Abstract
Abnormalities in Z-disc proteins cause hypertrophic (HCM), dilated (DCM) and/or restrictive cardiomyopathy (RCM), but disease-causing mechanisms are not fully understood. Myopalladin (MYPN) is a Z-disc protein expressed in striated muscle and functions as a structural, signaling and gene expression regulating molecule in response to muscle stress. MYPN was genetically screened in 900 patients with HCM, DCM and RCM, and disease-causing mechanisms were investigated using comparative immunohistochemical analysis of the patient myocardium and neonatal rat cardiomyocytes expressing mutant MYPN. Cardiac-restricted transgenic (Tg) mice were generated and protein–protein interactions were evaluated. Two nonsense and 13 missense MYPN variants were identified in subjects with DCM, HCM and RCM with the average cardiomyopathy prevalence of 1.66%. Functional studies were performed on two variants (Q529X and Y20C) associated with variable clinical phenotypes. Humans carrying the Y20C-MYPN variant developed HCM or DCM, whereas Q529X-MYPN was found in familial RCM. Disturbed myofibrillogenesis with disruption of α-actinin2, desmin and cardiac ankyrin repeat protein (CARP) was evident in rat cardiomyocytes expressing MYPNQ529X. Cardiac-restricted MYPNY20C Tg mice developed HCM and disrupted intercalated discs, with disturbed expression of desmin, desmoplakin, connexin43 and vinculin being evident. Failed nuclear translocation and reduced binding of Y20C-MYPN to CARP were demonstrated using in vitro and in vivo systems. MYPN mutations cause various forms of cardiomyopathy via different protein–protein interactions. Q529X-MYPN causes RCM via disturbed myofibrillogenesis, whereas Y20C-MYPN perturbs MYPN nuclear shuttling and leads to abnormal assembly of terminal Z-disc within the cardiac transitional junction and intercalated disc.
INTRODUCTION
Cardiomyopathies are heterogeneous myocardial diseases associated with cardiac dysfunction (1,2). Various cardiomyopathy phenotypes exist and are formally classified, including hypertrophic cardiomyopathy (HCM), characterized by ventricular hypertrophy with diastolic dysfunction (3); dilated cardiomyopathy (DCM) with ventricular dilation and systolic dysfunction (4); restrictive cardiomyopathy (RCM), accompanied by increased stiffness of the myocardium due to diastolic dysfunction without significant hypertrophy (5); arrhythmogenic right ventricular (RV) cardiomyopathy (ARVC), consisting of RV and/or left ventricular (LV) dilation and dysfunction with fibrofatty infiltration and arrhythmias (6,7) and LV non-compaction (LVNC), characterized by heavy trabeculations in the left ventricle (8). More than half of the patients with HCM and 20–35% of DCM patients have a family history of the disease, usually consistent with autosomal dominant inheritance (2,4). Familial occurrence is also noted in RCM, ARVC and LVNC (2,5–8). A variety of genes have been identified as causes of the various cardiomyopathy subtypes, and these generally encode proteins in a ‘final common pathway’ (9). In HCM and RCM, mutations in genes encoding sarcomeric proteins are associated with the clinical phenotype, although DCM also develops when genes encoding sarcomere and cytoskeletal proteins are mutated (10). In ARVC, the ‘final common pathway’ appears to cause disruption of desmosomal proteins by gene mutations (10,11).
Myopalladin (MYPN; NM_032578), localized at chromosome 10q21.3, encodes a 147 kDa multifunctional protein found at the sarcomeric Z- and I-bands, as well as in the nucleus, in cardiac and skeletal myocytes (12). Central and C-terminal domains of MYPN bind at the Z-disc to α-actinin and nebulette (NEBL), respectively. The α-actinin–MYPN–NEBL complex tethers actin and titin to the Z-disc and may play roles in the signaling and regulation of gene expression in response to muscle stress (12). The N-terminal domain of MYPN binds cardiac ankyrin repeat protein (CARP), which is involved in the control of muscle gene expression (13). MYPN contains five immunoglobulin (Ig) domains, consistent with being a member of the Ig-domain protein family of which abnormalities can be associated with inherited human disorders (14).
Mutations in MYPN have previously been reported in subjects with DCM (15) and HCM (16), suggesting a disease causation effect and therefore the importance of genetic screening of the MYPN gene in affected subjects and family members. However, no data exist regarding the prevalence of MYPN mutations and molecular pathophysiologic contributions to the development of different clinical phenotypes. In our study, a large cohort of 900 patients with DCM, HCM and RCM were screened for mutations and 15 MYPN variants were identified, of which 14 were novel, including 10 missense variants being predicted to have damaging consequences on the protein (17). The average cardiomyopathy prevalence was estimated to be 1.66%, including 1.86% for HCM, 1.72% for DCM and 1.45% for RCM. Two variants were associated with heterogeneous phenotypes in humans: a missense p.Y20C variant found in HCM and DCM patients, whereas a nonsense p.Q529X variant was found in familial RCM. To test the hypothesis that heterogeneous phenotypes occur with identical mutations via alteration of multifunctional features of MYPN and/or via its interacting partners in a distinct, domain-defined manner (14), in vitro and murine models of these two variants were developed, and functional studies were performed.
RESULTS
Mutational analysis
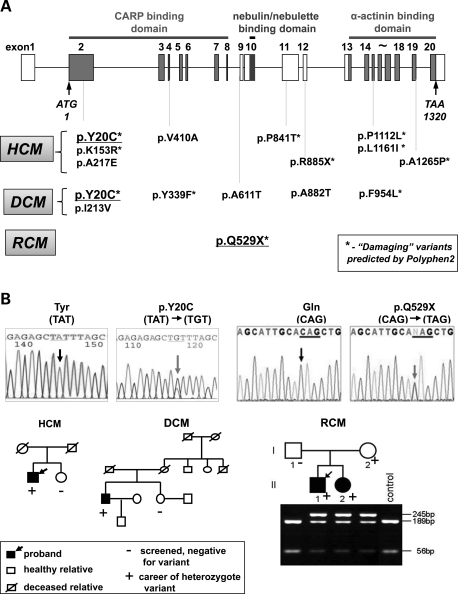
Fifteen rare sequence variations in the MYPN gene were identified in the 900 patients with heterogeneous forms of cardiomyopathy, with 14 being novel. A nonsense variant, p.R885X, and eight missense variants (p.Y20C, p.K153R, p.A217E, p.V410A, p.P841T, p.P1112L, p.L1161I and p.A1265P) were found in patients with HCM, resulting in an HCM prevalence of 1.86% (Table 1 and Fig. 1A). Six missense variants (p.Y20C, p.I213V, p.Y339F, p.A611T, p.A882T and p.F954L) were identified in patients with DCM, with the DCM prevalence being 1.72% (Table 2 and Fig. 1A). Ten variants (two nonsense and eight missense variants labeled with asterisks in Fig. 1A) are predicted to damage the protein function by the Polymorphism Phenotyping v2 (PolyPhen-2) prediction software. Variant p.P1112L, which was identified in an HCM patient, was previously reported in DCM (15) and HCM (16) cases. Patient III with HCM (Table 1) and patient MC31379 with DCM (Table 2) were noted to host compound heterozygous variants in MYPN. Interestingly, variant p.W816G (c.244T>G) in the MYH7 gene was identified in siblings MC31379 and JC31380 with DCM (Table 2).
Table 1.
Clinical characteristics of HCM and RCM patients carrying MYPN mutations
| ID | Mutation | Age, gender | Age at onset (years) | Clinical diagnosis | Family history | NYHA | LAD (mm) | LVEDD (mm) | LVESD (mm) | IVS (mm) | PW (mm) | %EF | Comments | Poly Phen-2 | SIFT | PANTHER |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VIII Caucasian | c.59A>G p.Y20C | 59 years, male | 45 | HCM | No | I | — | — | — | 22 | — | — | Pacemaker | Damaging: 0.998 | Intolerant: 0.15 | Deleterious: 0.5 |
| II Asian | c.458A>G p.K153R | 53 years, female | 24 | HCM | Yes | I | — | 36 | — | 33 | — | 65 | Damaging: 0.993 | 0.18 | Non | |
| III Asian | c.650C>A p.A217E + c.3793G> C p.A1265P | 44 years, female | 28 | HCM | Yes | I | 34 | 42 | 28 | 14 | 10 | 68 | Benign: 0.004 | 0.68 | Non | |
| Damaging: 0.975 | 0.27 | Non | ||||||||||||||
| I Asian | c.1229T>C p.V410A | 50 years, male | 29 | HCM | Yes | I | 50 | 50 | 31 | 20 | 12 | 80 | Myoectomy | Benign: 0.005 | 0.9 | Non |
| CM1023 Asian | c.1585C>T p.Q529X | 22 years, male | 7 | MR, TR, AF, RCM | Yes | IV | 62 | 43 | 39 | 10 | — | 50 | 11 years, heart transplant | Nonsense | ||
| CM1024 | c.1585C>T p.Q529X | 19 years, female | 8 | MR, RCM | Yes | III | 41 | 39 | 29 | 6 | — | 45 | 9 years, heart transplant | Nonsense | ||
| IV Caucasian | c.2521C>A p.P841T | 54 years, female | 47 | HCM | Yes | I | — | 48 | 29 | 19 | 12 | 73 | Damaging: 0.989 | 0.55 | Non | |
| V Caucasian | c.2653C>T p.R885X | 59 years, male | 50 | HCM | Yes | I | 47 | 54 | 35 | 18 | 25 | 68 | Nonsense | |||
| VI American Indian Alaskan | c.3335C>T p.L1161I | 63 years, male | 46 | HCM | No | I | — | — | — | 25 | 14 | 75 | Myoectomy | Damaging: 0.976 | 0 | Deleterious: −4.227 |
| VII Caucasian | c.3481C>A p.P1112L | 64 years, female | 55 | HCM | No | I | — | — | — | 15 | 15 | 80 | Damaging: 0.992 | 0.36 | Deleterious: −3.014 |
Ten variants were found. Variant p.Q529X was identified in two siblings with RCM, although healthy mother carried the same mutation. Patient III carried compound heterozygote variants, p.A217E and p.A1265P, in the MYPN gene. AF, atrial fibrillation; NYHA, New York Heart Association; LAD, left atrial dimension; LVEDD, left ventricular end-diastolic dimension; LVESD, left ventricular end-systolic dimension; IVS, interventricular septum; PW, posterior wall; %EF, percent ejection fraction; MR, mitral regurgitation; TR, tricuspid regurgitation; Non, non-deleterious.
Figure 1.
Mutational analysis. (A) Schematic representation of the MYPN gene and localization of variants identified in patients with HCM (upper panel), DCM and RCM (lower panel). Asterisks indicate nonsense and damaging missense variants evaluated by the PolyPhen-2 prediction software. (B) DNA sequence chromatogram and pedigrees of families carrying p.Y20C (A) and p.Q529X (B) MYPN gene mutations. Grey arrows indicate nucleotide substitutions. Black arrows indicate proband, a positive mark indicates career and a negative mark indicates a screened individual with no mutation. p.Q529X (B) was inherited in two affected children (II1 and II2, lower panel) from a clinically unaffected mother (I2).
Table 2.
Clinical characteristics of individuals with DCM carrying MYPN mutations
| ID | Variants | Age, gender | Age at onset | Clinical diagnosis | Family history | NYHA | %FS | %EF | Comments | PolyPhen-2 | SIFT | PANTHER |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H558307 Caucasian | c.59A>G Y20C | 58 years, male | 52 years | DCM, AF | No | I | 14 | Holter-salvo VT 7 bts | Damaging: 0.998 | 0.15 | Deleterious: 0.5 | |
| MC31379 Caucasian | c.637A>G p.I213V + c.2546G>A p.A882T | 35 years, female | 6 weeks | DCM | Yes, sibling of 31380 (+) MYH7 c.2447T>G, p.W816G | I | 25 | 47 | Treatment: lesinopril and metoprolol | Benign: 0.004 | 0.5 | Non |
| Benign: 0.025 | 0.39 | Non | ||||||||||
| JC31380 Caucasian | c.1016A>T p.Y339F | 38 years, female | 9 years | DCM | Yes, sibling of MC31379 (+) MYH7 c.2447T>G, p.W816G | I | — | 44 | Treatment: metoprolol | Damaging: 0.999 | 0.14 | Deleterious: −3.8114 |
| KA22974 Caucasian | c.1933A>G p.A611T | 10 years, male | At birth | DCM, LVNC | Yes | I | 28 | 46 | Hypertrophic LV | Benign: 0 | 0.54 | Non |
| 16017 Caucasian | c.2546G>Ap.A882T | 27 years, female | 11 years | DCM | Yes | III | 21 | Mother had heart transplant | Benign: 0.025 | 0.39 | Non | |
| 08V3294 Caucasian | c.2862C>A p.F954L | 17 years, female | 13 years | DCM | Yes | III | 9 | 15 | Damaging: 0.962 | 0.65 | Deleterious: −3.19 |
Seven variants were found in patients with DCM. Variant p.Y20C was associated with DCM and HCM phenotypes in humans. Siblings MC31379 and JC31380 were carriers of missense variant in MYH7. Patient MC31379 had compound heterozygote variants in the MYPN gene. AF, atrial fibrillation; LVNC, left ventricular non-compaction; LNSVT, non-sustained ventricular tachycardia; NYHA, New York Heart Association; %FS, percent fractional shortening; %EF, percent ejection fraction; Non, non-deleterious.
The p.Y20C (Y20C) variant was detected in one patient with DCM and another patient with HCM as a de novo variant (Fig. 1B). During the course of this study, the Y20C variant was described as a single-nucleotide polymorphism (SNP, rs140148105) in the dbSNP of the 1000 Genomes Pilot Database with a low genotype frequency of A/G = 0.001. Effects of the mutation on the affected protein were estimated by various prediction systems: PolyPhen-2, score = 0.998; SIFT, intolerant +0.15 and PANTHER, deleterious with score of P= 0.5. Y20C is located at the N-terminal domain of MYPN upstream to the coiled-coil domain, thus, affecting CARP binding (Supplementary Material, Fig. S1A).
In the RCM cohort, a nonsense variant, p.Q529X (Q529X), was identified in siblings with RCM (CM1023 and CM1024). Clinical examinations of the mother who carried the Q529X mutation revealed aortic regurgitation without RCM, although a possibility of mutation-associated regional pathologic changes around the aortic valve could not be excluded. The RCM prevalence was estimated to be 1.45% (Table 1 and Fig. 1). The Q529X variant truncated the three Ig-repeats and the NEBL and α-actinin-binding domains of MYPN (Supplementary Material, Fig. S1A).
None of the MYPN variants identified was found in 1020 ethnically matched control subjects and all were found to affect the evolutionarily conserved residues in MYPN from various species. There was no clinical evidence of skeletal muscle disease or other systemic involvement in any individuals carrying the MYPN variants. A complete list of additional genetic variants identified in the patients and the control cohort is provided (Supplementary Material, Table S2).
Myocardial findings in patients carrying the Q529X-MYPN mutation
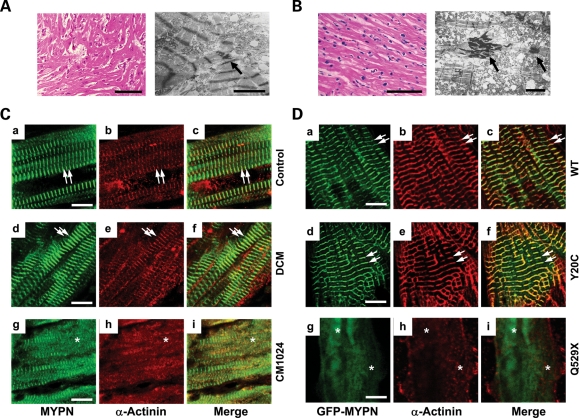
Histologic and transmisson electron microscopy (TEM) analyses of the hearts from siblings with RCM (CM1023 and CM1024) were performed in the 1990s, when these siblings underwent heart transplantation. Blood samples obtained from these subjects with DNA extracted at the time were recently screened for genetic mutations in a variety of candidate genes, and the Q529X-MYPN mutation was identified in these siblings. Since the CM1023 myocardial samples were not saved, only the myocardium from CM1024 was available for our molecular and cellular studies, although histologic analysis was available for both subjects.
Histologic analysis of the myocardium from sibling CM1023 (Fig. 2A) revealed multifocally disorganized cardiomyocytes with increased side-branching and frequent side-to-side junctions along with large, hyperchromatic and bizarrely shaped nuclei (Fig. 2A, left panel). TEM performed on this sample revealed myofibrillar disarray, side-to-side junctions and mitochondrial damage (Fig. 2A, right panel, arrow). The overlapping histologic and TEM features of RCM and HCM were consistent with the cardiac echocardiographic features of patient CM1023 in which a thickened interventricular septum (IVS) and dilated left atrium were notable (Table 1). Analysis of the CM1024 myocardium obtained from the other sibling (CM1024; Fig. 2B) demonstrated scattered myocardial degeneration, hypertrophied myocytes and small attenuated cells with interstitial fibrosis (Fig. 2B, left panel). Myofibrillar degeneration, swelling of the sarcotubular system, irregular spreading of Z-discs, increased tortuosity of the intercalated discs and electron dense material were noted by TEM (Fig. 2B, right panel, arrows).
Figure 2.
Histopathology and comparative immunohistology of an explanted human heart from siblings with RCM and NRCs. (A) Histology (left panel, bar = 100 μm) and electron micrograph (right panel, bar: 3 μm) of the LV myocardium from patient CM1023 with RCM. An arrow indicates electron-dense material accumulated in the intercellular junctions. (B) Histology (left panel, bar = 100 μm) and electron micrograph (right panel, bar: 3 μm) of the LV myocardium from patient CM1024 with RCM. Arrows indicate electron-dense material accumulated in the intercellular junctions similar to that seen in the sibling CM1023. (C) Expression of MYPN (a, d and g), α-actinin (b, e and h) and merged images (c, f and i) in human control heart (a–c), DCM (d–f) and RCM (CM1024, g–i) patients. Bar = 10 μm. Arrows indicate co-localized MYPN and α-actinin at the striated Z-discs. Asterisks indicate disrupted sarcomeric Z-discs with abnormal diffuse MYPN co-distributed focally with α-actinin in CM1024. (D) Expression of α-actinin (red) in NRCs transfected with GFP-WT-MYPN (a–c), GFP-Y20C-MYPN (d–f) or GFP-Q529X-MYPN (g–i). Right panels represent merged images. Arrows indicate GFP-WT-MYPN and GFP-Y20C-MYPN co-localized with α-actinin. Asterisks indicate disorganized GFP-Q529X-MYPN in the Z-discs and its disrupted co-localization with α-actinin. Bar = 5 μm.
Comparative analysis of human myocardium and rat neonatal cardiomyocytes
To further investigate the functional consequences of genetic mutations on the MYPN protein, two variants, Q529X and Y20C, were chosen due to the predicted deleterious effects on protein function and the heterogeneous cardiomyopathic phenotypes seen in the affected patients hosting these mutations. Comparative immunohistochemistry of human heart tissue was performed on specimens from normal control subjects (motor vehicle accident victims), and from subjects with DCM without MYPN mutations and in sibling CM1024, as well as in neonatal rat cardiomyocytes (NRCs) expressing green fluorescent protein (GFP) chimeras of WT-, Y20C- or Q529X-MYPN. Normal co-expression of MYPN and α-actinin was noted in myocardial samples obtained from control individuals and DCM patients (Fig. 2Ca–f, arrows). In contrast, disrupted sarcomeric Z-discs with abnormally diffuse MYPN co-distributed focally with abnormal sarcomeric α-actinin were seen in the specimen from sibling CM1024 with RCM (Fig. 2Cg–i, asterisks). Comparative immunocytochemistry revealed that the Z-disc assembly was disturbed in the Z-bodies in premature NRCs expressing Q529X-MYPN compared with control or Y20C-MYPN cells. The WT and Y20C-MYPN demonstrated diffuse distribution, forming patchy dense bodies (Supplementary Material, Fig. 2A, asterisks), although GFP signals were not observed at the borders of NRCs expressing Q529X-MYPN (Supplementary Material, Fig. 2A, arrowheads). Nuclear localization of both variants, Y20C- and Q529X-MYPN, was impaired (Supplementary Material, Fig. 2B, asterisks). In mature NRCs, WT-MYPN or Y20C-MYPN was assembled in the normal striated pattern at the Z-discs and co-localized with α-actinin similar to the expression patterns in control or DCM hearts (Fig. 2Da–f, arrows), whereas severe disruption of Z-disc assembly with disturbed co-expression of MYPN and α-actinin was noted in NRCs expressing Q529X-MYPN (Fig. 2Dg–i, asterisks).
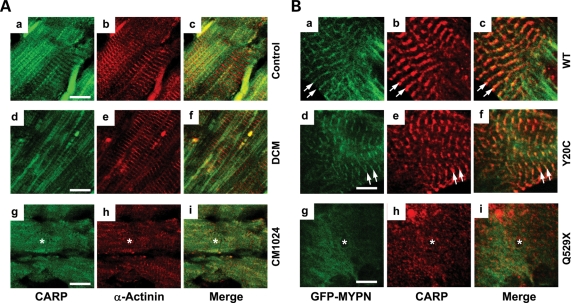
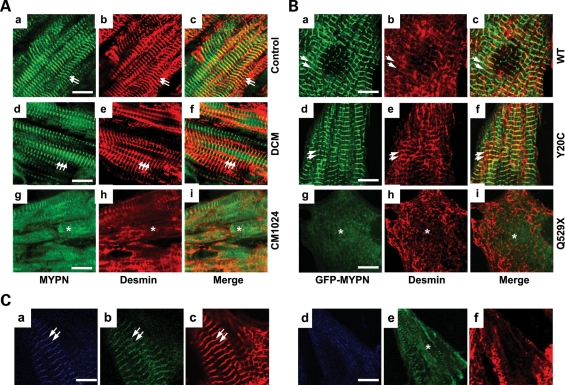
Abnormal diffuse CARP localization was also noted in the heart sample from subject CM1024 with RCM compared with control and DCM samples (Fig. 3Ag–i, asterisks). Despite normal co-distribution of endogenous CARP with WT-MYPN and Y20C-MYPN at the I-band neighboring the Z-disc (Fig. 3Ba–c and d–f, respectively, arrows), CARP was seen as small amorphous cytoplasmic aggregates in the Q529X-MYPN NRCs (Fig. 3Bg–i, asterisks). Loss of desmin at the Z-discs was also noted in the CM1024 myocardium and Q529X-MYPN NRCs (Fig. 4A and Bg–i, asterisks) as well.
Figure 3.
Comparative analysis of CARP in the human heart and NRCs. (A) Expression of CARP (a, d and g), α-actinin (b, e and h) and merged images (c, f and i) in the human control heart (a–c), DCM (d–f) or CM1024 (g–i) are shown. Expression of CARP is noted in Z/I bands in control and DCM hearts, whereas diffuse CARP expression in areas of disrupted sarcomere/Z-discs is indicated (asterisk) in CM1024. Bar = 10 μm. (B) Expression of CARP (red) in NRCs transfected with GFP-WT-MYPN (a–c), GFP-Y20C-MYPN (d–f) or GFP-Q529X-MYPN (g–i). Right panels represent merged images. Disrupted expression and localization of CARP and GFP-Q529X-MYPN (asterisks) is compared with normal localization of CARP at the I-bands and GFP-WT-MYPN and GFP-Y20C-MYPN at the Z-discs (arrows). Bar = 5 μm.
Figure 4.
Comparative analysis of desmin in human heart and NRCs. (A) Expression of MYPN (a, d and g), desmin (b, e and h) and merged images (c, f, and i) in control human heart (a–c), DCM (d–f) or CM1024 (g–i) heart samples are shown. The asterisk indicates loss of desmin in areas of disrupted sarcomere/Z-discs in the CM1024 heart. Bar = 10 μm. (B) Expression of desmin (red, b, e and h) in NRCs transfected with GFP-WT-MYPN (a–c), GFP-Y20C-MYPN (d–f) or GFP-Q529X-MYPN (g–i). Arrows indicate desmin and MYPN co-localization at the Z-discs; asterisks indicate disrupted desmin in Q529X cells. Bar = 5 μm. (C) Expression of WT and mutant MYPM fused with CFP- or GFP-containing constructs in NRCs. Cells co-transfected with CFP-MYPN-WT (a) or CFP-MYPN-Q529X (d) and GFP-MYPN-WT (b and e) 36 h after the transfection were stained with anti-α-actinin antibody (c and f). The CFP-MYPN-WT and GFP-MYPN-WT proteins were co-localized at the Z-disc (stained by anti-α-actinin antibody) in the NRCs (a–c). Localization of CFP-MYPN-Q529X protein at the Z-disc was completely disrupted in the NRCs co-transfected with CFP-MYPN-Q529X and GFP-MYPN-WT (d) and this abnormality was associated with that of GFP-MYPN-WT (e) and Z-disc itself (f); scale bar = 5 μm.
Dominant-negative effect on the Z-disc organization by the mutant Q529X-MYPN
To clarify whether deleterious effects of the Q529X mutation on Z-disc structure in NRCs were due to haploinsufficiency or a dominant-negative effect, co-expression studies were performed. Control NRCs co-expressing GFP alone with CFP (cyan fluorescence protein)-MYPN-WT or CFP alone with GFP-MYPN-WT showed co-localization of CFP- or GFP-MYPN and sarcomeric α-actinin at the Z-discs in a striated pattern (data not shown). Co-transfected CFP-MYPN-WT and GFP-MYPN-WT (Fig. 4Ca and b, respectively) of equal molar amounts were co-localized at the Z-disc overlapping with sarcomeric α-actinin (Fig. 4Cc). In contrast, when CFP-MYPN-Q529X and GFP-MYPN-WT (Fig. 4Cd and e, respectively) were co-transfected in NRCs, GFP-MYPN-WT was dissociated from the Z-discs diffusely expressing in almost all transfected cells. This was accompanied by diffuse distribution of both CFP-MYPN-Q529X and endogenous sarcomeric α-actinin (Fig. 4Cf). In addition, results of co-transfection of CFP-MYPN-WT and GFP-MYPN-Q529X were consistent (data not shown).
To determine the stability of mutant MYPN mRNA, the steady-state level of mutant mRNA was examined by RT-PCR analysis in the CM1024 myocardium. No significant difference in the expression of MYPN transcript levels between the WT and mutant alleles or no splicing abnormality of exon 9 in the mutant mRNA was observed in human hearts (Supplementary Material, Fig. S1B). In vitro, the stability of mutant MYPN at the cellular level was compared using western blotting at 12, 24 and 48 h after transfection of CFP-MYPN-Q529X and GFP-MYPN-WT constructs into HeLa cells. Relative intensity (RI) of mutant CFP-MYPN-Q529X to control GFP-MYPN-WT was gradually decreased from 12 to 24 h after transfection (0.77 ± 0.12-fold, Supplementary Material, Fig. S1C). At 48 h after transfection, RI was further decreased and it reached the statistical significance of 0.31 ± 0.06-fold, P< 0.001, suggesting that the nonsense mutation affected the stability of MYPN. These results were consistent in the cells co-transected with GFP-MYPN-Q529X and CFP-MYPN-WT (data not shown), indicating that the stability of mutant MYPN was not associated with fluorescent markers.
In vivo analysis of Y20C-MYPN
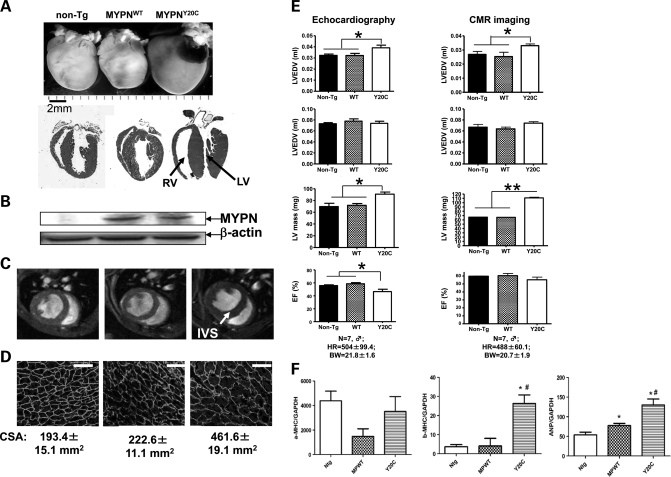
Based on the results of in vitro experiments showing deleterious effects of Q529X-MYPN on myofibrillogenesis of cardiomyocytes, we pursued further in vivo functional studies for the Y20C-MYPN mutant. We generated transgenic (Tg) mice with cardiac-restricted overexpression of WT- or Y20C-MYPN (MYPNWT or MYPNY20C, respectively). MYPNWT or MYPNY20C animals were born and they grew normally. By 6 months of age, MYPNY20C animals, unlike non-Tg and MYPNWT, developed significant cardiac enlargement with severe concentric hypertrophy and dilation of both the left and right ventricles (Fig. 5A). Expression of human MYPNWT or MYPNY20C in mouse hearts was confirmed by western blot analysis, although no expression of human MYPN was detected in non-Tg mice (Fig. 5B). The surface area of cardiomyocytes was significantly increased (P< 0.001) in MYPNY20C mice, measuring 461.6 ± 19.1 μm2 versus 193.4 ± 15.1 μm2 in non-Tg and 222.6 ± 11.1 μm2 in MYPNWT mice (Fig. 5D). The cardiac magnetic resonance (CMR) imaging confirmed LV hypertrophy and RV dilation (Fig. 5C). The IVS thickness and LV mass were significantly increased in MYPNY20C mice (P< 0.01), with an average IVS thickness of 1.26 mm and an average LV mass of 111.1 ± 2.2 mg versus those in non-Tg (0.93 mm and 66.3 ± 0.5 mg) and MYPNWT animals (0.88 mm and 65.9 ± 0.6 mg). Cardiac function evaluated by percentage ejection fraction (%EF) trended toward systolic dysfunction in MYPNY20C mice compared with non-Tg or MYPNWT mice. LV end-systolic volume (LVESV) was also significantly increased in MYPNY20C (P< 0.05, Fig. 5E). Real-time PCR of mouse heart tissue revealed up-regulation of hypertrophic genes such as β-MHC and ANP in MYPNY20C compared with MYPNWT littermates (Fig. 5F).
Figure 5.
Morphologic and functional assessment of MYPNY20C Tg mouse hearts. (A) Morphology (upper panel) and four-chamber H&E sections (lower panel) of non-Tg (left column), WT-Tg (middle column) and Y20C-Tg (right column) mouse hearts demonstrating concentric hypertrophy with severe thickening of the IVS, LV hypertrophy and RV dilation and thinning in MYPNY20C animals (arrows). (B) Protein analysis of human MYPN in non-Tg, MYPNWT and MYPNY20C hearts. (C) Short-axis end-diastolic gradient CMR imaging. Thickening of the IVS in the Y20C-Tg mouse is identified by the arrow. (D) Dystrophin staining of mouse cardiomyocytes (bar = 50 μm) demonstrating increase of cell surface area (CSA) in MYPNY20C mutants. (E) Mouse cardiac function analysis by CMR imaging and echocardiography. A significant increase of LVESV (*P< 0.05) and an increased LV mass (**P< 0.01) with reduced EF (%) were seen in MYPNY20C animals. HR, heart rate; BW, body weight. (F) Increase of hypertrophic marker genes expression in MYPNY20C mouse hearts compared with non-Tg and MYPNWT control littermates assessed by real-time PCR. The average value of increased folds measured by triplicate assays was used for quantification. *P< 0.05.
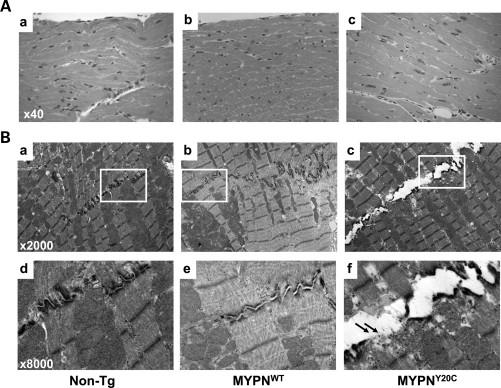
Histology demonstrated hypertrophied cardiomyocytes in MYPNY20C hearts (Fig. 6Ac). Masson trichrome staining revealed no cardiac fibrosis in any mice. TEM demonstrated no changes in sarcomeric Z-discs, mitochondria or other components of cardiomyocytes in control and MYPNWT or MYPNY20C animals (Fig. 6B). Unexpectedly, severe disruption of intercalated discs was observed in the MYPNY20C mouse hearts, including disappearance of terminal Z-discs adjacent to the intercalated discs and apparent pulling apart the cell–cell contacts (Fig. 6Bf, arrows) reminiscent of the typical findings observed in ARVC caused by desmosomal gene mutations (18).
Figure 6.
Histologic and ultrastructural assessment of MYPNY20C Tg mouse hearts. (A) Histology demonstrates hypertrophied cardiomyocytes with large rectangular nuclei in MYPNY20C (c) compared with non-Tg (a) and MYPNWT (b) animals. Resolution ×40. (B) Cardiac TEM demonstrates the structure of the intercalated disc in non-Tg (a), MYPNWT (b) and MYPNY20C animals (c) at a magnification of ×2000. Note the pulled apart intercalated disc in the mutant MYPNY20C heart. Arrows indicate disappearance of terminal Z-lines adjacent to intercalated discs in MYPNY20C (f, magnification ×8000).
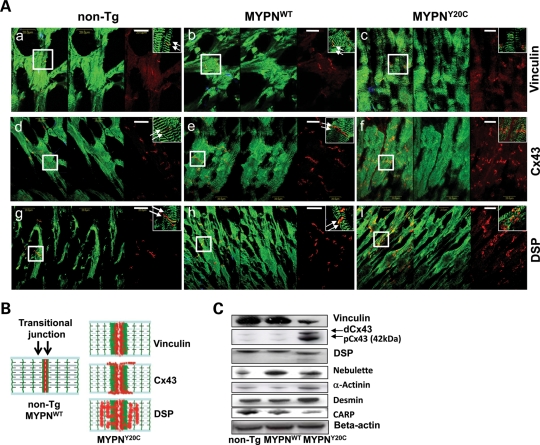
MYPN, binding partners and junctional proteins in mouse hearts
Owing to the discovery of disrupted intercalated discs in the MYPNY20C mouse hearts, we extended our protein–protein interaction studies to evaluate the interactions and co-localization of MYPN with intermediate filament, intercalated disc, gap junction and desmosomal proteins such as desmin, vinculin, connexin 43 (Cx43) and desmoplakin (DSP). Organized and two-sided expression of MYPN in Z-lines adjacent to intercalated discs (seen as a doublet) was demonstrated in the non-Tg and MYPNWT hearts (indicated by arrows; Fig. 7A, left and middle panels, respectively) and as schematically illustrated in Figure 7B (left panel). In contrast, expression of MYPN in the Z-lines adjacent to the intercalated discs was grossly abnormal and seen as a smear in MYPNY20C hearts (Fig. 7A, right panels). Expression of vinculin was abnormally expanded, smeared and overlaid with MYPN adjacent to the intercalated discs and Z-lines (Fig. 7Ac), and was down-regulated by quantitative analysis of RI compared with WT protein band intensity in western blotting (RI = 0.17, Fig. 7C). Expression of Cx43 was disorganized and expanded to the lateral surfaces of cardiomyocytes, presumably due to the redistribution of Cx43 from intercalated discs (Fig. 7Af) along with the up-regulation of both dephosphorylated (dCx43) and phosphorylated (pCx43) forms (RI = 1.59 and 3.62, respectively, Fig. 7C). Localization of DSP was expanded into the sarcoplasm, soaking into the sarcomeric I-bands and Z-lines, suggesting that the DSP disruption and aggregation also occurred in mutants (Fig. 7Ai). DSP expression levels were similar in all groups; however, immunoblotting revealed a lower molecular weight band for DSP in the MYPNY20C hearts, consistent with DSP cleavage, possibly occurring due to the formation of DSP aggregates (19,20). No significant changes were noted in the localization of NEBL, α-actinin and desmin in any of the mouse hearts by immunohistochemistry (data not shown). However, decreased CARP (RI = 0.6), increased α-actinin (RI = 1.9) and increased desmin (RI = 1.2) in the MYPNY20C hearts were notable, suggesting an imbalance in the expression of partner proteins due to mutated MYPN. Interestingly, expression was found to be increased for NEBL in the MYPNWT hearts (RI = 1.3, Fig. 7C).
Figure 7.
Expression of intercalated disc proteins in MYPNY20C Tg mouse hearts. (A) Co-immunolabeling of non-Tg (left panels), MYPNWT (middle panels) and MYPNY20C (right panels) hearts, with anti-MYPN in green and vinculin (a–c), Cx43 (d–f) or DSP (g–i) in red. Left column of each panel represents merged image. Bar = 20 μm. Right upper corner in each panel demonstrates high-resolution images (×100 digital magnification) of areas of interest indicated by squares. Arrows indicate normal expression of proteins in non-Tg and MYPNWT mice at intercalated discs. (B) Schematic illustration of MYPN (green), vinculin, Cx43 and DSP (red) expression in mouse hearts is shown. (C) Protein analysis in non-Tg, MYPNWT and MYPNY20C hearts. β-Actin was used as a loading control.
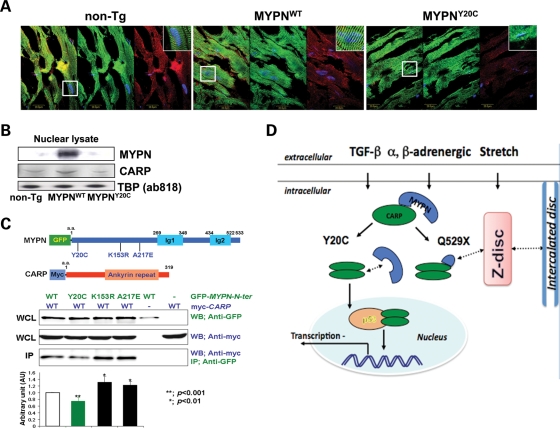
Distribution and interaction of Y20C-MYPN and CARP
In myocardial sections (Fig. 8A), only 24% of MYPNY20C cardiomyocytes (500 cells were counted in LV of five mice from each group) demonstrated nuclear localization (shuttling) of MYPN with Pearson values (PV) of −0.05 ± 0.05 (P< 0.05) compared with 55–60% in the MYPNWT (PV = 0.16 ± 0.02) and non-Tg animals (PV = 0.14 ± 0.01). These observations were supported by western blot analysis in which there was a notable absence of nuclear MYPN Y20C (Fig. 8B). Expression of CARP was decreased (RI = 0.6) in nuclear fractions (Fig. 8B, lower panel) of the MYPNY20C hearts despite the normal sarcoplasmic and nuclear distribution of CARP (Fig. 8A). These observations were associated with the significant up-regulation of CARP-specific target genes such as β-MHC, ANP and BNP in the MYPNY20C hearts compared with MYPNWT (Fig. 5F). Moreover, co-immunoprecipitation (co-IP) studies revealed the decreased binding of MYPN-Y20C and CARP compared with the binding of MYPN-WT and the other two variants, MYPN-K153R and MYPN-A217E (Fig. 8C).
Figure 8.
MYPN and CARP interactions in mouse hearts and in vitro. (A) Co-immunolabeling of non-Tg (left panels), MYPNWT (middle panels) and MYPNY20C (right panels) mouse hearts with MYPN (green); CARP (red); nuclei (blue; DAPI). Left column of each panel represents merged image. Right upper corner in each panel demonstrates high-resolution images (×100 digital magnification) of areas of interest indicated by squares showing localization of MYPN or CARP in nuclei. Bar = 20 μm. (B) Expression analysis of human MYPN and CARP in nuclear fractions of mouse hearts. Anti-TATA box protein (TBP, ab818, Abcam) was used as a nuclear protein-loading control. (C) Co-IP assay. Constructs of GFP-WT and -mutant MYPN and Myc-CARP shown in upper panels were transiently co-expressed in COS-7 cells and detected using anti-GFP and anti-Myc, respectively, in whole-cell lysate. The co-IP assay indicates significant impairment of GFP-MYPN-Y20C binding to Myc-CARP (**P< 0.001). (D) Schematic representation of pathogenic mechanisms involved in MYPN cardiomyopathies. MYPN binds to CARP (N-terminus), NEBL (rod domain) and α-actinin (C-terminus). MYPN Y20C mutant has reduced association with CARP, and lost ability to translocate to the nucleus. However, the level of nuclear CARP is reduced in the mutant MYPNY20C heart, thus, the function of CARP as a co-inhibitor of gene transcriptional regulation may be impaired. In addition, CARP also interacts with desmin at the I-band. At Z-discs, NEBL and α-actinin interact with desmin; thus, intermediate filament may impact the transitional junction/intercalated disc via DSP interaction. Maintenance of myofibrillogenesis is disturbed in MYPNQ529X mutants via truncation of MYPN binding to NEBL and α-actinin, the main molecules involved in Z disc/myofibrillar assembly. Hence, via a ‘domino effect’ of disturbance of multiple binding partners, MYPN mutations cause variable clinical forms of cardiomyopathy.
DISCUSSION
Pathophysiologic findings in affected subjects and animal models with cytoskeletal/Z-disc protein abnormalities suggest that the sarcomeric cytoskeleton not only provides structural support, but also acts as sensors for the mechanical, biochemical and electrical signals in striated muscle (21). MYPN is an Ig-domain family member protein that has been reported to be a key intermediate molecule at the Z-disc involved in sarcomere/Z-disc assembly and regulation of gene expression in cardiac cells (12,14). The goal of the genetic and functional studies presented herein was to determine whether MYPN gene mutations are common causes of cardiomyopathies and to explore the pathogenic mechanisms responsible for the development of heterogeneous cardiomyopathic phenotypes, as previously reported for MYPN in subjects with HCM and DCM. In our genetic screening, we identified 15 MYPN variants (of which 14 are novel), which were associated with HCM or DCM as previously reported by others, but was also found in familial RCM. The prevalence of 1.66% (DCM = 1.72%, HCM = 1.86% and RCM = 1.45%) in this large cardiomyopathic cohort is relatively high compared with other disease genes reported, and therefore suggests that it is likely to be clinically important. Two nonsense and 10 missense variants were predicted to have damaging consequences on the protein, using the protein function analysis software (17). Two of these MYPN variants, Y20C and Q529X, which disturb different functional domains of MYPN, resulted in variable clinical and pathologic features in the patients hosting these variants, as predicted to be deleterious by various protein prediction analyses. The Y20C variant was found in HCM and DCM patients, whereas the Q529X mutation was identified in an RCM family with variable penetrance and in whom the pathologic and echocardiographic signs consisted of overlapping features of HCM and RCM. The N-terminal variant, Y20C, affects the CARP-binding domain, whereas Q529X truncates the rod and C-terminus of MYPN, including both the NEBL- and α-actinin-binding domains. The portion of the protein truncated in Q529X-MYPN contains the last three Ig-domains of the five total domains that are found in MYPN (Fig. 8D). These Ig-domains are reported to be critically involved in sarcomere assembly and organization in cardiac muscle tissue (13,14,22).
Results of our biochemical, cell biologic and in vivo analyses of these variants have highlighted the divergent consequences of these gene abnormalities on protein function. The Q529X-MYPN mutant was shown to be involved in the pathogenesis of RCM by affecting and altering the recruitment of molecules, such as desmin, α-actinin and CARP during the myofibrillogenesis and Z-disc assembly. On the other hand, the Y20C-MYPN mutant altered the expression of MYPN-binding partners, including CARP, α-actinin and NEBL. Disruption of the Z-disc-transitional junction-intercalated disc assembly and integrity leading to HCM in vivo and its associated disturbance of desmin and intercalated disc molecules such as DSP, vinculin and Cx43 were also demonstrated along with TEM findings of disrupted cell–cell contacts, as reported in ARVC (18).
These results indicate that the MYPN variants, Y20C and Q529X, lead to different effects on MYPN function and its protein–protein interactions and that these differing effects cause variable phenotypes and possibly differing risks and severity of disease. We demonstrated that WT- and Y20C-MYPN form patchy, dense Z-bodies in the premature NRCs and incorporate with α-actinin at the Z-discs of mature myofibrils. Expression of Q529X-MYPN, in contrast to Y20C-MYPN, resulted in severely disrupted sarcomere/Z-disc assembly in NRCs with abnormal expression of α-actinin, desmin and CARP, similar to that seen in the human explanted heart hosting the Q529X mutation. Although the mechanisms responsible for the turnover and/or re-organization of the Z-bodies to the mature Z-discs remain unclear, we hypothesize that the severe disturbance of sarcomere/Z-disc assembly with the Q529X-MYPN is a result of losing the NEBL and α-actinin-binding domains, suggesting that the Q529X-MYPN mutation may have substantial impact on early myofibrillogenesis, with the clinical phenotype resulting in RCM in humans (Fig. 8D). The molecular mechanisms by which gene mutations cause cardiomyopathy can usually be explained by two alternative ways, dominant-negative or haploinsufficiency effects (23). Proteins with missense mutations expressed in cardiac muscle may affect its own structure and/or function through the dominant-negative effect. In contrast, the situation is more complex when the mutations generate C-terminal truncated proteins such as Q529X-MYPN. Truncated proteins are usually unstable due to the nonsense-mediated decay of mRNA and/or degradation through the lysosome or ubiquitin–proteasome system. Ultimately, insufficient quantities of the proteins may cause an imbalance of the function, termed as the haploinsufficiency effect. In the present study, we have showed that the steady-state expression of MYPN mRNA was not affected by the Q529X-MYPN mutation in the myocardium. Notably, mutant MYPN was relatively unstable compared with WT. This suggests that abnormal Z-disc assembly resulted from disturbed association of Q529X-MYPN with the Z-bodies/Z-discs during myofibrillogenesis due to the dominant-negative effect, although we cannot exclude the possibility of the haploinsufficiency effect by the progressive loss of unstable truncated protein expression.
Moreover, siblings with familial RCM had the Q529X-MYPN mutation, but their carrier mother (I-2 in Fig. 1B) did not suffer from RCM. The reason why she did not develop RCM remains unknown. There are several possibilities of explaining the difference of disease expressivity in mutation-prone individuals. First, the RCM patients carried other disease-associated mutations, which was absent from the mother. This possibility was evaluated by screening more than 20 known cardiomyopathy-associated genes in both patients, with no variants identified. Another possibility is that the RCM patients had other disease-modifier genes which were inherited from the father and not present in the mother, explaining the phenomenon of reduced penetrance of the p.Q529X mutation. For example, different amounts of mutant protein due to the difference in expression and/or degradation of protein in the lysosome and/or ubiquitin–proteasome system may interfere with the development of the phenotype. This might also explain the different overlapping pathologic findings between CM1023 (HCM) and CM1024 (DCM). Careful clinical follow-up is needed for the mother carrying the mutation.
Like CARP, a transcriptional co-activator of p53 and a co-inhibitor of gene transcription, MYPN has dual distribution in the cytoplasm and nucleus (24). Interestingly, we observed the occurrence of decreased binding of Y20C-MYPN to CARP and failure of Y20C-MYPN to translocate into nuclei (in vitro and in vivo). Results of in vivo studies demonstrating up-regulation of hypertrophic genes including CARP-specific target genes such as β-MHC and ANP (24) in MYPNY20C Tg animals suggest a decrease in CARP function. This may occur due to the down-regulation of CARP in MYPNY20C mice, which may subsequently cause decrease in the negative regulation of CARP-downstream genes, leading to the up-regulation of target hypertrophic genes and thus to the HCM phenotype in our model (Fig. 8D). Whether nuclear shuttling of MYPN modulates the CARP activity and whether nuclear MYPN is itself involved in cardiac gene regulation remain to be elucidated in future studies.
In addition to the altered expression of direct MYPN-binding partners such as CARP, α-actinin and NEBL, we also demonstrated that the intercalated discs in the MYPNY20C mice are disordered and disrupted with abnormal expression profiles of desmin, vinculin, DSP and Cx43. Desmosomal and intercalated disc proteins all play important roles in coordinating essential signaling events in the heart (25,26). Remarkably, the sarcomeric Z-discs maintained their ordered arrangement in the absence of terminal Z-discs adjacent to the intercalated discs. This suggests that MYPN may play a pivotal role at the ‘transitional junction’ (27), connecting the sarcomere to the intercalated disc. Supporting this notion, non-Tg mice showed two-sided expression of MYPN in the ‘transitional junction’ as schematically shown in Figure 7A. In contrast, MYPNY20C hearts demonstrated disrupted and smeared Y20C-MYPN expression in the terminal Z-discs or ‘transitional junctions’ adjacent to the intercalated discs. Owing to the lack of evidence for the direct binding of MYPN with any intercalated disc proteins, we hypothesize that the main target responsible for the disruption in the relationship of the sarcomere/Z-disc with the intercalated disc involves the intermediate filament protein desmin, which is overexpressed in MYPNY20C. Desmin binds to all direct MYPN partners, including NEBL, α-actinin and CARP, as well as to the desmosomal protein DSP, which are all disturbed. This would suggest that the desmin overexpression has downstream effects, including abnormal protein–protein interactions with the down-regulation of vinculin, and cleavage and aggregation of DSP, the protein complexes responsible for the function of the actin cytoskeleton and cell–cell adhesion, which in turn may cause lateralization of Cx43 distribution (26,28). Up-regulation of both phosphorylated and dephosphorylated forms of Cx43 noted in MYPNY20C suggests that this is a compensatory event that aims to balance the disturbed cell–cell uncoupling that occurs in the mutant heart. Notably, our results are consistent with the previously reported up-regulation of Cx43 that occurs in response to pulsatile stretch as seen in NRCs (29). These findings provide clues for understanding the interruption between myofibrillar filaments and intercalated discs in MYPNY20C animals as well as in the myocardium from the patients with the Q529X mutation. We have previously reported similar findings of disrupted and pulled apart intercalated discs in DSP-Tg mutant mice that were generated based on human DSP mutations in patients with ARVC (18). We also showed similar disruption of intercalated discs with disturbed desmin and MYPN expression in NEBL mutant mice (30), further supporting the role of desmin in the functional binding of the intercalated disc and sarcomere/Z-disc. In addition, conduction system abnormalities were seen in patients hosting the Y20C and Q529X variants (Table 1). As in patients with ARVC, these electrophysiologic abnormalities are likely due to disrupted gap junctions (Cx43) and desmosomes (DSP) (28), further supporting this hypothesis.
Finally, functional studies of Y20C and Q529X variants demonstrate that the specific form of cardiomyopathy appears to occur due to the disturbance of different binding partners of MYPN and downstream signaling pathways in a ‘domino effect’ pattern, causing overlapping cardiomyopathic phenotypes (9). Further supporting this hypothesis, the P1112L mutation in MYPN that was identified previously in patients with DCM (15) and HCM (16) was identified in another subject with HCM in this study. We also recently demonstrated that mutations in the ANKRD1 gene encoding CARP result in both the DCM and HCM phenotypes in patients (31,32), whereas novel mutations in the NEBL gene cause DCM and endocardial fibroelastosis (30). We suggest that the disturbed mechanosensation, one of the key features of Z-disc proteins, plays a substantial role in the development of the clinical phenotypes seen in heterogeneous cardiomyopathies.
In conclusion, we provide the first evidence that MYPN gene mutations have a relatively high prevalence in the development of heterogeneous cardiomyopathic phenotypes in humans, including DCM, HCM and RCM, with the suggestion that ARVC may also occur in some subjects. Although our primary genetic screening performed by denaturing high-performance liquid chromatography (DHPLC) or high-resolution melting (HRM) methods may miss rare variants, the relatively high frequency of MYPN mutations in patients with cardiomyopathy suggests that genetic screening of patients and family members with all forms of cardiomyopathy for mutations in the MYPN gene would be prudent. In addition to the reported multifunctional features of MYPN protein as a link between sarcomeric Z-discs and the nucleus, our studies suggest that MYPN forms a network linking sarcomeric proteins with the nucleus via CARP, as well as with intercalated discs and desmosomes, probably through the desmin intermediate filament network via its binding partners at the ‘transitional junction’. This work suggests that a combination of overlapping mechanisms which are dependent on the position of MYPN mutations and the protein-binding domains disturbed, as well as external forces (mechanical stress and stretch), leads to the disruption of key protein–protein interactions, resulting in heterogeneous clinical phenotypes. This study also demonstrates a possible basis for secondary disruption of cell–cell contacts due to Z-disc mutations and suggests a possible role for Z-disc mutations in ARVC.
MATERIALS AND METHODS
Mutational analysis
We analyzed 900 genetically unrelated patients with cardiomyopathy, including 484 (340 Caucasian and 144 Asian) HCM patients, 348 (280 Caucasian and 68 Asian) DCM patients and 68 (56 Caucasian, 6 African American and 6 Asian) RCM patients. The MYPN gene was amplified by polymerase chain reaction (PCR) (Supplementary Material, Table 1) in an exon-by-exon manner, screened by DHPLC or HRM curve and variants detected were confirmed by direct sequencing using an ABI3100 DNA Analyzer (Applied Biosystems, Foster, CA, USA). For each variant identified, available DNAs from family members and 1020 unrelated ethnic-matched (450 Asian, 370 Caucasian and 200 African American) healthy individuals were screened by DHPLC and direct sequencing to distinguish mutations from polymorphisms. Comparative analysis of a possible impact of an amino acid substitution on the structure and function of a human MYPN protein using various analyses including Polymorphism Phenotyping v2 (PolyPhen-2) at http://genetics.bwh.harvard.edu/pph2/ (17), Sorting Intolerant From Tolerant (SIFT) at http://blocks.fhcrc.org/sift/SIFT.html and PANTHER at http://www.pantherdb.org/tools/csnpScoreForm.jsp was performed.
mRNA expression, histology and immunohistochemical studies of human heart tissue
Paraffin-embedded LV myocardium from the explanted heart of a patient with RCM hosting a Q529X-MYPN mutation was available along with heart samples from two patients with DCM carrying no MYPN mutation and two control healthy subjects. Informed consents were obtained from all participating subjects and/or their guardians. Total RNA was extracted from paraffin-embedded heart tissues, using the RNeasy Fibrous Tissue Mini Kit (Qiagen, CA, USA) and first-strand cDNA was prepared using SuperscriptII (Invitrogen, CA, USA). To confirm the expression of both alleles, WT and c.1585C>T PCR products were digested using PvuII and then electrophoresed in a 2% agarose gel before being visualized by ethidium bromide. Histology and immunochemical analysis was performed as previously described (18,30).
Plasmid construction and transfection
Human MYPN mutant cDNA fragments with GFP (GFP-Y20C-MYPN and GFP-Q529X-MYPN) were constructed by primer-mediated mutagenesis from an entire WT-MYPN cDNA cloned into pEGFP-C1 (GFP-WT-MYPN) (Clontech, CA, USA). Each construct was transfected into NRCs isolated from hearts of 1- or 2-day-old Sprague–Dawley rats after multiple digestion with Liberase Blenzyme 3 (Roche) and Deoxyribonuclease II (Sigma) as described previously (31). For transient transfection, 0.4 μg of GFP-tagged construct was added into each well with 0.8 μl of TransFectin Lipid Reagent (Bio-Rad, CA, USA) according to the manufacturer's instructions. Eighteen, 36 or 48 h after transfection, cardiomyocytes were washed with PBS, fixed for 15 min in 100% ethanol at −20°C and stained with antibodies described in the following section. Over 200 transfected cells per each construct were analyzed.
Immuno- and co-IP, heart lysate fractioning and western blot analysis
MYPN-CARP binding was evaluated using co-IP studies. Human N-terminal-MYPN WT and mutant cDNA fragments (amino acids 1–533) with GFP-Y20C-MYPN, -K153R and -A217E were constructed by primer-mediated mutagenesis. cDNA encoding the CARP sequence was amplified from human cardiac mRNA (Ambion, Austin, TX, USA) and cloned into myc-tagged pCMV-Tag3 vector (Stratagene, La Jolla, CA, USA). Co-IP assay and western blot analyses were performed as described previously (18). Nuclear and cytoplasmic protein fractions were obtained from whole snap-frozen mouse hearts, using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL, USA) and 25 μg of protein was subjected to SDS–PAGE. Antibodies against α-actinin (Upstate), CARP, desmin (Abcam), DSP, Cx43 and vinculin (Santa Cruz) were purchased; human MYPN and NEBL antibodies were produced by Dr S. Labeit. Western blotting was quantified by density of bands, using the Kodak Image system. The level of protein in non-Tg hearts was referenced as a control and adjusted to RI = 1.0. The measured RI values compared with those of non-Tg hearts were expressed as mean ± SD.
To measure expression of CFP- or GFP-MYPN in transfected HeLa cells, nitrocellulose membrane (Invitrogen) with separated proteins was incubated with a primary BD Living Colors A.v. monoclonal antibody (Clontech) and a secondary rabbit anti-mouse Ig HRP-conjugated antibody (Dako A/S, Grostrup, Denmark). Recognized proteins were visualized by enhanced chemiluminescence (Pierce). The experiments were repeated seven times and the numerical densities were quantified as described earlier. Relative amount of CFP-MYPN-Q529X to GFP-MYPN-WT at 12 h after the transfection was set to 1.0 and those at 24 and 48 h after the transfection was calculated by assuming that the amount of GFP-MYPN was not changed during the experimental course.
Generation and characterization of α-MyHC-MYPN Tg mice
Cardiac-restricted MYPN-Tg mice were generated using a vector containing the mouse alpha-myosin heavy chain (α-MyHC) promoter (a gift from Dr Jeffrey Robbins, University of Cincinnati, OH, USA) and full-length WT- and Y20C-MYPN cDNA sequences. The linearized cDNAs were injected into fertilized oocytes (two-cell blastocyst stage) derived from C57/BL(6J) mice, and then the oocytes were transferred into the oviducts of pseudo-pregnant FVB mice. The genotypes of the offspring were determined by PCR of tail DNA. Two founders from each Tg line were expanded by crossing with non-Tg mice. Echocardiography was obtained using a 15 MHz linear transducer (Acuson Sequoia Cardiac System, Siemens Medical Solutions). To obtain the precise measurement of LV mass, three mice from each line underwent CMR imaging using the Bruker Biospin 7T horizontal MR scanner (Ettlingen, Germany) as described previously (17,18). Quantitative real-time PCR amplification of cDNA from total RNA extracted from mouse LV was performed using the 7500 Fast Real-time PCR system (Applied Biosystems). The average value of the triplicate assays was used for quantification. Final results are expressed as the ratio of a given gene/gene reference of GAPDH and/or beta-actin.
Histopathologic, immunohistochemical and ultrastructural assessment of mouse hearts
The mouse hearts were removed and perfused with relaxation buffer (25 mm KCl and 5% dextrose in PBS) containing heparin and nifedipine, using Langendorff system, for 10 min. Hearts were then fixed in 10% formalin and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin (H&E) and Masson trichrome to determine histologic and fibrotic changes in the hearts. Cryosections (5 μm) mounted on OCT (VWR, West Chester, PA, USA) were used for immunohistochemistry. To measure the myocyte area, cryosections were stained with C-terminal dystrophin to outline the sarcolemma. Multiple cross-sections from six mice per group with nearly circular capillary profiles were quantified. Quantitative image analysis was performed using the Image-Pro Plus software (Olympus). For TEM analysis, myocardial samples were processed according to the standard protocol. Images were taken using a Hitachi H-7500TEM.
Statistical analysis
Statistical analysis was performed using Student's t and ANOVA tests, using the GraphPad Prism 4.03 software. Data were reported as the mean with standard error mean, using Tukey's post hoc analysis. Values of P< 0.05 were considered to be significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by a Postdoctoral Fellowship and Beginning-Grant-in-Aid from the American Heart Association (E.P.), the Children's Cardiomyopathy Foundation (E.P., J.A.T.), the John Patrick Albright Foundation (J.A.T.) and NIH R01 HL53392, The Pediatric Cardiomyopathy Registry and R01 HL087000, The Pediatric Cardiomyopathy Specimen Repository (J.A.T.), Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology and grants from the Ministry of Health, Labour and Welfare, Japan; grants for Japan–France and Japan–Korea collaboration research from the Japan Society for the Promotion of Science; and grants from the Life Science Institute and Association Française contre les Myopathies, France (A.K., T.A.); the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program (M.J.A.).
Conflict of Interest statement. M.J.A. declared receiving royalties from Transgenomic as a consultant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the participating clinicians who contributed to the clinical evaluation and blood sampling of the patients, as well as the subjects and families.
REFERENCES
- 1.Maron B.J., Towbin J.A., Thiene G., Antzelevitch C., Corrado D., Arnett D., Marcus F.I., McKenna W.J., Sherrill D., Basso C., et al. Contemporary definitions and classification of the cardiomyopathies. An American Heart Association statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 2.Towbin J.A., Bowles N.E. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- 3.Keren A., Syrris P., McKenna W.J. Hypertrophic cardiomyopathy: the genetic determinants of clinical disease, expression. Nat. Clin. Pract. Cardiovasc. Med. 2008;5:158–168. doi: 10.1038/ncpcardio1110. [DOI] [PubMed] [Google Scholar]
- 4.Towbin J.A., Jefferies J.L. Dilated cardiomyopathy. Lancet. 2010;375:752–762. doi: 10.1016/S0140-6736(09)62023-7. [DOI] [PubMed] [Google Scholar]
- 5.Kushwaha S.S., Fallon J.T., Fuster V. Restrictive cardiomyopathy. N. Engl. J. Med. 1997;336:267–276. doi: 10.1056/NEJM199701233360407. [DOI] [PubMed] [Google Scholar]
- 6.Sen-Chowdhry S., Morgan R.D., Chambers J.C., McKenna W.J. Arrhythmogenic cardiomyopathy: etiology, diagnosis, and treatment. Ann. Rev. Med. 2010;61:233–253. doi: 10.1146/annurev.med.052208.130419. [DOI] [PubMed] [Google Scholar]
- 7.Marcus F.I., McKenna W.J., Sherrill D., Basso C., Bauce B., Bluemke D.A., Calkins H., Corrado C., Cox M.G.P.J., Daubert J.P., et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. Circulation. 2010;121:1533–1541. doi: 10.1161/CIRCULATIONAHA.108.840827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Towbin J.A. Left ventricular noncompaction: a new form of heart failure. Heart Fail. Clin. 2010;6:453–469. doi: 10.1016/j.hfc.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Bowles N.E., Bowles K.R., Towbin J.A. The ‘final common pathway’ hypothesis and inherited cardiovascular disease. The role of cytoskeletal proteins in dilated cardiomyopathy. Herz. 2000;25:168–175. doi: 10.1007/s000590050003. [DOI] [PubMed] [Google Scholar]
- 10.Vatta M., Marcus F., Towbin J.A. Arrhythmogenic right ventricular cardiomyopathy: a ‘final common pathway’ that defines clinical phenotype. Eur. Heart J. 2007;28:529–530. doi: 10.1093/eurheartj/ehl530. [DOI] [PubMed] [Google Scholar]
- 11.Towbin J.A. Desmosomal gene variants in patients with ‘possible ARVC’. Heart Rhythm. 2011;8:719–720. doi: 10.1016/j.hrthm.2011.03.040. [DOI] [PubMed] [Google Scholar]
- 12.Bang M.L., Mudry R.E., McElhinny A.S., Trombitás K., Geach A.J., Yamasaki R., Sorimachi H., Granzier H., Gregorio C.C., Labeit S. Myopalladin, a novel 145-kilodalton sarcomeric protein with multiple roles in Z-disc and I-band protein assemblies. J. Cell Biol. 2001;153:413–427. doi: 10.1083/jcb.153.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller M.K., Bang M.L., Witt C.C., Labeit D., Trombitas C., Watanabe K., Granzier H., McElhinny A.S., Gregorio C.C., Labeit S. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J. Mol. Biol. 2003;333:951–964. doi: 10.1016/j.jmb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Otey C.A., Dixon R., Stack C., Goicoechea S.M. Cytoplasmic Ig-domain proteins: cytoskeletal regulators with a role in human disease. Cell. Motil. Cytoskel. 2009;66:618–634. doi: 10.1002/cm.20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duboscq-Bidot L., Xu P., Charron P., Neyroud N., Dilanian G., Millaire A., Bors V., Komajda M., Villard E. Mutations in the Z-band protein myopalladin gene and idiopathic dilated cardiomyopathy. Cardiovasc. Res. 2007;77:118–125. doi: 10.1093/cvr/cvm015. [DOI] [PubMed] [Google Scholar]
- 16.Bagnall R.D., Yeates L., Semsarian C. Analysis of the Z-disc genes PDLIM3 and MYPN in patients with hypertrophic cardiomyopathy. Int. J. Cardiol. 2010;145:601–602. doi: 10.1016/j.ijcard.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z., Bowles N.E., Scherer S.E., Taylor M.D., Kearney D.L., Ge S., Nadvoretskiy V.V., DeFreitas G., Carabello B., Brandon L.I., et al. Desmosomal dysfunction due to mutations in desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ. Res. 2006;99:646–655. doi: 10.1161/01.RES.0000241482.19382.c6. [DOI] [PubMed] [Google Scholar]
- 19.Grossmann K.S., Grund C., Huelsken J., Behrend M., Erdmann B., Franke W.W., Birchmeier W. Requirement of plakophilin 2 for heart morphogenesis and cardiac junction formation. J. Cell Biol. 2004;167:149–160. doi: 10.1083/jcb.200402096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasile V.C., Edwards W.D., Ommen S.R., Ackerman M.J. Obstructive hypertrophic cardiomyopathy is associated with reduced expression of vinculin in the intercalated disc. Biochem. Biophys. Res. Commun. 2006;349:709–715. doi: 10.1016/j.bbrc.2006.08.106. [DOI] [PubMed] [Google Scholar]
- 21.Pyle W.G., Solaro R.J. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ. Res. 2004;94:296–305. doi: 10.1161/01.RES.0000116143.74830.A9. [DOI] [PubMed] [Google Scholar]
- 22.Granzier H.L., Labeit S. The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ. Res. 2004;94:284–295. doi: 10.1161/01.RES.0000117769.88862.F8. [DOI] [PubMed] [Google Scholar]
- 23.Vosberg H.P. The ubiquitin-proteasome system may be involved in the pathogenesis of hypertrophic cardiomyopathy. Cardiovasc. 2005;66:1–3. doi: 10.1016/j.cardiores.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Kojic S., Nestorovic A., Rakicevic L., Belgrano A., Stankovic M., Divac A., Faulkner G. A novel role for cardiac ankyrin repeat protein Ankrd1/CARP as a co-activator of the p53 tumor suppressor protein. Arch. Biochem. Biophys. 2010;502:60–67. doi: 10.1016/j.abb.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 25.Carisey A., Ballestrem C. Vinculin, an adapter protein in control of cell adhesion signaling. Eur. J. Cell Biol. 2011;90:157–163. doi: 10.1016/j.ejcb.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Severs N.J., Dupont E., Thomas N., Kaba R., Rothery S., Jain R., Sharpey K., Fry C.H. Alterations in cardiac connexin expression in cardiomyopathies. Adv. Cardiol. 2006;42:228–242. doi: 10.1159/000092572. [DOI] [PubMed] [Google Scholar]
- 27.Bennett P.M., Maggs A.M., Baines A.J., Pinder J.C. The transitional junction: a new functional subcellular domain at the intercalated disc. Mol. Biol. Cell. 2006;17:2091–2100. doi: 10.1091/mbc.E05-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saffitz J.E. Arrhythmogenic cardiomyopathy and abnormalities of cell-to-cell coupling. Heart Rhythm. 2009;6:62–65. doi: 10.1016/j.hrthm.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Zhuang J., Yamada K.A., Saffitz J.E., Kléber A.G. Pulsatile stretch remodels cell-to-cell communication in cultured myocytes. Circ. Res. 2000;87:316–322. doi: 10.1161/01.res.87.4.316. [DOI] [PubMed] [Google Scholar]
- 30.Purevjav E., Varela J., Morgado M., Kearney D.L., Li H., Taylor M.D., Arimura T., Moncman C.L., McKenna W., Murphy R.T., et al. Nebulette mutations are associated with dilated cardiomyopathy and endocardial fibroelastosis. J. Am. Coll. Cardiol. 2010;56:1493–1502. doi: 10.1016/j.jacc.2010.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arimura T., Bos J.M., Sato A., Kubo T., Okamoto H., Nishi H., Harada H., Koga Y., Moulik M., Doi Y.L., et al. Cardiac ankyrin repeat protein gene (ANKRD1) mutations in hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2009;54:334–342. doi: 10.1016/j.jacc.2008.12.082. [DOI] [PubMed] [Google Scholar]
- 32.Moulik M., Vatta M., Witt S.H., Arola A.M., Murphy R.T., McKenna W.J., Boriek A.M., Oka K., Labeit S., Bowles N.E., et al. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J. Am. Coll. Cardiol. 2009;54:325–333. doi: 10.1016/j.jacc.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.