This study identifies a novel chromatin remodeling complex, “ToRC,” comprising Drosophila Toutatis, ISWI, and CtBP. It is also found conserved in humans. CtBP is its key distinguishing component and is required for ToRC localization to euchromatic regions. ToRC catalyzes ATP-dependent nucleosomal assembly and provides an important new role for CtBP in directing remodeling activity to chromosomal arms.
Keywords: chromatin assembly and remodeling, ISWI, Toutatis, TIP5, CtBP, nucleolus, ac–sc complex (AS-C)
Abstract
SNF2-like motor proteins, such as ISWI, cooperate with histone chaperones in the assembly and remodeling of chromatin. Here we describe a novel, evolutionarily conserved, ISWI-containing complex termed ToRC (Toutatis-containing chromatin remodeling complex). ToRC comprises ISWI, Toutatis/TIP5 (TTF-I-interacting protein 5), and the transcriptional corepressor CtBP (C-terminal-binding protein). ToRC facilitates ATP-dependent nucleosome assembly in vitro. All three subunits are required for its maximal biochemical activity. The toutatis gene exhibits strong synthetic lethal interactions with CtBP. Thus, ToRC mediates, at least in part, biological activities of CtBP and Toutatis. ToRC subunits colocalize in euchromatic arms of polytene chromosomes. Furthermore, nuclear localization and precise distribution of ToRC in chromosomes are dependent on CtBP. ToRC is involved in CtBP-mediated regulation of transcription by RNA polymerase II in vivo. For instance, both Toutatis and CtBP are required for repression of genes of a proneural gene cluster, achaete–scute complex (AS-C), in Drosophila larvae. Intriguingly, native C-terminally truncated Toutatis isoforms do not associate with CtBP and localize predominantly to the nucleolus. Thus, Toutatis forms two alternative complexes that have differential distribution and can participate in distinct aspects of nuclear DNA metabolism.
Genomic activity in metazoans is regulated not only by transcription factors, but also by a host of coactivator and corepressor proteins in epigenetic pathways that use post-translational modification of histones in chromatin (Weake and Workman 2010; Li and Reinberg 2011). For instance, C-terminal-binding proteins (CtBPs) predominantly function as transcriptional corepressors (Chinnadurai 2002). More than 30 different transcription repressors have been reported to recruit CtBPs (Kuppuswamy et al. 2008). CtBPs have been shown to be incorporated in various multisubunit complexes and interface chromatin modification activities such as histone deacetylation, methylation, and demethylation with transcriptional repression (Chinnadurai 2007).
Chromosome structure and activity can also be modified through mobilization of nucleosomes in an ATP-dependent manner by chromatin remodeling factors (Flaus and Owen-Hughes 2001; Narlikar et al. 2002) that contain polypeptides from the SNF2 family of DNA-stimulated ATPases (Gorbalenya and Koonin 1993; Eisen et al. 1995). In addition to remodeling existing nucleosomes, SNF2-like motor proteins can facilitate the de novo assembly of nucleosomes (Fyodorov and Kadonaga 2001; Akey and Luger 2003; Haushalter and Kadonaga 2003) in a concerted reaction with core histone chaperones (Adams and Kamakaka 1999; Mello and Almouzni 2001; Ransom et al. 2010). Whereas histone chaperones mediate the formation of nascent prenucleosome structures, ATP-dependent enzymes are required to convert them into periodic arrays of mature nucleosomes (Torigoe et al. 2011). For instance, ACF (Acf1–ISWI complex) can mediate chromatin assembly in conjunction with histone chaperone NAP-1 (Ito et al. 1997). ACF consists of two subunits: the ATPase ISWI (Elfring et al. 1994) and a polypeptide termed Acf1, which is specific to ACF and a closely related complex, CHRAC (Varga-Weisz et al. 1997). Other known ATP-dependent chromatin assembly factors include human ISWI/SNF2H-containing RSF, CHD1, and ATRX (Loyola et al. 2001; Lusser et al. 2005; Lewis et al. 2010).
Despite similar biochemical activities, SNF2-like factors possess distinct and variable biological functions in vivo (Fyodorov and Kadonaga 2001). Consistently, they exhibit specific distribution patterns in the genome. Mechanisms that control this differential distribution are still not fully understood. It has been demonstrated, for instance, that motor proteins can be tethered to specific targets by direct physical interactions with transcription factors (Armstrong et al. 1998; Yudkovsky et al. 1999; de la Serna et al. 2001; Pedersen et al. 2001; Kadam and Emerson 2003; Memedula and Belmont 2003). In addition, locus-specific localization of remodeling factors can be modulated by the presence of specialized docking motifs (bromodomain and PHD finger, SANT, SLIDE, and HAND domains) that recognize post-translationally modified histone tails (Hassan et al. 2001, 2002; Li et al. 2006; Wysocka et al. 2006; Dang and Bartholomew 2007; Pinskaya et al. 2009; Ryan et al. 2011).
Mouse NoRC is a nucleolar-specific, SNF2H-containing chromatin remodeling factor (Strohner et al. 2001). Its large subunit, TIP5 (TTF-I-interacting protein 5), exhibits strong similarity to mammalian homologs of Acf1 and shares with it a number of domains that are important for the chromatin assembly activity of ACF (Fyodorov and Kadonaga 2002). The Drosophila ortholog of TIP5 is known as Toutatis (Tou) (Fauvarque et al. 2001). Here we describe purification and biochemical/biological characterization of a novel Drosophila ATP-dependent chromatin assembly factor termed ToRC (Toutatis-containing chromatin remodeling complex), which consists of TIP5/Tou, ISWI, and CtBP. Tou and CtBP exhibit strong genetic interactions, which suggests the existence of common biological functions that they share as subunits of ToRC. ToRC can assemble nucleosome arrays in an ATP-dependent manner and requires all three subunits to achieve its optimal biochemical activity. In vivo, ToRC is tethered to target sites of CtBP and requires CtBP for proper localization. These findings provide evidence for a transcriptional cofactor that forms a complex with SNF2-like ATPase and is dedicated to stimulation and recruitment of its enzymatic activity to specific target genes.
Results
The ToRC complex consists of Tou, ISWI, and CtBP
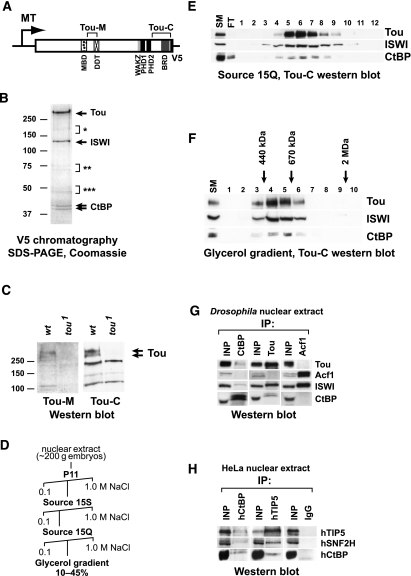
To identify the components of native complexes formed by Tou, we generated S2 cells with a stably integrated transgene that expresses V5-tagged full-length Tou under the control of inducible metallothionein promoter (Fig. 1A). After induction by copper sulfate, Tou-V5 was purified by anti-V5 immunoaffinity chromatography. The purified complex contained three major polypeptides (Fig. 1B) whose identities were examined by mass spectrometry and determined to be Tou, ISWI, and CtBP (Supplemental Table 1).
Figure 1.
Drosophila Toutatis (Tou) protein forms a complex with ISWI and CtBP in S2 cells. (A) Transgene construct for ectopic expression of Tou in S2 cells. Full-length Tou (1–2999) was cloned in-frame with a C-terminal V5 tag (black box). Evolutionarily conserved domains are indicated. (Dotted box) MBD, residues 953–1027; (crossed box) DDT, residues 1255–1308; (light-gray box) WAKZ, residues 2470–2508; (black boxes) PHD fingers, residues 2509–2556 and 2563–2613; (dark-gray box) bromodomain (BRD), residues 2896–2975. Brackets at the top indicate polypeptide antigen fragments as follows: Tou-M, 1033–1265; and Tou-C, 2670–2999. (MT) Metallothionein promoter. (B) The protein complex formed by Tou in S2 cells. The protein was purified by V5 chromatography from S2 cells that extopically express Tou-V5. Arrows point to complex components identified by sequencing (Tou, ISWI, and CtBP). Brackets indicate Tou degradation products (*) or nonspecific polypeptide bands ([**]heat-shock proteins; [***] α-tubulin and β-tubulin [CG1913, CG9277, and CG3401]), which are also present in mock purifications from S2 cells that do not express Tou. Molecular weight markers are shown on the left. (C) Western blot analysis of Tou in Drosophila embryos. Several Tou isoforms (between 250 and 350 kDa) that are recognized by Tou-M (left panel) and Tou-C (right panel) antibodies in lysates of wild-type (wt) embryos are not present in lysates of homozygous tou1 embryos. (D) Partial purification of the native complex of Tou from Drosophila embryos. Nuclear extracts were subjected to four steps of purification. Peak fractions were identified by Western blot with Tou-C, ISWI, and CtBP antibodies. (E) Fractionation of the native complex of Tou on anion exchange (Source 15Q) column. Tou, ISWI, and CtBP cofractionate in a narrow range of the salt gradient (320–400 mM NaCl). (SM) Starting material (peak Tou- and CtBP-containing fractions from Source 15S chromatography); (FT) flowthrough. Fraction numbers are shown at the top. (F) Glycerol gradient (10%–45%) sedimentation of the native Tou complex. The complex of Tou, ISWI, and CtBP cosediments with a molecular weight between 440 and 670 kDa according to sedimentation profiles of molecular weight markers. (SM) Starting material (fractions 5–7 from Source 15Q chromatography). (G) Co-IP analysis of Tou, ISWI, and CtBP. Drosophila nuclear extract was immunoprecipitated with CtBP, Tou-M, or Acf1 antibodies, and precipitated material was analyzed by Western blot for the presence of Tou, Acf1, ISWI, and CtBP. CtBP and ISWI coimmunoprecipitate with Tou, whereas CtBP does not coimmunoprecipitate with Acf1. (INP) Nuclear extract, 10% of immunoprecipitation input. (H) Co-IP analysis of TIP5, SNF2H, and hCtBP, human orthologs of Tou, ISWI, and dCtBP. HeLa nuclear extract was immunoprecipitated with hCtBP, TIP5 antibodies, or an equivalent amount of control IgGs, and precipitated material was analyzed by Western blot for the presence of TIP5, SNF2H, and hCtBP. Similar to their Drosophila orthologs, human TIP5, SNF2H, and CtBP form a complex in HeLa nuclear extract. (INP) Nuclear extract, 10% of immunoprecipitation input.
Whereas the physical association of Tou and ISWI could be predicted from the existence of an orthologous mammalian complex (NoRC), the presence of CtBP was unexpected. Thus, we decided to confirm that a similar complex is formed in vivo. We raised polyclonal antibodies to two distinct fragments of Tou—Tou-M and Tou-C—located in the middle and C-terminal portions of the protein, respectively (Fig. 1A). We then prepared whole-embryo lysates of wild-type and homozygous tou1 mutant alleles and analyzed the expression of Tou by Western blot (Fig. 1C). Several bands with apparent molecular masses between 250 and 350 kDa were recognized in wild-type lysates but were absent in tou1 lysates. We also prepared antibodies to full-length Drosophila CtBP, which specifically recognize two protein bands with apparent molecular masses of ∼39 and 40 kDa (data not shown). We then partially purified the native form of endogenous Tou from Drosophila embryos by a series of conventional chromatography steps and glycerol gradient sedimentation (Fig. 1D). In all chromatographic steps, Tou, ISWI, and CtBP showed precise copurification (Fig. 1E; data not shown). Furthermore, in the glycerol gradient, Tou, ISWI, and CtBP cosedimented in a single peak with an estimated molecular mass of ∼500 kDa (Fig. 1F), which approximately corresponds to the calculated mass of a trimeric Tou–ISWI–CtBP complex. Thus, in vivo in Drosophila, Tou forms a stable complex with ISWI and CtBP. We termed this novel complex ToRC.
ToRC is conserved in evolution
To further confirm the physical interactions among Tou, ISWI, and CtBP, we performed coimmunoprecipitation (co-IP) of Tou and CtBP from embryonic nuclear extracts. The material precipitated by either Tou or CtBP antibodies contained Tou, ISWI, and CtBP (Fig. 1G). However, it did not contain Acf1. Conversely, Acf1 did not coimmunoprecipitate Tou or CtBP. Thus, CtBP is a specific component of ToRC and does not interact with ACF.
To examine whether ToRC is conserved in evolution, we performed co-IP experiments with human (HeLa) nuclear extracts and antibodies to human orthologs of Tou (TIP5) and CtBP. Both antibodies specifically coimmunoprecipitated the components of putative human ToRC, including TIP5, SNF2H, and human CtBP (Fig. 1H). Thus, similar to Drosophila, in addition to the TIP5–SNF2H complex (NoRC), human cells contain a complex that includes CtBP (putative human ToRC). Alternatively, CtBP may be a previously unrecognized component of mammalian NoRC.
ToRC is an ATP-dependent chromatin assembly factor
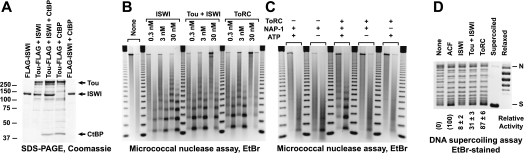
Drosophila ACF is one of the four known factors that can facilitate ATP-dependent nucleosome assembly in vitro. We decided to determine whether ToRC, too, can mediate nucleosome assembly in conjunction with the histone chaperone NAP-1. To this end, we expressed and purified various recombinant Tou-, ISWI-, and CtBP-containing complexes in baculovirus (Fig. 2A). Both ISWI and CtBP directly interact with Tou to form ToRC but do not associate with each other. We then tested recombinant Drosophila ISWI, NoRC (Tou + ISWI), and ToRC (Tou + ISWI + CtBP) in the defined nucleosome assembly system. Both NoRC and ToRC can assemble periodic nucleosome arrays more efficiently than ISWI when used in substoichiometric concentrations relative to nucleosomes/histones (Fig. 2B). Thus, Tou and/or CtBP can stimulate the intrinsic weak chromatin assembly activity of ISWI. This observation parallels a similar finding for Acf1 and ISWI (Ito et al. 1999; Fyodorov and Kadonaga 2002). Also similar to ACF, a stoichiometric excess of the NoRC or ToRC enzymes (when their molecular ratio to assembled nucleosomes approaches 1:1) inhibits the assembly of nucleosome arrays.
Figure 2.
ToRC is an ATP-dependent chromatin assembly factor. (A) Purification of recombinant ToRC. Various combinations of Flag-tagged and untagged Tou, ISWI, and CtBP were coexpressed in Sf9 cells, purified in two steps by Flag and anion exchange (Source 15Q) chromatography, and analyzed by SDS-PAGE and Coomassie staining. ISWI and CtBP directly interact with Tou to form ToRC but do not interact with each other. Molecular weight markers are shown on the left, and arrows on the right point to recombinant polypeptides. (B) ATP-dependent chromatin assembly activity of ISWI, Tou–ISWI complex, and ToRC. Recombinant Flag-ISWI, Tou-Flag + ISWI, or Tou-Flag + ISWI + CtBP was used in the defined ATP-dependent chromatin assembly system (Fyodorov and Kadonaga 2003). Each reaction contained 2.5 nM relaxed plasmid DNA (∼3 kb), 100 nM each core histone polypeptide, 500 nM NAP-1 polypeptide, and indicated concentrations of ATP-dependent factors. After 2 h at 27°C, the assembled nucleosome arrays in each reaction were analyzed by partial digestion with two distinct amounts of micrococcal nuclease and DNA agarose gel electrophoresis. A 123-bp DNA ladder (Invitrogen) was used as a molecular weight marker. (C) Cofactor requirements for ToRC in the chromatin assembly reaction. The chromatin assembly activity was assayed as described in B in the presence of all or in the absence of one or more of the following components: ToRC, NAP-1, and ATP. ToRC assembles periodic nucleosome arrays in an ATP- and NAP-1-dependent manner. (D) Stimulation of nucleosome assembly activity of ISWI by Tou and CtBP. Chromatin was assembled in vitro with 0.3 nM indicated ATP-dependent factor for 10 min as described above, and histone deposition was assayed by DNA supercoiling. Supercoiling activity of NAP-1 in the absence of remodeling factors was set to 0. Supercoiling activity of recombinant ACF was set to 100%. The activity of ISWI alone (∼8%) was stimulated approximately fourfold in a complex with Tou and further stimulated (approximately threefold) when the complex (ToRC) additionally contained CtBP. Quantitation is presented as an average of four independent experiments with two distinct preparations of each factor. Standard deviations are shown. The positions of nicked (N) and highly negatively supercoiled (S) DNA bands on the gel are shown on the right.
Motor proteins require both ATP and histone chaperones for the assembly of nucleosomes. Therefore, we tested cofactor requirements of ToRC in the reaction. We discovered that ToRC cannot assemble nucleosome arrays in the absence of NAP-1 or ATP (Fig. 2C). Thus, ToRC shares these properties with ACF, CHD1, and RSF. Finally, we wanted to examine quantitative contributions of ToRC subunits to the efficiency of nucleosome assembly. To this end, we used a DNA supercoiling assay for reactions with limiting amounts of ATP-dependent enzymes (Fyodorov and Kadonaga 2002). These measurements allow accurate quantitation of the rate of nucleosome formation. We discovered that successive additions of Tou and CtBP substantially stimulate (threefold to fourfold each) the intrinsic histone deposition activity of ISWI (Fig. 2D). Interestingly, the relative activity of NoRC (Tou + ISWI), although appreciably higher than that of ISWI alone, was less than one-third of that of ACF (Acf1 + ISWI). On the other hand, the activity of the complete ToRC complex was comparable with that of ACF. Therefore, the presence of all three subunits of ToRC is essential to achieve its full enzymatic activity.
tou and CtBP exhibit genetic interactions
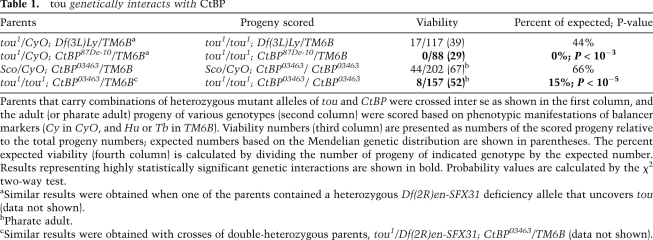
Since Tou and CtBP appear to function as subunits of a protein complex, we tested whether their cognate genes interact. To this end, we used existing alleles of tou and CtBP. tou1 contains a P-element insertion in the second intron of the gene (Fauvarque et al. 2001) and is a null or weak hypomorphic allele (Fig. 1C). In inter se crosses of heterozygous tou1 parents, the viability of homozygous progeny is only moderately reduced (Table 1), similar to null alleles of Acf1 (Fyodorov et al. 2004). CtBP86De-10 and CtBP03463 are amorphic and hypomorphic alleles, respectively. They are both recessive lethal. Whereas homozygous CtBP86De-10 animals die during early larval development, homozygous CtBP03463 larvae pupariate and survive to the adult pharate stage (Poortinga et al. 1998).
Table 1.
tou genetically interacts with CtBP
tou and CtBP alleles exhibited strong synthetic lethal interactions. For instance, reduced dosage of CtBP in trans-heterozygous tou1/CyO; CtBP87De-10/TM6B parents rendered their tou1/tou1; CtBP87De-10/TM6B progeny unviable (Table 1). Additionally, in crosses of tou1/tou1; CtBP03463/TM6B parents, the majority (>85%) of double-homozygous tou1/tou1; CtBP03463/CtBP03463 progeny failed to survive to the adult pharate stage and died prior to metamorphosis. Thus, Tou is required for the full biological activity of CtBP, and, conversely, maintaining the proper dosage of CtBP is necessary for the development of homozygous mutant tou animals.
ToRC is excluded from the nucleolus and depends on CtBP for its nuclear localization
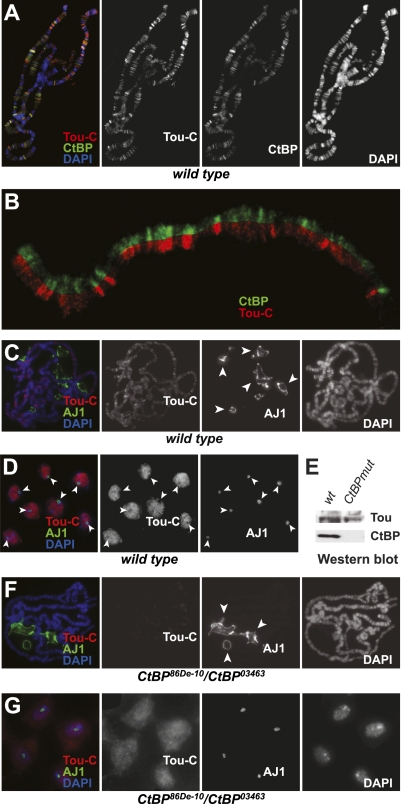
To further understand the function of ToRC in vivo, we used indirect immunofluorescence (IF) to examine distribution of Tou in polytene chromosomes. Tou-C antibody recognizes multiple (>100) bands in euchromatic arms of polytene chromosomes (Fig. 3A). As a control, Tou-C antibody does not produce discernable staining of polytene chromosomes of tou1 homozygous mutant larvae (Supplemental Fig. 1A). Furthermore, Tou colocalizes extensively with CtBP. In higher-resolution images, it is apparent that Tou is primarily present in CtBP-specific loci. However, a number of CtBP-positive bands did not contain Tou (Fig. 3B). This result is consistent with CtBP being a part of several distinct complexes (Chinnadurai 2007) in addition to ToRC.
Figure 3.
ToRC requires CtBP for proper localization on chromosome arms and is excluded from the nucleolus. (A) Genome-wide localization of ToRC. Localization patterns of Tou and CtBP in larval polytene chromosomes were analyzed by indirect IF staining with Tou-C and CtBP antibodies. Tou-C (red) and CtBP (green) signals extensively overlap in euchromatic arms of polytene chromosomes. DAPI staining is shown in blue. (B) Split image view of colocalization of CtBP and Tou. Tou-C-specific (red) and CtBP-specific (green) signals overlap in many but not all genomic loci on the distal 3L arm of the third chromosome. (C) Genome-wide localization of ToRC and nucleolus-specific antigen AJ1 in wild-type larvae. Localization patterns of Tou-C and AJ1 antigens in polytene chromosomes were analyzed by indirect IF staining with Tou-C and AJ1 antibodies. Whereas the AJ1 antibody (green) stains nucleolar structures in polytene spreads, the Tou-C antibody (red) stains multiple bands in euchromatic arms of polytene chromosomes but does not stain nucleoli. DAPI staining is shown in blue. (D) Subnuclear localization of ToRC in larval neuroblasts. Neuroblasts from wild-type L3 larvae were stained with Tou-C and AJ1 antibodies. AJ1 is present exclusively in nucleoli (arrowheads), whereas Tou-C signal is concentrated in nuclei but is weak or absent from nucleoli. (Red) Tou-C staining; (green) AJ1 staining; (blue) DAPI staining. (E) Expression of Tou and CtBP proteins in late L3 larvae. Whole wild-type and CtBP mutant (CtBP86De-10/CtBP03463) larvae were collected, and protein expression was analyzed by Western blot with Tou-C and CtBP antibodies. CtBP mutation eliminates detectable CtBP protein but does not substantially affect the expression of Tou. (F) Genome-wide localization of ToRC and nucleolus-specific antigen AJ1 in CtBP mutant larvae. Localization patterns of Tou-C and AJ1 antigens in CtBP86De-10/CtBP03463 polytene chromosomes were analyzed by indirect IF staining with Tou-C and AJ1 antibodies. AJ1 staining (green) is restricted to the nucleolus, similar to that of wild-type polytene spreads (Fig. 5C). No band-specific Tou-C staining (red) is observed in euchromatic arms of polytene chromosomes or elsewhere in polytene spreads. DAPI staining is shown in blue. (G) Abberrant distribution of ToRC in the neuroblasts of CtBP mutant larvae. Neuroblasts from CtBP86De-10/CtBP03463 L3 larvae were stained with Tou-C (red) and AJ1 (green) antibodies. The Tou-C signal does not localize in the nucleus, but rather is diffusely distributed throughout the cell. DAPI staining is shown in blue.
Mammalian TIP5 has been shown to localize predominantly to the nucleolus in NIH-3T3 cells (Strohner et al. 2001). However, we did not observe Tou-C staining in nucleoli of polytene spreads (Fig. 3A,C), in contrast to staining with AJ1 antibody raised against a nucleolus-specific antigen (Fig. 3C; Orihara-Ono et al. 2005). When we analyzed the distribution of TIP5/Tou in Drosophila larval neuroblasts by IF with Tou-C antibodies, we observed strong Tou-C signal in the nucleus (Fig. 3D), which could not be detected in tou1 mutant neuroblasts (Supplemental Fig. 1B). Similar to that in polytene chromosomes, IF staining patterns of Tou-C and AJ1 did not overlap. Rather, their distribution patterns appeared mutually exclusive (Fig. 3D). Thus, in contrast to mammalian NoRC, Drosophila ToRC is localized throughout the nucleus but is excluded from the nucleolus.
CtBP is recruited to its sites of action by interactions with transcriptional repressor factors (Kumar et al. 2004; Kuppuswamy et al. 2008). It is possible that ToRC tethering to its nuclear targets is in turn dependent on CtBP. Thus, we analyzed distribution of Tou in CtBP mutant animals in vivo. Heterozygous of CtBP86De-10/CtBP03463 animals do not express detectable CtBP at the late larval stage, but the expression of Tou in these animals is not affected (Fig. 3E). When we stained polytene chromosomes of CtBP mutant larvae with Tou-C antibodies, we could not detect Tou in polytene chromosome arms (Fig. 3F). Furthermore, Tou was no longer present exclusively in the neuroblast nuclei. Rather, the Tou-C signal appeared to be diffusely spread throughout the cell (Fig. 3G). Therefore, CtBP is required for nuclear localization and precise genomic distribution of ToRC. In a reciprocal experiment, the distribution of CtBP was not affected in homozygous mutant tou1 larvae (Supplemental Fig. 1C), which suggests that CtBP is translocated to the nucleus and targeted to its specific chromosomal loci independent of Tou or ISWI.
ToRC is involved in transcriptional repression of genes of achaete–scute complex (AS-C)
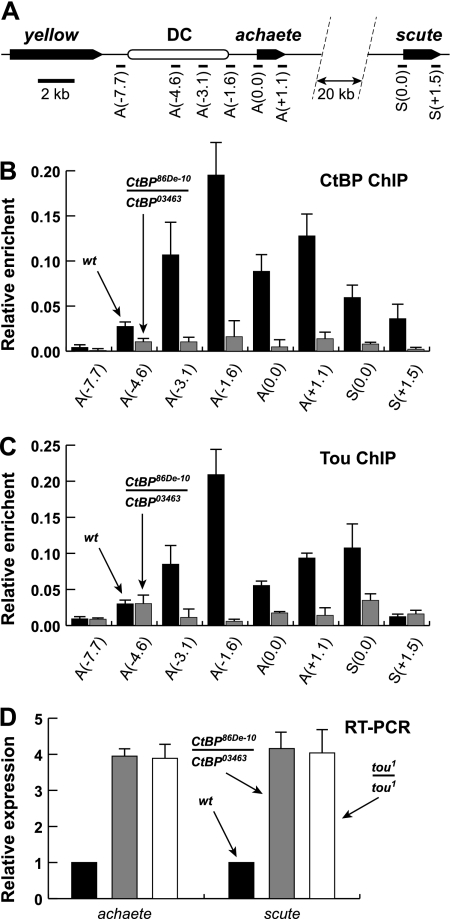
Drosophila proneural genes achaete (ac) and scute (sc) are members of the AS-C and are coregulated in larval imaginal discs by shared cis elements to establish the identity of precursor cells of adult sensory organs (Gómez-Skarmeta et al. 1995). For instance, proper regulation of ac and sc in the dorsocentral (DC) proneural cluster of wing disc requires an upstream 5.7-kb DC enhancer that mediates both transcriptional activation and repression in various parts of the wing disc (Garcia-Garcia et al. 1999).
Genetic experiments suggest that both CtBP (Biryukova and Heitzler 2008) and Tou (Vanolst et al. 2005) can regulate ac and sc in vivo. Thus, we decided to examine by chromatin immunoprecipitation (ChIP) whether CtBP and Tou colocalize at regulatory regions of AS-C in Drosophila larvae (Fig. 4A). Indeed, the loci with higher occupancy of CtBP contained Tou, whereas lower occupancy of CtBP correlated with lower occupancy of Tou (Fig. 4, cf. B and C), consistent with the recruitment of ToRC to these loci. Specifically, CtBP and Tou exhibited co-occupancy at promoters of ac and sc as well as the proximal part of the DC enhancer. When Tou and CtBP occupancy at these sites was examined in CtBP86De-10/CtBP03463 mutant larvae, which do not express detectable CtBP (Fig. 3E), near-complete elimination of occupancy for both CtBP and Tou was observed (Fig. 4B,C). Therefore, CtBP is required for tethering of ToRC to regulatory regions of AS-C in Drosophila larvae.
Figure 4.
ToRC is a repressor of achaete and scute in Drosophila larvae. (A) Schematic of the yellow–scute genomic interval in Drosophila. Black arrows indicate genes, and a white oval designates the 5.7-kb DC enhancer element that regulates the expression of ac and sc in larval wing discs (Garcia-Garcia et al. 1999). Bars at the bottom indicate approximate genomic intervals in kilobase pairs (kb). Positions of primer pairs are shown as black bars below the locus schematic. In probe names, numbers in parentheses indicate approximate positions (in kilobase pairs [kb]) of the primer pairs relative to the transcription start sites of ac (A) and sc (S), respectively. (B) ChIP analyses of CtBP occupancy at genomic loci within AS-C. ChIP was performed with CtBP antibodies in wild-type (wt; black bars) and CtBP86DE-10/CtBP03463 mutant (gray bars) larvae. Relative enrichment of CtBP at indicated sites was measured by real-time PCR. Error bars indicate standard deviation of six independent data points. In wild-type larvae, CtBP is enriched in the proximal part of the DC enhancer element and in transcription units of ac and sc. In CtBP mutant larvae, CtBP occupancy is strongly reduced at all sites. The w1118 allele was used as the wild-type control. (C) ChIP analyses of Tou occupancy at genomic loci within AS-C. ChIP was performed with Tou-M antibodies in wild-type (wt; black bars) and CtBP86DE-10/CtBP03463 mutant (gray bars) larvae. Relative enrichment of Tou was measured and presented as in B. Tou and CtBP colocalize in the proximal part of the DC enhancer element and at ac and sc promoters (cf. B). In CtBP mutant larvae, Tou occupancy at these sites is strongly reduced. (D) RT–PCR analysis of ac and sc expression in vivo in whole L3 larvae. Relative expression was measured by real-time RT–PCR on RNA prepared from wild-type (wt; black bars), CtBP86DE-10/CtBP03463 (gray bars), or tou1 (white bars) mutant larvae. CT values were normalized to the reference gene (rpL32), and the expression in wild-type was set to 1. Error bars indicate standard deviation of six independent data points. The mutation of either CtBP or tou results in an approximately fourfold activation of both ac and sc. The expression of control genes (actin5C or piwi) was not significantly affected by either mutation (data not shown).
Finally, we analyzed effects of CtBP and tou mutations on expression of AS-C genes in vivo. We performed RT–PCR analyses of ac and sc transcripts in wild-type and mutant larvae. In animals that do not express CtBP or Tou, both genes were strongly and reproducibly up-regulated (Fig. 4D). Thus, CtBP and Tou are required for repression of ac and sc in Drosophila larvae. Considering their association with known regulatory elements of these genes (Fig. 4B,C), the transcriptional repression is likely exerted through a direct action of ToRC at these sites.
Tou forms an alternative complex that does not contain CtBP and is targeted to the nucleolus in vivo
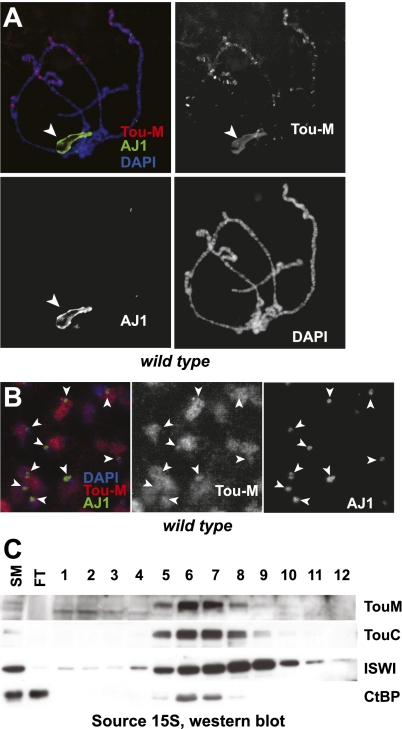
The existence of Tou polypeptides of various sizes is evident from Western blots of native Tou (Fig. 1). The presence of multiple variable expressed sequence tags (ESTs) of tou (FlyBase) indicates that it undergoes alternative splicing, which results in the formation of several protein isoforms. Proteolytic clipping of the C terminus similar to that of Acf1 (Ito et al. 1999) may also result in expression of truncated Tou polypeptides. Alternative Tou polypeptides may not associate with CtBP and could be distributed differentially. To test this hypothesis, we stained polytene chromosomes with antibodies to an antigen from the middle portion of Tou (Tou-M) (Fig. 1A). In addition to specific bands in chromosome arms that are detected by the Tou-C antibody, Tou-M antibody brightly stains the nucleoli of salivary gland cells, overlapping with AJ1 staining (cf. Figs. 3C and 5A). Similarly, nucleolus-specific signal was observed in the IF analysis of larval neuroblasts (Fig. 5B). Tou-M antibody specificity controls (staining of tou1 mutant polytene chromosomes and neuroblasts) are provided in Supplemental Figure 1, D and E.
Figure 5.
Alternative CtBP-free complex of Tou is localized in the nucleolus. (A) Genome-wide localization of Tou. The Tou-M antibody was used for IF staining of polytene chromosomes. In addition to multiple Tou-specific bands in chromosome arms that are also recognized by the Tou-C antibody, the Tou-M antibody produces a strong signal in the nucleolus of salivary gland cells (arrowhead). (Red) Tou-M antigen; (green) nucleolus-specific antigen AJ1. (B) Subnuclear localization of Tou in larval neuroblasts. Neuroblasts from wild-type L3 larvae were stained with Tou-M and AJ1 antibodies. AJ1 is present exclusively in nucleoli (arrowheads), whereas Tou-M signal is spread throughout the nuclei and is also present in nucleoli. (Red) Tou-M staining; (green) AJ1 staining; (blue) DAPI staining. (C) Alternative, CtBP-free native complex of Tou in Drosophila embryos: fractionation of the native complex of Tou on cation exchange (Source 15S) column. Full-length Tou and CtBP cofractionate in a narrow peak (280–350 mM NaCl; fractions 5–8) that corresponds to ToRC. An additional peak of Tou-specific material, which is recognized by Tou-M but not Tou-C antibody (200–260 mM NaCl; fractions 1–3), contains ISWI but not CtBP. (SM) Starting material (peak fractions of Tou-immunoreactive material from phosphocellulose P11 chromatography); (FT) flowthrough. Fraction numbers are shown at the top.
We originally used Tou-C antibody to follow partial purification of native embryonic ToRC (Fig. 1E,F). When we examined fractions from early chromatographic steps (Fig. 1D) with the Tou-M antibody, we discovered that, indeed, Tou is present in an additional protein complex. This complex contains ISWI but does not include CtBP and is largely separated from ToRC in the first two steps of purification (Fig. 5C). Importantly, Tou polypeptides in this alternative complex have a smaller apparent molecular mass. These shorter Tou polypeptides do not encompass the C terminus of Tou-PA, since they are not recognized by the Tou-C antibody. Therefore, in addition to the full-length polypeptide that forms the three-subunit ToRC (Tou + ISWI + CtBP) and is excluded from the nucleolus, native Drosophila TIP5/Tou exists in an alternative, C-terminally truncated form that forms a CtBP-free complex (putative Drosophila NoRC) and is present in the nucleolus.
Discussion
Mounting genetic and molecular evidence suggests that Toutatis, the Drosophila ortholog of mammalian TIP5, plays important roles in the regulation of transcription by RNA polymerase II. In fact, tou was originally characterized as a suppressor of a Polycomb group gene polyhomeotic (Fauvarque et al. 2001). Furthermore, tou mutants exhibit wing defects, suggesting that Tou may be involved in the regulation of wing development.
Data from a recent report (Liu et al. 2008) indicate that ISWI and CtBP colocalize at certain loci in the fly genome. For instance, a wingless target gene, hth, contains several sites where CtBP colocalizes with ISWI (Liu et al. 2008). Furthermore, hth is regulated by CtBP and ISWI in Kc cells and the developing wing (Fang et al. 2006; Liu et al. 2008). Thus, it is possible that Tou, ISWI, and CtBP may share genomic targets in vivo in Drosophila, where polymerase II-mediated transcription can be regulated by remodeling of chromatin structure by ToRC
The expression pattern of AS-C genes in the wing imaginal disc is maintained through antagonistic actions of transcriptional activators, such as GATA factor Pannier, and transcriptional repressors, such as zinc finger protein U-shaped, at the 5.7-kb DC enhancer element (Garcia-Garcia et al. 1999). Long-distance enhancer–promoter communications in AS-C are further facilitated by a LIM domain-binding protein, Chip (Ramain et al. 2000). Tou has been previously shown to be involved in AS-C regulation (Vanolst et al. 2005). tou genetically interacts with pnr and Chip, and Tou protein may physically associate with Pannier and Chip. Accordingly, it has been proposed that tou, Iswi, pnr, and Chip cooperate to establish long-distance enhancer–promoter interactions in AS-C, possibly through chromatin remodeling (Vanolst et al. 2005). Consistent with biochemical and genetic interactions of Tou and CtBP that we report here, loss-of-function mutants of CtBP exhibit similar genetic interactions with pnr and ac, as does tou, and CtBP physically interacts with Pannier and U-shaped (Biryukova and Heitzler 2008; Stern et al. 2009).
Here we demonstrate that Tou and CtBP function as repressors of genes of AS-C in vivo. Our data provide evidence that CtBP tethers Tou to regulatory regions of ac and sc and that Tou is required for the corepressor function of CtBP. In the future, it will be interesting to examine whether particular sequence-specific transcription factors such as U-shaped exhibit physical and genetic interactions with Tou. Furthermore, it will be important to understand how biochemical activities of ToRC mediate remodeling of nucleosome structure in AS-C and affect transcriptional regulation of ac and sc.
In previous studies, it has been assumed that mammalian TIP5 is localized exclusively to the nucleolus, where it regulates transcription by RNA polymerase I (Nemeth et al. 2004). NoRC is localized to the nucleolus by a complex mechanism that involves direct physical interactions of TIP5 with short RNA originating from the intergenic spacer in the rRNA gene cluster (IGS transcripts) and with TTF-I, a general transcription factor of RNA polymerase I (Strohner et al. 2001). Mammalian NoRC can also regulate replication of rDNA in cultured cells (Zhou et al. 2002; Li et al. 2004) through mechanisms that involve histone and DNA methylation (Santoro and Grummt 2005) and IGS transcripts (Mayer et al. 2006).
Our immunoprecipitation experiments indicate that in HeLa cells, human TIP5 associates with CtBP, which is recruited to and regulates genes that are transcribed by polymerase II. Thus, the existence of an alternative TIP5/Tou-containing complex (ToRC) is conserved in evolution from flies to mammals (Fig. 1H). Since NIH-3T3 cells exhibit exclusive nucleolus-specific IF staining with anti-TIP5 antibody, it is possible that these cells predominantly express the truncated form of TIP5 or limiting amounts of CtBP, which would largely abolish the formation of ToRC and tethering of TIP5 to sites outside of the nucleolus.
We discovered that Tou/TIP5 is localized at multiple genomic sites, where it is tethered by a polymerase II-specific corepressor, CtBP. Based on molecular and genetic interaction data, the stable complex of Tou, ISWI, and CtBP mediates at least some aspects of the regulatory function of CtBP. On the other hand, C-terminally truncated polypeptide isoforms of Tou fail to interact with CtBP and are instead recruited to nucleoli. Thus, depending on the primary structure and interaction partners, TIP5/Tou targets ATP-dependent nucleosome assembly/remodeling activity of ISWI to alternative genomic sites that undergo transcription by RNA polymerase I or II. In the future, it will be interesting to examine how the primary structure of Tou isoforms and, potentially, their association with CtBP affect the ability of Tou to interact with TTF-I and IGS transcripts and vice versa. These analyses will help to understand the differential distribution of NoRC/ToRC and the dual role of TIP5/Tou in regulation of transcription by RNA polymerases I and II.
Materials and methods
Ectopic expression of Tou-V5 and purification of protein complexes from S2 cells
Full-length Tou cDNA was prepared from three EST clones and two PCR fragments. The cDNA corresponds to the longest predicted product of tou (Tou-RA) and encodes a 2999-amino-acid protein (Tou-PA).
Drosophila S2 cells (Invitrogen) were grown in Schneider's Drosophila medium (Invitrogen) containing 10% FBS (Gemini Bioproducts). Cells (3 × 106) were cotransfected with pMT-Tou-V5 and pCoHygro vector (Invitrogen) by calcium phosphate transfection kit (Invitrogen) in the mass ratio of 19:1. After 2 d, standard growth medium was replaced by selective medium containing 300 mg/mL hygromycin-B (Invitrogen), and resistant cells were collected and replated after 4 wk of selection.
Stably transfected S2 cells were grown in 2 L of selective medium to mid-log phase (∼4 × 106 cells per milliliter), and the expression of recombinant Tou-V5 protein was induced by adding copper sulfate (II) to 0.5 mM. After 24 h of induction, the cells were harvested and washed with ice-cold PBS. The cell pellet (∼8 mL) was resuspended in 5 vol of buffer H (10 mM Tris-HCl at pH 7.9, 10 mM KCl, 0.75 mM spermidine, 0.15 mM spermine, 0.1 mM EDTA, 0.1 mM EGTA) that contained 2 mM DTT and protease inhibitors (0.2 mM PMSF, 1 mM benzamidine-HCl, 1 mM sodium metabisulfite, 2 mg/mL each aprotinin, leupeptin, and pepstatin).
The cells were disrupted by 20 strokes of a Wheaton Dounce homogenizer (pestle A), and nuclei were pelleted by centrifugation (8000 rpm for 10 min in SS-34 rotor; Beckman Coulter). The nuclear pellet (∼1 mL) was resuspended in a Dounce homogenizer (pestle A) in an equal volume of buffer L (50 mM Tris-HCl at pH 7.5, 20% glycerol, 10% sucrose, 0.6 M KCl, 5 mM MgCl2, 0.1 mM EDTA) that contained 2 mM DTT and protease inhibitors and was extracted for 30 min on ice. The nuclear extract was cleared by centrifugation (25,000 rpm for 1 h in TLA-120.1; Beckman Coulter) and dialyzed against two changes of 2 L of buffer TM (50 mM Tris-HCl at pH 7.9, 10% glycerol, 100 mM KCl, 2 mM MgCl2, 1 mM EDTA, 2 mM DTT) containing protease inhibitors for 2 h at 4°C.
The nuclear extract (∼2 mL) was mixed with 0.2 mL of V5-agarose slurry (Sigma). After incubation for 4 h, the resin was briefly spun in a microcentrifuge and washed five times with 1 mL of buffer W (25 mM HEPES at pH 7.6, 10% glycerol, 0.1 mM EDTA, 100 mM NaCl, 0.01% NP-40, 1 mM DTT, protease inhibitors). The proteins were eluted by four successive additions of 100 μL of buffer W additionally containing 0.4 mg/mL recombinant insulin and 0.8 mg/mL V5 peptide and were analyzed by SDS-PAGE and Coomassie or silver staining. The identities of protein bands in Coomassie-stained gels were determined by mass spectrometry. Full details of cloning and purification procedures are available on request.
Antibodies, Western blot, and immunoprecipitation
cDNA fragments corresponding to Acf1 amino acid residues 1–311, Tou residues 1033–1265 and 2670–2999, and full-length CtBP cDNA were amplified by PCR. The fragments were cloned in pET-29b vector. The respective C-terminally His-tagged polypeptides (Acf1-M17, Tou-M, Tou-C, and CtBP) were expressed in bacteria, purified by Ni-NTA chromatography in 8 M urea, and used as antigens to raise polyclonal antibodies. Tou-M antibody was raised in rabbits, and Tou-C antibodies were raised in rabbits and chickens, whereas Acf1-M17 and CtBP antibodies were raised in guinea pigs. The rabbit polyclonal Acf1 antibody recognizing the C-terminal portion of Acf1 (1290–1479 residues, Acf1-M23) is described elsewhere (Fyodorov et al. 2004). Rabbit polyclonal CtBP and ISWI antibodies were gifts of Ken Cadigan and Jim Kadonaga, respectively. Mouse monoclonal AJ1 antibody was obtained from the Developmental Studies Hybridoma Bank.
For whole-animal lysate analyses, ∼100 mg of fly embryos or L3 larvae was resuspended in 500 μL of 2× SDS-PAGE loading buffer, homogenized with a Roto-Dounce (Fisher Scientific), and boiled. Ten microliters of lysate per lane was assayed by Western blot. Equal protein loading was controlled by Ponceau S staining of Western membranes and Coumassie staining of equivalently loaded SDS-PAGE gels.
For immunoprecipitation, 300 μL of Drosophila embryos or HeLa nuclear extract (>5 mg of total protein) was used. The extracts were incubated for 3 h at 4°C with either of the following antibodies: 10 μL of guinea pig anti-CtBP, 20 μL of rabbit anti-Tou-M, 10 μL of guinea pig anti-Acf1-M17, 10 μL of rabbit anti-hTIP5 (Invitrogen), 10 μL of mouse anti-hCtBP (Santa Cruz Biotechnology), or 10 μL of normal IgG (Santa Cruz Biotechnology). Immunocomplexes were collected by addition of 25 μL of protein A-agarose (Sigma) for 2 h at 4°C. After washing four times with 1 mL of buffer HEG (25 mM HEPES at pH 7.6, 0.1 mM EDTA at pH 8.0, 10% glycerol) + 0.15 M NaCl, the immunoprecipitated proteins were eluted with 2× SDS-PAGE loading buffer and analyzed by SDS-PAGE and Western blot.
The following antibodies were used for Western detection: rabbit anti-Tou-C or Tou-M (1:10,000), rabbit anti-Acf1-M23 (1:10,000), rabbit anti-ISWI (1:25,000), guinea pig anti-CtBP (1:2000), rabbit anti-hTIP5 (1:1000; Invitrogen), goat anti-hSNF2H (1:20; Santa Cruz Biotechnology), and rabbit or mouse anti-hCtBP (1:1000; Santa Cruz Biotechnology). Chicken anti-Tou-C antibody (1:2000) was used for Western detection of Tou in material that was immunoprecipitated by rabbit anti-Tou-M (Fig. 1G, “Tou” and the adjacent “INP” lanes). Corresponding HRP-conjugated secondary antibodies (goat anti-rabbit, rabbit anti-goat, goat anti-mouse, rabbit anti-chicken, or donkey anti-guinea pig from Jackson Laboratories) were used at 1:5000 dilution.
Purification of the native form of Tou
Native Tou was purified from Drosophila embryos that were collected 0–12 h after egg deposition. Nuclear extract (∼200 g of embryos) was prepared as described (Kamakaka and Kadonaga 1994). The extract (∼25 mL) was applied to a 50-mL P11 phosphocellulose (Whatman) column equilibrated in HEG buffer containing 0.1 M NaCl, 0.01% NP-40, 1 mM DTT, 0.2 mM PMSF, 10 mM glycerophosphate, and 0.5 mM benzamidine. The column was washed with 5 column volumes (cv) of HEG + 0.1 M NaCl, which was followed by a linear gradient from 0.1 M to 1 M NaCl in HEG buffer (4.5 cv). Fractions were assayed for the presence of Tou by Western blot with Tou-C antibodies. Peak fractions of Tou-immunoreactive material (∼25 mL) were pooled and dialyzed against 4 L of buffer R (10 mM HEPES at pH 7.5, 10% glycerol, 1.5 mM MgCl2, 0.5 mM EGTA, 1 mM DTT, 0.2 mM PMSF, 10 mM glycerophosphate, 0.5 mM benzamidine) at 4°C until the sample conductivity was equivalent to that of Buffer R + 0.1 M NaCl.
The dialyzed sample was applied to a 4-mL Source 15S (GE Biosciences) column equilibrated to buffer R + 0.1 M NaCl. The column was washed with 5 cv of buffer R + 0.1 M NaCl, and the retained proteins were eluted by a linear gradient from 0.1 M to 1 M NaCl in buffer R (10 cv). Fractions were assayed for the presence of ToRC by Western blot with Tou-C, ISWI, and CtBP antibodies. Peak Tou-containing fractions (∼4 mL) were pooled and dialyzed against two changes of 2 L of buffer HEG + 0.1 M NaCl for 2 h at 4°C.
Dialyzed material was applied to a 0.5-mL Source 15Q (GE Biosciences) column equilibrated to HEG + 0.1 M NaCl. The column was washed with 5 cv of equilibration buffer, and the wash was followed by a linear gradient from 0.1 M to 1 M NaCl in HEG (8.5 cv). The fractions were assayed for the presence of ToRC as described above. Tou-containing fractions (1 mL) were pooled and applied to a preformed 10%–45% glycerol gradient in HEG + 0.15 M NaCl. After centrifugation in SW-41 (Beckman Coulter) at 40,000 rpm for 16 h, the fractions were collected and analyzed by Western blot.
Recombinant proteins and chromatin assembly in vitro
Tou cDNA was cloned from pMT-Tou-V5 into pFastBac1 vector (Invitrogen) in-frame with C-terminal His and Flag tags. Full-length CtBP cDNA was amplified by PCR from an EST clone (GH20987) and cloned into pFastBac1 vector. Specific details of cloning are available on request. Tou-Flag and CtBP (untagged) baculoviruses were prepared using BacToBac system (Invitrogen). Untagged ISWI, Flag-ISWI, and Acf1-Flag baculoviruses are described elsewhere (Ito et al. 1999; Fyodorov and Kadonaga 2003).
Recombinant ACF, ISWI, Tou, and its complexes with ISWI and/or CtBP were synthesized in Sf9 cells and purified by Flag immunoaffinity chromatography as described (Ito et al. 1999). Because of the low yield of recombinant Tou (<100 ng per 20-cm cell culture plate) and contamination with nucleic acids, all recombinant proteins were additionally purified and concentrated by Source 15Q chromatography. Protein concentrations were determined by SDS-PAGE and Coomassie staining along with BSA protein mass standards (Pierce).
Chromatin assembly was performed in a defined system (Fyodorov and Kadonaga 2003) that contained native Drosophila core histones, recombinant NAP-1 and ISWI, or ISWI-containing complexes. See the legend for Figure 2 for concentrations of components. Partial micrococcal nuclease digestion and DNA supercoiling assays were performed as described (Fyodorov and Levenstein 2002; Fyodorov and Kadonaga 2003).
Fly stocks and genetic experiments
Flies were reared, and crosses were performed at 25°C on standard cornmeal/molasses medium with dry yeast added to the surface. For the analyses of genetic interactions between tou and CtBP, the following fly strains were generated in a series of crosses: tou1/CyO; Df(3L)Ly/TM6B, Tb; tou1/CyO; CtBP87De-10/TM6B, Tb; Df(2R)en-SFX31/CyO; CtBP87De-10/TM6B, Tb; nocSco/CyO; CtBP03463/TM6B, Tb; tou1/tou1; CtBP03463/TM6B, Tb; and tou1/Df(2R)en-SFX31; CtBP03463/TM6B, Tb.
Parents with double mutations were mated inter se or with appropriate counterparts as shown in Table 1, and the appearance of double-homozygous mutant progeny (adult or pharate adult) was scored based on balancer markers.
Indirect IF
Antibody staining of polytene chromosomes was performed as described (Emelyanov et al. 2010). Salivary glands were dissected in PBS (pH 7.5) + 0.1% Triton X-100 solution and incubated for 30 sec (Tou-M and AJ1 antibodies) or for 3 min (Tou-C and CtBP antibodies) in fixing solution (3.7% formaldehyde, 1% Triton X-100 in PBS at pH 7.5). Fixed tissues were then transferred to 3.7% formaldehyde and 50% acetic acid for 2 min 45 sec, and the glands were squashed. Slides were frozen in liquid nitrogen and immersed in PBS. After washing twice in TBS (20 mM Tris-HCl at pH 7.5, 150 mM NaCl) for 15 min, the slides were incubated with blocking solution (TBS + 3% BSA, 0.05% Triton X-100) for 30 min. Primary antibodies were diluted in blocking solution supplemented with 2% normal goat serum and added to the slide. Affinity-purified rabbit Tou-C, Tou-M, and guinea pig CtBP antibodies were diluted 1:20, and monoclonal AJ1 antibody was used at 1:50 dilution. Overnight incubation in a humid chamber at 4°C followed. Slides were rinsed with TBS and washed three times for 15 min in blocking solution. After rinsing twice with TBS, the secondary antibody was added. Secondary antibodies (anti-rabbit Alexa Fluor 594, anti-mouse Alexa Fluor 488, or anti-guinea pig Alexa Fluor 488; Invitrogen) were diluted 1:200 in blocking solution containing 2% normal serum and incubated with the slides for 40 min at room temperature. The slides were washed as described for primary antibodies. After staining for 10 min in DAPI solution (0.5 μg/mL in PBS) and washing for 10 min in PBS, VectaShield (Vector Laboratories) was added before a coverslip was mounted to prevent bleaching.
For neuroblast staining, larval brains were dissected in PBS (pH 7.5) and 0.1% Triton X-100 solution and incubated for 30 sec in fixing solution (3.7% formaldehyde, 1% Triton X-100 in PBS at pH 7.5). Fixed brains were then transferred to 3.7% formaldehyde and 50% acetic acid for 2 min 45 sec and squashed. Immunostaining was performed exactly as described for polytene chromosomes staining.
Quantitative ChIP
ChIP was performed as described (Lu et al. 2009). Approximately 200 mg of L3 larvae was collected and homogenized with a Dounce glass homogenizer, type A pestle (10 strokes each), in 2 mL of cross-linking buffer containing 1.8% formaldehyde. The material was cross-linked for 15 min at room temperature, and the cross-linking was terminated by addition of 0.1 mL of 2.5 M glycine. The homogenate was washed twice with cold PBS, and nuclei were prepared as described (Takahashi et al. 2000). Chromatin was sonicated to an average size of ∼500 base pairs (bp) with Bioruptor (Diagenode, model UCD 200) in a dry ice–ethanol bath.
The following antibodies were used: normal rabbit immunoglobulin G (IgG) (Santa Cruz Biotechnology), guinea pig polyclonal anti-CtBP, and rabbit polyclonal anti-Tou-M. Approximately 6 μg of DNA and 4–10 μL of antibody were used per ChIP. After immunoprecipitation and cross-link reversal, the DNA was isolated by QIAquick PCR purification kit (Qiagen). Samples were analyzed quantitatively by real-time PCR (ABI Prism 7700, Applied Biosystems) using the method described in Garrett et al. (2005). Primer sequences are available in the Supplemental Material. Each ChIP was performed in duplicate, and each sample was analyzed in three independent real-time PCR reactions.
Quantitative real-time RT–PCR
Total RNA from 40 larvae of each genotype was isolated by RiboPure kit (Ambion) and quantitated with an Ultraspec 2100 spectrophotometer (GE Bioscience). One microgram of total RNA was treated with RNase-free DNase I (Promega), and oligo(dT)-primed cDNA was prepared using the SuperScript III kit (Invitrogen). Real-time quantitative PCR reactions were carried out in an ABI Prism 7700 sequence detection system (Applied Biosystems). One-step RT–PCR was done using a SYBR Green quantitative RT–PCR kit as per the manufacturer's instructions. To quantitate the expression levels, CT values of an endogenous reference gene, rpL32, were included. Each RNA sample was prepared in duplicate, and all reactions were carried out in triplicate, along with no-template controls. Primer sequences are available in the Supplemental Material.
Acknowledgments
We are grateful to Barbara Birshtein (AECOM) for HeLa nuclear extracts, and Jim Kadonaga (University of California at San Diego) and Kenneth Cadigan (University of Michigan) for antibodies. We thank Konstantin Beirit, Barbara Birshtein, Jason Ford, Andreas Jenny, Harsh Kavi, Michael Keogh, Alexandra Lusser, Arthur Skoultchi, and Na Xu for critical reading of the manuscript and valuable suggestions. We also thank Anna Makase and Lyudmila Kotlovanova for help with immunostaining techniques. Protein sequencing was performed by the Proteomics Resource Center at Rockefeller University. This work was supported by grants from the NIH (GM074233) to D.V.F. and from RFBR (09-04-01676-a) to A.Y.K. D.V.F. was a Scholar of the Sidney Kimmel Foundation for Cancer Research. A.Y.K was additionally supported by the “Genetic Diversity” Program of Presidium of the Russian Academy of Sciences.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.180604.111.
References
- Adams CR, Kamakaka RT 1999. Chromatin assembly: Biochemical identities and genetic redundancy. Curr Opin Genet Dev 9: 185–190 [DOI] [PubMed] [Google Scholar]
- Akey CW, Luger K 2003. Histone chaperones and nucleosome assembly. Curr Opin Struct Biol 13: 6–14 [DOI] [PubMed] [Google Scholar]
- Armstrong JA, Bieker JJ, Emerson BM 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95: 93–104 [DOI] [PubMed] [Google Scholar]
- Biryukova I, Heitzler P 2008. Drosophila C-terminal binding protein, dCtBP is required for sensory organ prepattern and sharpens proneural transcriptional activity of the GATA factor Pnr. Dev Biol 323: 64–75 [DOI] [PubMed] [Google Scholar]
- Chinnadurai G 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell 9: 213–224 [DOI] [PubMed] [Google Scholar]
- Chinnadurai G 2007. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol 39: 1593–1607 [DOI] [PubMed] [Google Scholar]
- Dang W, Bartholomew B 2007. Domain architecture of the catalytic subunit in the ISW2–nucleosome complex. Mol Cell Biol 27: 8306–8317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet 27: 187–190 [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC 1995. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res 23: 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW 1994. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol 14: 2225–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov AV, Konev AY, Vershilova E, Fyodorov DV 2010. Protein complex of Drosophila ATRX/XNP and HP1a is required for the formation of pericentric β-heterochromatin in vivo. J Biol Chem 285: 15027–15037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM, 2006. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J 25: 2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauvarque MO, Laurenti P, Boivin A, Bloyer S, Griffin-Shea R, Bourbon HM, Dura JM 2001. Dominant modifiers of the polyhomeotic extra-sex-combs phenotype induced by marked P element insertional mutagenesis in Drosophila. Genet Res 78: 137–148 [DOI] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T 2001. Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev 11: 148–154 [DOI] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT 2001. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106: 523–525 [DOI] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT 2002. Binding of Acf1 to DNA involves a WAC motif and is important for ACF-mediated chromatin assembly. Mol Cell Biol 22: 6344–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT 2003. Chromatin assembly in vitro with purified recombinant ACF and NAP-1. Methods Enzymol 371: 499–515 [DOI] [PubMed] [Google Scholar]
- Fyodorov DV, Levenstein ME 2002. Chromatin assembly in Drosophila systems. In Current protocols in molecular biology (ed. FM Ausubel et al.), pp. 21.27.21–21.27.27. Wiley & Sons, New York. doi: 10.1002/0471142727.mb2107s58 [Google Scholar]
- Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT 2004. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev 18: 170–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia MJ, Ramain P, Simpson P, Modolell J 1999. Different contributions of pannier and wingless to the patterning of the dorsal mesothorax of Drosophila. Development 126: 3523–3532 [DOI] [PubMed] [Google Scholar]
- Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK 2005. Chromatin architecture near a potential 3′ end of the igh locus involves modular regulation of histone modifications during B-cell development and in vivo occupancy at CTCF sites. Mol Cell Biol 25: 1511–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Skarmeta JL, Rodriguez I, Martinez C, Culi J, Ferres-Marcó D, Beamonte D, Modolell J 1995. Cis-regulation of achaete and scute shared enhancer-like elements drive their coexpression in proneural clusters of the imaginal discs. Genes Dev 9: 1869–1882 [DOI] [PubMed] [Google Scholar]
- Gorbalenya AW, Koonin EV 1993. Helicases: Amino acid sequence comparisons and structure–function relationships. Curr Opin Struct Biol 3: 419–429 [Google Scholar]
- Hassan AH, Neely KE, Workman JL 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104: 817–827 [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379 [DOI] [PubMed] [Google Scholar]
- Haushalter KA, Kadonaga JT 2003. Chromatin assembly by DNA-translocating motors. Nat Rev Mol Cell Biol 4: 613–620 [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90: 145–155 [DOI] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev 13: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell 11: 377–389 [DOI] [PubMed] [Google Scholar]
- Kamakaka RT, Kadonaga JT 1994. The soluble nuclear fraction, a highly efficient transcription extract from Drosophila embryos. Methods Cell Biol 44: 225–235 [DOI] [PubMed] [Google Scholar]
- Kumar R, Wang RA, Barnes CJ 2004. Coregulators and chromatin remodeling in transcriptional control. Mol Carcinog 41: 221–230 [DOI] [PubMed] [Google Scholar]
- Kuppuswamy M, Vijayalingam S, Zhao LJ, Zhou Y, Subramanian T, Ryerse J, Chinnadurai G 2008. Role of the PLDLS-binding cleft region of CtBP1 in recruitment of core and auxiliary components of the corepressor complex. Mol Cell Biol 28: 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci 107: 14075–14080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Reinberg D 2011. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev 21: 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Santoro R, Koberna K, Grummt I 2004. The chromatin remodeling complex NoRCcontrolsreplication timing of rRNA genes. EMBO J 24: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442: 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YI, Chang MV, Li HE, Barolo S, Chang JL, Blauwkamp TA, Cadigan KM 2008. The chromatin remodelers ISWI and ACF1 directly repress Wingless transcriptional targets. Dev Biol 323: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, LeRoy G, Wang YH, Reinberg D 2001. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev 15: 2837–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, Skoultchi AI 2009. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev 23: 452–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A, Urwin DL, Kadonaga JT 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 12: 160–166 [DOI] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R 2006. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361 [DOI] [PubMed] [Google Scholar]
- Mello JA, Almouzni G 2001. The ins and outs of nucleosome assembly. Curr Opin Genet Dev 11: 136–141 [DOI] [PubMed] [Google Scholar]
- Memedula S, Belmont AS 2003. Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr Biol 13: 241–246 [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487 [DOI] [PubMed] [Google Scholar]
- Nemeth A, Strohner R, Grummt I, Längst G 2004. The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res 32: 4091–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orihara-Ono M, Suzuki E, Saito M, Yoda Y, Aigaki T, Hama C 2005. The slender lobes gene, identified by retarded mushroom body development, is required for proper nucleolar organization in Drosophila. Dev Biol 281: 121–133 [DOI] [PubMed] [Google Scholar]
- Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C 2001. Cooperation between C/EBPα TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev 15: 3208–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya M, Nair A, Clynes D, Morillon A, Mellor J 2009. Nucleosome remodeling and transcriptional repression are distinct functions of Isw1 in Saccharomyces cerevisiae. Mol Cell Biol 29: 2419–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM 1998. Drosophila CtBP: A Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J 17: 2067–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramain P, Khechumian R, Khechumian K, Arbogast N, Ackermann C, Heitzler P 2000. Interactions between Chip and the Achaete/Scute-Daughterless heterodimers are required for Pannier-driven proneural patterning. Mol Cell 6: 781–790 [DOI] [PubMed] [Google Scholar]
- Ransom M, Dennehey BK, Tyler JK 2010. Chaperoning histones during DNA replication and repair. Cell 140: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T 2011. The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J 30: 2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Grummt I 2005. Epigenetic mechanism of rRNA gene silencing: Temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol 25: 2539–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MD, Aihara H, Roccaro GA, Cheung L, Zhang H, Negeri D, Nibu Y 2009. CtBP is required for proper development of peripheral nervous system in Drosophila. Mech Dev 126: 68–79 [DOI] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Längst G, Grummt I 2001. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 20: 4892–4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD 2000. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes Dev 14: 804–816 [PMC free article] [PubMed] [Google Scholar]
- Torigoe SE, Urwin DL, Ishii H, Smith DE, Kadonaga JT 2011. Identification of a rapidly formed nonnucleosomal histone–DNA intermediate that is converted into chromatin by ACF. Mol Cell 43: 638–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanolst L, Fromental-Ramain C, Ramain P 2005. Toutatis, a TIP5-related protein, positively regulates Pannier function during Drosophila neural development. Development 132: 4327–4338 [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388: 598–602 [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL 2010. Inducible gene expression: Diverse regulatory mechanisms. Nat Rev Genet 11: 426–437 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Yudkovsky N, Logie C, Hahn S, Peterson CL 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev 13: 2369–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Santoro R, Grummt I 2002. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 21: 4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]