Aging in plants is an intricate process that balances vegetative growth with flowering and reproductive success. This work describes the identification of JUNGBRUNNEN1, a NAC transcription factor that regulates this process in Arabidopsis thaliana and additionally affects abiotic stress tolerance by activating expression of the DREB2A transcription factor.
Abstract
The transition from juvenility through maturation to senescence is a complex process that involves the regulation of longevity. Here, we identify JUNGBRUNNEN1 (JUB1), a hydrogen peroxide (H2O2)-induced NAC transcription factor, as a central longevity regulator in Arabidopsis thaliana. JUB1 overexpression strongly delays senescence, dampens intracellular H2O2 levels, and enhances tolerance to various abiotic stresses, whereas in jub1-1 knockdown plants, precocious senescence and lowered abiotic stress tolerance are observed. A JUB1 binding site containing a RRYGCCGT core sequence is present in the promoter of DREB2A, which plays an important role in abiotic stress responses. JUB1 transactivates DREB2A expression in mesophyll cell protoplasts and transgenic plants and binds directly to the DREB2A promoter. Transcriptome profiling of JUB1 overexpressors revealed elevated expression of several reactive oxygen species–responsive genes, including heat shock protein and glutathione S-transferase genes, whose expression is further induced by H2O2 treatment. Metabolite profiling identified elevated Pro and trehalose levels in JUB1 overexpressors, in accordance with their enhanced abiotic stress tolerance. We suggest that JUB1 constitutes a central regulator of a finely tuned control system that modulates cellular H2O2 level and primes the plants for upcoming stress through a gene regulatory network that involves DREB2A.
INTRODUCTION
In plants, the transition from juvenility through maturation to senescence is a physiologically complex process that involves a large number of molecular and physiological events regulated by genetically determined and environmentally modified regulatory networks (e.g., Smart et al., 1995; Hinderhofer and Zentgraf, 2001; He and Gan, 2002; Gepstein et al., 2003; Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Lim et al., 2007). Senescence is triggered by adverse environmental conditions, such as high salinity, low light intensity, drought, pathogen attack, nutrient deficiency, and other stresses (Dwidedi et al., 1979; Dhindsa et al., 1981; Bohnert et al., 1995; Balazadeh et al., 2010a, 2011). Many genes, including those that control transcription, undergo expression changes during senescence (e.g., Gepstein et al., 2003; Guo et al., 2004; Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Balazadeh et al., 2008b; Parlitz et al., 2011). Recently, high-resolution temporal profiling of gene expression during leaf senescence in Arabidopsis thaliana revealed clusters of coexpressed genes and a distinct chronology of senescence-associated processes (Breeze et al., 2011). Among the transcription factors (TFs), the NAC (for NAM, ATAF1, 2, and CUC2) and WRKY families are particularly rich in senescence-regulated TFs in many plant species (Andersson et al., 2004; Guo et al., 2004; Lin and Wu, 2004; Buchanan-Wollaston et al., 2005; Gregersen and Holm, 2007; Balazadeh et al., 2008b), suggesting they play important roles in leaf senescence. Although more than 20 of the 106 known NAC genes in Arabidopsis exhibit senescence-dependent expression (and, thus, represent senescence-associated genes [SAGs]), a distinct regulatory function with respect to senescence has only been reported for some members so far, including At NAP (for NAC-LIKE, ACTIVATED BY APETALA3/PISTILLATA; also called Arabidopsis NAC [ANAC] 029; Guo and Gan, 2006), ORESARA1 (ORE1; ANAC092, At NAC2; Kim et al., 2009; Balazadeh et al., 2010a), and ORESARA1 SISTER1 (ORS1; ANAC059; Balazadeh et al., 2011). Inhibiting either NAC individually delays senescence, identifying them as nonredundant positive regulators of senescence (Guo and Gan, 2006; Kim et al., 2009; Balazadeh et al., 2010a, 2011). A dual-function NAC gene, VASCULAR-RELATED NAC-DOMAIN INTERACTING2 (VNI2; ANAC083), was recently reported to integrate abscisic acid (ABA) signaling with leaf senescence (Yang et al., 2011) and to negatively regulate xylem vessel formation (Yamaguchi et al., 2010). Genes downstream of ORE1 and ORS1 have been identified; from these studies, it became apparent that both TFs exert their function through regulatory networks that include many known SAGs (Balazadeh et al., 2010a, 2011). Of the 170 genes upregulated after expression of ANAC092 is induced, 102 genes were also enhanced by senescence in the time-course experiment reported by Breeze et al. (2011). The majority of these genes (75%) fell into clusters whose gene members were enriched for promoter-localized NAC binding sites. This observation supports the conclusion that ANAC092/ORE1 and most likely other NAC TFs play an important role in regulating senescence.
An apparent signaling element for the regulation of senescence is hydrogen peroxide (H2O2). Diverse environmental and developmental stimuli, such as heat stress, salinity, cold, and pathogen attack, are known to trigger an accumulation of intracellular H2O2 and through this regulate the expression of many genes, including TFs of, for example, the NAC, WRKY, and APETALA2 (AP2)/ethylene-responsive element binding protein (EREBP) families (Vanderauwera et al., 2005; Gadjev et al., 2006). Notably, the expression of at least 15 senescence-associated NAC TFs increases rapidly after H2O2 treatment, including, for example, At NAP, ANAC092/ORE1, ORS1, ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR1 (ATAF1), and ANAC032 (Balazadeh et al., 2010b, 2011). However, the H2O2 and senescence-dependent gene regulatory networks of these TFs are just emerging and are far from being fully understood.
DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) belongs to the EREBP family of TFs. Although DREB2A expression increases during senescence, its biological role in this process has not been analyzed. Expression of DREB2A is induced by dehydration, salinity, heat, and different oxidative stress treatments, including H2O2 stress (Gadjev et al., 2006; Sakuma et al., 2006a; Suzuki et al., 2011). DREB2A regulates the water deficit–inducible expression of target genes and requires posttranslational modification for its activation. Two C3HC4 RING domain-containing proteins, including DREB2A-INTERACTING PROTEIN1 (DRIP1) and DRIP2, have been discovered and shown to interact with DREB2A in the nucleus. DRIPs act as E3 ubiquitin ligases that mediate the ubiquitination of DREB2A, thereby targeting the TF to 26S proteasome degradation. DRIP1 and DRIP2 thus function as negative regulators of drought-responsive gene expression (Qin et al., 2008). Recently, Suzuki et al. (2011) identified a new heat stress regulon in Arabidopsis regulated by multiprotein bridging factor 1c (MBF1c). DREB2A is one of the 36 genes whose expression during heat stress is regulated by MBF1c, and genetic studies demonstrated that DREB2A is required for plant survival under heat stress (Suzuki et al., 2011). Overexpression of a constitutively active form of DREB2A (35S:DREB2A CA) resulted in the upregulation of nearly 500 genes, including drought- and salt-responsive genes, and also heat shock–related genes (Sakuma et al., 2006b). The heat shock TF gene HsfA3 is a known direct target of DREB2A (Schramm et al., 2008).
Here, we report the identification of JUNGBRUNNEN1 (JUB1), a NAC TF that negatively regulates senescence. JUB1 is rapidly and strongly induced by H2O2 treatment. Overexpression of JUB1 markedly extends leaf longevity and promotes tolerance to various abiotic stresses. We found that increased tolerance to abiotic stress correlates with reduced levels of H2O2 in JUB1 overexpressors, whereas the opposite is observed in jub1-1 knockdown plants, suggesting that JUB1 participates in regulating the cellular H2O2 homeostasis network. We further determined the binding site of the JUB1 TF by in vitro binding site selection and discovered DREB2A as one of its direct downstream target genes. Thus, JUB1 links H2O2 signaling to senescence regulation and the downstream activation of DREB2A and its direct target HsfA3.
RESULTS
JUB1 Promotes Leaf Longevity
To identify novel transcription regulators that modulate leaf senescence, we systematically screened NAC gene T-DNA insertion mutants and cauliflower mosaic virus (CaMV) 35S-driven overexpression lines for extended longevity compared with wild-type Arabidopsis plants kept under identical growth conditions. We found that plants overexpressing At2g43000 developed senescence considerably later than the wild type and retained fully green leaves for a much longer period (see below). By contrast, a knockdown mutant of At2g43000 (Salk ID 036474) developed leaf senescence earlier than the Columbia-0 (Col-0) wild type (see below). Similarly, we observed earlier senescence in transgenic plants upon suppression of At2g43000 transcript abundance by means of a genome-inserted artificial microRNA construct (see Supplemental Figure 1 online). These observations identify At2g43000 as a novel genetic regulator of plant senescence. As elevated expression of At2g43000 extended longevity, we designated it JUNGBRUNNEN1 (JUB1; German for “Fountain of Youth”).
JUB1 encodes a 275–amino acid protein of a calculated molecular mass of 31.5 kD. JUB1 contains a NAM domain (pfam02365) at its N terminus. Its coding region consists of three exons, interrupted by two introns. Phylogenetic analyses revealed the presence of JUB1 orthologs in other plant species, including rice (Oryza sativa; ONAC066; Ooka et al., 2003) and Populus trichocarpa (PNAC080-083; Hu et al., 2010). Expression of a JUB1–green fluorescence protein (GFP) fusion in Arabidopsis showed its predominant accumulation in the nucleus (see Supplemental Figure 2 online), consistent with its function as a transcription regulator.
RNA gel blot analysis and quantitative RT-PCR (qRT-PCR) confirmed elevated JUB1 transcript level in multiple independent 35S:JUB1 transformants compared with the wild type (Figures 1A and 1B). When grown in soil under long-day conditions, leaf senescence and bolting were strongly delayed in 35S:JUB1 plants compared with the wild type and empty vector (EV)–transformed control lines (Figure 1C). Overexpressors started bolting 10 to 14 d later than the EV lines. Chlorophyll levels of 12 individual rosette leaves of overexpression plants were higher than those in age-matched EV plants (Figure 1D). Mortality curves confirmed later senescence in overexpressors than in EV lines (Figure 1E). In accordance with this, ion leakage as an indicator of senescence was much less pronounced in overexpression plants than in EV controls (Figure 1F). We also expressed JUB1 from the RD29A promoter, which shows basal activity in nonstressed plants, but enhanced activity upon abiotic stress (Kasuga et al., 1999). We observed extended longevity, accompanied by delayed bolting, in RD29A:JUB1 plants, which was dependent on JUB1 expression level (see Supplemental Figure 3 online).
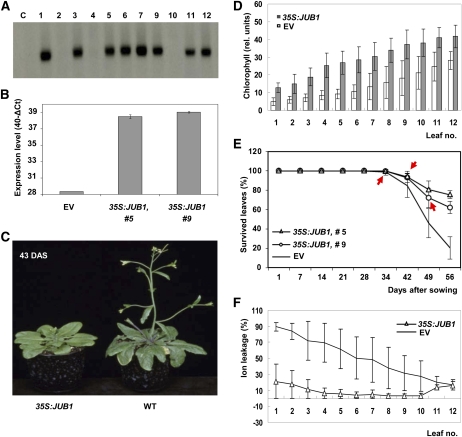
Figure 1.
Physiological and Molecular Characterization of JUB1 Overexpression Plants.
(A) RNA gel blot analysis of plants transformed with the 35S:JUB1 construct. Radiolabeled JUB1 cDNA was used as hybridization probe. Numbers indicate individual transformants; C, nontransformed wild-type (Col-0) control. Elevated JUB1 expression compared with wild-type plants is observed in transgenic lines 1, 3, 5, 6, 7, 9, 11, and 12.
(B) Increased JUB1 expression in lines 5 and 9, as confirmed by qRT-PCR, compared with EV control.
(C) Delayed bolting in 35S:JUB1 overexpression line compared with the wild type (WT) at 43 d after sowing (DAS).
(D) Elevated chlorophyll content in leaves 1 to 12 of 35S:JUB1 overexpressors compared with EV control plants at ~60 DAS (n = 6).
(E) Percentage of survived leaves at different plant ages (given as DAS). Bolting time points are indicated by red arrows. Note steeper slope for curve of EV plants compared with 35S:JUB1 overexpressors between days 34 and 56 (n = 14 to 17).
(F) Ion leakage of leaves 1 to 12 of 35S:JUB1 and EV control plants at ~60 DAS.
Data in (B) and (D) to (F) are the means of at least three biological replicates ± sd.
Next, we characterized a SALK T-DNA insertion line (Col-0 background). The T-DNA insertion in the second intron, 1034-bp downstream of the start codon (Figure 2A), was confirmed by PCR on genomic DNA of jub1-1 plants (see Supplemental Figure 4 online). RT-PCR analysis revealed a reduction in JUB1 transcript abundance in fully expanded jub1-1 mutant leaves compared with leaves of equivalently aged wild-type plants (Figure 2B). Phenotypic analysis of the jub1-1 mutant revealed earlier bolting than in the wild type (up to 3 d; Figure 2C). A >95% reduction of JUB1 transcript level was detected by qRT-PCR in jub1-1 plants (Figure 2D), which was accompanied by an earlier loss of chlorophyll (Figure 2E) and precocious senescence (Figures 2F).
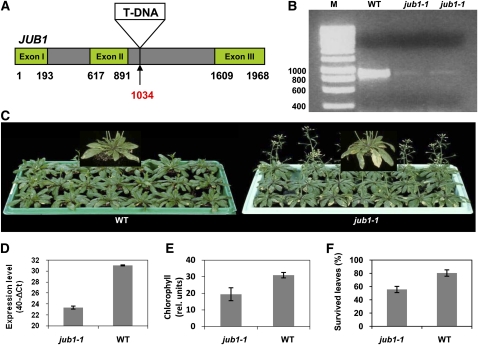
Figure 2.
Physiological and Molecular Characterization of the jub1-1 Mutant.
(A) T-DNA inserted at nucleotide position 1034 downstream of the start codon (indicated by arrow) in the second intron.
(B) Downregulation of JUB1 transcripts in jub1-1 mutant shown by RT-PCR with primers annealing to the start and stop regions of the coding sequence. M, molecular size marker (sizes in base pairs); WT, wild type.
(C) Comparison of jub1-1 plants with wild-type plants at 47 DAS. Note early bolting and early senescence in the mutant plants.
(D) Downregulation of JUB1 transcript abundance in jub1-1 line, as confirmed by qRT-PCR.
(E) The jub1-1 mutant contains less chlorophyll than the wild type in the five biggest leaves at 47 DAS.
(F) The jub1-1 mutant exhibits a lower percentage of survived leaves than the wild type at 47 DAS. Data in graphs are the means of at least three biological replicates ± sd.
As our data indicated a longevity-extending effect of elevated JUB1 expression, we also tested its role in dark-induced senescence. Leaves were detached from 39-d-old soil-grown plants and kept for up to 6 d on moist filter paper in the dark; we observed that 35S:JUB1 overexpressors retained chlorophyll better than the jub1-1 knockdown and EV control lines, reminiscent of a delay in senescence (see Supplemental Figure 5 online).
Expression Profiling of SAGs in JUB1 Transgenic Plants
To further substantiate the role of JUB1 for the regulation of leaf senescence, we analyzed the expression of 168 SAGs (including 49 TFs) in wild-type and JUB1-modified plants by qRT-PCR. The SAGs included in our expression profiling platform were previously shown to be highly upregulated during natural senescence in wild-type plants (see Supplemental Data Set 1 online; Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Balazadeh et al., 2008b; Parlitz et al., 2011). The platform included SAG12, a well-known senescence marker gene (Noh and Amasino, 1999). At a twofold cutoff, the expression of 89 SAGs (including 21 TFs and SAG12), representing ~53% of all SAGs tested, was downregulated in 35S:JUB1 plants compared with the wild type, while only six SAGs (including JUB1) were upregulated (see Supplemental Data Set 1 online; Figure 3A). By contrast, expression of 97 SAGs (including 19 TFs), representing ~58% of all SAGs tested here, were upregulated in jub1-1 compared with the wild type, and only two SAGs (including JUB1) were downregulated in jub1-1 plants (Figures 3B and 3C; see Supplemental Data Set 1 online). Collectively, our data support the model that JUB1 constitutes a negative regulator of leaf senescence and, hence, a driver of longevity.
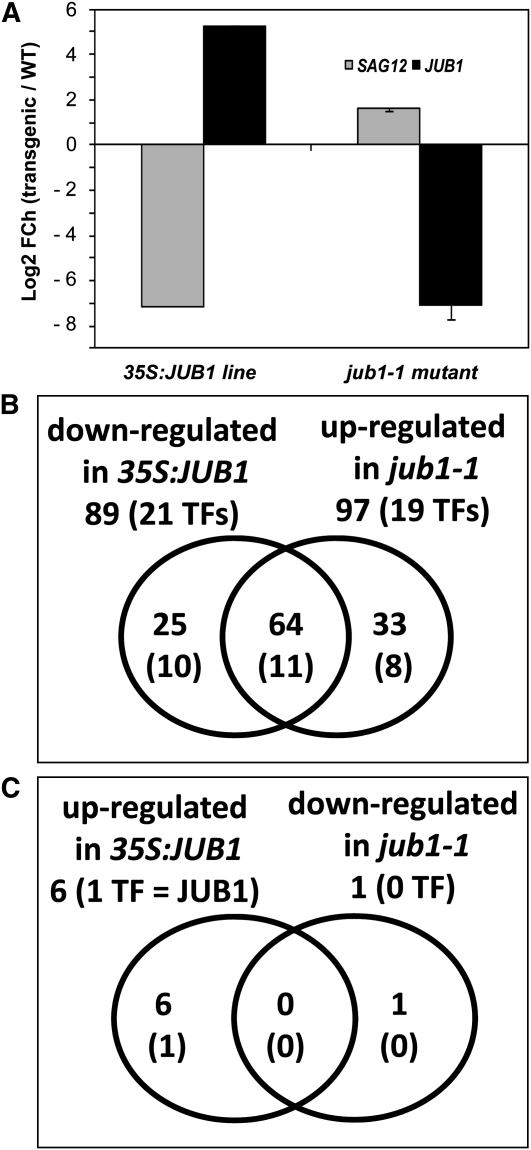
Figure 3.
Transcript Profiling of SAGs.
(A) Expression of JUB1 and the late-senescence marker gene SAG12 in 35S:JUB1 and jub1-1 plants compared with the wild type (WT) (numbers on the y axis indicate log2 fold-change (FCh) expression ratio compared with the wild type).
(B) and (C) Venn diagrams of SAGs differentially expressed in 35S:JUB1 and jub1-1 plants compared with the wild type at 47 DAS. Numbers in parentheses indicate senescence-associated TFs. See also Supplemental Data Set 1 online.
Estradiol-Inducible JUB1 Overexpression
We next tested the effect of estradiol (EST)-inducible JUB1 expression on bolting and leaf senescence using the system described by Zuo et al. (2000). qRT-PCR revealed induction of JUB1 expression in EST-treated JUB1-IOE seedlings already after 1 h of EST treatment and expression further increased at longer incubation times (Figure 4A). We sowed JUB1-IOE lines on half-strength Murashige and Skoog (MS) medium containing 1% Suc and 10 μM EST (or 0.1% ethanol in control experiments). After stratification at 4°C for 3 d, plates were transferred to a growth chamber at long-day conditions. Bolting was delayed in EST-treated JUB1-IOE plants compared with mock-treated controls (Figure 4B). When grown in liquid medium, JUB1-IOE seedlings remained green (nonsenescent) longer in the presence of EST (15 μM) than in mock-treated seedlings (Figure 4C). Thus, high JUB1 expression suppressed bolting and delayed senescence under in vitro conditions, similar to plants grown in soil.
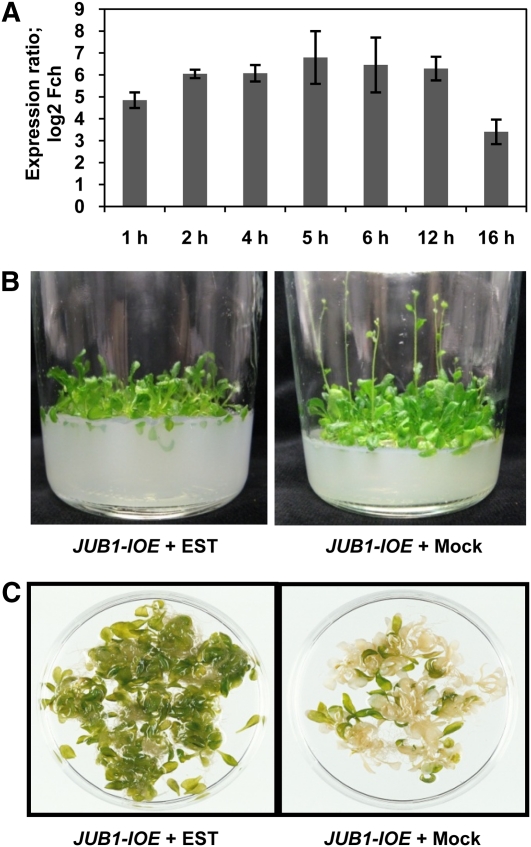
Figure 4.
Phenotypic Analysis of JUB1-IOE Lines.
(A) JUB1 expression is induced in leaves of JUB1-IOE seedlings after treatment with 10 μM EST compared with mock treatment (0.1% ethanol). Treatment times are indicated. Data are the means of three biological replicates ± sd. Fch, fold change.
(B) Induction of JUB1 expression by EST in JUB1-IOE plants delays bolting when grown in vitro. Plants were grown for 6 weeks in glass jars on medium containing 10 μM EST (0.1% ethanol for control experiment). In this experiment, five independent transgenic lines were tested; the photographs shown represent a typical result.
(C) Delayed senescence in JUB1-IOE plants grown in vitro. Two-week-old JUB1-IOE seedlings were transferred to flasks with liquid medium containing 15 μM EST, or 0.15% ethanol as control, and kept on a rotary shaker (slow motion) under continuous light for 1 week. Note the delayed senescence upon JUB1 induction. Three independent transgenic lines were used to confirm the observation made here.
Age-Dependent JUB1 Expression
We next analyzed the JUB1 expression pattern in Arabidopsis (Col-0) and tobacco (Nicotiana tabacum cv Samsum NN) plants transformed with a JUB1 promoter-β-glucuronidase (GUS) reporter construct (ProJUB1:GUS). JUB1 expression was observed in various tissues throughout plant development. In Arabidopsis seedlings, GUS activity was preferentially detected in roots, cotyledons, and the tips of young leaves. GUS staining was generally more pronounced in leaf tips and margins than the central part of the leaf blade (Figures 5A and 5B). Expression was also observed in floral tissues, preferentially in old sepals, petals, stamens, mature anthers, and pollen grains, while immature floral tissue did not show GUS activity (Figures 5C and 5D). GUS activity was also observed in the abscission zone of open flowers (data not shown). In the half expanded leaves of soil-grown plants, GUS staining was observed in the tip region only; in fully expanded leaves, strong GUS staining was observed in the senescent regions (Figures 5E and 5F). Moreover, GUS activity was detected in primary and lateral roots (Figures 5A, 5G, and 5H), including the root cap of extended roots (Figure 5I). However, JUB1 promoter activity was absent from cells of emerging lateral roots (Figure 5H) and from root meristematic zones that faded into the elongation zones (Figures 5G and 5I).
Figure 5.
GUS Activity in ProJUB1:GUS Lines.
(A) to (I) Arabidopsis.
(A) Ten-day-old seedling. GUS staining is mainly localized to cotyledons, the tip regions and margins of leaves representing the oldest but not yet senescent leaf regions, and primary and secondary roots.
(B) Ten-day-old seedling. Strong GUS staining located in tips of newly emerging leaves (arrows).
(C) and (D) Flowers at different development stages. GUS staining is virtually absent in unopened young flowers. In open flowers (D), GUS staining appears in mature anthers, filaments, and the stamen abscission zone.
(E) GUS staining is weak to absent in ~50% expanded leaves from soil-grown plants.
(F) Strong GUS staining is located in senescent regions of a partially senescent leaf from soil-grown plants.
(G) GUS staining in roots; note the absence of GUS activity in the meristematic zone.
(H) GUS staining is absent from emerging lateral roots (arrows).
(I) GUS staining in root cap (arrow). Staining was for ~1 h.
(J) to (P) Tobacco.
(J) Leaf, with more intense staining in the tip and margins. GUS activity is also visible around wound sites.
(K) and (L) Young flower without GUS staining.
(M) and (N) Open flower showing GUS staining in petal tips and anthers.
(O) Roots. Note the absence of GUS activity in the meristematic zone, whereas the root cap shows GUS staining (arrows).
(P) Absence of GUS activity from emerging lateral roots (arrows). Staining was for ~6 h.
In transgenic ProJUB1:GUS tobacco leaves, GUS staining was only observed in older parts (i.e., the tips and margins), consistent with an age-dependent upregulation of JUB1 expression (Figure 5J). No GUS staining was observed in young flowers when corolla limbs began to open (Figures 5K and 5L); however, in open flowers, intense GUS staining was evident in the tip regions of corolla limbs, but not corolla tubes. Additionally, JUB1 promoter activity was visible in anthers (Figures 5M and 5N). Moreover, GUS staining was observed in roots (Figures 5O and 5P), including the root tip of extended, but not young, emerging roots. As in Arabidopsis, no JUB1 promoter activity was detected in the root meristematic zone (Figure 5O).
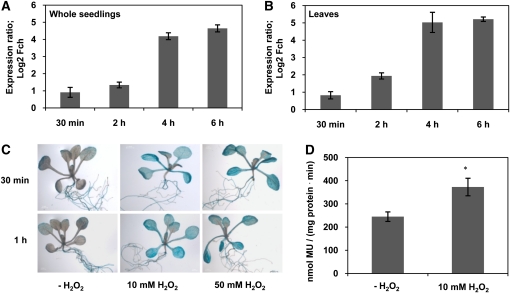
H2O2 Triggers JUB1 Expression
H2O2 plays a central role in plant signaling and stress responses. In microarray hybridization experiments, we previously observed strong induction (~25-fold) of JUB1 transcript level in Arabidopsis seedlings after 5 h of exposure to 10 mM H2O2 (Balazadeh et al., 2010b). Here, we confirmed H2O2-responsive JUB1 expression by qRT-PCR in 2-week-old Arabidopsis plants treated for 30 min or 2, 4, or 6 h with 10 mM external H2O2. JUB1 transcript abundance increased in both whole seedlings and leaves already 30 min after treatment and increased further thereafter (Figures 6A and 6B). Similarly, JUB1 expression increased approximately fivefold within 5 h of pharmacological inhibition of the H2O2-scavenging enzyme catalase in seedlings (see Supplemental Figure 6A online). In ProJUB1:GUS lines, enhanced GUS activity was observed 30 min or 1 h after treatment with 10 mM or 50 mM H2O2 (Figures 6C and 6D), indicating that the response of JUB1 to H2O2 is regulated at the promoter level and mediated by one or more currently unknown upstream transcription regulators.
Figure 6.
Effect of H2O2 on JUB1 Expression.
(A) and (B) JUB1 transcript level in whole seedlings (A) and leaves (B) of wild-type Arabidopsis plants as determined by qRT-PCR after treatment with 10 mM H2O2 for 30 min or 2, 4, and 6 h compared with nontreated samples.
(C) ProJUB1:GUS lines treated with H2O2. Two-week-old seedlings were transferred to medium containing 10 or 50 mM H2O2 and incubated for 30 min or 1 h, respectively. Elevated GUS activity was observed at both concentrations already 30 min after treatment.
(D) GUS activity of ProJUB1:GUS seedlings measured by a MUG assay after treatment with 10 mM H2O2 for 30 min. Asterisk indicates significant difference (P < 0.05, Student’s t test). Data are the means of three biological replicates ± sd. MU, methylumbelliferone.
It was previously reported that endogenous H2O2 concentration rises during bolting in Arabidopsis leaves, and downregulation of catalase (CAT2) activity was suggested to be the initial step of this rise (Zimmermann et al., 2006). To investigate a possible correlation between the level of endogenous H2O2 and JUB1 expression, we measured both H2O2 content and JUB1 expression in leaves of ~35-d-old Arabidopsis wild-type plants at bolting (1-cm main flower stalk). Leaves number 2 (second oldest rosette leaf) to 14 were sampled individually; younger leaves were collected in groups A (>25 mm leaf length), B (15 to 25 mm), and C (below 15 mm), respectively (leaf number 1 was too old and deteriorated for measurements). JUB1 expression was determined by qRT-PCR, and H2O2 level was quantified using an Amplex Red assay. We observed that endogenous H2O2 and JUB1 transcript abundance followed similar patterns with higher levels in older leaves (numbers 2 to 12) than in younger leaves (numbers 13 and 14, groups A to C); particularly low H2O2 and JUB1 transcript levels were present in the youngest rosette leaves (see Supplemental Figure 7 online). Thus, our data indicate that JUB1 expression follows the level of endogenous H2O2 dependent on leaf age.
Various abiotic stresses, including salinity, cold, and heat stress, cause the accumulation of endogenous H2O2. In a microarray hybridization experiment, we observed induction of JUB1 expression when hydroponically grown plants were subjected to salinity stress (150 mM NaCl) over a period of 4 d (see Supplemental Figure 6B online). Salt stress (150 and 200 mM NaCl) slightly induced GUS activity in ProJUB1:GUS plants after 24 h (4-methyl umbelliferyl β-d-glucuronide [MUG] assay; see Supplemental Figure 8A online). Several other treatments that cause an intracellular rise of H2O2 level induce JUB1 expression, including treatment with cellulase R-10 (see Supplemental Figures 8B and 8C online), paraquat (methyl viologen) (see Supplemental Figures 8B and 8C online), ozone (Gadjev et al., 2006), 3-aminotriazole, which blocks the activity of the H2O2 scavenging enzyme catalase (Gechev and Hille, 2005), and AAL (Alternaria alternata fungal toxin that induces H2O2 accumulation and cell death through perturbation of sphingolipid metabolism; Gechev and Hille, 2005). Collectively, these data indicate that JUB1 expression is triggered by an intracellular rise of H2O2 level.
Overexpression of JUB1 Enhances Tolerance to Salt Stress
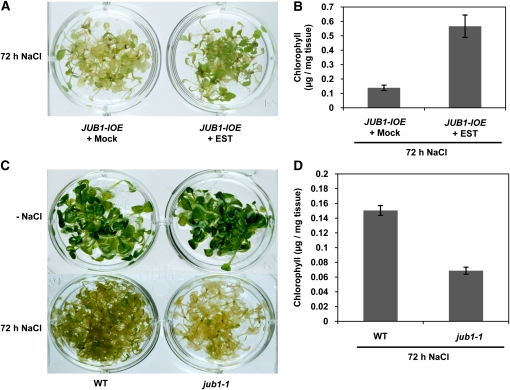
Salt stress triggers the accumulation of intracellular H2O2 (e.g., Chung et al., 2008). To investigate whether overexpression of JUB1 enhances tolerance to salt stress, the effect of 150 mM NaCl on JUB1-IOE and jub1-1 knockdown mutants was studied. To this end, 2-week-old jub1-1 mutant and JUB1-IOE seedlings were transferred from solid MS medium to liquid MS medium containing 150 mM NaCl. In the case of JUB1-IOE lines, 15 μM EST was added to induce JUB1 expression, and, as a control, 0.15% ethanol was used. All seedlings were incubated on a shaker in a growth chamber with continuous light; after 3 d of stress, the JUB1-IOE plants incubated in saline medium containing EST retained fivefold higher levels of chlorophyll than the plants incubated in saline medium in the absence of EST (Figures 7A and 7B). By contrast, chlorophyll content in jub1-1 seedlings was only half that of wild-type plants when stressed by salt (Figures 7C and 7D).
Figure 7.
EST-Treated JUB1-IOE Plants Exhibit Increased Tolerance to NaCl Stress, Whereas Tolerance to NaCl Is Reduced in the jub1-1 Mutant.
(A) Two-week-old JUB1-IOE seedlings were transferred from solid MS medium to liquid MS medium containing 150 mM NaCl, in the absence (mock; 0.15% ethanol) or presence of 15 μM EST added to induce JUB1 expression. Plants were incubated for 72 h. EST-treated JUB1-IOE seedlings are more tolerant to salt stress.
(B) Higher chlorophyll levels are retained in EST-treated JUB1-IOE seedlings after salt treatment.
(C) Seedlings of wild-type (WT) plants are less affected by 72 h salt treatment (150 mM NaCl) than those of the jub1-1 mutant.
(D) Chlorophyll content remains higher in the wild type (WT) than jub1-1 plants after stress treatment. Data in (B) and (D) are the means of three biological replicates ± sd.
JUB1 Enhances Tolerance to H2O2 by Regulating Its Cellular Concentration
Similar to findings in animals, extended life span in plants has been observed to be closely related to increased tolerance to oxidative stress. In particular, several Arabidopsis mutants with extended longevity have been shown to exhibit superior tolerance to oxidative stress. Examples include transgenic plants overexpressing the CBF2 and CBF3 TFs, the ore1, ore3, and ore9 mutants, and the very late flowering, long-living mutant gigantea (Kurepa et al., 1998; Woo et al., 2004; Sharabi-Schwager et al., 2010). The gigantea mutant exhibits enhanced tolerance to various types of oxidative stress, such as paraquat- and H2O2-induced oxidative stress (Kurepa et al., 1998).
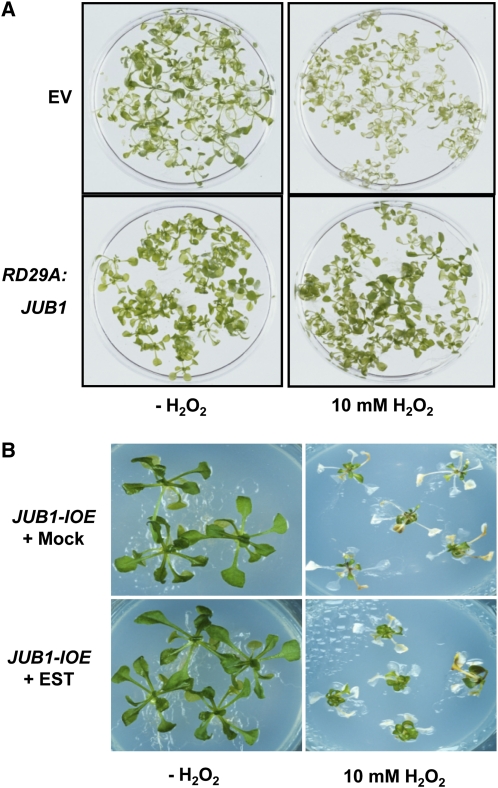
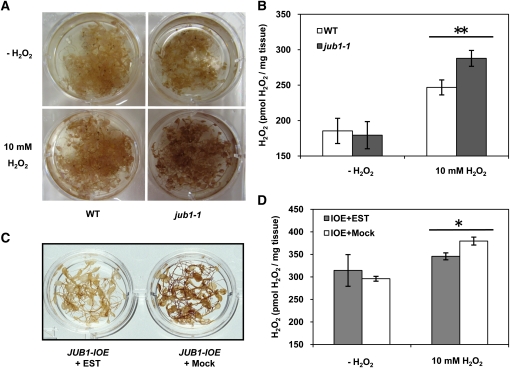
To determine whether there is a link between increased JUB1 expression and enhanced tolerance to oxidative stress, we first analyzed the effect of H2O2 treatment on JUB1 transgenic plants. Two-week-old RD29A:JUB1 seedlings were treated with 10 mM H2O2. As shown in Figure 8A, seedlings subjected to treatment for 24 h retained chlorophyll more efficiently than EV-transformed control plants. Additionally, we cultured 35S:JUB1 and EV seedlings on sterile medium containing 0 or 10 mM H2O2. 35S:JUB1 plants remained healthier and showed less leaf bleaching in the presence of H2O2 than the control line (data not shown). We next tested H2O2 tolerance of JUB1-IOE lines by transferring 3-week-old seedlings from half-strength solid MS medium to fresh medium containing 15 μM EST and 10 mM H2O2; control seedlings were similarly treated with H2O2, but EST was omitted (mock treatment: 0.15% ethanol). After 6 d, the JUB1-IOE plants treated with H2O2 and EST were more vital than plants treated with H2O2 alone (Figure 8B). We next tested the effect of reduced JUB1 expression on H2O2 tolerance. Leaves detached from 35-d-old soil-grown jub1-1 and EV plants were incubated in 10 mM H2O2 for 5 d; EV leaves remained greener than those of the jub1-1 mutant (see Supplemental Figures 9A and 9B online). Moreover, when 12-d-old seedlings were incubated in MS medium with 10 mM H2O2 for 6 h tissue damage was more prominent in jub1-1 than wild-type seedlings. Accordingly, diaminobenzidine (DAB) staining revealed a higher H2O2 accumulation in jub1-1 than wild-type plants (Figure 9A). We confirmed a slight (~14%) but significant increase in H2O2 concentration in 12-d-old jub1-1 seedlings compared with the wild type using an Amplex Red assay (Figure 9B). Next, we tested the effect of elevated JUB1 expression on cellular H2O2 level. To this end, we treated JUB1-IOE lines in the absence (control) or presence of 15 μM EST (to induce JUB1 expression) with 10 mM H2O2 for 6 h and observed that EST-treated seedlings accumulated slightly less (~10%) H2O2 than the controls (Figures 9C and 9D). Jointly these data indicate that JUB1 counteracts the cellular accumulation of H2O2.
Figure 8.
Overexpression of JUB1 Confers Tolerance to H2O2.
(A) Two-week-old RD29A:JUB1 and EV seedlings grown on solid MS medium were transferred to liquid medium containing 10 mM H2O2 and incubated for 24 h. RD29A:JUB1 seedlings remained green, whereas EV lines bleached in the presence of H2O2.
(B) JUB1-IOE lines treated with 15 μM EST survived better than plants treated with 0.15% ethanol (mock treatment) after transferring 3-week-old plants to fresh medium containing 10 mM H2O2 for 6 d.
Figure 9.
Overexpression of JUB1 Reduces Endogenous H2O2 Content, While the Opposite Is Observed in the jub1-1 Mutant.
(A) DAB staining of wild-type (WT) and jub1-1 seedlings treated with 10 mM H2O2 for 6 h. Note the stronger DAB staining in jub1-1 seedlings in the presence of H2O2.
(B) Amplex Red assay. Note the higher H2O2 level in H2O2-treated jub1-1 seedlings compared with the wild type. Asterisks indicate significant difference (P < 0.001).
(C) DAB staining. JUB1-IOE seedlings treated with EST for 6 h accumulate less H2O2 than mock-treated seedlings.
(D) Amplex Red assay. Note the reduced H2O2 level in EST-treated JUB1-IOE seedlings compared with mock-treated plants (asterisk indicates significant difference; P < 0.02). Data in (B) and (D) are the means of three independent biological replicates ± sd.
A Heat Shock Protein Response Network Regulated by JUB1
To start unraveling the H2O2 regulatory network regulated by JUB1, we tested the expression of 187 reactive oxygen species (ROS)–responsive genes in 2-week-old seedlings of RD29A:JUB1 and jub1-1 plants before and after treatment with H2O2 (10 mM, 6 h) and compared results with data from wild-type seedlings. Expression analysis was performed by qRT-PCR. Genes included in the ROS expression platform are known to be induced by H2O2, superoxide anion, and/or singlet oxygen (for details, see Methods) and encode TFs, heat shock proteins (HSPs), protein kinases, glutathione S-transferases (GSTs), and others (see Supplemental Data Set 2 online).
Our data revealed detectable expression of 179 ROS genes in all samples at both control and treatment conditions. We observed that expression of 36 genes was more strongly induced by H2O2 in RD29A:JUB1 than wild-type seedlings, while expression of these genes was either not or only marginally affected by H2O2 treatment in jub1-1 seedlings compared with the wild type (Table 1). These genes therefore represent prime candidates for ROS-responsive genes regulated by JUB1 during H2O2 signaling. Genes encoding HSPs constitute the largest fraction of the genes in the regulatory network. Upon H2O2 treatment, 14 HSP genes were more strongly induced in RD29A:JUB1 seedlings compared with the wild type, whereas their expression was not or only marginally induced in jub1-1 plants (Table 1). In addition to HSPs, several GST genes (including GST10, 24, and 25), TFs (e.g., DREB2A, DREB2B, ANAC047, and ANAC055), and some heat shock binding proteins were also among the ROS genes induced by H2O2 treatment in RD29A:JUB1 transgenic plants. In accordance with the reduced induction of HSPs after stress in jub1-1 seedlings, we observed a reduced heat stress tolerance (45°C, 5 h) of jub1-1 seedlings relative to the wild type (see Supplemental Figure 10 online), whereas overexpression of JUB1 enhanced tolerance to heat stress (see Supplemental Figure 11 online).
Table 1.
Genes Affected by H2O2 Treatment
| H2O2 Treatment versus Control | ||||
| RD29A:JUB1/Wild Type | jub1-1/Wild Type | |||
| log2 FCh | ||||
| AGI | Description | A | B | A-B |
| AT1G74310 | HOT1_HSP101; ATP binding/ATPase/ nucleoside-triphosphatase/nucleotide binding | 4.69 | 0.99 | 3.70 |
| AT2G29500 | 17.6-kD Class I small HSP (HSP17.6B-CI) | 3.57 | 0.05 | 3.52 |
| AT4G25200 | ATHSP23.6-mitochondrial small HSP23.6 | 3.62 | 0.45 | 3.17 |
| AT5G12030 | HSP17.6; unfolded protein binding | 3.98 | 0.97 | 3.01 |
| AT1G74590 | GST TAU10 (ATGSTU10_GSTU10) | 2.69 | −0.22 | 2.90 |
| AT1G53540 | 17.6-kD Class I small HSP (HSP17.6C-CI) (AA 1-156) | 3.16 | 0.26 | 2.89 |
| AT1G71000 | HSP binding | 3.87 | 1.09 | 2.78 |
| AT2G28210 | ALPHA CARBONIC ANHYDRASE2 (ACA2) | 2.82 | 0.29 | 2.53 |
| AT3G09350 | Armadillo/β-catenin repeat family protein | 3.36 | 0.83 | 2.53 |
| AT5G51440 | 23.5-kD mitochondrial small HSP (HSP23.5-M) | 3.54 | 1.03 | 2.51 |
| AT5G12020 | 17.6 KDA CLASS II HSP (HSP17.6II) | 2.94 | 0.42 | 2.51 |
| AT3G12580 | HSP70; ATP binding | 3.43 | 0.92 | 2.51 |
| AT5G52640 | ATHS83_HSP81-1_HSP81.1_HSP83__ATHSP90.1; ATP binding/unfolded protein binding | 3.55 | 1.05 | 2.50 |
| AT3G24500 | ATMBF1C__MBF1C; DNA binding/transcription coactivator/TF | 3.48 | 1.04 | 2.44 |
| AT5G64510 | Unknown protein | 3.19 | 0.77 | 2.42 |
| AT3G46230 | ATHSP17.4 | 2.54 | 0.13 | 2.41 |
| AT1G17170 | GST TAU24 (GST_ATGSTU24) | 2.34 | −0.05 | 2.39 |
| AT5G48570 | Peptidyl-prolyl cis-trans isomerase, putative/FK506 binding protein | 3.26 | 0.91 | 2.35 |
| AT1G54050 | 17.4-kD Class III HSP (HSP17.4-CIII) | 3.27 | 0.92 | 2.35 |
| AT4G12400 | Stress-inducible protein, putative | 3.09 | 0.83 | 2.26 |
| AT1G07160 | Protein phosphatase 2C, putative/PP2C, putative | 1.50 | −0.70 | 2.20 |
| AT2G46240 | ATBAG6__BAG6; calmodulin binding/protein binding | 2.61 | 0.43 | 2.18 |
| AT2G38340 | AP2 domain-containing TF, putative (DRE2B) | 1.83 | −0.34 | 2.17 |
| AT2G26150 | HSFA2__ATHSFA2; DNA binding/TF | 3.73 | 1.59 | 2.14 |
| AT5G05410 | DREB2A; DNA binding/transcription activator/TF | 2.28 | 0.15 | 2.13 |
| AT2G32120 | HSP70T-2; ATP binding | 2.47 | 0.34 | 2.13 |
| AT3G04070 | ANAC047; TF | 1.96 | −0.14 | 2.10 |
| AT2G20560 | DNAJ heat shock family protein | 3.23 | 1.19 | 2.04 |
| AT4G37370 | CYP81D8; electron carrier/heme binding/iron ion binding/monooxygenase/oxygen binding | 1.93 | −0.02 | 1.94 |
| AT4G34410 | REDOX RESPONSIVE TRANSCRIPTION FACTOR1 (RRTF1) | 1.41 | −0.53 | 1.94 |
| AT1G17180 | GST TAU25 (ATGSTU25) | 2.71 | 0.91 | 1.80 |
| AT1G16030 | Hsp70b; ATP binding | 2.27 | 0.48 | 1.79 |
| AT3G15500 | ATNAC3__ANAC055; TF | 1.51 | −0.12 | 1.63 |
| AT3G28210 | PMZ; zinc ion binding | 1.66 | 0.06 | 1.60 |
| AT4G37990 | ATCAD8_CAD-B2__ELICITOR-ACTIVATED GENE 3-2 (ELI3-2); aryl-alcohol dehydrogenase/mannitol dehydrogenase | 0.30 | −1.29 | 1.58 |
| AT1G14200 | Zinc finger (C3HC4-type RING finger) family protein | 1.77 | 0.21 | 1.56 |
Genes highly induced by H2O2 treatment (6 h) in the RD29A:JUB1 line but either not or marginally affected in the jub1-1 mutant when compared to wild-type plants. A threefold change (log2 = 1.5) was selected as threshold. HSPs are indicated in bold. AGI, Arabidopsis Genome Initiative; FCh, fold change.
Collectively, our data indicate that JUB1 is a key regulator of plant stress responses. Under stress, JUB1 regulates expression of a set of stress tolerance genes in transgenic Arabidopsis, thereby leading to improved stress tolerance.
JUB1 Binding Site
To identify cis-elements recognized by the JUB1 TF, we performed in vitro binding site selection using the CELD-TF fusion method (Xue, 2002, 2005). Thirty-six unique double-stranded oligonucleotides bound by JUB1-CELD fusion protein were recovered after five rounds of selection using biotin-labeled double-stranded oligonucleotides containing a 30-nucleotide random sequence and were analyzed for binding activity (Table 2). Alignment of the target sequences identified RRYGCCGT as the JUB1 consensus core binding sequence. Notably, all JUB1-selected motifs, except for pf17d127, had a fixed distance to the primer sequence flanking the 30-nucleotide random sequence, indicative of primer sequences establishing part of the JUB1 recognition site. To identify nucleotide positions required for efficient JUB1 binding, and to discover potential secondary motifs contributing to JUB1 binding, nucleotide substitution experiments were performed using the sequences of selected oligonucleotides, F15d64 and F17d127, as a starting point. This analysis demonstrated that the JUB1 binding sequence consists of two elements (Table 3). We observed at least two types of sequences that differ in their 3′ parts: TGCCGT(7N)ACG and TGCCGT(7N)CCGC (N, any nucleotide). However, the second element is not essential for JUB1 binding but increases binding affinity.
Table 2.
JUB1-Selected Binding Sequences
| JUB1-Selected Oligonucleotide (5′/3′) | JUB1 RBA | |
| Bio-RS-Oligo 1 | ||
| Pf15d21 | TCCCAATAGGATTCGTAAAGTGCCGTGTTCcgtccgccagcgcacc | 0.82 |
| Pf15d30 | AGCGAAGGATCAATTGAAGACGCCGTGATCcgtccgccagcgcacc | 0.74 |
| Pf15d25 | AATTTGACGTCATATTCTAACGCCGTAGTCcgtccgccagcgcacc | 0.83 |
| Pf15d64 | CAACATGAAGCTAGATGCCGTAGACcgtccgccagcgcacc | 1.18 |
| Bio-RS-Oligo 2 | ||
| Pf16d41 | TCACCCCCCCTTCTGAGGAACTCGGTGCCGTTCCTTTCcgtccacctgcag | 0.85 |
| Pf16d43 | CCTGTGACTTTCTCGAATCATGAGTGCCGTGCTCTCTCcgtccacctgcag | 0.48 |
| Pf16d44 | AGCGTATTCCCACTCCCGCTATGAGTGCCGTGCCCCCCcgtccacctgcag | 0.87 |
| Pf16d48 | TGCGAACCTTGTAGTGCTCCAGGATGCCGTACACCCCcgtccacctgcag | 0.84 |
| Pf16d82 | ATACTTTCCCCGAGTGTGATCGGGTGCCGTGCTCCCCcgtccacctgcag | 0.96 |
| Pf16d84 | CCATTTCGCCTTGCTGATTGCGCGGTGCCGTGTATCTCcgtccacctgcag | 0.96 |
| Pf16d88 | AAGCATTATCGTTGTTAAATACGGTGCCGTGTTCTGGCcgtccacctgcag | 0.66 |
| Pf16d98 | GGGCGGGCTGGTCTCGTATTGAGATGCCGTACTTGCCcgtccacctgcag | 0.92 |
| Pf16d100 | CCGATATCCTGTGAACTCAGCAAGATGCCGTCGTCCCCcgtccacctgcag | 0.89 |
| Pf16d102 | TTGGTGGTGCACGTATTTGATAGGGTGCCGTGTGTTCCcgtccacctgcag | 0.90 |
| Pf16d104a | ATGTTCGGCTGGATCTATATCACGATGCCGTGCGTTGCcgtccacctgcag | 1.20 |
| Pf16d106 | CCAATTCCCTTTTGCTGTTTAGTAGTGCCGTGCTGTCCcgtccacctgcag | 1.01 |
| Bio-RS-Oligo 3 | ||
| Pf17d51 | GGGACTTGTATACCTGTAAGGTGCCGTACCtcatgcggtacccacgtc | 0.87 |
| Pf17d52b | GGGAGGCCTCGTGCCAACCAGTGCCGTACGtcatgcggtacccacgtc | 1.07 |
| Pf17d54 | ACGTGATACACGCTCTATCAGTGCCGTGCCtcatgcggtacccacgtc | 0.85 |
| Pf17d57 | AGGCCGTTAAACATACATGAGTGCCGTACGtcatgcggtacccacgtc | 1.06 |
| Pf17d58 | ACACAATTGTGACGCGAAAGGTGCCGTACAtcatgcggtacccacgtc | 1.02 |
| Pf17d112 | CATCGGTTTCGGCCTTGTAGGTGCCGTACCtcatgcggtacccacgtc | 0.94 |
| Pf17d114 | TCCGTCCTCCGAGGATCATGATGCCGTACGtcatgcggtacccacgtc | 0.77 |
| Pf17d115 | TCAGGTACAACTCTGATGCAGTGCCGTACCtcatgcggtacccacgtc | 0.91 |
| Pf17d116 | CGAGCGTGGCCCAAAACACGGTGCCGTACCtcatgcggtacccacgtc | 0.94 |
| Pf17d117 | TCAGCTTGGCTGGAGCTAGGATGCCGTGGGtcatgcggtacccacgtc | 0.72 |
| Pf17d118 | GATCCCCCTCCTTGCCTCTAGTGCCGTACCtcatgcggtacccacgtc | 0.86 |
| Pf17d120 | TTCCCAGAACCTCTAACTGGATGCCGTACCtcatgcggtacccacgtc | 0.86 |
| Pf17d121 | GTACTAGATGCCGTACGtcatgcggtacccacgtc | 0.84 |
| Pf17d124 | AATGTCACTGTCCCCCTACAGTGCCGTGGCtcatgcggtacccacgtc | 0.91 |
| Pf17d126 | GGCTTAACCCGACAGCACAGACGCCGTGCCtcatgcggtacccacgtc | 0.94 |
| Pf17d127 | TGCCCAATGCCGTGTGTAGCACGCTGCCCA | 1.00 |
| Pf17d128 | AAATCCTTGTAAATCCCTAGATGCCGTACTtcatgcggtacccacgtc | 0.86 |
| Pf17d129 | GGTCGCACATCTCATCATGGATGCCGTACCtcatgcggtacccacgtc | 0.81 |
| Pf17d130 | ACGCACGTGTCTAGTATTGAGTGCCGTGCAtcatgcggtacccacgtc | 0.99 |
Thirty-six JUB1-selected oligonucleotides were obtained after five rounds of in vitro DNA binding site selection. Relative binding activity (RBA) of JUB1 to oligonucleotide Pf17d127 is set to 1. Values are based on a single assay. Nucleotides in lowercase letters are from flanking primer sequences. The JUB1 core binding sequence is in bold.
Table 3.
Base Substitution or Insertion Analysis of JUB1 Binding Motifs
| Synthetic Oligonucleotide Probe | JUB1 RBA | |
| F17d127 | TGCCCAATGCCGTGTGTAGCACGCTGCCCA | 1.00 ± 0.03 |
| F17d127m8 | TGCCCTTTGCCGTGTGTAGCACGCTGCCCA | 0.82 ± 0.05 |
| F17d127m7 | TGCCCAAAACCGTGTGTAGCACGCTGCCCA | 0.46 ± 0.02 |
| F17d127m1 | TGCCCAATGAAGTGTGTAGCACGCTGCCCA | 0.23 ± 0.01 |
| F17d127m3 | TGCCCAATGCCAAGTGTAGCACGCTGCCCA | 0.07 ± 0.01 |
| F17d127m9 | TGCCCAATGCCGTTTGTAGCACGCTGCCCA | 0.96 ± 0.02 |
| F17d127m13 | TGCCCAATGCCGTGAATAGCACGCTGCCCA | 1.00 ± 0.07 |
| F17d127m14 | TGCCCAATGCCGTGTGAAGCACGCTGCCCA | 1.02 ± 0.04 |
| F17d127m10 | TGCCCAATGCCGTGTGTTTCACGCTGCCCA | 1.10 ± 0.02 |
| F17d127m17 | TGCCCAATGCCGTGTGTAGAACGCTGCCCA | 1.02 ± 0.07 |
| F17d127m4 | TGCCCAATGCCGTGTGTAGTTCGCTGCCCA | 0.49 ± 0.01 |
| F17d127m2 | TGCCCAATGCCGTGTGTAGCAAACTGCCCA | 0.56 ± 0.02 |
| F17d127m5 | TGCCCAATGCCGTGTGTAGCACGAAGCCCA | 0.92 ± 0.05 |
| F17d127m15 | TGCCCAATGCCGTGTGTAGCACGCTAACCA | 1.05 ± 0.06 |
| F17d127m6f | TGCCCAATGCCGTGTGTAGCACGCTGTTCA | 1.06 ± 0.04 |
| F17d127m11 | TGCCCAATGCCGTGTGAAATAGCACGCTGCCCA | 0.08 ± 0.01 |
| F17d127m12 | TGCCCAATGCCGTGTGTACAAAACAAACCA | 0.19 ± 0.02 |
| F17d127m18a | TGCCCAATGCCGTGTGAAAACCGCCAGCCA | 1.01 ± 0.02 |
| F15d64 | AGCTAGATGCCGTAGACCGTCCGCCAGCGC | 0.84 ± 0.02 |
| F15d64m1 | AGCTAGATGCCGTAGACCGTAAGCCAGCGC | 0.45 ± 0.05 |
| F15d64m2 | AGCTAGATGCCGTAGACCGTCCAACAGCGC | 0.35 ± 0.02 |
| F15d64m3 | AGCTAGATGCCGTAGACCGTCCGCATGCGC | 0.59 ± 0.01 |
| F15d64m4 | AGCTAGATGCCGTAGACCGTCCGCCAAAGC | 0.73 ± 0.04 |
Nucleotides of motif 1 and motif 2 of the JUB1 binding site are shown in bold. Mutated nucleotides are underlined. Values are means ± sd of three replicated assays. RBA, relative binding activity.
Nucleotides shown in italics in F17d127m18 are derived from the flanking primer sequence of Pf15d64 (see Table 2).
DREB2A Is a Direct Target of JUB1
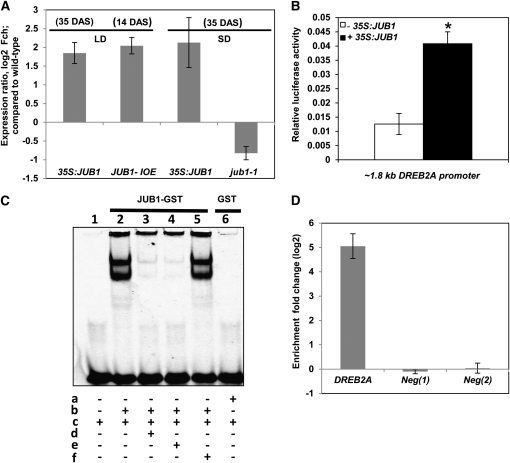
One of the genes induced by H2O2 in RD29A:JUB1 seedlings that attracted our attention was DREB2A, a member of the AP2/EREBP TF family whose function with respect to plant responses to various abiotic stresses, such as cold and drought, is well established (Sakuma et al., 2006a, 2006b). DREB2A was approximately fivefold more induced by H2O2 in the RD29A:JUB1 overexpressor than wild-type seedlings (Table 1). However, enhanced expression of DREB2A in JUB1 overexpressors was also observed in the absence of H2O2 treatment. We found that DREB2A expression was elevated by more than threefold in 35-d-old 35S:JUB1 plants compared with wild-type plants (Figure 10A). By contrast, reduced DREB2A expression (~1.7-fold decrease) was observed in the jub1-1 mutant when compared with the wild type. We also tested DREB2A expression in 2-week-old JUB1-IOE seedlings shortly (~3 h) after EST treatment. DREB2A expression was almost fourfold induced in the EST-treated lines, suggesting DREB2A as a JUB1 target gene (Figure 10A). Notably, the DREB2A promoter harbors a perfect match to the full JUB1 binding sequence (TGCCGTNNNNNNNACG) ~1 kb upstream of its translation start site.
Figure 10.
DREB2A Is a Direct Target of JUB1.
(A) Expression of DREB2A in 35S:JUB1, JUB1-IOE, and jub1-1 lines compared with the wild type. Plant ages are indicated in days after sowing (DAS). LD, long day; SD, short day. Numbers on the y axis indicate expression fold change (log2 basis) compared with the wild type. Data represent means ± sd of five (LD) or three (SD) independent experiments.
(B) Transactivation of DREB2A expression (from its ~1.8-kb promoter) by JUB1 in Arabidopsis mesophyll cell protoplasts. The ProDREB2A:FLuc construct harboring the DREB2A promoter upstream of the firefly (Photinus pyralis) luciferase (FLuc) open reading frame was cotransformed with the 35S:JUB1 plasmid (omitted in control experiments). The 35S:RLuc vector was used for transformation efficiency normalization. Bars indicate the sd of at least four biological replicates. The asterisk indicates significant difference to control at P < 0.05.
(C) EMSA. Purified JUB1-GST protein binds specifically to the JUB1 binding site within the DREB2A promoter. In vitro DNA binding reactions were performed with the 40-bp wild-type fragment of the DREB2A promoter containing the JUB1 motif (5′-GATGCCGTTAGAGACACG-3′). a, GST protein; b, JUB1-GST protein; c, 5′-DY682 double-stranded oligonucleotide containing the perfect JUB1 binding site; d, 100× competitor (unlabeled oligonucleotide containing perfect JUB1 binding site); e, 200× competitor (unlabeled oligonucleotide containing perfect JUB1 binding site); f, 200× mutated oligonucleotide (unlabeled with mutation in JUB1 binding site where 5′-GATGCCGTTAGAGACACG-3′ was replaced by 5′-GATGCCAATAGAGACACG-3′).
(D) ChIP-qPCR. Whole shoots of 35-d-old Arabidopsis plants expressing GFP-tagged JUB1 under the control of the CaMV 35S promoter (35S:JUB1-GFP) and wild-type plants were harvested for the ChIP experiment. qPCR was used to quantify enrichment of the DREB2A promoter. As negative controls, primers annealing to promoter regions of two Arabidopsis genes lacking a JUB1 binding site, At3g18040 (Neg 1) and At2g22180 (Neg 2), were used. Data represent means ± sd of three independent experiments.
To provide further evidence for regulation of DREB2A by JUB1, we performed luciferase-based transactivation assays in Arabidopsis mesophyll cell protoplasts using the ~1.8-kb DREB2A promoter (including the JUB1 binding site) fused to the firefly luciferase reporter. JUB1 significantly transactivated the 1.8-kb DREB2A promoter (Figure 10B).
We next performed an electrophoretic mobility shift assay (EMSA) to test the physical interaction of JUB1 with the DREB2A promoter. As shown in Figure 10C, JUB1 interacts with a 40-bp DREB2A promoter fragment harboring the JUB1 binding site. Finally, we used chromatin immunoprecipitation–quantitative PCR (ChIP-qPCR) to demonstrate that JUB1 binds also in vivo to the DREB2A promoter (Figure 10D). Thus, our data demonstrate that JUB1 is an upstream transcriptional regulator of DREB2A.
Metabolite Profiling Reveals Accumulation of Trehalose and Pro in JUB1 Overexpressors
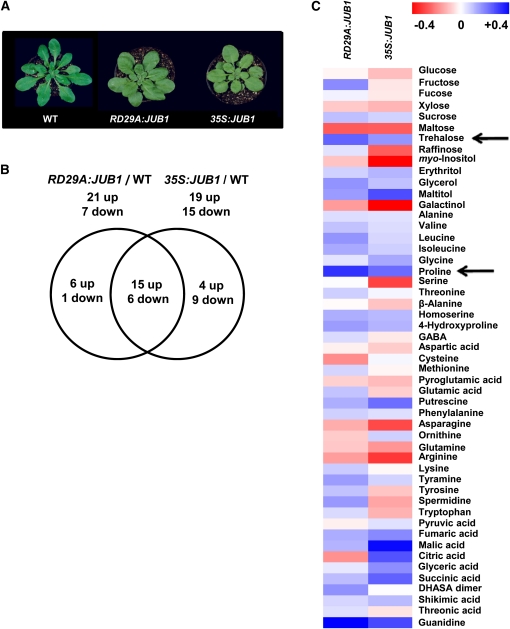
Next, we were interested to know how overexpression of JUB1 affects metabolism of the corresponding plants. Therefore, the profile of primary metabolites in rosette leaves was compared between wild-type and JUB1 overexpression lines (35S:JUB1 and RD29A:JUB1). Metabolic profiling by gas chromatography coupled to mass spectrometry (GC-MS) was performed on extracts from rosette leaves of 35-d-old plants (Figure 11A) and metabolic profiles were compared (Figure 11B). A total of 51 metabolites of known chemical structure were accurately quantified in every chromatogram. These compounds mostly included amino acids, carbohydrates (sugars and sugar alcohols), and organic acids. Overall, 67% (34 out of 51) and 55% (28 out of 51), respectively, of the identified metabolites revealed a significant difference (*P < 0.05, Student’s t test) when 35S:JUB1 and RD29A:JUB1 metabolite profiles were compared with the wild type (for an overview, see Figure 11C and Supplemental Data Set 3 online). Metabolite profiles of the two types of overexpression plants were highly similar to each other: of the 19 metabolites upregulated in 35S:JUB1 plants, 15 were also elevated in RD29A:JUB1 plants. Similarly, of the 15 metabolites downregulated in 35S:JUB1 plants, six were also reduced in RD29A:JUB1 plants compared with the wild type (Figure 11B). In general, the content of the major organic acids increased in the overexpression lines compared with the wild type. Among these, the level of glyceric acid and various tricarboxylic acid cycle intermediates, fumaric acid, malic acid, citric acid, and succinic acid, significantly increased in the 35S:JUB1 line compared with wild-type plants. Most of these components were just slightly increased in RD29A:JUB1 plants. Shikimic acid content was significantly higher in both types of overexpression plants. Our data revealed that the most drastic changes were detectable in amino acids and carbohydrates. For example, among amino acids, a significant increase was observed for Pro and 4-hydroxy-Pro levels in both overexpression plants. Polyamines and compatible osmolytes, such as Pro, are known to be involved in the plant’s responses to various environmental stresses, including osmotic and salt stress, heat stress, drought, cold, and pathogen infection (e.g., Yoshiba et al., 1997; Bhatnagar-Mathur et al., 2008; Verbruggen and Hermans, 2008; Gill and Tuteja, 2010). Among the detected disaccharides, the proportion of Suc significantly increased in both overexpression lines compared with the wild type. Moreover, a significant increase in the level of trehalose was observed in both overexpression lines. Trehalose is a nonreducing disaccharide of Glc that functions as an osmoprotectant (Müller et al., 1995) and stabilizes biological structures under abiotic stress conditions in bacteria, fungi, and invertebrates (Djilianov et al., 2005). Among the sugar alcohols, the levels of maltitol, glycerol (a compatible osmolyte), and erythritol were significantly higher in both overexpression lines, and the level of myo-inositol was significantly decreased in these lines compared with the wild type. In general, it appears that JUB1 overexpressors accumulate higher levels of trehalose, Pro, and polyols (glycerol) than wild-type plants, which is in accordance with their enhanced tolerance to abiotic stress.
Figure 11.
Primary Metabolite Profiling of JUB1 Overexpression Plants.
(A) Phenotype of 35-d-old wild-type (WT), RD29A:JUB1, and 35S:JUB1 plants subjected to metabolite profiling.
(B) Venn diagram showing an overview of metabolites that are significantly different (P < 0.05, Student’s t test) in 35S:JUB1 and RD29A:JUB1 lines compared with the wild type.
(C) Hierarchical average linkage clustering of all detected primary metabolites. For every metabolite, the metabolic content of the wild type was considered as 1 and the metabolic content of overexpression lines was normalized to that. Metabolic ratios: red, minimum (between 0 and −0.4); blue, maximum (between 0 and + 0.4); see also Supplemental Data Set 3 online. Arrows indicate increased trehalose and Pro content in JUB1 overexpressors compared with the wild type.
Secondary Metabolite Profile of JUB1 Transgenics
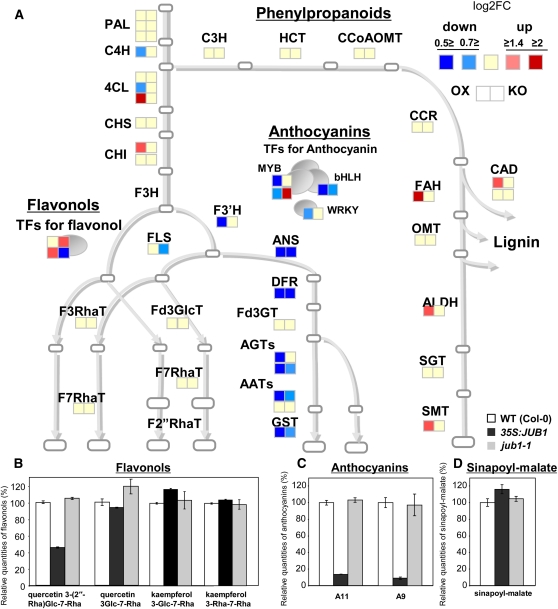
Secondary metabolites confer various advantages to the plants, such as regulation of development and response to biotic and abiotic stresses. To study secondary metabolite profiles of JUB1 transgenics, liquid chromatography–mass spectrometry (LC-MS) analysis was performed on extracts from rosette leaves of 35-d-old JUB1 overexpression and jub1-1 lines. Moreover, to identify JUB1-regulated genes involved in secondary metabolism, we tested by qRT-PCR the expression of 94 genes encoding enzymes and TFs that regulate the biosynthetic pathway from shikimate to phenylpropanoids, including anthocyanins, flavonols, and sinapoyl derivatives (see Supplemental Data Set 4 online). Our metabolite analysis revealed the most drastic changes for cyanidin derivatives (Figure 12). Cyanidin derivative (A11 and A9; Tohge et al., 2005) levels were significantly decreased in 35S:JUB1 plants, whereas no significant change was observed for the level of anthocyanidins in the jub1-1 mutant. Accordingly, expression of genes encoding dihydroflavonol 4-reductase (DFR; At5g42800), a key enzyme in shunting flavonols into the anthocyanins, leucoanthocyanidin dioxygenase (LDOX; At4g22880), anthocyanin glycosyltransferases (A5GT, At4g14090; A3G2”XT, At5g54060), anthocyanin acyltransferases (A5GMaT, At3g29590; A3GCouT, At1g03940), and GST (TT19, At5g17220) were significantly downregulated in JUB1 overexpression plants. Similarly, expression of anthocyanin regulatory TFs of the MYB (PAP1 and PAP2), bHLH (TT8), and WRKY (TTG2) families was also reduced in JUB1 overexpression plants compared with the wild type. Expression of PRODUCTION OF ANTHOCYANIN PIGMENT2 (PAP2) was significantly induced in jub1-1 knockdown plants (see Supplemental Figure 12 online). Apart from PAP2, expression of anthocyanin biosynthesis genes was either not affected or only slightly reduced in jub1-1 knockdown plants. These data indicate that JUB1, most probably in conjunction with other TFs, negatively regulates the expression of the anthocyanin biosynthesis genes. Anthocyanins represent a group of flavonoids that accumulate under conditions of various types of environmental stresses. Moreover, they accumulate in senescing leaves preceding chlorophyll breakdown and play a photoprotective role against strong light in combination with coolness that may occur during autumn (Hoch et al., 2003; Diaz et al., 2006). The reduced level of anthocyanins in JUB1 overexpression plants is consistent with their prolonged longevity.
Figure 12.
Gene Expression Profiling of Phenylpropanoids, Flavonols, and Anthocyanins, and Metabolite Profiling of Related Secondary Metabolites in 35S:JUB1, jub1-1, and Wild-Type Plants.
(A) Expression of 33 enzymatic genes and six TFs as measured by qRT-PCR. Intensity of fold change against wild-type (WT) expression level (log2FC) is indicated by color. Abbreviations are given in Supplemental Table 2 online.
(B) to (D) The content of flavonols [quercetin-3-O-(2′′-O-Rha)Glc-7-O-Rha, quercetin-3-O-Glc-7-O-Rha, kaempferol-3-O-Glc-7-O-Rha, and kaempferol-3-O-Rha-7-O-Rha], anthocyanins (A11 and A9; Tohge et al., 2005), and sinapoyl-malate in 35S:JUB1, jub1-1, and wild-type plants was analyzed by LC-MS. Average of two biological replicates ± sd.
Of the other secondary metabolites, one of the major phenylpropanoid compounds, sinapoylmalate, was significantly induced in the JUB1 overexpression plants (Figure 12). Similarly, transcript levels of several genes encoding enzymes of phenylpropanoid metabolism, such as FAH1 (encodes ferulate-5-hydroxylase), ALDH (aldehyde dehydrogenase 2C4), and SMT (sinapoylglucose:malate sinapoyltransferase, which catalyzes the formation of sinapoylmalate from sinapoylglucose), were induced in JUB1 overexpression plants (see Supplemental Figure 12 online).
Hormonal Adjustments in JUB1 Transgenics
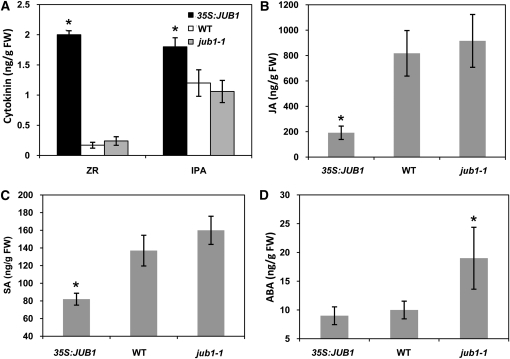
Plant hormones play an important role in regulating senescence (in particular cytokinins) and the response to stress (including ABA, salicylic acid [SA], and jasmonic acid [JA]). We determined the concentrations of these hormones in rosette leaves of 43-d-old 35S:JUB1, jub1-1, and wild-type plants. Significantly higher levels of isopentenyladenosine (IPA) and zeatin riboside (ZR) were detected in leaves of 35S:JUB1 transgenics compared with jub1-1 and wild-type plants (Figure 13A), while levels of zeatin (Z), dihydrozeatin (DHZ), and dihydrozeatin riboside (DHZR) were not significantly altered (see Supplemental Figure 6C online). By contrast, levels of the biotic stress hormones JA and SA were significantly reduced in 35S:JUB1 lines compared with the wild type (Figures 13B and 13C), while ABA was not affected (Figure 13D). However, ABA was increased by ~94% in jub1-1 mutant plants compared with the wild type (Figure 13D).
Figure 13.
Hormone Contents in 35S:JUB1, jub1-1, and Wild-Type Plants.
Determination of ZR and IPA (A), JA (B), SA (C), and ABA (D) in 43-d-old 35S:JUB1, jub1-1, and wild-type plants grown at long-day conditions (16 h light/8 h dark). Values represent the means ± sd from five independent sets of samples. Asterisks indicate significant differences compared with the wild type (WT) (P < 0.05, Student’s t test). FW, fresh weight.
DISCUSSION
To identify novel regulators of plant senescence, we screened NAC overexpression and T-DNA insertion lines for changes in leaf senescence. We previously reported the identification of ORS1 as a TF that positively regulates senescence (Balazadeh et al., 2011). Similarly, At NAP and ORE1 (ANAC092) have been shown to act as positive regulators of senescence in Arabidopsis (Guo and Gan, 2006; Kim et al., 2009; Balazadeh et al., 2010a). Here, we discovered another member of the NAC gene family, designated JUB1, which in contrast with these previously characterized NAC factors, strongly delays senescence when overexpressed in transgenic plants and triggers precocious senescence at low expression level (in the jub1-1 mutant and artificial microRNA lines). Thus, JUB1 represents a strong negative regulator of senescence whose molecular function may differ from those of the positively acting NAC factors. Notably, the expression of all four NACs is triggered by H2O2, although the H2O2-dependent induction is slightly less pronounced for ORE1 compared with At NAP, ORS1, and JUB1 (Balazadeh et al., 2010b). Additionally, the expression of several other senescence-regulated NAC genes, such as ANAC032, ATAF1, and ANAC102, is triggered by H2O2 (Balazadeh et al., 2010b). This observation is interesting and suggests a close regulatory node connecting the accumulation of cellular H2O2 to the regulation of senescence and possibly bolting, a developmental process often tightly linked with the onset of leaf senescence (Levey and Wingler, 2005; Balazadeh et al., 2008a). However, a distinct role during senescence has not been reported for the other NACs so far. Recently, VNI2 (ANAC083; At5g13180) was found to regulate senescence by integrating ABA signaling (Yang et al., 2011); it also regulates xylem vessel specification (Yamaguchi et al., 2010). Currently, however, it remains largely unknown how these diverse cellular functions are integrated by VNI2.
Similar to observations in animals, extended longevity in plants is known to be correlated with increased tolerance to oxidative stress (Finkel and Holbrook, 2000; Muller et al., 2007). The correlation between stress tolerance and the onset of senescence and determination of life span in plants is supported by experimental evidence (Jing et al., 2003). Additionally, increased stress tolerance was also observed for late-flowering/long-living gigantea, ore1, ore3, and ore9 mutants (Kurepa et al., 1998; Woo et al., 2004). It has been suggested that aging is triggered by oxidative stress as a result of an imbalance between production and scavenging of oxygen radicals. In plants, this hypothesis is in part supported by the observation that timing of senescence is altered in mutants with a decreased level of the antioxidant l-ascorbic acid (vitamin C). The vtc1 mutant enters senescence prematurely and is more sensitive to various oxidative stresses than the corresponding wild type (Barth et al., 2004). Compared with other ROSs, H2O2 has a relatively long half-life of ~1 ms, although its stability is influenced by the cellular pH and redox equilibrium (Reth, 2002). H2O2 acts as a signaling molecule that regulates plant development and adaptation to various stresses. It has been observed that a decrease of catalase (CAT2) and cytosolic ascorbate peroxidase 1 (APX1) activities during bolting time is followed by an accumulation of H2O2 and an enhanced expression of the senescence-associated TF WRKY53, suggesting H2O2 functions as a signal to promote senescence (Ye et al., 2000; Miao et al., 2004; Zimmermann et al., 2006).
Although the precise molecular pathways through which JUB1 regulates longevity and abiotic stress tolerance are not known at present, one possible scenario is that it does so by affecting a gene regulatory network that possibly involves DREB2A, its direct downstream target. DREB2A is an important transcription regulator acting in response to various abiotic stresses (e.g., Sakuma et al., 2006a; Kant et al., 2008). Transcriptome studies have shown that DREB2A activates a large number of abiotic stress–responsive genes involved in drought and heat stress responses (Sakuma et al., 2006b). The heat shock TF gene HsfA3 has been identified as a direct downstream target of DREB2A during heat stress (Schramm et al., 2008; Yoshida et al., 2008), and DREB2A itself is a heat shock–responsive gene (Suzuki et al., 2011). Another direct target of DREB2A is RESPONSIVE TO DESSICATION29A (RD29A; also referred to as COR78; Liu et al., 1998). Although we have not experimentally tested heat stress–dependent JUB1 expression here, global transcriptome data from Swindell (2006) identified JUB1 as a heat stress–responsive gene. Another observation of interest is that all three genes are significantly upregulated by H2O2 treatment (10 mM, 5 h) in Arabidopsis seedlings, although induction levels varied between the three TF genes (JUB1, ~25-fold; DREB2A, ~90-fold; HsfA3, approximately threefold; see Supplemental Figure 6D online). Taken together, our data in conjunction with published reports (Schramm et al., 2008; Yoshida et al., 2008) establish an extended transcriptional cascade involving three consecutive positive regulators (JUB1–DREB2A–HsfA3), with JUB1 adopting an upstream position. Furthermore, HsfA3 has been suggested to be part of an expression amplification loop (Nishizawa-Yokoi et al., 2011) involving two additional heat shock TFs (i.e., HsfA1e and HsfA2), where HsfA1e activates HsfA2 expression (likely by direct binding to heat shock cis-elements present in its promoter; Nishizawa-Yokoi et al., 2011), HsfA2 activates HsfA3 expression (Schramm et al., 2006), and HsfA3 stimulates expression of HsfA1e (Yoshida et al., 2008). HsfA2 has been shown to directly regulate the expression of APX2, which encodes a key cytosolic enzyme for the detoxification of H2O2 (Shigeoka et al., 2002; Nishizawa et al., 2006; Schramm et al., 2006), consistent with our observation of reduced H2O2 level in JUB1 overexpressors. The control network linking H2O2 signaling with this Hsf activation loop most likely involves additional transcriptional regulators, including MBF1c, which was shown to be required for enhanced expression of DREB2A during heat stress (Suzuki et al., 2011). Notably, MBF1c is highly responsive to abiotic stresses, including heat stress (Suzuki et al., 2011), and is also rapidly and strongly upregulated by H2O2 treatment (~30-fold up already after 1 h at 10 mM H2O2; see Supplemental Figure 6E online). As we have shown here (Table 1), MBF1c is significantly more upregulated after H2O2 challenge in JUB1 overexpressors than in the jub1-1 mutant, further supporting the model of a regulatory link between JUB1 and MBF1c upstream of DREB2A and HsfA3. In accordance with this model is the observation that MBF1c contains a JUB1 binding site (CGCCGT) in its promoter at around 640 bp upstream of the transcription start site; however, we have not yet tested its functional relevance.
Our analysis presented here also identified changes in primary and secondary metabolism in JUB1 transgenic lines. In general, our analysis revealed an accumulation of various compatible solutes, including trehalose, Pro, and various sugar alcohols (maltitol, glycerol, and erythritol) in JUB1 overexpressors compared with wild-type plants, which may contribute to the enhanced abiotic stress tolerance of such lines. Although the role of these metabolites in senescence is not well established, examples indicate that trehalose and sugar alcohols (mannitol and inositol) delay senescence of cut flowers in some species (reviewed in van Doorn and Woltering, 2008). We also noticed a significant increase in Suc concentration due to JUB1 overexpression. At the level of secondary metabolites, we observed a decrease of cyanidin derivatives upon JUB1 overexpression, which was accompanied by reduced expression of anthocyanin biosynthesis genes (including DFR, LDOX, A5GT, and others) and known anthocyanin regulatory TFs (i.e., PAP1, PAP2, TTS, and TTG2). By contrast, expression of PAP2 was significantly induced in jub1-1 knockdown plants. Our data thus indicate that JUB1, possibly jointly with other TFs, has a negative effect on the expression of anthocyanin biosynthesis genes. Anthocyanins accumulate as a response to various types of environmental stresses and in senescing leaves, where they play a photoprotective role against high light stress in combination with low temperature, which occurs, for instance, in autumn (Hoch et al., 2003; Diaz et al., 2006). The reduced anthocyanin level in JUB1 overexpression plants is consistent with their prolonged longevity. Furthermore, minor flavonol glycosides, such as quercetin glycosides, were slightly decreased in leaves of JUB1 overexpressors. This may be due to the downregulated expression of F3′H (TT7) caused by the suppression of PAP1. Despite this fact, the increased levels of other effective phenolic antioxidants, like sinapoyl-malate and major glycosides of kaempferol, may contribute to the enhanced oxidative stress tolerance (Figure 12).
At the hormone level, we found higher levels of cytokinins (ZR and IPA) in JUB1 overexpressors. Cytokinins are important regulators of senescence, and leaf senescence and the induction of SAGs can only be initiated when cytokinin levels are below a threshold (Gan and Amasino, 1995; Noodén et al., 1997). The high cytokinin level in JUB1 overexpressors is thus in accordance with their extended lifespan. On the contrary, ABA level increased in the jub1-1 mutant, indicating cellular stress, possibly due to disturbed H2O2 homeostasis in these plants.
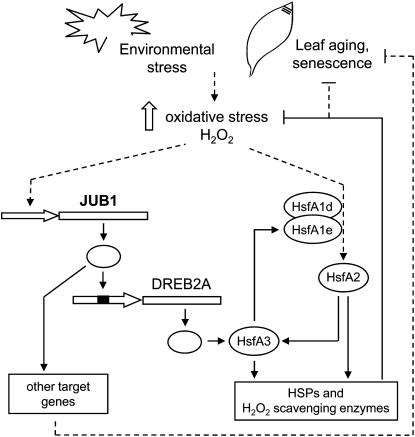
Model for JUB1 Action
Based on the available experimental data, we propose the following model for JUB1 action (Figure 14). JUB1 transcription is activated by a rise of endogenous H2O2 concentration that is triggered by developmental input or environmental stress, including wounding, salinity stress, or cellulase treatment. Notably, JUB1 expression follows the cellular H2O2 concentration that changes with plant development, showing a peak during bolting (Zimmermann et al., 2006). The upstream TF(s) regulating H2O2-dependent or senescence-associated JUB1 transcription remains unknown at present. JUB1 directly targets DREB2A, which functions as a positive regulator of HsfA3 and RD29A. HsfA3 itself regulates the expression of HSP genes and is part of a positive feedback loop together with HsfA1e and HsfA2 (Nishizawa-Yokoi et al., 2011), whereas the molecular function of RD29A is not known. Elevation of JUB1 expression lowers H2O2 concentration in plant tissues, possibly through the Hsf amplification loop, while the opposite effect (i.e., increased H2O2 concentration) is observed in the jub1-1 mutant, suggesting that JUB1 assists in regulating cellular H2O2 homeostasis. Although the precise molecular mechanism through which JUB1 regulates intracellular H2O2 concentration remains to be established, the current model proposes an involvement of HSPs and H2O2 scavenging enzymes. Notably, several GST genes (including GST10, 24, and 25) were induced in JUB1 overexpressors after H2O2 treatment, while expression of these genes remained either unchanged or was slightly reduced in the jub1-1 mutant (Table 1).
Figure 14.
Model of JUB1 Action.
JUB1 is activated by H2O2 and during leaf senescence. JUB1 TF binds to the DREB2A promoter, thereby activating its expression. DREB2A positively regulates the expression of HsfA3 (Schramm et al., 2008; Yoshida et al., 2008), thus establishing a transcriptional cascade. As suggested by Nishizawa-Yokoi et al. (2011), HsfA3 together with Hsf1A1e and HsfA2 form an expression amplification loop. HsfA3 and HsfA2 regulate the expression of HSPs and H2O2 scavenging enzymes, leading to reduced intracellular H2O2 levels, extended longevity, and increased stress tolerance. Increased longevity may also be regulated through other JUB1 target genes.
Concomitant with enhanced JUB1 expression, we observed reduced expression of many SAGs, which is in accordance with the delayed senescence observed in these lines. Notably, expression of SAGs is increased in the jub1-1 mutant, constituting JUB1 as a negative regulator of senescence. A possible explanation of the reduced expression of SAGs in JUB1 overexpressors may be derived from the observation that expression of many SAGs is enhanced by H2O2 (e.g., Navabpour et al., 2003; Balazadeh et al., 2010b; Genevestigator at http://www.genevestigator.com). In particular, expression of the majority of the senescence-associated NAC TFs is also triggered by H2O2 treatment (15 NACs in total), including At NAP, ORS1, and ORE1 (Balazadeh et al., 2010b), all of which have been shown to affect senescence positively. Thus, our model suggests that JUB1 lowers the cellular H2O2 level, thereby minimizing the stimulatory effect on NAC gene expression and, hence, senescence. On the contrary, reduced JUB1 expression (such as in the jub1-1 mutant) would favor the accumulation of cellular H2O2 that drives NAC gene expression and through this supports precocious senescence. However, there is also the additional possibility that JUB1 regulates senescence through other target genes of currently unknown molecular function (Figure 14). Future work will have to address the intricacies of the underlying regulatory network in greater detail.
METHODS
General
Standard molecular techniques were performed as described (Sambrook et al., 2001; Skirycz et al., 2006). Oligonucleotide sequences are given in Supplemental Data Set 5 online. Chemicals and reagents for GC-MS analysis were obtained from Sigma-Aldrich, Fluka, or Merck with the exception of N-methyl-N-(trimethylsilyl) trifluoroacetamide, which was obtained from Macherey-Nagel. For sequence analyses, the tools provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), MIPS (http://mips.gsf.de/), The Arabidopsis Information Resource (http://www.arabidopsis.org/), and the Plant Transcription Factor Database (http://plntfdb.bio.uni-potsdam.de/v3.0/) were used.
Plants
Seeds of Arabidopsis thaliana accession Col-0 were obtained from the Arabidopsis thaliana Resource Centre for Genomics (Institut National de la Recherche Agronomique, France; http://dbsgap.versailles.inra.fr/publiclines/). For growth under long-day conditions, seedlings were grown in soil (Einheitserde GS90; Gebrüder Patzer) in a climate-controlled chamber with a 16-h daylength provided by fluorescent light at ~100 μmol m−2 s−1 and a day/night temperature of 20/16°C and a RH of 60/75%. After 2 weeks, seedlings were transferred to a growth chamber with a 16-h day (80 or 120 μmol m−2 s−1) and a day/night temperature of 22/16°C and 60/75% RH. For growth under short-day conditions, the light period was reduced to 8 h. Growth in hydroponic culture, salinity treatment, and sample preparation were done as described using stage 1 plants (28 d old) (Balazadeh et al., 2010a). T-DNA insertion lines screened for extended longevity (see Supplemental Table 1 online) were obtained from the European Arabidopsis Stock Centre (http://Arabidopsis.info/). Homozygous plants were identified by PCR using the T-DNA left border primer, as well as the gene-specific primers LP and RP.
Constructs
Constructs were generated by PCR- and restriction enzyme-mediated cloning. Primer sequences are given in Supplemental Data Set 5 online. PCR-generated amplicons were checked by DNA sequence analysis (MWG). Constructs were transformed into Arabidopsis Col-0 via Agrobacterium tumefaciens–mediated transformation.
For 35S:JUB1, the JUB1 open reading frame was amplified by PCR from Arabidopsis Col-0 leaf cDNA and inserted into pUni/V5-His-TOPO (Invitrogen). The cDNA was cloned via added PmeI-PacI sites into a modified pGreen0229-35S plant transformation vector (Skirycz et al., 2006). For RD29A:JUB1, the RD29A promoter (1 kb upstream of translation start site) was amplified from Arabidopsis (Col-0) genomic DNA, cloned into pCR2.1 (Invitrogen), and then transferred via BamHI and NcoI sites into pCAMBIA1305.1-hygromycin, giving rise to plasmid RD29A:pCAMBIA. The JUB1 coding region was amplified by PCR from leaf cDNA using primers JUB1-forward and JUB1-reverse and cloned downstream of the RD29A promoter in plasmid RD29A:pCAMBIA via primer-added NcoI and PmlI sites. For 35S:JUB1-GFP, the full-length JUB1 open reading frame was amplified without its stop codon. The PCR product was cloned into the pENTR/D-TOPO vector using the pENTR Directional TOPO cloning kit (Invitrogen). The sequence-verified entry clone was then transferred to the pK7FWG2 vector (Ghent University) by LR recombination (Invitrogen). For JUB1-IOE, the JUB1 coding region was amplified by PCR from Arabidopsis leaf cDNA using primers JUB1-IOE-fwd and JUB1-IOE-rev, inserted into pBluescript SK+, and then cloned via XhoI and SpeI sites into the pER8 vector (Zuo et al., 2000). For the ProJUB1:GUS fusion, an ~1.8-kb 5′ genomic fragment upstream of the translation initiation codon was amplified by PCR from Arabidopsis Col-0 genomic DNA, inserted into plasmid pGEM-T Easy (Promega), and fused via HindIII (present in pGEM-T Easy) and NcoI restriction sites to the GUS reporter gene in pCAMBIA1305.1-hygromycin (CAMBIA). For JUB1-amiRNA, the Web MicroRNA Designer platform (http://wmd2.weigelworld.org/cgi-bin/mirnatools.pl?page=1) was used to design amiRNA sequences (21-mers). For ProDREB2A:FLuc, the ~1.8-kb DREB2A promoter containing the JUB1 binding site was amplified by PCR from Arabidopsis genomic DNA and inserted into the pENTR/D-TOPO vector (Invitrogen). The sequence-verified promoter was then transferred to the p2GWL7.0 vector harboring the firefly (Photinus pyralis) luciferase (FLuc) coding region (Licausi et al., 2011) by LR recombination (Invitrogen).
Expression Profiling by qRT-PCR
Total RNA extraction, synthesis of cDNA, and qRT-PCR were performed as described (Caldana et al., 2007; Balazadeh et al., 2008b). Expression analysis platforms contained primer pairs for 168 SAGs (Parlitz et al., 2011), 179 ROS-responsive genes, and 94 phenolic secondary metabolite biosynthetic genes. Genes included in the SAG platform are highly upregulated during natural senescence in wild-type Arabidopsis plants (Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Balazadeh et al., 2008b). ROS-responsive genes were extracted from the literature (Gechev et al., 2004, 2005; Davletova et al., 2005a, 2005b; Gadjev et al., 2006) and in-house experiments. Primers for the metabolite platform were designed for the biosynthetic genes from primary metabolism to flavonoid production; the platform also included primers for other phenolic secondary metabolite biosynthetic genes and TFs that control anthocyanin biosynthesis. Genes included in the qRT-PCR platforms, including primer sequences, are given in Supplemental Data Set 5 online. Primers were designed using QuantPrime (Arvidsson et al., 2008). PCR reactions were run on an ABI PRISM 7900HT sequence detection system (Applied Biosystems Applera), and amplification products were visualized using SYBR Green (Applied Biosystems). ACTIN2 served as reference gene; primers were Actin2-F (5′-TCCCTCAGCACATTCCAGCAGAT-3′) and Actin2-R (5′-AACGATTCCTGGACCTGCCTCATC-3′).
DNA Binding Site Selection
In vitro binding site selection was performed using the CELD system with the pTacJUB1-LCELD6XHis construct, employing three biotin-labeled double-stranded oligonucleotides (i.e., Bio-RS-Oligo 1, RS-Oligo 2, and Bio-RS-Oligo 3), which contained 30-nucleotide random sequences that differed in flanking primer sequence (Xue, 2005). JUB1-selected oligonucleotides were cloned and sequenced. The DNA binding activity of JUB1-CELD was measured using methylumbelliferyl β-d-cellobioside as substrate (Xue, 2002). DNA binding assays with a biotin-labeled single-stranded oligonucleotide or a biotin-labeled double-stranded oligonucleotide without a target binding site were used as controls.
Transactivation Assays
Arabidopsis mesophyll cell protoplasts were prepared according to the protocol of Sheen (2002). The construct containing the ~1.8-kb DREB2A promoter fragment in front of the FLuc coding region (ProDREB2A:FLuc) was cotransformed in the presence or absence of the 35S:JUB1 plasmid. The 35S:RLuc vector (Licausi et al., 2011) was used for normalization against transformation efficiency. Firefly luciferase and Renilla luciferase (RLuc) were assayed using the Dual Luciferase Reporter Assay System (Promega). Six micrograms of DNA were used for transient transformation of protoplasts according to Yoo et al. (2007); 16 h after incubation, protoplasts were lysed by adding 400 μL of passive lysis buffer, and the resulting suspension was briefly vortexed. Forty microliters of luciferase assay buffer was added to the same volume of crude extract and FLuc activity was measured. Forty microliters of Stop and Glow buffer was then added and Renilla chemiluminescence was measured. Relative light units were determined in a GloMax 20/20 luminometer (Promega) using a 10-s measurement. Data were collected as ratio (FLuc activity:RLuc activity). Protoplasts transformed with only the promoter-FLuc, 35S:RLuc reporter plasmid (no TF), were analyzed as background controls.
EMSA
JUB1-GST fusion protein was purified from Escherichia coli expression strain BL21 Star (DE3) pRARE, which was generated by transforming the pRARE plasmid isolated from Rosetta (DE3) pRARE cells (Merck) into E. coli BL21 Star (DE3) (Invitrogen). Protein expression was induced in a 100-mL expression culture using 1 mM isopropyl thio-β-d-galactoside, and cells were harvested 4 h after induction at 30°C. Cells were sonicated in lysis buffer (20 mM sodium phosphate buffer, pH 7.3, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 1 mM phenylmethanesulfonyl fluoride). Supernatant of centrifuged sample was used for purification using a 1-mL GSTrap HP column (GE Healthcare) coupled to the Äkta-Purifier FPLC system (GE Healthcare). Aliquots of the flow-through fractions were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. One-milliliter elution fractions containing the purified JUB1-GST fusion protein were pooled and dialyzed against PBS buffer (20 mM Na-phosphate, pH 7.4, and 150 mM NaCl). Protein concentration was determined by the Bradford assay (Bradford, 1976). 5′-DY682-labeled DNA fragments were ordered from MWG. Sequences of labeled DNA fragments, unlabeled competitors, and mutated fragments are given in Supplemental Data Set 5 online. Annealing was performed by heating the primers to 100°C followed by slow cooling to room temperature. The binding reaction was performed at room temperature for 20 min as described in the Odyssey Infrared EMSA kit instruction manual. DNA-protein complexes were separated on 6% retardation gel, while DY682 signal was detected using the Odyssey Infrared Imaging System from LI-COR Biosciences.
In Vivo Binding of JUB1 to the DREB2A Promoter
To investigate in vivo binding of JUB1 to its DNA binding site in the DREB2A promoter, we used ChIP-qPCR, using whole shoots from long day–grown, 35-d-old Arabidopsis plants expressing GFP-tagged JUB1 protein from the CaMV 35S promoter (35S:JUB1-GFP). Wild-type plants were used as negative control. For the ChIP, we followed a protocol previously described by Kaufmann et al. (2010) employing anti-GFP antibody to immunoprecipitate protein-DNA complexes. The ChIP experiment was run in three independent replications. qPCR was used to test binding of JUB1 to its binding site within the DREB2A promoter; the primers flanked the JUB1 binding site. As a negative control, we used primers annealing to promoter regions of two other Arabidopsis genes (At3g18040 and At2g22180) lacking a JUB1 binding site. Primer sequences are given in Supplemental Data Set 5 online. We analyzed ChIP-qPCR data relative to input, as this includes normalization for both background levels and input chromatin going into the ChIP. The amount of genomic DNA coprecipitated by GFP antibody (ChIP signal) was calculated in comparison to the total input DNA used for each immunoprecipitation in the following way: cycle threshold (CT) = CT(ChIP) − CT(Input). To calculate fold enrichment, normalized ChIP signals were compared between 35S:JUB1-GFP and wild-type plants, where the ChIP signal is given as the fold increase in signal relative to the background signal.
H2O2 Measurements
The H2O2 staining agent, DAB (D5637, Sigma-Aldrich), was dissolved in water and adjusted to pH 3.8 with KOH. To avoid auto-oxidation, the DAB solution was freshly prepared (Fryer et al., 2002). Whole seedlings were infiltrated under vacuum with 0.5 mg mL−1 DAB staining solution and further incubated for 12 h in medium; chlorophyll was removed by incubating seedlings in 90% ethanol at 70°C for 10 min. H2O2 was visualized as brown color due to DAB polymerization.
Quantitative measurement of H2O2 production was performed using the Amplex Red hydrogen peroxide/peroxidase assay kit (Molecular Probes) following the manufacturer’s instructions. Briefly, samples were ground in liquid nitrogen, and 30 mg of ground frozen tissue from each sample was placed in an Eppendorf tube and kept frozen. Four hundred milliliters of 20 mM sodium phosphate buffer, pH 6.5, was immediately added into the tube and mixed. The extraction was centrifuged at 10,000g for 10 min at 4°C, and the supernatant was used for the assay. Measurements were performed at excitation and emission wavelengths of 560 and 590 nm, respectively, using a 96-well LS55 luminescence spectrometer (PerkinElmer). H2O2 levels are given in pmol/mg frozen tissue.
Primary Metabolite Profiling by GC-MS
To perform this study, we grew all plants alongside each other under carefully controlled conditions. Metabolite extraction, derivatization, and relative metabolite levels were determined using an established GC-MS protocol as described previously (Roessner et al., 2001; Lisec et al., 2006). Metabolites were identified in comparison to database entries of authentic standards (Kopka et al., 2005; Schauer et al., 2005).
Secondary Metabolite Profiling by LC-MS
Secondary metabolite analysis by LC-MS was performed as described by Tohge and Fernie (2010). All data were processed using Xcalibur 2.1 software (Thermo Fisher Scientific). The obtained data matrix was normalized using an internal standard (Isovitexin; CAS 29702-25-8). Metabolites were identified and annotated based on comparisons with data in our previous publications (Tohge et al., 2005, 2007; Hirai et al., 2007; Yonekura-Sakakibara et al., 2008), metabolite databases (reviewed in Tohge and Fernie, 2009), and standard compounds (Yonekura-Sakakibara et al., 2008; Nakabayashi et al., 2009).
Hormone Analyses
The extraction and analysis of ABA, SA, JA, and the cytokinins Z, ZR, DHZ, DHZR, and IPA were performed as described previously (Abreu and Munné-Bosch, 2009), except that internal deuterated standards and ultraperformance liquid chromatography–MS/MS (instead of HPLC-MS/MS) were used for the analysis. Briefly, 100 mg of leaf samples was ground in liquid nitrogen and extracted with 1.5 mL methanol using sonication. After centrifugation, the supernatant was collected and the pellet was reextracted with isopropanol:glacial acetic acid (99:1) to fully extract cytokinins. The two supernatants were dried completely under a nitrogen stream and redissolved in 150 μL methanol. Then, supernatants were combined, filtered through a 0.22-μm polytetrafluoroethylene filter (Waters), and injected into the LC-MS/MS system. MS/MS analyses were performed on an API 3000 triple quadrupole mass spectrometer (PE Sciex). All analyses were performed using the Turbo Ion Spray source in negative ion mode for ABA, SA, and JA and in positive ion mode for cytokinins. Internal standards (deuterium-labeled hormone analogs, including d4-SA, d6-ABA, d5-JA, d6-IPA, d5-Z, and d5-ZR purchased from OlChemIm) were added to each sample immediately after grinding, thus allowing the calculation of specific recovery rates for each compound. Quantification by MS/MS using the Multiple Reaction Monitoring method was performed as described by Abreu and Munné-Bosch (2009). Multiple Reaction Monitoring acquisition was done monitoring the following transitions: ABA, 263/153; SA, 137/93; JA, 209/59; IPA, 336/204; Z, 220/136; ZR, 352/220; DHZ, 222/136; DHZR, 354/222.2; d6-ABA, 269/159; d4-SA, 141/97.2; d5-JA, 241/64; d6-IPA, 342/210; d5-Z, 225/137; and d5-ZR, 357/225. The declustering potential and collision energy were optimized for each compound.
Treatments
For EST induction, 12-d-old seedlings were incubated in liquid MS medium containing 15 μM EST (control treatment: 0.15% ethanol). The seedlings were kept on a rotary shaker for 30 min or 2, 6, or 24 h, harvested, and, after removal of the roots, immediately frozen in liquid nitrogen. JUB1 expression level was determined by qRT-PCR. For EST induction on plates, MS medium was supplemented with 10 μM EST (control: 0.1% ethanol). Cellulase treatment was performed as described (Rentel et al., 2004). Briefly, 12-d-old Col-0 seedlings grown on agar plates were transferred to liquid medium supplied with 0.1% cellulase R-10 (Onozuka R-10; Yakult). Seedlings were incubated for 5 h on a rotary shaker at continuous light, and expression of JUB1 was determined by qRT-PCR. For histochemical GUS assays, ProJUB1:GUS seedlings were treated with 0.5% cellulase R-10 for 3 h. For methyl viologen (MV) treatment, 12 d-old Col-0 seedlings grown on agar plates were transferred to liquid medium, supplied with 10 μM MV (Sigma-Aldrich). Seedlings were incubated for 5 h on a rotary shaker, and expression of JUB1 was determined by qRT-PCR. For histochemical GUS assays, ProJUB1:GUS seedlings were treated with 50 μM MV for 3 h. For salt treatment, seedlings grown on MS medium were transferred to liquid MS medium containing 150 or 200 mM NaCl, followed by incubation for the indicated times; alternatively, seeds were sown on MS medium containing 100 mM NaCl.
Microscopy
Distribution of JUB1-GFP fusion protein was analyzed by confocal fluorescence microscopy using an Eclipse E600 microscope (Nikon).
Other Methods
Histochemical GUS assays was performed as described by Plesch et al. (2001). Fluorometric determination of GUS activity was done using 4-MUG (Sigma-Aldrich) as substrate (Jefferson et al., 1987). Chlorophyll content was determined using a SPAD analyzer (N-tester; Hydro Agri). Alternatively, frozen Arabidopsis leaves were ground in liquid nitrogen, resuspended in 1 mL of 96% (v/v) ethanol, and homogenized for 1 min. The soluble fraction, which contains chlorophyll, was separated by centrifugation (5 min, 13,000 rpm), and the amount of chlorophyll in the extract was determined spectrophotometrically at 650 nm. Ion leakage in the first six leaves was determined as described (Guo and Gan, 2006).
Statistical Analyses
Unless otherwise specified, statistical analyses were performed using Student’s t test embedded in Microsoft Excel. Only the return of P < 0.05 was designated as statistically significant.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACTIN2 (At3g18780), ALDH (At3g24503), At NAP (At1g69490), DREB2A (At5g05410), FAH1 (At4g36220), JUB1 (At2g43000), MBF1c (At3g24500), ORE1 (At5g39610), ORS1 (At3g29035), SMT (At2g22990), TT7 (At5g07990), and VNI2 (At5g13180). Additional accession numbers are given in Supplemental Tables 1 and 2 online and Supplemental Data Sets 1, 2, 4, and 5 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Early Leaf Senescence in JUB1-amiRNA Lines.
Supplemental Figure 2. Localization of JUB1-GFP Fusion Protein in Guard Cells of Transgenic Arabidopsis.
Supplemental Figure 3. Phenotypic and Physiological Analyses of Transgenic Plants Overexpressing JUB1 under Control of Stress-Inducible Promoter RD29A.
Supplemental Figure 4. Confirmation of T-DNA Insertion in jub1-1 Mutant.
Supplemental Figure 5. Dark-Induced Senescence Is Affected in Detached Leaves of JUB1 Overexpressors.
Supplemental Figure 6. Gene Expression and Hormone Levels in Arabidopsis Plants Subjected to Different Treatments.
Supplemental Figure 7. H2O2 Content and JUB1 Expression Level in Individual Leaves of 35-d-Old Wild-Type Plants.
Supplemental Figure 8. Effect of NaCl, Cellulase, and Methyl Viologen on JUB1 Expression.
Supplemental Figure 9. Effect of H2O2 on Detached Leaves of the jub1-1 Mutant.
Supplemental Figure 10. The jub1-1 Mutant Exhibits Decreased Heat Stress Tolerance Compared with the Wild Type.
Supplemental Figure 11. Enhanced Heat Stress Tolerance of JUB1 Overexpressor.
Supplemental Figure 12. Expression of Secondary Metabolite-Associated Genes in 35S:JUB1, jub1-1, and Wild-Type Plants, Determined by qRT-PCR.
Supplemental Table 1. NAC Transcription Factor T-DNA Insertion Lines Included in the Screen for Extended Longevity.
Supplemental Table 2. Abbreviations of Enzyme Names.
Supplemental Data Set 1. Expression of 168 Senescence-Associated Genes in 47-d-Old 35S:JUB1, jub1-1, and Wild-Type Plants.
Supplemental Data Set 2. Expression of 179 ROS Genes in 2-Week-Old, H2O2-Treated (10 mM, 6 h) and Nontreated RD29A:JUB1, jub1-1, and Wild-type Plants.
Supplemental Data Set 3. Metabolite Composition of Rosette Leaves of 35-d-Old Wild-type, 35S:JUB1, and RD29A:JUB1 Plants.
Supplemental Data Set 4. Expression of 94 Secondary Metabolite-Associated Genes in 35-d-Old 35S:JUB1, jub1-1, and Wild-Type Plants.
Supplemental Data Set 5. Oligonucleotide Sequences.
Supplementary Material
Acknowledgments
B.M.-R. thanks the Deutsche Forschungsgemeinschaft (FOR 948; MU 1199/14-1) for funding and the Bundeministerium für Bildung und Forschung for funding the GoFORSYS Research Unit for Systems Biology (FKZ 0313924). A.W. was supported by the Chinese Academy of Sciences as a member of the Joint Doctoral Promotion Program and the National Basic Research Program of China (2009CB118604). We thank the Pakistan Higher Education Commission and the Deutscher Akademischer Austauschdienst for providing a scholarship to H.S. (DAAD A/02/37115). Prashanth Garapati is supported by funds from the International Max-Planck Research School of the Max-Planck Society and the University of Potsdam. S.M.-B. was supported by the Institució Catalana de Recerca i Estudis Avançats Academia prize funded by the Generalitat de Catalunya and the BFU2009-07294 grant funded by the Ministry of Science and Innovation of the Spanish Government. We also thank Sara Shahnejat for performing some of the heat stress experiments, Josef Bergstein for photographic work, Karin Koehl and the Green Team for expert plant care, Eugenia Maximova for help with microscopy, Isabell Witt for great support during the initiation of the project, Maren Müller for hormone analyses, and Tsanko Gechev for assistance in selecting genes for the ROS qRT-PCR expression profiling platform. We thank the European Arabidopsis Stock Centre for providing seeds of T-DNA insertion lines.
AUTHOR CONTRIBUTIONS
B.M.-R. and S.B. designed the research and supervised the group. A.D.A., A.W., H.S., P.G., and S.B. performed the research. M.-I.Z. generated the 35S:JUB1 lines. H.D. produced recombinant JUB1-GST protein. M.A.A.-F. and S.M.-B. performed the hormone analysis. C.A., T.T., and A.R.F. did the metabolite profiling. G.-P.X. performed the CELD experiment. K.K. and S.B. performed the ChIP experiment. B.M.-R. and S.B. wrote the article with contributions from the other authors.
References
- Abreu M.E., Munné-Bosch S. (2009). Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J. Exp. Bot. 60: 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson A., et al. (2004). A transcriptional timetable of autumn senescence. Genome Biol. 5: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S., Kwasniewski M., Riaño-Pachón D.M., Mueller-Roeber B. (2008). QuantPrime—A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S., Kwasniewski M., Caldana C., Mehrnia M., Zanor M.I., Xue G.P., Mueller-Roeber B. (2011). ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 4: 346–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S., Parlitz S., Mueller-Roeber B., Meyer R.C. (2008a). Natural developmental variations in leaf and plant senescence in Arabidopsis thaliana. Plant Biol. (Stuttg.) 10(suppl. 1): 136–147 [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Riaño-Pachón D.M., Mueller-Roeber B. (2008b). Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. (Stuttg.) 10(suppl. 1): 63–75 [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Siddiqui H., Allu A.D., Matallana-Ramirez L.P., Caldana C., Mehrnia M., Zanor M.-I., Köhler B., Mueller-Roeber B. (2010a). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Wu A.H., Mueller-Roeber B. (2010b). Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal. Behav. 5: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C., Moeder W., Klessig D.F., Conklin P.L. (2004). The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin c-1. Plant Physiol. 134: 1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar-Mathur P., Vadez V., Sharma K.K. (2008). Transgenic approaches for abiotic stress tolerance in plants: Retrospect and prospects. Plant Cell Rep. 27: 411–424 [DOI] [PubMed] [Google Scholar]
- Bohnert H.J., Nelson D.E., Jensen R.G. (1995). Adaption to environmental stresses. Plant Cell 7: 1099–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Breeze E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.F., Wu S.H., Swidzinski J., Ishizaki K., Leaver C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Caldana C., Scheible W.R., Mueller-Roeber B., Ruzicic S. (2007). A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.S., Zhu J.K., Bressan R.A., Hasegawa P.M., Shi H.Z. (2008). Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 53: 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D.J., Coutu J., Shulaev V., Schlauch K., Mittler R. (2005a). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S., Schlauch K., Coutu J., Mittler R. (2005b). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa R.S., Dhindsa P.P., Thorpe T.A. (1981). Leaf senescence: Correlated with increased levels membrane permeability and lipid peroxidation and decreased levels of SOD and CAT. J. Exp. Bot. 32: 93–101 [Google Scholar]
- Diaz C., Saliba-Colombani V., Loudet O., Belluomo P., Moreau L., Daniel-Vedele F., Morot-Gaudry J.F., Masclaux-Daubresse C. (2006). Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol. 47: 74–83 [DOI] [PubMed] [Google Scholar]
- Djilianov D., Georgieva T., Moyankova D., Atanassov A., Shinozaki K., Smeeken S.C.M., Verma D.P.S., Murata N. (2005). Improved abiotic stress tolerance in plants by accumulation of osmoprotectants - Gene transfer approach. Biotechnol. Biotechnol. Equip. 19(Special Issue): 63–71 [Google Scholar]
- Dwidedi S., Kar M., Mishra D. (1979). Biochemical changes in excised leaves of Oryza sativa subjected to water stress. Physiol. Plant. 45: 35–40 [Google Scholar]
- Finkel T., Holbrook N.-J. (2000). Oxidants, oxidative stress and the biology of ageing. Nature 408: 239–247 [DOI] [PubMed] [Google Scholar]
- Fryer M.J., Oxborough K., Mullineaux P.M., Baker N.R. (2002). Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 53: 1249–1254 [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S., Amasino R.M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988 [DOI] [PubMed] [Google Scholar]
- Gechev T.S., Gadjev I.Z., Hille J. (2004). An extensive microarray analysis of AAL-toxin-induced cell death in Arabidopsis thaliana brings new insights into the complexity of programmed cell death in plants. Cell. Mol. Life Sci. 61: 1185–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev T.S., Hille J. (2005). Hydrogen peroxide as a signal controlling plant programmed cell death. J. Cell Biol. 168: 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev T.S., Minkov I.N., Hille J. (2005). Hydrogen peroxide-induced cell death in Arabidopsis: Transcriptional and mutant analysis reveals a role of an oxoglutarate-dependent dioxygenase gene in the cell death process. IUBMB Life 57: 181–188 [DOI] [PubMed] [Google Scholar]
- Gepstein S., Sabehi G., Carp M.J., Hajouj T., Nesher M.F., Yariv I., Dor C., Bassani M. (2003). Large-scale identification of leaf senescence-associated genes. Plant J. 36: 629–642 [DOI] [PubMed] [Google Scholar]
- Gill S.S., Tuteja N. (2010). Polyamines and abiotic stress tolerance in plants. Plant Signal. Behav. 5: 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen P.L., Holm P.B. (2007). Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol. J. 5: 192–206 [DOI] [PubMed] [Google Scholar]
- Guo Y., Cai Z., Gan S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27: 521–549 [Google Scholar]
- Guo Y., Gan S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46: 601–612 [DOI] [PubMed] [Google Scholar]
- He Y., Gan S. (2002). A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderhofer K., Zentgraf U. (2001). Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213: 469–473 [DOI] [PubMed] [Google Scholar]
- Hirai M.Y., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 104: 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch W.A., Singsaas E.L., McCown B.H. (2003). Resorption protection. Anthocyanins facilitate nutrient recovery in autumn by shielding leaves from potentially damaging light levels. Plant Physiol. 133: 1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R., Qi G., Kong Y., Kong D., Gao Q., Zhou G. (2010). Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 10: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H.-C., Hille J., Dijkwel P.P. (2003). Ageing in plants: Conserved strategies and novel pathways. Plant Biol. 5: 455–464 [Google Scholar]
- Kant P., Gordon M., Kant S., Zolla G., Davydov O., Heimer Y.M., Chalifa-Caspi V., Shaked R., Barak S. (2008). Functional-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses. Plant Cell Environ. 31: 697–714 [DOI] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Kim J.H., Woo H.R., Kim J., Lim P.O., Lee I.C., Choi S.H., Hwang D., Nam H.G. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kopka J., et al. (2005). GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kurepa J., Smalle J., Van Montagu M., Inzé D. (1998). Oxidative stress tolerance and longevity in Arabidopsis: The late-flowering mutant gigantea is tolerant to paraquat. Plant J. 14: 759–764 [DOI] [PubMed] [Google Scholar]
- Levey S., Wingler A. (2005). Natural variation in the regulation of leaf senescence and relation to other traits in Arabidopsis. Plant Cell Environ. 28: 223–231 [Google Scholar]
- Licausi F., Weits D.A., Pant B.D., Scheible W.R., Geigenberger P., van Dongen J.T. (2011). Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol. 190: 442–456 [DOI] [PubMed] [Google Scholar]
- Lim P.O., Kim H.J., Nam H.G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lin J.F., Wu S.H. (2004). Molecular events in senescing Arabidopsis leaves. Plant J. 39: 612–628 [DOI] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Laun T., Zimmermann P., Zentgraf U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Muller F.L., Lustgarten M.S., Jang Y., Richardson A., Van Remmen H. (2007). Trends in oxidative aging theories. Free Radic. Biol. Med. 43: 477–503 [DOI] [PubMed] [Google Scholar]
- Müller J., Boller T., Wiemken A. (1995). Trehalose and trehalase in plants: recent developments. Plant Sci. 112: 1–9 [Google Scholar]
- Nakabayashi R., Kusano M., Kobayashi M., Tohge T., Yonekura-Sakakibara K., Kogure N., Yamazaki M., Kitajima M., Saito K., Takayama H. (2009). Metabolomics-oriented isolation and structure elucidation of 37 compounds including two anthocyanins from Arabidopsis thaliana. Phytochemistry 70: 1017–1029 [DOI] [PubMed] [Google Scholar]
- Navabpour S., Morris K., Allen R., Harrison E., A-H-Mackerness S., Buchanan-Wollaston V. (2003). Expression of senescence-enhanced genes in response to oxidative stress. J. Exp. Bot. 54: 2285–2292 [DOI] [PubMed] [Google Scholar]
- Nishizawa A., Yabuta Y., Yoshida E., Maruta T., Yoshimura K., Shigeoka S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A., Nosaka R., Hayashi H., Tainaka H., Maruta T., Tamoi M., Ikeda M., Ohme-Takagi M., Yoshimura K., Yabuta Y., Shigeoka S. (2011). HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 52: 933–945 [DOI] [PubMed] [Google Scholar]
- Noh Y.-S., Amasino R.M. (1999). Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 41: 181–194 [DOI] [PubMed] [Google Scholar]
- Noodén L.D., Guiamét J.J., John I. (1997). Senescence mechanisms. Physiol. Plant. 101: 746–753 [Google Scholar]
- Ooka H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Parlitz S., Kunze R., Mueller-Roeber B., Balazadeh S. (2011). Regulation of photosynthesis and transcription factor expression by leaf shading and re-illumination in Arabidopsis thaliana leaves. J. Plant Physiol. 168: 1311–1319 [DOI] [PubMed] [Google Scholar]
- Plesch G., Ehrhardt T., Mueller-Roeber B. (2001). Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J. 28: 455–464 [DOI] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel M.C., Lecourieux D., Ouaked F., Usher S.L., Petersen L., Okamoto H., Knight H., Peck S.C., Grierson C.S., Hirt H., Knight M.R. (2004). OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427: 858–861 [DOI] [PubMed] [Google Scholar]
- Reth M. (2002). Hydrogen peroxide as second messenger in lymphocyte activation. Nat. Immunol. 3: 1129–1134 [DOI] [PubMed] [Google Scholar]
- Roessner U., Luedemann A., Brust D., Fiehn O., Linke T., Willmitzer L., Fernie A.R. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Osakabe Y., Qin F., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006a). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2006b). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsche E.F., Maniatis T. (2001) Molecular Cloning: A Laboratory Manual, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press) [Google Scholar]
- Schauer N., Steinhauser D., Strelkov S., Schomburg D., Allison G., Moritz T., Lundgren K., Roessner-Tunali U., Forbes M.G., Willmitzer L., Fernie A.R., Kopka J. (2005). GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Schramm F., Ganguli A., Kiehlmann E., Englich G., Walch D., von Koskull-Döring P. (2006). The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 60: 759–772 [DOI] [PubMed] [Google Scholar]
- Schramm F., Larkindale J., Kiehlmann E., Ganguli A., Englich G., Vierling E., von Koskull-Döring P. (2008). A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 53: 264–274 [DOI] [PubMed] [Google Scholar]
- Sharabi-Schwager M., Lers A., Samach A., Guy C.L., Porat R. (2010). Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J. Exp. Bot. 61: 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2002). A transient expression assay using Arabidopsis mesophyll protoplasts. http://genetics.mgh.harvard.edu/sheenweb/
- Shigeoka S., Ishikawa T., Tamoi M., Miyagawa Y., Takeda T., Yabuta Y., Yoshimura K. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53: 1305–1319 [PubMed] [Google Scholar]
- Skirycz A., Reichelt M., Burow M., Birkemeyer C., Rolcik J., Kopka J., Zanor M.I., Gershenzon J., Strnad M., Szopa J., Mueller-Roeber B., Witt I. (2006). DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 47: 10–24 [DOI] [PubMed] [Google Scholar]
- Smart C.M., Hosken S.E., Thomas H., Greaves J.A., Blair B.G., Schuch W. (1995). The timing of maize leaf senescence and characterization of senescence-related cDNAs. Physiol. Plant. 93: 673–682 [Google Scholar]
- Suzuki N., Sejima H., Tam R., Schlauch K., Mittler R. (2011). Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. 66: 844–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell W.R. (2006). The association among gene expression responses to nine abiotic stress treatments in Arabidopsis thaliana. Genetics 174: 1811–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Fernie A.R. (2009). Web-based resources for mass-spectrometry-based metabolomics: A user’s guide. Phytochemistry 70: 450–456 [DOI] [PubMed] [Google Scholar]
- Tohge T., Fernie A.R. (2010). Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 5: 1210–1227 [DOI] [PubMed] [Google Scholar]
- Tohge T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Tohge T., Yonekura-Sakakibara K., Niida R., Watanabe-Takahashi A., Saito K. (2007). Phytochemical genomics in Arabidopsis thaliana: A case study for functional identification of flavonoid biosynthsis genes. Pure Appl. Chem. 79: 811–823 [Google Scholar]
- Vanderauwera S., Zimmermann P., Rombauts S., Vandenabeele S., Langebartels C., Gruissem W., Inzé D., Van Breusegem F. (2005). Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E., Schwacke R., Schneider A., Desimone M., Flügge U.I., Kunze R. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn W.G., Woltering E.J. (2008). Physiology and molecular biology of petal senescence. J. Exp. Bot. 59: 453–480 [DOI] [PubMed] [Google Scholar]
- Verbruggen N., Hermans C. (2008). Proline accumulation in plants: A review. Amino Acids 35: 753–759 [DOI] [PubMed] [Google Scholar]
- Woo H.R., Kim J.H., Nam H.G., Lim P.O. (2004). The delayed leaf senescence mutants of Arabidopsis, ore1, ore3, and ore9, are tolerant to oxidative stress. Plant Cell Physiol. 45: 923–932 [DOI] [PubMed] [Google Scholar]
- Xue G.P. (2002). Characterisation of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res. 30: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G.P. (2005). A CELD-fusion method for rapid determination of the DNA-binding sequence specificity of novel plant DNA-binding proteins. Plant J. 41: 638–649 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Ohtani M., Mitsuda N., Kubo M., Ohme-Takagi M., Fukuda H., Demura T. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22: 1249–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.D., Seo P.J., Yoon H.K., Park C.M. (2011). The arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Rodriguez R., Tran A., Hoang H., de los Santos D., Brown S., Vellanoweth R.L. (2000). The developmental transition to flowering represses ascorbate peroxidase activity and induces enzymatic lipid peroxidation in leaf tissue in Arabidopsis thaliana. Plant Sci. 158: 115–127 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., Watanabe-Takahashi A., Inoue E., Saito K. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshiba Y., Kiyosue T., Nakashima K., Yamaguchi-Shinozaki K., Shinozaki K. (1997). Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 38: 1095–1102 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Sakuma Y., Todaka D., Maruyama K., Qin F., Mizoi J., Kidokoro S., Fujita Y., Shinozaki K., Yamaguchi-Shinozaki K. (2008). Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 368: 515–521 [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Heinlein C., Orendi G., Zentgraf U. (2006). Senescence-specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ. 29: 1049–1060 [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.-W., Chua N.-H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.