Abstract
Pluripotent stem cells are derived from culture of early embryos or the germline and can be induced by reprogramming of somatic cells. Barriers to reprogramming that stabilize the differentiated state and have tumor suppression functions are expected to exist. However, we have a limited understanding of what such barriers might be. To find novel barriers to reprogramming to pluripotency, we compared the transcriptional profiles of the mouse germline with pluripotent and somatic cells, in vivo and in vitro. There is a remarkable global expression of the transcriptional program for pluripotency in primordial germ cells (PGCs). We identify parallels between PGC reprogramming to pluripotency and human germ cell tumorigenesis, including the loss of LATS2, a tumor suppressor kinase of the Hippo pathway. We show that knockdown of LATS2 increases the efficiency of induction of pluripotency in human cells. LATS2 RNAi, unlike p53 RNAi, specifically enhances the generation of fully reprogrammed iPS cells without accelerating cell proliferation. We further show that LATS2 represses reprogramming in human cells by post-transcriptionally antagonizing TAZ but not YAP, two downstream effectors of the Hippo pathway. These results reveal transcriptional parallels between germ cell transformation and the generation of iPS cells and indicate that the Hippo pathway constitutes a barrier to cellular reprogramming.
INTRODUCTION
Pluripotent stem cells can be propagated almost indefinitely without undergoing senescence and can give rise to all cell types of the body, both in vitro and in vivo. Because of these properties, pluripotent stem cells are an excellent system to study cellular differentiation in normal and diseased states and may contribute to the development of cell-replacement therapies (1–4). Embryonic stem (ES) cells are the prototypical pluripotent stem cells and are derived from in vitro culture of the inner cell mass (ICM) of the blastocyst (5–7). Remarkably, pluripotent stem cells can be generated by over-expressing particular key transcription factors or microRNAs in somatic cells (8–18). This approach allows the generation of disease-specific induced pluripotent stem (iPS) cells (19–21) and holds enormous promise in Regenerative Medicine. However, the efficiency of iPS cell generation is very low, and this is likely due to genes or pathways that act as barriers to reprogramming to pluripotency. Senescence has been reported as a barrier to reprogramming. Preventing senescence by over-expressing SV40T antigen or hTERT (15), or down-regulating p53 or p21 (22–29), can significantly increase the efficiency of iPS cell generation. However, these manipulations appear to facilitate reprogramming largely by inducing a higher rate of cell proliferation, and thereby increasing the probability of stochastic events that may underlie reprogramming (23). Targets of the ES cell-specific cell cycle-regulating (ESCC) family of miRNA have also been shown to antagonize reprogramming (17). In addition, lineage-specific transcription factors may also act as barriers to reprogramming (30,31). Therefore, the assay of iPS cell generation provides an opportunity to dissect the mechanisms that act as barriers to reprogramming and antagonize cellular transformation (32).
A cell lineage where barriers to reprogramming may be of particular importance is the germline. Primordial germ cells (PGCs) are the embryonic precursors to the gametes, which re-establish the totipotent zygote upon fertilization. When PGCs are cultured in vitro they give rise to pluripotent stem cells very similar to ES cells, called embryonic germ (EG) cells (33–35). Unlike the reprogramming of somatic cells to iPS cells, reprogramming of PGCs to EG cells does not require introduction of exogenous genes. This is largely due to the fact that critical regulators of ES cell pluripotency and reprogramming, such as the transcription factors Oct4 and Nanog, are highly expressed in PGCs and indeed are essential for their development (36–38). However, important differences between PGCs and pluripotent stem cells must exist. PGCs, unlike ES cells or EG cells, proliferate for only a short period of time and do not contribute to chimeras when injected into blastocysts (39). Germ cell tumors are thought to arise from loss of tumor suppressor mechanisms that are active in PGCs but not in pluripotent stem cells (40). A direct comparison of transcriptional profiles between PGCs and other pluripotent cell types would therefore be expected to shed light on the mechanisms that protect PGCs against cellular transformation, and potentially also reveal novel barriers to reprogramming of somatic cells to pluripotency. While several recent studies have described transcriptional analyses of PGCs (41–47), no study to date has directly compared the transcriptome of the ICM, ES cells, PGCs and EG cells, and no insights into potential barriers to reprogramming have been reported.
We report a comparative study of the gene-expression profiles of mouse pluripotent stem cells and the cells in the embryo from which they are derived, including PGCs. Our results reveal a core transcriptional program present in all pluripotent cells analyzed, including a remarkable global expression of the transcriptional program for pluripotency in PGCs. We find that reprogramming of PGCs to the pluripotent stem cell state involves transcriptional changes that parallel both human germ cell tumorigenesis and the generation of iPS cells. The tumor suppressor Lats2 is highly expressed in PGCs but not in pluripotent stem cells or human germ cell tumors. Lats2 is a kinase of the Hippo pathway, a signaling cascade that regulates cell growth and tumorigenesis in both Drosophila and mammals (48–50). We show that LATS2 acts as a barrier to induction of pluripotency in human cells, and that this effect is mediated by suppression of TAZ, a downstream target of the Hippo pathway. We discuss the potential implications of our results for the parallels between germ cell transformation and the generation of iPS cells.
RESULTS
Identification of the transcriptional profiles of pluripotent cells
We first used microarrays to identify and compare the transcriptional profiles of mouse pluripotency associated cells in vivo, the ICM and PGCs, as well as the pluripotent stem cells they give rise to when cultured: ES cells and EG cells, respectively (Fig. 1A). ICMs were isolated by immunosurgery from embryonic day E3.5 blastocysts (51). PGCs were purified from mouse embryos by fluorescence-activated cell sorting (FACS) using the Oct4/GFP transgenic mouse line GOF18/delta PE/GFP (52–54) (Fig. 1A). These mice express GFP under the control of the Oct4 promoter specifically in PGCs (52,53). We initially focused our analysis on PGCs isolated from E11.5 mouse embryos. PGCs at this stage are still sexually indifferent and are capable of giving rise to EG cells (55). ES cells and EG cells were cultured in vitro and removed from feeder cells before analysis. In order to identify the transcriptional program of the various pluripotency associated cells analyzed, we used the following non-pluripotent cells as controls: freshly isolated somatic cells of the genital ridge/mesonephros area at E11.5 (SGM, in vivo control) and E13.5 cultured mouse embryonic fibroblasts (MEFs, in vitro control; Fig. 1A). All cell types were analyzed with a minimum of three and a maximum of six biological replicates. In addition, our analysis included our previous data on the transcriptional profiles of two adult stem cells: hematopoietic stem cells (HSCs) and neural stem/precursor cells (NSCs) (56). We analyzed all samples in C57Bl/6 background, with the only exception of ICM samples (C57Bl/6xC3H F1), which were controlled for as described in the Materials and Methods section. Detailed information on all samples used for microarray studies is provided in Supplementary Material, File S1. The raw data can be obtained from GEO (http://www.ncbi.nlm.nih.gov/geo/, GSE35416). The full normalized log 2-transformed expression data can be found in Supplementary Material, File S2.
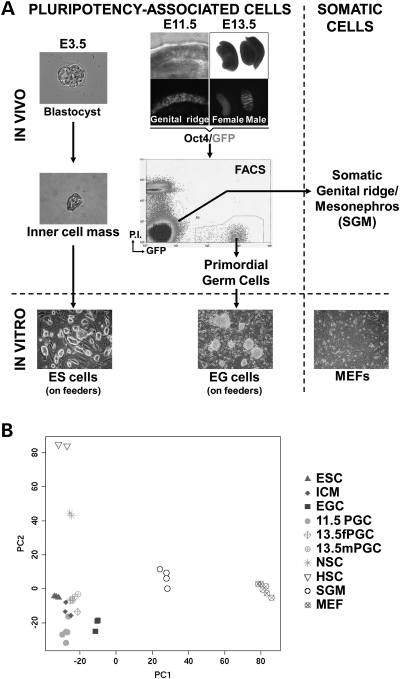
Figure 1.
Pluripotent cells in vivo and in vitro express a shared transcriptional program. (A) The transcriptional profiles of mouse inner cell mass (ICM) of the blastocyst, embryonic stem (ES) cells, primordial germ cells (PGCs) and embryonic germ (EG) cells were determined. ICMs were isolated by immunosurgery and PGCs by FACS using Oct4/GFP transgenic embryos. GFP fluorescence images in transgenic E11.5 and E13.5 embryos are shown. Note that E11.5 PGCs, but not E13.5 PGCs, give rise to EG cells when cultured in vitro. Controls used were: Somatic cells of the genital ridge/mesonephros (SGM) and mouse embryonic fibroblasts (MEFs). All cell types were analyzed with three to six replicas per cell type using Affymetrix microarrays. Also analyzed were previously collected data on gene-expression profiles of adult hematopoietic and NSCs (56). (B) Principal component analysis (PCA) of the transcriptional profiles of pluripotent and somatic cells, freshly isolated or in vitro cultured.
We analyzed the broad similarities and differences between the various cell types using principal component analysis (PCA) of the entire expression data (Fig. 1B). Figure 1B shows that pluripotent cells, regardless of whether they are freshly isolated from embryos (ICM and PGCs) or cultured in vitro (ES and EG cells), cluster closely together. Adult stem cells (HSCs and NSCs) or other somatic cells (SGM, MEFs) are clearly distant from pluripotent cells. These results indicate that pluripotent cells share similarities in their transcriptional programs that distinguish them from somatic cells.
We addressed in more detail the relative transcriptional similarities between all pluripotent cells. Hierarchical clustering indicates that the similarities between the transcriptional profiles of the various pluripotent cells are very high, to the point that sample clustering changes depending on the statistical method used (Supplementary Material, Fig. S1). We also determined the number of genes differentially expressed between ES cells and various other cell types, quantified along a continuum of fold-change cutoff, as a measure of the relative similarities between these samples. The reasoning behind this method is that if the transcriptional profiles of two cell types are similar, there will be few genes that are differentially expressed between them. As expected, there are large numbers of genes whose expression changes between ES cells and the somatic cell controls, SGM and MEFs (Supplementary Material, Fig. S2). ES cells would be predicted to have high transcriptional similarities to the ICM, the cells from which they are derived, or to EG cells, which are also cultured pluripotent stem cells. In agreement with these predictions, there are few differentially expressed genes when ES cells are compared with EG cells or the ICM (Supplementary Material, Fig. S2). Interestingly, there are also few differentially expressed genes when ES cells are compared with PGCs (Supplementary Material, Fig. S2). The similarities between ES cells and PGCs are validated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Out of 34 comparisons by qRT-PCR, 33 (97%) confirmed the microarray data, and one was ambiguous (Supplementary Material, Fig. S3). Taken together the PCA (Fig. 1B), hierarchical clustering results (Supplementary Material, Fig. S1), differential gene-expression data (Supplementary Material, Fig. S2) and qRT-PCR analysis (Supplementary Material, Fig. S3) indicate that there are high transcriptional similarities between ES cells and E11.5 PGCs, comparable with the similarities between ES cells and the ICM or EG cells. We find that PGCs downregulate the pluripotency program as they progress to sexual differentiation from E11.5 and E13.5 (Supplementary Material, Fig. S4), in agreement with a previous report (42). Importantly, E11.5 PGCs are functionally distinct from ES cells and do not contribute to chimeras (39). Nevertheless, these results suggest that the transcriptional program of pluripotency is globally maintained in E11.5 PGCs.
Transcriptional differences between E11.5 PGCs and pluripotent stem cells
The global similarities between the transcriptional profiles of E11.5 PGCs and pluripotent stem cells raise the question of what are the specific differences that may underlie the distinct biology and tumorigenic potential of these two cell types. In particular, the expression profile of PGCs is expected to differ from pluripotent stem cells in ways that protect against cellular transformation (39,40). One possibility would be that, while there are overall transcriptional similarities between E11.5 PGCs and ES cells, E11.5 PGCs do not express the critical core regulators of ES cell pluripotency, or express them at inappropriate levels. We observed that this is not the case: both E11.5 PGCs and ES cells express Oct4, Sox2, Nanog, Sall4, Utf1, Rex1, Fbx15 and Dppa4, among other pluripotency regulators or markers, at similarly high levels (Fig. 2A). The similarity in expression levels between E11.5 PGCs and ES cells for these genes is striking: there is no significant difference in gene levels between the two cell types (Fig. 2A). These observations are validated by qRT-PCR (Supplementary Material, Table S1). This is the case even for Oct4, the levels of which have to be very tightly regulated in ES cells to avoid differentiation (57). Therefore, E11.5 PGCs express the core transcriptional regulators of pluripotency at levels very similar to those in ES cells.
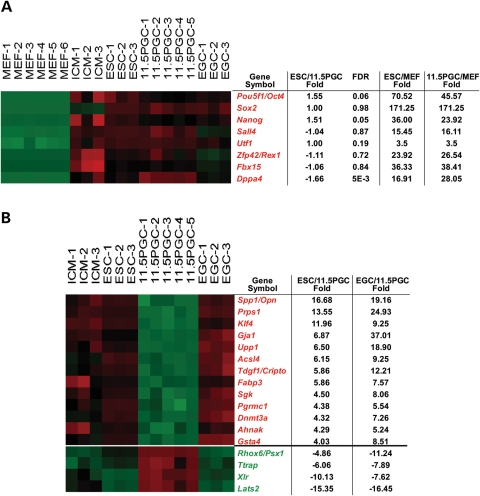
Figure 2.
Few genes are highly differentially expressed between E11.5 PGCs and pluripotent stem cells. (A) Expression of core transcriptional regulators and markers of pluripotency of ES cells and E11.5 PGCs. ES cells and E11.5 PGCs express similarly high levels of Oct4, Sox2, Nanog, Sall4, Utf1, Rex1, Fbx15 and Dppa4. The table shows fold changes in the expression of these genes in ES cells versus E11.5 PGCs, ES cells versus MEFs and E11.5 PGCs versus MEFs. The data are validated by qRT-PCR (Supplementary Material, Table S1). (B) Genes showing high differential expression between E11.5 PGCs and ICM, ESC cells and EG cells. Genes shown were identified as follows: they do not change between ES cells and ICM by more than 4-fold but are differentially expressed between ES cells and E11.5 PGCs and between EG cells and E11.5 PGCs by greater than 4-fold. Differential gene expression in the samples indicated is color-coded: red represents up-regulation, green represents down-regulation. Note the high differential expression of Klf4 and Lats2. The absent/present calls in the microarray data (not shown) and qRT-PCR (Supplementary Material, Table S1) confirm that Klf4 is either expressed at very low levels or not at all in E11.5 PGCs.
We next sought to identify genes expressed at similar levels in the ICM, ES cells and EG cells, but highly differentially expressed (by greater than 4-fold) between each of these cell types and E11.5 PGCs. Only 17 genes fulfilled these criteria (Fig. 2B and Supplementary Material, File S3), further highlighting the global transcriptional similarities between E11.5 PGCs and pluripotent stem cells. These results suggest that relatively few transcriptional changes may underlie transformation of germ cells to the tumorigenic pluripotent stem cell state.
Transcriptional differences between PGCs and pluripotent stem cells are recapitulated in human germ cell tumors
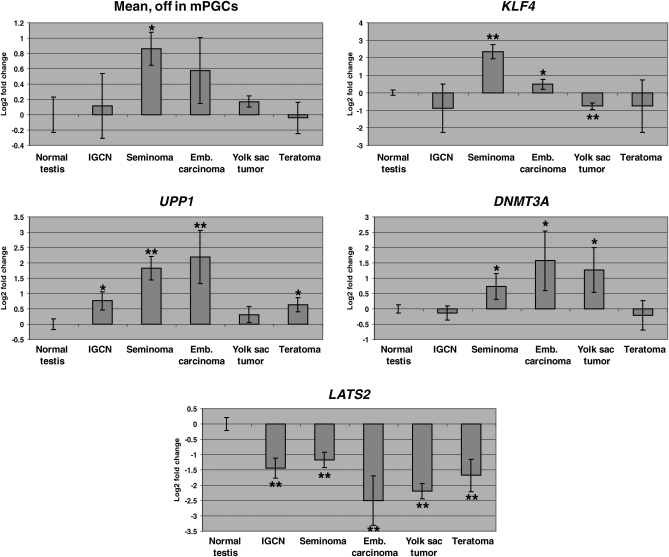
Given that germ cell tumors are thought to arise from transformation of PGCs (40), we then tested whether genes highly differentially expressed between PGCs and pluripotent stem cells (Fig. 2B and Supplementary Material, File S3) also show differential expression in germ cell tumors. We analyzed the expression of human orthologs of the genes in Figure 2B, making use of microarray data on the transcriptional profiles of human germ cell tumors (58). Interestingly, the average expression of the genes not expressed in PGCs (top part of Fig. 2B), without any bias in gene selection other than the existence of a human ortholog and its detection by the human microarray used, shows a significant up-regulation in seminomas (t-test P-value = 0.009) and a slight up-regulation in embryonic carcinomas (P = 0.054), but not in yolk sac tumors and teratomas (Fig. 3). Seminomas and embryonic carcinomas are two types of germ cell tumors with transcriptional similarities to pluripotent stem cells (58). Yolk sac tumors and teratomas, on the other hand, have transcriptional similarities to differentiated extra-embryonic and somatic tissues, respectively (58). This trend is more pronounced in the expression of specific genes such as UPP1, DNMT3A and KLF4 (Fig. 3). UPP1 is a uridine phosphorylase that has been proposed to be a prognostic factor in breast cancer (59), pancreatic cancer (60) and oral squamous cell carcinoma (61). DNMT3A is a de novo DNA methyl-transferase that regulates imprinting and gene silencing, which when dysregulated may lead to cancer (62). Of note, the pluripotency-inducing factor Klf4 (9–16) is highly upregulated in pluripotent stem cells relative to PGCs (Fig. 2B) and is induced in human germ cell tumors (Fig. 3). These data suggest that there may be molecular similarities between transformation of PGCs to the tumorigenic pluripotent stem cell state and induction of pluripotency in somatic cells.
Figure 3.
Transcriptional differences between PGCs and pluripotent stem cells are recapitulated in human germ cell tumors. The expression of genes differentially expressed between mouse E11.5 PGCs and ICM, ES and EG cells was analyzed in the transcriptional profiles of normal human testis and human germ cell tumors (58). ‘Mean’ depicts the average expression pattern (averaged Log2 of signal ratio using as normalizer Universal Human Reference RNA) in human germ cell tumor samples of the orthologs of genes not expressed in mouse PGCs but highly expressed in pluripotent stem cells (top part of Fig. 2B). KLF4, UPP1 and DNMT3A are upregulated in seminomas and/or embryonic carcinomas. LATS2 shows the reverse pattern of expression: it is highly expressed in mouse PGCs but not in pluripotent cells (bottom part of Fig. 2B), and it is strongly downregulated in all types of germ cell tumors analyzed. Note that Y-axes represent Log2 transformations of the fold change relative to normal testis. IGCN, intratubular germ cell neoplasia. Data points are averages from three to four independent tissue/tumor samples. Error bars depict standard deviation. *P < 0.05; **P < 0.005.
Knockdown of LATS2 facilitates the generation of human iPS cells
We found that Rhox6/Psx1, Trap, Xlr and Lats2 have the expression pattern opposite to Klf4, i.e. they are highly expressed in PGCs but not in pluripotent stem cells (Fig. 2B). In particular, the tumor suppressor Lats2 is the most differentially expressed gene in this comparison (Fig. 2B and Supplementary Material, File S3) and shows a striking down-regulation in all human germ cell tumor types (Fig. 3). We therefore focused on its role in reprogramming for our next experiments. Loss of Lats2 in Drosophila leads to tumorigenesis (63) and silencing of LATS2 has been associated with a variety of human cancers (64). Interestingly, Lats2 is a target of miRNAs that are highly expressed in germ cell tumors (65) and that can induce pluripotency in somatic cells (8). Lats2 is also strongly downregulated when MEFs are reprogrammed to iPS cells (66). We therefore hypothesized that Lats2 may represent a barrier to reprogramming to pluripotency. We tested the effect of knockdown of LATS2 in induction of pluripotency in human fibroblasts by four factors (4F: OCT4, SOX2, KLF4 and C-MYC). Indeed, three different shRNAs against LATS2 all significantly increase the number of human iPS cell colonies positive for the pluripotency marker Tra-1-81, as compared to infection with a non-targeting shRNA control (Fig. 4A). Interestingly, LATS2 RNAi decreases the number of non-iPS cell colonies, which do not express TRA-1-81, and include both partially reprogrammed colonies and transformed fibroblasts (Fig. 4A). We also found that LATS2 RNAi leads to the appearance of iPS cell colonies 2–5 days earlier than controls, and probably because of this it increases the size of iPS cell colony (Fig. 4B). Efficient knockdown of LATS2 mRNA was confirmed by qRT-PCR (Supplementary Material, Fig. S5A). While LATS2 RNAi enhances reprogramming efficiency, it does not replace any of the reprogramming factors (data not shown).
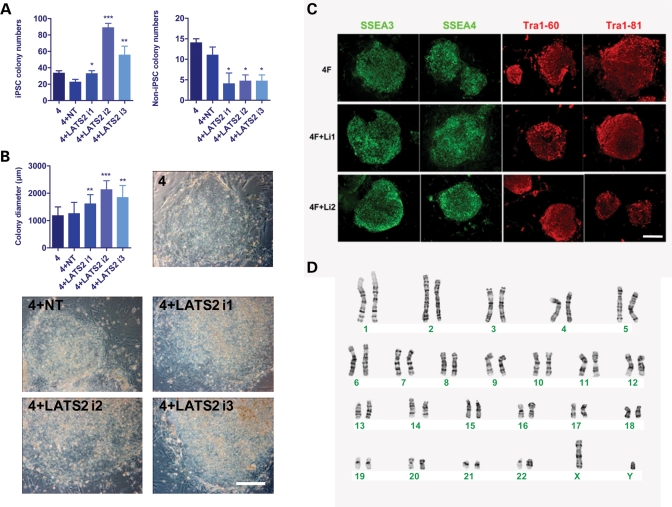
Figure 4.
Knockdown of LATS2 increases the efficiency of human iPS cell generation. (A) The number of Tra1-81-positive iPS cell and Tra-1-81-negative non-iPS cell colonies was counted on d20 after infection of human BJ foreskin fibroblasts with 4F alone (4), 4F + non-targeting shRNA (4 + NT) and 4F + LATS2 shRNA (three different short hairpins targeting LATS2 were independently tested, 4 + LATS2 i1, 4 + LATS2 i2 and 4 + LATS2 i3). Infections were performed in triplicate. Knockdown of LATS2 resulted in a significant increase in the number of Tra1-81-positive iPS cell colonies, and in a significant reduction in the number of Tra1-81-negative iPS cell colonies when compared with 4F + NT. (B) The diameter of iPS cell colonies was measured on d24 after infection of BJ foreskin fibroblasts with 4F alone (4), 4 + NT and 4F + LATS2 i1/2/3. For each condition 10 iPS cell colonies were randomly picked. Knockdown of LATS2 resulted in a significant increase in the diameter of iPS cell colonies. Phase-contrast representative images of a colony for each condition are also shown. (C) The iPS cell clones (P5) generated by 4F alone and 4F + LATS2 i1/2 showed strong, positive staining for all human ES cell-specific markers analyzed by immunostaining. (D) iPS cells (P10) generated by 4F + LATS2 i2 showed a normal male karyotype (46, XY). In all relevant panels, error bars represent standard deviation, and scale bars represent 300 μm. *P < 0.05; **P < 0.01; ***P < 0.001.
Once human iPS cell lines generated with LATS2 RNAi are established, they have normal growth rates (data not shown) and express human ES/iPS cell-specific surface markers including SSEA3, SSEA4, TRA-1-60 and TRA-1-81 (Fig. 4C). RT-PCR showed that these cells activate the endogenous expression of pluripotency markers OCT4, SOX2 and NANOG, and silence the viral transgenes (Supplementary Material, Fig. S5B), all of which are indicative of faithful reprogramming. These results indicate that, as suggested by our expression-profiling studies (Figs 2B and 3), LATS2 constitutes a novel barrier to reprogramming to the pluripotent stem cell state.
LATS2 represses human iPS cell generation by antagonizing TAZ
We sought to understand the mechanism by which LATS2 represses reprogramming. The reported role for Lats2 in genomic stability of mouse cells (67) prompted us to analyze the karyotypes of human iPS cells generated with LATS2 RNAi. All of these lines were found to be karyotypically normal (Fig. 4D and Supplementary Material, Fig. S5C), excluding chromosomal abnormalities as a potential underlying cause of increased reprogramming. We next considered whether LATS2 RNAi might increase reprogramming efficiency by accelerating cell proliferation, because Lats2−/− MEFs display increased proliferation rates (68). In addition, LATS2 has been shown to negatively regulate CDK2 and cooperate with p53 at the G2/M checkpoint (69,70). Accelerated cell proliferation, such as that caused by p53 RNAi, has been shown to increase the efficiency of iPS cell generation (22–29). p53 RNAi in human fibroblasts leads to a 2–5-fold increase in the efficiency of iPS cell generation, which is comparable to LATS2 RNAi (22,29). However, we found that, unlike p53 RNAi, LATS2 RNAi neither leads to increased proliferation in four factor-induced reprogramming nor in fibroblasts alone. If anything, LATS2 RNAi may globally reduce cell proliferation (Fig. 5A and Supplementary Material, Figs S6A and B). A further important difference between p53 RNAi and LATS2 RNAi is that in p53 RNAi a general cellular overgrowth is observed and all types of colonies, including partially reprogrammed non-iPS cell colonies, are increased in number (Supplementary Material, Fig. S6A and data not shown). LATS2 RNAi, in contrast, leads to a specific increase in iPS cell colony numbers and a concomitant decrease in non-iPS cell colony numbers (Fig. 4A). Taken together, these results indicate that LATS2 RNAi enhances the efficiency of human iPS cell generation by mechanisms that are distinct from p53 RNAi-driven cellular over-proliferation.
Figure 5.
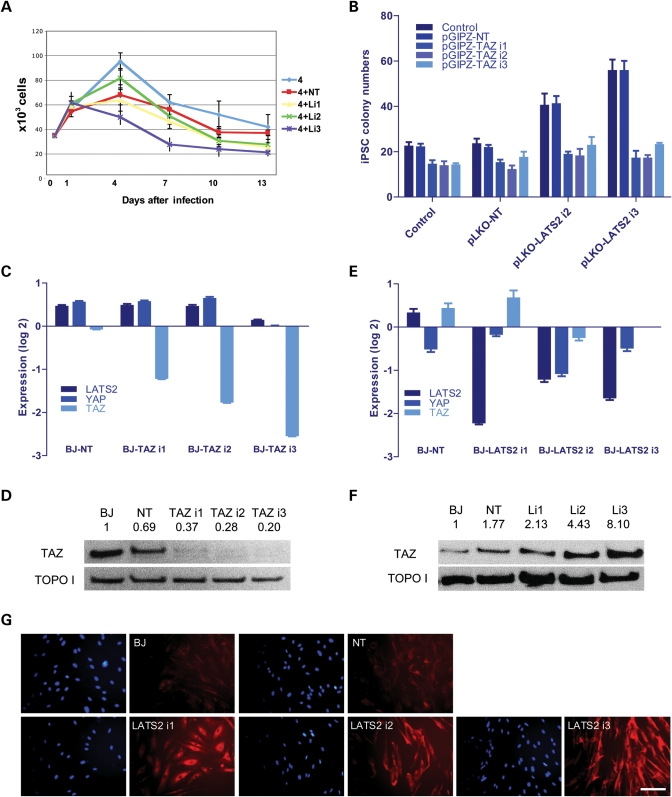
LATS2 antagonizes human cell reprogramming by repressing TAZ. (A) Growth curves of human fibroblasts infected with 4 factor, 4F + non-targeting shRNA (4 + NT), 4F + LATS2 shRNA (4 + Li1/2/3), counted on d0, d1, d4, d7, d10 and d13 post-infection. Infections were performed in triplicates. LATS2 RNAi did not increase total cell numbers during the first 13 days of reprogramming. Data shown are representative of two independent experiments, and error bars represent standard deviations. (B) TAZ knockdown suppresses the LATS2 RNAi-mediated increase in efficiency of iPS cell generation. The number of iPS cell colonies was counted on d21 after infection of BJ foreskin fibroblasts with 4F alone (control), 4F + non-targeting shRNA (pLKO-NT) and 4F + LATS2 shRNA (pLKO-LATS2 i2/3). For each condition, TAZ was also knocked down by pGIPZ lentivirus infection (pGIPZ-TAZ i1/2/3, with pGIPZ-NT used as a negative control). Infections were performed in triplicates and error bars represent standard deviation. (C) Reduction in the levels of TAZ expression achieved by each of the three shRNA constructs (TAZ i1/2/3) was confirmed by qRT-PCR. The expression of LATS2 and YAP showed no significant change upon TAZ RNAi. (D) Reduction in the levels of TAZ protein expression achieved by each of the three shRNA constructs (TAZ i1/2/3) was confirmed by western blotting. Topoisomerase I (TOPO I) was used as loading control. Numbers indicate densitometry analysis of the TAZ expression level standardized to TOPOI. (E) Reduction in the levels of LATS2 expression achieved by each of the three shRNA constructs was confirmed by qRT-PCR. The mRNA level of TAZ and YAP showed no significant change upon LATS2 RNAi. For (C,E), values were standardized to GAPDH and UBB, and then normalized to uninfected BJ fibroblasts. Note log 2 scale in y-axis: e.g. −2 equals down 4×, −3 equals down 8×, etc. Data are from triplicate PCR reactions, and error bars represent standard deviation. (F) Western blotting shows that LATS2 RNAi (Li1/2/3) increases TAZ protein expression level in human fibroblasts. TOPO I was used as loading control. Numbers indicate densitometry analysis of the TAZ expression level standardized to TOPOI. (G) Immunofluorescence shows that LATS2 RNAi (LATS2 i1/2/3) increases TAZ protein expression level in human fibroblasts. Immunostaining was performed 5 days after infection with lentiviruses. Blue, Dapi; red, TAZ. Scale bars represent 80 μm.
LATS2 also functions as a member of the Hippo signaling pathway, which regulates important developmental processes including apoptosis, stem cell maintenance, differentiation and organ size control (48–50). We therefore explored whether the effect of LATS2 RNAi on reprogramming may be mediated by dysregulation of the Hippo pathway. The paralog transcriptional regulators YAP and TAZ are downstream effectors of the Hippo pathway, and are negatively regulated by LATS2. Activated LATS2 can phosphorylate both YAP and TAZ, which leads to their cytoplasmic retention and protein degradation (49). YAP over-expression was recently reported to increase the efficiency of mouse iPS cell generation (71). However, we found that, unlike in mouse, YAP over-expression or RNAi has no effect on human fibroblast reprogramming to iPS cells, whether on its own or combined with LATS2 RNAi (Supplementary Material, Fig. S6C). Efficient knockdown of YAP mRNA and over-expression of YAP protein was confirmed by qRT-PCR (Supplementary Material, Fig. S6D) and western blotting (Supplementary Material, Fig. S6E), respectively. These results indicate that the increase in the efficiency of human iPS cell generation upon LATS2 RNAi is not mediated by de-repression of YAP.
While mouse ES cells are not affected by knockdown of Taz, TAZ RNAi leads to self-renewal defects and differentiation in human ES cells (72). Interestingly, we found that knockdown of TAZ completely suppresses the enhancement in human iPS cell generation seen with LATS2 RNAi (Fig. 5B). Moreover, TAZ RNAi on its own leads only to a slight decrease in the efficiency of human iPS cell generation (Fig. 5B). Specific knockdown of TAZ mRNA and protein was confirmed by qRT-PCR (Fig. 5C) and western blotting (Fig. 5D), respectively. These results suggest that de-repression of TAZ is an essential downstream effect of LATS2 RNAi in human cell reprogramming. We tested whether LATS2 directly regulates TAZ expression, using qRT-PCR, western blotting and immunofluorescence. We found that although LATS2 RNAi has no effect on TAZ mRNA levels (Fig. 5E), it significantly increases TAZ protein expression in both the nucleus and cytoplasm of human fibroblasts (Fig. 5F and G and Supplementary Material, Fig. S6F). Thus, LATS2 appears to primarily regulate total levels of TAZ, rather than differential nuclear import/export. Taken together, these results indicate that LATS2 acts as a barrier to reprogramming of human cells by post-transcriptional regulation of TAZ.
DISCUSSION
We sought to identify barriers to reprogramming to pluripotency by analyzing the transcriptional profiles of PGCs, pluripotent stem cells and somatic cells. We found that the transcriptional program of pluripotent stem cells is extensively maintained in PGCs. We identified specific differences between the transcriptional profiles of PGCs and pluripotent stem cells, and propose that these differences may protect PGCs against germ cell tumorigenesis. We focused on the tumor suppressor Lats2, which is highly expressed in PGCs but not in pluripotent stem cells. We showed that LATS2 is strongly downregulated in all types of human germ cell tumors. We tested the role of LATS2 in human iPS cell generation and found that it acts as a barrier to reprogramming. We further showed that LATS2 antagonizes human cell reprogramming via post-transcriptional regulation of TAZ but not YAP, two downstream effectors of the Hippo pathway. These data suggest that there may be parallels between germ cell transformation and the generation of iPS cells, and indicate that the Hippo pathway constitutes a barrier to cellular reprogramming.
Relationship between PGCs, germ cell tumorigenesis and induction of pluripotency
Our data reveal transcriptional parallels between reprogramming of PGCs to the pluripotent stem cell state, germ cell tumorigenesis and the generation of iPS cells. It is generally assumed that iPS cells represent an artificial manipulation of somatic cells in vitro that leads to the re-acquisition of the ES cell state, which in turn represents a ‘freezing in time’ of the late ICM of the blastocyst (73). An alternative and not mutually exclusive possibility suggested by our data is that iPS cells can represent the implementation in somatic cells of a recipe for germ cell transformation to a self-renewing tumorigenic state, which occurs naturally at low frequencies in both mice and humans. Similar alternative routes to achieving the ES cell state upon blastocyst culture, one involving a direct ICM–ES cell transition and another involving an intermediate PGC-like state, have recently been hypothesized and proposed to be dependent on culture conditions (74,75).
Possible role for Lats2 in the suppression of germ cell tumors
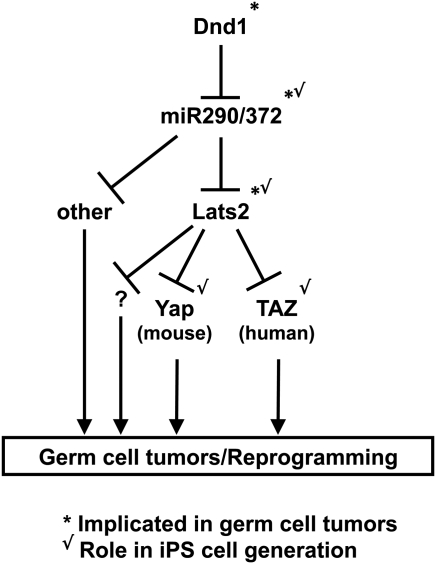
Our data suggest that Lats2 may protect PGCs from transformation. The high expression of Lats2 in PGCs relative to pluripotent stem cells (Fig. 2B) and its downregulation in germ cell tumors (Fig. 3) are consistent with a potential tumor-suppressive role for Lats2 in the germline, although this remains to be tested. Considering the high global transcriptional similarities between PGCs and pluripotent stem cells, it is interesting to speculate as to how Lats2 is so strikingly differentially expressed between these cell types (Fig. 6). Lats2 is a target of the 290 family of miRNAs, which are highly expressed in pluripotent stem cells (76) and germ cell tumors (65), and can induce pluripotency in somatic cells (8,18). These miRNAs are also expressed in PGCs (77), but their activity is inhibited by the RNA-binding protein Dead end 1, Dnd1 (78). Dnd1 mutant mice have a high incidence of germ cell tumors (79), and it will be of interest to determine if loss of Lats2 contributes to the Dnd−/− phenotype. A major unresolved question about this model concerns how the expression of Dnd1 in PGCs is regulated. It is important to emphasize the speculative and simplistic nature of this model, which is intended as a framework for future studies of a potential role for the Hippo pathway in suppressing germ cell tumorigenesis. In addition, this model highlights some of the potential parallels identified in this study between germ cell tumorigenesis and reprogramming to the iPS cell state (Fig. 6).
Figure 6.
Speculative model for the role of the Hippo pathway in germ cell tumorigenesis and reprogramming. Lats2 may be under tight control in PGCs via Dnd1-mediated inhibition of miRNAs of the 290 family. Loss of Dnd1 in PGCs may allow these miRNAs to inhibit their targets, including Lats2. ‘Other’ represents targets of the miRNA 290 family other than Lats2. Question mark (‘?’) represents functions of Lats2 independent of the Hippo pathway effectors Yap and Taz, such as in cell cycle and mitotic stability. Loss of Lats2 de-represses Yap or Taz, which promote reprogramming to the tumorigenic pluripotent stem cell state. Checkmarks indicate cases where a gene has been implicated in germ cell tumorigenesis and/or iPS cell generation in the literature (Dnd1, mir290, Yap) or in this study (Lats2, Taz). The different role for Yap and Taz in mouse versus human cell reprogramming may be due to species- or stage-specific differences. See Discussion section for details.
Role of the Hippo pathway in reprogramming
We show that LATS2 acts as a novel barrier to human iPS cell generation. We consistently find that intermediate levels of knockdown of LATS2 and TAZ protein induction have the best effects on reprogramming efficiency. This might be because such levels reduce the Hippo pathway arm of LATS2 while leaving its independent roles in cytokinesis/ploidy intact. Our data indicate that LATS2 antagonizes human cell reprogramming by repressing TAZ, but not YAP, two conserved downstream targets of the Hippo pathway that are directly regulated by LATS2. The Hippo pathway is emerging as a global regulator of progenitor cells, organ size and tumor suppression (48–50). We had found Yap to be upregulated in multiple mouse stem cell populations (56), and Yap has since been shown to regulate stem and progenitor compartments in the intestine, liver, skin and brain (80–82). There is evidence for shared as well as specific roles for Yap and Taz. Yap−/− mice display embryonic lethality at E8.5, whereas Taz−/− mice can survive to adulthood but with defects in the kidneys and lungs (83–86). However, Yap and Taz likely act redundantly during pre-implantation because Yap−/−;Taz−/− embryos arrest prior to morula development (87). Yap and Taz are thought to regulate commitment to the trophectoderm lineage in the morula and early blastocyst and are repressed by Lats1/2 in the ICM (87). These findings are compatible with ours because we detect Lats2 in the ICM, albeit at much lower levels than in PGCs (Fig. 2B), and Lats2 is further downregulated by ∼3-fold in the conversion ICM–ES cells (Supplementary Material, File S3). Indeed, Lats2 is downregulated in various settings of reprogramming to the pluripotent stem cell state, such as ICM–ES cells, PGC–EG cells (this work), MEF to iPS cells (66) and epiblast stem cells to ES cells (88). These results suggest that suppression of the Hippo pathway may be a common feature of reprogramming and, more broadly, cellular transformation.
Interestingly, TAZ and YAP have divergent roles in mouse and human cells. YAP regulates mouse ES cell self-renewal (89) and can increase the efficiency of mouse iPS cell generation (71), but has no role in human ES cells (90) or in iPS cell generation (this work). The reverse is the case for TAZ: it is important for human ES cell self-renewal (72) and iPS cell generation (this work), but has no role in mouse ES cells (72). We speculate that this difference may be due to the distinct signaling requirements of mouse versus human pluripotent stem cells. YAP can act as a co-activator of the BMP signaling pathway, while TAZ is important for TGFβ/Activin signaling (72,91). Both YAP and TAZ have, in addition, been shown to regulate the Wnt signaling pathway (92,93). The BMP pathway, with which Yap interacts, is critical for mouse ES/iPS cell self-renewal, but not for human. On the other hand, the TGFβ/Activin pathway, with which TAZ interacts, is essential for human ES/iPS cell self-renewal, but not for mouse (94,95). In addition, human ES cells correspond to a developmental stage that is more similar to mouse Epiblast stem cells than to ES cells (88,96), and therefore the distinct roles for TAZ and YAP in mouse versus human may be related to stage-specific differences in Hippo pathway signaling. Further work will be needed to dissect the distinct signaling interactions mediated by TAZ and YAP in different types of pluripotent stem cells.
MATERIALS AND METHODS
Culture of mouse ES cells and EG cells
C57Bl/6 (B6) ES cells were cultured in standard conditions in the presence of fetal bovine serum (FBS, Hyclone), MEFs and leukemia inhibitory factor (LIF) as previously described (97). ES cells were removed from MEFs by serial re-plating (56) or serial re-plating followed by culture for one passage in the absence of MEFs in gelatin-coated dishes. B6 EG cells (Patricia Labosky, Vanderbilt U.), derived from E12.5 male PGCs (55), were cultured in the presence of FBS, STO feeder fibroblasts and LIF, as described (98). Feeders were removed by serial re-plating (56) followed by culture for one passage in gelatin-coated dishes.
Isolation of ICMs
Throughout this study, we collected mouse samples on an inbred B6 background. The only exception was that B6 inbred mice (100% B6), upon super-ovulation, yielded few properly staged blastocysts for ICM isolation. We therefore collected 50% B6–50% C3H (B6C3H) ICMs. We tested the effect of reducing the B6 background on the transcriptional profiles of ICMs. We collected one ICM sample that is only 25% B6 (ICM-1) and compared its transcriptional profile with the other two ICM replicates (ICM-2 and ICM-3), which are 50% B6. If the genetic background was a major factor in our analysis, we would have expected the two replicates with 50% B6 (ICM-2 and ICM-3) to be the most closely correlated of the three replicates, with ICM-1 being an outlier. We observed that not to be the case. Analysis of the correlation coefficients (CC) between the ICM datasets indicates that reducing the B6 contribution by half did not significantly bias the data: CC ICM-1/2 = 0.9840; CC ICM-1/3 = 0.9775; CC ICM-2/3 = 0.9799. B6C3H (B6 × C3H F1) females were super-ovulated between 5–8 weeks of age with 10 IU PMS (Calbiochem) followed by 10 IU HCG (Calbiochem) 46 h later and mated to B6C3H males overnight. Embryos were flushed at E3.5 with M2 (Sigma), washed with Dulbecco's modified Eagle's medium (DMEM)/10% FBS before being transferred to a droplet of rabbit anti-mouse serum (Sigma) under oil. Embryos were incubated in serum for 30 min at 37°C before being washed three times in M2. They were then transferred to a droplet of guinea pig serum (Sigma) under oil and incubated at 37°C for 30–60 min. Embryos were examined for trophectoderm lysis and washed three times in DMEM/FBS before being transferred into a droplet of 0.5% pronase (Sigma) at 37°C. When the zona pellucida started to disintegrate, embryos were pipetted vigorously to strip them of the zonas as well as lysed trophectoderm cells. ICMs were then washed in M2 before being transferred into RNA lysis buffer (RLT, Qiagen).
Isolation of primordial germ cells
Male mice of the Oct4/EGFP transgenic line (53), kept on a B6 background, were crossed to B6 females. The morning of the day of vaginal plug was considered E0.5. PGCs were isolated at E11.5 and E13.5. E11.5 PGCs are still sexually indifferent, so a mixture of male and female embryos was used. E13.5 PGCs have initiated sexual differentiation, and at this stage male and female embryos were processed separately. Embryo fragments were dissected and dissociated in the presence of 0.25% trypsin (Invitrogen) and 1 mg/ml DNAse (Worthington) in phosphate-buffered saline (PBS, Invitrogen) at 37°C for 10 min, with occasional vortexing and pipetting. Trypsinization was stopped with the addition of FBS to 5%. Cells were pelleted at 2000 rpm for 4 min, re-suspended in PBS with 1% fetal calf serum and 1 μg/ml propidum iodide (PI, Invitrogen) and passed through a 40 μm cell strainer. PGCs (PI−; EGFP+) and E11.5 SGM (PI−;EGFP−) fractions were purified in the UCSF Diabetes Center Cell Sorting Facility using a MoFlo cell sorter (Cytomation). Cell fractions were collected directly into RLT.
RNA amplification and microarray hybridization
We collected three to six replicates per cell type. The cell numbers used for the following cell types were:
| ICM | ESC | E11.5 PGC | EGC | E13.5F PGC | E13.5M PGC | E11.5 SGM | |
| Cell number | 50–75 #(no. of ICMs) | 500–15 000 | 1500–25 400 | 1000 | 2100–11 900 | 5400–7000 | 20 000–41 000 |
RNA was isolated using the RNeasy kit (Qiagen) with in-column DNAse digestion. mRNA was amplified using a two-round in vitro transcription protocol as described (56). Alternatively, mRNA was amplified using the RiboAmp HS kit (Arcturus), which is a modified two-round in vitro transcription protocol. Samples amplified using the RiboAmp HS kit were treated as a separate batch and, after batch effect calculation (see below), clustered correctly with replicates of the same tissues amplified using our protocol (56). For example, the mRNA for ES-1 and ES-2 were amplified using our protocol and had previously been reported (56), whereas ES-3 was amplified using the RiboAmp HS kit (Figs 1B and 2). Twelve micrograms of biotin-labeled amplified RNA was hybridized to Affymetrix U74Av2 arrays at the UCSF Gladstone Genomics Core Facility, according to the manufacturer's instructions. The raw data can be downloaded from GEO (GSE35416). Data on the transcriptional profiles of mouse adult stem cells have been previously described (56).
Statistical analyses
All the statistical analyses were performed using custom scripts and packages in the free statistical computing environment R/Bioconductor (www.r-project.org) (99). The probe intensities of the Affymetrix CEL files were background subtracted, quantile normalized and summarized for each probe set into a logarithm base 2 intensity value using the robust multi-array average method implemented in the Affymetrix package (100). Subsequently, we applied the mean center global adjustment to remove apparent batch effects across the experiments to facilitate between-experiment comparisons (101). Differential expression analysis between conditions were performed using the moderated t-statistics as computed by the limma package (102). P-values were adjusted to control for false discovery rate (FDR) using the Benjamini–Hochberg method (103) to account for multiple testing. PCA was done as described (104,105). The expression of genes highly differentially expressed between E11.5 PGCs and ICM, ES cells and EG cells (Supplementary Material, File S3 and Fig. 2B) was analyzed in human germ cell tumor data obtained from GEO dataset GDS1742 (58). Genes with human orthologs whose expression was assayed for in the microarrays used (58) were the following: PPRS1, KLF4, UPP1, ACSL4, TDGF1, FABP3, SGK, PGRMC1, DNMT3A, GSTA4, TTRAP and LATS2. P-values were calculated using a two-tailed t-test.
Quantitative real-time RT-PCR
Independent samples containing approximately the same number of ES cells, PGCs and ICM cells were obtained as described above. RNA was isolated using the RNeasy Mini RNA Isolation kit (Qiagen) and reverse-transcribed using the iScript first strand cDNA synthesis kit (BioRad) or the High-Capacity cDNA Reverse Transcription kit (Applied BioSystems). The cDNA reaction was diluted 1:5 in TE (10 mm Tris–Cl/1 mm EDTA, pH 7.6) and used in Sybr Green real-time PCR reactions (BioRad or Applied BioSystems). PCR primers were designed to amplify 100–200 bp fragments spanning exons. Housekeeping genes used were Ubiquitin-b and Ribosomal protein L7, which were determined from the microarray data to not be differentially expressed in the samples analyzed, or as indicated. Reactions were run in duplicates or triplicates on a MyiQ qPCR machine (BioRad) or a 7900HT machine (Applied BioSystems) according to the manufacturer's instructions. Only samples with single and matching end-point melting curve peaks were used for subsequent analysis. Cycle threshold values were imported into the REST software (106) for fold-change calculations of ES or PGC relative to ICM, using the housekeeping genes as controls, or as indicated. When a gene was detected in one tissue but not another, no fold change was calculated and instead the Present/Absent (P/A) notation was used. Primer sequences are listed in Supplementary Material, Table S2.
Immunostaining and western blotting
For immunofluorescence, cells were fixed directly in culturing plates with 4% paraformaldehyde or cold methanol, and permeabilized with 0.1% Triton X-100. Cells were then stained with primary antibodies against SSEA-3 (MAB4303, Millipore, 1:100), SSEA-4 (MAB4304, Millipore, 1:100), Tra1–60 (ab16288, Abcam, 1:100), Tra1-81 (MAB4381, Millipore, 1:100), V5 (46-0705, Invitrogen, 1:5000 for western blotting), TAZ (no.4883, Cell Signaling, 1:200; no.560235, BD, 1:1000 for western blotting), Topoisomerase I (ab85038, Abcam, 1:800 for western blotting), Bex1/Rex3 (Frank Margolis, U. Maryland, 1:20 000) and DMRT1 (Silvana Guioli, NIMR, London, 1:1000). Respective secondary antibodies were conjugated to either Alexa Fluor 594 or Alexa Fluor 488 (Invitrogen) and used at 1:500. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blotting using the same primary antibodies and respective secondary antibodies conjugated with horse radish peroxidase were performed according to standard protocols.
Lentivirus production
Lentiviral vectors that lead to the expression of shRNA were obtained from OpenBiosystems. shRNA for LATS2 (RHS3979–9569292, RHS3979–9569293, RHS3979–9569294) is in pLKO.1 backbone, and shRNA for YAP (RHS4430–98525388, RHS4430–98818907, RHS4430–98893379) and TAZ (RHS4430–98514207, RHS4430–99293240, RHS4430–101097950) is in pGIPZ. For virus production, 293T cells at 60–70% confluency were transfected in 10 cm plates with 4 μg of the lentiviral vectors together with 1 μg each of the packaging plasmids VSV-G, MDL-RRE and RSVr using Fugene 6 (Roche). After 72 h, viral supernatants were harvested, filtered and stored at −80°C.
Generation of human iPS cells
Human primary newborn foreskin (BJ) fibroblasts were obtained from ATCC (reference no.: CRL-2522) and cultured in DMEM with 10% FBS, 1× glutamine, 1× non-essential amino acids, 1× sodium pyruvate, 2× penicillin/streptomycin and 0.06 mm β-mercaptoethanol (fibroblast medium). Fibroblasts were seeded at 60 000 cells per well of a six-well plate the day before infection. Cells were infected with 0.5 μl each of concentrated retroviruses (obtained from the Harvard Gene Therapy Initiative) leading to the over-expression of OCT4, SOX2 and KLF4 and 0.05 μl in the case of c-MYC, alone or in combination with 20 μl (for LATS2) or 100 μl (for YAP and TAZ) of non-concentrated lentivirus for shRNA. Cells were infected in 1 ml human ES cell medium (DMEM/F12 with 20% KSR, 0.5× glutamine, 1× non-essential amino acids, 2× penicillin/streptomycin, 0.1 mm β-mercaptoethanol, 10 ng/ml bFGF) and 8 μg/ml polybrene. Cells remained in the presence of virus for 48 h and on the day after virus addition, 1 ml of fibroblast medium was added. Forty-eight hours after infection, virus was removed and cells were cultured in human ES cell medium. On d20–d28 after infection, live Tra1-81 (MAB4381, Millipore) staining was performed in order to identify fully reprogrammed iPS cell colonies.
Cytogenetic analysis
Human iPS cells were treated with 10 ng/ml of Colcemid (Invitrogen) overnight at 37°C. Cells were harvested and G-banded according to standard cytogenetic protocols (107). Metaphase cells were analyzed under the microscope and karyotyped according to an International System for Human Cytogenetic Nomenclature (108) using CytoVision system (Applied Imaging).
SUPPLEMENTARY MATERIAL
FUNDING
Work in M.H.'s laboratory is supported by grants from the JDRF and the Leona M. and Harry B. Helmsley Charitable Trust. Work in the Santos laboratory is supported by an NIH Director's New Innovator Award, the Leona M. and Harry B. Helmsley Charitable Trust and CIRM.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Hans Scholer for Oct4/GFP mice, Trish Labosky for EG cells, Frank Margolis and Silvana Guioli for antibodies, Kevin Eggan for pMXs-4F plasmids and Deepa Subramanyam for advice on human iPS cell generation. We thank the Santos laboratory and Matt Cook for discussions and Robert Blelloch, Marco Conti, Susan Fisher, Renee Reijo Pera, Diana Laird, Marica Grskovic, Connie Wong and Christina Chaivorapol for critical reading of the manuscript.
Conflict of Interest statement. The authors declare no competing financial interests.
REFERENCES
- 1.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 2.Smith A.G. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- 3.Stadtfeld M., Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2011;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiskinis E., Eggan K. Progress toward the clinical application of patient-specific pluripotent stem cells. J. Clin. Invest. 2010;120:51–59. doi: 10.1172/JCI40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 6.Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 8.Anokye-Danso F., Trivedi C.M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P.J., Epstein J.A., et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 11.Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B.E., Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 12.Maherali M., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 15.Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 16.Lowry W.E., Richter L., Yachechko R., Pyle A.D., Tchieu J., Sridharan R., Clark A.T., Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl Acad. Sci. USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanyam D., Lamouille S., Judson R.L., Liu J.Y., Bucay N., Derynck R., Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyoshi N., Ishii H., Nagano H., Haraguchi N., Dewi D.L., Kano Y., Nishikawa S., Tanemura M., Mimori K., Tanaka F., et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Brennand K.J., Simone A., Jou J., Gelboin-Burkhart C., Tran N., Sangar S., Li Y., Mu Y., Chen G., Yu D., et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howden S.E., Gore A., Li Z., Fung H.L., Nisler B.S., Nie J., Chen G., McIntosh B.E., Gulbranson D.R., Diol N.R., et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc. Natl Acad. Sci. USA. 2011;108:6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G.H., Barkho B.Z., Ruiz S., Diep D., Qu J., Yang S.L., Panopoulos A.D., Suzuki K., Kurian L., Walsh C., et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y., Yin X., Qin H., Zhu F., Liu H., Yang W., Zhang Q., Xiang C., Hou P., Song Z., et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Hanna J., Saha K., Pando B., van Zon J., Lengner C.J., Creyghton M.P., van Oudenaarden A., Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banito A., Rashid S.T., Acosta J.C., Li S., Pereira C.F., Geti I., Pinho S., Silva J.C., Azuara V., Walsh M., et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Utikal J., Polo J.M., Stadtfeld M., Maherali N., Kulalert W., Walsh R.M., Khalil A., Rheinwald J.G., Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marion R.M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Collado M., Villasante A., Strati K., Ortega S., Canamero M., Blasco M.A., Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A., Wahl G.M., Belmonte J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanna J., Markoulaki S., Schorderet P., Carey B.W., Beard C., Wernig M., Creyghton M.P., Steine E.J., Cassady J.P., Foreman R., et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikkelsen T.S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B.E., Jaenisch R., Lander E.S., Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramalho-Santos M. iPS cells: insights into basic biology. Cell. 2009;138:616–618. doi: 10.1016/j.cell.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsui Y., Zsebo K., Hogan B.L. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- 34.Resnick J.L., Bixler L.S., Cheng L., Donovan P.J. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- 35.Shamblott M.J., Axelman J., Wang S., Bugg E.M., Littlefield J.W., Donovan P.J., Blumenthal P.D., Huggins G.R., Gearhart J.D. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc. Natl Acad. Sci. USA. 1998;95:13726–13731. doi: 10.1073/pnas.95.23.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okamura D., Tokitake Y., Niwa H., Matsui Y. Requirement of Oct3/4 function for germ cell specification. Dev Biol. 2008;317:576–584. doi: 10.1016/j.ydbio.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Kehler J., Tolkunova E., Koschorz B., Pesce M., Gentile L., Boiani M., Lomeli H., Nagy A., McLaughlin K.J., Scholer H.R., et al. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 39.Donovan P.J. Growth factor regulation of mouse primordial germ cell development. Curr. Top Dev. Biol. 1994;29:189–225. doi: 10.1016/s0070-2153(08)60551-7. [DOI] [PubMed] [Google Scholar]
- 40.Stevens L.C. Origin of testicular teratomas from primordial germ cells in mice. J. Natl Cancer Inst. 1967;38:549–552. [PubMed] [Google Scholar]
- 41.Kurimoto K., Yabuta Y., Ohinata Y., Shigeta M., Yamanaka K., Saitou M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabour D., Arauzo-Bravo M.J., Hubner K., Ko K., Greber B., Gentile L., Stehling M., Scholer H.R. Identification of genes specific to mouse primordial germ cells through dynamic global gene expression. Hum. Mol. Genet. 2011;20:115–125. doi: 10.1093/hmg/ddq450. [DOI] [PubMed] [Google Scholar]
- 43.Mise N., Fuchikami T., Sugimoto M., Kobayakawa S., Ike F., Ogawa T., Tada T., Kanaya S., Noce T., Abe K. Differences and similarities in the developmental status of embryo-derived stem cells and primordial germ cells revealed by global expression profiling. Genes Cells. 2008;13:863–877. doi: 10.1111/j.1365-2443.2008.01211.x. [DOI] [PubMed] [Google Scholar]
- 44.Grskovic M., Chaivorapol C., Gaspar-Maia A., Li H., Ramalho-Santos M. Systematic identification of cis-regulatory sequences active in mouse and human embryonic stem cells. PLoS Genet. 2007;3:e145. doi: 10.1371/journal.pgen.0030145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharov A.A., Piao Y., Matoba R., Dudekula D.B., Qian Y., VanBuren V., Falco G., Martin P.R., Stagg C.A., Bassey U.C., et al. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 2003;1:E74. doi: 10.1371/journal.pbio.0000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharova L.V., Sharov A.A., Piao Y., Shaik N., Sullivan T., Stewart C.L., Hogan B.L., Ko M.S. Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev. Biol. 2007;307:446–459. doi: 10.1016/j.ydbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo G., Huss M., Tong G.Q., Wang C., Li Sun L., Clarke N.D., Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 48.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 49.Zhao B., Li L., Lei Q., Guan K.L. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan S.W., Lim C.J., Chen L., Chong Y.F., Huang C., Song H., Hong W. The Hippo pathway in biological control and cancer development. J. Cell Physiol. 2011;226:928–939. doi: 10.1002/jcp.22435. [DOI] [PubMed] [Google Scholar]
- 51.Solter D., Knowles B.B. Immunosurgery of mouse blastocyst. Proc. Natl Acad. Sci. USA. 1975;72:5099–5102. doi: 10.1073/pnas.72.12.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeom Y.I., Fuhrmann G., Ovitt C.E., Brehm A., Ohbo K., Gross M., Hubner K., Scholer H.R. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 53.Yoshimizu T., Sugiyama N., De Felice M., Yeom Y.I., Ohbo K., Masuko K., Obinata M., Abe K., Scholer H.R., Matsui Y. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 1999;41:675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- 54.Szabo P.E., Hubner K., Scholer H., Mann J.R. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 2002;115:157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- 55.Labosky P.A., Barlow D.P., Hogan B.L. Mouse embryonic germ (EG) cell lines: transmission through the germline and differences in the methylation imprint of insulin-like growth factor 2 receptor (Igf2r) gene compared with embryonic stem (ES) cell lines. Development. 1994;120:3197–3204. doi: 10.1242/dev.120.11.3197. [DOI] [PubMed] [Google Scholar]
- 56.Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R.C., Melton D.A. ‘Stemness’: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 57.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 58.Skotheim R.I., Lind G.E., Monni O., Nesland J.M., Abeler V.M., Fossa S.D., Duale N., Brunborg G., Kallioniemi O., Andrews P.W., et al. Differentiation of human embryonal carcinomas in vitro and in vivo reveals expression profiles relevant to normal development. Cancer Res. 2005;65:5588–5598. doi: 10.1158/0008-5472.CAN-05-0153. [DOI] [PubMed] [Google Scholar]
- 59.Kanzaki A., Takebayashi Y., Bando H., Eliason J.F., Watanabe Si S., Miyashita H., Fukumoto M., Toi M., Uchida T. Expression of uridine and thymidine phosphorylase genes in human breast carcinoma. Int. J. Cancer. 2002;97:631–635. doi: 10.1002/ijc.10105. [DOI] [PubMed] [Google Scholar]
- 60.Sahin F., Qiu W., Wilentz R.E., Iacobuzio-Donahue C.A., Grosmark A., Su G.H. RPL38, FOSL1, and UPP1 are predominantly expressed in the pancreatic ductal epithelium. Pancreas. 2005;30:158–167. doi: 10.1097/01.mpa.0000151581.45156.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyashita H., Takebayashi Y., Eliason J.F., Fujimori F., Nitta Y., Sato A., Morikawa H., Ohashi A., Motegi K., Fukumoto M., et al. Uridine phosphorylase is a potential prognostic factor in patients with oral squamous cell carcinoma. Cancer. 2002;94:2959–2966. doi: 10.1002/cncr.10568. [DOI] [PubMed] [Google Scholar]
- 62.Baylin S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2(Suppl. 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 63.Xu T., Wang W., Zhang S., Stewart R.A., Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 64.Visser S., Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 2010;9:3892–3903. doi: 10.4161/cc.9.19.13386. [DOI] [PubMed] [Google Scholar]
- 65.Voorhoeve P.M., le Sage C., Schrier M., Gillis A.J., Stoop H., Nagel R., Liu Y.P., van Duijse J., Drost J., Griekspoor A., et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 66.Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 67.McPherson J.P., Tamblyn L., Elia A., Migon E., Shehabeldin A., Matysiak-Zablocki E., Lemmers B., Salmena L., Hakem A., Fish J., et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 2004;23:3677–3688. doi: 10.1038/sj.emboj.7600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yabuta N., Okada N., Ito A., Hosomi T., Nishihara S., Sasayama Y., Fujimori A., Okuzaki D., Zhao H., Ikawa M., et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. J. Biol. Chem. 2007;282:19259–19271. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- 69.Li Y., Pei J., Xia H., Ke H., Wang H., Tao W. Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene. 2003;22:4398–4405. doi: 10.1038/sj.onc.1206603. [DOI] [PubMed] [Google Scholar]
- 70.Aylon Y., Michael D., Shmueli A., Yabuta N., Nojima H., Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lian I., Kim J., Okazawa H., Zhao J., Zhao B., Yu J., Chinnaiyan A., Israel M.A., Goldstein L.S., Abujarour R., et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varelas X., Sakuma R., Samavarchi-Tehrani P., Peerani R., Rao B.M., Dembowy J., Yaffe M.B., Zandstra P.W., Wrana J.L. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 73.Nichols J., Silva J., Roode M., Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nichols J., Smith A. The origin and identity of embryonic stem cells. Development. 2011;138:3–8. doi: 10.1242/dev.050831. [DOI] [PubMed] [Google Scholar]
- 75.Zwaka T.P., Thomson J.A. A germ cell origin of embryonic stem cells? Development. 2005;132:227–233. doi: 10.1242/dev.01586. [DOI] [PubMed] [Google Scholar]
- 76.Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J., et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hayashi K., Chuva de Sousa Lopes S.M., Kaneda M., Tang F., Hajkova P., Lao K., O'Carroll D., Das P.P., Tarakhovsky A., Miska E.A., et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kedde M., Strasser M.J., Boldajipour B., Oude Vrielink J.A., Slanchev K., le Sage C., Nagel R., Voorhoeve P.M., van Duijse J., Orom U.A., et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 79.Youngren K.K., Coveney D., Peng X., Bhattacharya C., Schmidt L.S., Nickerson M.L., Lamb B.T., Deng J.M., Behringer R.R., Capel B., et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao X., Pfaff S.L., Gage F.H. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Avruch J., Zhou D., Fitamant J., Bardeesy N. Mst1/2 signalling to Yap: gatekeeper for liver size and tumour development. Br. J. Cancer. 2011;104:24–32. doi: 10.1038/sj.bjc.6606011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Camargo F.D., Gokhale S., Johnnidis J.B., Fu D., Bell G.W., Jaenisch R., Brummelkamp T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 83.Hossain Z., Ali S.M., Ko H.L., Xu J., Ng C.P., Guo K., Qi Z., Ponniah S., Hong W., Hunziker W. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc. Natl Acad. Sci. USA. 2007;104:1631–1636. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Makita R., Uchijima Y., Nishiyama K., Amano T., Chen Q., Takeuchi T., Mitani A., Nagase T., Yatomi Y., Aburatani H., et al. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am. J. Physiol. Renal Physiol. 2008;294:F542–F553. doi: 10.1152/ajprenal.00201.2007. [DOI] [PubMed] [Google Scholar]
- 85.Tian Y., Kolb R., Hong J.H., Carroll J., Li D., You J., Bronson R., Yaffe M.B., Zhou J., Benjamin T. TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex. Mol. Cell Biol. 2007;27:6383–6395. doi: 10.1128/MCB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morin-Kensicki E.M., Boone B.N., Howell M., Stonebraker J.R., Teed J., Alb J.G., Magnuson T.R., O'Neal W., Milgram S.L. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol. Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R.O., Ogonuki N., et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 88.Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 89.Tamm C., Bower N., Anneren C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J. Cell Sci. 2011;124:1136–1144. doi: 10.1242/jcs.075796. [DOI] [PubMed] [Google Scholar]
- 90.Chia N.Y., Chan Y.S., Feng B., Lu X., Orlov Y.L., Moreau D., Kumar P., Yang L., Jiang J., Lau M.S., et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 91.Alarcon C., Zaromytidou A.I., Xi Q., Gao S., Yu J., Fujisawa S., Barlas A., Miller A.N., Manova-Todorova K., Macias M.J., et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Varelas X., Miller B.W., Sopko R., Song S., Gregorieff A., Fellouse F.A., Sakuma R., Pawson T., Hunziker W., McNeill H., et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev. Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 93.Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R.L., Martin J.F. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hyslop L.A., Armstrong L., Stojkovic M., Lako M. Human embryonic stem cells: biology and clinical implications. Expert Rev. Mol. Med. 2005;7:1–21. doi: 10.1017/S1462399405009804. [DOI] [PubMed] [Google Scholar]
- 95.Zhang H., Wang Z.Z. Mechanisms that mediate stem cell self-renewal and differentiation. J. Cell Biochem. 2008;103:709–718. doi: 10.1002/jcb.21460. [DOI] [PubMed] [Google Scholar]
- 96.Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A., et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 97.Joyner A. The practical approach series. In: Alexandra L.J., editor. Gene Targeting, a practical approach. Vol. 212. Oxford: Oxford University Press; 2000. p. 293. [Google Scholar]
- 98.Labosky P.A., Hogan B.L. Mouse primordial germ cells. Isolation and in vitro culture. Methods Mol. Biol. 1999;97:201–212. doi: 10.1385/1-59259-270-8:201. [DOI] [PubMed] [Google Scholar]
- 99.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 101.Johnson W.E., Rabinovic A., Li C. Adjusting batch effects in microarray expression data using Empirical Bayes methods. Biostatistics. 2006;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 102.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 103.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 104.Mardia K.V., Kent J.T., Bibby J.M. Multivariate analysis. London/New York: Academic Press; 1979. [Google Scholar]
- 105.Venables W.N., Ripley B.D., Venables W.N. Modern applied statistics with S. New York: Springer; 2002. [Google Scholar]
- 106.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rooney D.E. Human cytogenetics: constitutional analysis, a practical approach. New York: Oxford University Press; 2001. [Google Scholar]
- 108.Shaffer L.G., Slovak M.L., Campbell L.J. An international system for human cytogenetic nomenclature. Switzerland: Karger Publishers, Inc; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.