LST8 is a member of the widely conserved TOR kinase complex. This work shows that LST8 is important for plant growth as well as metabolic and developmental processes linked to changes in light conditions, probably by influencing the activity of the TOR complex.
Abstract
The conserved Target of Rapamycin (TOR) kinase forms high molecular mass complexes and is a major regulator of cellular adaptations to environmental cues. The Lethal with Sec Thirteen 8/G protein β subunit-like (LST8/GβL) protein is a member of the TOR complexes, and two putative LST8 genes are present in Arabidopsis thaliana, of which only one (LST8-1) is significantly expressed. The Arabidopsis LST8-1 protein is able to complement yeast lst8 mutations and interacts with the TOR kinase. Mutations in the LST8-1 gene resulted in reduced vegetative growth and apical dominance with abnormal development of flowers. Mutant plants were also highly sensitive to long days and accumulated, like TOR RNA interference lines, higher amounts of starch and amino acids, including proline and glutamine, while showing reduced concentrations of inositol and raffinose. Accordingly, transcriptomic and enzymatic analyses revealed a higher expression of genes involved in nitrate assimilation when lst8-1 mutants were shifted to long days. The transcriptome of lst8-1 mutants in long days was found to share similarities with that of a myo-inositol 1 phosphate synthase mutant that is also sensitive to the extension of the light period. It thus appears that the LST8-1 protein has an important role in regulating amino acid accumulation and the synthesis of myo-inositol and raffinose during plant adaptation to long days.
INTRODUCTION
Cell growth is a fundamental energy-consuming process that needs to be tightly coordinated with nutrient availability and other environmental stimuli to preserve cellular homeostasis. In plants, identification of central key regulators that integrate exogenous signals, such as light, water, stress, or the presence of nutrient, and adjust plant metabolism and morphogenesis to optimize development and enhance survival is a challenging goal. The Target of Rapamycin (TOR) pathway, which is conserved among all eukaryotes, is now well known in animals and yeast as a central regulator of growth in response to environmental cues (reviewed in Wullschleger et al., 2006; Shaw, 2008; Soulard et al., 2009; Moreau et al., 2010). Indeed, this signaling pathway integrates hormonal and nutritional information to translate them into growth, developmental, and metabolic decisions. The TOR kinase is a large protein (molecular mass of ~250 kD) that belongs to the phosphatidyl inositol kinase-related kinase family and is essential in all eukaryotic organisms. TOR associates in complexes with other protein partners to regulate, in response to environmental signals (nutrient availability, stress, or growth factors), various cellular processes like translation, transcription, ribosome biogenesis, autophagy, actin organization, and metabolic adaptation (Wullschleger et al., 2006). In yeast and animals, there are two TOR complexes (TORC1 and TORC2) influencing different functions in cells. TORC1 is composed of three major proteins: TOR, KOG1/RAPTOR, and LST8/GβL (for Lethal with Sec Thirteen 8/G protein β subunit-like), whereas TORC2 is composed of TOR, LST8/GβL, and AVO3/RICTOR (Hara et al., 2002; Kim et al., 2002; Loewith et al., 2002). Other than TOR, the small 34-kD LST8/GβL protein is the only one that is common to both complexes.
LST8 was first identified in yeast through a screen for mutations that show synthetic lethality with alleles of sec13, a gene involved in endoplasmic reticulum–to–Golgi transport and also required for the regulated transport from the Golgi to the plasma membrane of Gap1, a general amino acid permease (Roberg et al., 1997). The LST8 protein contains seven WD 40 repeats, which are implicated in a wide range of functions like signal transduction and vesicular trafficking (Neer et al., 1994), and its primary sequence shows similarity with those of heterotrimeric G protein β-subunits (Kim et al., 2003). These WD repeats result in the formation of a stable, propeller-like platform allowing interactions with several protein partners (Smith et al., 1999). At the same time, LST8 was identified in fission yeast as Wat1p, a protein partner of Prp2, the large subunit of the U2AF essential splicing factor (Kemp et al., 1997). Mutations in Wat1p caused diploidization, genome instability, and decreased level of α-tubulin transcripts (Ochotorena et al., 2001). In yeast, LST8 is a negative regulator of the retrograde (RTG) signaling pathway that mediates responses to mitochondrial dysfunction and to the presence of poor nitrogen sources by readjusting carbon and nitrogen metabolism through nuclear translocation of the heterodimeric transcription factors Rtg1/3 (Liu et al., 2001). Interestingly, the inactivation of TOR activity by rapamycin also resulted in nuclear accumulation of Rtg1/3 (Komeili et al., 2000). More recently the LST8 protein was involved in connecting the TOR and RTG signaling pathways, and some aspects of the LST8-dependent RTG responses could be separated from TOR activity (Giannattasio et al., 2005). In yeast, some of the effects of lst8 mutations were ascribed to an intracellular accumulation of amino acids and to a partial inhibition of the TOR signaling pathway (Chen and Kaiser, 2003). Indeed, lst8 mutants were found to be hypersensitive to rapamycin, an inhibitor of TORC1 functions, and had cell wall defects. In animal cells, the mLST8 protein binds and activates the TOR kinase domain and seems to be required to maintain the TOR–RAPTOR interaction of the TORC1 complex in a nutrient-dependent manner (Kim et al., 2003; Adami et al., 2007). Recently, it has also been shown that mLST8 interacts with IkappaB kinase and that it inhibits the phosphorylation of this kinase by recruiting PP2A and PP6 phosphatases (You et al., 2010). So far, little is known about the impact of LST8 mutations in multicellular organisms. In mouse, it has been demonstrated that embryos devoid of mLST8 (GβL) expression survive for some time and die at a period corresponding to an increase in the vasculature (Guertin et al., 2006). These embryos resembled RICTOR-deficient ones, and the lack of mLST8 protein appeared to affect only the functions of the TORC2 complex without any visible effects on the readouts of TORC1 activity.
In plants there are no clear homologs of the TOR complex 2 (TORC2)–specific components like AVO1/hSIN1 or AVO3/RICTOR. Conversely, there is strong evidence for the existence of a TORC1 complex in both land plants and algae (Mahfouz et al., 2006; Moreau et al., 2010). In Chlamydomonas reinhardtii, the LST8 protein (Cr-LST8), as well as Cr-TOR, were found in high molecular mass complexes that are associated with internal membranes (Díaz-Troya et al., 2008). The same report showed that Cr-LST8 was also localized around the nucleus and near peribasal bodies, close to the flagella. The fact that the expression of the Cr-LST8 coding sequence in yeast complemented lst8 mutations suggests that Cr-LST8 is a functional homolog of the yeast LST8 protein. In C. reinhardtii, the Cr-LST8 protein was shown to copurify with the Cr-TOR protein and to interact, as in other eukaryotes, with its kinase domain (Díaz-Troya et al., 2008). Taken together, these data all support the assumption that a conserved TORC1 protein complex is formed in plants and algae with the TOR, RAPTOR, and LST8 partners.
The loss of TOR or RAPTOR expression is embryo lethal in plants (Menand et al., 2002; Deprost et al., 2005) even if the penetrance of raptor mutation is not complete, allowing the recovery of homozygous mutants under some conditions (Anderson et al., 2005). By studying Arabidopsis thaliana lines either over- or underexpressing the Arabidopsis TOR gene, we previously showed that the level of TOR expression was very well correlated with the size of the plants, the amount of seed produced, and the abundance of polysomes (Deprost et al., 2007). Ethanol-inducible RNA interference (RNAi) lines were also obtained that allow a conditional silencing of TOR (Deprost et al., 2007). When the expression of TOR was abolished by ethanol induction, plants growth was arrested and senescence-linked markers (genes and metabolites) became upregulated. As for RAPTOR, there are two genes in Arabidopsis coding for proteins showing high sequence identities when compared with the LST8 protein sequences from other organisms (At3g18140 and At2g22040 for LST8-1 and LST8-2 genes, respectively).
In this article, we investigated the role and properties of the LST8 proteins in Arabidopsis. We showed that the LST8 protein interacts with the TOR kinase domain and that mutations in the most highly expressed LST8 gene affect Arabidopsis growth and development while impeding plant transcriptomic and metabolic adaptations to long-day (LD) conditions. Our results provide insight into the important role of LST8 in adapting plant metabolism and development to external conditions.
RESULTS
Only One of the Two Arabidopsis LST8 Genes Is Significantly Expressed
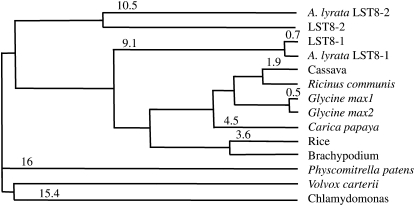
A similarity search of the Arabidopsis translated genome with the yeast LST8 protein sequence revealed that it contains two genes potentially coding for homologs of yeast and animal LST8 proteins, as described earlier (Mahfouz et al., 2006). An LST8-like sequence was also previously reported in cotton (Gossypium hirsutum; Duan et al., 2006). These genes correspond to the predicted At3g18140 and At2g22040 loci in Arabidopsis, which we will name throughout this study LST8-1 and LST8-2, respectively. Both LST8-1 and LST8-2 genes are ~2 kb long and are composed of 10 exons and nine introns. The LST8-1 gene encodes a protein of 305 amino acids with a predicted molecular mass of 34 kD, whereas LST8-2 encodes a 35-kD protein of 312 amino acids. The two proteins are slightly more similar to yeast LST8 (51% sequence identity) than to animal LST8 protein sequences (45% sequence identity with mouse LST8). As in other organisms, both Arabidopsis LST8 proteins contain seven predicted WD 40 repeats, which can form a propeller-like platform structure composed of β-strands (see Supplemental Figure 1 online; Díaz-Troya et al., 2008). The two Arabidopsis LST8 proteins have a high percentage of sequence identity to each other (75%), but, interestingly, other angiosperm LST8 protein sequences are more similar to LST8-1 than is LST8-2 (Figure 1). For example, the rice (Oryza sativa) LST8 protein sequence displays an 80% sequence identity to LST8-1, whereas LST8-2 is only 77% identical. It seems that the LST8 gene was duplicated in the ancestor of Arabidopsis and Arabidopsis lyrata since a sequence closely related to LST8-2 was found only in A. lyrata (Figure 1). The protein corresponding to LST8-2 in A. lyrata seems to also have diverged from other plant LST8 sequences. Numerous ESTs corresponding to the LST8-1 gene are available in databases, but we have not been able to identify specific ESTs or complete cDNAs that would correspond to LST8-2. Moreover, the last exon of the LST8-2 gene has been labeled as nonconfirmed in the TAIR 10 Arabidopsis genome release (www.Arabidopsis.org). Affymetrix microarrays contain only one oligonucleotide set for both LST8 genes, which makes it difficult to assess their expression levels. Conversely, CATMA microarrays (Crowe et al., 2003) carry tags that are specific for each LST8 gene. A survey of all publicly available CATMA chip results (CatDB; Gagnot et al., 2008) did not reveal any detectable expression of LST8-2. It is thus likely, given the divergence of the LST8-2 from other plant LST8 proteins and the lack of detectable expression of its coding sequence, that LST8-2 is a nonfunctional gene. This implies that expression data from Affymetrix microarrays probably mainly reflect the level of LST8-1–derived transcripts. Data from the Genevestigator website (www.genevestigator.com) reveal that LST8-1 is expressed throughout plant development and in all plant organs with a higher level in micropylar and chalazal endosperm (see Supplemental Figure 2 online). Furthermore, LST8-1 expression seems to be higher in aerial parts than in roots.
Figure 1.
Distances between LST8 Protein Sequences.
Tree showing the average distances based on sequence identities between plant and algae LST8 protein sequences using the COBALT multiple alignment tool (Papadopoulos and Agarwala, 2007). The alignment is available as Supplemental Data Set 1 online.
Subcellular Localization and Expression Pattern of the LST8-1 Protein
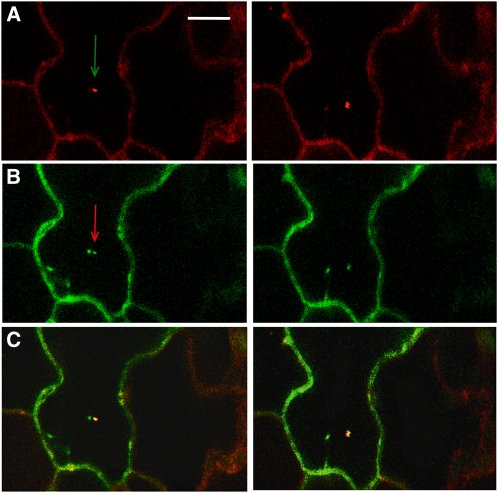
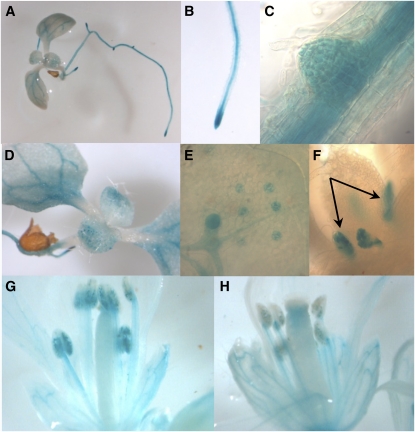
First, we investigated the subcellular localization of the LST8-1 protein by transiently expressing a 35S:LST8-1-green fluorescent protein (GFP) construct in Arabidopsis cotyledons. The fusion protein was mainly detected in mobile dots (Figure 2). To establish the nature of the mobile fluorescent dots, the 35S:LST8-1-GFP construct was introduced in cotyledons of Arabidopsis seedlings expressing a RabC1-red fluorescent protein (RFP) fusion that specifically label the endosomes (Rutherford and Moore, 2002). In most of the cases, the GFP- and RFP-labeled dots were colocalized or at least very close (Figure 2C), indicating that the LST8-1 protein could be associated with endosomes. Next, the LST8-1 expression pattern in Arabidopsis organs was investigated using plants expressing the β-glucuronidase (GUS) reporter gene driven by the LST8-1 promoter and 5′-untranslated region (UTR; 1 kb upstream of ATG). Staining was detected in plantlets at the level of the root central cylinder, the root tip, in emerging lateral roots (Figures 3A to 3C), in cotyledon vasculature (Figures 3A and 3D), and in stomata (Figure 3E). Promoter activity was also observed in leaf stipules (Figure 3F). In flowers, LST8-1 seemed to be also strongly expressed in anthers, in pollen, and in the filament (Figures 3G and 3H; see Supplemental Figure 3 online), as well as in the vasculature of petals and sepals (Figure 3H).
Figure 2.
Localization of a LST8-GFP Fusion Protein after Transient Transformation of Cotyledons from a RabC1-RFP–Expressing Arabidopsis Line.
Each row is derived from a time-lapse series (5-s intervals). The arrows indicate mobile dots. Scale bar = 10 μm.
(A) RFP-specific fluorescence from the RabC1-RFP construct that labels the endosomes.
(B) GFP-specific fluorescence from the 35S:LST8-GFP construct.
(C) RFP and GFP signals were merged.
Figure 3.
GUS Staining of Transformed Arabidopsis Plants Carrying a pLst8:GUS Construct Containing 1 kb of LST8-1 Promoter.
Plantlet (A), primary root tip (B), emerging secondary root (C), aerial part (D), close-up on a leaf showing staining of stomatal guard cells (E), emerging leaves and stipules ([F], indicated by arrows), and flowers ([G] and [H]).
The Arabidopsis LST8-1 Gene Can Complement Yeast lst8 Mutants
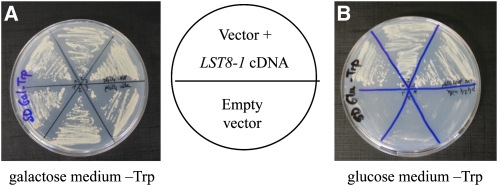
To test the conservation of LST8 function between yeast and Arabidopsis, we tried to complement a yeast lst8 mutant strain. Since LST8 is an essential gene in yeast, the mutant strain we used expressed the yeast LST8 gene under the control of a Gal-inducible promoter (Loewith et al., 2002). This strain can grow only on a Gal-containing medium, and, on Glc, a rapid growth arrest of the lst8 mutant is observed. We expressed the Arabidopsis LST8-1 coding sequence in this strain under the control of a constitutive promoter (see Methods for details) and tested the ability of transformants to grow on Glc as well as on the Gal control medium (Figure 4). Yeast expressing the LST8-1 coding sequence was able to grow normally on Glc medium, whereas yeast transformed with the empty expression vector alone grew only on Gal medium. This demonstrates that the Arabidopsis LST8 protein is a functional homolog of yeast LST8 and that it can perform the same functions as the yeast protein. This is of interest given the known role of this protein in the regulation of the TOR complexes.
Figure 4.
Complementation of a Yeast lst8 Mutant with the Arabidopsis LST8-1 cDNA.
A yeast lst8 mutant strain expressing the Saccharomyces cerevisiae LST8 cDNA under the control of an inducible Gal promoter was used for complementation studies. On a permissive, Gal-containing medium, the yeast lst8 mutant strain is able to grow (A), but on selective, Glc-containing medium, the yeast lst8 mutant strain containing an empty transformation vector fails to grow ([B], bottom part). The expression of the Arabidopsis LST8-1 cDNA in the yeast lst8 mutant strain fully restores the ability to grow on Glc medium ([B], top part).
[See online article for color version of this figure.]
Arabidopsis LST8-1 Interacts with the TOR FRB and Kinase Domains
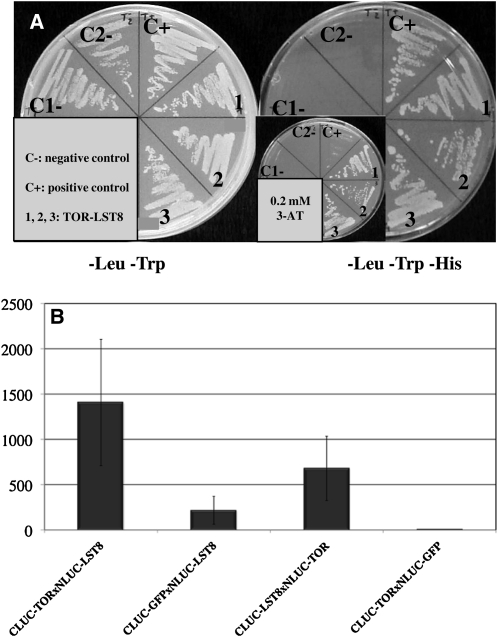
The yeast two-hybrid system was first used to test whether Arabidopsis LST8-1 can interact with the TOR FRB and kinase domain (FK domain) as described in animal cells (Kim et al., 2003). A yeast strain expressing the Arabidopsis LST8-1 and TOR FK proteins as prey and bait, respectively, was able to grow under selective conditions lacking His (Figure 5A). Control strains expressing either the bait or the prey protein together with the corresponding empty vector failed to grow on the same medium.
Figure 5.
Arabidopsis LST8-1 Interacts with the C-Terminal FRB-Kinase Domain of TOR.
(A) Yeast two-hybrid assay with the TOR FRB-kinase domain as bait and the LST8-1 protein as prey. C1-, pADH::GAL4BD pADH::GAL4AD-LST8; C2-, pADH::GAL4BD-FRBK pADH::GAL4AD; C+, pADH::GAL4BD-HSD1pADH::GAL4AD-GAPC2; 1, 2, 3, independent double yeast transformants with pADH:GAL4BD-TOR/FRB and pADH:GAL4AD-LST8-1.
(B) Split-luciferase assay in Arabidopsis cotyledons after transient expression. Relative light emission of the different split-luciferase protein pairs. Luciferase (LUC) activity was monitored with at least two independent infiltration experiments per tested interactions. The mean of the experiments is shown together with the corresponding sd values.
To confirm this interaction in planta, we used a split-luciferase system in which the two proteins to be tested are fused to either the N- or C-terminal parts of firefly luciferase as we described previously (Van Leene et al., 2010). After transient expression in Arabidopsis cotyledons of the two fusion proteins, we measured light emissions from 6 to 10 plantlets grown in vitro in multiwell plates (Figure 5B). Although light levels were quite weak, a reproducible signal was observed when the LST8-1 protein was fused to luciferase N- or C-terminal parts and expressed together with the TOR FK domain. Negative controls expressing the LST8-1 or TOR FK proteins together with GFP gave much lower light signals (Figure 5B). Collectively, these results strongly suggest that the LST8-1 protein interacts with the TOR FK domain in Arabidopsis.
Disruption of the LST8-1 Gene Affects Plant Growth and Development
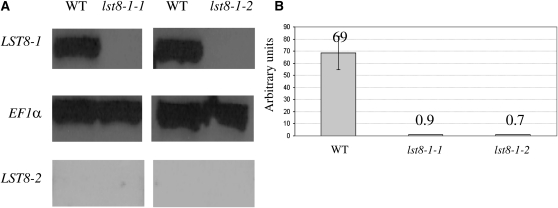
Next, we investigated further the role of Arabidopsis LST8-1 by studying the consequences of mutations in the corresponding gene. We isolated two LST8-1 homozygous mutants named lst8-1-1 (Salk collection; Alonso et al., 2003) and lst8-1-2 (SAIL collection) with T-DNA insertions in the 4th exon and 5th intron of the gene, respectively (see Supplemental Figure 4 online). No full-length LST8-1 transcripts were detected in these mutants by RT-PCR (Figure 6). Homozygous null mutants were also obtained for the LST8-2 gene, which displayed no visible phenotype. Expression of the LST8-2 gene was not detected by RT-PCR in the wild type or in the mutant lst8-1 background (Figure 6A). This confirms that LST8-2 is not, or is only very weakly, expressed in Arabidopsis plants.
Figure 6.
Impact of T-DNA Insertions on Transcription of the LST8 Genes.
(A) Analysis of LST8-1 and LST8-2 expression levels in lst8-1-1 and lst8-1-2 mutants by RT-PCR. The reference constitutive gene is EF1a (Elongation factor 1a). See Methods for details. WT, wild type.
(B) Analysis of LST8-1 expression level in lst8-1-1 and lst8-1-2 mutants by quantitative real-time RT-PCR. Arbitrary units are calculated relative to the EF1a expression level. Values are the mean of at least three independent repetitions ± sd.
Under controlled short-day (SD) conditions (8 h of light), lst8-1-1 and lst8-1-2 homozygous mutants showed a reduction in growth compared with the wild type and did not flower (Figure 7A). By contrast, the mutant plants did not display defects in rosette development. When plants were transferred to LD conditions (16 h of light), growth of the mutants was much more retarded compared with the wild type than under control SD conditions both in vitro (Figure 7B) and when grown in soil (Figures 7C and 7D). In the most severe cases, lst8-1 mutants stopped growing and started to yellow and died after 2 weeks, without producing flowers (Figure 7D). When the plants survived, they became bushy and developed multiple apical meristems (Figures 7E and 7F). The severity of the phenotype depended on the developmental stage of the plant when transferred to LD conditions and on the quantity of light received by the plants. Indeed, an early transfer of SD-grown plants resulted in decreased survival and a stronger phenotype in LDs. Both lst8-1 mutants exhibited the same growth defects under LD conditions. In the growth chamber, or in the greenhouse in summer or spring with high levels of outside light, mutant plants were particularly affected by LD periods. In winter, even if the LD conditions were maintained with artificial light in the greenhouse, lst8-1 mutants showed a milder response to extended light periods. Indeed, mutant plants displayed defects in leaf development but continued to grow, and after a few weeks under permissive LD conditions, lst8-1 mutants became bushy and developed multiple apical meristems (Figures 7E and 7F). Under these growth conditions, half of the plants stayed at the vegetative stage for several months without developing inflorescences. The other half of the mutant plants started to flower while producing a higher number of stems from the rosette than the control wild type (Figure 7). However, the organization of the stem was affected (revealing a loss of apical dominance) with axillary buds larger than the wild-type ones (see Supplemental Figure 5 online). The emerging flower buds also presented an abnormal development (see Supplemental Figure 5 online). Indeed, the flowers remained closed, with no emergence of petals as the carpel started to elongate. Afterwards, the flowers often generated very small siliques containing aborted seeds. In one case, we were able to obtain viable seeds from one lst8-1-1 mutant plant grown in a growth chamber. These seeds germinated normally and produced plants similar to the parent homozygous mutant. It thus seems that growth conditions are important for seed production in the lst8-1 mutants.
Figure 7.
Phenotype of the lst8-1 Insertion Mutants.
(A) to (D) The control wild-type plants are on the left, and the lst8-1-1 mutant plants are on the right.
(A) Plants cultivated in growth chambers for 4 weeks under SD conditions.
(B) Plants grown in vitro for 7 d under LD conditions.
(C) Plants cultivated in the greenhouse for 6 weeks under LD conditions (winter).
(D) Plants cultivated for 4 weeks under SD conditions as in (A) followed by 1 week under LD conditions.
(E) and (F) lst8-1-1 mutant grown in the greenhouse under LD conditions. lst8-1 mutants develop multiple meristems ([E], indicated by red arrows), become bushy, and produce several stems (F).
The lst8-1-1 and lst8-1-2 mutants were transformed with a pLST8-1:LST8-1 construct, where the LST8-1 genomic DNA was driven by 1 kb of its own promoter. The wild-type phenotype could be restored in the obtained transformants, and this construct complemented the mutant phenotype under both SD and LD conditions (see Supplemental Figure 6 online).
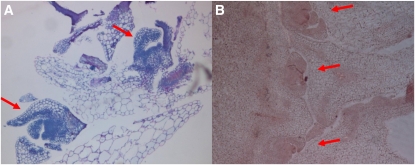
We then investigated in more detail the cause of the lst8-1 mutants’ bushy phenotype. lst8-1 meristems were embedded in resin or paraffin, cut in sections, and observed by microscopy. This revealed that lst8-1 mutants presented not one but multiple apical meristems that appeared correctly organized and functional (Figure 8). These meristematic structures derive most probably from activated axillary meristems, which could explain the production of multiple stems and the bushy phenotype that was observed in these mutants.
Figure 8.
Development of Multiple Meristems in lst8-1-1 Mutant Plants.
Sections of the apical meristem zone were performed and observed after resin embedding and NBB staining (A) or paraffin embedding and Schiff reagent staining (B). Arrows indicate multiple meristems.
[See online article for color version of this figure.]
Metabolite and Enzyme Analysis of lst8-1-1 and lst8-1-2 Mutants Reveals an Impaired Adaptation to LD Conditions
The LST8 and TOR proteins have been implicated in the regulation of several metabolic pathways. In yeast, it has been demonstrated that under nitrogen-rich conditions, LST8 represses transcription factors that promote the expression of genes implicated in amino acid synthesis. Accordingly, these genes are constitutively expressed in yeast lst8 mutants, resulting in an accumulation of amino acids (Liu et al., 2001; Chen and Kaiser, 2003).
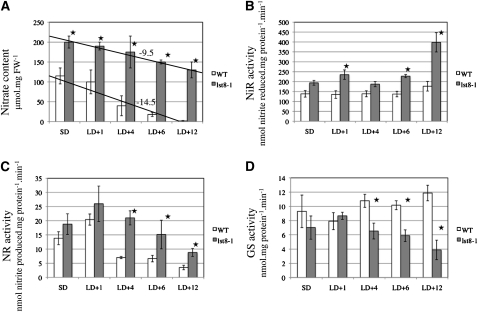
This prompted us to investigate the nitrate assimilation pathway in lst8-1 mutants under LD conditions. After transfer from SDs to LDs, a decrease in nitrate contents was observed in the leaves of wild-type plants with almost no nitrate left after 12 d of growth in LDs (Figure 9A). By contrast, the amount of nitrate stored in lst8-1 mutants was already higher in SDs and declined only slightly in LDs. Enzymatic activities of the two first enzymes involved in nitrate assimilation, namely, nitrate and nitrite reductases (NR and NiR, respectively), were always higher in leaves of the mutant plants when compared with the wild type (Figures 9B and 9C). NR activity showed a transient increase just after transfer to LD conditions and decreased afterwards, whereas NiR activity was more constant, except for in the mutant plants 12 d after transfer, where NiR activity was increased. Interestingly, Gln synthetase activity diminished in the mutant plants, whereas it increased in wild-type leaves (Figure 9D).
Figure 9.
Influence of Long-Day Conditions on Nitrogen Assimilation in lst8 Mutants.
Nitrate content (A), NiR (B), total NR (C), and Gln synthetase (GS; [D]) activities after transfer to LD conditions of wild-type and lst8-1-2 mutant plants. Plants were grown under controlled conditions. Values are the mean of at least three independent repetitions ± sd. The values for nitrate concentrations (A) have been fitted to a regression line, and the corresponding slope is indicated. Statistically different values between the wild type and lst8-1-2 are indicated by a star (Student’s t test). FW, fresh weight; LD+n, number of days under LD conditions.
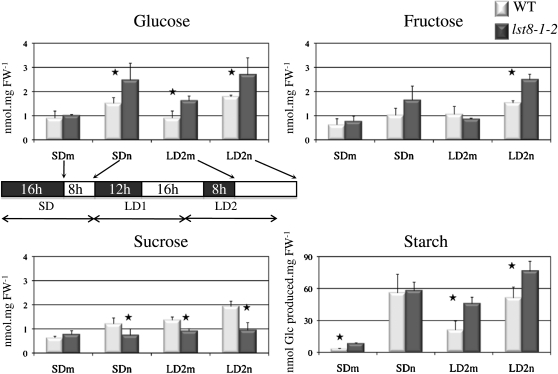
Sugar concentrations were shown to vary after exposure of Arabidopsis to LDs (Corbesier et al., 1998). Therefore, we measured concentrations of soluble sugars and starch at the beginning and at the end of the day during an SD-to-LD transition. Glc and Fru concentrations in leaves did not show major changes after transfer to LDs but were significantly higher in the lst8-1-2 mutant at the end of the day (Figure 10). Conversely, Suc was less abundant in the mutant at the end of SDs or at the beginning of LDs. Suc concentrations increased in LDs, but the difference between the mutant and the wild type was unchanged (Figure 10). Although the amount of starch was similar in wild-type and mutant lines at the end of a SD, it significantly increased in the lst8-1-2 mutant under LD conditions. Similar results were obtained with the lst8-1-1 mutant (see Supplemental Figure 7 online). This suggests that the lst8-1 mutants are unable to adjust sugar and starch metabolism properly in response to an abrupt transition to LDs.
Figure 10.
Diurnal Variations in Soluble Sugar and Starch Content during the Transition from SDs to LDs in the Wild Type and the lst8-1-2 Mutant.
Plants were first grown under controlled SD conditions and harvested at the beginning (morning [m]) and end (night [n]) of the day preceding the shift to LD. Plants were again harvested at day 2 after the start of LD conditions. Results are mean of at least three different samples ± sd. Statistically different values between the wild type (WT) and lst8-1-2 are indicated by a star (Student’s t test). FW, fresh weight.
lst8-1 Mutants Show Faster Movements of a Phloem Tracer
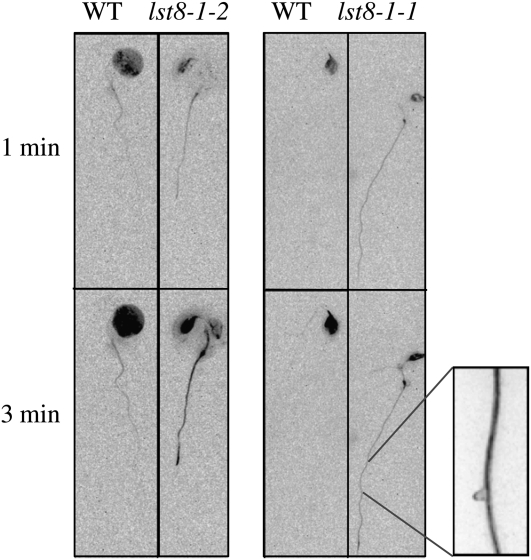
Sugar concentration in leaves is influenced by sugar export through the phloem to sink organs. Moreover, it has been shown previously that phloem transport is affected by transitions to LDs (Gisel et al., 1999, 2002). Several fluorescent tracers can be used to monitor phloem fluxes. For instance 6(5)-carboxyfluorescein (CF) diacetate can be loaded into leaf cells and the impermeant CF moiety can move only after loading into the phloem (Roberts et al., 1997). Loading of one cotyledon from in vitro–grown plants with CF allowed us to follow phloem flux and tracer unloading in roots (Figure 11). We used a segregating population from lst8-1 heterozygous mutants to compare the appearance of CF fluorescence in the phloem of connected organs. The genotype of the plants was determined by PCR analysis. A kinetic experiment after CF labeling revealed that fluorescence appeared more rapidly in the roots and cotyledons of lst8-1 mutants than in the wild type (Figure 11). This suggests that phloem loading and/or transport is more active in lst8-1 mutants.
Figure 11.
Kinetic Analysis of Phloem Labeling Using CF Diacetate.
CF was applied on cotyledons of plants grown in vitro under LDs. Fluorescence was recorded every 3 s. Magnification shows the labeling of conductive tissues inside the root. WT, wild type.
Global Metabolite Profiling of lst8-1 Mutants
To explore more globally the impact of lst8-1 mutation on metabolism, we performed an analysis of metabolites by gas chromatography coupled to mass spectrometry (GC-MS) using leaves from the lst8-1-1 and lst8-1-2 mutant plants as well as wild-type plants grown under SD or LD conditions. Under SD growth conditions, lst8-1-1 and lst8-1-2 presented a higher level of several amino acids, such as Gln, Pro, and Ala (see Supplemental Figure 8A online). The quantity of soluble protein was found to be similar between lst8-1 mutants and wild-type plants (11.8 and 12 μg·mg−1 fresh weight, respectively). By contrast, it was found that lst8-1 mutants accumulated almost twofold more ammonium than wild-type plants (2.8 versus 1.6 μmol·mg−1 fresh weight, respectively).
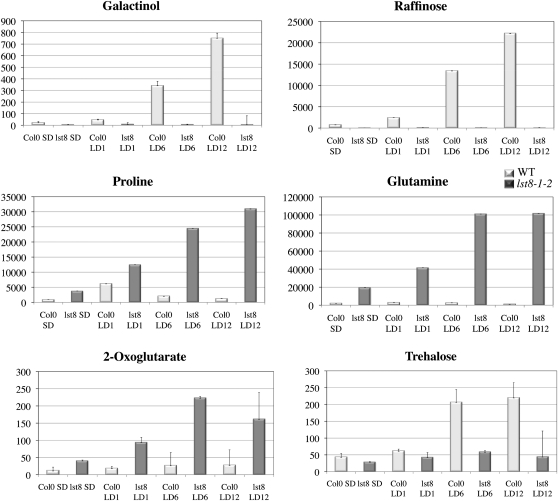
Metabolite profiling analysis also showed that lst8-1-1 and lst8-1-2 mutants underwent profound metabolic perturbations when plants were transferred from SD to LD conditions. The increase in amino acid content that was detected in mutants when grown under SD conditions was dramatically amplified under LD growth conditions in lst8-1-1 and lst8-1-2 compared with the wild type (see Supplemental Figures 8B, 9, and 10 online). In particular, Pro, γ amino-butyric acid, and Gln levels increased after transfer to LDs to more than 20, 25, and 100 times the amount measured in wild-type plants, respectively (Figure 12; see Supplemental Figure 9 online). There was also an increase in Gln and other amino acid levels when we compared lst8-1 mutants grown in LDs to mutants grown in SDs (Figure 12; see Supplemental Figure 10 online). Wild-type plants presented a smaller increase in amino acids (see Supplemental Figure 11 online). For example, there was a transient increase in Pro after transfer to LDs, but the Pro concentration later returned to the level found in SDs. Conversely, lst8-1 mutants seem unable to control this accumulation of Pro under LD conditions, which continued progressively as most mutant plants were starting to senesce (Figure 12). Similar accumulations of Gln, Pro, and γ amino-butyric acid but also of Leu and Ala were observed in both the lst8-1 mutants and in TOR-inducible RNAi lines (Deprost et al., 2007; see Supplemental Figure 12 online), which supports the role of LST8 in regulating TOR kinase activity.
Figure 12.
Leaf Metabolite Contents after Transfer to LD Conditions of Wild-Type and lst8-1-2 Mutant Plants.
Values are derived from normalized areas of specific peaks after GC-MS experiments (see Methods for details). Plants were grown in controlled growth chambers. Values are the means of three independent repetitions ± sd. Darker bars correspond to the mutant plants. LDn, number of days under LD conditions (16 h light); SD, 8 h light.
Concerning organic acids, there was also a strong increase in the levels of malate, succinate (see Supplemental Figure 9 online), and 2-oxoglutarate (Figure 12) in lst8-1 mutants and in TOR RNAi lines (see Supplemental Figure 12 online). The higher level of 2-oxoglutarate, the C-skeleton used for N assimilation, is consistent with an increased amount of Gln. Finally, after 12 LDs, nearly all metabolites were increased in lst8-1 mutant, which is often indicative of cell death.
When wild-type plants were transferred to LDs, we observed a dramatic increase in the amounts of trehalose, galactinol, and raffinose stored in the leaves (Figure 12; see Supplemental Figure 11 online). There was also an increase in myo-inositol but to a lower extent (see Supplemental Figure 11 online). Galactinol is made from UDP-Gal by addition of myo-inositol. Raffinose is a trisaccharide synthesized by the addition of a Gal moiety donated by galactinol to Suc. Raffinose and galactinol accumulate in response to various stresses, including high light irradiance (Nishizawa et al., 2008). Metabolite profiling clearly reveals a lack of myo-inositol, galactinol, and raffinose accumulation in lst8-1 mutants when shifted to LD conditions (see Supplemental Figure 8 online; Figure 12). Under SD conditions, the myo-inositol level in wild-type plants was already 3 to 4 times higher than those in lst8-1-1 and lst8-1-2 mutants (see Supplemental Figure 8A online). However, upon shifting to LD conditions, myo-inositol (see Supplemental Figures 8B, 9, and 10 online), galactinol, and raffinose (Figure 12) remained very low in the lst8-1 mutants, whereas it strongly increased in wild-type plants. In LDs, wild-type plants also accumulated minor sugars like trehalose, whereas lst8-1 mutants did not (Figure 12). Interestingly, dark-treated seedlings of the TOR-inducible RNAi lines also show a reduced level of both galactinol and raffinose (see Supplemental Figure 12 online). It thus appears that in the absence of LST8-1, Arabidopsis plants exposed to LDs are unable to induce the raffinose biosynthetic pathway and the production of myo-inositol, which is needed for important signaling and metabolic pathways.
Transcriptomic Analyses of lst8-1 Mutants in SD and LD Conditions
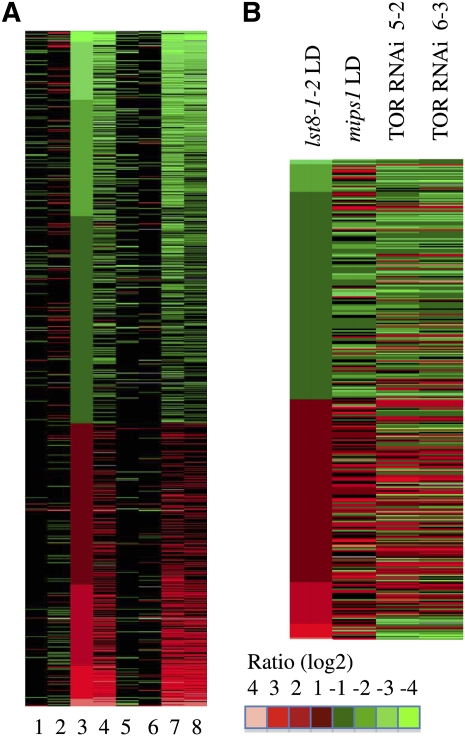
To analyze variations in global gene expressions in the lst8-1 mutants upon transfer to selective LD conditions, both lst8-1-1 and lst8-1-2 mutant as well as wild-type plants were grown in SD conditions until they reached the seven- to eight-leaf stage. Subsequently, some plants were transferred to LD conditions for 2 d, and three plants were harvested for each condition and each genotype to compare transcriptome variations. Global expression profiles were determined using RNA isolated from either SD or LD conditions, and each lst8-1 mutant was compared with the wild type grown in the same light regime. Columbia-8 (Col-8) was used as a reference for the lst8-1-1 mutant (Salk library) and Col-0 for lst8-1-2 (SAIL library). mRNAs from each sample were extracted, amplified, and hybridized on CATMA microarrays (Lurin et al., 2004; see Methods for details). There were fewer up- and downregulated genes in mutant plants grown in SDs than there were under LD conditions (Figure 13A). The genes that are downregulated in the mutants grown in SDs were mainly categorized in abiotic and biotic stress responses (see Supplemental Figure 13 online). The two lst8-1 mutant lines together with the corresponding wild type were grown in independent experiments. Transcriptome comparisons between the two wild-type ecotypes cultivated in either SDs or LDs reveals a good match between the two experiments; indeed, ~70% of differentially expressed genes were in common between the two wild-type samples (401 out of 561 genes for Col-0 and 624 for Col-8; Figure 13A). Similarly, 50% of the differentially regulated genes in the lst8-1-1 mutant grown in SDs were also detected in the other mutant line. After transfer to LDs, only 25% of the differentially expressed genes were conserved between the two mutant lines, which probably reflects the wide range of phenotypes displayed by the mutants in these conditions and possibly the different genetic background of the two mutant lines. Therefore, we decided to focus our analysis on genes that were found to be statistically differentially expressed in both lst8-1 mutant lines. We also verified that, for a few genes, quantitative RT-PCR would confirm the microarrays data (see Supplemental Figure 14 online). Furthermore, ~20% of differentially expressed genes in LD-grown lst8-1 mutants were also differentially expressed after induction of TOR silencing (Figure 13B). Genes that were found to be downregulated in lst8-1 mutants transferred to LDs mainly belong to stress response genes (see Supplemental Figure 15 online). Accordingly, we found that the lst8-1-1 mutant showed a higher sensitivity to osmotic stress, similar to the TOR RNAi line (Deprost et al., 2007; see Supplemental Figure 6 online). By contrast, there was no significant effect of moderate salt stress. Upregulated genes corresponded to genes involved in energy generating pathways or coding for ribosomal proteins (see Supplemental Figure 16 online).
Figure 13.
Differentially Expressed Genes in the Transcriptomic Analysis of lst8-1 Mutants Using CATMA Arrays.
For each condition, gene expression in the mutant samples was compared with that in wild-type samples grown under the same light regimes as references. 1, lst8-1-2 to wild type in SD; 2, lst8-1-2 to wild type in LD for 2 d; 3, wild type in LD to wild type in SD (reference); 4, lst8-1-2 in LD to lst8-1-2 in SD; 5 to 8, same as 1 to 4, except with the lst8-1-1 mutant.
(A) Differentially expressed genes were ordered from the lowest to the highest ratio with the wild type LDs (LD after 2 d) to SDs comparison as reference (see Methods for the definition of differentially expressed genes).
(B) Differentially expressed genes in the comparison between lst8-1-2 and wild-type grown under LD conditions, which were also differentially expressed in a mips1 mutant compared with wild-type plants grown in LD and in TOR ethanol-inducible RNAi lines induced by ethanol for 24 h (see Methods for details). Only genes that are found in common between at least two comparisons were retained for this analysis. Data were obtained from the CatDB database and from Meng et al. (2009).
Mutants affected in the MYO-INOSITOL1 PHOSPHATE SYNTHASE (MIPS1) gene were also previously described as lacking galactinol accumulation and being sensitive to LD conditions (Meng et al., 2009). This prompted us to compare the transcriptome of mips1 and lst8-1 mutants. Interestingly, out of 317 genes differentially expressed in the lst8-1-2 mutant grown in LDs, 217 (68%) were also found to be either down- or upregulated in the mips1 mutant in LDs when compared with wild-type plants (Figure 13B). This indicates that a large proportion of the impact of lst8-1 mutations on the transcriptome of LD-grown plants can be explained by a default in MIPS1 activity. This gene has been identified as being closely correlated with biomass accumulation and carbon perturbations (Sulpice et al., 2009).
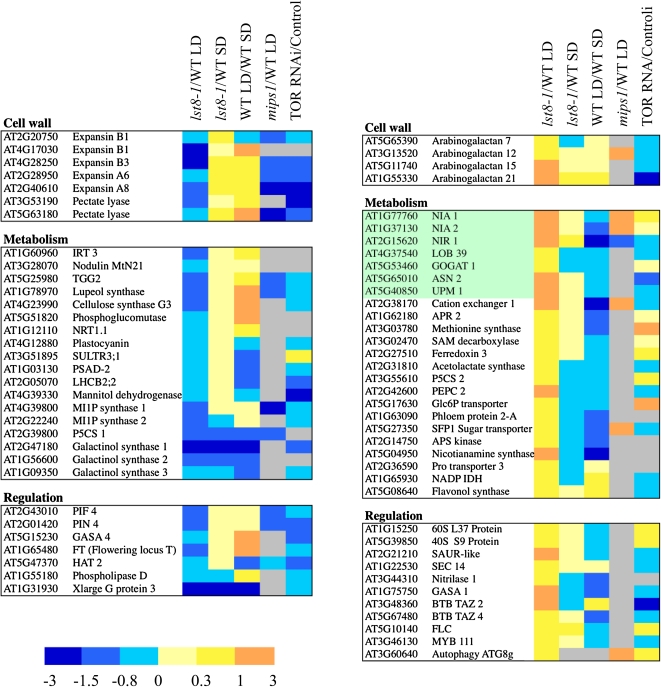
Next, we investigated the genes that are either up- or downregulated in both lst8-1 mutants after transfer to LDs. To better explain the increased penetrance of the lst8-1 mutation in LD conditions, we focused our analysis on differentially expressed genes that show different trends in SDs or LDs (Figure 14). Among genes that were downregulated in the mutants, we found several genes involved in cell wall formation, such as expansin genes and CELLULOSE SYNTHASE-LIKE G3. The gene encoding phospholipase D, an enzyme involved in the synthesis of phosphatidic acid, was also downregulated in lst8-1 mutants (Figure 14). Interestingly, it was shown that the overexpression of phospholipase Dα3 resulted in increased TOR activity in Arabidopsis (Hong et al., 2008). As expected, MIPS1 and MIPS2 were specifically repressed in the mutants grown in LDs, whereas GALACTINOL SYNTHASE1, 2, and 3 were downregulated in lst8-1 mutants in SDs and LDs. These results are in agreement with the observed lack of galactinol and raffinose accumulation in mutant leaves. The downregulation of the PIF4, a phytochrome-interacting transcription factor involved in photoperiodic regulations, and PIN4 proteins is also noteworthy (Figure 14). Interestingly, these genes were also repressed in the mips1 mutant and in inducible TOR RNAi lines but were found to be upregulated when wild-type plants were transferred to LDs, which suggests that they may be involved in the adaptation to these conditions (Figure 14).
Figure 14.
Differentially Expressed Genes in lst8-1 Mutants.
Transcriptome comparisons were performed between leaves of lst8-1 mutants and wild-type plants grown either in SDs (8 h) or transferred to LDs (16 h) between wild-type plants grown in SDs and transferred to LDs and between mips1 mutants and the corresponding wild type in LDs, and between the TOR ethanol-inducible RNAi lines and the corresponding control line (mean of 6-3 and 5-2 RNAi lines induced for 24 h with ethanol). Genes showing opposite variations when compared with the wild type in SDs or LDs, or of special interest, were selected among differentially expressed transcripts in the lst8-1 mutants. The results are the mean of the intensity ratios for lst8-1-1 and lst8-1-2 mutants and are presented as log2 ratios. Experiments were run in duplicate. A color code was used to visualize the data.
A substantial number of genes that were upregulated in the mutants compared with the wild type after transfer to LDs were involved in either nitrate or sulfur assimilation (Figure 14). These genes include adenosine 5′-phosphosulfate kinase and reductase 2, NR, NiR, the nitrate-specific transcription factor LOB39, ASPARAGINE SYNTHETASE2, and uroporphyrin methylases (UPM1), which are involved in biosynthesis of siroheme, a cofactor found in nitrite and sulfite reductases. The P5CS2 gene involved in Pro biosynthesis was also upregulated. In addition, the expression of PEPC2 and IDH, which are involved in the synthesis of organic acids and of carbon skeletons needed for the production of organic nitrogen, was also clearly induced (Figure 14). This transcriptome analysis is in line with the observed accumulation of amino acids, especially Gln and Pro, after transfer to LDs and with the higher levels of NR and NiR activities that were observed in lst8-1 mutants. Most of these genes were found to be repressed in wild-type plants transferred to LDs, which is in agreement with the observed decrease in nitrate assimilation and N metabolism. We also observed a specific increase in the mRNA level of the BTB-TAZ2 (for BRIC-À-BRAC, TRAMTRACK AND BROAD-TRANSCRIPTIONAL ADAPTOR ZINC FINGER2) and BTB-TAZ4 genes, which were previously shown to be induced by nitrate but repressed by sugars (Mandadi et al., 2009). The inhibition of the TOR kinase results in the induction of autophagy (Wullschleger et al., 2006). Interestingly, the expression of a known marker for the formation of autophagic vesicles (ATG8) is induced in the lst8-1 mutant, in the TOR RNAi lines, and in the mips1 mutant.
It is known that LDs induce flowering in Arabidopsis (Bernier and Périlleux, 2005). Indeed, expression of the flowering inducer gene Flowering Locus T (FT) is augmented after exposure of Arabidopsis plants to LD growth conditions. As expected, FT expression increased fourfold in wild-type plants after transfer from SD to LD conditions but much less in lst8-1 mutants (log2 ratio of 0.3 and 0.1 for, respectively, lst8-1-1 and lst8-1-2 mutants as determined by quantitative RT-PCR). Accordingly, Flowering Locus C, which represses flowering by regulating FT (Greenup et al., 2009), shows the opposite trend. These results could contribute to the delayed onset of flowering in LD-grown lst8-1 mutants.
DISCUSSION
In this article, we show that Arabidopsis LST8 plays an important role in growth and organ development as well as metabolic regulation and flowering in response to LDs. This protein is known to be a component of the two TOR complexes in animals and yeast (Wullschleger et al., 2006). In C. reinhardtii, it was also demonstrated that LST8 interacts with the TOR kinase domain (Díaz-Troya et al., 2008). In Arabidopsis, there are two genes potentially coding for homologs of the LST8 proteins, and only one of them (LST8-1, At3g18140) seems to be expressed at significant levels. Interrogation of public EST, transcriptome, and protein databases gave no indication of expression of LST8-2 (At2g22040). In addition, the LST8-2 sequence seems to have diverged from other plant LST8 sequences (Figure 1). It could thus be the result of a recent duplication that is no longer expressed and is therefore relieved from evolutionary pressure. Moreover, we were not able to amplify a cDNA corresponding to LST8-2 by RT-PCR in all tested organs and conditions (Figure 6). Therefore, the vast majority of LST8 activity in Arabidopsis is likely to derive from the expression of the sole LST8-1 gene.
The Arabidopsis LST8 protein, like in other organisms, displays seven conserved WD 40 repeats that can form the typical β-propeller fold of WD 40 domains (Smith et al., 1999). More than 200 proteins in Arabidopsis contain WD 40 repeats, which are thought to be mainly involved in protein–protein interactions, and among these are several important signaling components like Cop1, the major repressor of photomorphogenesis, and the floral regulator FY (van Nocker and Ludwig, 2003).
The localization of a LST8-GFP fusion protein indicates that it is associated with endosomes and mobile vesicles. In yeast, the LST8 protein, together with the TOR kinase, was also found to copurify with endosomes and Golgi particles (Chen and Kaiser, 2003). Similarly, in C. reinhardtii, the LST8 protein was associated with microsomes (Díaz-Troya et al., 2008). Therefore, it seems that the association of LST8 with intracellular vesicles is a common trend in eukaryotes and could be linked to the regulation of trafficking by the TOR kinase (Shaw, 2008).
The analysis of transformed Arabidopsis lines harboring a translational fusion of the LST8-1 promoter and 5′-UTR to the GUS reporter gene shows that LST8-1 is mainly expressed in root meristems and vascular tissues as well as in developing organs like emerging root or leaf primordia. There was also a strong GUS activity in flowers. This is similar to the GUS staining pattern resulting from the expression of a TOR-GUS fusion in Arabidopsis (Menand et al., 2002) and implies that LST8 and TOR have some overlapping domains of expression. Interestingly, strong GUS staining was also detected in stomatal guard cells, which are known to be central regulators of gas exchange processes and were submitted to many metabolic and environmental regulations (Casson and Gray, 2008). Like the C. reinhardtii LST8 gene (Díaz-Troya et al., 2008), the Arabidopsis LST8-1 coding sequence can fully replace the lethal depletion of LST8 activity in a yeast mutant strain. This result shows that LST8 is functionally conserved between Arabidopsis and yeast. Moreover, we have shown that, like in animal cells (Kim et al., 2002), the LST8-1 protein interacts with the Arabidopsis TOR FRB and kinase domains both in yeast and in planta (Figure 5). Unlike yeast (Loewith et al., 2002) and mouse (Guertin et al., 2006), mutations in the LST8-1 gene are not lethal in Arabidopsis. A similar result has been observed in fission yeast (Kemp et al., 1997). Nevertheless, the growth and development of lst8-1 mutants was severely affected with a delay in plant growth, sterility of most flowers, and high sensitivity to LDs. Interestingly, deficiency in the RAPTOR protein, the other component of the TORC1 complex, is also lethal in yeast and mice (Guertin et al., 2006), whereas viable raptor mutants can be obtained in Arabidopsis (Anderson et al., 2005; Moreau et al., 2010). The lethality of lst8 mutations in other eukaryotes could be due to TOR-independent activities, which were recently described (You et al., 2010). The reduced growth of lst8-1 mutants is reminiscent of the effect of partial silencing of the expression of Arabidopsis TOR (Deprost et al., 2007). The lst8-1 mutants have altered flower development, and a recent report linked the overexpression of the RIBOSOMAL S6 KINASE gene, a major readout of TOR kinase activity, to abnormal flower development (Tzeng et al., 2009). The bushy phenotype and the development of multiple meristems in lst8-1 mutants could indicate a role for LST8 in the regulation of apical dominance and meristem cell proliferation. It is noteworthy that a knockdown of the ribosomal protein gene RPL23aA in tobacco (Nicotiana tabacum) produced a pleiotropic phenotype characterized by altered and retarded growth, abnormal phyllotaxy, and loss of apical dominance (Degenhardt and Bonham-Smith, 2008). Indeed, the TORC1 complex has a central regulatory role in ribosome biogenesis and mRNA translation (Wullschleger et al., 2006).
The LD sensitivity and the delay in flowering in lst8-1 mutants may be the results of perturbations in cell adaptation to the length of the light period. One well-documented signal transduction pathway responding to light level is the so-called RTG signaling from chloroplast to nucleus (Leister, 2005; Queval et al., 2007; Fernández and Strand, 2008). Arabidopsis plants normally respond to extension of the light period by adjusting the metabolic and energetic status of the cell and also by inducing flowering (Corbesier et al., 2002; Bernier and Périlleux, 2005; Greenup et al., 2009). In yeast, LST8 has been implicated in vesicular trafficking and in RTG responses linking mitochondrial dysfunction to nuclear gene transcription (Liu et al., 2001; Chen and Kaiser, 2003). In response to upstream signals, LST8 negatively regulates the transcription factors Rtg1/3 responsible for the activation of amino acid synthesis genes in response to environmental conditions. Indeed, yeast lst8 mutants show a strong accumulation of amino acids due to an abolished repression of RTG1/3 transcription factors by Glu (Liu et al., 2001). It was suggested that Glu is sensed by the Ssy1-Ptr3 signaling pathway and that this signal is conveyed to RTG1/3 through the action of TOR and LST8. Interestingly, we observed the same accumulation of amino acids, mainly Gln, in LD-grown lst8-1 mutant plants but also in Arabidopsis plants silenced for TOR expression (Figure 12; see Supplemental Figure 9 online). This accumulation of amino acids could be the result of a stimulation of nitrate assimilation and amino acid synthesis. Indeed, it is known that overexpression of NR causes ammonium and Gln accumulation like that observed in lst8-1 mutants (Lea et al., 2006). Accordingly, transcriptome analysis of lst8-1 mutants transferred to LDs showed a strong induction of several genes involved in the nitrate assimilation pathway, including NR and NiR as well as Asn synthetase (Meyer and Stitt, 2001; Lillo, 2008; Figure 14). In Arabidopsis, silencing of the genes encoding either TOR or TAP46, a target of TOR interacting with PP2A phosphatase, resulted in a decreased NR activity (Deprost et al., 2007; Ahn et al., 2011), whereas there was an accumulation of Gln. It thus seems that inactivation of LST8-1 has the opposite effect on NR activity than on Gln level. It has been previously shown that a shift to LD conditions causes in Arabidopsis a substantial decrease in nitrate concentration and in the expression of genes involved in its assimilation (Corbesier et al., 1998, 2002). Thus, it appears that lst8-1 mutants do not sense properly the shift to LD conditions and do not adjust their nitrogen metabolism accordingly. Furthermore, the induction of the flowering locus FT did not occur in lst8-1 mutants when transferred to LDs. FT is regulated by the expression of the CONSTANS gene and represents one of the most potent activators of flowering, integrating several signaling pathways (Searle and Coupland, 2004). Taken together, these data show that in the absence of LST8, Arabidopsis plants show a delay in the onset of flowering and appear to be impaired in the perception of the extension of light period. Arabidopsis LST8 also seems to be implicated in flower development. Indeed, lst8-1 mutants displayed abnormal and often sterile flowers, with a lack of petal and anther extension and altered flower insertion on the stems (see Supplemental Figure 5 online). Moreover, we showed that LST8-1 is expressed in petal and sepal conducting tissues, in pollen, and in stamen filaments. It was previously described that a decrease in leaf-to-shoot trafficking of phloem symplastic tracers like CF occurs during the transition to flowering in Arabidopsis (Gisel et al., 1999, 2002). The higher phloem loading and/or flux detected in lst8-1 mutants could thus impede the onset of flowering by altering the transport of signaling molecules.
When wild-type plants were transferred to LD conditions, we noticed an accumulation of Suc and of osmoprotectants like raffinose and galactinol, which did not occur at all in lst8-1-1 and lst8-1-2 mutants (Figure 12). Raffinose and galactinol have been shown to be involved in resistance to various stresses, including high light conditions, and to protect plants by detoxifying reactive oxygen species (Taji et al., 2002; Nishizawa et al., 2008; Usadel et al., 2008). When shifted to LDs, wild-type plants displayed a transient increase in Pro concentration, followed by raffinose and galactinol accumulation. It is possible that these sugars replace Pro as osmoprotectants during adaptation to LDs. By contrast, lst8-1 mutants fail to synthesize galactinol and raffinose while showing a continuous increase in Pro. Interestingly, TOR-silenced lines also showed a decrease in the accumulation of galactinol and raffinose when germinated in the dark (see Supplemental Figure 12 online). Therefore, the sensitivity of lst8-1 mutants to LDs may be partly due to the absence of these molecules. The low concentration of myo-inositol found in lst8-1 mutants can explain the lack of galactinol and raffinose production because myo-inositol is needed for the synthesis of these compounds from UDP-Gal (Nishizawa et al., 2008). Interestingly, Arabidopsis mutants affected in the MIPS1 gene had lower levels of myo-inositol and galactinol and showed lesion formation in LD conditions, like lst8-1 mutants (Meng et al., 2009). Furthermore, we found a remarkable overlap between the transcriptome of lst8-1 and mips1 mutants (Figure 13), which suggests that a significant part of the consequences of mutations in the LST8-1 gene can be explained by a lack of myo-inositol synthesis. This is in agreement with the previous identification of MIPS1 as a candidate gene highly correlated with carbon perturbations and growth (Sulpice et al., 2009). Apart from the production of galactinol and raffinose, the lack of myo-inositol also affects signal transduction via a decrease in phosphorylated forms of inositol like inositol 1,4,5-trisphosphate (IP3) and phosphatidylinositol-(4,5)-bisphosphate (PI[4,5]P2) (Xu et al., 2005). lst8-1 mutants share several aspects of their phenotype, like accumulation of amino acid or sensitivity to osmotic stress, with TOR RNAi lines. Moreover, the absence of TOR or LST8 activity results in the differential expression of a common set of genes (Figure 13B). Collectively, these results suggest that the LST8-1 protein, probably by interacting with the TOR kinase complex and regulating its activity, makes a connection between external cues and the regulation of growth and carbon metabolism by affecting the expression of MIPS1.
In conclusion, we have shown that Arabidopsis LST8 is important for growth and developmental processes linked to changes in light conditions probably by influencing the activity of the TOR complex. LST8-1 is needed for the adaptation of primary metabolism to changes in daylength by inducing the synthesis of myo-inositol and of the osmoprotectants galactinol and raffinose and by restraining nitrate assimilation and amino acid accumulation that would exhaust cellular energy stores. This is similar to the role of yeast LST8, the function of which is to lower amino acid production under stress conditions by modulating the TOR kinase activity. Our goal is now to identify the targets and protein partners of LST8 in plants.
METHODS
Plant Material
The lst8-1-1 (SALK_02459) and lst8-1-2 (SAIL_641D10) mutants were obtained from the Nottingham Arabidopsis Stock Centre in a T-DNA–mutagenized population of the Col-8 Arabidopsis thaliana ecotype and from the Syngenta Arabidopsis Insertion Library (SAIL) T-DNA–mutagenized population in the Col-0 Arabidopsis ecotype, respectively. Homozygous mutant plants were identified by PCR using the primers listed in Supplemental Table 1 online. A DNA fragment comprising 1 kb of LST8-1 promoter and the 5′-UTR (1 kb upstream of the gene initiation codon) were amplified with forward and reverse primers containing EcoRI and NotI restriction sites, respectively (primers are listed in Supplemental Table 1 online). The resulting PCR product was cloned into the pENTR4 (Invitrogen) vector after digestion with the same restriction enzymes. The pLst8:GUS construct was then obtained after cloning into Gateway technology–compatible (Invitrogen) pGWB3 binary plasmid. Transgenic plants were obtained by floral dipping (Clough and Bent, 1998) of Col-0 Arabidopsis plants. To complement the lst8-1 mutant, a genomic DNA fragment containing the LST8-1 gene encompassing 1 kb of promoter sequence was amplified by PCR with forward and reverse primers containing a BamHI and a NotI restriction site, respectively (primers are listed in Supplemental Table 1 online) and checked by sequencing. The resulting PCR product was cloned into the pENTR4 vector after digestion with the same restriction enzymes. The pLst8:LST8 construct was then obtained after cloning into Gateway technology–compatible (Invitrogen) pGWB1 binary plasmid. Transgenic plants were obtained by floral dipping of lst8-1-1 and lst8-1-2 heterozygous mutant plants. The ethanol-inducible TOR RNAi lines 5-2 and 6-3, the control alcA:GUS line, as well as the constitutive TOR RNAi line 35-7 were previously described by Deprost et al. (2007). Plants used for either global metabolite profiling or transcriptome analysis were harvested at the beginning of the light period.
Plant Growth Conditions
Seeds were sown in vitro on half-strength Murashige and Skoog medium containing 1% Suc and transferred to soil 7 d after germination. SD and LD conditions were 8 h light/16 h night and 16 h light/8 h night, respectively, in controlled growth chambers (70% relative humidity) with fluorescent tubes and a light intensity of respectively 150 μmol·m−2·s−1 and 130 μmol·m−2·s−1. The greenhouse was used for LDs with natural light, supplemented, according to the season, with artificial light bulbs. For in vitro studies, surface-sterilized seeds were sown on half-strength Murashige and Skoog medium containing 1% Suc. Ethanol-inducible TOR RNAi lines were induced either by direct sowing on half-strength Murashige and Skoog medium containing 0.3% Suc and 50 mM ethanol or by adding an Eppendorf cap containing 50% ethanol for 1 or 2 d on the solid medium. Other details were as described by Deprost et al. (2007).
Subcellular Localization
The LST8-GFP fusion protein was obtained and expressed transiently in Arabidopsis cotyledons as described earlier (Marion et al., 2008). Confocal microscopy was performed on a Zeiss Lsm710 spectral laser scanning microscope. GFP and mRFP1 fluorescence were detected with an emission band of 495 to 540 nm and 600 to 645 nm and excited with an argon laser line at 488 nm and a HeNe laser at 594 nm, respectively.
GUS Staining
Transgenic plants carrying a pLST8-1:GUS construct were grown on horizontal plates for 10 d at 25°C under LD conditions (16 h light/8 h dark). Some of the plants were transferred to soil in growth chamber under LD conditions (16 h light/8 h dark) until flowering. Staining of GUS activity with X-Gluc was performed as described (Menand et al., 2002) with 2 mM ferricyanure (potassium hexacyanoferrate III) and 2 mM ferrocyanure (potassium hexacyanoferrate II).
Yeast Complementation
The LST8-1 coding sequence was amplified by RT-PCR with forward and reverse primers containing SalI and NotI restriction sites, respectively (primers are listed in Supplemental Table 1 online) and checked by sequencing. The resulting PCR product was cloned into the p424GPD yeast expression vector under control of the constitutive glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter (Mumberg et al., 1995). lst8 mutant yeast strain was then transformed with the empty p424GPD vector or with the p424GPD-LST8-1 construct. Transformants were plated on SD Glc-Trp medium or SD Gal-Trp medium and scored for growth. Plates were incubated at 30°C for 3 d.
Two-Hybrid Experiments in Yeast and Split-Luciferase Assays in Arabidopsis Cotyledons
The Lst8-1 full-length cDNA was amplified by PCR as described above. The resulting cDNA was digested with these enzymes and cloned by ligation in a SalI- and NotI-digested pEXPAD-502 plasmid (Invitrogen) containing a GAL4 activation domain. A 2.5-kb DNA fragment containing the TOR FRB and kinase domains was amplified by PCR as described previously (Deprost et al., 2007) and introduced in the pENTR4 plasmid after digestion with EcoRI and NotI. This partial cDNA was then inserted by recombination in the two-hybrid pDEST32 vector (Invitrogen) that contains the GAL4 DNA binding domain. Two-hybrid experiments were performed in yeast using the AH109 strain, and interaction between LST8-1 and TOR proteins was tested on a complete medium lacking Leu, Trp, and His with increasing concentrations of 3-amino triazole as described by the supplier (Matchmaker; Clontech).
Split-luciferase experiments were performed with the same LST8-1 and TOR proteins after cloning of the corresponding DNA fragments in Gateway vectors carrying either the N- or C-terminal parts of firefly luciferase as previously described (Van Leene et al., 2010). This way, we obtained four different vector combinations. Transient transformation of Landsberg erecta Arabidopsis seedlings and detection of luciferase activity were performed as previously described (Van Leene et al., 2010) except that luciferase emission was normalized according to the number of infiltrated plants. Levels of light emissions were measured with an ultra-amplified charge-coupled device camera (Photonic Science) and obtained after integrating 2000 images (Photolite 32 software).
Paraffin or Resin Embedding and Chloro-Naphtol Staining
lst8-1 mutant meristems were dissected, fixed, and embedded in paraffin or resin as described by Macquet et al. (2007). Tissue sections were cut at 8 μm for paraffin-embedded samples and at 4 μm for resin-embedded samples.
Enzymatic Activities
Rosettes of three plants of each genotype were harvested for each time point. NiR, NR, and Gln synthetase activities were measured on leaf tissue as previously described by Lea et al. (2006) and Deprost et al. (2007).
Phloem Labeling
To image phloem transport, the adaxial surface of cotyledons was gently abraded with carborundum, and ~5 μL of CF diacetate (60 μg·mL−1) was applied to the cotyledon surface following the protocol of Roberts et al. (1997). Roots were examined at 3-s intervals between 10 s and 5 min after labeling using a Nikon SMZ 1500 binocular equipped with a fluorescence excitation and detection module.
RNA Extraction
Total RNAs were prepared from shoots and roots using the Trizol reagent (Invitrogen) following the manufacturer’s protocol.
Quantitative RT-PCR
Total RNA (1 μg) was used as a template to perform RT reactions using Moloney murine leukemia virus reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Quantitative RT-PCR reactions were achieved using 2× Mesa Fast qPCR MasterMix Plus for SYBR assay (Eurogentec) following the manufacturer’s protocols. The expression of the EF1α gene (At5g60390) was used as a constitutive reference with primers described previously (Deprost et al., 2007).
Transcriptome Studies
Microarray analysis was performed at the Unité de Recherche en Génomique Végétale (Evry, France) using the CATMA arrays containing 24,576 GSTs corresponding to 22,089 genes from Arabidopsis (Crowe et al., 2003; Hilson et al., 2004). Two independent biological replicates were produced. For each biological repetition and each point, RNA samples were obtained by pooling RNAs from three plants. Leaves were collected from plants at the rosette 3.1 developmental growth stage (Boyes et al., 2001) and cultivated in growth chamber conditions. Total RNA was extracted using leaves according to the supplier’s instructions. For each comparison, one technical replicate with fluorochrome reversal was performed for each biological replicate (i.e., four hybridizations per comparison). The labeling of cRNAs with Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products), the hybridization to the slides, and the scanning were performed as described by Lurin et al. (2004).
Statistical Analysis of Microarray Data
Statistical analysis of each comparison was based on two dye swaps (i.e., four arrays, each containing 24,576 GSTs and 384 controls) and followed the analysis described by Gagnot et al. (2008). For each array, the raw data comprised the logarithm of median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green), and no background was subtracted. An array-by-array normalization was performed to remove systematic biases. First, spots considered as badly formed features were excluded. Then, a global intensity-dependent normalization using the loess procedure was performed to correct the dye bias. Finally, for each block, the log ratio median calculated over the values for the entire block was subtracted from each individual log ratio value to correct print tip effects. To determine differentially expressed genes, we performed a paired t test on the log ratios averaged on the dye swap. A trimmed variance was then calculated from spots that did not display extreme variance. The spots that were excluded were those with a specific variance/common variance ratio smaller than the α-quantile of a χ2 distribution of one degree of liberty or greater than the 1-α-quantile of a χ2 distribution of one degree of liberty with α = 0.0001. The raw P values were adjusted by the Bonferroni method, which controls the family-wise error rate to keep a strong control of the false positives in a multiple-comparison context. We considered as being differentially expressed the probes with a Bonferroni P value ≤ 0.05, as described by Gagnot et al. (2008). The P values corresponding to the differentially expressed genes are available online in the CatDB database (http://urgv.evry.inra.fr/cgi-bin/projects/CATdb/cons_diff.pl?project_id=197andexperiment_id=303).
Metabolite Profiling and Analysis
Rosettes of three plants (seven- to eight-leaf stage) of each genotype in each growth condition were harvested, and 20 μg of powder of each sample was used for extraction. Nitrate concentrations were measured as described previously by Lea et al. (2006). Extraction, derivatization, analysis, and data processing were performed according to Fiehn (2006). Metabolites were analyzed by GC-MS 3 h and 20 min after derivatization. One microliter of the derivatized samples was injected in splitless mode on an Agilent 7890A gas chromatograph coupled to an Agilent 5975C mass spectrometer. The column was an Rtx-5SilMS from Restek (30 m with 10 m Integraguard column). The liner (Restek 20994) was changed before each series of analysis and 10 cm of column was cut. Oven temperature ramp was 70°C for 7 min then 10°C/min to 325°C for 4 min (run length 36.5 min). Helium constant flow was 1.5231 mL/min. Temperatures were as follows: injector, 250°C; transfer line, 290°C; source: 250°C; and quadripole, 150°C. Samples and blanks were randomized. Amino acid standards were injected at the beginning and end of the analysis for monitoring of the derivatization stability. An alkane mix (C10, C12, C15, C19, C22, C28, C32, and C36) was injected in the middle of the queue for external calibration. Five scans per second were acquired.
Raw Agilent data files were converted in NetCDF format and analyzed with AMDIS (http://chemdata.nist.gov/mass-spc/amdis/). An in-house retention indices/mass spectra library built from the National Institute of Standards and Technology, Golm, and Fiehn databases and standard compounds were used for metabolite identification. Peak areas were then determined using the Quanlynx software (Waters) after conversion of the NetCDF file in Masslynx format. Statistical analysis was made with TMEV (http://www.tm4.org/mev.html): Univariate analysis by permutation (one-way and two-way analysis of variance) was first used to select the significant metabolites. Multivariate analysis (hierarchical clustering and principal component analysis) was then performed on them. MapMan (http://www.gabipd.org/projects/MapMan/) was used for graphical representation of the metabolic changes after log2 transformation of the mean of the three replicates.
Bioinformatics
Gene expression data were analyzed using the Genevestigator (www.genevestigator.com), CatDB (Gagnot et al., 2008; urgv.evry.inra.fr/CATdb), or BAR (bar.utoronto.ca) website. Metabolomic data were analyzed using MapMan (Usadel et al., 2005; mapman.mpimp-golm.mpg.de).
Multiple alignment of protein sequences was performed using the COBALT tool (Papadopoulos and Agarwala, 2007) with default settings. This method uses progressive multiple alignment to combine pairwise aligned sequences. It ultimately produces an optimal alignment and a tree based on sequence identities and not on evolution rates. The resulting alignment is available as Supplemental Data Set 1 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or Genbank/EMBL databases under the following accession numbers: LST8-1 (At3g18140), LST8-2 (At2g22040), lst8-1-1 (SALK_02459), and lst8-1-2 (SAIL_641D10) mutants. The following protein sequences were used for multiple alignments: XP_003542977 and XP_002880414 (Arabidopsis lyrata); 012143m (Manihot esculenta); XP_002523640 (Ricinus communis); Gm15g09170 and Gm13g29940 (Glycine max); CP00092G00040 (Carica papaya); Os03g0681700 (rice [Oryza sativa]); XP_003565062 (Brachypodium distachyon); PP00138G00630 (Physcomitrella patens); VC00031G01430 (Volvox carteri); CR17G03790 (Chlamydomonas reinhardtii); NP_001186102 (Homo sapiens); and P41318 (Saccharomyces cerevisiae). Microarray normalized data are available in the Gene Expression Omnibus database under accession numbers GSE25731 and GSE25721 for the analysis of lst8-1 mutants and TOR RNAi lines, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Plants, Yeast, and Human LST8 Protein Sequences.
Supplemental Figure 2. Expression of the Arabidopsis LST8 Genes in Various Organs.
Supplemental Figure 3. GUS Staining of Transformed Arabidopsis Plants Carrying a pLst8:GUS Construct.
Supplemental Figure 4. Position of the T-DNA Insertions in the lst8-1 Mutants.
Supplemental Figure 5. Mutations in LST8-1 Gene Affect Stem and Flower Bud Morphology and Organization.
Supplemental Figure 6. Germinating Plantlets from the lst8-1 Mutant Line Display the Same Sensitivity to High Sugar Concentrations as a TOR RNAi Line.
Supplemental Figure 7. Diurnal Variations in Sugar and Starch Content during the Transition from Short Days to Long Days in the Wild Type and in the lst8-1-1 Mutant.
Supplemental Figure 8. MapMan Representations of Metabolite Variations in lst8-1-2 Mutants Using Global GC-MS Analysis.
Supplemental Figure 9. Variations in Metabolite Levels in lst8-1-2 Mutants and Wild-Type Plants Exposed to Long Days.
Supplemental Figure 10. MapMan Representations of Global Metabolite Analysis Using GC-MS in lst8-1 Mutants.
Supplemental Figure 11. MapMan Representations of Global Metabolite Analysis Using GC-MS in Wild-Type Plants.
Supplemental Figure 12. Changes in Metabolite Accumulations Determined by GC-MS in the lst8-1-2 Mutant and in TOR-Inducible RNAi Lines.
Supplemental Figure 13. Frequency of Functional Classes in the Genes Downregulated in lst8-1 Mutants Grown in Short-Day Conditions and Compared with Wild-Type Plants Grown in Short Days.
Supplemental Figure 14. Changes in Transcript Abundance Determined by Either Quantitative Real-Time RT-PCR or Microarray Hybridizations.
Supplemental Figure 15. Frequency of Functional Classes in the Genes Downregulated in lst8-1 Mutants after 2 d under Long-Day Conditions.
Supplemental Figure 16. Frequency of Functional Classes in the Genes Upregulated in lst8-1 Mutants.
Supplemental Table 1. Primer Sequences.
Supplemental Data Set 1. Text File of the Alignment Corresponding to the Tree in Figure 1.
Supplementary Material
Acknowledgments
We thank Halima Morin from the Institut Jean-Pierre Bourgin Plant Imaging Platform (Unité Mixte de Recherche 1318, Institut National de la Recherche Agronomique, AgroParisTech, Versailles, France) for help producing and imaging meristem sections, Lionel Gissot for help with confocal microscopy and for providing the RabC1-RFP construct, Sylvie Dinant for help with phloem labeling, Magali Bedu for performing enzyme activity measurements, Jessica Marion for help with transient transformation of Arabidopsis, and Michael Hall (Biozentrum, University of Basel, Switzerland) for providing the yeast lst8 mutants. We thank Joël Talbotec, François Gosse, and Philippe Maréchal for taking care of the plants and Jean-Christophe Palauqui, Patrick Laufs, Jean-Denis Faure, and Hoai-Nam Truong for helpful discussions. This work was partly supported by an Agence Nationale de la Recherche grant (ANR Blanc06-3-135436) to J.-P.R., C.M., and C.R. and by a European Union 6th framework project (Agronomics) to C.M. M.M. was supported by a joint PhD grant from Institut National de la Recherche Agronomique (Plant Biology Department) and Direction des Sciences du Vivant-Commissariat à l’Energie Atomique.
AUTHOR CONTRIBUTIONS
M.M. designed and performed research, analyzed data, and cowrote the article. M.A., T.D., C. Renne, L.T., and C. Marchive designed and performed research. G.C. designed and performed research, contributed new analytic/computational tools, and analyzed data. J.-P.R. and M.-L.M.-M. analyzed data and contributed new analytic/computational tools. C. Robaglia and C. Meyer designed research, analyzed data, and cowrote the article.
References
- Adami A., García-Alvarez B., Arias-Palomo E., Barford D., Llorca O. (2007). Structure of TOR and its complex with KOG1. Mol. Cell 27: 509–516 [DOI] [PubMed] [Google Scholar]
- Ahn C.S., Han J.A., Lee H.S., Lee S., Pai H.S. (2011). The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23: 185–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson G.H., Veit B., Hanson M.R. (2005). The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier G., Périlleux C. (2005). A physiological overview of the genetics of flowering time control. Plant Biotechnol. J. 3: 3–16 [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S., Gray J.E. (2008). Influence of environmental factors on stomatal development. New Phytol. 178: 9–23 [DOI] [PubMed] [Google Scholar]
- Chen E.J., Kaiser C.A. (2003). LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 161: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Bernier G., Périlleux C. (2002). C:N ratio increases in the phloem sap during floral transition of the long-day plants Sinapis alba and Arabidopsis thaliana. Plant Cell Physiol. 43: 684–688 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Lejeune P., Bernier G. (1998). The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: Comparison between the wild type and a starchless mutant. Planta 206: 131–137 [DOI] [PubMed] [Google Scholar]
- Crowe M.L., et al. (2003). CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 31: 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt R.F., Bonham-Smith P.C. (2008). Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol. 147: 128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D., Truong H.N., Robaglia C., Meyer C. (2005). An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem. Biophys. Res. Commun. 326: 844–850 [DOI] [PubMed] [Google Scholar]
- Deprost D., Yao L., Sormani R., Moreau M., Leterreux G., Nicolaï M., Bedu M., Robaglia C., Meyer C. (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H.Y., Li F.G., Wu X.D., Ma D.M., Wang M., Hou Y.X. (2006). The cloning and sequencing of a cDNA encoding a WD repeat protein in cotton (Gossypium hirsutum L.). DNA Seq. 17: 49–55 [DOI] [PubMed] [Google Scholar]
- Díaz-Troya S., Florencio F.J., Crespo J.L. (2008). Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii. Eukaryot. Cell 7: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández A.P., Strand A. (2008). Retrograde signaling and plant stress: Plastid signals initiate cellular stress responses. Curr. Opin. Plant Biol. 11: 509–513 [DOI] [PubMed] [Google Scholar]
- Fiehn O. (2006). Metabolite profiling in Arabidopsis. Methods Mol. Biol. 323: 439–447 [DOI] [PubMed] [Google Scholar]
- Gagnot S., Tamby J.P., Martin-Magniette M.L., Bitton F., Taconnat L., Balzergue S., Aubourg S., Renou J.P., Lecharny A., Brunaud V. (2008). CATdb: A public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 36(Database issue): D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio S., Liu Z., Thornton J., Butow R.A. (2005). Retrograde response to mitochondrial dysfunction is separable from TOR1/2 regulation of retrograde gene expression. J. Biol. Chem. 280: 42528–42535 [DOI] [PubMed] [Google Scholar]
- Gisel A., Barella S., Hempel F.D., Zambryski P.C. (1999). Temporal and spatial regulation of symplastic trafficking during development in Arabidopsis thaliana apices. Development 126: 1879–1889 [DOI] [PubMed] [Google Scholar]
- Gisel A., Hempel F.D., Barella S., Zambryski P. (2002). Leaf-to-shoot apex movement of symplastic tracer is restricted coincident with flowering in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 1713–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup A., Peacock W.J., Dennis E.S., Trevaskis B. (2009). The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. (Lond.) 103: 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D.A., Stevens D.M., Thoreen C.C., Burds A.A., Kalaany N.Y., Moffat J., Brown M., Fitzgerald K.J., Sabatini D.M. (2006). Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell 11: 859–871 [DOI] [PubMed] [Google Scholar]
- Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002). Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110: 177–189 [DOI] [PubMed] [Google Scholar]
- Hilson P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Pan X., Welti R., Wang X. (2008). Phospholipase Dalpha3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp J.T., Balasubramanian M.K., Gould K.L. (1997). A wat1 mutant of fission yeast is defective in cell morphology. Mol. Gen. Genet. 254: 127–138 [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sarbassov D.D., Ali S.M., King J.E., Latek R.R., Erdjument-Bromage H., Tempst P., Sabatini D.M. (2002). mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175 [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sarbassov D.D., Ali S.M., Latek R.R., Guntur K.V., Erdjument-Bromage H., Tempst P., Sabatini D.M. (2003). GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11: 895–904 [DOI] [PubMed] [Google Scholar]
- Komeili A., Wedaman K.P., O’Shea E.K., Powers T. (2000). Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151: 863–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea U.S., Leydecker M.T., Quilleré I., Meyer C., Lillo C. (2006). Posttranslational regulation of nitrate reductase strongly affects the levels of free amino acids and nitrate, whereas transcriptional regulation has only minor influence. Plant Physiol. 140: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. (2005). Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene 354: 110–116 [DOI] [PubMed] [Google Scholar]
- Lillo C. (2008). Signalling cascades integrating light-enhanced nitrate metabolism. Biochem. J. 415: 11–19 [DOI] [PubMed] [Google Scholar]
- Liu Z., Sekito T., Epstein C.B., Butow R.A. (2001). RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J. 20: 7209–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., Oppliger W., Jenoe P., Hall M.N. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A., Ralet M.C., Loudet O., Kronenberger J., Mouille G., Marion-Poll A., North H.M. (2007). A naturally occurring mutation in an Arabidopsis accession affects a beta-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz M.M., Kim S., Delauney A.J., Verma D.P. (2006). Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi K.K., Misra A., Ren S., McKnight T.D. (2009). BT2, a BTB protein, mediates multiple responses to nutrients, stresses, and hormones in Arabidopsis. Plant Physiol. 150: 1930–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion J., Bach L., Bellec Y., Meyer C., Gissot L., Faure J.D. (2008). Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 56: 169–179 [DOI] [PubMed] [Google Scholar]
- Menand B., Desnos T., Nussaume L., Berger F., Bouchez D., Meyer C., Robaglia C. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng P.H., Raynaud C., Tcherkez G., Blanchet S., Massoud K., Domenichini S., Henry Y., Soubigou-Taconnat L., Lelarge-Trouverie C., Saindrenan P., Renou J.P., Bergounioux C. (2009). Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS ONE 4: e7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C., Stitt M. (2001). Nitrate reduction and signalling. In Plant Nitrogen, P.J. Lea and J.-F. Morot-Gaudry, eds (Berlin: Springer), pp, 37–59
- Moreau M., Sormani R., Menand B., Veit B., Robaglia C., Meyer C. (2010). The TOR complex and signaling pathway in plants. In The TOR Complexes, Enzyme Series 27, M. Hall and F. Tamanoi, eds (Oxford, UK: Academic Press/Elsevier), pp. 285–302
- Mumberg D., Müller R., Funk M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Neer E.J., Schmidt C.J., Nambudripad R., Smith T.F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371: 297–300 [DOI] [PubMed] [Google Scholar]
- Nishizawa A., Yabuta Y., Shigeoka S. (2008). Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochotorena I.L., Hirata D., Kominami K., Potashkin J., Sahin F., Wentz-Hunter K., Gould K.L., Sato K., Yoshida Y., Vardy L., Toda T. (2001). Conserved Wat1/Pop3 WD-repeat protein of fission yeast secures genome stability through microtubule integrity and may be involved in mRNA maturation. J. Cell Sci. 114: 2911–2920 [DOI] [PubMed] [Google Scholar]
- Papadopoulos J.S., Agarwala R. (2007). COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 23: 1073–1079 [DOI] [PubMed] [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F.A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52: 640–657 [DOI] [PubMed] [Google Scholar]
- Roberg K.J., Bickel S., Rowley N., Kaiser C.A. (1997). Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics 147: 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.G., Cruz S.S., Roberts I.M., Prior D., Turgeon R., Oparka K.J. (1997). Phloem unloading in sink leaves of Nicotiana benthamiana: Comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9: 1381–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S., Moore I. (2002). The Arabidopsis Rab GTPase family: Another enigma variation. Curr. Opin. Plant Biol. 5: 518–528 [DOI] [PubMed] [Google Scholar]
- Searle I., Coupland G. (2004). Induction of flowering by seasonal changes in photoperiod. EMBO J. 23: 1217–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R.J. (2008). mTOR signaling: RAG GTPases transmit the amino acid signal. Trends Biochem. Sci. 33: 565–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., Neer E.J. (1999). The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 24: 181–185 [DOI] [PubMed] [Google Scholar]
- Soulard A., Cohen A., Hall M.N. (2009). TOR signaling in invertebrates. Curr. Opin. Cell Biol. 21: 825–836 [DOI] [PubMed] [Google Scholar]
- Sulpice R., et al. (2009). Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T., Ohsumi C., Iuchi S., Seki M., Kasuga M., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. (2002). Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Tzeng T.Y., Kong L.R., Chen C.H., Shaw C.C., Yang C.H. (2009). Overexpression of the lily p70(s6k) gene in Arabidopsis affects elongation of flower organs and indicates TOR-dependent regulation of AP3, PI and SUP translation. Plant Cell Physiol. 50: 1695–1709 [DOI] [PubMed] [Google Scholar]
- Usadel B., Bläsing O.E., Gibon Y., Poree F., Höhne M., Günter M., Trethewey R., Kamlage B., Poorter H., Stitt M. (2008). Multilevel genomic analysis of the response of transcripts, enzyme activities and metabolites in Arabidopsis rosettes to a progressive decrease of temperature in the non-freezing range. Plant Cell Environ. 31: 518–547 [DOI] [PubMed] [Google Scholar]
- Usadel B., et al. (2005). Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S., Ludwig P. (2003). The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M.N. (2006). TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Xu J., Brearley C.A., Lin W.H., Wang Y., Ye R., Mueller-Roeber B., Xu Z.H., Xue H.W. (2005). A role of Arabidopsis inositol polyphosphate kinase, AtIPK2alpha, in pollen germination and root growth. Plant Physiol. 137: 94–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D.J., Kim Y.L., Park C.R., Kim D.K., Yeom J., Lee C., Ahn C., Seong J.Y., Hwang J.I. (2010). Regulation of IκB kinase by GβL through recruitment of the protein phosphatases. Mol. Cells 30: 527–532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.