotech, San Leandro, CA).
Chromatin Immunoprecipitation.
Cells were washed and scraped off of culture dishes in PBS, and nuclei were prepared in ice-cold hypotonic buffer (10 mM Tris⋅HCl, pH 7.4/10 mM NaCl/5 mM MgCl2). Each step was performed on ice and in the presence of a mixture of protease inhibitors (Complete, Roche Molecular Biochemicals). After centrifugation, nuclei were resuspended in hypotonic buffer containing 0.1% Nonidet P-40 and centrifuged at 850 × g. Proteins were then cross-linked to DNA with 0.3% formaldehyde. The cells were lysed and cross-linking was performed on nuclei. Samples were washed and lysed in SDS buffer (1% SDS/10 mM EDTA/50 mM Tris⋅HCl, pH 8.1) for 10 min on ice. Nucleoprotein complexes were sonicated to reduce DNA fragments to 400–600 bp. Debris were removed, and the supernatant was diluted 1:10 in chromatin immunoprecipitation (ChIP) dilution buffer (0.01% SDS/1.1% Triton X-100/1.2 mM EDTA/16.7 mM Tris base, pH 8.1/167 mM NaCl). The chromatin solution was precleared with 80 μl of salmon sperm DNA/protein A-agarose beads (Upstate Biotechnology, Lake Placid, NY). The soluble fraction was collected, and 5 μl of anti-acetyl-histone H3 (residues 1–21) or H4 (residues 2–19) antibodies (Upstate Biotechnology) was added. To identify the methyl-CpG binding proteins, 5 μl of the TRD-MeCP2 (provided by Peter L. Jones, Laboratory of Molecular Embryology) or MBD2 (provided by Paul Wade, Laboratory of Molecular Embryology) antibody was added. After immunoprecipitation, immune complexes were collected by adding 60 μl of salmon sperm DNA/protein A-agarose beads. After washing, complexes were eluted in 1% SDS/0.1 M NaHCO3 and cross-links were reversed by heating. Samples were heated at 65°C to reverse DNA-proteins cross linking. DNA was recovered by proteinase K digestion, phenol extraction, and ethanol precipitation.
PCR Amplification.
PCR amplification was performed in 50 μl as described (11, 16) with the following primers: Fp16-island, 5′-GGGCTCTCACAACTAGGAA; Rp16-island, 5′-CGGAGGAGGTGCTATTAACTC; FAlu, 5′-GTAATAGTGATAATTCTATCCAAAGCA; RAlu, 5′-GAATCTTGTTTTGAGGCATATAA; Fp14-island, 5′-TGCGACTCCACCTACCTAGTC; Rp14-island, 5′-ACTTTCCCGCCCTGTGTGC. Ten microliters of the PCR products was size-fractionated on a 2% ethidium bromide/agarose gel and quantitated under UV transillumination.
Results
Use of ChIP to Study the in Vivo Binding of Methyl-CpG Binding Proteins.
To determine whether the silencing of hypermethylated p16 and p14 genes observed in human cancer is consistent with a model involving methyl-CpG binding proteins, we used the in vivo formaldehyde cross-linking technique to study the occupancy of methylated promoters by methyl-CpG binding proteins in three colon carcinoma cell lines with methylated p16 (HCT116, HCT15, and SW48) and p14 (HCT15) genes and a cervix cell line, HeLa, in which neither gene is methylated (16, 18). In HCT116, the methylation level of the p16 promoter is about 45% (28) but p16 expression is still detectable (11, 18) because of the expression of the mutated allele concomitant with the silencing of the wild-type allele (19).
A large component of the human genome consists of Alus and long interspersed elements (LINES), interspersed in coding and regulatory regions that contain functional promoters. In human tissues, most of these repetitive elements are methylated, probably, as a result of a host defense system (20, 21). In cell lines, the methylation of the two Alu elements located in the intergenic region between the p16 and p14 genes (Fig. 1A) apparently does not interfere with expression of these genes (22). Thus, to determine whether the formation of a repressive chromatin structure was targeted to the regulatory regions, the putative binding of methyl-CpG binding proteins also was investigated with primers specific for the Alu element located upstream of p16 (22).
We first evaluated the sensitivity of the PCR amplification on serial dilutions (0–100 ng) of total DNA collected after sonication but before the preclearing step (input DNA) with specific primers for the promoter region of p16 and Alu (Fig. 1B) or p14 (Fig. 1C). Data indicate that the signal is proportional to the amount of input DNA from 0 to 25 ng of DNA for the Alu element (Fig. 1B) and from 0 to 50 ng of DNA for p16 and p14 promoters (Fig. 1 B and C). For further experiments on DNA collected after incubation of immune complexes with salmon sperm DNA/agarose-protein A beads or on the corresponding unbound fractions, PCRs were performed under linear dose–response conditions for each sequence analyzed.
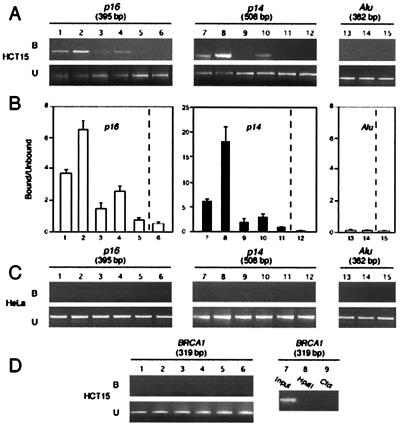
A second quantitative assay was performed for each set of primers to determine the efficiency of amplification of the target DNA sequence in the bound chromatin fraction after ChIP. Each experiment was repeated at least three times in the different cell lines and representative experiments with HCT15 (p16 and p14 M+) and HeLa (p16 and p14 M−) cells are shown in Fig. 2. To assess whether the recovery of PCR products was proportional to the amount of antibody or dependent on the stringency of the reaction, immunoprecipitations were done with 2 or 5 μl of antibody, in the presence or absence of SDS. The band intensity of bound DNA (Fig. 2A Upper) and unbound DNA (Fig. 2A Lower) were measured with CHEMIIMAGER 4400 software and normalized to the value for the input fraction. The ratios (normalized values for bound vs. unbound fractions) were calculated for p16 and p14 primers. The enrichment of the bound fraction for p16 and p14 was calculated from the above ratio and the value was obtained when the antibody was omitted (Fig. 2B, lanes 6, 12, and 15).
Figure 2.
ChIP analysis of the occupancy of p16 and p14 promoters by methyl-CpG binding proteins. (A) A quantitative analysis of PCR products was performed on chromatin immunoprecipitated with various amounts of antibodies against MBD2 and MeCP2 in HCT 15 cells. For the different conditions, the bound (B) and unbound (U) fractions were amplified with primers specific for p16, p14, and Alu. (B) Histograms of the ratio of the bound DNA fraction vs. the unbound DNA fraction normalized to the no antibody value (lane 6, p16; lane 12, p14; lane 15, Alu). Data are the mean ± SD for at least three experiments. (C) As a control experiment, amplification of the bound and unbound fractions was performed in HeLa where p16 and p14 are unmethylated. (D) DNA corresponding to the transcription start site of the BRCA1 gene was amplified with primers for exon 1a (16), after digestion of genomic DNA with HpaII or CfoI (lanes 8 and 9), or after ChIP, on input (lane 7), bound (lanes 1–6, Upper) and unbound (lanes 1–6, Lower) fractions.
When sonicated chromatin was incubated with 2 μl of anti-MBD2 antibodies, we observed a 3.7-fold enrichment of p16 promoter in the bound DNA (Fig. 2 A and B, lanes 1) and a 6-fold enrichment for p14 (Fig. 2 A and B, lanes 7). This enrichment was improved when samples were incubated with 5 μl of anti-MBD2 for p16 (Fig. 2 A and B, lanes 2) and p14 (Fig. 2 A and B, lanes 8). When 0.1% SDS and 2 μl of anti-MBD2 was added, low (Fig. 2 A and B, p16, lanes 3) or negligible amounts of chromatin were precipitated (Fig. 2 A and B, p14, lanes 9), but significant amounts of DNA was amplified when 5 μl of antibody and detergent was added (Fig. 2 A and B, p16, lanes 4, and p14, lanes 10). MBD2 protein binds to both p16 and p14 promoters, but we failed to detect a significant enrichment for the Alu element, located 1.1–1.5 kb upstream of p16, with anti-MBD2 (Fig. 2 A and B, lanes 13) compared with a similar experiment without the antibody (Fig. 2 A and B, lanes 15) or with preimmune serum (data not shown). Moreover, negligible chromatin was collected when a polyclonal anti-MeCP2 was used (Fig. 2 A and B, p16, lanes 5; p14, lanes 11; Alu, lanes 14). Specificity of binding was validated by detecting no enrichment for any sequence analyzed in HeLa cells where neither gene is methylated (Fig. 2C). As a control experiment, we used ChIP to investigate the association of MBD2 and MeCP2 to an unmethylated CpG island in HCT15 cells. In HCT15 cells, the promoter region and first exon of the breast cancer predisposition gene BRCA1 are unmethylated as shown by a PCR-based methylation analysis after digestion of DNA with HpaII or CfoI (Fig. 2D, lanes 8 and 9) and as expected for unmethylated CpG sites, no significant amplification of this region was noticed in the DNA fraction corresponding to bound chromatin (Fig. 2D, lanes 1–6, Upper) compared with the unbound sample (Fig. 2D, lanes 1–6, Lower) or input DNA (Fig. 2D, lane 7).
Thus, the values obtained indicate that immunoprecipitation yielded larger quantities of DNA when 5 μl of antibody was used and that the transcriptional repressor MBD2 binds to the methylated p16 and p14 regulatory regions but not to the Alu element upstream of p16 in colon cancer cell lines where genes are silenced.
Inverse Correlation Between the Presence of MBD2 and Gene Expression or Histone Acetylation.
Transcriptional repression by methyl-CpG binding proteins appears to be a dynamic and complex process involving nuclease-resistant chromatin assembly and histone deacetylation (15). However, acetylation of nucleosomal histones H3 and H4 correlates very closely with transcriptional activity, and many proteins that are transcriptional coactivators are also histone acetyltransferases. To address whether p16 and p14 expression correlates with changes in the status of histone acetylation, we have analyzed the acetylation of histones H3 and H4 and the presence of MBD2 on p14/p16 promoters and the Alu element located nearby p16.
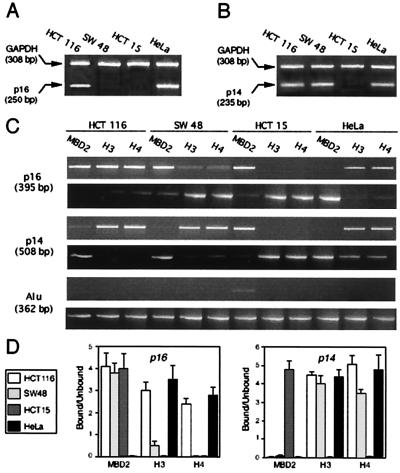
The level of p14 or p16 transcripts was evaluated in the different cell lines by a semiquantitative RT-PCR assay after coamplification of the ubiquitous glycolytic enzyme GAPDH as an internal control. p16 expression was detected in HCT116 cells, where one allele is methylated, and in the HeLa cells, where p16 is unmethylated, but p16 expression was not detected in SW48 and HCT15 cells, where the gene is methylated (Fig. 3A). p14 expression was detected in HCT116, SW48, and HeLa cells but was not detected in HCT15 cells (Fig. 3B). Promoter occupancy by transcriptional coactivators or corepressors also was analyzed by ChIP with antibodies to MBD2 or acetylated H3 and H4 (Fig. 3C). The intensity of the bands in the bound and unbound fractions (Fig. 3C) was measured by densitometry, and the enrichment value for the bound fraction vs. unbound fraction is presented for each cell line (Fig. 3D). Results obtained with ChIPs and RT-PCR were compared, and the expression of p16 (HCT116 and HeLa; Fig. 3A) or p14 (HCT116, SW48, and HeLa; Fig. 3B) correlates with the occupancy of promoters by acetylated histones H3 or H4 (Fig. 3 C and D). Alternatively, the presence of bound MBD2 was associated with DNA hypermethylation and the absence of gene (SW48 and HCT15) or allele (HCT116) expression for p16 or p14 (HCT15). In contrast, we detected no acetylation of histones H3 and H4 for the intergenic Alu element (Fig. 3C). Thus, MBD2 appears to be associated with promoter regions of hypermethylated and silenced p16 or p14 genes and excludes histone hyperacetylation.
Figure 3.
Expression of p16 and p14 and ChIP analysis of MBD2 and acetylated histones H3 and H4 in colon cancer cell lines and HeLa. (A) p16 transcripts were coamplified by RT-PCR, with the GAPDH transcripts as an internal control; 1 μg of total RNA from HCT116, SW48, HCT15, and HeLa cells was used. PCR products (5 μl) were separated by electrophoresis on a 2% agarose gel, and positions of the bands for p16 (250 bp) and GAPDH (308 bp) are indicated on the left. (B) Expression of p14 was analyzed by RT-PCR after coamplification of p14 transcript (235 bp) with GAPDH (308 bp). (C) PCR analysis of DNA in chromatin immunoprecipitated with anti-MBD2 and anti-acetylated histone H3 and H4 for p16 (395 bp), p14 (508 bp), and Alu (362 bp). Experiments were performed on the following cell lines: HCT116 with one allele methylated (M+) for p16 and p14 M−; SW48 with p16 M+ and p14 M−; HCT15 with p16 M+, p14 M+, and HeLa cells with p16 and p14 M−. (D) Statistical analysis (mean ± SD of at least three experiments) of the enrichment of the bound DNA fraction compared with the unbound DNA fraction for p16 and p14, for each antibody and in each cell line.
Chemical Treatment with 5aza-dC and TSA Induces Histone Deacetylation and Chromatin Changes on Hypermethylated Promoters.
DNA methylation and histone deacetylation appear to act synergistically to silence genes in cancer cell lines (13), presumably through a complex chromatin structure where methyl-CpG binding proteins are associated with chromatin-modifying factors. Therefore, the observed association between MBD2 and the methylated p16 and p14 promoters led us to search for chromatin rearrangement, such as changes in acetylation in response to chemical stimuli. Cell lines were treated with 5aza-dC in the presence or in the absence of TSA, and gene expression was compared between untreated and treated cells. Signals for p16 (Fig. 4A) or p14 (Fig. 4C) were normalized to the intensity of the amplification product for GAPDH. The distribution of MBD2 and acetylated histones H3 and H4 on p16 and p14 promoter regions (Fig. 4 B and D) were compared with that for untreated cells (Fig. 3).
Figure 4.
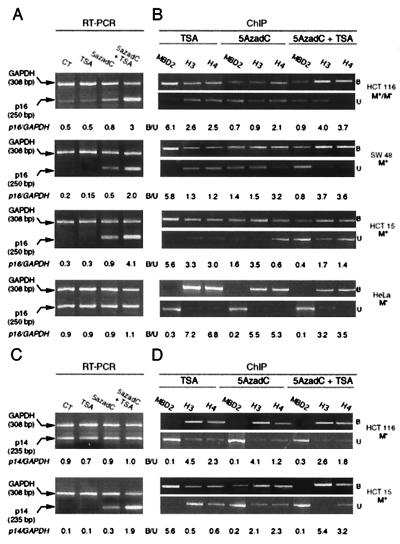
Changes in p16 and p14 promoter occupancy and gene expression after drug treatments. (A) For each cell line, p16 expression was monitored by RT-PCR after coamplification with the GAPDH. The position of the bands is indicated to the right. For each condition, the ratio for p16 vs. GAPDH bands is indicated. (B) ChIP was performed with antibodies for MBD2 or acetylated histones H3 and H4, as indicated, in HCT116, SW48, HCT15, and HeLa cells after treatment with TSA, 5aza-dC, or a combination of both. Bound DNA (B) and unbound DNA (U) fractions were amplified by PCR with primers for the p16 promoter. Ratios (B/U) are below the gel. (C) For each conditions, p14 expression was monitored by RT-PCR (235 bp) after coamplification with the GAPDH (308 bp). The ratio for p14 expression vs. GAPDH is indicated below the gel for each cell line. (D) Bound and unbound DNA–chromatin samples immunoprecipitated with anti-MBD2 or anti-acetylated histones H3 or H4 were amplified with primers for the 3′ end of the p14 CpG island (508 bp) after treatment with TSA, 5aza-dC, or both. Ratios (B/U) are indicated below the gel for each cell line.
In this experiment, TSA alone did not enhance p16 gene expression, but chemically induced demethylation resulted in a 2- to 3-fold increase in expression (Fig. 4A) dependent on the demethylation rate (data not shown). The combination of 5aza-dC and TSA exhibits a 6- to 13-fold increase, suggesting synergistic gene activation. Moreover, we have analyzed the p16 promoter occupancy by MBD2 and acetylated histones H3 and H4 under various conditions for each cell line (Fig. 4B). As expected, TSA treatment can hyperacetylate histones H3 and H4 but did not significantly change promoter occupancy by MBD2. However, the enrichment in hyperacetylated histones was not sufficient to induce gene expression when DNA was methylated (Fig. 4 A and B). Treatment of the colon cell lines with 5aza-dC reduced the levels of MBD2 bound to the p16 promoter and was associated with a slight increase in hyperacetylated histones. Exposure to both drugs increases the recruitment of acetylated histones H3 and H4 and the depletion of MBD2 proteins leading presumably in a complete remodeling of the chromatin architecture at the p16 locus (Fig. 4B).
As observed for p16, demethylation by 5aza-dC increased p14 expression slightly (3-fold) in HCT15 cells and the effects of demethylation became more apparent when 5aza-dC was combined with TSA (18-fold, Fig. 4C). Moreover, by using ChIP, we observed results similar to those described for p16—i.e., the loss of MBD2 binding and an enrichment of acetylated histones H3 and H4 after demethylation and inhibition of histone deacetylases (Fig. 4D).
Thus CpG methylation appears to be the dominant mechanism, compared with histone posttranslational modification, for gene silencing, and DNA demethylation appears to be a prerequisite event for gene reexpression. Moreover, the increase in gene expression associated with the inhibition of deacetylation, demethylation, and the absence of binding of MBD2 suggests that this protein MBD2 operates by recruiting a histone deacetylase corepressor complex to strengthen gene silencing.
Discussion
CpG islands originally were defined as regions of DNA with a G + C content of >0.5 and a frequency of CpG observed versus CpG expected of >0.6 (23). Most CpG islands are associated with promoter regions of housekeeping genes or tissue-specific genes (24). In cancer, inhibition of transcription by hypermethylation contributes to the functional inactivation of growth regulatory (25) or DNA repair (26) genes. Among the genes involved in the control of the cell cycle, aberrant methylation of p16 is well documented in various human neoplasms (12, 22). However, hypermethylation of the CpG island of the p14/ARF gene, initiated close to the p16 gene, is a less common event (11, 12). Two mechanisms could be involved in silencing genes by DNA methylation, a mechanism that prevents the binding of transcription factors to their recognition sequence (27) or a mechanism through which repressor molecules bind to methylated DNA. Several proteins can recognize methylated CpG islands, including the MeCP1 complex (28), MeCP2 (29, 30), and MBD1, 2, 3, and 4 (31). Although all of these proteins can recruit corepressors and histone deacetylases to remodel chromatin (32–36), the precise ways by which they exert their repressure effect in vivo remain unclear.
Using the formaldehyde cross-linking technique, we show in this report that the MBD2 protein binds in vivo to the methylated regulatory regions of p16 and p14 and could thereby contribute to gene silencing in colon carcinoma cell lines. However, we did not detect significant binding of MBD2 to methylated Alu elements that are located between p16 and p14. Although MBD2 and MeCP2 antibodies can recognize their respective antigens in whole-cell extracts or after immunoprecipitation of DNA–protein complexes and deplete the unbound fraction of most target proteins (data not shown), it appears that MeCP2 is not recruited to methylated promoters of p16 or p14. Results from our laboratory using ChIP, demonstrating the binding of MeCP2 to different sequences in vivo (A. El Osta and A.P.W., unpublished results), suggest that different repressive complexes involving different methyl-CpG binding proteins might be targeted to a methylated regulatory region depending on the cell type, sequence specificity, or the presence of other transacting factors.
MBD2 has been identified as a component of the MeCP1 complex (28) and can immunoprecipitate material with histone deacetylase activity and histone binding proteins (35). MBD2 can also interact with nucleosome remodeling histone deacetylase complex, NuRD, thereby, recruiting the Mi2–NuRD complex to methylated DNA (34, 36). With our model, it remains to be established whether MBD2 exerts its repressive effects by interacting with MeCP1, the Mi2–NuRD complex, or another unidentified complex, depending on the cell type, the differentiation status, or proliferation status. In this report, we examined the effects of the hypomethylating agent 5aza-dC, the histone deacetylase inhibitor TSA, or both drugs on MBD2 binding and acetylation of histones H3 and H4 in the p16 and p14 regulatory region. Silenced genes are not reactivated by TSA treatment, as observed for the β polymerase gene (35), whereas the synergic action of 5aza-dC and TSA suggests cooperation between methylation and deacetylation as described (13). These data strongly suggest that the repressive action of the MBD2 complex operates through a histone deacetylase-dependent pathway, where DNA methylation plays a dominant function in gene silencing. For p16 and/or p14, CpG island methylation presumably leads to a localized repressive nucleosomal structure that prevents the initiation of transcription (Fig. 5). However, the presence downstream of a highly methylated p16 promoter has little effect on the expression of p14, indicating that the repressive structure does not affect the elongation of transcription, as observed (37).
Figure 5.
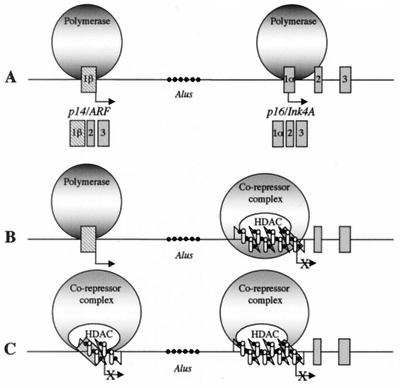
Model for the methylation-dependent silencing of p14 and p16 in colon cancer. (A) Normally, p14/ARF and p16/Ink4A are transcribed from different promoters and different first exons (exon 1β for p14 and 1α for p16) but share exons 2 and 3. The presence of methylated Alu elements between the two genes does not inhibit the elongation of transcription. ●, Methyl-CpG sites. (B) In cancer cells where p16 is hypermethylated and silenced, methyl-CpG binding proteins MBD2 (open cylinders) are recruited to the methylated CpG island containing the transcription start site and remodel the chromatin structure into an inactive state. Transcriptional silencing can be relieved by the histone deacetylase inhibitor TSA, suggesting a mechanism of gene repression that involves methyl-CpG binding proteins and a multisubunit chromatin-remodeling complex containing histone deacetylases (HDAC). The condensed region does not inhibit the expression of the p14/ARF gene initiated 20 kb upstream of p16/Ink4A. (C) p14/ARF also can be methylated in colon cancer cells. MBD2 interacts with the methylated region and directs the chromatin-remodeling complex to methylated promoters. The absence of MBD2 proteins associated with the methylated Alu elements indicates that repressor complexes containing the methyl-CpG binding protein are targeted to regulatory regions, suggesting a in vivo role in the silencing of genes by DNA methylation.
In HCT15 cells, the hypermethylated area is restricted to a small region at the 3′ end of the p14/ARF CpG island that do not extend beyond position −450 relative to the transcription start site (11). Moreover, the CpG island of p15 located upstream of the p14 promoter remains unmethylated in colorectal tumors but is methylated in hematological malignancies (5). The description of these localized hypermethylated areas at the p15/p14/p16 locus and our results on the specific distribution of the transcriptional repressor MBD2 demonstrate that the methylation-dependent silencing is a selective epigenetic alteration in human cancers; however, the mechanisms that turn off the genes remain obscure. Three functional DNA methyltransferases have been identified in mammalian cells, but little is known of their role in the changes in methylation observed in cancer. DNA methyltransferase 1 (DNMT1) interacts with corepressors and histone deacetylases (38–40). In malignant cells, the loss of function of transacting factors, the artificial presence of DNMT1 at CpG islands, and the addition of methyl groups could be one of the mechanisms that recruits methyl-CpG binding proteins to translate this aberrant epigenetic signal into a repressive chromatin structure.
To date, only a few examples of gene repression by methyl-CpG binding proteins have been reported in vitro (35, 41, 42), and, consequently, identification in vivo of such proteins binding to regulatory sequences of specific chromatin modifiers is a major task in the understanding of gene silencing. Therefore, further studies are needed to investigate this mode of regulation for other tumor suppressor genes and to identify the proteins involved in the corresponding repressive complex. Furthermore, the dynamic of histone acetylation could provide an attractive model for the reversible repression of genes hypermethylated in human pathologies.
Acknowledgments
The antibodies against MeCP2 were provided by Dr. Peter L. Jones. We thank Dr. P. Wade for providing MBD2 antibodies and for his advice on ChIPs experiments and Timur Yusufzai for his critical reading of the manuscript. F.M. is the recipient of a fellowship from L'Association pour la Recherche sur le Cancer (France).
Abbreviations
- ChIP
chromatin immunoprecipitation
- 5aza-dC
5-aza-2′-deoxycytosine
- TSA
trichostatin A
- RT-PCR
reverse transcriptase–PCR
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 3.Kamb A, Gruis N A, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian S V, Stockert E, Day R S, 3rd, Johnson B E, Skolnick M H. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 4.Stone S, Jiang P, Dayananth P, Tavtigian S V, Katcher H, Parry D, Peters G, Kamb A. Cancer Res. 1995;55:2988–2994. [PubMed] [Google Scholar]
- 5.Ruas M, Peters G. Biochim Biophys Acta. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 6.Nobori T, Miura K, Wu D J, Lois A, Takabayashi K, Carson D A. Nature (London) 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- 7.Duro D, Bernard O, Della Valle V, Berger R, Larsen C J. Oncogene. 1995;11:21–29. [PubMed] [Google Scholar]
- 8.Gardie B, Cayuela J M, Martini S, Sigaux F. Blood. 1998;91:1016–1020. [PubMed] [Google Scholar]
- 9.Foster S A, Wong D J, Barrett M T, Galloway D A. Mol Cell Biol. 1998;18:1793–1801. doi: 10.1128/mcb.18.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nuovo G J, Plaia T W, Bellinsky S A, Baylin S A, Herman J G. Proc Natl Acad Sci USA. 1999;96:12754–12759. doi: 10.1073/pnas.96.22.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson K D, Jones P A. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteller M, Tortola S, Toyota M, Capella G, Peinado M A, Baylin S B, Herman J G. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- 13.Cameron E E, Bachman K E, Myohanen S K, Herman J G, Baylin S B. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 14.Coffee B, Zhang F, Warren S T, Reines D. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 15.Bird A P, Wollfe A P. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 16.Magdinier F, Ribieras S, Lenoir G M, Frappart L, Dante R. Oncogene. 1998;17:3169–3176. doi: 10.1038/sj.onc.1202248. [DOI] [PubMed] [Google Scholar]
- 17.Dante R, Ribieras S, Baldassini S, Martin V, Benzerara O, Bouteille C, Bremond A, Frappart L, Rio M C, Lasne Y. Lab Invest. 1994;71:188–192. [PubMed] [Google Scholar]
- 18.Pao M M, Liang G, Tsai Y C, Xiong Z, Laird P W, Jones P A. Oncogene. 2000;19:943–952. doi: 10.1038/sj.onc.1203414. [DOI] [PubMed] [Google Scholar]
- 19.Myohanen S K, Baylin S B, Herman J G. Cancer Res. 1998;58:591–593. [PubMed] [Google Scholar]
- 20.Yoder J A, Walsh C P, Bestor T H. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 21.Wolffe A P, Matze M A. Science. 1999;282:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalgo M L, Hayashida T, Bender C M, Pao M M, Tsai Y C, Gonzales F A, Nguyen H D, Nguyen T T, Jones P A. Cancer Res. 1998;58:1245–1252. [PubMed] [Google Scholar]
- 23.Gardiner-Garden M, Frommer M. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 24.Bird A P. Nature (London) 1986;21:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 25.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J P. Adv Cancer Res. 1997;72:141–196. [PubMed] [Google Scholar]
- 26.Herman J G, Umar A, Polyak K, Graff J R, Ahuja N, Issa J P, Markowitz S, Willson J K, Hamilton S R, Kinzler K W, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tate P H, Bird A P. Curr Biol. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 28.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Cell. 1991;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 29.Lewis J D, Meehan R R, Henzel W J, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 30.Nan X, Campoy J, Bird A. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 31.Hendrich B, Bird A. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 33.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 34.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Nat Genet. 1999;1:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 35.Ng H H, Zhang Y, Hendrich B, Johnson C A, Turner B M, Erdjument-Bromage H E, Tempest P, Reinberg D, Bird A. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones P A. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 38.Fuks F, Burgers W A, Brehm A, Hughes-Davies L, Kouzarides T. Nat Genet. 2000;1:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 39.Rountree M R, Bachman K E, Baylin S B. Nat Genet. 2000;3:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 40.Robertson K D, Ait-Si-Ali S, Yokochi T, Wade P A, Jones P L, Wolffe A P. Nat Genet. 2000;3:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 41.Fujita N, Shimotake N, Ohki I, Chiba T, Saya H, Shirakawa M, Nakao M. Mol Cell Biol. 2000;14:5107–5118. doi: 10.1128/mcb.20.14.5107-5118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magdinier F, Billard L M, Wittmann G, Frappart L, Benchaïb M, Lenoir G M, Guérin J F, Dante R. FASEB J. 2000;14:1585–1594. doi: 10.1096/fj.14.11.1585. [DOI] [PubMed] [Google Scholar]