This work presents a technology for effectively silencing endogenous small RNAs by expressing a small tandem target mimic (STTM) composed of two noncleavable small RNA binding sites linked by an empirically determined spacer. Expression of STTM in Arabidopsis thaliana leads to the specific degradation of endogenous small RNAs by small RNA degrading nuclease family enzymes.
Abstract
MicroRNAs (miRNAs) and other endogenous small RNAs act as sequence-specific regulators of the genome, transcriptome, and proteome in eukaryotes. The interrogation of small RNA functions requires an effective, widely applicable method to specifically block small RNA function. Here, we report the development of a highly effective technology that targets specific endogenous miRNAs or small interfering RNAs for destruction in Arabidopsis thaliana. We show that the expression of a short tandem target mimic (STTM), which is composed of two short sequences mimicking small RNA target sites, separated by a linker of an empirically determined optimal size, leads to the degradation of targeted small RNAs by small RNA degrading nucleases. The efficacy of the technology was demonstrated by the strong and specific developmental defects triggered by STTMs targeting three miRNAs and an endogenous siRNA. In summary, we developed an effective approach for the destruction of endogenous small RNAs, thereby providing a powerful tool for functional genomics of small RNA molecules in plants and potentially animals.
INTRODUCTION
Small RNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs), play central roles in growth and development, epigenetics, genome integrity, defense against viral infection, and responses to environmental changes in plants (Brosnan and Voinnet, 2009; Ghildiyal and Zamore, 2009; Matzke et al., 2009; Simon and Meyers, 2010). miRNAs are especially important in controlling plant development by negatively regulating many transcription factor genes at the posttranscriptional level (Jones-Rhoades et al., 2006). siRNAs play a predominant role in RNA interference and in transcriptional gene silencing through DNA and histone methylation (Matzke et al., 2009). To date, hundreds of miRNAs and hundreds of thousands of endogenous siRNAs have been identified from dozens of plant species (Griffiths-Jones et al., 2008). With increasing efforts in small RNA discovery by high-throughput sequencing and the completion of whole-genome sequences of more plant species, there will be a growing need for functional genomics of small RNAs (Nobuta et al., 2010). Understanding the functions of individual small RNAs in vivo requires an effective means to block their production or activity.
Traditional approaches to the interrogation of gene function rely on the generation and characterization of genetic mutants (Till et al., 2003). Such genetic approaches are not easily applicable to small RNAs due to their small size and the fact that many miRNA families are composed of multiple members with potentially overlapping functions. A major, indirect approach to investigating miRNA functions relies on the generation and analysis of transgenic lines expressing miRNA-resistant targets containing silent mutations (Baker et al., 2005; Mallory et al., 2005). However, given that a miRNA usually regulates multiple target genes with either redundant or distinct functions (Bartel, 2009), this approach could only partially reveal the functions of a particular miRNA in vivo.
The ideal approach to exploring the functions of small RNAs is the simultaneous blockage of all the members of the small RNA family so as to reveal the effects of derepressing all the target genes through a single genetic transformation event. An endogenous regulatory mechanism that modulates miR399 activity, termed target mimicry (TM), has been uncovered in Arabidopsis thaliana (Franco-Zorrilla et al., 2007). miR399 is normally induced by phosphate starvation and targets the ubiquitin E2 conjugation enzyme UBC24 (PHO2), which in turn enhances phosphorus acquisition by activating specific phosphate transporters in plants (Franco-Zorrilla et al., 2007). Interestingly, phosphate starvation also induces the transcription of the non-protein-coding gene INDUCED BY PHOSPHATE STARVATION1 (IPS1). IPS1 transcripts are partially complementary to miR399, so that the miR399/IPS1 RNA duplex forms a central bulge that effectively blocks the cleavage of IPS1 RNA by miR399 (Franco-Zorrilla et al., 2007). Since IPS1 does not affect the abundance of miR399 but reduces its ability to regulate PHO2, it is thought that IPS1 inhibits the activity of miR399 by sequestering the miRNA through TM. This endogenous TM via IPS1 has been adapted to block the functions of several other miRNAs in Arabidopsis (Franco-Zorrilla et al., 2007; Wu et al., 2009). However, IPS1 functions mainly in sequestering miR399 rather than effectively destroying the small RNA, leaving a possibility of incomplete inactivation of the small RNA (Franco-Zorrilla et al., 2007; Todesco et al., 2010). Furthermore, a recent study examined the broad applicability of IPS1-based TM and found that only a small set of miRNAs was blocked by TM to give rise to phenotypes comparable to those caused by expressing individual miRNA-resistant target genes (Todesco et al., 2010); thus, exploring new approaches that complement the IPS1-based TM technology in the functional genomics of small RNAs is necessary.
Here, we report the development of a powerful technology that induces the degradation of specific small RNAs in vivo. This technology is based on the expression of a short tandem target mimic (STTM), which harbors two copies of small RNA partially complementary sequences linked by a short spacer. We show that STTM triggers efficient degradation of targeted small RNAs, and this degradation requires the activities of the SMALL RNA DEGRADING NUCLEASEs (SDNs). By testing parameters that contribute to STTM efficacy, we derived a general rule for optimal STTM design. By comparing STTM and IPS1-based TM, we show that STTM is more effective in reducing the levels of targeted small RNAs. STTM is a powerful approach for the interrogation of small RNA functions in vivo.
RESULTS
Design of an STTM for Functional Blockage of the miR165/166 Family
To test if shorter noncoding RNAs (sncRNA) (compared with IPS1) can effectively block the functions of endogenous small RNAs, we designed and expressed an artificial 108-nucleotide sncRNA, termed STTM, to block the functions of the miR165/166 family. We chose the miR165/166 family because it has a clearly defined set of target mRNAs, which encode class III homeodomain/Leu zipper (HD-ZIP III) transcription factors, including PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA (REV), ATHB8, and ATHB15 (Mallory et al., 2004; Reinhart et al., 2002; Zhong and Ye, 2007). Previous studies demonstrated that these genes are regulated by miR165/166 through target cleavage (Tang et al., 2003). Loss of miR165/166 regulation of these target mRNAs is predicted to produce clear phenotypes; for example, the gain-of-function allele phb-1d is a dominant mutation that disrupts the miR165/166 binding site in the PHB gene and produces a loss of apical dominance and changes in leaf symmetry (McConnell and Barton, 1998).
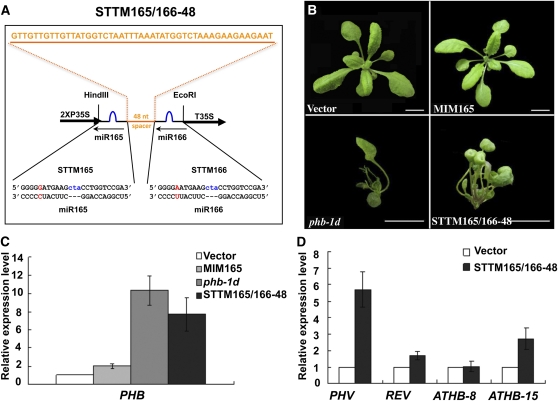
The STTM construct, which we named STTM165/166-48 (Figure 1A), contained two copies of imperfect miR165/166 binding sites (24 nucleotides), one for miR165 and the other for miR166, that were linked by a 48-nucleotide RNA spacer. To trap miR165/166 without being cleaved by it, STTM165/166-48 was designed to contain three additional nucleotides (CTA) so that a cleavage-preventive bulge could be formed between STTM165/166-48 and miR165/166 around positions 10 to 11 of the miRNA. STTM165/166-48 was constitutively expressed in transgenic Arabidopsis plants using a 2×35S promoter and a 35S terminator from Cauliflower mosaic virus.
Figure 1.
STTM165/166-48 Induced Dramatic Alterations in Arabidopsis Development.
(A) Diagram of STTM165/166-48 structure showing the design strategy. Orange indicates the spacer region and the spacer sequence. Blue indicates the bulge sequences in the miRNA binding sites. Red indicates the nucleotides that are different between miR165 and miR166. nt, nucleotides.
(B) Phenotypes of 3-week-old STTM165/166-48 transformants compared with vector control (Columbia-0), MIM165, and phb-1d plants. phb-1d is a dominant genetic mutant of the PHB (AT2G34710) gene in the Landsberg erecta background, and this mutation abolishes the binding of miR165/166 to PHB mRNA. Bars = 1.0 cm.
(C) qRT-PCR analysis of the target gene PHB in STTM165/166-48 transformants compared with vector control, MIM165, and phb-1d plants. Bars show se.
(D) qRT-PCR analysis of selected targets of miR165/166 in STTM165/166-48 transformants. Bars show se.
Compared with transgenic plants harboring miR165 target mimic (MIM165), an IPS1-based TM construct targeting miR165/166 (Todesco et al., 2010) (Figure 1B), STTM165/166-48 transgenic plants exhibited more severe, pleiotropic developmental defects, which were comparable to those displayed by the phb-1d mutant (Figure 1B). In addition to the loss of apical dominance, the transgenic plants displayed a typical loss of leaf asymmetry (Figure 1B), as observed in mutants in which one miR165/166 target, such as PHB, PHV, or REV, is resistant to miR165/166 (Emery et al., 2003; Mallory et al., 2004; McConnell et al., 2001). About 90% of more than 30 T1 STTM165/166-48 transgenic plants, selected for resistance to the herbicide BASTA, exhibited such phenotypes, and none of those transformed with the vector alone showed any phenotypes, indicating that these phenotypic changes were caused by STTM165/166-48. The phenotypes were stably inherited from generation to generation. These initial results suggest that the 108-nucleotide STTM165/166-48 can effectively inhibit miRNA165/166 activity when expressed in Arabidopsis.
To determine whether the STTM was able to block the function of miR165/166, we examined the levels of the miR165/166 target mRNAs. Consistent with our expectations, all five targets (PHB, PHV, REV, ATHB8, and ATHB15) were upregulated to different extents in STTM165/166-48 transgenic plants but not in the empty vector control transgenic plants (Figures 1C and 1D). We specifically compared PHB expression levels in vector control transgenic plants, MIM165, phb-1d, and STTM165/166-48, and found that PHB expression in STTM165/166-48 was a little lower than that in phb-1d but much higher than that in MIM165 (Figure 1C). In summary, the above data strongly support the conclusion that STTM165/166-48 inhibited the activities of miR165/166 and caused an increase in the levels of five HD-ZIP III transcription factor genes, leading to a series of developmental alterations. The effect of STTM165/166-48 is comparable to the gain-of-function phb-1d mutation, but much stronger than the IPS1-based MIM165 in both phenotypic severity and the upregulation of the miRNA target genes.
The Optimal Length and the Secondary Structure of the RNA Spacer between the Two miRNA Binding Sites
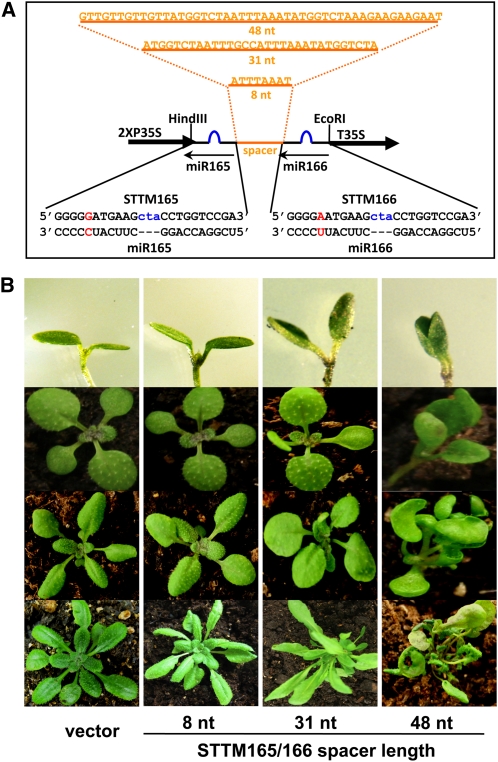
In animals, mRNAs targeted by miRNAs usually contain, within their 3′ untranslated regions, multiple miRNA binding sites that are bound by either one miRNA or different miRNA species (Bartel, 2009). The distance between the miRNA binding sites is important in let-7–mediated translational repression of lin-41 mRNA in Caenorhabditis elegans (Vella et al., 2004). To explore how the length of the spacer could affect the functional blockage of miR165/166 by STTM165/166, we designed spacers with different lengths (8, 31, to 48 nucleotides) between the two miR165/166 binding sites and introduced them into plants (Figure 2A). The severity of phenotypic alterations was used to score the transgenic plants for the effectiveness of the STTM.
Figure 2.
The Length of the RNA Spacer between the miR165 and miR166 Complementary Regions Is Crucial for STTM165/166 Function.
(A) Diagrams of the STTM structures with varying lengths of the spacer region. Orange indicates the spacer region and the spacer sequence. Blue indicates the bulge sequences in the miRNA binding sites. Red indicates the nucleotides that are different between miR165 and miR166.
(B) The phenotypes of representative STTM165/166 transgenic plants with different lengths of the RNA spacers at different developmental stages. Rows 1 to 4 represent plants at days 4, 8, 15, and 30, respectively. nt, nucleotides.
We mainly examined the morphology of cotyledons, true leaves, and overall plant architecture. After germination, the STTM165/166 transgenic plants with a 48-nucleotide spacer (STTM165/166-48) exhibited altered cotyledon morphology such that the cotyledons were spoon-shaped (Figure 2B). During later stages, the STTM165/166-48 transgenic plants always showed the strongest phenotypic alterations in the shape and polarity of the true leaves as well as overall plant architecture (Figure 2B). STTM165/166 plants with 31-nucleotide spacers (STTM165/166-31) exhibited less severe phenotypes than those of the STTM165/166-48 transgenic plants, while the transgenic plants with an 8-nucleotide spacer resembled plants transformed with the empty vector.
Given that STTM with a longer (48 nucleotide) spacer had better efficacy than that with a shorter (31 nucleotide) spacer, we further extended the 48-nucleotide spacer of STTM165/166-48 to 88 and 96 nucleotides (see Supplemental Figure 1A online) while retaining the stem structures (see Supplemental Figures 2D and 2E online) formed by the spacers between the two miRNA binding sites. Compared with STTM165/166-48, STTM165/166-88 had increased efficacy (see Supplemental Figure 1B online), but a further increase in the spacer length to 96 nucleotides did not improve the efficiency significantly (see Supplemental Figure 1B online).
One potential function for the spacers used for STTM might be in the stability of the expressed STTM transcripts. We thus performed a thermodynamic stability analysis of the potential RNA secondary structures for STTM165/166 RNAs with different lengths of spacers (see Supplemental Figure 2 online). As suggested by RNA folding analyses, the structure of STTM165/166-8 RNA was predicted to be rather unstable with a dG of −11.7 kcal/mol. STTM165/166 structures with longer spacers were predicted to be more stable with a dG lower than −20 kcal/mol. The lack of efficacy of STTM165/166-8 might be attributed to its low stability. However, the difference in effectiveness between STTM165/166-31 and STTM165/166-48 could not be simply explained by their thermostabilities because the dG values of STTM165/166-31 and -48 were even slightly opposite (see Supplemental Figure 2 online). Further analysis of the folded RNA structures showed that STTM165/166-48 formed a stable stem region that did not exist in STTM165/166-31, leaving the two STTM modules as dangling ends that perhaps would make it possible for miR165/166 to dock onto the STTM (see Supplemental Figure 2 online). The stem region of STTM165/166-48 may play a role in stabilizing the STTM transcripts and explain the difference in effectiveness between STTM165/166-31 and STTM165/166-48. Extending the spacer length from 48 to 88 or 96 nucleotides while retaining the stem structure increased the STTM thermostability and consequently the efficiency in the reduction of the target miRNAs (see Supplemental Figures 1 and 2 online).
Additionally, we randomly mutated the STTM165/166-48 in the spacer region without changing the spacer length to generate STTM165/166-48mut, in which the stem structure in the spacer was altered (see Supplemental Figure 3 online). Compared with STTM165/166-48, the proportion of STTM165/166-48mut transgenic plants with strong abnormal phenotypes was reduced approximately from 30 to 20%. Taken together, these analyses suggest that the thermostabilities and the secondary structures of STTM transcripts, especially the stem region formed by the RNA spacer between the two STTM modules, might contribute to STTM efficacy.
STTM-Directed Functional Blockage of miR165/166 Occurs through the Reduction of miR165/166 Levels
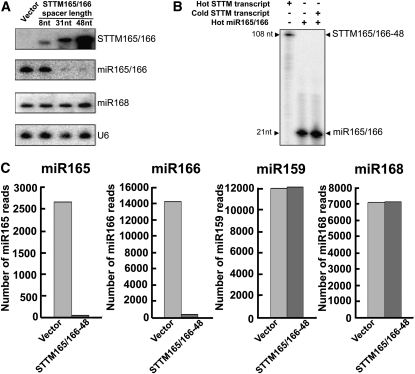
Recent studies indicated that TM also triggered reduction of the targeted miRNAs (Gu et al., 2010; Todesco et al., 2010). To investigate the specific mechanism and the effectiveness by which the STTM inhibited miR165/166 functions, we conducted miRNA gel blotting analysis to determine the levels of miR165/166. MIM165 plants, which were included as a control, showed a modest reduction in miR165/166 levels as observed before (see Supplemental Figure 4 online; Todesco et al., 2010). Consistent with the stronger developmental defects caused by STTM165/166-31 or -48, miR165/166 was almost undetectable from STTM165/166-31 and STTM165/166-48 transgenic plants (see Supplemental Figure 4 online). Plants transformed with vector alone or STTM165/166-8 had much higher levels of miR165/166 than STTM165/166-31 and STTM165/166-48 plants (Figure 3A). The reduction in miR165/166 levels by STTM165/166 was very specific: miR168 was not affected by STTM165/166 (Figure 3A). Further examination of miR165/166 target genes in the corresponding transgenic plants revealed that all five targets were upregulated significantly in STTM165/166-31 and STTM165/166-48 transgenic plants but not in the empty vector control or the STTM165/166-8 transgenic plants (see Supplemental Figure 5 online). The above data strongly support the conclusion that STTM165/166 triggered a reduction in miR165/166 levels and activated five HD-ZIP III transcription factors in the transgenic plants, leading to a series of developmental alterations.
Figure 3.
STTM165/166 Triggered Drastic Reduction of miR165/166 Levels.
(A) Representative RNA gel blotting to detect STTM165/166, miR165/166, and miR168. U6 served as an internal control. nt, nucleotides.
(B) STTM165/166-48 was unable to affect the migration of miR165/166 on a denaturing polyacrylamide gel.
(C) Copy numbers of small RNA reads in vector control and STTM165/166-48 by deep sequencing.
To determine whether the failure to detect miR165/166 by RNA gel blotting was due to the artifact of in vitro sequestration of miR165/166 by the 108-nucleotide STTM structure during electrophoresis, we generated the 108-nucleotide STTM165/166-48 RNA transcripts by in vitro RNA transcription and subjected them to in vitro miR165/166 binding. After ensuring full annealing between radiolabeled synthetic miR165/166 and nonradiolabeled STTM165/166-48 transcripts by examining the altered migration on a native gel, we resolved the annealing products on a denaturing polyacrylamide gel without a heat-denaturing step prior to sample loading. It was clear that the STTM165/166-48 RNA transcripts were unable to sequester the radiolabeled miR165/166 to the expected position (~108 nucleotides) or significantly alter the migration of miR165/166 in the denaturing gel (Figure 3B). Therefore, our results suggest that the observed reduction in miR165/166 levels by STTM165/166-48 was not likely due to the sequestration of miR165/166 during gel electrophoresis.
To further confirm the reduction of miR165/166 levels in vivo in STTM165/166-48 plants, we constructed small RNA libraries using the small RNA fraction isolated from the aerial parts of 3-week-old seedlings of vector control and STTM165/166-48. The abundance of small RNA reads of each sample was normalized to 9 million, and the read numbers of miR165, miR166, miR168, and miR159 from vector control and STTM165/166-48 were calculated (Figure 3C). Although the read numbers of both miR168 and miR159 were nearly at the same level between vector control and STTM-165/166-48, the reads of miR165 and miR166 were dramatically reduced in STTM165/166-48 compared with vector control. This further demonstrates that STTM triggers a drastic and highly specific reduction of the targeted small RNAs in vivo.
Contribution of Spacer Length and STTM Expression Level to STTM Efficacy.
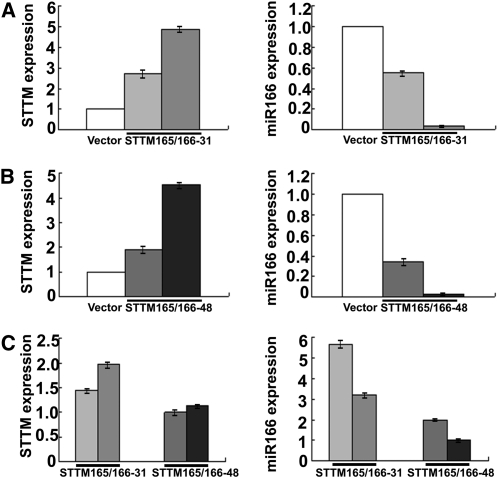
Both STTM165/166-31 and STTM165/166-48 independent transgenic lines displayed variable phenotypes. These phenotypic variations, likely due to positional or transgene copy effects, allowed us to examine the potential relationship among the phenotypic severity, the STTM transcript level, and the degree of reduction in miR166 levels in independent transgenic plants. For each population of the STTM165/166-31 and the STTM165/166-48 transgenic plants, we analyzed two types of independent transgenic lines with either weak or strong phenotypes. For either STTM165/166-31 or STTM165/166-48, much higher levels of STTM transcripts and significantly lower levels of miR166 were detected in transgenic plants displaying severe phenotypic defects than in transgenic plants displaying less severe phenotypes (Figures 4A and 4B). This indicates that for both 31-nucleotide-spacer and 48-nucleotide-spacer STTMs, the expression level of STTMs plays critical roles in the downregulation of the targeted miRNA. The higher the expression of the STTM, the more the reduction in miRNA levels and the more severe the phenotypic alterations.
Figure 4.
Comparisons of STTM and miR166 levels in STTM165/166 Transformants with Different Spacer Lengths.
(A) qRT-PCR analysis of STTM and miR166 levels in independent STTM165/166-31 transgenic plants. Bars show se.
(B) qRT-PCR analysis of STTM and miR166 levels in independent STTM165/166-48 transgenic plants. Bars show se.
(C) Comparison of STTM and miR166 levels between independent STTM165/166-31 transgenic plants and independent STTM165/166-48 transgenic plants.
Transgenic plants used in (A) to (C) were independent lines. Actin mRNA (for STTM) or SnoR101 (for miRNA) was used as an internal control. Values were obtained by normalizing to Actin or SnoR101 and then comparing the normalized values to those of control plants. Different shades of gray are used to indicate different independent lines to allow for the easy comparison between STTM and miR165/166 levels in the same lines. Bars show se. Note: The vector controls also contain 2XP35S and T35S (see Supplemental Figure 7 online), which bind the common qRT-PCR primers for the quantification of STTM expression and thus gave a value by qRT-PCR.
We further compared the efficacy of the 31-nucleotide-spacer and the 48-nucleotide-spacer STTMs in the reduction of miR166 levels. Although in some cases the STTM expression level in STTM165/166-31 transgenic plants was higher than that in STTM165/166-48 transgenic plants, STTM165/166-31 was not as effective as STTM165/166-48 in triggering the reduction of miR166 levels (Figure 4C). Together, these data demonstrated that both the expression level of STTM and the length of the linker inside the STTM are important in the downregulation of miRNAs in Arabidopsis.
STTM Efficacy Requires Both the Spacer and the Two miRNA Binding Sites
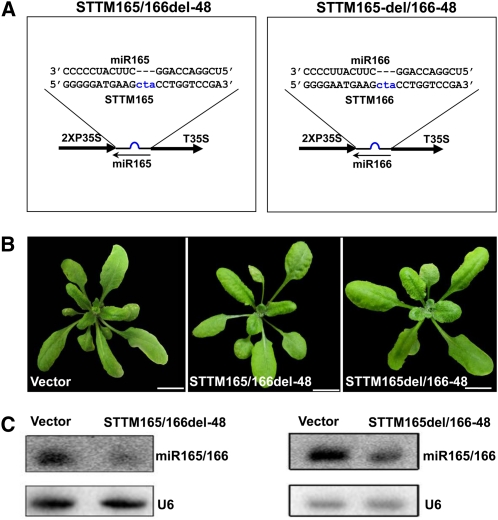
Compared with the IPS1-based TM, a featured distinction of STTM is that it contains one spacer and two target miRNA binding sites. To further evaluate the contribution of the spacer and the two binding sites to the efficiency of STTM, a set of STTMs was engineered to have different variations in their structures based on the STTM165del/166-48 structure. First, we examined the effects of STTM165del/166-48 and STTM165/166del-48 constructs (Figure 5A), in which we kept one binding site of miR165 or miR166 and deleted the spacer and the other binding site in the STTM165/166-48. Both kinds of transgenic plants showed reduced levels of miR165/166 (Figure 5C) but displayed no obvious phenotypic alterations (Figure 5B). Therefore, constructs containing one miRNA binding site were not as effective as STTMs in blocking the function of miRNAs, even when expressed by a 2X35S promoter.
Figure 5.
Comparison of the Efficacy of STTM Constructs with One miRNA Binding Site.
(A) Diagram of STTM with one miR165 binding site or one miR166 binding site. Blue indicates the bulge sequences in the miRNA binding sites.
(B) Phenotypes of transgenic plants containing STTM constructs with one miR165 binding site or one miR166 binding site. Bars = 1.0 cm.
(C) RNA gel blotting to determine the levels of miR165/166 in transgenic plants containing STTM with one miR165 binding site or one miR166 binding site. Total RNAs were prepared from multiple primary transformants.
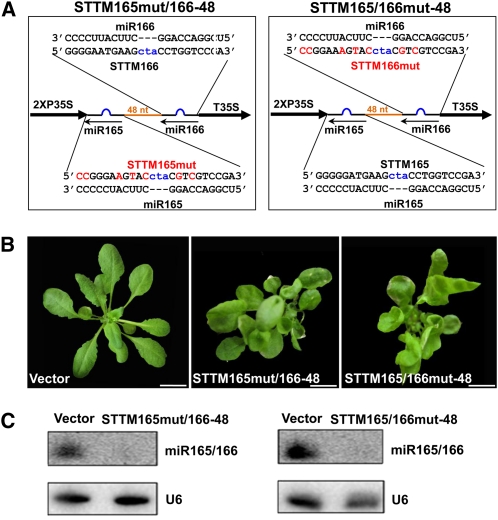
We generated two additional constructs, STTM165mut/166-48 and STTM165/166mut-48, by mutating one miRNA binding site of STTM165/166-48 without changing the overall length of the STTM and the spacer. In both cases, miR165/166 was expected to interact only with the nonmutated miRNA binding site (Figure 6A). In contrast with the STTM165del/166-48 and STTM165/166del-48, both STTM165mut/166-48 and STTM165/166mut-48 transgenic plants displayed obvious morphological defects (Figure 6B), but the phenotypes were not as severe as those observed in the STTM165/166-48 transgenic plants.
Figure 6.
Comparison of the Efficacy of STTM with One Mutated Binding Site.
(A) Diagram of STTM with mutated miR165 binding site or mutated miR166 binding site. Orange indicates the spacer region. Blue indicates the bulge sequences in the miRNA binding sites. Red indicates mutations in the miRNA binding sites.
(B) Phenotypes of transgenic plants containing STTM with mutated miR165 binding site or mutated miR166 binding site. Bars = 1.0 cm.
(C) RNA gel blotting to determine the levels of miR165/166 in transgenic plants containing STTM constructs with mutated miR165 binding site or mutated miR166 binding site. Total RNAs were prepared from multiple primary transformants.
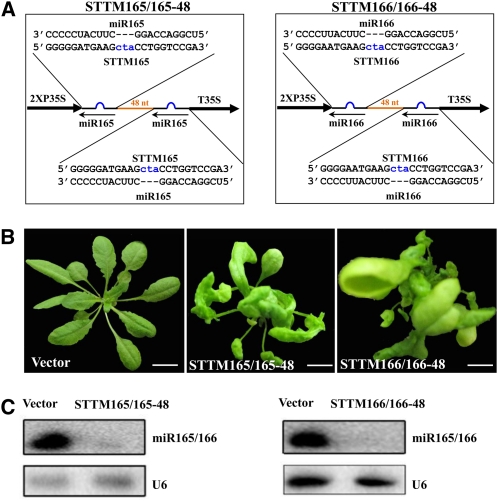
To gain more insights into the relationship between STTM structure and function, we analyzed the effects of two additional constructs: STTM165/165-48 and STTM166/166-48 (Figure 7A), which have two identical miR165 or miR166 binding sites rather than one binding to miR165 and the other to miR166. We found that these STTMs resulted in strong phenotypes with drastically decreased miR165/166 accumulation (Figures 7B and 7C), which resembled those of STTM165/166-48.
Figure 7.
Comparison of the Efficacy of STTMs with Two Identical Binding Sites.
(A) Diagram of STTMs with two miR165 binding sites or two miR166 binding sites. Orange indicates the spacer region. Blue indicates the bulge sequences in the miRNA binding sites.
(B) Phenotypes of transgenic plants containing STTMs with two miR165 binding sites or two miR166 binding sites. Bars = 1.0 cm.
(C) RNA gel blotting to determine the levels of miR165/166 in transgenic plants containing STTMs with two miR165 binding sites or two miR166 binding sites. Total RNAs were prepared from multiple primary transformants.
We further compared the efficacy of these different STTM constructs in the reduction of miR166 levels. We found that although the expression level of STTM in STTM165/165-48 or STTM165/166-48 transgenic plants is slightly lower than that in the STTM165/166del-48 or STTM165/166mut-48 transgenic plants in some cases, the extent of reduction of miR166 is higher (see Supplemental Figure 6 online).
In summary, among many STTM165/166del-48 or STTM165del/166-48 transgenic plants, none of them displayed obvious abnormal phenotypes. In the case of STTM165mut/166-48 or STTM165/166mut-48 transgenic plants, ~20% of the transformants displayed abnormal phenotypes, but the phenotypes were not comparable to the strong abnormal phenotypes of STTM165/165-48 or STTM165/166-48 transgenic plants. Overall, ~30% of STTM165/165-48 or STTM165/166-48 transformants displayed strong abnormal phenotypes. These data are in agreement with the expression levels of miR166, the corresponding levels of STTM transcripts, and the contributions from the spacer and the miRNA binding sites in each category of modified STTM constructs. By dissecting and comparing the effects of differently modified STTM constructs, we conclude that both the spacer and the two miRNA binding sites are necessary for the effectiveness of STTM in the functional blockage of small RNAs.
STTM-Directed Functional Blockage of Other miRNAs and Endogenous siRNAs
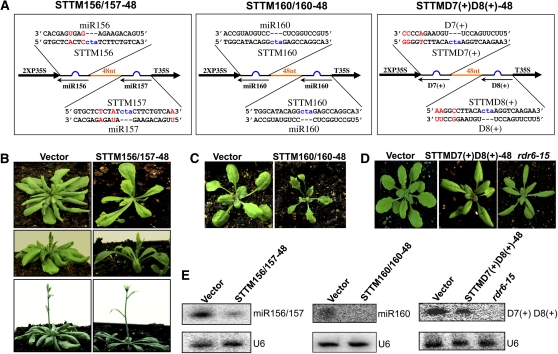
To evaluate the general applicability of STTM to the functional studies of miRNAs and siRNAs in plants, we used the STTM-48 structure to target two additional miRNA families, miR156/157 and miR160, and one family of endogenous trans-acting siRNAs (tasiRNAs), D7(+) and D8(+), in Arabidopsis (Figure 8A). Similar to the miR165/166 family composed of highly homologous but not identical miRNAs, miR156 and miR157 are different in only three nucleotides (Figure 8A) and both target the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factor genes, including SPL3, SPL4, and SPL5, which are involved in the promotion of vegetative phase transitions as well as flowering (Wu and Poethig, 2006). Functional blockage of miR156/157 is predicted to trigger an early vegetative phase change and early flowering. Indeed, STTM156/157-48 transgenic plants appeared delayed in leaf initiation and advanced in transition from juvenile to adult leaves following the production of the two abnormal, turbine-blade shaped cotyledons (Figure 8B). STTM156/157-48 transgenic plants showed early production of adult leaves that were characterized by marked serration and trichomes on both the abaxial and adaxial surfaces (Figure 8B). Flowering was accelerated in STTM156/157-48 plants as they bolted with six to seven leaves, while the empty vector control plants bolted after making over 10 true leaves (Figure 8B). To determine whether STTM156/157-48 similarly induced the reduction of miR156/157 levels, we performed miRNA gel blotting to determine miR156/157 levels. As expected, the abundance of miR156/157 was reduced by 90% in the STTM156/157-48 transgenic plants compared with the empty vector control plants (Figure 8E).
Figure 8.
STTM Is Effective in Triggering the Reduction of miRNAs and siRNAs.
(A) Diagram of STTMs containing miR156/157, miR160, or tasiRNAs (D7[+] and D8[+]) binding sites. Orange indicates the spacer region. Blue indicates the bulge sequences in the miRNA binding sites. Red indicates the nucleotides that are different between the two miRNAs or the two tasiRNAs.
(B) to (D) Phenotypes of STTM156/157, STTM160/160, and STTMD7(+)D8(+) transgenic plants.
(E) RNA gel blotting to determine the levels of miR156/157, miR160, or tasiRNAs (D7[+] and D8[+]) in STTM156/157, STTM160/160, or STTMD7(+)D8(+) transgenic plants. Total RNAs were prepared from multiple primary transformants.
We next used the STTM-48 structure to target miR160, which negatively regulates auxin response factor genes, ARF10, 16, and 17, and plays important roles in many stages of plant growth and development. The STTM160/160-48 transgenic plants exhibited hyponastic and serrated leaves (Figure 8C); these phenotypes were also observed in transgenic plants harboring miR160-resistant ARF10 (Liu et al., 2007). Compared with MIM160, STTM160/160-48 transgenic plants displayed deeper leaf serrations, and the derepression of the miR160 target gene, ARF17, was also higher (see Supplemental Figure 8 online), suggesting that STTM160/160-48 was more effective than MIM160. As expected, miR160 was decreased to an undetectable level in these transgenic plants (Figure 8E).
To assess the effectiveness of STTM against endogenous siRNAs, we sought to target the D7(+) and D8(+) tasiRNAs from tasiRNA precursor3 (TAS3) using STTM (Figure 8A). D7(+) and D8(+) are different only in three nucleotides at their 3′ ends, and both are generated through the tasiRNA pathway composed of ARGONAUTE7 (AGO7), RNA-DEPENDENT RNA POLYMERASE6 (RDR6), SUPPRESSOR OF GENE SILENCING3 (SGS3), and DICER-LIKE4 (DCL4). D7(+) and D8(+) target ARF2, 3, and 4 mRNAs to regulate developmental timing and patterning. Mutations in tasiRNA pathway components abrogate the production of D7(+) and D8(+) and result in precocious vegetative phase transitions such that juvenile leaves adopt characteristics of adult leaves, including a higher length/width ratio. Indeed, the leaf morphology of STTM D7(+)D8(+)-48 transgenic plants resembled that of the rdr6-15 mutant lacking the TAS3 tasiRNAs (Figure 8D). Small RNA gel blotting analysis revealed that D7(+) and D8(+) accumulated to a lower level compared with the empty vector control plants (Figure 8E).
Degradation of Small RNAs by SDNs Contributes to the Reduction of Small RNAs Triggered by STTM
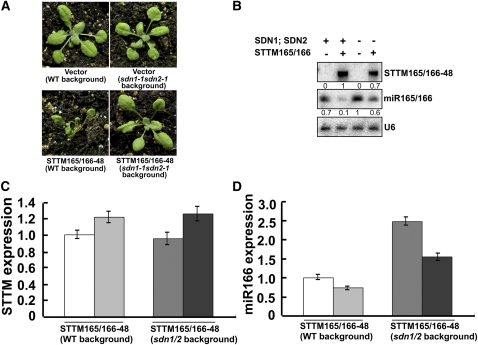
The SDN family of exonucleases was found to turnover small RNAs in vivo (Ramachandran and Chen, 2008). To test if the reduction of small RNAs triggered by STTM may be attributed to the degradation of small RNAs by SDNs, we sought in vivo genetic evidence in Arabidopsis. We generated STTM165/166-48 transgenic plants in the background of the sdn1-1 sdn2-1 double mutant. The sdn1-1 sdn2-1 double mutant exhibited no obvious morphological phenotypes (Ramachandran and Chen, 2008). The great majority (~95%) of the STTM165/166-48 transgenic plants in the sdn1sdn2 mutant background showed no phenotypes, and a small portion (~5%) showed mild phenotypic alterations in the cotyledons at an early developmental stage, whereas at later stages the plants developed normally (Figure 9A). By contrast, STTM165/166-48 transgenic plants in the wild-type background showed visible phenotypes at a high frequency (~90%). We analyzed the levels of STTM165/166-48 transcripts and miR165/166 from the STTM165/166-48 transgenic plants in the wild-type and sdn1-1 sdn2-1 backgrounds. Although the STTM165/166-48 transcripts were only slightly lower in the sdn1-1 sdn2-1 background than in the wild-type background (Figure 9B), the levels of miR165/166 differed significantly between these two genotypes. miR165/166 accumulated to higher levels in the transgenic plants in the sdn1-1 sdn2-1 background (Figure 9B).
Figure 9.
Degradation of miR165/166 by the SDNs Contributes to the Reduction of miR165/166 Triggered by STTM in Arabidopsis.
(A) Phenotypes of vector and STTM165/166-48 transgenic plants in the sdn1-1 sdn2-1 background (at right) compared with those in the wild type (WT) background (at left).
(B) RNA gel blotting to determine the levels of STTM165/166-48 RNA, miR165/166, and the U6 control RNA in the wild type (SDN1; SDN2 +) or sdn1-1 sdn2-1 (SDN1; SDN2 −) background. The numbers indicate the relative levels of the RNAs.
(C) Comparison of STTM expression in STTM165/166-48 transgenic plants in sdn1-1 sdn2-1 background and in wild-type background. Bars show se.
(D) Comparison of miR166 levels in STTM165/166-48 transgenic plants in sdn1-1 sdn2-1 background and in wild-type background. Bars show se. (C) and (D) Different shades of gray are used to indicate different independent lines to allow for the easy comparison between STTM and miR165/166 levels in the same lines. [See online article for color version of this figure.]
To exclude the possibility that the slightly lower level of STTM165/166-48 transcripts in sdn1-1 sdn2-1 may compromise the degradation of miR165/166 in the mutant plants, we identified two independent STTM165/166-48 transgenic plants in the sdn1-1 sdn2-1 background, in which the levels of STTM165/166-48 transcripts were comparable to those of two independent STTM165/166-48 transgenic plants in the wild-type background (Figure 9C). Using quantitative RT-PCR (qRT-PCR), we found that although the two kinds of transgenic plants had similar levels of STTM165/166-48 transcripts, the STTM165/166-48 transgenic plants in the sdn1-1 sdn2-1 background accumulated a higher level of miR166 than that of STTM165/166-48 transgenic plants in the wild-type background (Figure 9D). The derepressed expression of miR166 caused by sdn mutations in the STTM165/166-48 transgenic plants in sdn1-1 sdn2-1 background might account for the weakened phenotype. In summary, we found that mutations in SDN1 and SDN2 compromised the degradation of miR165/166 triggered by STTM165/166.
Similarly, we generated STTM156/157-48 transgenic plants in the sdn1-1 sdn2-1 background. We found that the proportion of abnormal transgenic plants was reduced from 30% in the wild type to 3.5% in the sdn1-1 sdn2-1 background. Based on the above observations, we conclude that STTM-triggered target miRNA reduction is at least partially through SDNs.
General Rules for STTM Design in Application
Based on this study, we recommend the following general rules for STTM design. First, the STTM should be designed to have two noncleavable miRNA binding sites linked by a spacer. The two noncleavable miRNA binding sites can be either identical, to target one specific miRNA, or slightly different to target two slightly different miRNA members of the same family. To reach a maximal effect, we do not recommend using the current STTM structure to target two distinct miRNAs from different families due to insufficient degradation of miRNAs by STTM with only one miRNA binding site. Second, the miRNA binding sites should have a CTA trinucleotide bulge corresponding to positions 10 and 11 from the 5′ end of the mature miRNAs, so that the STTM can effectively bind but will not be cleaved by the miRNAs. Some small RNAs may coincidently have the sequence TAG after the 10th nucleotide, which complements the introduced CTA in the STTM. This may result in a shift of the position of the introduced bulge and a subsequent cleavage of the STTM. In these rare cases, a different trinucleotide bulge should be used instead to prevent the cleavage of the STTM. Third, the optimal length of the spacer is from 48 to 88 nucleotides. The sequences of the spacer should be relatively AT rich and able to form a stable stem, such as those used in this study.
DISCUSSION
STTM as an Effective Approach for Functional Genomics of Small RNAs
In this study, we explored the use of sncRNAs, STTM, to block the functions of endogenous miRNAs and siRNAs, two major types of small RNAs in plants. Our results show that STTM is highly effective and specific in reducing the levels of targeted small RNAs in Arabidopsis. As such, this approach can be employed to explore the functions of small RNAs in planta. Although we only examined the effects of STTM in Arabidopsis in this study, STTM is also effective in other tested plant species, such as tobacco (Nicotiana tabacum) and rice (Oryza sativa). For plant species that are recalcitrant to transformation, it is possible that STTM can be transiently expressed through agroinfiltration or particle bombardment. Therefore, STTM may prove to be a widely applicable tool for the functional genomics of small RNAs.
The STTM approach has advantages over the IPS1-based approach in dissecting small RNA functions. First, STTM is more effective than IPS1 in that it induced marked phenotypic alterations (Figures 1B and 2B) in the case of STTM165/166-48. IPS1-based MIM165/166 Arabidopsis plants had only a mild phenotype (Todesco et al., 2010) that is far weaker than those of the STTM165/166-48 plants in this study and the gain-of-function HD-ZIP-III mutants published previously (Figures 1B and 2B). Additionally, lower expression of STTM165/166-48 than MIM165 (i.e., IPS1 backbone) could induce more reduction of miR165/166 (see Supplemental Figure 9 online). We noticed that STTM was particularly more successful than MIM in targeting miR165/166 for degradation (Figure 1; see Supplemental Figure 4 online). In the case of miR160, MIM was also very successful although STTM was slightly more effective (see Supplemental Figure 8 online). Thus, STTM is an effective approach that complements MIM in the functional dissection of small RNAs in plants.
Second, the STTM transgene is small, fewer than 100 nucleotides, and thus is easy to construct from synthetic oligonucleotides. Third, STTM induces small RNA degradation, a process that could not be reversed; thus, the blockage of small RNAs is effective and stable in transgenic plants. Fourth, because of its small size, multiple STTMs can be introduced together as concatamers to block multiple small RNAs simultaneously, with each kind of small RNA having two noncleavable binding sites. STTM is therefore useful for the functional dissection of small RNA networks and the interactions among small RNAs. We expect that STTM will be widely adopted in the functional dissection of small RNAs in plants and that the technology may be adapted for similar purposes in animals. In fact, two recent reports have shown that expression of RNAs with complementary target sites to specific miRNAs triggers the degradation of the miRNAs in animals (Ameres et al., 2010; Cazalla et al., 2010).
Transgenerational Stability of STTM
The successful application of STTM technology to the interrogation of miRNA functions requires its transgenerational stability. We followed the phenotypes of STTM165/166 transgenic plants for three generations. The phenotypes of STTM165/166-31 and -48 transgenic plants all remained unchanged and stable for all three generations, and STTM165/166-48 transgenic plants consistently displayed stronger phenotypes than STTM165/166-31 plants over the generations. The transgenerational stability indicates that STTM is a reliable and powerful tool for the study of small RNA functions.
Stability and Secondary Structure of STTM RNAs
The success of STTM should depend on the ability to stably express high levels of the noncoding STTM RNAs. We observed that highly expressed STTMs triggered more effective degradation of the targeted small RNAs, leading to greater derepression of the small RNA target gene expression (Figures 3A and 4). Not all noncoding transcripts are stable in cells. Because of their potential resemblance to nonsense codon-containing RNAs, cells are likely to consider them as aberrant RNAs and actively clear them away through the nonsense-mediated decay (NMD) pathway (Kurihara et al., 2009). We have been able to stably express STTMs by including two STTM submodules that pair with miRNAs and by varying the lengths of the spacer region between the two submodules. STTM with only one miR165/166 binding site was not effective enough in the blockage of miR165/166 (Figure 5). Two submodules with an 8-nucleotide spacer failed to effectively block miRNA functions (Figure 2B). A shorter (31 nucleotide) spacer was also less effective (Figure 2B). A spacer of ~48 to 88 nucleotides was optimal for STTM-mediated small RNA degradation. A spacer longer than 88 nucleotides did not dramatically improve the small RNA degradation efficiency further (see Supplemental Figure 1 online). A previous study showed that the stem region provides unusual stability to the IS10 antisense RNA and is critical for its function (Case et al., 1989). RNA folding analysis (see Supplemental Figure 2 online) shows that a stable stem region was formed by the RNA spacer in the cases of STTMs with 48-, 88-, and 96-nucleotide spacers, and the stem structure may provide stability to the STTM transcripts. The expression level of the STTM transcripts with different RNA spacers correlated overall with the ability to induce the degradation of the target small RNAs. Although the thermodynamic stability and the secondary structure of STTM transcripts may play a role in the expression level of STTM, other unidentified factors may also affect the stable expression of STTM transcript in plants. In addition to contributing to the stability of the STTM transcripts, the spacer may also affect the interaction between the STTM transcript and the targeted small RNA by influencing the structure of the STTM transcript, hence contributing to the efficiency of SDN-mediated degradation of the small RNA.
Potential Mode of Action of STTM
It cannot be absolutely excluded that STTM might partially function in sequestering the targeted small RNAs, but the mode of action of STTM is likely through the induction of degradation of small RNAs. This conclusion is based on several observations. First, four STTMs targeting four different small RNA families all lead to drastic and specific reduction of the steady state levels of the targeted small RNAs (Figures 3 and 8). Second, the levels of the STTM transcripts inversely correlate with the levels of the targeted small RNAs (Figures 3A and 4). Third, no reduction in miRNA primary precursors was observed for at least two miR156/157 family members, pri-miR156A and pri-miR156B (see Supplemental Figure 10 online), suggesting that STTM downregulates mature miRNAs but has no apparent negative effect on their precursors. Fourth, the effectiveness of STTM-165/166 is largely compromised in the sdn1-2 sdn2-1 double mutant, in which the levels of miR165/166 were increased (Figure 9).
These pieces of evidence point to a model whereby an STTM transcript induces SDN-based small RNA degradation through pairing with the small RNA. The underlying molecular mechanism of action is unknown but can be speculated based on structural insights into small RNA/AGO interactions. Structural studies on AGO in complex with a guide strand in the presence of a short or long complementary target strand reveal that the target strand induces conformational changes in AGO (Wang et al., 2008). While the 5′ and 3′ ends of a small RNA are anchored by different AGO domains in the absence of the target strand, the presence of a complementary target strand causes the release of the 3′ end of the small RNA from the PAZ domain. We predict that STTM causes the release of the 3′ end of the targeted small RNA from AGO1 to allow SDN1 to access the 3′ end of the small RNA for degradation. The requirement for single strandedness of the small RNA substrate by SDN1 implies that the 3′ end of the small RNA is not tightly bound by STTM. Perhaps the bulge in the middle or the structure of the STTM transcript allows the 3′ end of the targeted small RNA to exist, at least transiently, in an unpaired state to allow access by SDN1.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines used were in the Columbia-0 background except the phb-1d mutant, which was in the Landsberg erecta accession. Plants were grown in long days (16 h light/8 h dark) at 23°C. The sdn1-1 sdn2-1 mutant has been described (Ramachandran and Chen, 2008).
Plasmid Construction and Transgenes
All STTM modules, such as STTM165/166-8, STTM165/166-31, STTM165/166-48, STTM165/166-88, STTM165/166-96, STTM156/157-48, STTM160/160-48, and STTM D7(+)D8(+)-48, were engineered to be from 68 to 156 bp depending on the size of the spacer. The STTM modules were first inserted between the 2X35S promoter and the 35S terminator in a small-sized (~1.5 kb) pOT2 vector. This was done by PCR amplification of the vector with a proofreading Taq polymerase and a pair of long primers (~36 to 104 nucleotides; see Supplemental Table 1 online) that were designed to contain a SwaI site in each primer. The sequences of the two primers covered the entire STTM sequences to minimize errors in STTM regions during the PCR reaction. The PCR product that includes the pOT2 backbone (~3.6 kb) was purified and cleaved by SwaI, followed by purification and self-ligation. The ligated product was transformed into XL1-blue. Single colonies were propagated for plasmid isolation. The recombinant constructs were verified by linearization of the plasmids by SwaI, and the noncleavable template plasmids were abandoned. The recombinant plasmids (~3.6 kb) were further amplified by a pair of primers that contained PacI sites (see Supplemental Table 1 online) to delete the plasmid replication origin. The PCR products that contained the STTM and a chloramphenicol selection marker were introduced into a modified pFGC5941 binary vector through the unique PacI site. Recombinant binary plasmids were selected on Luria-Bertani plates containing the antibiotics chloramphenicol and kanamycin. The final constructs were verified by DNA sequencing before being used for plant transformation.
For STTM variants, such as STTM165del/166-48, STTM165/166del-48, STTM165mut/166-48, STTM165/166mut-48, and STTM165/166-48mut, mutated reverse or forward primers that covered the deletion or mutations were used to replace the primers used for the construction of nonmutated STTMs. The mutation of miR165 or miR166 binding sites to result in STTM165mut/166-48 and STTM165/166mut-48 (Figure 6A) followed the same strategy for the mutation of the miR399 binding site on IPS1 described previously (Franco-Zorrilla et al., 2007). All the primers used for plasmid construction are listed in Supplemental Table 1 online. Transgenic plants were generated by Agrobacterium tumefaciens–mediated floral dip transformation (Clough and Bent, 1998). Transgenic plants were selected by resistance to the herbicide BASTA.
Total RNA Isolation and Small RNA Gel Blot Analysis
Total RNA was isolated from leaves of pooled T1 or later-generation plants using Trizol (Invitrogen), and the total RNA was resolved by 15% PAGE under denaturing conditions (8 M urea). End-labeled, synthetic, 21-nucleotide RNA oligonucleotides were used as size standards. Blots were hybridized using end-labeled oligonucleotide probes complementary to the small RNAs (Haley et al., 2003; Tang and Zamore, 2004). The sequences of the small RNA probes are listed in Supplemental Table 2 online. The reverse primers, such as 165-166-STTMSwa8ntlink-PR, 156-157-STTMSwa48ntlink-PR, and D7D8-STTMSwa48ntlink-PR (see Supplemental Table 1 online), were used as probes in RNA gel blots to detect STTM RNAs from the transgenic plants.
mRNA Analysis by qRT-PCR
For examination of the mRNA levels of the genes of interest, total RNA was extracted using an RNeasy mini kit (Qiagen). Purified RNA was first treated with DNase I to remove any potential genomic DNA contamination and then used for RT with a High-Capacity cDNA Archive Kit (Applied Biosystems). qRT-PCR was performed with an Applied Biosystems step one instrument using the SYBR Green PCR master mix kit (Applied Biosystems) according to the manufacturer’s instructions. Actin mRNA was used as an internal control. The sequences of primers are listed in Supplemental Table 3 online. Values were obtained by normalizing to Actin and then comparing the normalized values to those of control plants. The relative levels of gene expression were calculated using the 2-ΔΔ cycle threshold method (Livak and Schmittgen, 2001). Three biological replicates were examined to ensure reproducibility.
miRNA qRT-PCR
RT and the qRT-PCR for the quantification of miR166 were performed according to TaqMan Small RNA Assays protocol (Applied Biosystems) The TaqMan Gene Expression Master Mix (Applied Biosystems) was used for qRT-PCR. Arabidopsis SnoR101 was used as an internal control. The sequences of primers are listed in Supplemental Table 3 online.
In Vitro Assay for miR165/166 Sequestration by STTM165/166-48
In vitro production of STTM165/166-48 RNAs was conducted by in vitro RNA transcription using T7 RNA polymerase and the DNA templates made by PCR with a pair of primers named T7-STTM165/166-48-PF and T7-STTM165/166-48-PR as well as a DNA oligo template named STTM-Spacer-48nt-template (see Supplemental Table 1 online). Transcription was conducted as described previously (Haley et al., 2003; Tang and Zamore, 2004). Synthetic miR165/166 was produced in Dharmacon RNAi Technologies at Thermo Fisher Scientific (see Supplemental Table 2 online). Hot miR165/166 and STTM165/166 were made by 5′ end labeling as previously described (Haley et al., 2003; Tang and Zamore, 2004). Annealing of STTM165/166-48 with miR165/166 was conducted in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) buffer first by heating the RNAs at 95°C for 5 min and then by cooling the mixture to room temperature for an additional 2 h of hybridization. The hybridized STTM-miRNA complex was first evaluated on native gel and then subjected to sequestering assay using 15% denatured PAGE. The sequestering results were evaluated using a phosphor imager.
Small RNA Deep Sequencing
RNA samples from the aerial parts of 3-week-old seedlings were isolated. Small RNAs were isolated from total RNAs following size selection using polyacrylamide/urea gel electrophoresis. Purified small RNAs were ligated to a 3′ Solexa DNA adaptor first. The 3′ ligated product was then purified and ligated to a 5′ Solexa RNA adaptor. The small RNA library was reverse transcribed and then amplified by PCR. Small RNA sequencing was conducted using Genome Analyzer II (Illumina) as previously described (Ghildiyal et al., 2008).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: miR165/166 (AT1G01183, AT4G00885, AT2G46685, AT3G61897, AT5G08712, AT5G08717, AT5G41905, AT5G43603, and AT5G63715), miR156/157 (AT2G19425, AT2G25095, AT4G30972, AT4G31877, AT5G10945, AT5G11977, AT5G26147, AT5G55835, AT1G48742, AT1G66783, AT1G66795, and AT3G18217), miR160 (AT2G39175, AT4G17788, and AT5G46845), miR168 (AT4G19395 and AT5G45307), tasiRNA D7(+) and D8(+) (JA643511 and JA643515), PHB (AT2G34710), PHV (AT1G30490), REV (AT5G60690), ATHB8 (AT4G32880), ATHB15 (AT1G52150), IPS1 (AT3G09922), ARF17 (AT1G77850), and ACTIN2 (AT3G18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Optimal Length of the Spacer between the Two Small RNA Binding Sites in STTM.
Supplemental Figure 2. The Secondary Structures and Thermodynamic Stabilities of Various STTM165/166s with Different Spacer Lengths (8, 31, 48, 88, and 96 Nucleotides).
Supplemental Figure 3. Mutation in the Spacer Region Changed the Stem Structure and Reduced the Efficacy in the Functional Blockage of miR165/166.
Supplemental Figure 4. Comparison of the Expression Levels of miR165/166 in STTM165/166 and MIM165 Transformants and Vector Control Plants.
Supplemental Figure 5. Comparison of the Expression Levels of miR165/166 Targets in STTM165/166 Transformants with Different Spacer Lengths.
Supplemental Figure 6. Comparison of the Expression Levels of STTM and miR166 in STTM165/166del-48, STTM165/166mut-48, STTM165/165-48, and STTM165/166-48 Transformants.
Supplemental Figure 7. Diagrams of the STTM-31/48 Plasmids and Their Vector Control as Well as the Real-Time Primer Locations.
Supplemental Figure 8. Comparison of the Phenotypes and the Expression Levels of the miR160 Target, ARF17, in Vector Control, MIM160, and STTM160/160-48.
Supplemental Figure 9. Comparison of the Expression Levels of MIM, STTM, and miR165/166 in MIM165 and STTM165/166-48 Transformants.
Supplemental Figure 10. The Expression Levels of miR156 Primary Transcripts (pri-miR156A and pri-miR156B) as Well as the Mature miR156 in STTM156/157-48.
Supplemental Table 1. Primers Used for Plasmid Construction in This Study.
Supplemental Table 2. Synthetic miRNAs and Small RNA Probes Used in This Study.
Supplemental Table 3. Real-Time PCR Primers Used in This Study.
Supplementary Material
Acknowledgments
This work was supported by the Kentucky Tobacco Research and Development Center and the USDA–National Research Initiative Grants 2006-35301-17115 and 2006-35100-17433, National Science Foundation grants (MCB-0718029: Subaward S-00000260 and IOS-1048216), an award from the Kentucky Science and Technology Corporation under Contract KSTC-144-401-08-029, startup funds from Michigan Tech University to G.T., and grants from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (K01 DK078648-01 and R03 DK084166-01) to X.T. Detlef Weigel and Zhixin Xie kindly provided the MIM165, MIM160, and rdr6-15 mutant, respectively. Chengjian Li and Phillip Zamore provided the training and guidance in small RNA library construction and the practice for small RNA deep sequencing at the University of Massachusetts Medical School. Yu Tian and Xuguo (Joe) Zhou performed the bioinformatic analysis of the small RNA deep sequencing data.
AUTHOR CONTRIBUTIONS
G.T. conceived and directed the project. G.T. and J.Y. designed the research strategies. J.Y. and Y.G. conducted the majority of experiments. X.J. conducted the in vitro assay for miR165/166 sequestration experiment. W.K. conducted some RNA gel blotting experiments. S.P. constructed some STTMs for their expression in Arabidopsis. X.T. conducted some data analysis and guided radioactive and real-time experiments. X.C. conceived the experiments with the sdn1 sdn2 double mutant. Y.G. did the framework and J.Y. conducted the deep work; thus, both contributed equally to this article. G.T., J.Y., and X.C. wrote the article. All authors contributed to writing the article.
References
- Ameres S.L., Horwich M.D., Hung J.H., Xu J., Ghildiyal M., Weng Z., Zamore P.D. (2010). Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.C., Sieber P., Wellmer F., Meyerowitz E.M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15: 303–315 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan C.A., Voinnet O. (2009). The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 21: 416–425 [DOI] [PubMed] [Google Scholar]
- Case C.C., Roels S.M., Jensen P.D., Lee J., Kleckner N., Simons R.W. (1989). The unusual stability of the IS10 anti-sense RNA is critical for its function and is determined by the structure of its stem-domain. EMBO J. 8: 4297–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D., Yario T., Steitz J.A. (2010). Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328: 1563–1566 Erratum. Science 329: 1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M., Seitz H., Horwich M.D., Li C., Du T., Lee S., Xu J., Kittler E.L., Zapp M.L., Weng Z., Zamore P.D. (2008). Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M., Zamore P.D. (2009). Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. (2008). miRBase: Tools for microRNA genomics. Nucleic Acids Res. 36(Database issue): D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Jia X., Tang X., Tang G. (2010). Small RNA destruction by a novel short target mimic in Arabidopsis thaliana. In RNA Silencing Mechanisms in Plants, 2010 Keystone Symposia. (Silverthorne, CO: Keystone Symposia on Molecular and Cellular Biology), p. 86
- Haley B., Tang G., Zamore P.D. (2003). In vitro analysis of RNA interference in Drosophila melanogaster. Methods 30: 330–336 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Kurihara Y., et al. (2009). Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 2453–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.P., Montgomery T.A., Fahlgren N., Kasschau K.D., Nonogaki H., Carrington J.C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52: 133–146 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Bartel D.P., Bartel B. (2005). MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17: 1360–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. (2004). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B., Matzke A.J. (2009). RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21: 367–376 [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Barton M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Nobuta K., McCormick K., Nakano M., Meyers B.C. (2010). Bioinformatics analysis of small RNAs in plants using next generation sequencing technologies. Methods Mol. Biol. 592: 89–106 [DOI] [PubMed] [Google Scholar]
- Ramachandran V., Chen X. (2008). Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321: 1490–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. (2002). MicroRNAs in plants. Genes Dev. 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S.A., Meyers B.C. (2010). Small RNA-mediated epigenetic modifications in plants. Curr. Opin. Plant Biol. 14: 148–155
- Tang G., Reinhart B.J., Bartel D.P., Zamore P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Zamore P.D. (2004). Biochemical dissection of RNA silencing in plants. Methods Mol. Biol. 257: 223–244 [DOI] [PubMed] [Google Scholar]
- Till B.J., et al. (2003). Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13: 524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella M.C., Reinert K., Slack F.J. (2004). Architecture of a validated microRNA:target interaction. Chem. Biol. 11: 1619–1623 [DOI] [PubMed] [Google Scholar]
- Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J. (2008). Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456: 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Poethig R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ye Z.H. (2007). Regulation of HD-ZIP III genes by microRNA 165. Plant Signal. Behav. 2: 351–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.