Laser microdissection combined with Solexa sequencing of cDNA was used to analyze the transcriptome of the shoot meristem during the floral transition. Activated genes were placed in pathways downstream or parallel to the inductive signal encoded by FLOWERING LOCUS T.
Abstract
Flowering of Arabidopsis thaliana is induced by exposure to long days (LDs). During this process, the shoot apical meristem is converted to an inflorescence meristem that forms flowers, and this transition is maintained even if plants are returned to short days (SDs). We show that exposure to five LDs is sufficient to commit the meristem of SD-grown plants to flower as if they were exposed to continuous LDs. The MADS box proteins SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) and FRUITFULL (FUL) play essential roles in this commitment process and in the induction of flowering downstream of the transmissible FLOWERING LOCUS T (FT) signal. We exploited laser microdissection and Solexa sequencing to identify 202 genes whose transcripts increase in the meristem during floral commitment. Expression of six of these transcripts was tested in different mutants, allowing them to be assigned to FT-dependent or FT-independent pathways. Most, but not all, of those dependent on FT and its paralog TWIN SISTER OF FT (TSF) also relied on SOC1 and FUL. However, this dependency on FT and TSF or SOC1 and FUL was often bypassed in the presence of the short vegetative phase mutation. FLOR1, which encodes a leucine-rich repeat protein, was induced in the early inflorescence meristem, and flor1 mutations delayed flowering. Our data contribute to the definition of LD-dependent pathways downstream and in parallel to FT.
INTRODUCTION
In annual plants, such as Arabidopsis thaliana, floral transition initiates reproduction and the end of vegetative development. The timing of this transition is crucial for reproductive success and is therefore stringently controlled by developmental and environmental response pathways. In Arabidopsis, six genetic pathways contribute to the regulation of flowering (Fornara et al., 2010). Here, we focus on the photoperiod pathway that strongly promotes flowering under long days (LDs), while under short days (SDs), flowering is delayed. Photoperiod is perceived in the leaves, whereas flowers develop at the shoot apical meristem.
FLOWERING LOCUS T (FT) and its close homolog TWIN SISTER OF FT (TSF) are transcribed in the vascular tissue in response to LDs (Kardailsky et al., 1999; Kobayashi et al., 1999; Yamaguchi et al., 2005; Jang et al., 2009). Activation of these genes in response to LDs requires the CONSTANS (CO) transcription factor, which is stabilized specifically under these conditions (Turck et al., 2008). FT and TSF are small proteins related to phosphatidyl ethanolamine binding proteins. FT, and probably TSF, is a mobile signal that can move from the leaf to the meristem where it activates the floral transition (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Tamaki et al., 2007). FT and TSF interact with FD, a bZIP transcription factor (Abe et al., 2005; Wigge et al., 2005). FD is strongly and broadly expressed in the shoot apical meristem before the floral transition, and mutations in FD reduce the early flowering phenotype of plants overexpressing FT. Comparison of gene expression patterns in wild-type, fd, and ft mutants identified genes that are proposed to be activated in response to FT arrival at the meristem (Schmid et al., 2003; Searle et al., 2006; Wang et al., 2009). SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), which encodes a MADS box transcription factor, is of particular interest as this is the earliest activated gene in the meristem that has so far been identified after transferring plants from SDs to LDs (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000). Early activation of SOC1 depends on FT and TSF and is delayed by mutations in FD (Searle et al., 2006). After activation, SOC1 plays an important role as a floral promoter at the meristem. FRUITFULL (FUL), another MADS box gene closely related to SOC1, is also activated in response to FT (Schmid et al., 2003; Teper-Bamnolker and Samach, 2005). Although loss-of-function mutations of this gene only weakly delay flowering (Ferrándiz et al., 2000), ful mutations enhance the late-flowering phenotype of soc1 mutants, and soc1-3 ful-2 double mutants are strongly attenuated in their response to FT overexpression from the viral 35S promoter (Melzer et al., 2008).
Several members of the SQUAMOSA PROMOTER BINDING LIKE (SPL) gene family, particularly SPL3, SPL4, SPL5, SPL9, and SPL15, also play important roles in the floral transition (Cardon et al., 1997; Schwarz et al., 2008; Wang et al., 2009; Yamaguchi et al., 2009). Most of these genes are increased in expression in the shoot apex in response to LDs based on Affymetrix microarray analysis (Schmid et al., 2003). Additionally, the temporal and spatial patterns of SPL3, SPL4, SPL5, and SPL9 mRNA expression have been described by in situ hybridization during or after the floral transition (Cardon et al., 1999, 1997; Wang et al., 2009). These SPL factors directly bind to and activate transcription of SOC1, APETALA1 (AP1), LEAFY (LFY), and FUL (Wang et al., 2009; Yamaguchi et al., 2009). The SPL genes proposed to have roles in floral transition belong to those negatively regulated by microRNA156 and microRNA157 (Schwab et al., 2005).
Repressors of flowering are also present in the meristem prior to floral transition. Of these, the MADS box proteins encoded by FLOWERING LOCUS C (FLC) and SHORT VEGETATIVE PHASE (SVP) strongly delay flowering (Michaels and Amasino, 1999; Sheldon et al., 1999; Hartmann et al., 2000). These proteins interact to form a heterodimer proposed to repress SOC1 transcription (Fujiwara et al., 2008; Li et al., 2008). However, genetic analysis demonstrated that flc svp double mutants are earlier flowering than either single mutant, suggesting that these proteins likely have distinct as well as overlapping functions (Fujiwara et al., 2008; Li et al., 2008). Both proteins act in the leaf and meristem to delay flowering by repressing FT and TSF or SOC1 transcription, respectively (Searle et al., 2006; Lee et al., 2007; Jang et al., 2009).
The process of floral induction ends with the initiation of flower development. Expression of AP1 defines this stage, and by the time AP1 is expressed, floral determination has occurred, so that plants continue to flower independently of environmental cues, such as light quality and daylength (Hempel et al., 1997). AP1 encodes a MADS box transcription factor that together with LFY specifies floral identity, and during the early stages of floral development, its expression is restricted to the floral primordium.
Present genetic data as well as temporal and spatial expression patterns are consistent with a largely linear process during the early stages of floral induction by photoperiod. According to this representation, FT/TSF acting through FD activates SOC1 and FUL transcription in the meristem, leading to downstream events, such as the activation of SPL genes. SPLs would then activate LFY and AP1 transcription. However, arguing against such a simple representation, the soc1-3 ful-2 double mutant retains a response to photoperiod; indeed, those plants are only moderately late flowering in LDs compared with wild-type plants and much earlier flowering than ft-10 tsf-1 double mutants (Melzer et al., 2008). Furthermore, downregulation of expression of several SPL genes by 35S:miR156 does not strongly delay flowering under LDs (Schwab et al., 2005). Such observations suggest that additional genes promote the floral transition in response to photoperiod through FT and TSF. Consistent with this proposal, expression profiling of apices during floral induction identified several hundred genes that changed in expression within 7 d of exposure to LDs and a few hundred prior to AP1 induction (Schmid et al., 2003). Such data suggest a complex transcriptional reprogramming of the apex during floral induction.
In this study, we employed a highly tissue-specific approach using laser microdissection to collect specifically shoot apical meristems of plants shifted from SDs to LDs and studied global gene expression changes in this material by sequencing cDNA. We identified ~200 genes that are upregulated in the shoot meristem prior to induction of AP1 and placed them in hierarchies relative to FT/TSF, SOC1/FUL, and SVP. Inactivation of one of the newly identified genes encoding a leucine-rich repeat (LRR) protein caused later flowering and enhanced the late flowering of soc1 ful double mutants.
RESULTS
Contribution of SOC1, FUL, and SVP to Floral Commitment in Response to Long Photoperiods
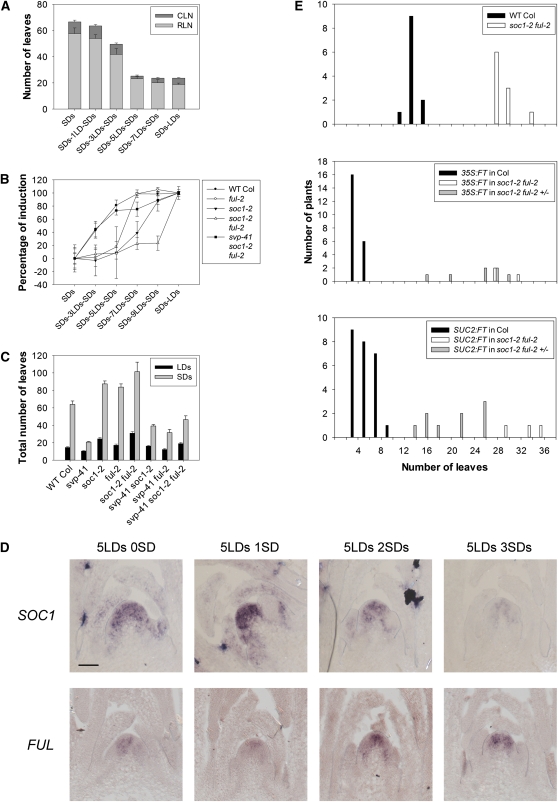
Arabidopsis plants are induced to flower by exposure to LDs and when exposed to sufficient LDs can maintain the floral transition even if returned to SDs, a process called commitment. To better characterize this process, the minimum number of LDs required to stably commit Arabidopsis Columbia (Col) to flowering was determined. Two-week-old SD-grown plants were transferred to LDs for various lengths of time and then returned to SDs. After exposure to five LDs, 80 to 100% of plants flowered with a similar number of leaves to plants transferred to continuous LDs (Figure 1A). Therefore, five LDs were sufficient to commit the plants to flower. In agreement with this conclusion AP1 mRNA was already expressed in floral primordia after exposure to five LDs (see Supplemental Figure 1A online). Exposure to only one LD did not influence flowering time, whereas three LDs accelerated flowering, but not to the same extent as plants exposed to continuous LDs.
Figure 1.
The Influence of SOC1 and FUL on the Commitment to Flower.
(A) Commitment of wild-type Col to flower after exposure to LDs. Flowering time expressed as leaf number. Plants were grown for 2 weeks in SDs and then transferred to different numbers of LDs, as indicated in the axis legend, before returning to SDs. CLN, cauline leaf number; RLN, rosette leaf number. Error bars represent sd. Sixteen plants were used to score flowering time under each condition.
(B) Commitment of soc1-2 ful-2 and svp-41 soc1-2 ful-2 as well as control genotypes after SD-grown plants were exposed to various numbers of LDs. Percentage of induction is calculated according to the formula indicated in Methods. Error bars represent sd. At least eight plants were used to score flowering time under each condition and genotype. WT, wild type.
(C) Flowering time of soc1-2 ful-2 and svp-41 soc1-2 ful-2 as well as control genotypes grown continuously under LDs (black columns) or SDs (gray columns). The number of leaves is total leaves (rosette plus cauline). Error bars represent sd. At least eight plants were used to score flowering time under each genotype.
(D) Analysis of SOC1 and FUL expression after transient exposure of SD-grown plants to LDs. Plants were grown for 2 weeks in SDs, transferred to LDs for 5 d, and transferred back to SDs as indicated above each panel. Bar = 50 μm.
(E) Effects of Pro35S:FT and ProSUC2:FT on flowering time in Col and soc1-2 ful-2 genotypes. Number of leaves is rosette leaf number in individual T1 plants. The ful-2 homozygote is difficult to transform so that soc1-2 homozygous ful-2 heterozygous plants were transformed. T1 plants are therefore all soc1-2 homozygous but segregate for the ful-2 mutation. Only soc1-2 ful-2 and soc1-2 ful-2/+ individuals are shown.
[See online article for color version of this figure.]
The effect of varying the duration of SD exposure prior to transfer to LDs was also tested. In addition to 2 weeks in SD conditions, plants were grown for 1, 3, or 4 weeks in SDs before transfer to LDs (see Supplemental Figure 1B online). Generally, plants grown for longer in SDs became more sensitive to LD exposure. Two weeks in SDs was chosen as the period of vegetative growth prior to transfer to LDs for all of the subsequent experiments because under these conditions, the floral transition takes place within a relatively extended time window of five LDs, allowing processes to be temporally separated at the meristem during the transition.
The MADS box proteins SOC1 and FUL are both required to maintain the transition to flowering when plants are exposed continuously to LDs (Melzer et al., 2008). To test the contributions of SOC1 and FUL to floral commitment, 2-week-old SD-grown soc1-2 and ful-2 mutants, as well as the soc1-2 ful-2 double mutant, were transiently exposed to LDs. The soc1-2 and ful-2 single mutants required nine and seven LDs, respectively, to become fully committed to flowering, and unlike wild-type plants were almost insensitive to shorter exposures of three and five LDs (Figure 1B). Therefore, soc1-2 and ful-2 mutants required significantly longer exposure to LDs than Col to become committed to flowering, although when grown continuously under LDs, the ful-2 mutant flowered only after forming two to three more leaves than Col (Figure 1C). The double mutant soc1-2 ful-2 required longer exposure to LDs than either single mutant to become committed, and even after 9 d exposure only 20% of plants were committed, supporting the idea that SOC1 and FUL are partially redundant (Figure 1B; see Supplemental Figure 1C online). Despite this dramatic increase in LD exposure required for commitment, soc1-2 ful-2 plants were not dramatically late flowering under continuous LDs, as previously reported (Melzer et al., 2008), and flowered after forming around 12 leaves more than the wild type (Figure 1C).
In response to LDs, FT and TSF expression in the leaves is necessary for activation of SOC1 and FUL transcription in the meristem. However, FT transcription is rapidly repressed after return of plants from LDs to SDs (Corbesier et al., 2007). Therefore, SOC1 and FUL might maintain the floral state because after transfer to SDs their mRNA expression persists in the meristem even after reduction of FT transcription in the leaves or because after exposure to several LDs, commitment becomes independent of SOC1 and FUL expression. To test these possibilities, in situ hybridizations were performed on apices of plants exposed to one, three, or five LDs and then transferred back to SDs (Figure 1D; see Supplemental Figure 2 online). Both SOC1 and FUL were reduced in expression on return to SDs when plants were exposed to three or fewer LDs (see Supplemental Figure 2 online), consistent with these plants not being committed to flower. SOC1 expression was also gradually reduced in the shoot apical meristem after return to SDs when plants were exposed to five LDs. By contrast, FUL expression was maintained under SDs after exposure to five LDs (Figure 1D). Consistent with the commitment step, AP1 mRNA was visible by in situ hybridization at the floral meristem after five LDs and remained expressed after the committed plants were moved to SDs (see Supplemental Figure 2 online). These results suggest that maintenance of high levels of SOC1 mRNA requires continued exposure to LDs and is not required for commitment to flowering. By contrast, FUL becomes independent of LDs once plants are committed to flower, and its expression does not subside after return to SDs, suggesting that mechanisms independent of FT expression or continued exposure to LDs maintain FUL expression.
Previously, SVP was shown to repress FT and TSF expression in the leaves and SOC1 expression at the meristem (Lee et al., 2007; Li et al., 2008; Jang et al., 2009). To better characterize the function of SVP in the meristem, svp-41 soc1-2 ful-2 triple mutants were constructed and compared with soc1-2 ful-2. When exposed continuously to LDs or SDs, the soc1-2 ful-2 double mutant flowered later than either single mutant, as described previously (Melzer et al., 2008), and was extremely late flowering under SDs. By contrast, the triple mutant svp-41 soc1-2 ful-2 flowered only slightly later than Col under LDs and much earlier than Col under SDs, although the triple mutant was later flowering than svp-41 mutants under both conditions (Figure 1C). In addition, soc1-2 ful-2 double mutants underwent reversion to vegetative growth after transition to flowering, as previously observed for soc1-3 ful-2 (Melzer et al., 2008), and the svp-41 mutation also partially suppressed this aspect of the phenotype. The svp-41 soc1-2 ful-2 triple mutant still formed an additional set of leaves growing on the secondary inflorescences that does not occur in wild-type plants (see Supplemental Figure 3 online), but did not produce the typical aerial rosette leaves on the senescent inflorescences of the soc1-2 ful-2 double mutant. Therefore, svp-41 can almost completely suppress the effect of soc1-2 ful-2 on flowering time and inflorescence development. These data suggest that additional targets of SVP at the apex can promote flowering in the absence of SOC1 and FUL. Alternatively, FT and TSF overexpression in leaves of the svp-41 mutant (Jang et al., 2009) might be responsible for activating floral promoters other than SOC1 and FUL.
These possibilities were first tested by exposing SD-grown soc1-2 ful-2 and svp-41 soc1-2 ful-2 plants to LDs and then returning them to SDs (Figure 1C; see Supplemental Figure 1C online). The svp-41 mutant grown for 2 weeks in SDs was fully committed to flower after exposure to one LD, while if grown for only 1 week in SDs was fully committed after three LDs and is therefore more sensitive to LDs than Col. Furthermore, the reduction of sensitivity to LDs in the soc1-2 ful-2 double mutant was almost completely overcome in the svp-41 soc1-2 ful-2 triple mutant. These results indicate that in addition to repressing SOC1, SVP likely represses genes that can bypass the effect of soc1-2 ful-2 on floral commitment and flowering.
The strong early flowering phenotype conferred by FT in a single Pro35S:FT transformant was suppressed in the soc1-3 ful-2 double mutant (Melzer et al., 2008), suggesting that increased expression of FT in svp-41 soc1-2 ful-2 plants is unlikely to be the cause of suppression of the soc1-2 ful-2 phenotype. To test this observation further in a wider range of Pro35S:FT transformants and in ProSUC2:FT plants, in which FT is overexpressed directly in the companion cells where FT is normally expressed, Pro35S:FT and ProSUC2:FT transgenes were introduced into soc1-2 ful-2 and Col plants by Agrobacterium tumefaciens–mediated transformation (see Methods). The effect of overexpressing FT from Pro35S:FT or ProSUC2:FT was strongly suppressed in soc1-2 ful-2 plants and did not result in a significant acceleration of flowering of the double mutants (Figure 1E). Therefore, the suppression of soc1-2 ful-2 by svp-41 is unlikely to be due to increased FT expression and suggests that SVP represses additional genes at the meristem that are capable of suppressing soc1-2 ful-2 when increased in expression.
Taken together, the transient exposure of plants to LDs reveals a role for SOC1 and FUL in committing the plant to flower in response to LDs, and this effect is markedly stronger than the delay in flowering observed under continuous LDs. SOC1 and FUL are functionally important downstream of FT, and LD induction of SOC1 is not maintained when plants are returned to SDs, suggesting that SOC1 is highly responsive to FT/TSF expression levels and that maintenance of a high level of SOC1 expression is not required for commitment. The requirement for SOC1 and FUL to commit plants to flowering can be overcome in an svp mutant background, suggesting that SVP represses further redundant genes.
RNA-Seq Analysis of Laser-Dissected Meristems to Identify Further Genes Expressed during the Floral Transition
The analysis of svp-41 soc1-2 ful-2 triple mutants suggested that additional genes are likely to be active in the meristem and that some of these can overcome the requirement for SOC1 and FUL during floral induction. To identify a wider range of genes increased in expression in the meristem during the transition to flowering, RNA-seq was performed on laser-dissected meristems. Use of this approach excluded young leaves that are present in hand-dissected apical samples and therefore further enriched for meristem-expressed mRNAs. Meristems were dissected from plants 8 h after dawn (Zeitgeber time [ZT] 8) after 2 weeks in SDs, and 1 and 3 d after transfer to LDs (see Supplemental Figure 4 online). This procedure was repeated in triplicate. RNA was extracted from these meristems, and the resulting cDNA from the nine samples was sequenced using Solexa/Illumina technology. The sequences obtained were then aligned to cDNAs of Arabidopsis using Megablast (see Methods). The number of sequence reads obtained in all nine samples that aligned to each Arabidopsis cDNA are shown in Supplemental Data Set 1 online.
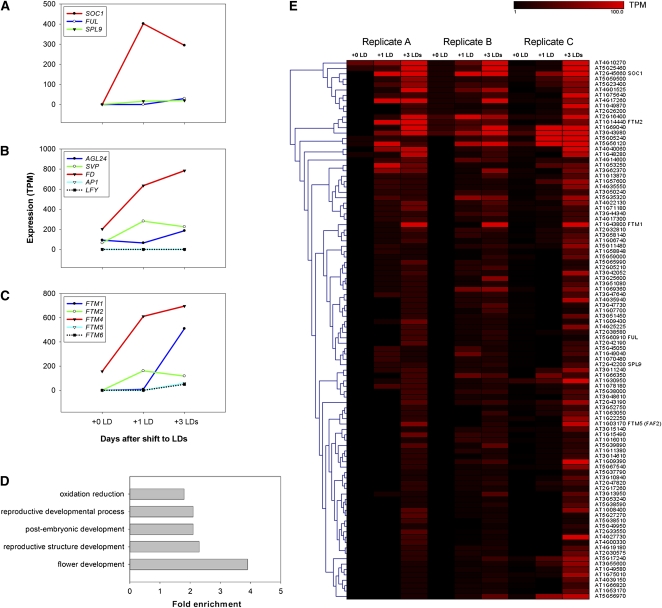
Genes that increased in expression during the floral transition after exposure to LDs were then identified from the sequence reads by screening for cDNAs to which more sequence reads aligned from the 2 weeks of SDs plus LDs sample than the 2 weeks of SDs sample (see Methods). The initial comparison was done using the 2 weeks of SDs plus three LDs sample because LD-induced genes were often more highly expressed as the exposure to LDs was extended. Only cDNAs that aligned to more sequence reads from the 2 weeks of SDs plus three LDs sample in all three experiments with a P value of ≤0.05 were initially considered. Although genes that exhibited reduced expression during this period were present in the reads, these were not studied in detail. In total, 202 upregulated genes were identified using the criteria described, and these genes are listed in Supplemental Data Set 2 online. The list included previously characterized flowering time genes that increase in expression in the meristem during floral transition, such as SOC1, FUL, and SPL9. The number of reads obtained for each of these genes at different time points is illustrated graphically in Figure 2A. No reads were detected for the AP1 or LFY genes in any of the samples, consistent with the idea that the mRNA was extracted early in the flowering process before these genes are expressed (Figure 2B). The mRNAs of other flowering genes known to be expressed in the shoot apical meristem, such as AGL24, SVP, and FD, were detected in the samples (Figure 2B). These genes are expressed in the SDs sample, as observed previously (Yu et al., 2002; Wigge et al., 2005; Jang et al., 2009) and therefore were not consistently detected as increased in abundance in the 2 weeks of SDs plus three LDs samples and thus do not appear in Supplemental Data Set 2 online.
Figure 2.
Transcriptomics of Shoot Apical Meristems Undergoing the Floral Transition.
(A) to (C) Analysis of expression of specific genes in the Solexa sequence reads (for replicate A). Expression level is indicated on the y axis as transcripts per million (TPM) reads. Time points are represented on the x axis: 2 weeks in SDs (+0 LDs); 2 weeks in SDs plus 1 LDs (+1 LDs); 2 weeks in SDs plus 3 LDs (+ 3 LDs).
(A) Known flowering time genes not expressed in the meristem prior to LD exposure.
(B) Floral meristem identity genes and flowering time genes (FD and AGL24) expressed prior to exposure to LDs.
(C) FTM genes identified and studied here in detail. Data for FTM3 are shown in Supplemental Figure 5 online.
(D) GO terms of classes of genes enriched among the 202 upregulated genes shown in Supplemental Data Set 3 online. These classes are enriched in the 202 genes compared with the whole genome.
(E) Heat map showing a clustering of genes among the 202 upregulated genes. FTM genes and flowering time genes are indicated on the right side along with all gene codes.
A Gene Ontology (GO) enrichment analysis was performed on the 202 upregulated genes compared with the whole genome (Figure 2D; see Supplemental Data Set 3 online; see Methods). Some terms expected to be enriched were indeed overrepresented, such as genes involved in postembryonic development, reproductive development, and flower development. In addition, enzymes involved in oxidation/reduction reactions were also unexpectedly overrepresented.
The Genesis program was used to cluster and identify coregulated genes among the 202 candidate genes proposed to be upregulated in the meristem during the floral transition (see Methods; Figure 2E; see Supplemental Figure 5 online). This approach identified classes of genes for further analysis, some of which are studied in detail in the following section.
Spatial and Temporal Patterns of Expression of Selected Genes during the Transition to Flowering
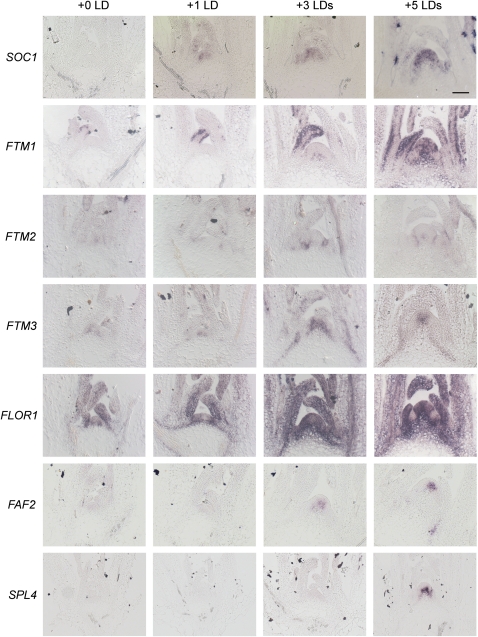
The temporal and spatial patterns of expression of a subset of the candidate genes were characterized in detail by in situ hybridization. These genes increased in expression during the time course and were initially named FLORAL TRANSITION AT THE MERISTEM1 (FTM1) to FTM6 (Figure 2C; see Supplemental Figure 6A online). Three of the candidate genes were chosen because they belonged to different groups in the cluster analysis (FTM1, FTM2, and FTM5 [FAF2]; illustrated in Figure 2E and described in Table 1). In addition, FTM4 was chosen because, although it was excluded by the stringent criteria applied to assemble Supplemental Data Set 2 online, it appeared to be expressed in the meristem of plants exposed to three LDs, was clearly upregulated in two experiments, and little data on its expression during floral induction is available because it is not present on the ATH1 Affymetrix array. FTM6 was upregulated after three LDs in the Illumina sequence data. Also, because it is a member of the SPL family of transcription factor–encoding genes that are important in the floral transition, it was important to define its temporal expression pattern during floral induction. Finally, FTM3 was included because it is upregulated in the sequence data, but it was not included in Supplemental Data Set 2 online because its complex annotation led to an ambiguous assignment of reads to transcripts and therefore prevented its detection in the Megablast pipeline (see Methods; see Supplemental Methods 1, Supplemental References 1, Supplemental Data Set 4, and Supplemental Figure 6B online). These six genes also encode proteins with diverse biochemical functions, including enzymes (FTM1), transcription factors (FTM2, FTM3, and FTM6), and novel proteins of unknown functions (FTM4 and FTM5). Some of these genes have been studied previously (see below), but their temporal and spatial expression patterns in the early stages of floral transition have not been described. In situ hybridizations were performed on the meristems of wild-type plants after growth for 2 weeks under SDs and then 1, 3, and 5 d after transfer to LDs (Figure 3). SOC1 was used as a control.
Table 1.
Summary of Genes Identified as Being Upregulated and Examined by in Situ Hybridization in Different Genetic Backgrounds
| At Gene No. | Gene Isolation Nos. | Other Names | Predicted Gene Product |
| AT1G43800 | FTM1 | S-ACP-DES6 | Stearoyl-ACP-desaturase |
| AT1G14440 | FTM2 | ATHB31 | Zinc finger-homeodomain protein. |
| AT2G18160 | FTM3 | GBF5, BZIP2 | bZIP transcription factor |
| AT3G12145 | FTM4 | FLOR1 | LRR protein |
| AT1G03170 | FTM5 | FAF2 | Unknown protein |
| AT1G53160 | FTM6 | SPL4 | SQUAMOSA PROMOTER BINDING LIKE transcription factor |
Figure 3.
Spatial Patterns of Expression in the Shoot Apex of Candidate Genes during Floral Transition Induced by Transfer from SDs to LDs.
In situ hybridization on apices of wild-type Col grown for 2 weeks in SDs (0 LD) and then transferred to LDs for one LD, three LDs, or five LDs. The genes used as probe are shown on the left of each time course. Samples were harvested at ZT8. SOC1 was used as a control. The remaining six genes were extracted from the Solexa reads, as described in the text. Bar = 50 μm.
[See online article for color version of this figure.]
SOC1 (Figure 3) was not expressed after 2 weeks in SDs, and expression of this gene increased progressively during exposure from one to five LDs, as described previously (Borner et al., 2000; Samach et al., 2000; Searle et al., 2006).
FTM1 encodes the enzyme stearoyl-ACP desaturase, and expression of this member of the gene family could not be detected previously by RT-PCR (Kachroo et al., 2007). However, FTM1 mRNA is present in the epidermis of young leaves of SD-grown plants by in situ hybridization (Figure 3). As the floral transition proceeds, FTM1 mRNA also becomes expressed in the apex, in the region of the rib meristem, of plants exposed to three and five LDs.
FTM2 encodes the zinc finger homeodomain transcription factor ATHB31 (Tan and Irish, 2006). The mRNA of FTM2 was not detected in the meristem prior to exposure to LDs, but after exposure to three LDs, it was present on the flanks of the meristem adjacent to floral primordia (Figure 3). The mRNA persisted in a similar pattern after five LDs.
FTM3 encodes the bZIP transcription factor AtbZIP2, related to ATB2, which is involved in Suc signaling (Rook et al., 1998; Jakoby et al., 2002). The mRNA was expressed in SD apices, specifically in a region in the center of the SAM; however, after three LDs, its abundance increased strongly. After five LDs, FTM3 mRNA was also detected in regions extending into the vascular tissue adjacent to the shoot apical meristem.
FTM4 is FLOR1, which encodes an intracellular LRR protein described to interact with the MADS box transcription factor AGAMOUS (Gamboa et al., 2001). It was weakly expressed prior to floral induction but increased strongly in expression in the meristem after exposure to three LDs, as the meristem changes shape, becoming more domed (Figure 3). After five LDs, this gene was strongly expressed in the meristem but appeared to be excluded from the developing floral primordia (see also below).
FTM5 is FANTASTIC FOUR2 (FAF2), which encodes a protein of unknown function and was previously shown to be increased in expression during the floral transition by analysis of Affymetrix array data (Wahl et al., 2010), although its spatial expression was not followed through an inductive time course as shown in Figure 3. FAF2 mRNA was only faintly detected in the center of the meristem in 2-week-old SD-grown plants but increased in the meristem after exposure to three and five LDs.
FTM6 is SPL4 and its mRNA was not detected prior to LD induction. However, it was detected by in situ hybridization after exposure to five LDs in the rib meristem, in a distinct spatial pattern to that described at a similar stage for SPL3 and SPL9 (Wang et al., 2009). This pattern of SPL4 mRNA expression likely persists after floral induction into the mature inflorescence meristem (Cardon et al., 1999).
These data demonstrate that the mRNAs of genes identified in the expression profiling are increased in abundance in the meristem early during the floral transition, within three to five LDs after transfer from SDs and that they accumulate in different spatial patterns within the meristem and shoot apex. Once expressed, their mRNAs persist throughout the period that the meristem undergoes commitment to flowering.
Roles of Known Floral Regulators in Induction of FTM Genes during the Floral Transition
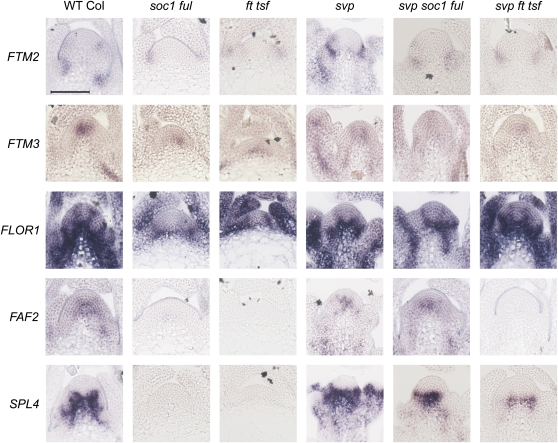
Several mutations that impair early changes in gene expression at the shoot apical meristem during photoperiodic floral induction have been characterized (Turck et al., 2008). To place the induced genes within this regulatory hierarchy, in situ hybridizations were performed for the subset of induced genes described above on apices of ft-10 tsf-1, soc1-2 ful-2, svp-41, svp-41 ft-10 tsf-1, and svp-41 soc1-2 ful-2 plants transferred from SDs to LDs. The results are shown in Figure 4 for plants exposed to five LDs, and the complete time course of in situ hybridizations is shown in Supplemental Figure 7 online.
Figure 4.
Expression of Candidate Genes in the Shoot Apices of Mutants Impaired in Various Genes of the Photoperiodic Pathway after Exposure to Five LDs.
In situ hybridization on apices of wild-type Col and illustrated mutant genotypes grown for 2 weeks in SDs and then transferred to five LDs. The genes used as probes are shown on the left. The entire time course after transfer is shown in Supplemental Figure 7 online. The svp mutant flowers early under SDs; therefore, these plants were grown for only 10 SDs before transfer. Samples were harvested at ZT8. FTM1 is not shown as its expression did not change significantly in these genotypes, and it is shown in Figure 5. Bar = 50 μm.
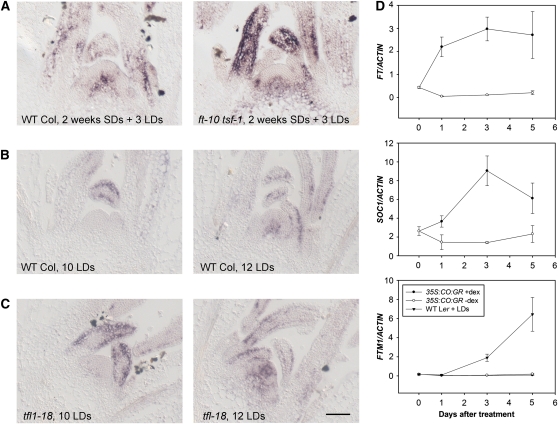
Flowering of the ft-10 tsf-1 double mutant is insensitive to daylength due to loss of photoperiodic signaling between the leaf and the meristem; therefore, in this double mutant, the expression of many genes induced in the meristem on transfer to LDs is expected to be impaired (Yamaguchi et al., 2005; Jang et al., 2009). As previously shown, SOC1 expression is delayed in ft-10 tsf-1 double mutants (Jang et al., 2009), and it is not induced in those mutants after transferring plants to five LDs (see Supplemental Figure 8 online). Similarly, FLOR1, FTM2, FAF2, and SPL4 were not induced in ft-10 tsf-1 double mutants after exposure to five LDs (Figure 4). These experiments confirm that FT and TSF ultimately regulate the expression of a wide range of genes expressed in diverse spatial patterns in the meristem in response to photoperiod, as previously proposed based on Affymetrix array analysis of apical RNA samples extracted from ft mutants (Schmid et al., 2003). However, surprisingly, some genes were induced in the meristem in response to photoperiod even in the absence of FT and TSF. In ft-10 tsf-1 double mutants, an increase in expression of FTM3 still occurs in the meristem and vascular tissue after transfer to five LDs, although this increase is reduced compared with Col (Figure 4). Similarly, FTM1 is induced in the center of the meristem of ft-10 tsf-1 double mutants around three LDs after transfer in a similar way as it is in Col plants (Figure 5A). These data demonstrate that some genes are induced in the meristem in response to LDs at least partly independently of FT and TSF.
Figure 5.
Relationship between Induction of FTM1 and Activity of FT.
(A) In situ hybridization analysis of FTM1 expression in apices of Col (left panel) and ft-10 tsf-1 (right panel) grown for 2 weeks in SDs and then induced for three LDs. WT, wild type.
(B) In situ hybridization analysis of FTM1 in apices of Col grown from germination under LDs for 10 (left panel) or 12 (right panel) d.
(C) In situ hybridization analysis of FTM1 in apices of tfl1-18 mutants grown from germination under LDs for 10 (left panel) or 12 (right panel) d. Bar = 50 μm.
(D) Activation of FT by CO does not lead to FTM1 activation under SDs. Pro35S:CO:GR plants were grown under SDs and exposed to dex. Ler plants were transferred from SDs to LDs. RNA was extracted from apices. Top panel: Analysis of FT mRNA by RT-PCR in Pro35S:CO:GR plants treated with dex or a mock solution . Middle panel: Analysis of SOC1 mRNA by RT-PCR in Pro35S:CO:GR plants treated with dex or a mock solution. Bottom panel: Analysis of FTM1 mRNA by RT-PCR in Pro35S:CO:GR plants treated with dex or a mock solution. Ler plants transferred from SDs to LDs act as a control. Two biological replicates of the experiment were performed, and for each of them three technical replicates were compared in the PCR reaction. Error bars represent sd in one experiment. In all panels, x axis is days after treatment. Closed circles, Pro35S:CO:GR treated with dex; open circles, Pro35S:CO:GR treated with mock. In the bottom panel, closed triangles indicate Col transferred from SDs to LDs.
[See online article for color version of this figure.]
FT and TSF are proposed to activate flowering through the rapid induction of SOC1 in the shoot apical meristem. Therefore, we tested whether the expression of the analyzed genes in the meristem in response to LDs depends on SOC1 and the partially redundant gene FUL. The abundance of FLOR1, FTM2, and SPL4 mRNA was not increased in response to five LDs in soc1-2 ful-2 plants (Figure 4); therefore, the strong response of these genes to five LDs in Col plants depends on SOC1 and FUL. By contrast, FAF2 expression was increased in the soc1-2 ful-2 double mutant 5 d after transfer to LDs, but to a lesser extent than in Col (Figure 4; see Supplemental Figure 7 online). Furthermore, in agreement with what was observed in ft-10 tsf-1 double mutants, FTM3 was induced in soc1-2 ful-2 double mutants after transfer to LDs (Figure 4).
The svp-41 mutation strongly suppressed the effect of ft-10 tsf-1 and soc1-2 ful-2 on flowering time (Jang et al., 2009) (Figure 1). Therefore, to test whether the effect of ft-10 tsf-1 and soc1-2 ful-1 on induction of gene expression could be overcome by the svp-41 mutation, in situ hybridizations were performed on svp-41 soc1-2 ful-2 and svp-41 ft-10 tsf-1 plants transferred from SDs to LDs. SPL4 was induced in svp-41 soc1-2 ful-2 and, to a lesser extent, in svp-41 ft-10 tsf-1 after five LDs (Figure 4), whereas no SPL4 mRNA was detected in soc1-2 ful-2 or ft-10 tsf-1 meristems (Figure 4). Therefore, in svp-41 mutants, SOC1 and FUL are not required for SPL4 activation in response to LDs, and the requirement for FT and TSF is reduced. By contrast, FAF2 mRNA expression was dependent on FT and TSF, even in svp-41 mutants. In svp-41 ft-10 tsf-1 triple mutant plants, FAF2 mRNA was not detected in the meristem after exposure to five LDs, but in svp-41 soc1-2 ful-2 plants, FAF2 mRNA was expressed in the meristem (Figure 4). This result indicates that svp-41 can overcome the requirement for SOC1 and FUL for activation of FAF2 in response to LDs but not the requirement for FT and TSF. FLOR1, on the other hand, seemed to show a higher dependency on SOC1 and FUL than FT and TSF in an svp-41 mutant background. In svp-41 ft-10 tsf-1 plants, FLOR1 mRNA was expressed after exposure to five LDs and in a similar spatial pattern to wild-type plants (Figure 4). However, in svp-41 soc1-2 ful-2 plants, FLOR1 mRNA level was not as highly or broadly expressed after five LDs as in wild-type or svp-41 ft-10 tsf-1 plants. These results suggest that svp-41 can overcome the requirement for FT and TSF in FLOR1 induction but can only partially do so for SOC1 and FUL.
Taken together, these results suggest that early induction of gene expression in the meristem in response to LDs is regulated to a large extent by FT and TSF as well as by SOC1 and FUL. Generally, FTM2, FLOR1, and SPL4 expression in the meristem correlates with flowering in all genotypes. However, exposure to LDs does lead to the activation of some genes in the meristem independently of flowering and of SOC1 and FUL or FT and TSF. Also, the capacity of loss of SVP to overcome the impairment of SOC1 and FUL or FT and TSF function differs depending on the downstream gene, so that FAF2 does not respond to svp-41 in the absence of FT and TSF, whereas SPL4 responds more strongly to svp-41 in the absence of SOC1 and FUL than in the absence of FT and TSF.
FTM1 Is Induced in the Shoot Meristem during Floral Induction but Is Not Activated by the Photoperiodic Flowering Pathway
FTM1 expression was increased in the meristem of Col and ft-10 tsf-1 plants transferred from SDs to LDs (Figure 5A). To test whether this induction is associated with floral induction or with another process related to transfer from SDs to LDs, the pattern of FTM1 mRNA was followed in plants of different ages continuously grown in LDs. FTM1 mRNA was not detected in the shoot meristem of 10-d-old wild-type plants, which are still vegetative, but was present in the meristems of 12-d-old plants, which are in the early stages of floral induction (Figure 5B). Therefore, in these plants, FTM1 expression in the meristem correlates with floral induction. The expression pattern of FTM1 was also tested in tfl1-18 mutants grown under LDs, which flower earlier than the wild type (Shannon and Meeks-Wagner, 1991; Alvarez et al., 1992). FTM1 mRNA was already weakly detected in 10-d-old tfl1-18 plants and was strongly detected in the meristem of 12-d-old plants (Figure 5C) and so is expressed earlier than in wild-type plants, again correlating with floral induction. These experiments indicate that FTM1 expression in the meristem is associated with the floral transition even in plants grown continuously in LDs.
FT and TSF were not required to induce FTM1 expression in the meristem of plants transferred from SDs to LDs. However, FT and TSF might still activate FTM1, but do so redundantly with another LD-regulated signal. To test whether induction of FT mRNA is able to activate FTM1 expression, transgenic plants in which FT expression can be induced in SDs due to activation of CO function were used (Simon et al., 1996). Pro35S:CO:GR plants grown in SDs were treated with dexamethasone (dex) to induce CO activity. RT-PCR analysis demonstrated that expression of FT and SOC1 mRNA was induced rapidly after treatment with dex (Figure 5D). However, no increase in abundance of FTM1 mRNA was detected in these plants. By contrast, RT-PCR analysis did detect an increase in FTM1 expression after transferring Landsberg erecta (Ler) plants from SDs to LDs, as shown above by in situ hybridization for wild-type Col. Therefore, activation of CO and FT activities is not sufficient to induce expression of FTM1 under SDs, and FT is neither required nor sufficient to induce FTM1 expression. Thus, the induction of FTM1 in the shoot apical meristem during floral induction in LDs is independent of CO, FT, and TSF.
FLOR1 Contributes to Early Flowering
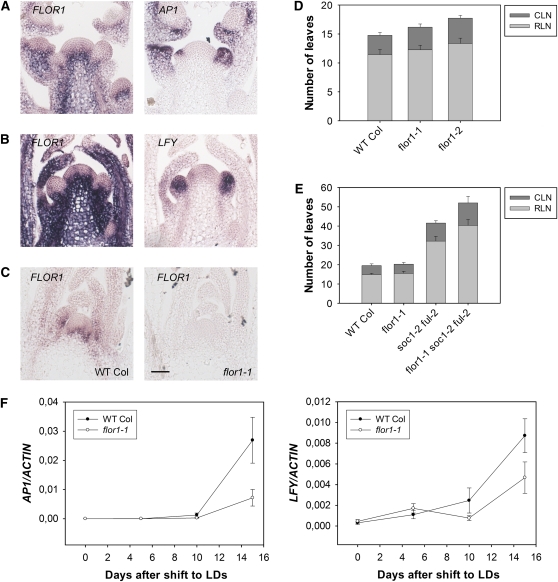
FLOR1 is weakly expressed in the rib meristem before floral transition in plants grown under SDs and strongly increased in expression after transfer to LDs (Figure 3). The strong expression in the inflorescence rib meristem persists after floral induction and is excluded from floral primordia that express LFY and AP1 mRNA (Figures 6A and 6B). To test whether FLOR1 contributes to floral induction, the gene was overexpressed and insertion mutations were identified. Overexpression of FLOR1 from the cauliflower mosaic virus 35S promoter had no consistent effect on flowering, indicating that FLOR1 expression is not limiting on flowering. In addition, two T-DNA insertion mutations were identified, flor1-1 and flor1-2, both of which carry a T-DNA insertion in the single intron of FLOR1. No expression of the second exon was detected by RT-PCR in either mutant (see Supplemental Figure 9A online). Furthermore, in the flor1-1 mutant, no FLOR1 mRNA was detected in the meristem region by in situ hybridization (Figure 6C). These results suggest that both mutant alleles reduce FLOR1 activity.
Figure 6.
Genetic Analysis of Role of Candidate Genes in the Floral Transition.
(A) In situ hybridization of FLOR1 and AP1 probes on consecutive sections of apices of 22-d-old Col plants grown under LDs.
(B) In situ hybridization of FLOR1 and LFY probes on consecutive sections of apices of 22-d-old Col plants grown under LDs.
(C) In situ hybridization of FLOR1 probe on apices of Col and flor1-1 mutant plants grown for 12 d under LDs. WT, wild type. Bar = 50 μm.
(D) Flowering time in LD of flor1-1 and flor1-2 plants compared with wild-type Col. The flor1-2 mutant was later flowering than Col (P < 0.05 in two experiments). flor1-1 was significantly late flowering (P < 0.05) in terms of total leaf number in only two of five experiments. CLN, cauline leaf number; RLN, rosette leaf number. Error bars represent sd. At least eight plants were used to score flowering time of each genotype in each experiment.
(E) flor1-1 enhances the late-flowering phenotype of soc1-2 ful-2 double mutants (P < 0.05 in two experiments). Plants were grown under LDs. CLN, cauline leaf number; RLN, rosette leaf number. Error bars represent sd. At least eight plants were used to score flowering time of each genotype in each experiment.
(F) The flor1-1 mutation delays activation of AP1 and LFY. RNA was extracted from apices of plants grown under SDs for 2 weeks and transferred to LDs. Left panel: AP1 expression, as determined by qRT-PCR. Right panel: LFY expression. x axis: days after transfer from SDs to LDs. Two biological replicates of the experiment were performed, and for each of them three technical replicates were compared in the PCR. Error bars represent sd in one experiment, and the other biological replicate produced similar results.
[See online article for color version of this figure.]
The flowering time of flor1-1 and flor1-2 mutants was measured under LDs and SDs. Both mutants flowered slightly late under LDs, producing two to three more leaves than wild-type plants (Figure 6D). Under SDs, no effect on flowering time was detected. To test whether flor1 mutations enhance the effect of soc1-2 and ful1-2 on flowering time, the double and triple mutant combinations were constructed. The flor1-1 soc1-2 and flor1-1 ful-2 double mutants flowered at similar times to soc1-2 and ful-2 single mutants (see Supplemental Figure 9B online). However, the triple mutant flor1-1 soc1-2 ful-2 flowered significantly later than the soc1-2 ful-2 control, producing ~10 more leaves than soc1-2 ful-2 double mutants (Figure 6E). Consistent with this conclusion, quantitative RT-PCR analysis of LFY and AP1 mRNAs in flor1-1 mutants transferred from SDs to LDs showed that these genes are more rapidly induced in Col plants than flor1-1 mutants (Figure 6F). Thus, FLOR1 promotes flowering and its role is partially redundant with SOC1 and FUL.
DISCUSSION
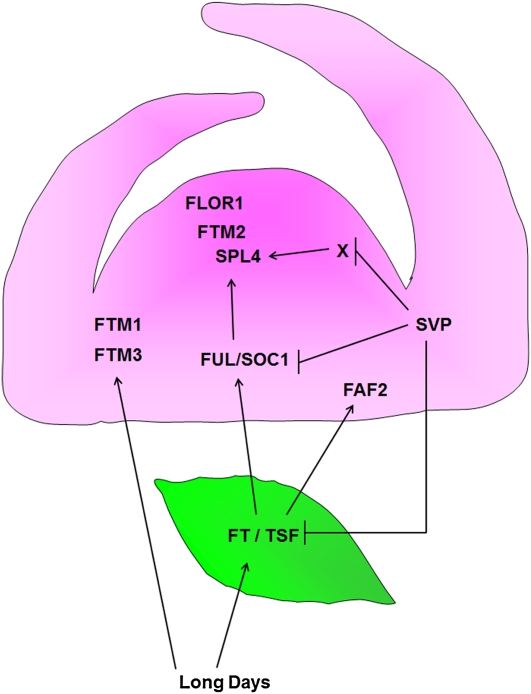
Analysis of the meristem during the five LDs required to commit the plant to flowering demonstrated the complexity of the inductive pathways involved in the early stages of floral induction within the shoot apical meristem. Pathways downstream of FT and TSF were found to act in a nonlinear way, and pathways acting in parallel to FT and TSF were identified. Mutations in one of the genes strongly induced in the inflorescence meristem, FLOR1, that encodes a LRR protein enhanced the late-flowering phenotype of soc1-2 ful-2 plants. Our conclusions are summarized in the diagram shown in Figure 7.
Figure 7.
Summary of the Major Gene Interactions during Floral Induction Identified by Genetic and Experimental Analyses Described in the Text.
Exposure of plants to LDs activates FT and TSF transcription in leaves. FT and TSF are required for activation of SOC1 and FUL in the shoot apical meristem. Expression of FLOR1, FTM2, and SPL4 in the meristem of wild-type plants during floral induction depends on FT and TSF as well as SOC1 and FUL. FAF2 can be activated independently of SOC1 and FUL but depends on FT and TSF. FTM1 and FTM3 are activated independently of FT and TSF and SOC1 and FUL. Mutation of SVP increases expression of FLOR1, FTM2, and SPL4 even in the absence of FUL and SOC1 or FT and TSF. Leaf shown in green; meristem shown in pink.
Transient Exposure to LDs and Commitment of the Meristem to Flowering
Commitment to flowering after transient exposure to LDs indicates that the inflorescence meristem can stably maintain its identity in noninductive conditions. Expression of AP1, which only occurs in floral primordia, was detected after five LDs and defines floral commitment. AP1 expression was previously shown to mark the time of commitment, or determination, after exposure to different wavelengths of light (Hempel et al., 1997). The duration of this commitment period likely depends on the age of the plants and the precise environmental conditions, such as light intensity, wavelength of light, and temperature (Corbesier et al., 1996; Hempel et al., 1997; King et al., 2008).
The related MADS box transcription factors SOC1 and FUL have partially redundant roles in flowering time and maintenance of the inflorescence meristem in plants continuously exposed to LDs (Melzer et al., 2008). The expression of both genes is induced in the meristem after exposure of SD-grown plants to one to three LDs (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000). Failure of soc1-2 ful-2 to commit to flowering in response to five LDs is probably due to a severe delay in activation in the meristem of many flowering genes. The failure of the soc1-2 ful-2 double mutant to maintain the inflorescence under continuous LDs (Melzer et al., 2008) may be related to the commitment to flower phenotype described here; however, the maintenance phenotype was observed in plants that had undergone the transition to flowering and then reverted to vegetative growth, which cannot easily be explained by delayed flowering. The inflorescence maintenance phenotype may be a consequence of impairment of a later function of SOC1 in the inflorescence meristem or floral primordia (Lee and Lee, 2010).
Commitment of the meristem to continue forming flowers after return to SDs is not due to continued expression of FT in the leaves (Corbesier et al., 2007). SOC1 mRNA level in the meristem fell substantially when plants exposed to five LDs were returned to SDs, suggesting that SOC1 mRNA level is highly responsive to FT. The reduction in SOC1 mRNA after return of plants to SDs suggests that its contribution to floral commitment has already occurred during the first five LDs. By contrast, FUL mRNA level was maintained in the meristem after return to SDs and indeed appeared to continue to increase, perhaps allowing it to continue to contribute to floral commitment. This expression pattern suggests the presence of a positive feedback loop that maintains FUL expression even after FT and SOC1 mRNA levels have declined. Such feedback loops mark developmental shifts in other systems (Adrian et al., 2009). SPL transcription factors, whose mRNAs rise in expression after induction of SOC1 and FUL, might be involved in maintaining FUL expression because SPL3, for example, has been shown to directly activate FUL (Wang et al., 2009; Yamaguchi et al., 2009). Furthermore, we found that SOC1 and FUL are required for activation of SPL4. These observations support the existence of a loop in which FUL is involved in activation of SPL genes, which then maintain FUL expression.
The svp-41 mutation can overcome the strong impairment of floral commitment and delay of flowering time in soc1-2 ful-2 double mutants. SVP directly represses SOC1 expression in the meristem, but the observation that svp-41 can overcome the phenotype of soc1-2 ful-2 argues that SVP has additional targets in the meristem. If it represses expression of such genes, then when derepressed in an svp-41 mutant their increased expression might overcome the late flowering phenotype of the soc1-2 ful-2 double mutant. These additional genes are unlikely to be the known targets of SVP in the leaves, FT and TSF, because overexpression of FT from the phloem-specific SUC2 promoter had little effect on the flowering time of soc1-2 ful-2 plants, as shown previously for the 35S promoter (Melzer et al., 2008). Understanding the multiple roles of SVP in the meristem in delaying flowering time would contribute to unraveling the layers of genetic redundancy that exist during the early stages of floral induction.
Transcriptional Reprogramming of the Shoot Apical Meristem during Floral Induction
Laser microdissection combined with next-generation sequencing or microarrays was applied to plant tissues previously, for example, the shoot apical meristem and leaves of maize (Zea mays) or the female gametophyte of Arabidopsis (Jones-Rhoades et al., 2007; Li et al., 2010; Matias-Hernandez et al., 2010). The spatial resolution of the method allowed us to focus specifically on meristem tissue, excluding the leaf primordia, young leaves, and vascular tissue that are present in hand-dissected apical samples. Such enrichment of meristem tissue likely also increases the sensitivity with which meristem transcripts are detected. GO analysis demonstrated that genes involved in processes such as reproductive development were indeed enriched in upregulated genes compared with the whole transcriptome. However, enzymes involved in oxidation-reduction reactions were also unexpectedly highly overrepresented. The function of most of these enzymes is unknown, but the expression pattern of one of them, FTM1, was confirmed by in situ hybridization.
Previously, Affymetrix ATH1 microarray hybridizations were used to analyze gene expression changes in apical samples during floral induction (Schmid et al., 2003). Of the 202 genes identified here as being upregulated in the shoot apical meristem, only 174 were present on the ATH1 microarray. Of these 174, three were detected in the Affymetrix analysis as being upregulated by at least twofold in the apical samples after 3 d and nine after 7 d. Of the six genes analyzed in detail in this study, one, FLOR1, was not annotated on the ATH1 array. Only FAF2 of the remaining five was upregulated by at least twofold in the three LD apical sample. At seven LD, SPL4 and FTM1 were also upregulated twofold. FTM3 and FTM2 never reached twofold upregulation in the apical samples, even after exposure to seven LDs. Therefore, there is an overlap between genes upregulated in the laser microdissection Solexa data and the Affymetrix apical samples. However, genes were also identified in the laser microdissection data that were not present in the apical Affymetrix data, partly because of the limited annotation of the ATH1 array and presumably also due to the higher sensitivity conferred by enriching for meristems with the laser microdissection method. Furthermore, in our experiments, plants were grown for a shorter time in SD conditions prior to transfer to LDs, and this may have helped to reduce the expression level in the SD control samples of genes associated with flowering.
Induced Genes Differentially Respond to Known Floral Regulators Active in the Meristem
A prediction of the current model of photoperiod response is that many or all of the genes induced in wild-type meristems by exposure to LDs would not be induced in ft-10 tsf-1 double mutants. Indeed, this was the case for SOC1, FLOR1, FTM2, FAF2, and SPL4. Under SDs, some of these genes might respond to other inputs as has been shown for SOC1 (Moon et al., 2003). Complex mechanisms must be required to interpret the FT signal transmitted from the leaves to ultimately activate distinct pathways leading to expression of genes in such diverse spatial patterns.
Surprisingly, meristem induction of FTM1 and FTM3 still responded to daylength in ft-10 tsf-1 double mutants with similar kinetics and amplitude observed in wild-type plants. FTM3 encodes a bZIP transcription factor in Group S (Jakoby et al., 2002). The best-characterized member of this group, ATBZIP11, is expressed in sink tissues and responds to Suc levels (Rook et al., 1998). During the transition to flowering, the shoot apical meristem becomes a stronger sink tissue and levels of Suc increase at the meristem (Bernier et al., 1993). Therefore, the increase in FTM3 mRNA observed in the meristem might be a response to increasing carbohydrate levels. If so, this would represent an FT- and TSF-independent mechanism by which expression of regulatory genes can increase at the meristem during floral induction. FTM1 is also involved in metabolism, encoding a putative stearoyl-acyl carrier protein-desaturase (S-ACP-DES) enzyme. This gene family is composed in Arabidopsis of seven members (Kachroo et al., 2007). The best characterized member is SUPPRESSOR OF SA INSENSITIVITY2, and mutants for this gene contain low levels of the monosaturated fatty acid and are impaired in salicylic acid as well as jasmonic acid signaling (Kachroo et al., 2001; Shah et al., 2001). Although other members of the S-ACP-DES gene family also seem to regulate lipid profiles, it is not clear to what extent they regulate SA or JA signaling (Kachroo et al., 2007). The specific expression pattern of FTM1 during floral induction could indicate involvement in the meristem during floral induction of SA signaling, which has been reported to alter flowering time (Martínez et al., 2004). FTM1 is not regulated by FT, and the mechanism by which it is induced in the meristem during floral induction is not known.
Genes that require FT and TSF for their induction in the meristem generally also require SOC1 and FUL. Thus, FLOR1, SPL4, and FTM2 expression patterns are similarly affected in ft-10 tsf-1 and soc1-2 ful-2 backgrounds. However, the spatial patterns of SPL4 and FTM2 do not overlap, indicating that SOC1 and FUL somehow activate spatially restricted pathways within the meristem. Also, ft-10 tsf-1 and soc1-2 ful-2 double mutants are not identical with respect to meristem gene expression patterns. FAF2 mRNA is faintly detected in soc1-2 ful-2, but absent from ft-10 tsf-1, suggesting a stronger requirement for FT and TSF. Also, these genes can be divided into distinct regulatory hierarchies based upon their regulation by SVP, as described below. In addition, the late flowering phenotype of ft-10 tsf-1 is more extreme than that of soc1-2 ful-2, excluding the possibility that FT and TSF act only through SOC1 and FUL.
SPL4 expression is restored in the meristems of svp-41 soc1-2 ful-2 or svp-41 ft-10 tsf-1 plants exposed to five LDs. Therefore, genes deregulated in the meristems of svp-41 mutants are able to induce SPL4 expression in ft-10 tsf-1 or soc1-2 ful-2. This expression of SPL4 correlates with earlier flowering of svp-41 ft-10 tsf-1 and svp-41 soc1-2 ful-2 compared with ft-10 tsf-1 or soc1-2 ful-2. By contrast, induction of FAF2 during floral induction showed an absolute requirement for FT and TSF even in the presence of svp-41. FAF2 is a member of a gene family comprising four related genes (Wahl et al., 2010). Based on analysis of Pro35S:FAF plants, the FAF genes were proposed to regulate meristem function by modulating the WUSCHEL (WUS)-CLAVATA3 (CLV3) feedback loop that maintains the meristem. These results of Wahl et al. (2010) together with our observations that FAF2 expression depends on FT and TSF suggest a link between FT (and TSF) signaling and the WUS-CLV3 feedback loop that maintains meristem function.
The PENNYWISE (PNY) and POUNDFOOLISH (PNF) homeobox genes are also strongly required for expression of SPL4 and SPL5 at the apex and suppress the early flowering caused by FT overexpression (Kanrar et al., 2008; Lal et al., 2011). These effects are similar to those described above for soc1-2 ful-2. However, the mechanism by which PNY and PNF affect SPL4 and SPL5 mRNA levels is unlikely to be through SOC1, as this gene is expressed at almost normal levels in pny pnf plants (Smith et al., 2004). How the function of the PNY and PNF genes intersects with that of FT and TSF as well as SOC1 and FUL remains unclear.
Role of FLOR1 in Regulating Flowering Time
FLOR1 is a member of the plant-specific LRR protein subfamily (Gamboa et al., 2001; Acevedo et al., 2004). Unlike most plant-specific LRR proteins that have been characterized, FLOR1 has an intracellular location in the cytoplasm and nucleus (Acevedo et al., 2004). FLOR1 was initially isolated because it interacts in vitro and in yeast with AGAMOUS, a MADS box transcription factor involved in the development of stamens and carpels. In situ hybridizations demonstrated that FLOR1 mRNA is present in mature stamens and carpels and therefore overlaps with AG expression (Acevedo et al., 2004). Our in situ analyses showed that FLOR1 is weakly expressed in the vegetative shoot meristem and then upregulated in the inflorescence meristem within 3 d of induction. Subsequently, it is excluded from young floral primordia marked with AP1 and LFY mRNAs. This inflorescence pattern is similar to that of SOC1 mRNA, which is excluded from the domain in which AP1 is expressed but is expressed later in the center of the flower. As observed for AG, FLOR1 might interact with other MADS box proteins expressed in the inflorescence meristem during floral transition. Such proteins are unlikely to be SOC1 or FUL because flor1 mutations delayed the flowering phenotype of soc1-2 ful-2 double mutants. However, several other MADS box proteins are expressed in the inflorescence meristem (Michaels et al., 2003; Dorca-Fornell et al., 2011). The biochemical significance of the interaction between FLOR1 and MADS box proteins has not been established but is likely to represent a function not previously implicated in floral induction.
METHODS
Plant Material and Growth Conditions
Plants used in this study were Arabidopsis thaliana accession Col or Ler. Mutant alleles in the Col background were previously described: soc1-2 (Lee et al., 2000), ft-10 tsf-1 (Jang et al., 2009), ful-2 (Ferrándiz et al., 2000), svp-41 (Hartmann et al., 2000), and tfl1-18 (Conti and Bradley, 2007). Lines carrying T-DNA insertions in the FLOR1 locus were SALK_093764 for flor1-1 and SALK_120575 for flor1-2.
The Pro35S:CO:GR line is in the co-2 background in Ler (Simon et al., 1996).
Plants were genotyped using specific primers listed in Supplemental Table 1 online.
To promote germination, seeds were stratified on soil at 4°C for 3 d in the dark. Plants for the experiments were grown in growth cabinets. LDs were 16 h of light and 8 h of dark, and SDs were 8 h of light and 16 h of dark. The temperature in the growth cabinet was 18°C.
Flowering Time Measurements and Percentage of Induction
Flowering time was scored as number of leaves at bolting. The number of rosette leaves was counted until the bolting shoot reached around 1 cm in length. Cauline leaves were counted when they were all visible on the shoot. At least eight genetically identical plants were used to score flowering time of each genotype. The Student’s t test was used to test the significance of flowering time differences.
For each population X (a distinct genotype or a population with a certain time of vegetative growth in SDs), the following formula was used to calculate the degree of induction in the double shift experiments:
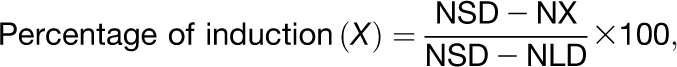 |
where NSD is the number of leaves at flowering for the plants in SDs, NLD is the number of leaves at flowering for the plants shifted from SDs to LDs, and NX is the number of leaves at flowering of the plants for which the percentage of induction is calculated. NSD, NLD, and NX are total leaf numbers (rosette plus cauline leaves), calculated as an average for the population exposed to a particular condition. sd was calculated according to the classical propagation of errors for a division.
Plant Transformation
Plants were transformed with Agrobacterium tumefaciens strain GV3101 pMP90 RK by floral dipping (Clough and Bent, 1998). Plasmids containing ProSUC2:FT and Pro35S:FT in Agrobacterium were used to stably transform wild-type Col and soc1-2 ful-2 double mutants and they were described (An et al., 2004).
Plants carrying the transgene were selected by spraying with glufosinate (BASTA) at 100 mg/L, and then the insertion of the transgene was checked with specific primers by PCR on genomic DNA extracted from single leaves from the independent lines. Plants at the T1 generation were scored for flowering time. To make transgenic lines of soc1-2 ful-2 double mutants, plants homozygous for the soc1 mutation and heterozygous for the ful-2 mutation were transformed with Agrobacterium. The ful-2 mutation at the T1 generation was followed by sequencing the PCR product with specific primers (see Supplemental Table 1 online) for the heterozygotes and by the silique phenotype for the ful homozygotes.
In Situ Hybridizations
The methods used for digoxigenin labeling of mRNA probes, tissue preparation, and in situ hybridization were previously described (Bradley et al., 1993) with small modifications. Protease treatment was performed with Proteinase K (1 μg/mL in 100 mM Tris, pH 8, and 50 mM EDTA) at 37°C for 30 min, instead of Pronase. The posthybridization washes were performed in 0.1× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate).
For AP1, a previously described plasmid (Mandel et al., 1992) was used to synthesize a probe of 720 bp.
For all the other genes, templates for probes to detect their transcripts were PCR amplified from cDNA using specific primer pairs (see Supplemental Table 1 online) that amplify part of the cDNA, with T7 RNA polymerase binding sites attached to the reverse primers. Hybridization experiments for each gene were generally repeated three times on each set of samples using at least two biological replicates.
Sample Collection and Preparation for Laser Microdissection
The procedure used to prepare the samples followed the one used for the in situ hybridization with specific modifications.
Seedlings were collected and fixed with ethanol:acetic acid in a 3:1 ratio and continuously kept on ice during the harvesting to preserve the RNA. To allow penetration of the fixative, the tissue was vacuum infiltrated using a pump, fixative was replaced, and the samples left at 4°C on ice overnight. The following day, the fixative was replaced with a stepwise ethanol:water series at 4°C (85% ethanol, 4 h; 95% ethanol, 4 h; 100% ethanol, overnight; 100% ethanol, fresh). The samples were stored at 4°C in 100% ethanol until embedding.
Samples were stained with eosin (0.1% Eosin Y in 100% ethanol) prior to embedding in paraffin. Embedding in Paraplast Plus (McCormick) paraffin was performed with the automated system ASP300 tissue processor (Leica). Wax blocks with eosin-stained samples were stored at 4°C until sectioning.
The slides used for laser microdissection were PALM MembraneSlides (PEN-membrane, 1 mm) from P.A.L.M. Microlaser Technologies. They were treated to remove possible RNase contamination with dry heat at 180°C for 4 h. This was followed by UV treatment by irradiation with UV light at 254 nm for 30 min using a cross-linker UV Stratalinker 1800 (Stratagene). This provides further sterilization and allows the membranes to become more hydrophilic.
Embedded plants were sectioned using a rotary microtome (Leitz 1512) at 10-μm thickness and collected on the laser microdissection slides.
Laser Microdissection
To dissolve the paraffin, the slides were exposed to Histo-clear solvent (National Diagnostics) and then to a series of ethanol/water solutions with increasing concentrations of water (100% Histo-clear, 2 min; 100% Histo-clear, 2 min; 100% ethanol, 1 min; 96% ethanol, 1 min; 70% ethanol, 1 min; 50% ethanol, 1 min; water, 1 min) and allowed to dry.
Laser microdissection optionally coupled to laser pressure catapulting was performed with the HAL 100 model (230 VZ) from P.A.L.M., equipped with an Axiovert 200 M from Zeiss. After microdissection of the tissue, the sample was collected into PALM AdhesiveCaps (from P.A.L.M.).
RNA Extraction and Amplification after Laser Microdissection
The samples were dissolved from the caps of the collection tubes, to extract the RNA, with 100 μL RLT buffer (from the RNeasy kit; Qiagen) + β-mercapto-ethanol (10 μL for 1 mL of buffer).
After 10 min at room temperature, tubes were vortexed for 10 min and spun in a bench-top centrifuge at 13,400 relative centrifugal force for 5 min. Samples were stored at −80°C to avoid RNA degradation. Total RNA was extracted using a PicoPure extraction kit (Arcturus).
RNA amplification was performed using a RiboAmp HS amplification kit (Arcturus). The procedure was followed according to the manufacturer’s manual.
The RNA quality tests were performed with the Agilent 2100 bioanalyzer (Agilent Technologies) using an RNA 6000 Pico assay kit (Agilent Technologies).
RT-PCR Methods
Total RNA was extracted using an RNAeasy kit (Qiagen). Any DNA contamination was removed by DNaseI treatment (Ambion). cDNA synthesis was performed with SuperScript II (Invitrogen) using oligo(dT) primers. Three micrograms of RNA per sample was used to synthesize cDNA for quantitative real-time PCR. The synthesized cDNA was diluted to a final volume of 150 μL with water, and 3 μL was used for the PCR amplification.
For real-time PCR, three replicates were used for each sample. Amplified products were detected using SyBR green I in an IQ5 (Bio-Rad) thermal cycler. ACTIN2 was used as a housekeeping gene to normalize the expression of the genes investigated.
Illumina-Solexa Sequencing
To evaluate gene expression in the samples collected by laser microdissection, the RNA was converted into double-stranded cDNA with two methods depending on the samples. For replicates A and B, the cDNA synthesis was done using the RiboAmp kit (Arcturus), and the synthesis was initiated using polyT-based primers. Samples were then sequenced as “genomic sample preparation.” For the samples of replicate C, the mRNA-Seq sequencing protocol derived from Illumina was followed and random primers were used to initiate the cDNA synthesis. The sequencing was performed by FASTERIS Life Sciences. Short sequence tags of 35 bp were obtained as single-end for replicates A and B and 36 bp were obtained as paired-end for replicate C.
Analysis of Short-Sequence Reads from Illumina-Solexa Sequencing
The reads can be accessed from the National Center for Biotechnology Information (NCBI), as described below under Accession Numbers.
Trimming and Filtering
The data were initially filtered using Seqclean (release dated August 18, 2005). This program trims matches against user-specified target sequences, which here were the primer sequences (5′-GACGGCCAGTGAATTGTAATACGACTCACTATAGGGAGATCTGTATGCTGG-3′ and 5′-CCAGCATACAGATCTCCCTATAGTGAGTCGTATTACAATTCACTGGCCGTC-3′), as well as the UniVec_Core database (dated October 8, 2008), poly(A) tails, and ends rich in undetermined bases. After trimming, a read may be removed entirely for one of three reasons: (1) the sequence is shorter than the minimum length specified via the “−l” parameter (here, 30), (2) the percentage of undetermined bases is >3%, and (3) <40 nucleotides of the sequence is left unmasked by the “dust” low-complexity filter. Further details on analysis of sequences is provided in the Supplemental Methods 1 online.
Mapping the Reads
Each data set of reads was converted into a blast database using “formatdb.” To identify matches to known genes, the TAIR8 cDNA collection (TAIR8_cdna_20080325) was then compared with the read databases using Megablast (settings: -v 2000 -b 500 -a 4 -W16 -p 0.6 -e 1 -D3).
The initial runs were performed using the Megablast version BLASTN 2.2.13 (November 27, 2005); the last runs were done with BLASTN 2.2.21 (June 14, 2009).
Determining Raw Expression Counts of Genes (Loci)
The Megablast output was converted to an expression count by the following four steps, in this order: (1) Discard a match if its bit score is 5 or more below the best bit score that is reached by the respective read. (2) Discard a match if it is shorter than 20 bp or if its edit distance (number of gaps + number of mismatches) is 4 or more, or if the match does not start within the first three bases of the read (rationale for the last condition: sequencing quality is best at the 5′ end; therefore, a true match should cover the 5′ end). (3) Discard all matches involving reads that map to more than a single locus (note that a locus can encompass more than a single transcript [cDNA]). (4) For each locus, count the number of different reads that map to it (a single read can map multiple times to a locus if the locus has multiple transcripts; yet, the read will be counted only once at this step). The count of step (4) is output as a raw expression count.
Identifying Differentially Expressed Genes
The count data were normalized with edgeR (Robinson et al., 2010), and the package's exact test for the negative binomial distribution was used to compute P and false discovery rate values.
Clustering and Heat Map
Clustering and heat map of the normalized count data were performed using Genesis software (Sturn et al., 2002). Genes were clustered by hierarchical clustering (parameters: distance = mismatch distance; agglomeration rule = average linkage clustering).
Gene Functional Classification
The DAVID gene functional annotation tool (Huang et al., 2009) was used for the gene-GO term enrichment analysis. The 202 upregulated genes were classified into the different GO FAT terms (Biological Process). The following thresholds were used: minimum number of counts (2) and EASE score (0.05).
ATH1 Microarray Comparison
Archives corresponding to the processed data (ID: E-GEOD-576) were downloaded from the Array Express repository (http://www.ebi.ac.uk/arrayexpress/).
Accession Numbers
The Solexa sequence reads are available from NCBI with the series entry GSE34476. Access to these sequences is provided at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE34476.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Floral Commitment in Arabidopsis Wild Type and Mutants.
Supplemental Figure 2. In Situ Hybridization on Wild-Type Col Apices in Double Shift Experiments Using Probes for SOC1, FUL, SVP, and AP1.
Supplemental Figure 3. Comparison of Inflorescences of soc1-2 ful-2 and svp-41 soc1-2 ful-2 Plants.
Supplemental Figure 4. Laser Microdissection Performed on Wild-Type Col Apices during the Floral Transition.
Supplemental Figure 5. K-Means Clustering of the Upregulated Genes.
Supplemental Figure 6. Annotation and Sequence Reads of FTM3 (At2G18160).
Supplemental Figure 7. Time Course of in Situ Hybridizations in Different Genotypes for Genes Identified as Being Upregulated during Floral Induction.
Supplemental Figure 8. Time-Course Analysis of SOC1 Expression in Col and ft-10 tsf-1.
Supplemental Figure 9. Genetic and Molecular Analysis of flor1 Mutations.
Supplemental Table 1. List of Primers Used in This Study.
Supplemental Data Set 1. Number of Reads Aligned to Each Arabidopsis Gene in Each Experiment.
Supplemental Data Set 2. List of 202 Genes Upregulated in the Shoot Apical Meristem during Floral Induction According to the Criteria Described in the Text.
Supplemental Data Set 3. Gene Ontology Analysis.
Supplemental Data Set 4. Number of Reads Aligned to Each Arabidopsis Gene in Each Experiment Taking into Account the Promiscuous Counts.
Supplemental Methods 1. Methods for the Supplemental Data.
Supplemental References 1. References for the Supplemental Data.
Supplementary Material
Acknowledgments
We thank Doris Falkenhan for her technical assistance. We also thank Laurent Farinelli at Fasteris for advice and assistance on the processing of the RNA samples. Aimone Porri kindly provided primers for in situ hybridization probe of LFY. We also thank Emiel Ver Loren van Themaat for help in data analysis. This work was partly funded by a Marie Curie Training Fellowship to S.T. through the TRANSISTOR program, by the European Research Area–Plant Genomics grant BLOOMNET through the Deutsche Forschungsgemeinshaft to G.C., and by the Max Planck Society via a core grant to G.C.
AUTHOR CONTRIBUTIONS
S.T., F.F., and G.C. designed the experiments. S.T., F.F., C.V., and F.A. performed the experiments. K.N., U.G., D.K., and H.S. performed the bioinformatics analysis. S.T., F.F., C.V., F.A., and G.C. analyzed the data. S.T. and G.C. wrote the article.
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Acevedo F.G., Gamboa A., Paez-Valencia J., Jimenez-Garcia L.F., Izaguirre-Sierra M., Alvarez-Buylla E.R. (2004). FLOR1, a putative interaction partner of the floral homeotic protein AGAMOUS, is a plant-specific intracellular LRR. Plant Sci. 167: 225–231 [Google Scholar]
- Adrian J., Torti S., Turck F. (2009). From decision to commitment: The molecular memory of flowering. Mol. Plant 2: 628–642 [DOI] [PubMed] [Google Scholar]
- Alvarez J., Guli C.L., Yu X.H., Smyth D.R. (1992). Terminal flower: A gene affecting inflorescence development in Arabidopsis thaliana. Plant J. 2: 103–116 [Google Scholar]
- An H.L., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Bernier G., Havelange A., Houssa C., Petitjean A., Lejeune P. (1993). Physiological signals that induce flowering. Plant Cell 5: 1147–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner R., Kampmann G., Chandler J., Gleissner R., Wisman E., Apel K., Melzer S. (2000). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24: 591–599 [DOI] [PubMed] [Google Scholar]
- Bradley D., Carpenter R., Sommer H., Hartley N., Coen E. (1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72: 85–95 [DOI] [PubMed] [Google Scholar]
- Cardon G., Höhmann S., Klein J., Nettesheim K., Saedler H., Huijser P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237: 91–104 [DOI] [PubMed] [Google Scholar]
- Cardon G.H., Höhmann S., Nettesheim K., Saedler H., Huijser P. (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 12: 367–377 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conti L., Bradley D. (2007). TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 19: 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Gadisseur I., Silvestre G., Jacqmard A., Bernier G. (1996). Design in Arabidopsis thaliana of a synchronous system of floral induction by one long day. Plant J. 9: 947–952 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S.H., Fornara F., Fan Q.Z., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dorca-Fornell C., Gregis V., Grandi V., Coupland G., Colombo L., Kater M.M. (2011). The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 67: 1006–1017 [DOI] [PubMed]
- Ferrándiz C., Gu Q., Martienssen R., Yanofsky M.F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Fornara F., de Montaigu A., Coupland G. (2010). SnapShot: Control of flowering in Arabidopsis. Cell 141: 550–, 550, e1–e2 [DOI] [PubMed] [Google Scholar]
- Fujiwara S., Oda A., Yoshida R., Niinuma K., Miyata K., Tomozoe Y., Tajima T., Nakagawa M., Hayashi K., Coupland G., Mizoguchi T. (2008). Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 20: 2960–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamboa A., Paéz-Valencia J., Acevedo G.F., Vázquez-Moreno L., Alvarez-Buylla R.E. (2001). Floral transcription factor AGAMOUS interacts in vitro with a leucine-rich repeat and an acid phosphatase protein complex. Biochem. Biophys. Res. Commun. 288: 1018–1026 [DOI] [PubMed] [Google Scholar]
- Hartmann U., Höhmann S., Nettesheim K., Wisman E., Saedler H., Huijser P. (2000). Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Hempel F.D., Weigel D., Mandel M.A., Ditta G., Zambryski P.C., Feldman L.J., Yanofsky M.F. (1997). Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124: 3845–3853 [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Wigge P.A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F.; bZIP Research Group (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Jang S., Torti S., Coupland G. (2009). Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 60: 614–625 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Borevitz J.O., Preuss D. (2007). Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet. 3: 1848–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A., Shanklin J., Whittle E., Lapchyk L., Hildebrand D., Kachroo P. (2007). The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol. Biol. 63: 257–271 [DOI] [PubMed] [Google Scholar]
- Kachroo P., Shanklin J., Shah J., Whittle E.J., Klessig D.F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 98: 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar S., Bhattacharya M., Arthur B., Courtier J., Smith H.M.S. (2008). Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J. 54: 924–937 [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- King R.W., Hisamatsu T., Goldschmidt E.E., Blundell C. (2008). The nature of floral signals in Arabidopsis. I. Photosynthesis and a far-red photoresponse independently regulate flowering by increasing expression of FLOWERING LOCUS T (FT). J. Exp. Bot. 59: 3811–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Lal S., Pacis L.B., Smith H.M. (2011). Regulation of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE genes/microRNA156 Module by the Homeodomain Proteins PENNYWISE and POUND-FOOLISH in Arabidopsis. Mol. Plant 4: 1123–1132 [DOI] [PubMed]
- Lee H., Suh S.S., Park E., Cho E., Ahn J.H., Kim S.G., Lee J.S., Kwon Y.M., Lee I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Lee I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61: 2247–2254 [DOI] [PubMed] [Google Scholar]
- Lee J.H., Yoo S.J., Park S.H., Hwang I., Lee J.S., Ahn J.H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., Helliwell C.A., Ito T., Meyerowitz E., Yu H. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Li P.H., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Mandel M.A., Gustafson-Brown C., Savidge B., Yanofsky M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Martínez C., Pons E., Prats G., León J. (2004). Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. 37: 209–217 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Matias-Hernandez L., Battaglia R., Galbiati F., Rubes M., Eichenberger C., Grossniklaus U., Kater M.M., Colombo L. (2010). VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell 22: 1702–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer S., Lens F., Gennen J., Vanneste S., Rohde A., Beeckman T. (2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40: 1489–1492 [DOI] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Ditta G., Gustafson-Brown C., Pelaz S., Yanofsky M., Amasino R.M. (2003). AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 33: 867–874 [DOI] [PubMed] [Google Scholar]
- Moon J., Suh S.S., Lee H., Choi K.R., Hong C.B., Paek N.C., Kim S.G., Lee I. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F., Gerrits N., Kortstee A., van Kampen M., Borrias M., Weisbeek P., Smeekens S. (1998). Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 15: 253–263 [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Schwarz S., Grande A.V., Bujdoso N., Saedler H., Huijser P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I., He Y.H., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Kachroo P., Nandi A., Klessig D.F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25: 563–574 [DOI] [PubMed] [Google Scholar]
- Shannon S., Meeks-Wagner D.R. (1991). A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3: 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Burn J.E., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Igeño M.I., Coupland G. (1996). Activation of floral meristem identity genes in Arabidopsis. Nature 384: 59–62 [DOI] [PubMed] [Google Scholar]
- Smith H.M., Campbell B.C., Hake S. (2004). Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr. Biol. 14: 812–817 [DOI] [PubMed] [Google Scholar]
- Sturn A., Quackenbush J., Trajanoski Z. (2002). Genesis: Cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tan Q.K.G., Irish V.F. (2006). The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 140: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper-Bamnolker P., Samach A. (2005). The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Wahl V., Brand L.H., Guo Y.L., Schmid M. (2010). The FANTASTIC FOUR proteins influence shoot meristem size in Arabidopsis thaliana. BMC Plant Biol. 10: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T. (2005). TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46: 1175–1189 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Wu M.F., Yang L., Wu G., Poethig R.S., Wagner D. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Xu Y.F., Tan E.L., Kumar P.P. (2002). AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. USA 99: 16336–16341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.