Abstract
The importance and diversity of calcium signaling in the brain is mirrored by the expression of a multitude of voltage-activated calcium channel (CaV) isoforms. Whereas the overall distributions of α1 subunits are well established, the expression patterns of distinct channel isoforms in specific brain regions and neurons, as well as those of the auxiliary β and α2δ subunits are still incompletely characterized. Further it is unknown whether neuronal differentiation and activity induce changes of CaV subunit composition. Here we combined absolute and relative quantitative TaqMan reverse transcription PCR (RT-PCR) to analyze mRNA expression of all high voltage-activated CaV α1 subunits and all β and α2δ subunits. This allowed for the first time the direct comparison of complete CaV expression profiles of mouse cortex, hippocampus, cerebellum, and cultured hippocampal neurons. All brain regions expressed characteristic profiles of the full set of isoforms, except CaV1.1 and CaV1.4. In cortex development was accompanied by a general down regulation of α1 and α2δ subunits and a shift from β1/β3 to β2/β4. The most abundant CaV isoforms in cerebellum were CaV2.1, β4, and α2δ-2, and in hippocampus CaV2.3, β2, and α2δ-1. Interestingly, cultured hippocampal neurons also expressed the same CaV complement as adult hippocampus. During differentiation specific CaV isoforms experienced up- or down-regulation; however blocking electrical activity did not affect CaV expression patterns. Correlation analysis of α1, β and α2δ subunit expression throughout all examined preparations revealed a strong preference of CaV2.1 for β4 and α2δ-2 and vice versa, whereas the other α1 isoforms were non-selectively expressed together with each of the other β and α2δ isoforms. Together our results revealed a remarkably stable overall Ca2+ channel complement as well as tissue specific differences in expression levels. Developmental changes are likely determined by an intrinsic program and not regulated by changes in neuronal activity.
Keywords: VGCC, Ca2+, realtime RT-PCR, beta, alpha(2)delta, mRNA distribution
Voltage-activated Ca2+ channels (CaV) control multiple neuronal functions including transmitter release, gene transcription, and synaptic plasticity (Catterall, 2000). High voltage-activated CaVs are heteromultimers consisting of a pore-forming α1 and the auxiliary α2δ and β subunits (Catterall et al., 2005). Seven genes encode for α1 subunits of L-type (CaV1.1 to CaV1.4) and non L-type (CaV2.1 to CaV2.3) channels, and four genes for each of the auxiliary β and α2δ subunits (Dolphin, 2003; Davies et al., 2007). Most of the subunit isoforms are expressed in the central nervous system (Ludwig et al., 1997; Dolphin, 2003; Davies et al., 2007).
The L-type channels CaV1.2 and CaV1.3 perform primarily postsynaptic functions in transcriptional regulation and synaptic plasticity, whereas the non-L-type channels (CaV2.1, CaV2.2, CaV2.3) are responsible for neurotransmitter release. While some peripheral neurons, like superior cervical ganglion cells, are known to express only one presynaptic channel (Mochida et al., 2003), it is evident that the majority of brain regions and neurons express the whole plethora of CaVs (Vacher et al., 2008). Considering the additional diversity of the auxiliary subunits and the fact that all α1 subunits seem to be capable of assembling with all β and α2δ isoforms, the complexity of possible subunit compositions becomes enormous. For example three distinct presynaptic CaV2 α1 isoforms plus three α2δ and four β subunits already give 36 possible channel compositions; and that is without including the splice variants existing for all of the isoforms.
In light of this subunit diversity specific mechanisms must exist to assemble distinct α1/β/α2δ complexes in neurons. The simplest possible mechanism is to limit the number of isoforms expressed in a single cell at a given time. This is the case in skeletal muscle (CaV1.1/β1a/α2δ-1), cardiac myocytes (CaV1.2/β2/α2δ-1), and retina photoreceptor cells (CaV1.4/β2/α2δ-4) (Mori et al., 1991; Ball et al., 2002; Barnes and Kelly, 2002; Arikkath and Campbell, 2003; Wycisk et al., 2006). Similarly, the cerebellum shows a strong preference towards one subunit combination (CaV2.1/β4/α2δ-2) (Ludwig et al., 1997; Brodbeck et al., 2002). In contrast, the cerebral cortex shows a more heterogenous expression of CaV isoforms (Ludwig et al., 1997; Klugbauer et al., 1999). Existing evidence from electrophysiological, pharmacological, and immunostaining experiments indicates that these narrow and broad expression patterns in cerebellum and cortex respectively, are also reflected in the α1 subunit expression of specific types of neurons, like cerebellar granule cells and hippocampal pyramidal cells (Hell et al., 1993; Lorenzon and Foehring, 1995; Randall and Tsien, 1995; Westenbroek et al., 1995). However, little to no quantitative comparable data exist on the expression of CaV isoforms in different brain tissues and cells, and information on expression patterns of auxiliary subunits is sparse.
To bring more clarity into this complex situation we employed TaqMan quantitative RT-PCR (qRT-PCR) to measure the mRNA expression of all seven high voltage-activated CaV α1, and each of the four β and α2δ subunit genes. The generation of standard curves enabled the quantitative comparison of the transcript levels between the individual genes, and a rigorous normalization to endogenous reference genes allowed the direct comparison of the expression levels in mouse cortex, hippocampus, cerebellum, and cultured hippocampal neurons. All examined tissues and cells expressed the full complement of subunit isoforms, with the exception of CaV1.1 and CaV1.4. Characteristic developmental changes in the CaV subunit expression were evident in brain regions and cultured neurons. However, alteration of the electrical activity of cultured hippocampal neurons did not affect the CaV expression patterns. Together these data emphasize the great complexity of CaV expression in brain as well as in hippocampal pyramidal cells, and indicate a limited role of differential expression in controlling the subunit and isoform composition in neurons.
EXPERIMENTAL PROCEDURES
RNA isolation from cultured hippocampal neurons
Low-density cultures of hippocampal neurons were prepared from 16.5-day-old embryonic BALB/c mice as described (Obermair et al., 2003; Kaech and Banker, 2006). Briefly, dissected hippocampi were dissociated by trypsin treatment and trituration. Neurons were plated on poly-l-lysine-coated glass coverslips at a density of 7000 cells/cm2. After plating, cells were allowed to attach for 3–4 h before transferring the coverslips neuron-side-down into 60-mm culture dishes with a glial feeder layer. Neurons and glial feeder layer were cultured in serum-free neurobasal medium (Invitrogen GmbH, Karlsruhe, Germany) supplemented with Glutamax and B27 supplements (Invitrogen GmbH). Five or 24 days after plating coverglasses with neurons were removed from the dishes with glia cells, harvested by trypsin treatment, and homogenized using QiaShredder columns (Qiagen, GmbH, Hilden, Germany). Total RNA was extracted using the RNeasy Protect Mini Kit (Qiagen, GmbH, Hilden, Germany). Reverse transcription was performed on 5 μl of RNA using Superscript II reverse transcriptase (Invitrogen, Carlsbad, USA) and random hexamer primers (Promega, Madison WI, USA); the RT mix was incubated at 37 °C for 60 min.
RNA isolation from tissue
BALB/c mice of different age (E16, PD1, 2 and 8 weeks) were euthanized by CO2 exposure and decapitated. From each mouse total RNA was isolated from the entire cerebral hemispheres (referred to as cortex), the hippocampus, and the cerebellum. To this end the skull was opened from caudal to rostral and the brain was carefully removed and placed in ice cold Hank’s buffered salt solution. The entire cerebral hemispheres were isolated by separation from the diencephalon. The hippocampus was removed from one hemisphere using fine scissors. Next the entire cerebellum was cut from the brainstem. The respective tissues were further cut in four to six pieces and immediately transferred into RNAlater RNA Stabilization Reagent (Qiagen, GmbH, Hilden, Germany). Tissue samples were disrupted by using a rotor-stator homogenizer (Ultraturrax T8, IKA, Staufen, Germany) and QiaShredder columns. Total RNA was extracted from homogenized brain tissue using the RNeasy Protect Mini Kit (Qiagen, GmbH, Hilden, Germany). RNA concentrations were determined photometrically. Reverse transcription was performed on 1 μg of RNA using Superscript II reverse transcriptase (Invitrogen, Carlsbad CA, USA) and random hexamer primers (Promega, Madison WI, USA); RT mix was incubated for 60 min at 37 °C. Animal handling was in accordance with national and international standards of animal welfare.
Quantitative real time PCR
The relative abundance of different CaV subunit transcripts was assessed by TaqMan qRT-PCR using a standard curve method based on PCR products of known concentration. TaqMan gene expression assays (Table 1), designed to span exon–exon boundaries, were purchased from Applied Biosystems (Foster City, CA, USA). For each assay, flanking primer pairs (Eurofins MWG Operon, Ebersberg, Germany) were designed to amplify templates for the standard curves using cDNA from mouse whole brain (Table 2). PCR products were separated on 1.5% low melting point agarose gels (Amresco, Solon, OH, USA). Bands were excised, DNA was extracted using Nucleospin Extract II columns (Macherey-Nagel, Düren, Germany) and sequenced (Eurofins MWG Operon Sequencing Department, Martinsried, Germany) to confirm the integrity of the obtained fragments. Concentrations of PCR products were determined using Quant-IT PicoGreen dsDNA Assay Kit (Invitrogen). Standard curve dilution series ranging from 101 to 107 DNA molecules were generated in water containing 1 μg/ml of poly-dC-DNA (Midland, TX, USA). qRT-PCRs of the standard curve samples were performed in triplicates and samples without template (background) served as negative controls. In order to determine standard reliability all standard curves were repeated two to three times (including all steps, see above) over the course of 2 years. Finally, average linear regressions were calculated for the combined results of all standard curve replicates. Only data points in the logarithmic amplification range of the respective standard curve were included for regression analysis. The limits of detection (LOD) and quantification (LOQ) of each TaqMan assay were obtained by subtracting 3 and 10 times, respectively, the root mean squares of the residuals, including background values, from the cycle threshold (Ct) value of the Y-intercept (Table 3) (Corley, 2003). In cases where assays did not show background Ct values or the spread of the background Ct values was large, LOQ was determined by subtracting 10 times the root mean squares of the residuals, excluding background, from the Ct value of the Y-intercept (Table 3). All standard curve data were included for this analysis.
Table 1.
TaqMan gene expression assays for all high-voltage activated Ca2+ channel α1, β, and α2δ subunits
| Subunit | GenBank accession |
Assay ID | Exon boundary |
Intron length (bp) |
|---|---|---|---|---|
| CaV1.1 | NM_014193 | Mm00489257_m1 | 9–10 | 1768 |
| CaV1.2 | NM_009781 | Mm00437917_m1 | 8a–9 | 7241 |
| CaV1.3 | NM_028981 | Mm01209919_m1 | 29–30 | 5412 |
| CaV1.4 | NM_019582 | Mm00490443_m1 | 18–19 | 541 |
| CaV2.1 | NM_007578 | Mm00432190_m1 | 40–41 | 3281 |
| CaV2.2 | NM_007579 | Mm00432226_m1 | 38–39 | 2530 |
| CaV2.3 | NM_009782 | Mm00494444_m1 | 43–44 | 4240 |
| β 1 | NM_031173 | Mm00518940_m1 | 1–2 | 2575 |
| β 2 | NM_023116 | Mm00659092_m1 | 13–14 | 1148 |
| β 3 | NM_007581 | Mm00432233_m1 | 1–2 | 4274 |
| β 4 | NM_146123 | Mm00521623_m1 | 11–12 | 12,878 |
| α2δ-1 | NM_009784 | Mm00486607_m1 | 33–34 | 2583 |
| α2δ-2 | NM_020263 | Mm00457825_m1 | 1–2 | 25,769 |
| α2δ-3 | NM_009785 | Mm00486613_m1 | 5–6 | 44,477 |
| α2δ-4 | NM_001033382 | Mm01190105_m1 | 8–9 | 2407 |
Table 2.
cDNA specific primer sequences for standard template amplification
| Forward primer | Reverse primer | Fragment size (bp) | |
|---|---|---|---|
| CaV1.1 | 5′-gttacatgagctggatcacacag-3′ | 5′-atgagcatttcgatggtgaag-3′ | 349 |
| CaV1.2 | 5′-atgcaagacgctatgggctat-3′ | 5′-caggtagcctttgagatcttcttc-3′ | 201 |
| CaV1.3 | 5′-acattctgaacatggtcttcacag-3′ | 5′-aggacttgatgaaggtccacag-3′ | 327 |
| CaV1.4 | 5′-ctcttcatctgtggcaactacatc-3′ | 5′-gtaccaccttctccttgggtacta-3′ | 324 |
| CaV2.1 | 5′-ggtcacacctcacaagtccac-3′ | 5′ -ccagtcttctggaacatctcttg-3′ | 306 |
| CaV2.2 | 5′-cacttagacgaattcattcgagtct-3′ | 5′-tatcatgagagcagcatagacctt-3′ | 408 |
| CaV2.3 | 5′-aaggtaaagaaacagagacagcag-3′ | 5′ -gtctgttaccaccagagattgttg-3′ | 267 |
| β1 | 5′-gatcctctccatggtccagaa-3′ | 5′ -ctgcctccttccttaaggcttc-3′ | 266 |
| β2 | 5′-gactatctggaggcatactggaag-3′ | 5′-ctctcttgggtttcagagtcaaa-3′ | 317 |
| β3 | 5′-cccatgtatgacgactcctacg-3′ | 5′ -acagtagctgacattggtcctcac-3′ | 216 |
| β4 | 5′-gctgattaagtccagaggaaagtc-3′ | 5′ -tgtctcattcgctgactctgtaat-3′ | 288 |
| α2δ-1 | 5′-gcatgatgagacacctggttaata-3′ | 5′ -acagtccagtaaaccactgaatga-3′ | 347 |
| α2δ-2 | 5′ -ccgctcttgctcttgctg-3′ | 5′-cttcctgtccagcaggctct-3′ | 273 |
| α2δ-3 | 5′-gtatgaatacttcaatgctgtgctg-3′ | 5′-atttaatccctgggtactgtctga-3′ | 305 |
| α2δ-4 | 5′-cacatctcccaaagacatcgt-3′ | 5′-caaggaagtctctgcaaccag-3′ | 337 |
Table 3.
Properties of standard curves and limits of detection (LOD) and quantification (LOQ)
| B | SE-Ba | Y-int.b | SE-Ya | R2 | LODc | LOQd | |
|---|---|---|---|---|---|---|---|
| CaV1.1 | −3.449 | 0.028 | 38.02 | 0.13 | 0.998 | 3 | 15 |
| CaV1.2 | −3.648 | 0.023 | 38.54 | 0.11 | 0.998 | 37 | 142 |
| CaV1.3 | −3.692 | 0.043 | 41.14 | 0.19 | 0.995 | 60 | 78 |
| CaV1.4 | −3.420 | 0.029 | 37.41 | 0.13 | 0.997 | 5 | 34 |
| CaV2.1 | −3.556 | 0.043 | 38.36 | 0.20 | 0.995 | 4 | 102 |
| CaV2.2 | −3.497 | 0.053 | 37.86 | 0.25 | 0.993 | 5 | 155 |
| CaV2.3 | −3.506 | 0.040 | 37.23 | 0.18 | 0.995 | 4 | 59 |
| β1 | −3.474 | 0.037 | 36.34 | 0.17 | 0.995 | 3 | 34 |
| β2 | −3.491 | 0.048 | 39.48 | 0.22 | 0.989 | 6 | 190 |
| β3 | −3.520 | 0.039 | 37.58 | 0.18 | 0.993 | 12 | 114 |
| β4 | −3.535 | 0.050 | 38.01 | 0.23 | 0.992 | 5 | 137 |
| α2δ-1 | −3.436 | 0.058 | 36.80 | 0.27 | 0.990 | 5 | 127 |
| α2δ-2 | −3.572 | 0.036 | 38.07 | 0.17 | 0.997 | 22 | 117 |
| α2δ-3 | −3.533 | 0.035 | 37.88 | 0.16 | 0.994 | 4 | 59 |
| α2δ-4 | −3.412 | 0.028 | 37.70 | 0.13 | 0.996 | 3 | 27 |
SE-B, -Y, standard error of B and Y intercept, respectively.
Y-int., Y-intercept (Ct value).
LOD, limit of detection (number of transcripts).
LOQ, limit of quantification (number of transcripts).
qRT-PCR (50 cycles) was performed in duplicates using 20 ng total RNA equivalents of cDNA and the specific TaqMan gene expression assay for each 20 μl reaction in TaqMan Universal PCR Master Mix (Applied Biosystems). Measurements were performed on at least three independent RNA preparations from each tissue and developmental stage. Analyses were performed using the 7500 Fast System (Applied Biosystems).
Endogenous controls and data normalization
To compare the relative expression of distinct CaV subunits in different preparations, data were normalized based on the most stable control gene determined as previously described (Vandesompele et al., 2002). The endogenous control genes included were [name (gene symbol), assay ID (Applied Biosystems)]: β-cytoplasmic actin (ACTB), Mm00607939_s1; beta-2-microglobulin (B2M), Mm00437762_m1; glyceraldehyde-3-phosphate dehydrogenase (GAPD), Mm99999915_g1; hypoxanthine phosphoribosyl-transferase 1 (HPRT1), Mm00446968_m1; succinate dehydrogenase complex, subunit A (SDHA), Mm01352363_m1; tata box binding protein (TBP), Mm00446973_m1; transferrin receptor (TFRC), Mm00441941_m1. HPRT1 and SDHA were determined to be the most stably expressed reference genes when comparing all preparations (Suppl. 1). The Ct values for each CaV gene expression assay were recorded for each individual preparation. To allow a direct comparison between the expression levels in different tissues, we normalized all experiments to the Ct value of HPRT1 in the RNA preparation yielding the highest HPRT1 Ct value. Subsequently, normalized molecule numbers were calculated for each CaV subunit from their respective standard curve. For determining the number of molecules per neuron (see below), molecule numbers were calculated from the raw Ct values.
Estimating the number of transcript molecules per cultured neuron
To estimate the number of transcript molecules of each CaV subunit we first determined the number of neurons per coverslip. Coverslips of four 24 DIV hippocampal cultures were stained with Hoechst dye to distinguish nuclei and analyzed on an Axiovert 200M microscope (Carl Zeiss GmbH, Wien, Austria) using a 10× objective. The number of neurons per coverslip was extrapolated to the total number of neurons per RNA preparation and RT reaction and, finally, to the amount of RNA equivalents used for the qRT-PCR reaction.
Data analysis and statistics
Data were organized and analyzed using MS Excel and SPSS statistical software (SPSS Inc., Chicago, IL, USA) as indicated. Statistical significance was determined by Student’s t-test and ANOVA. To correct for multiplicity in analyses involving many pairwise comparisons the Holm procedure (Bonferroni step-down correction) was applied (Bender and Lange, 2001). After Holm correction P-values <0.05 were considered as statistically significant. All data are presented as mean±SE for the indicated number of experiments. Graphs and figures were generated using MS Excel, Origin 7, and Adobe Photoshop 8.0 software.
RESULTS
CaV expression profiles of mouse hippocampus, cortex, and cerebellum
Previous studies demonstrated the existence of mRNA and protein of CaV α1 (CaV1.2, CaV1.3, CaV2.1, CaV2.2, and CaV2.3), α2δ (α2δ-1, α2δ-2, α2δ-3), and of all four β subunits in the mammalian brain (for review see Catterall, 2000; Arikkath and Campbell, 2003). Nevertheless, quantitatively comparable expression levels of the distinct CaV subunit isoforms in different brain regions of a single mammalian species were missing. One reason being the inherent difficulty to quantitatively compare the outcomes of methods based on different antibodies, riboprobes, or PCR primers. Quantitative TaqMan RT-PCR analysis is the state-of-the art approach to analyze relative mRNA amounts in distinct probes, provided a suitable and stably expressed reference gene has been identified. Moreover, the quantitative comparison of the expression level of different genes is hampered by variations in the sensitivities and amplification kinetics of the various qRT-PCR assays. Therefore we combined relative quantification based on endogenous control genes with absolute quantification based on individual standard curves. This allowed us to determine the amount of each CaV isoform transcript in different mouse brain regions and cultured hippocampal neurons, and to get an accurate estimate on the quantities of each transcript relative to each other.
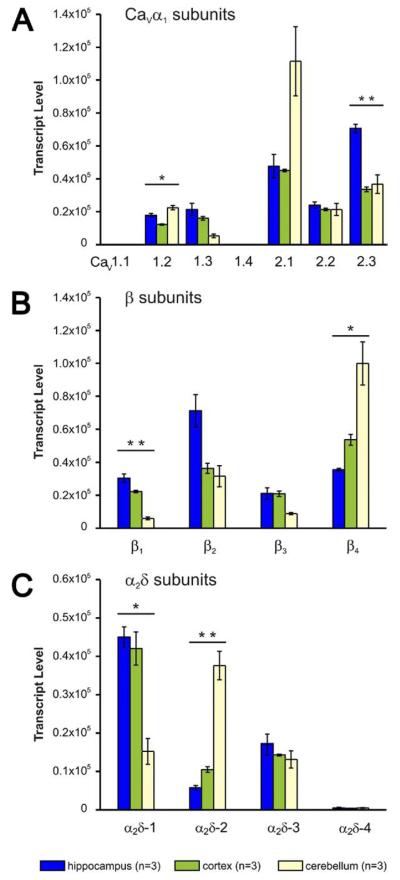
Hippocampus, cortex, and cerebellum of adult (8 weeks old) mice expressed all high voltage-activated CaV α1 subunits, except the skeletal muscle CaV1.1 and the retinal CaV1.4, which were expressed at ~1000-fold lower levels, around the limit of quantification (Fig. 1A). Furthermore, all four β subunit genes and three of the four α2δ subunit genes were robustly expressed in these brain tissues (Fig. 1B, C). α2δ-4 molecule numbers were above the limit of quantification in all qRT-PCR runs. However, when compared to the other auxiliary subunits its expression levels were negligible. Overall, the L-type channels (CaV1.2 and CaV1.3) were expressed at lower levels than each of the non-L-type channels (CaV2.1, CaV2.2, and CaV2.3). Whereas the expression profile was fairly similar between cortex and hippocampus, cerebellum showed some striking differences. CaV2.1, β4, and α2δ-2 were the dominating isoforms in cerebellum and were expressed two to threefold higher compared to cortex and hippocampus (Fig. 1A–C). The high expression of these three CaV isoforms in the cerebellum confirms previous findings (Ludwig et al., 1997; Hobom et al., 2000; Barclay et al., 2001; Cole et al., 2005) and thus strengthens the confidence in our new quantification method. Cerebellar expression levels of L-type channels also differed from those in the other two brain regions in that the ratio of CaV1.2 to CaV1.3 was 4:1 in cerebellum, whereas it was ~1:1 in cortex and hippocampus. Our observation that CaV1.3 transcripts accounted for only ~20% of cerebellar L-type channels is again consistent with previous observations (Koschak et al., 2007; Sinnegger-Brauns et al., 2009). Moreover, whereas α2δ-1 was the dominating α2δ subunit isoform in cortex and hippocampus, this isoform was markedly reduced in the cerebellum (Fig. 1C). Unexpectedly the CaV2.3 and β2 mRNAs were twofold higher in the hippocampus than in cortex and cerebellum. Actually they were the highest expressed α1 and β isoforms in the hippocampus. Finally, the expression of CaV2.2 and α2δ-3 was remarkably uniform throughout the three brain regions.
Fig. 1.
Expression profile of the high voltage-activated Ca2+ channel α1, β, and α2δ subunits in mouse hippocampus, cortex, and cerebellum. (A) Hippocampus (blue), cortex (green), and cerebellum (yellow) express all CaV α1 subunits except CaV1.1 and CaV1.4. Subunit expression levels in hippocampus and cortex are similar with the exception of CaV2.3, which is highest expressed in hippocampus. In cerebellum CaV2.1 is the most abundant isoform, CaV1.2 is higher and CaV1.3 lower than in hippocampus and cortex. CaV2.2 is uniformly expressed in all three brain regions. (B) mRNA of all four β subunits is present in hippocampus, cortex, and cerebellum. β2 and β4 are the dominant isoforms in hippocampus and cerebellum, respectively. In cortex β subunits are expressed at similar levels. (C) α2δ-1 is the major α2δ isoform in hippocampus and cortex, whereas in cerebellum it is α2δ-2. Levels of α2δ-3 are uniform throughout the brain regions tested. Compared to the other auxiliary subunits α2δ-4 expression levels were negligible, although above the limit of quantification in all qRT-PCR runs. * P<0.05; ** P<0.01; 2-way ANOVA plus post hoc ANOVA with Holm correction; error bars: ±SEM.
CaV expression patterns change during development
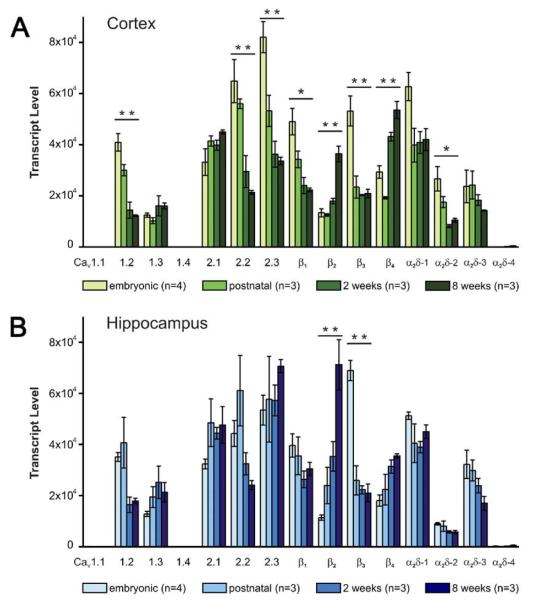
Because CaVs have been previously demonstrated to be important for neuronal maturation and development (Pravettoni et al., 2000; Splawski et al., 2004), we next sought to investigate whether and how their expression patterns change from late embryonic until adult stages. To this end we analyzed the expression profiles on embryonic day 16 (E16), postnatal day 1 (PD1), and at 2 weeks and 8 weeks of age (Fig. 2). In line with the expression profiles of 8 weeks old mice (Fig. 1), CaV1.1 and CaV1.4 were absent from both tissues at all time points (Fig. 2A, B). In cortex three different patterns of developmental changes could be observed (Fig. 2A): the majority of isoforms showed decreasing expression levels (CaV1.2, CaV2.2, CaV2.3, β1, β3, α2δ-2), four isoforms showed stable expression levels (CaV1.3, CaV2.1, α2δ-1 and α2δ-3), and the levels of two isoforms (β2 and β4) showed a significant increase with development. Interestingly, the total number of CaV α1 transcripts was about twofold larger in developing than in mature brain. A similar trend was observed for the α2δ subunits, but not for total β subunits. Individually, the β subunits showed very pronounced developmental changes. Whereas β1 and β3 levels declined as the majority of the α1 subunits, β2 and β4 expression levels significantly increased. Thus, in contrast to the α1 subunits, the total number of β subunit transcripts remained fairly constant during development. However, their ratio shifted from predominantly β1 and β3 in embryonic cortex, to mostly β2 and β4 in the mature cortex.
Fig. 2.
Developmental changes of CaV subunit mRNA expression in cortex and hippocampus. CaV subunit expression profiles were determined in embryonic (E16), postnatal (PD1), and 2 and 8 weeks old BALB/c mice. (A) During development cortical mRNA levels of CaV1.2, CaV2.2, CaV2.3, β1, β3, and α2δ-2 significantly decline, whereas levels of β2 and β4 increase. Levels of CaV1.3, CaV2.1, and α2δ-1 and α2δ-3 remain stable. (B) In hippocampus the overall developmental changes are less striking than in cortex. However, the increase in β2 and the significant drop of β3 levels between E16 and PD1 are more pronounced. In contrast to whole cortex, levels of CaV2.3 did not decline. Although total mRNA levels are negligible in comparison with the other α2δ subunits, expression of α2δ-4 increases ~20-fold during development. * P<0.05; ** P<0.01; 2-way ANOVA plus post hoc ANOVA with Holm correction; error bars: ±SEM.
In the developing hippocampus (Fig. 2B) expression levels of many subunits showed the same tendency as in whole cortex. The most striking difference was the fact that CaV2.3 and β1 levels did not show the developmental decline observed in cortex. The developmental increase of β2 was even more pronounced whereas β3 showed a dramatic drop within the short period between E16 and PD1. This drop was also observed in cortex (Fig. 2A). Surprisingly, in hippocampus expression of α2δ-4 increased ~20 fold during development, although still at a very low level.
Cultured hippocampal neurons express the same subunit isoforms as adult hippocampus
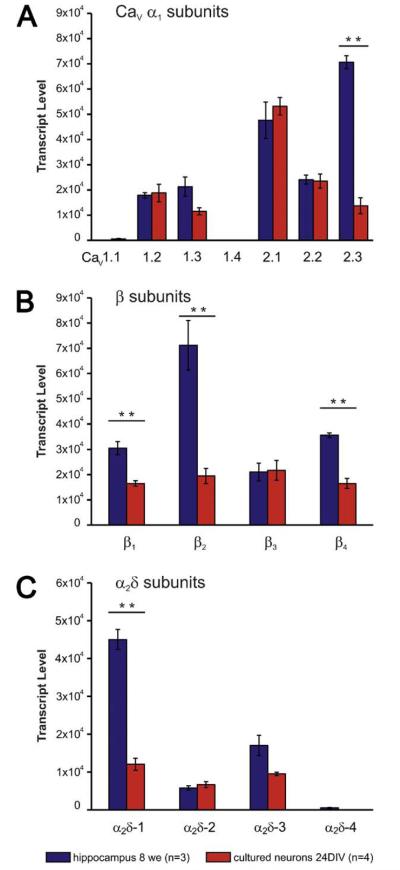
The overall CaV expression profile in hippocampus, cortex, and cerebellum represents the sum of RNAs derived from a large number of different neuronal and non-neuronal cell types. As a consequence, developmental changes may indicate an overall change in all cells, or it may result from manifold greater changes in subpopulations of cells. On the other hand, a decline of a specific transcript in one cell type might coincide with an increase of the same transcript in another cell type and thus mask the developmental change. Therefore, in addition to understanding the overall CaV channel expression patterns in brain and distinct brain regions, it is vitally important to understand the subunit expression in single types of neurons. Low density cultured hippocampal neurons serve as an ideal model to address this question. First, this cell culture system is a pure neuronal culture without glia. Second, it represents a highly homogenous culture with >90% glutamatergic pyramidal cells (Benson et al., 1994; Obermair et al., 2003). In light of this it is remarkable that qRT-PCR analysis revealed the same set of CaV subunit isoforms in differentiated cultured hippocampal neurons (24 DIV) as in adult hippocampus expressed (Fig. 3). This indicates that the complexity of CaV expression pattern exists in individual neuron types also and does not only arise from a mix of different cell types with distinct sets of calcium channels. Interestingly, with one exception (CaV2.3) the expression levels of the α1 isoforms were similar to those in hippocampus (Fig. 3A). Transcript levels of CaV2.3 in the culture were much lower than in hippocampus. In the culture CaV2.1 was the most abundant α1 subunit. Similar to CaV2.3, β2 and α2δ-1 (and to a lesser degree also β1 and β4) showed substantially lower expression levels in hippocampal neurons than in whole hippocampus. Consequently the hippocampus-specific expression profile of the auxiliary subunits was not observed in the cultured neurons, but β1 to β4 and α2δ-1 to α2δ-3 were all expressed at approximately equal amounts (Fig. 3B, C).
Fig. 3.
Expression profile of high voltage-activated Ca2+ channel α1, β, and α2δ subunits in differentiated cultured hippocampal neurons (24 DIV). (A) The majority of α1 subunits show similar expression levels in the cultured neurons as in 8 weeks old hippocampus. However, expression of CaV2.3 was 5-fold lower in cultured neurons. (B) In cultured neurons all β isoforms are expressed at equal amounts, but at significantly lower levels than in hippocampus. (C) α2δ subunits are expressed at equal amounts and generally lower levels than in hippocampal tissue. α2δ-4 levels were analyzed separately in five DIV and 24 DIV old neurons (cf. Fig. 4 and Suppl. Fig. 2), but always below detectability. ** P<0.01; 2-way ANOVA plus post hoc t-test with Holm correction; error bars: ±SEM.
Specific developmental upregulation of CaV2.1 and α2δ-2 in cultured hippocampal neurons
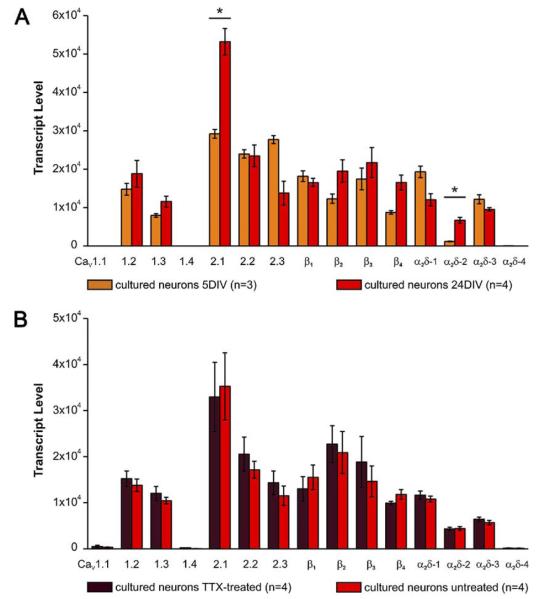
Cultured hippocampal neurons undergo dramatic morphological changes during growth and differentiation in vitro. The first week is characterized by massive neurite outgrowth. While first synapses appear around 3 DIV, the maturation of the dendritic tree starts around 5–7 DIV. In the following weeks the neurons develop a large and elaborate dendritic tree with numerous dendritic spines and an increasing number of synapses (Fletcher et al., 1994; Obermair et al., 2003). Interestingly, during this period of continuous differentiation from 5 DIV to 24 DIV most CaV subunit isoforms showed only a slight increase or remained expressed at constant levels (Fig. 4A). The notable exceptions were the P/Q-type channel CaV2.1 and the α2δ-2 subunit, both of which experienced a significant up-regulation. Among the β subunits β4 showed the strongest increase. In contrast, expression levels of CaV2.3 and α2δ-1 decreased during the same time frame.
Fig. 4.
Effects of development and neuronal activity on CaV subunit expression in cultured hippocampal neurons. (A) The majority of CaV isoforms shows a slight increase in transcript levels between 5 and 24 DIV. This increase is most obvious for CaV2.1, α2δ-2, and β4. In contrast to the overall trend, amounts of CaV2.3 and α2δ-1 transcripts decrease during in vitro development. (B) Blocking electrical activity by TTX for 24 h did not alter the expression level of any CaV subunit isoform. * P<0.05; 2-way ANOVA plus post hoc t-test with Holm correction; error bars: ±SEM.
Blocking neuronal activity does not alter the CaV expression pattern in cultured hippocampal neurons
Neurons adapt to alterations in the activity status and synaptic transmission by homeostatic plasticity (Turrigiano, 2008). Decreasing overall activity generally leads to an increase in the cellular excitability, whereas increased activity leads to a decrease therein. These homeostatic alterations are caused by both presynaptic and postsynaptic adaptations also involving gene transcription. Because CaVs are major constituents of both pre- and postsynaptic compartments and are indirectly (Deisseroth et al., 2003; Dolmetsch, 2003) and likely also directly (Gomez-Ospina et al., 2006; Subramanyam et al., 2009) involved in the regulation of transcription, we tested whether their expression patterns change upon blocking neuronal activity. Well differentiated cultured hippocampal neurons display robust spontaneous activity, which can be suppressed by tetrodotoxin (TTX) or further increased by blocking inhibitory input with bicuculline (Harms and Craig, 2005; Turrigiano and Nelson, 2004). Surprisingly, overnight application of TTX did not affect the expression level of any CaV subunit isoforms (Fig. 4B) although the same experimental paradigm induced the nuclear translocation of the CaV β4b subunit (Subramanyam et al., 2009). N-methyl-d-aspartate (NMDA) receptors are critically involved in the control of plasticity-related gene expression and long-term memory formation (e.g. Jordan and Kreutz, 2009), and blockage by dl-2-amino-5-phosphonopentanoic acid (dl-AP5) leads to a local increase of synaptic NMDA receptors without inducing overall homeostatic changes (Rao and Craig, 1997; Obermair et al., 2003; Turrigiano, 2008). However, also chronic blockage of NMDA receptors did not change the CaV expression profile (Suppl. 2).
DISCUSSION
This is the first study providing a comprehensive calcium channel expression profile of mouse brain and cultured hippocampal neurons. This study is unique in that it quantifies the expression of the full complement of high voltage-activated calcium channels α1 together with all α2δ and β subunits, and in that for the first time this quantification is thoroughly based on a quantitative assessment of the underlying assay variability, thus allowing a direct comparison of expression levels between all isoforms and across multiple preparations.
The CaV transcriptome of mouse brain regions and cultured hippocampal neurons
Comparing the calcium channel expression profiles of cortex, hippocampus, and cerebellum revealed expression of the same complement of CaV isoforms. Overall, expression of L-type calcium channels was lower than that of non-L-type channels, both individually and in sum. CaV1.2 and CaV1.3 were the only L-type calcium channels expressed in mouse brain. In the cerebellum they were found at a ratio of 4:1 (CaV1.2: CaV1.3), which is consistent with results from previous dihydropyridine binding studies and qRT-PCR analysis (Koschak et al., 2007; Sinnegger-Brauns et al., 2009). In cortex and hippocampus CaV1.2 and CaV1.3 were expressed at equal levels. Generally this is in agreement with published data on the relative L-type CaV distribution in different rat and mouse brain regions (Qin et al., 2002; Doering et al., 2007; Sinnegger-Brauns et al., 2009). However, we found no evidence for the expression relatively high levels of Cav1.1 mRNA that has been reported in a study on human brain (Takahashi et al., 2003). Thus, our expression profile confirms previous data on the expression of L-type calcium channels in the brain, and it adds the new finding that, at least in some species or mouse strains, levels of CaV1.3 mRNA in the cortex and hippocampus can be as high as those of CaV1.2. This is important in light of their distinct roles in the formation of long term memory (Moosmang et al., 2005) and the involvement of CaV1.3 in depression (Busquet et al., 2009).
In the family of the non-L-type calcium channels, we observed the expected high levels of CaV2.1 in the cerebellum. CaV2.1 was also the highest expressed α1 subunit in the cortex. In hippocampus however, CaV2.3 was expressed at twice the levels found in cortex and cerebellum, and thus was the dominant CaV isoform. CaV2.3 has been shown to be involved in presynaptic transmitter release in calyx-type terminals in the trapezoid body (Wu et al., 1998, 1999) and in presynaptic LTP in mossy fiber synapses (Dietrich et al., 2003). Postsynaptically, CaV2.3 is a major contributor to action potential evoked calcium transients in dendritic spines and is involved in postsynaptic plasticity (Yasuda et al., 2003). The high expression levels observed here may correspond to this twofold role in hippocampal neurons. Interestingly, hippocampal neurons in culture, which consist of mostly pyramidal cells, did not show similarly prominent CaV2.3 expression levels (cf. Fig. 3A).
Each of the four β subunits and three of the four α2δ subunits were expressed in all examined brain regions. The tissue-specific prevalence of CaV2.1 in cerebellum was paralleled by high expression of β4 and α2δ-2. This is consistent with previous results of a Western blot and in situ hybridization study (Ludwig et al., 1997), as well as with the similarity of phenotypes reported for mutant mice of CaV2.1 (tottering, leaner), β4 (lethargic), and α2δ-2 (ducky and entla) (Burgess et al., 1997; Doyle et al., 1997; Barclay et al., 2001; Brill et al., 2004). In hippocampus β2 and α2δ-1 were the highest expressed auxiliary subunits. Based on experiments involving specific β2 riboprobes and antibodies (Ludwig et al., 1997; Day et al., 1998; Pichler et al., 1997) the overall high levels of β2 transcripts in all brain regions was not to be expected. Because our β2 TaqMan assay showed a comparably low sensitivity (high Ct values and standard curves), we reanalyzed selected samples with a second β2 assay. This confirmed the high β2 expression levels (data not shown). Apparently specific properties of the β2 transcript, like the secondary structure, may render it less accessible to riboprobes and PCR primers. This would explain both, the previously reported low signals of in situ hybridization and the high raw Ct values observed in our own. Nevertheless, previous Western Blot experiments performed on rat and rabbit tissues still indicate a low β2 subunit abundance in brain (Ludwig et al., 1997; Pichler et al., 1997).
The CaV expression profile in hippocampus reflects the sum of expression patterns in a variety of different neuronal and non-neuronal cells. Thus CaV expression of individual neuron types may differ greatly from the overall hippocampal profile. Therefore it was quite surprising to find that cultured hippocampal pyramidal cells express all the same CaV subunit isoforms as hippocampus, although the expression levels of all four β subunits and α2δ-1 through α2δ-3 were fairly uniform. Nevertheless, this expression profile is characteristic for cultured hippocampal neurons and distinct from CaV expression patterns in other differentiated neurons, like dorsal root ganglion or cerebellar granule neurons (Obermair et al., unpublished results). This suggests that in cultured hippocampal pyramidal cells, a restricted expression of auxiliary subunit isoforms is not the strategy to achieve specific CaV subunit compositions. Consequently specific targeting properties and interactions with anchoring proteins in pre- and postsynaptic compartments must be responsible for assembling channels with distinct subunit compositions (Obermair et al., 2010). One striking difference to hippocampus tissue was the reduction of CaV2.3, β2, and α2δ-1 levels in hippocampal neurons. Either neurons other than pyramidal cells account for the prominent expression of these isoforms in the hippocampus, or transcriptional regulation in the hippocampus is strongly dependent on cell-cell interactions or trophic factors missing in the culture system.
Developmental changes of selected isoforms in brain and hippocampal neurons
In the weeks following birth expression of CaV1.2, CaV2.2, and CaV2.3 in the cortex experiences a dramatic decline, while transcript levels of CaV1.3 and CaV2.1 remain constant. Thus, 8 weeks after birth the quantity of total α1 subunit mRNA is reduced to about half of that expressed in late embryonic development. Considering the numerous important functions of voltage-gated calcium channels in mature neurons this developmental drop is remarkable. Either CaVs play more important roles in developing neurons than so far anticipated, or upon differentiation the stabilization of CaVs in pre- and postsynaptic compartments reduces their turnover rate and thus high levels of protein expression may be maintained, even though transcription rates are reduced.
The fact that during this critical period, in which electrical activity sets in and synaptic connections are established, none of the α1 subunits is up-regulated indicates that the pore-forming subunits do not undergo a developmental isoform switch. The same is true for the α2δ subunits. In contrast, developmental expression of the β subunits shows a marked isoform switch. Whereas β1 and β3 decline in parallel to the α subunits, expression of β2 and β4 experiences a significant increase, suggesting that calcium channels change their β subunit composition upon neuronal differentiation. The β isoform shift during neuronal differentiation is expected to result in a functional switch of calcium channels without actually changing the α1 subunit. Furthermore, β subunits are essential for membrane expression of CaVs in heterologous cells as well as in neurons (Dolphin, 2003; Leroy et al., 2005; Obermair et al., 2008, 2010). At the late embryonic stage total α1 transcripts are in excess of total β transcripts and around birth total expression of α1 subunits declines, while total expression of β subunits remains constant. In fact, at 8 weeks after birth total α1 and β transcript levels are nearly balanced. If the available amount of β subunits is limiting membrane expression of CaVs, the overall decline of functionally expressed CaV proteins may be smaller than indicated by the numbers of α1 subunit transcripts.
In hippocampus developmental changes of CaV isoforms mirror those of cortex with one notable exception. CaV2.3 remains expressed at constantly high levels, thus becoming the predominant α1 subunit isoform in mature hippocampus. Furthermore the developmental up-regulation of β2 is more pronounced than that of β4. Together these changes give rise to the predominant expression levels of CaV2.3 and β2 in the 8 weeks old hippocampus when compared to cortex (see Fig. 1A, B). In vitro differentiating cultured hippocampal neurons do not reflect the developmental changes observed in whole hippocampus. During the period when the cultured pyramidal cells differentiate into axons and dendrites and form numerous synaptic contacts, CaV2.3 declines, whereas expression of mainly presynaptic isoforms CaV2.1, α2δ-2, and β4 increases. This observation is consistent with the dramatic increase in the synapse number (Obermair et al., 2003) and is consistent with a change in the channels coupled to glutamate release from mainly N-type channels to P/Q-type channels during in vitro development (Scholz and Miller, 1995).
Global changes in activity quickly induce synaptic scaling mechanisms involving gene-expression (Ibata et al., 2008), and activity-dependent regulation of membrane turnover of CaV1.2 expression has been suggested (Green et al., 2007). Therefore we examined whether the specific changes in CaV isoform expression observed during differentiation are activity dependent. Contrary to our expectation treating cultured hippocampal neurons with TTX or DL-AP5 did not result in any changes in CaV isoform expression. This indicates that the observed developmental changes in CaV expression in neurons may be intrinsically regulated, independent of electrical or synaptic activity.
Correlated expression of specific subunit isoforms
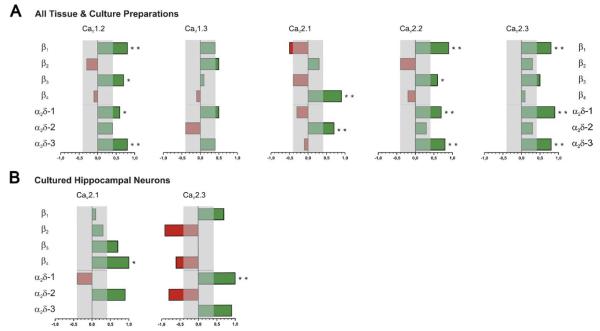
Recurring α1, β, and α2δ expression patterns in different samples or their coordinated up- and down-regulation can be indicative of the existence of preferential subunit combinations in neurons throughout the brain. Thus correlation analysis may be useful to suggest or exclude the existence of such preferential CaV complexes. Our correlation analysis is based on the following assumptions: (1) if CaV α1 isoforms have a preference for specific auxiliary isoforms, or (2) if the expression of certain isoforms is co-regulated, and (3) if this preference or co-regulation is constant in different brain regions, cells and conditions, then this should be revealed in a correlation analysis. For example the correlation analysis (Fig. 5A) clearly identified the preference of CaV2.1 for β4 and α2δ-2 subunits, reflecting their disproportional high expression in cerebellum and their concomitant up-regulation during development of hippocampal pyramidal cells. In tissues such a correlation could also arise from parallel but independent up- and down-regulation of these subunits in different cells. Therefore it is remarkable that a correlated up-regulation of the same subunits (CaV2.1, β4, and α2δ-2) could also be observed during the differentiation of cultured neurons (Fig. 5B). Within one cell type the most likely interpretation of a correlated up-regulation of CaV2.1, β4, and α2δ-2 is indeed that these subunit isoforms actually form a channel together.
Fig. 5.
Correlation analysis of CaV α1 subunit expression with individual β and α2δ isoforms. Correlation coefficients were calculated between the transcript amounts of the individual α1 and the auxiliary β and α2δ subunits including measurements from all tissue and culture preparations. The abscissa represents the size and direction of the correlation coefficients whereby direct (positive) and indirect (negative) coefficients are indicated by green and red bars, respectively. Asterisks mark significant correlations and the gray area indicates the cut-off (r=0.4) for weak correlations. (A) Correlation analysis clearly identified the previously demonstrated association of the CaV2.1 α1 subunit with β4 and α2δ-2. Interestingly, CaV1.2, CaV2.2, and CaV2.3 showed similar degrees of correlations with the same set of β and α2δ subunits. Expression of CaV1.3 did not reveal a strong correlation with any auxiliary subunit. (B) Similar correlation coefficients were identified for CaV2.1 and CaV2.3 by including only the measurements from cultured hippocampal neurons. * P<0.05; ** P<0.01; Pearson correlation.
Another striking upshot of the correlation analysis is the similarity of correlations between CaV1.2, CaV2.2 and CaV2.3. Expression of these α1 subunits correlates positively with β1, β3, α2δ-1, and α2δ-3 (Fig. 5A). Interestingly, the same four auxiliary subunit isoforms correlate negatively with CaV2.1. During early development of the cortex all these channels and auxiliary subunits undergo a strong and concomitant down-regulation. The preference for the same auxiliary subunits may lead to a functional redundancy of CaV2.2 and CaV2.3 in supporting synaptic transmission. However, their differential expression in cortex and hippocampus suggests that CaV2.2 and CaV2.3 may be dominating in distinct cell types. The specific subunit combination CaV2.1/β4/α2δ-2 on one side and a promiscuous association of CaV2.2 and CaV2.3 with any combination of β1, β3, α2δ-1, and α2δ-3 on the other side may explain why loss of CaV2.1 in null-mutant mice causes severe neurological disease (Jun et al., 1999), whereas loss of CaV2.2 or 2.3 results in little to no neurological phenotype (Saegusa et al., 2000; Ino et al., 2001).
Transcript numbers of individual neurons
Estimated transcript numbers of the different CaV subunit isoforms per cultured hippocampal neuron ranged from 0 to 12 (Table 4). The total number of transcripts per cell was 26 α1, 16 β, and 6 α2δs. However, depending on the length of the reading frame a single transcript will yield different numbers of proteins per unit time. Assuming a maximal translation rate of 5 amino acids per second (Boehlke and Friesen, 1975) a single ribosome yields approximately 10 α1, 20 α2δ, and 40 β proteins per hour. If we further assume that every transcript is occupied by 10 ribosomes (Mata et al., 2005), then our present data predict the translation of ~2600 α1, ~6400 β, and ~1200 α2δ molecules per hour and neuron. On the protein level our previous quantitative immunofluorescence analysis revealed that an average cultured hippocampal neuron may contain approximately 4000 CaV1.2 clusters, each consisting of eight channels on average (Obermair et al., 2004), in total ~32,000 proteins. With the translation rates estimated above, this corresponds to a complete turnover of the entire CaV1.2 complement in 64 h. Such protein turnover rates are slower than previously suggested membrane turnover rates based on TIRF analysis (Green et al., 2007), but are in line with estimated turnover rates of β subunits (Berrow et al., 1995) and the high stability of membrane expressed CaV1.2 clusters observed in hippocampal neurons (Di Biase et al., 2008). If CaV subunits in the neurons are associated at a 1:1:1 (α1:β:α2δ) stoichiometry, these numbers suggest two- and fourfold higher β protein turnover rates compared with α1 and α2δ, respectively. On the other hand, additional free auxiliary subunits might also be involved in functions independent of the CaV complex. This possibility is supported by the existence of a fraction of α2δ-2 subunits not associated with the channel complex (Davies et al., 2006), and the recent findings of a β4 subunit located in nuclei of cerebellar neurons (Subramanyam et al., 2009).
Table 4.
Number of mRNA molecules in a single cultured hippocampal neuron
| CaV subunit | Number of transcript molecules |
|---|---|
| CaV1.1 | 0.0 |
| CaV1.2 | 5.0 |
| CaV1.3 | 2.2 |
| CaV1.4 | 0.0 |
| CaV2.1 | 11.2 |
| CaV2.2 | 5.4 |
| CaV2.3 | 2.3 |
| β1 | 3.6 |
| β2 | 4.2 |
| β3 | 4.4 |
| β4 | 3.8 |
| α2δ-1 | 2.3 |
| α2δ-2 | 1.8 |
| α2δ-3 | 2.1 |
| α2δ-4a | 0.0 |
Calculations are based on the qRT-PCR experiment on cultured hippocampal neurons (24 DIV) with highest RNA yield.
The assay for α2δ-4 was not included in the same qRT-PCR experiments. However, in other qRT-PCR runs on cultured hippocampal neurons α2δ-4 levels were always below detectability (cf. Fig. 4 and Suppl. Fig. 2).
CONCLUSION
Combining relative and absolute qRT-PCR quantification for the first time allowed the direct quantitative comparison of the CaV expression profile in different brain regions, cultured neurons, and treatment conditions. Our results clearly revealed a remarkably stable overall Ca2+ channel complement as well as tissue specific differences in expression levels. Furthermore we could show that low-density cultured hippocampal neurons, a widely used neuronal model system, express all the same Ca2+ channel subunit isoforms as adult hippocampus. Developmental changes are likely determined by an intrinsic program and not regulated by changes in neuronal activity. Interestingly, whereas correlation of expression patterns indicated a great permissiveness of interactions between α1 and auxiliary subunits, CaV2.1 was different in that it showed a strong preference for β4 and α2δ-2 subunits not only in cerebellum but also in hippocampal neurons.
Supplementary Material
Acknowledgments
We thank Gilda Pelster for technical assistance. This work was supported by grants from the Austrian Science Fund and the Austrian National Bank P17806-B05, P17807-B05, P20059-B05. This work is part of the PhD thesis of B.S.
Abbreviations
- CaV
voltage-activated calcium channel
- cDNA
complementary DNA
- Ct
cycle threshold
- DIV
days in vitro
- dl-AP5
dl-2-amino-5-phosphonopentanoic acid
- E16
embryonic day 16
- HPRT1
hypoxanthine phosphoribosyl-transferase 1
- LOD
limit of detection
- LOQ
limit of quantification
- mRNA
messenger RNA
- NMDA
N-methyl-d-aspartate
- PD1
postnatal day 1
- qRT-PCR
quantitative reverse transcription PCR
- TTX
tetrodotoxin
APPENDIX
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.neuroscience.2010.02.037.
REFERENCES
- Arikkath J, Campbell KP. Auxiliary subunits: essential components of the voltage-gated calcium channel complex. Curr Opin Neurobiol. 2003;13:298–307. doi: 10.1016/s0959-4388(03)00066-7. [DOI] [PubMed] [Google Scholar]
- Ball SL, Powers PA, Shin HS, Morgans CW, Peachey NS, Gregg RG. Role of the beta(2) subunit of voltage-dependent calcium channels in the retinal outer plexiform layer. Invest Ophthalmol Vis Sci. 2002;43:1595–1603. [PubMed] [Google Scholar]
- Barclay J, Balaguero N, Mione M, Ackerman SL, Letts VA, Brodbeck J, Canti C, Meir A, Page KM, Kusumi K, Perez-Reyes E, Lander ES, Frankel WN, Gardiner RM, Dolphin AC, Rees M. Ducky mouse phenotype of epilepsy and ataxia is associated with mutations in the Cacna2d2 gene and decreased calcium channel current in cerebellar Purkinje cells. J Neurosci. 2001;21:6095–6104. doi: 10.1523/JNEUROSCI.21-16-06095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes S, Kelly ME. Calcium channels at the photoreceptor synapse. Adv Exp Med Biol. 2002;514:465–476. doi: 10.1007/978-1-4615-0121-3_28. [DOI] [PubMed] [Google Scholar]
- Bender R, Lange S. Adjusting for multiple testing—when and how? J Clin Epidemiol. 2001;54:343–349. doi: 10.1016/s0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- Benson DL, Watkins FH, Steward O, Banker G. Characterization of GABAergic neurons in hippocampal cell cultures. J Neurocytol. 1994;23:279–295. doi: 10.1007/BF01188497. [DOI] [PubMed] [Google Scholar]
- Berrow NS, Campbell V, Fitzgerald EM, Brickley K, Dolphin AC. Antisense depletion of beta-subunits modulates the biophysical and pharmacological properties of neuronal calcium channels. J Physiol. 1995;482:481–491. doi: 10.1113/jphysiol.1995.sp020534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehlke KW, Friesen JD. Cellular content of ribonucleic acid and protein in Saccharomyces cerevisiae as a function of exponential growth rate: calculation of the apparent peptide chain elongation rate. J Bacteriol. 1975;121:429–433. doi: 10.1128/jb.121.2.429-433.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill J, Klocke R, Paul D, Boison D, Gouder N, Klugbauer N, Hofmann F, Becker CM, Becker K. entla, a novel epileptic and ataxic Cacna2d2 mutant of the mouse. J Biol Chem. 2004;279:7322–7330. doi: 10.1074/jbc.M308778200. [DOI] [PubMed] [Google Scholar]
- Brodbeck J, Davies A, Courtney JM, Meir A, Balaguero N, Canti C, Moss FJ, Page KM, Pratt WS, Hunt SP, Barclay J, Rees M, Dolphin AC. The ducky mutation in Cacna2d2 results in altered Purkinje cell morphology and is associated with the expression of a truncated alpha 2 delta-2 protein with abnormal function. J Biol Chem. 2002;277:7684–7693. doi: 10.1074/jbc.M109404200. [DOI] [PubMed] [Google Scholar]
- Burgess DL, Jones JM, Meisler MH, Noebels JL. Mutation of the Ca2+ channel beta subunit gene Cchb4 is associated with ataxia and seizures in the lethargic (lh) mouse. Cell. 1997;88:385–392. doi: 10.1016/s0092-8674(00)81877-2. [DOI] [PubMed] [Google Scholar]
- Busquet P, Khoi Nguyen N, Schmid E, Tanimoto N, Seeliger MW, Ben-Yosef T, Mizuno F, Akopian A, Striessnig J, Singewald N. CaV1.3 L-type Ca2+ channels modulate depression-like behaviour in mice independent of deaf phenotype. Int J Neuropsychopharmacol. 2009;11:1–15. doi: 10.1017/S1461145709990368. [DOI] [PubMed] [Google Scholar]
- Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- Cole RL, Lechner SM, Williams ME, Prodanovich P, Bleicher L, Varney MA, Gu G. Differential distribution of voltage-gated calcium channel alpha-2 delta (alpha2delta) subunit mRNA-containing cells in the rat central nervous system and the dorsal root ganglia. J Comp Neurol. 2005;491:246–269. doi: 10.1002/cne.20693. [DOI] [PubMed] [Google Scholar]
- Corley J. Best practices in establishing detection and quantification limits for pesticide residues in foods. In: Lee PW, editor. Handbook of residue analytical methods for agrochemicals. Vol. 1. John Wiley & Sons, Ltd; East Sussex, United Kingdom: 2003. pp. 1–18. LS0203. [Google Scholar]
- Davies A, Douglas L, Hendrich J, Wratten J, Tran Van Minh A, Foucault I, Koch D, Pratt WS, Saibil HR, Dolphin AC. The calcium channel alpha2delta-2 subunit partitions with CaV2.1 into lipid rafts in cerebellum: implications for localization and function. J Neurosci. 2006;26:8748–8757. doi: 10.1523/JNEUROSCI.2764-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A, Hendrich J, Van Minh AT, Wratten J, Douglas L, Dolphin AC. Functional biology of the alpha(2)delta subunits of voltage-gated calcium channels. Trends Pharmacol Sci. 2007;28:220–228. doi: 10.1016/j.tips.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Day NC, Volsen SG, McCormack AL, Craig PJ, Smith W, Beattie RE, Shaw PJ, Ellis SB, Harpold MM, Ince PG. The expression of voltage-dependent calcium channel beta subunits in human hippocampus. Brain Res Mol Brain Res. 1998;60:259–269. doi: 10.1016/s0169-328x(98)00186-7. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Mermelstein PG, Xia H, Tsien RW. Signaling from synapse to nucleus: the logic behind the mechanisms. Curr Opin Neurobiol. 2003;13:354–365. doi: 10.1016/s0959-4388(03)00076-x. [DOI] [PubMed] [Google Scholar]
- Di Biase V, Obermair GJ, Szabo Z, Altier C, Sanguesa J, Bourinet E, Flucher BE. Stable membrane expression of postsynaptic CaV1.2 calcium channel clusters is independent of interactions with AKAP79/150 and PDZ proteins. J Neurosci. 2008;28:13845–13855. doi: 10.1523/JNEUROSCI.3213-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich D, Kirschstein T, Kukley M, Pereverzev A, von der Brelie C, Schneider T, Beck H. Functional specialization of presynaptic Cav2.3 Ca2+ channels. Neuron. 2003;39:483–496. doi: 10.1016/s0896-6273(03)00430-6. [DOI] [PubMed] [Google Scholar]
- Doering CJ, Peloquin JB, McRory JE. The Ca(v)1.4 calcium channel: more than meets the eye. Channels. 2007;1:3–10. [PubMed] [Google Scholar]
- Dolmetsch R. Excitation-transcription coupling: signaling by ion channels to the nucleus. Sci STKE. 2003;2003:PE4. doi: 10.1126/stke.2003.166.pe4. [DOI] [PubMed] [Google Scholar]
- Dolphin AC. Beta subunits of voltage-gated calcium channels. J Bioenerg Biomembr. 2003;35:599–620. doi: 10.1023/b:jobb.0000008026.37790.5a. [DOI] [PubMed] [Google Scholar]
- Doyle J, Ren X, Lennon G, Stubbs L. Mutations in the Cacnl1a4 calcium channel gene are associated with seizures, cerebellar degeneration, and ataxia in tottering and leaner mutant mice. Mamm Genome. 1997;8:113–120. doi: 10.1007/s003359900369. [DOI] [PubMed] [Google Scholar]
- Fletcher TL, De Camilli P, Banker G. Synaptogenesis in hippocampal cultures: evidence indicating that axons and dendrites become competent to form synapses at different stages of neuronal development. J Neurosci. 1994;14:6695–6706. doi: 10.1523/JNEUROSCI.14-11-06695.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EM, Barrett CF, Bultynck G, Shamah SM, Dolmetsch RE. The tumor suppressor eIF3e mediates calcium-dependent internalization of the L-type calcium channel CaV1.2. Neuron. 2007;55:615–632. doi: 10.1016/j.neuron.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms KJ, Craig AM. Synapse composition and organization following chronic activity blockade in cultured hippocampal neurons. J Comp Neurol. 2005;490:72–84. doi: 10.1002/cne.20635. [DOI] [PubMed] [Google Scholar]
- Hell JW, Westenbroek RE, Warner C, Ahlijanian MK, Prystay W, Gilbert MM, Snutch TP, Catterall WA. Identification and differential subcellular localization of the neuronal class C and class D L-type calcium channel alpha 1 subunits. J Cell Biol. 1993;123:949–962. doi: 10.1083/jcb.123.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobom M, Dai S, Marais E, Lacinova L, Hofmann F, Klugbauer N. Neuronal distribution and functional characterization of the calcium channel alpha2delta-2 subunit. Eur J Neurosci. 2000;12:1217–1226. doi: 10.1046/j.1460-9568.2000.01009.x. [DOI] [PubMed] [Google Scholar]
- Ibata K, Sun Q, Turrigiano GG. Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron. 2008;57:819–826. doi: 10.1016/j.neuron.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Ino M, Yoshinaga T, Wakamori M, Miyamoto N, Takahashi E, Sonoda J, Kagaya T, Oki T, Nagasu T, Nishizawa Y, Tanaka I, Imoto K, Aizawa S, Koch S, Schwartz A, Niidome T, Sawada K, Mori Y. Functional disorders of the sympathetic nervous system in mice lacking the alpha 1B subunit (Cav 2.2) of N-type calcium channels. Proc Natl Acad Sci U S A. 2001;98:5323–5328. doi: 10.1073/pnas.081089398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan BA, Kreutz MR. Nucleocytoplasmic protein shuttling: the direct route in synapse-to-nucleus signaling. Trends Neurosci. 2009;32:392–401. doi: 10.1016/j.tins.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Jun K, Piedras-Renteria ES, Smith SM, Wheeler DB, Lee SB, Lee TG, Chin H, Adams ME, Scheller RH, Tsien RW, Shin HS. Ablation of P/Q-type Ca(2+) channel currents, altered synaptic transmission, and progressive ataxia in mice lacking the alpha(1A)-subunit. Proc Natl Acad Sci U S A. 1999;96:15245–15250. doi: 10.1073/pnas.96.26.15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Klugbauer N, Lacinova L, Marais E, Hobom M, Hofmann F. Molecular diversity of the calcium channel alpha2delta subunit. J Neurosci. 1999;19:684–691. doi: 10.1523/JNEUROSCI.19-02-00684.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschak A, Obermair GJ, Pivotto F, Sinnegger-Brauns MJ, Striessnig J, Pietrobon D. Molecular nature of anomalous L-type calcium channels in mouse cerebellar granule cells. J Neurosci. 2007;27:3855–3863. doi: 10.1523/JNEUROSCI.4028-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy J, Richards MW, Butcher AJ, Nieto-Rostro M, Pratt WS, Davies A, Dolphin AC. Interaction via a key tryptophan in the I–II linker of N-type calcium channels is required for beta1 but not for palmitoylated beta2, implicating an additional binding site in the regulation of channel voltage-dependent properties. J Neurosci. 2005;25:6984–6996. doi: 10.1523/JNEUROSCI.1137-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzon NM, Foehring RC. Characterization of pharmacologically identified voltage-gated calcium channel currents in acutely isolated rat neocortical neurons. I. Adult neurons. J Neurophysiol. 1995;73:1430–1442. doi: 10.1152/jn.1995.73.4.1430. [DOI] [PubMed] [Google Scholar]
- Ludwig A, Flockerzi V, Hofmann F. Regional expression and cellular localization of the alpha1 and beta subunit of high voltage-activated calcium channels in rat brain. J Neurosci. 1997;17:1339–1349. doi: 10.1523/JNEUROSCI.17-04-01339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Marguerat S, Bahler J. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem Sci. 2005;30:506–514. doi: 10.1016/j.tibs.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Mochida S, Westenbroek RE, Yokoyama CT, Itoh K, Catterall WA. Subtype-selective reconstitution of synaptic transmission in sympathetic ganglion neurons by expression of exogenous calcium channels. Proc Natl Acad Sci U S A. 2003;100:2813–2818. doi: 10.1073/pnas.262787299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmang S, Haider N, Klugbauer N, Adelsberger H, Langwieser N, Muller J, Stiess M, Marais E, Schulla V, Lacinova L, Goebbels S, Nave KA, Storm DR, Hofmann F, Kleppisch T. Role of hippocampal Cav1.2 Ca2+ channels in NMDA receptor-independent synaptic plasticity and spatial memory. J Neurosci. 2005;25:9883–9892. doi: 10.1523/JNEUROSCI.1531-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Friedrich T, Kim MS, Mikami A, Nakai J, Ruth P, Bosse E, Hofmann F, Flockerzi V, Furuichi T, et al. Primary structure and functional expression from complementary DNA of a brain calcium channel. Nature. 1991;350:398–402. doi: 10.1038/350398a0. [DOI] [PubMed] [Google Scholar]
- Obermair GJ, Kaufmann WA, Knaus HG, Flucher BE. The small conductance Ca2+-activated K+ channel SK3 is localized in nerve terminals of excitatory synapses of cultured mouse hippocampal neurons. Eur J Neurosci. 2003;17:721–731. doi: 10.1046/j.1460-9568.2003.02488.x. [DOI] [PubMed] [Google Scholar]
- Obermair GJ, Schlick B, Di Biase V, Subramanyam P, Gebhart M, Baumgartner S, Flucher BE. Reciprocal interactions regulate targeting of calcium channel β subunits and membrane expression of α1 subunits in cultured hippocampal neurons. J Biol Chem. 2010;285:5776–5791. doi: 10.1074/jbc.M109.044271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermair GJ, Szabo Z, Bourinet E, Flucher BE. Differential targeting of the L-type Ca2+ channel alpha 1C (CaV1.2) to synaptic and extrasynaptic compartments in hippocampal neurons. Eur J Neurosci. 2004;19:2109–2122. doi: 10.1111/j.0953-816X.2004.03272.x. [DOI] [PubMed] [Google Scholar]
- Obermair GJ, Tuluc P, Flucher BE. Auxiliary Ca(2+) channel subunits: lessons learned from muscle. Curr Opin Pharmacol. 2008;8:311–318. doi: 10.1016/j.coph.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Pichler M, Cassidy TN, Reimer D, Haase H, Kraus R, Ostler D, Striessnig J. Beta subunit heterogeneity in neuronal L-type Ca2+ channels. J Biol Chem. 1997;272:13877–13882. doi: 10.1074/jbc.272.21.13877. [DOI] [PubMed] [Google Scholar]
- Pravettoni E, Bacci A, Coco S, Forbicini P, Matteoli M, Verderio C. Different localizations and functions of L-type and N-type calcium channels during development of hippocampal neurons. Dev Biol. 2000;227:581–594. doi: 10.1006/dbio.2000.9872. [DOI] [PubMed] [Google Scholar]
- Qin N, Yagel S, Momplaisir ML, Codd EE, D’Andrea MR. Molecular cloning and characterization of the human voltage-gated calcium channel alpha(2)delta-4 subunit. Mol Pharmacol. 2002;62:485–496. doi: 10.1124/mol.62.3.485. [DOI] [PubMed] [Google Scholar]
- Randall A, Tsien RW. Pharmacological dissection of multiple types of Ca2+ channel currents in rat cerebellar granule neurons. J Neurosci. 1995;15:2995–3012. doi: 10.1523/JNEUROSCI.15-04-02995.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Craig AM. Activity regulates the synaptic localization of the NMDA receptor in hippocampal neurons. Neuron. 1997;19:801–812. doi: 10.1016/s0896-6273(00)80962-9. [DOI] [PubMed] [Google Scholar]
- Saegusa H, Kurihara T, Zong S, Minowa O, Kazuno A, Han W, Matsuda Y, Yamanaka H, Osanai M, Noda T, Tanabe T. Altered pain responses in mice lacking alpha 1E subunit of the voltage-dependent Ca2+ channel. Proc Natl Acad Sci U S A. 2000;97:6132–6137. doi: 10.1073/pnas.100124197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz KP, Miller RJ. Developmental changes in presynaptic calcium channels coupled to glutamate release in cultured rat hippocampal neurons. J Neurosci. 1995;15:4612–4617. doi: 10.1523/JNEUROSCI.15-06-04612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnegger-Brauns MJ, Huber IG, Koschak A, Wild C, Obermair GJ, Einzinger U, Hoda JC, Sartori SB, Striessnig J. Expression and 1,4-dihydropyridine-binding properties of brain L-type calcium channel isoforms. Mol Pharmacol. 2009;75:407–414. doi: 10.1124/mol.108.049981. [DOI] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Subramanyam P, Obermair GJ, Baumgartner S, Gebhart M, Striessnig J, Kaufmann WA, Geley S, Flucher BE. Activity and calcium regulate nuclear targeting of the calcium channel beta(4b) subunit in nerve and muscle cells. Channels. 2009;3:343–355. doi: 10.4161/chan.3.5.9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Jeong SY, Ogata K, Goto J, Hashida H, Isahara K, Uchiyama Y, Kanazawa I. Human skeletal muscle calcium channel alpha1S is expressed in the basal ganglia: distinctive expression pattern among L-type Ca2+ channels. Neurosci Res. 2003;45:129–137. doi: 10.1016/s0168-0102(02)00204-3. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- Vacher H, Mohapatra DP, Trimmer JS. Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev. 2008;88:1407–1447. doi: 10.1152/physrev.00002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenbroek RE, Sakurai T, Elliott EM, Hell JW, Starr TV, Snutch TP, Catterall WA. Immunochemical identification and subcellular distribution of the alpha 1A subunits of brain calcium channels. J Neurosci. 1995;15:6403–6418. doi: 10.1523/JNEUROSCI.15-10-06403.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LG, Borst JG, Sakmann B. R-type Ca2+ currents evoke transmitter release at a rat central synapse. Proc Natl Acad Sci U S A. 1998;95:4720–4725. doi: 10.1073/pnas.95.8.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LG, Westenbroek RE, Borst JG, Catterall WA, Sakmann B. Calcium channel types with distinct presynaptic localization couple differentially to transmitter release in single calyx-type synapses. J Neurosci. 1999;19:726–736. doi: 10.1523/JNEUROSCI.19-02-00726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wycisk KA, Budde B, Feil S, Skosyrski S, Buzzi F, Neidhardt J, Glaus E, Nurnberg P, Ruether K, Berger W. Structural and functional abnormalities of retinal ribbon synapses due to Cacna2d4 mutation. Invest Ophthalmol Vis Sci. 2006;47:3523–3530. doi: 10.1167/iovs.06-0271. [DOI] [PubMed] [Google Scholar]
- Yasuda R, Sabatini BL, Svoboda K. Plasticity of calcium channels in dendritic spines. Nat Neurosci. 2003;6:948–955. doi: 10.1038/nn1112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.