Like mammals, S. pombe has just one member of the Pif1 family of DNA helicases. Zakian and colleagues now report that this S. pombe member, Pfh1, is required for efficient replication fork progression through sites that are naturally difficult to replicate. This includes highly transcribed Pol III and also, interestingly, Pol II genes.
Keywords: Pif1 family, DNA helicase, DNA replication, Schizosaccharomyces pombe, swi1, natural fork barriers
Abstract
Replication forks encounter impediments as they move through the genome, including natural barriers due to stable protein complexes and highly transcribed genes. Unlike lesions generated by exogenous damage, natural barriers are encountered in every S phase. Like humans, Schizosaccharomyces pombe encodes a single Pif1 family DNA helicase, Pfh1. Here, we show that Pfh1 is required for efficient fork movement in the ribosomal DNA, the mating type locus, tRNA, 5S ribosomal RNA genes, and genes that are highly transcribed by RNA polymerase II. In addition, converged replication forks accumulated at all of these sites in the absence of Pfh1. The effects of Pfh1 on DNA replication are likely direct, as it had high binding to sites whose replication was impaired in its absence. Replication in the absence of Pfh1 resulted in DNA damage specifically at those sites that bound high levels of Pfh1 in wild-type cells and whose replication was slowed in its absence. Cells depleted of Pfh1 were inviable if they also lacked the human TIMELESS homolog Swi1, a replisome component that stabilizes stalled forks. Thus, Pfh1 promotes DNA replication and separation of converged replication forks and suppresses DNA damage at hard-to-replicate sites.
DNA replication is a fundamental process that must occur with high efficiency and precision. Schizosaccharomyces pombe is an excellent model for replication studies because its genome organization is very similar to higher eukaryotes. In eukaryotes, replication initiates at multiple origins along the chromosome, with replication proceeding bidirectionally from these origins. However, a few chromosomal regions are replicated unidirectionally due to naturally occurring replication fork barriers (RFBs) that pause or stall replication forks moving in one direction through the site. The ribosomal DNA (rDNA) and the mating type locus are two S. pombe genomic locations that replicate unidirectionally (Sanchez et al. 1998; Dalgaard and Klar 2000; Krings and Bastia 2004). Both regions contain cis-acting sequences that are maintained by the nonnucleosomal Swi1/Swi3 (human TIMELESS/TIPIN) complex and function as natural RFBs (Dalgaard and Klar 2000; Krings and Bastia 2004). The S. pombe Swi1/Swi3 complex migrates with the replisome, and the human TIMELESS/TIPIN homologs interact with replisome components (Noguchi et al. 2004; Gotter et al. 2007).
S. pombe rDNA, which is located in two clusters, with one at each end of chromosome III, consists of 100–150 copies of 10.9-kb rDNA repeats. The RFBs in each rDNA repeat pause replication forks moving in the direction opposite of transcription, ensuring that transcription and replication move in the same direction through these highly transcribed genes (Sanchez et al. 1998). Sap1 and Reb1 are two additional rDNA RFB-binding proteins (Sanchez-Gorostiaga et al. 2004; Krings and Bastia 2005; Mejia-Ramirez et al. 2005). There are four RFBs in each rDNA repeat: Ter1, Ter2, Ter3, and RFB4. Reb1 binds Ter2–3, and Sap1 binds Ter1. sap1+, but not reb1+, is required for cell viability (Arcangioli et al. 1994). The pausing at Ter1–3 is abolished in swi1+ or swi3+ mutant cells (Krings and Bastia 2004), but the requirements for fork pausing at RFB4 have not been characterized. Like Xenopus laevis and human cells (Little et al. 1993; Wiesendanger et al. 1994), the barrier activity at rDNA in S. pombe (Sanchez et al. 1998) is not as efficient as in Saccharomyces cerevisiae (Brewer and Fangman 1988; Linskens and Huberman 1988). In addition, similar to human cells, S. pombe 5S ribosomal RNA (rRNA) genes are not located in the rDNA cluster but are dispersed throughout all three chromosomes (Wood et al. 2002).
The mat1 locus, located on chromosome II, encodes the mating type of the cell. At this locus, replication progresses from the telomeric side of mat1 toward the centromere. Replication from the opposite direction is blocked by the Swi1, Swi3, Rtf1, and Rtf2 complex bound to the replication termination sequence (RTS1), which allows proper imprinting during mating type switching (Dalgaard and Klar 2001). In S. cerevisiae, replication forks also show modest fork pausing at tRNA genes (Deshpande and Newlon 1996) and highly transcribed RNA polymerase II (Pol II) genes (Azvolinsky et al. 2009). All of these replication pause sites—the rDNA RFB, the 5S rRNA genes, the mating type RTS1, tRNA genes, and highly transcribed Pol II genes—are associated with stable protein complexes.
Pif1 enzymes comprise a 5′–3′ DNA helicase family that is found in essentially all eukaryotes and some prokaryotes (for review, see Bochman et al. 2010, 2011). These helicases perform major roles in both nuclear and mitochondrial DNA maintenance. S. cerevisiae and several other fungi encode two Pif1 members. In contrast, S. pombe, like humans and most other metazoans, encodes a single Pif1 family helicase, Pfh1. So far, most of the information on Pif1 family helicases comes from studies on the two S. cerevisiae Pif1 proteins, ScPif1 and ScRrm3. Although ScPif1 and ScRrm3 affect replication of many of the same substrates, their effects on these substrates are different and sometimes even opposing. For example, at the rDNA, ScPif1 impedes, while ScRrm3 promotes, fork progression through the RFB (Ivessa et al. 2000). Likewise, ScPif1, but not ScRrm3, is important for maintenance of mitochondrial DNA (Foury and Kolodynski 1983; O'Rourke et al. 2005; Cheng et al. 2007, 2009). However, deletion of RRM3 in pif1Δ cells partially suppresses the high loss rate of mitochondrial DNA that occurs in the absence of ScPif1 (O'Rourke et al. 2005). ScRrm3 promotes semiconservative replication at telomeres, while ScPif1 inhibits telomerase (Zhou et al. 2000; Ivessa et al. 2002). ScRrm3 also promotes fork progression at other sites where ScPif1 is not known to act, such as at tRNA genes, inactive origins, and the silent mating type loci, while ScPif1 is important for replication through G-rich motifs that can form G-quadruplex structures in vitro (Ivessa et al. 2003; Paeschke et al. 2011). Human Pif1 (hPif1) also unwinds G-quadruplex structures in vitro, as well as DNA structures that resemble stalled replication forks (George et al. 2009; Sanders 2010). Although S. cerevisiae Pif1 family helicases have important roles in DNA replication, pif1Δ rrm3Δ cells are viable (Ivessa et al. 2000). In contrast, S. pombe pfhl+ is essential in both mitochondria and nuclei because of its key function in replication of both mitochondrial and nuclear DNA (Tanaka et al. 2002; Zhou et al. 2002; Pinter et al. 2008). Thus, an outstanding question in the field is how a single Pif1 family helicase performs roles that have been separated and often function in opposition to each other in S. cerevisiae. This question is compelling given the recent finding that several families with a high incidence of breast cancer carry hPIF1 mutations in a highly conserved residue (Chisholm et al. 2012). Moreover, the same mutation in Pfh1 results in a stable but inactive protein that is unable to provide either its nuclear or mitochondrial functions (Chisholm et al. 2012).
Although Pfh1 is required for chromosome replication (Pinter et al. 2008), its mechanism of action is unknown. Here, we tested the hypothesis that Pfh1 promotes chromosomal DNA replication at specific hard-to-replicate sites, such as RFBs and highly transcribed RNA Pol II and Pol III genes. We show that replication at all of these sites was impaired in Pfh1-depleted cells. We provide evidence that it is the stable protein complexes at the RFBs within the mating type locus and rDNA that made replication Pfh1-sensitive. Moreover, in the absence of Pfh1, genome integrity was compromised at those sites where its activity was needed for efficient replication, as DNA damage increased at these sites in Pfh1-depleted cells but not at sites that did not show Pfh1-sensitive DNA replication. These data support the hypothesis that Pfh1 has a ScRrm3-like role in promoting fork movement past stable protein complexes, thereby facilitating semiconservative DNA replication. However, Pfh1 is so far unique among eukaryotic DNA helicases in being required for efficient replication fork progression through highly transcribed RNA Pol II genes.
Results
Pfh1 binds to all tested DNA sequences but shows particularly high binding at sites known to inhibit fork progression
Because ScRrm3 travels with the replication fork (Azvolinsky et al. 2006), it binds to all sites in nuclear DNA, but its binding is enriched at the subset of sites where replication is slowed in its absence (Azvolinsky et al. 2009). In contrast, ScPif1 does not move with the fork but, rather, is recruited to its sites of action (Paeschke et al. 2011). To determine the pattern of Pfh1 binding in a quantitative manner, we epitope-tagged Pfh1, cross-linked asynchronously growing cells with formaldehyde, and performed chromatin immunoprecipitations (ChIP) in combination with quantitative PCR (qPCR). As a control, we carried out the same experiment in a strain that did not contain an epitope-tagged protein (no-tag control).
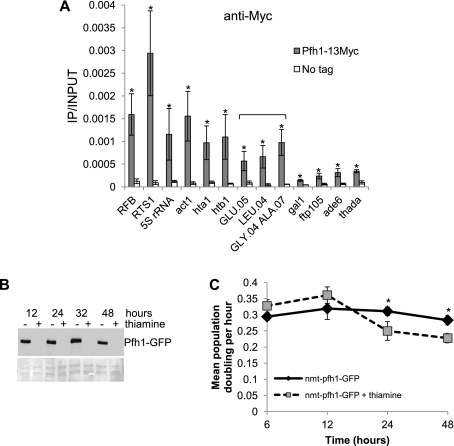
Because S. pombe replication forks slow at rDNA, mating type loci, and telomeres (Sanchez et al. 1998; Dalgaard and Klar 2000; Krings and Bastia 2004; Miller et al. 2006), and at a tRNA gene inserted within ade6+ (Pryce et al. 2009), we speculated that Pfh1 might affect replication at these sites. Therefore, we examined Pfh1 binding to the rDNA RFB, 5S rRNA genes, the mating type locus RFB (RTS1), and four tRNA genes (the role of Pfh1 at telomeres will be described elsewhere) (Fig. 1A). We also examined Pfh1 binding to seven loci (act1+, gal1+, hta1+, htb1+, thada, ade6+, and ftp105+) that are not known to be replication fork pause sites in S. pombe (Fig. 1A). Compared with the no-tag control, Pfh1 was significantly enriched at each of the tested sites. However, Pfh1 binding was particularly high at the RTS1 and low at the gal1+ gene (Fig. 1A). Four of the anticipated negative controls (gal1+, thada, ade6+, and ftp105+) had low binding compared with loci that are known or predicted to cause fork pausing. However, three of these sites (act1+ and the two histone genes hta1+ and htb1+) had high binding. All three of these genes are highly transcribed by RNA Pol II (Wilhelm et al. 2008).
Figure 1.
Pfh1 binds preferably to natural RFBs. (A) Samples from asynchronous cells from an untagged control and Pfh1-13Myc strain were chromatin-immunoprecipitated using an anti-Myc antibody. The immunoprecipitated DNA was analyzed using qPCR and is presented as immunoprecipitated DNA divided by input DNA. The immunoprecipitated DNA for tRNA genes (noted by bracket) was analyzed using qPCR with primers specific for just outside each tRNA gene. Primers for 5S rRNA genes recognize both 5S rRNA.03 and 5S rRNA.04 genes. Data represent the mean of three independent cultures; error bars indicate one standard deviation. (*) P < 0.05 was determined by two-tailed Student's t-test. thada is the name for SPCC1494.07. (B) TCA-precipitated protein samples from nmt-pfh1-GFP collected after 12, 24, 32, and 48 h growth in the presence (+) or absence (−) of thiamine. Samples were loaded on a 6% polyacrylamide gel. The gels were immunoblotted with anti-Pfh1 antibody (top) and later stained with Ponceau S to examine loading and transfer of protein samples (bottom). (C) Growth curves showing population doublings in cultures growing on EMMS (black solid line) or EMMS containing 30 μM thiamine (gray dashed line). (*) P < 0.05 determined by two-tailed Student's t-test for 24-h and 48-h growth.
The S. pombe genome encodes 169 tRNA genes (Wood et al. 2002, 2011). Because there are multiple copies of many tRNA genes, we amplified the unique DNA immediately adjacent to the tRNA gene rather than the genes themselves. Each of the four tRNA genes had robust Pfh1 binding compared with the no-tag control strain and the gal1+ gene (Fig. 1A), but Pfh1 binding to tRNA genes was not as high as to the RFBs, RTS1, or act1+ loci (Fig. 1A). We conclude that Pfh1 binds all of the tested sites, although some sites such as the rDNA, the mating type locus, act1+, histone, 5S, and tRNA genes have particularly high binding.
Replication at the rDNA RFBs is hindered in Pfh1-depleted cells due to stable DNA–protein complexes
Sites with high Pfh1 binding are candidates for loci that require Pfh1 for efficient replication. We examined replication through these sites in Pfh1-depleted and wild-type cells using two-dimensional (2D) gel analysis, which separates replication intermediates by size in the first dimension and by shape (and size) in the second dimension (Brewer and Fangman 1987). DNA samples were restriction enzyme-digested using information on the positions of ARS elements in the region being examined (Heichinger et al. 2006). This genome-wide study identified 401 strong origins and 503 weaker origins and calculated their firing efficiency. However, origins in S. pombe and higher eukaryotes do not have a clear consensus sequence and are difficult to map (Dubey et al. 1996; Feng et al. 2006; Heichinger et al. 2006). In some cases, the positions of active ARS elements in our experiments, as inferred from the pattern of replication intermediates in 2D gels, did not agree with results from the published study. These differences might be due to inherent difficulties mapping S. pombe origins or the different growth conditions used in our experiments, as in the other study, cells were grown at 25°C with or without hydroxyurea.
In addition to Pfh1's enrichment at the RFB in rDNA (Fig. 1A), Pfh1 is concentrated in the nucleolus (Pinter et al. 2008). Because pfh1Δ cells are inviable, in this and subsequent experiments, we used the repressible nmt81 promoter to regulate Pfh1 expression (Pinter et al. 2008). Addition of thiamine to a nmt-pfh1-GFP culture represses Pfh1 expression, so that after 12 h, Pfh1 was no longer detected by Western blot analysis (Fig. 1B; Pinter et al. 2008). In addition, population doubling time was increased after 24 h of Pfh1 depletion (Fig. 1C; Pinter et al. 2008).
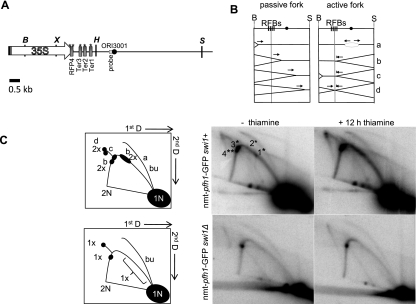
To determine the pattern of replication fork progression in cells that were not yet growth-arrested but had reduced Pfh1, we carried out 2D gel analysis on genomic DNA isolated from the nmt-pfh1-GFP strain grown for 12 h in minimal medium with or without thiamine. The pattern of replication intermediates seen in cells grown without thiamine (Pfh1 expressed) was similar to published results (Krings and Bastia 2004), except that pausing at Ter2, which is the weakest RFB in the rDNA (Krings and Bastia 2004), was barely detectable (Fig. 2). In Pfh1-depleted cells, pausing in the RFB region was increased twofold, and pausing at Ter1 and Ter2 appeared as a smear on the arc of replication instead of as discrete pause sites (Fig. 2C). Converged forks (X-shaped structures) also increased twofold compared with nondepleted cells (Fig. 2). These data suggest that Pfh1 activity is needed for normal fork progression and for resolving converged forks within rDNA.
Figure 2.
Pfh1 facilitates replication fork progression through rDNA in a Swi1-dependent manner. (A) Schematic of the rDNA region used for 2D gel analysis (here and elsewhere, schematics are drawn to scale). (X) XhoI; (H) HindIII; (B) BglII; (S) SalI recognition sites. Each repeat contains an origin of replication (ORI3001) (black circle). The RFB region is located between the XhoI and HindII sites. Swi1-dependent (*) and Swi1-independent (**) pause sites are indicated above each RFB. (B) Panels represent different replication intermediates during replication of rDNA. Replication forks migrating leftward from the origin are blocked by the RFB, while replication forks migrating in the same direction as transcription are not affected. (C, left) Cartoon representation of 2D gel analysis for DNA isolated from nmt-pfh1-GFP or nmt-pfh1-GFP swi1Δ cells. Letters on the arc indicate the replication intermediates shown in the panels in B. Here and in other figures: (1N) Nonreplicating DNA fragments; (2N) almost fully replicated DNA fragments; (bu) bubble arc; brackets mark quantified regions outside the pause sites. The BglII (B) SalI (S) fragment used for the 2D gel analysis is 7.4 kb. (Right) 2D gel analysis of nmt-pfh1-GFP or nmt-pfh1-GFP swi1Δ cells grown in the presence of thiamine for 0 or 12 h. Genomic DNA was digested with BglII and SalI. (*) Position of Swi1-dependent RFBs; (**) Swi1-independent RFBs. Numbers indicate RFBs ([1] Ter1; [2] Ter2; [3] Ter3; [4] RFP4). The gels were quantified in ImageQuant, and the numbers in the cartoon represent the fold differences between the 0-h and 12-h thiamine conditions. The fold difference is the average of two independent biological replicates.
To investigate whether the increased pausing at the RFBs is protein-dependent, we isolated DNA and carried out 2D gel analysis on DNA from nmt-pfh1-GFP swi1Δ cells grown in minimal medium with thiamine for 0 or 12 h. Swi1 stabilizes Ter1, Ter2, and Ter3 but not RFP4 (Fig. 2A; Krings and Bastia 2004). Pausing at Ter1–Ter3 was absent in swi1Δ cells, while pausing at RFP4 was not affected (Fig. 2C). In addition, pausing at the other RFBs and converged forks was not increased in swi1Δ Pfh1-depleted cells (Fig. 2C). These data suggest that the protein complexes bound to the RFBs, rather than the RFB sequence, confer Pfh1 sensitivity to replication through these sites. In addition, replication intermediates were equally abundant in other regions of the rDNA fragment (denoted by bracket in schematic, Fig. 2C) in the presence or absence of Pfh1, suggesting that Pfh1 does not promote replication through all regions of the genome.
Pfh1 facilitates replication fork movement at 5S rRNA genes
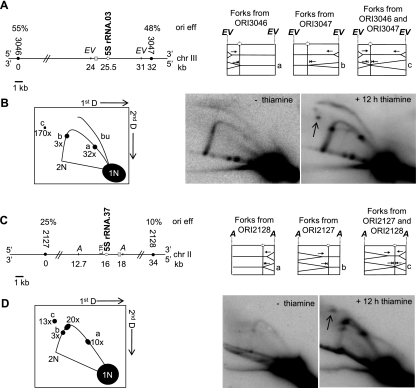
There are 33 5S rRNA genes dispersed throughout the S. pombe genome (Wood et al. 2011). To study replication fork progression through these genes, we examined EcoRV-digested DNA from nmt-pfh1-GFP cells grown in the presence or absence of thiamine and subjected it to 2D gel analysis. We used a hybridization probe close to 5S rRNA.03 located on chromosome III, which is reported to be ∼7 kb away from an efficient origin (ORI3046, 48% firing efficiency) (Fig. 3A). However, we detected a bubble arc indicative of an active origin within the fragment being examined, which was not reported in the origin mapping study (Fig. 3B; Heichinger et al. 2006). Thus, either ORI3047 is located in the fragment, or a previously unidentified origin is located in this EcoRV DNA fragment.
Figure 3.
Pfh1's activity is needed at 5S rRNA genes. (A, left) Schematic image of 5S rRNA.03 on chromosome III. Here and in other figures, origins of replication (closed circles) and origin efficiencies (ori eff) are from Heichinger et al. (2006). Here and in other figures, gray boxes indicate hybridization probes. (Right) Panels to the right illustrate replication intermediates formed during replication of 5S rRNA.03. Here and in other figures, lowercase letters mark these intermediates. (B, left) Cartoon of 2D gels analysis of 6-kb DNA EcoRV fragment. Letters indicate replication intermediates in A. (Right) 2D gel analysis from DNA isolated from nmt-pfh1-GFP cells grown in the presence of thiamine for 0 or 12 h. (Arrow) Converged forks. The fold change shown below each letter in the cartoon ([a] 32-fold; [b] threefold; [c] 170-fold) is the average difference between the 0-h and 12-h time points and was calculated from two independent biological replicates. (C, left) Schematic of 5.3-kb AgeI fragment containing the 5S rRNA.37 region from chromosome II. (A) AgeI site. (Right) Schematic showing expected replication intermediates during replication of the 5S rRNA.37 gene. (D, left) Cartoon of 2D gel analysis indicates replication intermediates shown in the panels on the top. (Right) 2D gel analysis of nmt-pfh1-GFP cells grown in the presence of thiamine for 0 or 12 h. Three discrete pause sites were visualized in Pfh1-depleted cells. We suggest that two of these correspond to either paused leftward-moving forks from ORI2128 or paused rightward-moving forks from ORI2127 at the 5S rRNA gene. The third one could be a pause site at the LTR positioned 600 bp downstream from the 5S rRNA gene, almost in the middle of the AgeI fragment. There were 10-fold (a) and threefold (b) increased pausing at the 5S rRNA gene after 12 h of Pfh1 depletion compared with the nondepleted conditions. The level of converged forks was 13-fold higher (c) in Pfhl-depleted cells versus nondepleted cells.
Although there was little evidence for pausing at the 5S rRNA gene in the presence of Pfh1 (−thiamine), there were two sites of discrete pausing in Pfh1-depleted cells. One pause mapped to the site expected if the gene was replicated by forks initiating at ORI3047 (Fig. 3A,B, position marked with b), in which case, replication moves through the gene in the same direction as transcription. The signal at this site was about threefold higher in Pfh1-depleted (+12 h thiamine) compared with nondepleted cells. We also detected pausing in Pfh1-depleted cells at the position expected for the 5S rRNA gene if it is replicated from forks initiating at ORI3046 (Fig. 3A,B, position marked with a). In this case, forks move in the opposite direction as transcription through the 5S gene. At this site, pausing was 32-fold higher compared with nondepleted cells. In addition, we detected converged forks (Fig. 3B, indicated with an arrow and c) that were much higher (170-fold higher) in the absence of Pfh1 (Fig. 3B). We also observed Pfh1-dependent replication fork progression through a second 5S gene, 5S rRNA.37, located on chromosome II (Fig. 3C,D). These data suggest that Pfh1-sensitive replication is a general feature of 5S rRNA genes.
Replication forks slow as they move through tRNA genes, and this pausing is increased in the absence of Pfh1
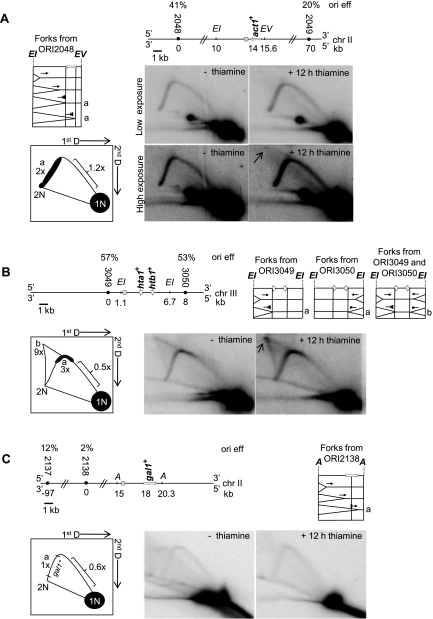
We also examined replication of five tRNA genes in Pfh1-depleted cells (Fig. 4; Supplemental Fig. S1). The tRNAGLU.05 gene, which is on chromosome II, is ∼1 kb from an efficient origin (ORI2026, 65% efficiency) (Fig. 4A; Heichinger et al. 2006). Because the next nearest origin (ORI2025) is 67 kb away and has a firing frequency of only 32%, this tRNA gene will almost always be replicated from a fork initiated downstream from the gene. Thus, replication and transcription move in opposite directions through tRNAGLU.05, an arrangement that causes fork pausing at tRNA genes in wild-type S. cerevisiae (Deshpande and Newlon 1996; Ivessa et al. 2003). Indeed, pausing was detected at tRNAGLU.05 even in the presence of Pfh1 (Fig. 4B). However, this pausing was increased threefold in Pfh1-depleted cells (Fig. 4B). Structures with the mobility of broken (Fig. 4B, fb) and converged (Fig. 4B, arrow) forks were also detected, but only in the absence of Pfh1 (Fig. 4B). We did not detect a bubble arc, suggesting that ORI2026 is either dormant or located outside the fragment being studied (Fig. 4B). Alternatively, this fragment could be replicated from forks initiating at ORI2027 (efficiency 37%).
Figure 4.
Pfh1 promotes fork progression through tRNA genes. (A, left) Schematic of genomic region surrounding the tRNAGLU.05 gene on chromosome II. (EV) EcoRV. (Right) Schematic of replication intermediates formed during replication of tRNAGLU.05. (B, left) Cartoon of 2D gel analysis. The letters on the cartoon illustrate replication intermediates shown in A. DNA digested with EcoRV yields a 6.2-kb DNA fragment. (fb) Broken forks. (Right) The 2D gel analysis of tRNAGLU.05 from nmt-pfh1-GFP cells grown in the presence of thiamine for 0 (−thiamine) or 12 h (+12 h thiamine). (Arrow) Converged forks. (C) tRNAASN.05 on chromosome III shown as in A. (EI) EcoRI; (EV) EcoRV. EcoRI/EcoRV-digested DNA yields a 6.1-kb fragment. (D) 2D gel analysis of tRNAASN.05 as in B.
tRNAASN.05 is located on chromosome III between ORI3035 (efficiency 52%, 12 kb upstream of the gene) and ORI3036 (efficiency 45%, 23 kb downstream from the gene) (Fig. 4C). Therefore, replication through tRNAASN.05 is expected to occur more frequently from “left to right” or in the same direction as transcription. Again, we detected two pause sites in the arc of replication intermediates that were both enhanced in the absence of Pfh1 (Fig. 4D). Forks migrating from ORI3036 and proceeding through the gene in the opposite direction of transcription had ninefold higher pausing (Fig. 4D, indicated by b), while forks from ORI3035 that move in the same direction as transcription increased threefold (Fig. 4D, indicated by a), compared with wild-type cells. Additionally, converged forks were increased eightfold in the absence of Pfh1 (Fig. 4D, marked as c in schematic 2D gel and by arrow in 2D gel). We observed similar results for three other tRNA genes (Supplemental Fig. S1). Together, these data indicate that Pfh1 affects replication fork progression through tRNA genes regardless of whether replication proceeds in the same or opposite direction as transcription, although tRNA genes that are replicated and transcribed in opposite directions are more Pfh1-dependent. In addition, the presence of Pfh1 did not affect the abundance of replication intermediates in regions of the restriction fragments that did not contain tRNA genes (Fig. 3B,D, denoted by brackets in schematic), again supporting the interpretation that Pfh1 acts in a locus-specific manner to affect replication fork progression.
Highly expressed RNA Pol II transcribed genes slow replication forks, and this slowing is increased in Pfh1-depleted cells
In S. cerevisiae, highly transcribed RNA Pol II genes slow progression of replication forks (Prado and Aguilera 2005; Azvolinsky et al. 2009), but this slowing is not enhanced in pif1Δ or rrm3Δ cells. act1+ is a highly expressed gene (Wilhelm et al. 2008) and displayed high Pfh1 binding by ChIP (Fig. 1A). Therefore, we used 2D gel analysis to determine whether replication forks pause within act1+. In the presence of Pfh1, there were more replication intermediates within the portion of the DNA fragment containing the act1+ ORF than in the rest of the fragment. Moreover, in the absence of Pfh1, this pausing was increased twofold (Fig. 5A, marked as a), while replication forks were not increased in the rest of the fragment (1.2-fold change in region marked by bracket in Fig. 5A). Converged forks were also detected (indicated by arrow in Fig. 5A).
Figure 5.
Pfh1 facilitates fork progression at highly transcribed RNA Pol II genes. (A, top) Schematic of act1+ gene. (EI) EcoRI; (EV) EcoRV sites. (Middle left) Schematic of replication intermediates migrating from the closest and most efficient origin toward the act1+ gene. (Bottom left) Cartoon of the 2D gels showing forks slowed within act1+ (marked as a). Pausing in this region was twofold higher after 12 h of Pfh1 depletion compared with nondepleted cells. (Middle and bottom right) 2D gel analysis of EcoRI/EcoRV-digested DNA, which produces a 5.6-kb fragment containing the act1+ ORF prepared from nmt-pfh1-GFP cells grown in thiamine for 0 or 12 h, hybridized to a probe for that region. Both a low and a high exposure are shown. (Arrow) Converged forks that were visualized at higher exposure. (B, top left) Schematic of hta1+ and htb1+ genes on chromosome III. (Top right) Panels showing expected replication intermediates, pause sites, and converged forks are marked with a or b. (Bottom left) Cartoon of 2D gel analysis of hta1+ and htb1+ digested with EcoRI, which produces a 5.6-kb fragment with hta1+ and htb1+. (b) Converged forks; (a) pause sites; (arrow) converged forks. The fold differences after 12 h of Pfh1 depletion compared with the nondepleted conditions are stated below each letter in the cartoon. (C, top left) Schematic of gal1+ region on chromosome II. (A) AgeI. AgeI digestion generates a 5.3-kb fragment containing the gal1+ ORF. (Top right) Schematic showing expected replication intermediates during replication of the gal1+ gene. (Bottom left) Cartoon of 2D gel analysis. (Bottom right) 2D gel analysis of nmt-pfh1-GFP cells grown in the presence of thiamine for 0 or 12 h. The gels were visualized with a gal1+ probe.
To test whether other highly transcribed genes show replication pausing, we performed 2D gels on the highly transcribed histone genes hta1+ and htb1+ (Fig. 5B), whose transcription is highest in S phase (Peng et al. 2005). Compared with wild-type cells, replication intermediates and converged forks were, respectively, three and nine times more abundant within the hta1+ and/or htb1+ genes in Pfh1-depleted cells (Fig. 5B). As with the act1+ gene, the increase in replication intermediates in the absence of Pfh1 was seen only in the portion of the fragment that contained the histone genes. Thus, Pfh1 is needed for replication progression through three of three highly transcribed genes. Consistent with our findings that Pfh1 binding was low at the gal1+ gene (Fig. 1A), a gene that is not expected to be highly transcribed in our growth conditions, pausing and converged forks were not detected by 2D gel analysis within the gal1+ ORF in wild-type or Pfh1-depleted cells (Fig. 5C).
Reduced Pfh1 results in slower growth in the presence of extra RTS1 sites
When the RFB from the mating type locus (RTS1) is placed on both sides of the ura4+ gene (RuraR cassette), the region between the RTS1 sites is particularly hard to replicate, owing to forks stalling at the two RTS1 sites (Lambert et al. 2005). Because our ChIP data (Fig. 1A) showed that RTS1 had particularly high Pfh1 binding, we determined whether Pfh1 is required to replicate through a RuraR cassette in the nmt-pfh1-GFP strain. We plated fivefold serial dilutions of the strain on minimal medium lacking (Pfh1 expressed) or containing (Pfh1 repressed) thiamine (Fig. 6A). By this assay, cell viability in Pfh1-depleted cells was more than fivefold lower in cells containing the RuraR cassette compared with cells without it. We also measured the doubling time of liquid-grown cells. Pfh1-depleted cells without the RuraR cassettes doubled 2.3 times faster than the same cells containing the RuraR cassette (Fig. 6B, time point 48 h).
Figure 6.
Pfh1 enhances the viability of cells with the RuraR cassette, and its depletion causes fork pausing and converged forks, which are eliminated in swi1Δ cells. (A) Spot assay using fivefold serial dilutions of cells on EMMS or EMMS containing 5 μg/mL thiamine. (B) Growth curves showing population doublings in cultures growing on EMMS or EMMS + 30 μM thiamine (data for nmt-pfh1-GFP+thiamine were also shown in Fig. 1C). (*) P < 0.05 was determined by two-tailed Student's t-test for 48 h of growth in thiamine for all three strains. For 24-h-growth time points, P < 0.05 was determined for RuraR-nmt-pfh1-GFP (**) and RuraR-nmt-pfh1-GFP+thiamine (*), as well as for RuraR-nmt-pfh1-GFP (**) and nmt-pfh1-GFP+thiamine (*). (C, top) Protein samples from RuraR nmt-pfh1-GFP collected after 0, 6, 9, 12, and 24 h in the presence of thiamine and analyzed by immunoblotting with anti-Pfh1 antibody. (Bottom) Coomassie Blue staining of the same membrane. (D, top left) A schematic of the RuraR region on chromosome III. (E) EaeI. (Top right) Panels showing expected replication intermediates during replication of the RuraR cassette are marked with letters a, b, and c. (Bottom left) Cartoon of 2D gel analysis indicates the replication intermediates shown in the panels on top. The fold differences after 6 h of Pfh1 depletion compared with the nondepleted conditions are stated below each letter in the cartoon. (Bottom right) 2D gel analysis of EaeI-digested DNA from RuraR nmt-pfh1-GFP or RuraR nmt-pfh1-GFP swi1Δ cells grown in the presence of thiamine for 0 or 6 h. Digestion by EaeI results in a 6.8-kb fragment. (Arrows) 2N spots. (E) Spot assay using fivefold serial dilutions of cells on EMMS or EMMS+5 μg/mL thiamine. Three independent isolates of nmt-pfh1-GFP swi1Δ cells, in addition to nmt-pfh1-GFP, nmt-pfh1-GFP RuraR swi1Δ, and RuraR swi1Δ cells, were tested.
Replication forks arrest at RuraR in Pfh1-depleted cells, and Swi1 enhances the viability of Pfh1-depleted cells
To evaluate whether the growth defect in the Pfh1-depleted strains was due to fork arrest at RTS1, Pfh1 was depleted by addition of thiamine for 0, 6, 9, and 12 h in the nmt-pfh1-GFP RuraR strain (Fig. 6C). DNA was prepared from each time point and analyzed by 2D gels (Fig. 6D). As expected, strong fork pausing was detected at both RTS1 sites even in the 0-h DNA sample (Fig. 6D). As shown previously (Lambert et al. 2005), structures consistent with forks converged at the RTS1 sites were also observed (Fig. 6D). Fork arrest at the RTS1 sites increased twofold to threefold (marked a and b in Fig. 6D) when Pfh1 was depleted for 6 h and even more (threefold to fourfold) after 9 h (Fig. 6D; data not shown). Structures interpreted as converged forks (Lambert et al. 2005) increased fourfold (marked c in Fig. 6D) and eightfold at these time points. By 12 h of Pfh1 depletion, genomic DNA was too degraded for 2D gel analysis (data not shown).
Swi1 is required for fork arrest at RTS1 (Dalgaard and Klar 2000; Lambert et al. 2005). As expected, forks did not pause at the RuraR cassette in a swi1Δ strain (Fig. 6D, bottom panel). Likewise, there was no fork arrest at the RTS1 cassette in RuraR swi1Δ cells depleted of Pfh1 for 6 h (Fig. 6D, bottom panel). Moreover, the fraction of DNA in replication intermediates was not higher in Pfh1-depleted swi1Δ cells compared with swi1Δ cells alone (Fig. 6D, region analyzed indicated by bracket), indicating that Pfh1 does not promote replication genome-wide. These data also indicate that it is the Swi1-dependent protein complex at RTS1 that makes replication through this site Pfh1-sensitive.
As controls for these experiments, we made swi1Δ variants of each strain. Unexpectedly, regardless of the presence or absence of the RuraR cassette, swi1Δ cells that were continuously depleted of Pfh1 by growth on thiamine plates were inviable (Fig. 6E; see the Discussion).
Pfh1 suppresses accumulation of phosphorylated histone H2A at sites of fork stalling
Stalled replication forks are particularly susceptible to breakage. Phosphorylated H2A is an early response to double-strand breaks (DSBs) in diverse eukaryotes. The increased formation of γ-H2A surrounding a break has been shown previously in both S. pombe and S. cerevisiae (Shroff et al. 2004; Rozenzhak et al. 2010). In S. pombe, H2A is phosphorylated in regions of ≥40 kb around a DSB but is lower at the break site itself (Rozenzhak et al. 2010). To confirm the presence of stalled and/or broken replication forks by an independent metric, we used ChIP-qPCR to measure levels of γ-H2A at sites of Pfh1-dependent fork arrest in nondepleted and Pfh1-depleted cells. As a control to establish the specificity of the antibody (a kind gift from C. Redon), we used an S. pombe strain in which the two genes that encode histone H2A, hta1+ and hta2+, were mutated so H2A can no longer be phosphorylated in response to DNA damage (hta1-S129A hta2-S128A) (Nakamura et al. 2004). We carried out ChIP in both wild-type and hta1-S129A hta2-S128A cells in the presence or absence of camptothecin (CPT), a drug that inhibits topoisomerase I and causes DNA breaks (Nakamura et al. 2004), which demonstrated the specificity of the antibody (see the Supplemental Material; Supplemental Fig. S2).
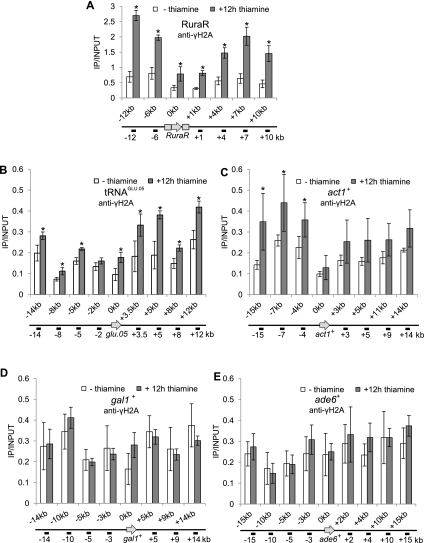
To determine whether Pfh1 depletion caused increased γ-H2A formation at the RuraR cassette and the surrounding region, we grew the nmt-pfh1-GFP RuraR strain in the presence or absence of thiamine and carried out ChIP-qPCR after precipitation with the anti-γ-H2A antibody. We detected a twofold to fourfold increase in γ-H2A in the region surrounding the RuraR cassette in Pfh1-depleted cells compared with nondepleted cells, with the lowest γ-H2A levels at the break site (Fig. 7A). Likewise, γ-H2A was approximately twofold higher in a 25-kb region surrounding the tRNAGLU.05 gene in Pfh1-depleted cells (Fig. 7B). This increase was especially evident in the region downstream from the gene. In addition, γ-H2A levels at the act1+ gene were higher in Pfh1-depleted cells (Fig. 7C). Although this increase was evident both upstream of and downstream from the act1+ gene, only the increase at the upstream sites was significant (P-value<0.05). In contrast, Pfh1 depletion did not result in higher γ-H2A levels in the regions surrounding the gal1+ or ade6+ genes (Fig. 7D,E). Thus, reduced Pfh1 results in high γ-H2A levels specifically in those regions where replication fork progression is slowed in its absence. The fact that Pfh1 depletion did not increase γ-H2A levels at gal1+ or ade6+ provides additional support for our interpretation that Pfh1 acts at a subset of genomic loci.
Figure 7.
Pfh1 protects hard-to-replicate sites against fork breakage. (A) Samples from asynchronous RuraR nmt-pfh1-GFP cell cultures growing in minimal medium supplemented with thiamine for 0 or 12 h were chromatin-immunoprecipitated using an anti-γH2A antibody. Immunoprecipitated DNA was analyzed with qPCR. The region upstream of the gene is marked with a minus sign, and the region downstream from the gene is marked with a plus sign. Here and in B–E, data are presented as immunoprecipitated DNA divided by input DNA. Data are the mean of three or four (shown in C–E) independent cultures, with error bars indicating one standard deviation. (*) P < 0.05 was determined by two-tailed Student's t-test. (B) nmt-pfh1-GFP cells were grown and treated as in A, but note the scale difference on the Y-axis. qPCR primers were specific for the region surrounding the tRNAGLU.05 gene. In nondepleted cells, γ-H2A also increased approximately twofold 12 kb downstream from the tRNAGLU.05 gene. (C) DNA from nmt-pfh1-GFP cells was quantified by qPCR with primers specific for the act1+ gene and its surrounding region, as described in A. (D) nmt-pfh1-GFP cells were grown and treated as in A. qPCR primers were designed for the surrounding regions of the gal1+ ORF. (E) ChIP and qPCR as in A for nmt-pfh1-GFP cells. qPCR primers were for surrounding regions of ade6+.
Discussion
DNA replication is a damage-prone process, as each DNA sequence is transiently single-stranded during S phase, making replicating DNA particularly vulnerable to DSBs. Induced lesions or natural impediments that slow fork progression increase this vulnerability by allowing forks to persist in a susceptible state. Most studies on replication fork progression examine the consequences of exogenous DNA damage. However, there are a large number of sites throughout eukaryotic genomes, such as highly transcribed genes and stable protein complexes, that impede fork progression, and unlike lesions induced by exogenous damage, these obstacles are encountered in every S phase. Despite their importance, systematic identification of hard-to-replicate sites and the mechanisms that allow cells to cope with them have so far been addressed only in S. cerevisiae.
Here we demonstrate that the sole S. pombe Pif1 family DNA helicase, Pfh1, has profound effects on replication fork progression at naturally occurring hard-to-replicate sites. Based on Pfh1 binding, these effects on replication are likely direct. Although all tested DNA sequences had significant Pfh1 binding, Pfh1 binding was particularly high at sites where replication fork progression was slowed in its absence (Fig. 1A). In fact, high Pfh1 binding was a good predictor of Pfh1-sensitive replication, as two “control” sites, act1+ and hta1+/htb1+, unexpectedly had high Pfh1 binding. As a result, we tested these sites for replication defects and found that their replication was indeed Pfh1-sensitive (Fig. 5A,B). Moreover, all sites with high Pfh1 binding and Pfh1-dependent replication also had elevated γ-H2A levels in Pfh1-depleted cells, while Pfh1-insensitive sites did not (Fig. 7). Pfh1 does not appear to promote fork progression genome-wide, as the increased fraction of DNA in replication intermediates in Pfh1-depleted cells was only seen at hard-to-replicate sites (Figs. 2C, 4B,D, 5A–C, 6D). These results demonstrate that replication through Pfh1-sensitive regions in the absence of Pfh1 results in DNA damage that is normally suppressed by Pfh1. Higher Pfh1 binding at Pfh1-sensitive sites can be explained if Pfh1 is recruited to its sites of action in response to replication fork stalling. Alternatively, Pfh1, like ScRrm3 (Azvolinsky et al. 2006), might be a replisome component, and its higher occupancy at Pfh1-sensitive sites would then reflect the increased occupancy of the entire replisome at hard-to-replicate sites (Azvolinsky et al. 2009).
Three types of sequences are known to cause replication fork pausing in S. pombe in their native context: the RFB in rDNA (Sanchez et al. 1998), RTS1 in the mating type locus (Dalgaard and Klar 2001), and telomeres (Miller et al. 2006). Replication through each of these was Pfh1-sensitive: rDNA (Fig. 2), RTS1 (Fig. 6D), and telomeres (KR McDonald, N Sabouri, VA Zakian, and IM Cristea, in prep.). Within the rDNA, Pfh1 depletion resulted in a twofold increase in fork pausing at the RFB and a similar increase in converged forks. The estimates of the impact of Pfh1 on fork progression at the rDNA and elsewhere are almost surely underestimates, as the DNA for 2D gels was prepared from cycling cells (Fig. 1C). Because pfh1+ is essential (Tanaka et al. 2002; Zhou et al. 2002), these cells must still have had residual Pfh1, even though it was not detectable by Western blotting (Fig. 1B). Indeed, by genetic criteria, it is extremely difficult to eliminate nuclear Pfh1 (Pinter et al. 2008). Another study also found replication defects in rDNA in Pfh1-depleted cells (Steinacher et al. 2012). However, there are differences between the two studies. For example, Steinacher et al. (2012) saw heightened pausing at only one of the four rDNA Ter sites, while our data are best explained by increased pausing at all four Ter sites (Fig. 2C). These and other inconsistencies can be explained by methodological differences, as Steinacher et al. (2012) used the pfh1-mt* allele, which at 30°C provides more Pfh1 function than thiamine-grown nmt1-pfh1-GFP cells (Pinter et al. 2008). In addition, their 2D gels examined sequences carried on multicopy plasmids (Steinacher et al. 2012), while, except for RTS1, we examined replication at sites in their normal chromosomal context.
Replication through RTS1 was also Pfh1-sensitive. For these experiments, we used a cassette with two head-to-head RTS1 sites separated by the ura4+ gene (Fig. 6D, called RuraR; Lambert et al. 2005). Cell viability (Fig. 6A) and growth rate (Fig. 6B) were severely compromised in Pfh1-depleted cells carrying the RuraR construct. We infer that these growth and viability problems were due to replication defects because 2D gels revealed a twofold to fourfold increase in both fork arrest and converged forks at RTS1 in Pfh1-depleted cells (Fig. 6D). Another group saw increased converged forks but not increased fork arrest at RTS1 upon Pfh1 depletion (Steinacher et al. 2012). Again, the differences between the two groups can be explained by there being more Pfh1 in pfh1-mt* cells than in thiamine-grown nmt-pfh1-GFP cells (Pinter et al. 2008). These results suggest the interesting interpretation that resolving converged forks is more Pfh1-dependent than fork movement through RTS1. The finding by Steinacher et al. (2012) of increased converged forks in the absence of increased pausing indicates that the elevated number of converged forks is not simply a consequence of increased pausing. Thus, Pfh1 likely has a direct role in resolving converged forks, as does ScRrm3 (Ivessa et al. 2000; Fachinetti et al. 2010).
In otherwise wild-type cells, fork arrest at both the rDNA RFB and the mating type RTS1 is Swi1-dependent (Dalgaard and Klar 2000; Krings and Bastia 2004), presumably because binding of the stable protein complexes at these sites is lost in this mutant. Likewise, Pfh1-sensitive replication both in the rDNA (Fig. 2C) and at RuraR (Fig. 6D) was lost in swi1Δ cells. These findings suggest that it is the stable multiprotein complexes at these sites that make their replication Pfh1-sensitive. Unexpectedly, swi1Δ cells were inviable upon long-term growth with reduced Pfh1 (Fig. 6E). Swi1 is thought to protect stalled replication forks (Noguchi et al. 2003, 2004). Therefore, the increase in stalled forks throughout the genome in Pfh1-depleted cells might lead to widespread fork collapse and death when Swi1 is not there to protect them. Alternatively, death may arise from additive effects of DNA damage, as γ-H2A levels are elevated in both Pfh1-depleted (Fig. 7A–C) and swi1Δ cells (Rozenzhak et al. 2010). The synergism of Pfh1 and Swi1 depletion emphasizes the important role of Pfh1, as similarly severe growth effects are only seen in checkpoint-defective swi1Δ cells that are treated with a DNA-damaging agent (Noguchi et al. 2003). Because both Swi1 and its human homolog, TIMELESS, interact with the replisome (Noguchi et al. 2004; Gotter et al. 2007), the conclusions reached here for Swi1/Pfh1 cooperation during DNA replication may also be relevant to mammals.
An earlier report presaged our finding that tRNA genes affect fork progression in S. pombe by showing pausing at an ectopic tRNA gene inserted within the ade6+ ORF (Pryce et al. 2009). Here, we demonstrate that five of five tRNA genes caused fork pausing in their normal chromosomal context in wild-type cells, and this pausing was evident regardless of the direction of replication vis-á-vis transcription in these genes (Fig. 4; Supplemental Fig. S1). Pausing at each of the five tRNA genes increased in Pfh1-depleted cells, with the magnitude of the increase (twofold to ninefold) depending on the gene and its direction of replication. Although Pfh1 was important for replication of all tested tRNA genes, those where replication and transcription move in opposite directions through the gene were particularly Pfh1-sensitive. Converged and broken replication forks were also seen at four of the tRNA genes, but only in Pfh1-depleted cells (Fig. 4; Supplemental Fig. S1). Pfh1-dependent increases in pausing and fork convergence are also seen at a plasmid-borne tRNA gene (Steinacher et al. 2012). Replication through another class of RNA Pol III transcribed genes, 5S rRNA genes, was also Pfh1-sensitive (Fig. 3). Neither pausing nor converged forks were detected at two of two 5S rRNA genes in wild-type cells, but both were dramatically increased in Pfh1-depleted cells (Fig. 3). As with tRNA genes, 5S rRNA genes were Pfh1-dependent regardless of the direction of replication through the gene. We propose that replication of all 169 tRNA and 33 5S rRNA genes is Pfh1-dependent. Based on the data presented here and in S. cerevisiae (Deshpande and Newlon 1996; Ivessa et al. 2000, 2003; Azvolinsky et al. 2009), we anticipate that RNA Pol III genes are hard to replicate in all eukaryotes, and Pif1 family helicases may also be important for their replication in more complex eukaryotes.
Finally, we present the first evidence that highly transcribed RNA Pol II genes slow fork progression in S. pombe, a slowing that was exacerbated in Pfh1-depleted cells (Fig. 5). Within the highly transcribed act1+ gene, this Pfh1-exacerbated replication defect was also associated with elevated γ-H2A, especially in the region upstream of the gene (Fig. 7C). This localized increase suggests that the negative supercoiling that accumulates behind the transcribing polymerase is relieved by Pfh1 because in its absence, there was DNA damage specifically in this region. Although replication forks in wild-type S. cerevisiae cells pause within highly transcribed RNA Pol II ORFs, pausing is not increased within these genes in either pif1 or rrm3 cells, even though both proteins bind robustly to these genes (Azvolinsky et al. 2009; Paeschke et al. 2011). However, this function has not yet been examined in the double mutant. Together with findings in S. cerevisiae (Azvolinsky et al. 2009; Bermejo et al. 2011), the demonstration that highly transcribed RNA Pol II genes slow replication forks in S. pombe suggests that this phenomenon is widespread. We predict that Pif1 family helicases promote replication through RNA Pol II transcribed genes in organisms other than S. pombe, a role that may have been missed in S. cerevisiae because ScPif1 and ScRrm3 have overlapping functions.
Our studies show that many of the same protein–DNA complexes that impede replication in S. cerevisiae also do so in S. pombe and that Pif1 family helicases, ScRrm3 in S. cerevisiae and Pfh1 in S. pombe, are critical for fork progression and suppression of DNA damage at these sites. In addition, like ScRrm3 (Ivessa et al. 2000; Fachinetti et al. 2010), Pfh1 appears to have a general role in completing DNA replication by resolving converged forks. This conclusion is strengthened by physical analysis of the X-shaped structures that accumulate in Pfh1-depleted cells (Steinacher et al. 2012). Because ScPif1 does not affect replication fork progression at stable protein complexes (Ivessa et al. 2000, 2002), Pfh1's role in chromosomal replication is more ScRrm3-like than ScPif1-like. In addition, Pfh1, unlike ScPif1, does not appear to inhibit telomerase (Pinter et al. 2008). However, Pfh1, like ScPif1 but unlike ScRrm3, is critical for maintenance of mitochondrial DNA.
So far, ScRrm3 and Pfh1 are the only eukaryotic DNA helicases known to promote fork progression at protein complexes and to resolve converged forks, although these problems are faced by all organisms in every S phase. S. cerevisiae and S. pombe diverged from each other more than a billion years ago (Heckman et al. 2001) and are as divergent from each other as each is from humans (Sipiczki 2000). Our findings that Pif1 family helicases promote fork progression at stable protein complexes and DNA termination in such divergent organisms raise the strong possibility that Pif1 family helicases in higher eukaryotes have similar functions. We think this hypothesis is particularly likely given that humans and most other eukaryotes, like S. pombe, encode a single Pif1 family helicase.
Materials and methods
See the Supplemental Material for more details.
Cells were grown in either yeast extract medium (YES) or synthetic minimal medium (EMM) in the presence or absence of 30 μM thiamine at 30°C. See Supplemental Table 1 for strains and Supplemental Table 2 for primers. To monitor Pfh1 binding by ChIP, we epitope-tagged the C terminus of Pfh1 with 13 Myc epitopes. The Pfh1-13Myc was inserted at the leu1+ locus in a pfh1+ strain.
Procedures for ChIP were as described (Webb and Zakian 2012), except that shearing to ∼400 base pairs (bp) was done with a Covaris E220 for the Pfh1-ChIP. For the γ-H2A ChIP, DNA was immunoprecipitated with anti-γ-H2A antibody (from C. Redon). For 2D gels, DNA was prepared from 1000 mL of 1×107 cells per milliliter. Lysis was performed by vortexing cells with glass beads, and genomic DNA was isolated by Qiagen Genomic-tip 500/G. The gels were run as described in Brewer and Fangman (1987). Hybridization probes used for each gel are indicated as a gray box in the restriction maps. The blot was exposed to a PhosphorImager screen, scanned with Typhoon 9410, and quantified by ImageQuant 5.2 software. Each indicated site on the thiamine-added 2D gel image was divided by the signal in the 1N spot in the same gel and then divided by the corresponding ratio for the “no thiamine” gel, setting the 0-h time point values as 1×. Quantitation is an average of at least two independent DNA preparations.
Acknowledgments
We are grateful to S. Pinter and S. Aubert for initiating this project, C. Redon for the kind gift of the γ-H2A antibody, and P. Russell and T. Carr for sending us strains. We also thank M. Bochman for valuable comments on the manuscript and M. Whitby for sharing results prior to publication. This work was supported by National Institutes of Health grants GM26938 (to V.A.Z.) and DP1DA026192 (to I.M.C.), Wenner-Gren foundation (to N.S.), Swedish Society for Medical Research (to N.S.), and New Jersey Commission on Cancer Research (to K.R.M.). C.J.W. was supported by an ARRA supplement to NIH grant GM43265.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.184697.111.
References
- Arcangioli B, Copeland TD, Klar AJ 1994. Sap1, a protein that binds to sequences required for mating-type switching, is essential for viability in Schizosaccharomyces pombe. Mol Cell Biol 14: 2058–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Dunaway S, Torres J, Bessler J, Zakian VA 2006. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev 20: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi P, Lieb J, Zakian V 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gomez-Gonzalez B, et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Sabouri N, Zakian VA 2010. Unwinding the functions of the Pif1 family helicases. DNA Repair (Amst) 9: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Judge CP, Zakian VA 2011. The Pif1 family in prokaryotes: What are our helicases doing in your bacteria? Mol Biol Cell 22: 1955–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643 [DOI] [PubMed] [Google Scholar]
- Cheng X, Dunaway S, Ivessa AS 2007. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion 7: 211–222 [DOI] [PubMed] [Google Scholar]
- Cheng X, Qin Y, Ivessa AS 2009. Loss of mitochondrial DNA under genotoxic stress conditions in the absence of the yeast DNA helicase Pif1p occurs independently of the DNA helicase Rrm3p. Mol Genet Genomics 281: 635–645 [DOI] [PubMed] [Google Scholar]
- Chisholm K, Aubert SD, Freese KP, Zakian VA, King M-C, Welcsh PL 2012. A genomewide screen for suppressors of Alu-mediated rearrangements reveals a role for PIF1. PLoS ONE (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102: 745–751 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ 2001. A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev 15: 2060–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS 1996. DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dubey DD, Kim SM, Todorov IT, Huberman JA 1996. Large, complex modular structure of a fission yeast DNA replication origin. Curr Biol 6: 467–473 [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, Kanoh Y, Shirahige K, Azvolinsky A, Zakian VA, Foiani M 2010. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell 39: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ 2006. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol 8: 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F, Kolodynski J 1983. pif mutation blocks recombination between mitochondrial rho+ and rho− genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc Natl Acad Sci 80: 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George T, Wen Q, Griffiths R, Ganesh A, Meuth M, Sanders CM 2009. Human Pif1 helicase unwinds synthetic DNA structures resembling stalled DNA replication forks. Nucleic Acids Res 37: 6491–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter AL, Suppa C, Emanuel BS 2007. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol 366: 36–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293: 1129–1133 [DOI] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P 2006. Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25: 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Zhou J-Q, Zakian VA 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100: 479–489 [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Zhou J-Q, Schulz VP, Monson EM, Zakian VA 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and sub-telomeric DNA. Genes Dev 16: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Krawchuk MD, Wahls WP 1999. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15: 1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings G, Bastia D 2004. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci 101: 14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings G, Bastia D 2005. Sap1p binds to Ter1 at the ribosomal DNA of Schizosaccharomyces pombe and causes polar replication fork arrest. J Biol Chem 280: 39135–39142 [DOI] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM 2005. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689–702 [DOI] [PubMed] [Google Scholar]
- Linskens MHK, Huberman JA 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol 8: 4927–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RD, Platt TH, Schildkraut CL 1993. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol 13: 6600–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia-Ramirez E, Sanchez-Gorostiaga A, Krimer DB, Schvartzman JB, Hernandez P 2005. The mating type switch-activating protein Sap1 Is required for replication fork arrest at the rRNA genes of fission yeast. Mol Cell Biol 25: 8755–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Du LL, Redon C, Russell P 2004. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol 24: 6215–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, Du LL, Russell P 2003. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol 23: 7861–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, McDonald WH, Yates JR III, Russell P 2004. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol 24: 8342–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke TW, Doudican NA, Zhang H, Eaton JS, Doetsch PW, Shadel GS 2005. Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability. Gene 354: 86–92 [DOI] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA 2011. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Karuturi RK, Miller LD, Lin K, Jia Y, Kondu P, Wang L, Wong LS, Liu ET, Balasubramanian MK, et al. 2005. Identification of cell cycle-regulated genes in fission yeast. Mol Biol Cell 16: 1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter SF, Aubert SD, Zakian VA 2008. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol 28: 6594–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A 2005. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J 24: 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce DW, Ramayah S, Jaendling A, McFarlane RJ 2009. Recombination at DNA replication fork barriers is not universal and is differentially regulated by Swi1. Proc Natl Acad Sci 106: 4770–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Brewer BJ, Fangman WL 1994. Activation of a yeast replication origin near a double-stranded DNA break. Genes Dev 8: 554–562 [DOI] [PubMed] [Google Scholar]
- Rozenzhak S, Mejia-Ramirez E, Williams JS, Schaffer L, Hammond JA, Head SR, Russell P 2010. Rad3ATR decorates critical chromosomal domains with γH2A to protect genome integrity during S-phase in fission yeast. PLoS Genet 6: e1001032 doi: 10.1371/journal.pgen.1001032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JA, Kim SM, Huberman JA 1998. Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp Cell Res 238: 220–230 [DOI] [PubMed] [Google Scholar]
- Sanchez-Gorostiaga A, Lopez-Estrano C, Krimer DB, Schvartzman JB, Hernandez P 2004. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol Cell Biol 24: 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders CM 2010. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem J 430: 119–128 [DOI] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 14: 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M 2000. Where does fission yeast sit on the tree of life? Genome Biol 1: reviews1011–reviews1011.4 doi: 10.1186/gb-2000-1-2-reviews1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinacher R, Osman F, Dalgaard JZ, Lorenz A, Whitby MC 2012. The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev (this issue). doi: 10.1101/gad.184663.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Ryu GH, Seo YS, Tanaka K, Okayama H, MacNeill SA, Yuasa Y 2002. The fission yeast pfh1+ gene encodes an essential 5′ to 3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res 30: 4728–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA 2012. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase–telomere association. Genes Dev 26: 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesendanger B, Lucchini R, Koller T, Sogo JM 1994. Replication fork barriers in the Xenopus rDNA. Nucleic Acids Res 22: 5038–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 [DOI] [PubMed] [Google Scholar]
- Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, et al. 2011. PomBase: A comprehensive online resource for fission yeast. Nucleic Acids Res 40: D695–D699 doi: 10.1093/nar/gkr853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J-Q, Monson EM, Teng S-C, Schulz VP, Zakian VA 2000. The Pif1p helicase, a catalytic inhibitor of telomerase lengthening of yeast telomeres. Science 289: 771–774 [DOI] [PubMed] [Google Scholar]
- Zhou J-Q, Qi H, Schulz V, Mateyak M, Monson E, Zakian V 2002. Schizosaccharomyces pombe pfh1+ encodes an essential 5′ to 3′ DNA helicase that is a member of the PIF1 sub-family of DNA helicases. Mol Biol Cell 13: 2180–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]