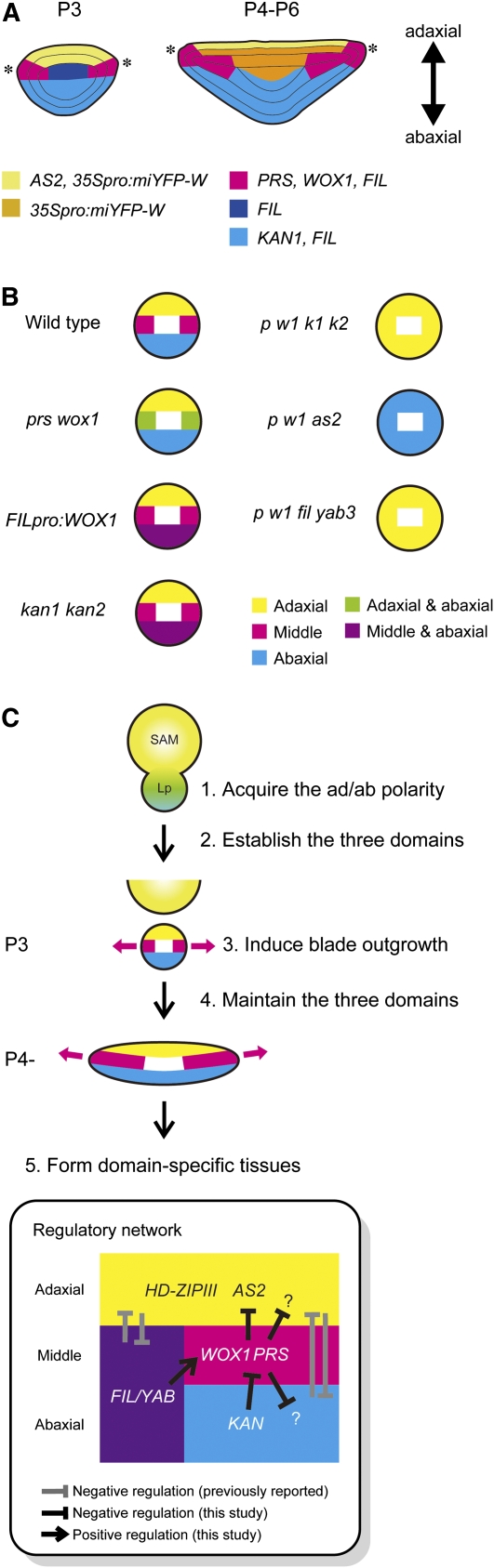
This work proposes that the middle domain, which is distinct from the adaxial (upper) and abaxial (lower) domains, plays a key role in coordinating two important processes in early leaf development, blade outgrowth, and adaxial/abaxial patterning, through the actions of the middle domain–specific WOX genes, PRS and WOX1, in concert with the adaxial- and abaxial-specific genes.
Abstract
During leaf development in flowering plants, adaxial (upper) and abaxial (lower) side–specific genes are responsible for blade outgrowth, which takes places predominantly in the lateral direction, and for margin development as well as differentiation of adaxial and abaxial tissues. However, the underlying mechanisms are poorly understood. Here, we show that two WUSCHEL-RELATED HOMEOBOX (WOX) genes, PRESSED FLOWER (PRS)/WOX3 and WOX1, encoding homeobox transcription factors, act in blade outgrowth and margin development downstream of adaxial/abaxial polarity establishment. The expression of PRS and WOX1 defines a hitherto undescribed middle domain, including two middle mesophyll layers and the margin, as a center that organizes the outgrowth of leaf blades. The expression of PRS and WOX1 is repressed in the abaxial leaf domain by the abaxial-specific transcription factor KANADI. Furthermore, PRS and WOX1 coordinate adaxial/abaxial patterning together with adaxial- and abaxial-specific genes. Our data suggest a model of blade outgrowth and adaxial/abaxial patterning via the middle domain–specific WOX genes in Arabidopsis thaliana leaves.
INTRODUCTION
Plant leaves have flat and broad blades containing region-specific tissues. The outgrowth of the leaf blade begins with the divisions of cells that are located at the lateral end of the primordium (Hagemann and Gleissberg, 1996; Donnelly et al., 1999; Dengler and Tsukaya, 2001). Subsequently, cell division continues throughout the blade predominantly in the lateral direction, making it flat and broad in shape. During blade expansion, differentiation of adaxial (upper) (adaxial epidermis and palisade cells) and abaxial (lower) tissues (abaxial epidermis, stomata, and spongy cells) occurs on the adaxial side close to the shoot apical meristem (SAM) and the abaxial side far from the SAM, respectively (Chitwood et al., 2007). In addition, margin-specific cell types (e.g., rectangular margin cells) are formed at the boundary between the adaxial and abaxial surfaces (Poethig and Sussex, 1985; McHale, 1993).
Based on the leaf phenotypes of the Antirrhinum majus phantastica mutant that displays reduced adaxial tissues, it was proposed that blade outgrowth depends on the interaction of adaxial and abaxial cells (Waites and Hudson, 1995). This hypothesis is supported by studies involving mutants of flowering plants that have defects in adaxial/abaxial patterning of leaves (Waites and Hudson, 1995; Timmermans et al., 1998; Bowman et al., 2002; Chitwood et al., 2007). Recent studies have identified adaxial- and abaxial-specific genes that encode transcription factors; the ASYMMETRIC LEAVES2 (AS2) gene and HD-ZIPIII family genes are adaxial-specific regulators (McConnell et al., 2001; Iwakawa et al., 2002), whereas FILAMENTOUS FLOWER (FIL)/YABBY (YAB) family genes and KANADI (KAN) family genes are abaxial specific (Sawa et al., 1999; Siegfried et al., 1999; Kerstetter et al., 2001). Mutations in these regulatory genes lead to abnormal phenotypes of the leaf blade; for example, the fil yab3 double mutant and the fil yab3 yab5 triple mutant plants have narrower or smaller leaves than those of the wild type (Siegfried et al., 1999; Stahle et al., 2009; Sarojam et al., 2010), whereas the kan1 kan2 double mutant displays adventitious outgrowths on the abaxial side of the leaf (Eshed et al., 2001, 2004). These findings suggest that the adaxial- and abaxial-specific genes regulate leaf blade outgrowth, but the mechanisms by which these genes influence cells that are located at the adaxial/abaxial boundary to promote blade outgrowth have not yet been established.
Previous studies have identified regulatory genes that are expressed at the margin of the lateral organ primordia. The Arabidopsis thaliana PRESSED FLOWER (PRS)/WUSCHEL-RELATED HOMEOBOX3 (WOX3) gene and its ortholog in maize (Zea mays; NARROW SHEATH [NS]) encode WOX transcription factors that are expressed at the margins of leaf primordia and of floral organ primordia (Matsumoto and Okada, 2001; Nardmann et al., 2004). PRS activity is required for the formation of lateral sepals and sepal margins in flowers and of stipules at the lateral leaf base (Matsumoto and Okada, 2001; Nardmann et al., 2004), whereas maize NS genes are required for the growth of the leaf sheath and of the proximal blade region (Scanlon et al., 1996). The Arabidopsis WOX1 gene and its orthologous genes in petunia (Petunia hybrida; MAEWEST), tobacco (Nicotiana sylvestris; LAMINA1 [LAM1]), and Medicago truncatula (STENOFOLIA [STF]) belong to the same clade of the WOX family as PRS (Haecker et al., 2004) and are expressed at the margins and in the provascular regions of leaf primordia and floral organ primordia (Vandenbussche et al., 2009; Tadege et al., 2011). The WOX1 orthologous genes are required for leaf growth, the development of margin cells, and patterning along the adaxial-abaxial axis at the leaf margin (McHale, 1993; Vandenbussche et al., 2009; Tadege et al., 2011). It has also been reported that PRS and WOX1 are redundantly required for leaf growth and adaxial/abaxial patterning in Arabidopsis (Vandenbussche et al., 2009).
In this study, we show that the specific expression of PRS and WOX1 in a domain between the adaxial and abaxial domains is important for lateral-specific blade outgrowth and margin-specific cell fate in Arabidopsis. We also show that the expression domain of these genes is restricted by the abaxial-specific KAN genes, while the two WOX genes coordinate adaxial/abaxial patterning in cooperation with adaxial- and abaxial-specific regulators.
RESULTS
PRS and WOX1 Act in Lateral-Specific Blade Outgrowth from the Middle Domain of the Leaf Primordia
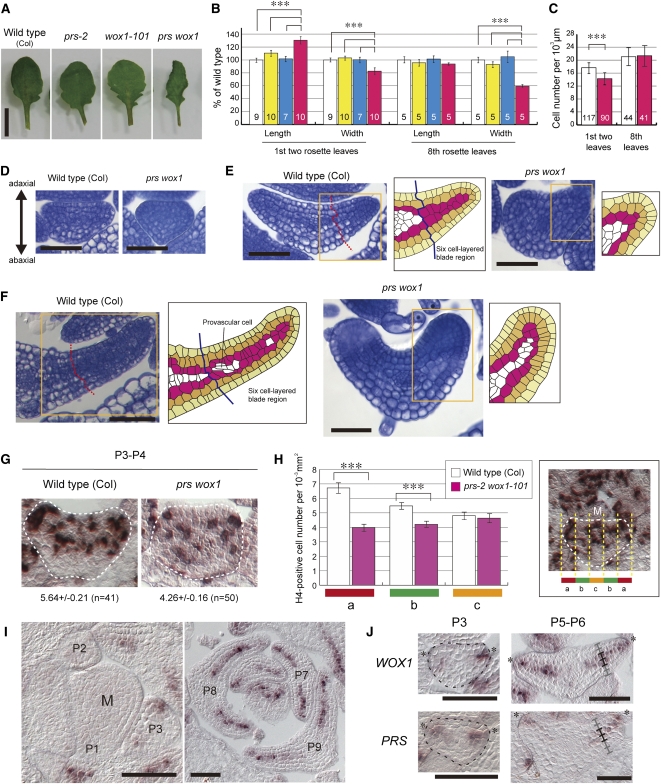
To reveal how PRS and WOX1 regulate the development of leaves and floral organs, we studied prs, wox1, and prs wox1 mutant phenotypes. Similar to the prs-1 mutant (Matsumoto and Okada, 2001), the leaves of prs-2, which is a putative null allele of PRS (see Supplemental Figure 1A online), were indistinguishable in size and shape from those of the wild type (Figures 1A and 1B), yet the sepals and petals were slightly narrower (see Supplemental Figures 2A and 2B online). The leaves of wox1-101, which is a putative null allele of WOX1 (see Supplemental Figures 1B and 1C online), were similar in size to those of the wild type (Figures 1A and 1B) but had deeper serrations (Figure 1A). The floral organs of wox1-101 looked normal (see Supplemental Figures 2A and 2B online). By contrast, the leaves of the prs-2 wox1-101 double mutant were curled upward and narrower than those of the wild type or either single mutant (Figures 1A and 1B). The number of the palisade cells per 1000 μm of width did not increase (Figure 1C), indicating that the narrow leaf phenotype of prs wox1 is due to a reduction in the number of cells. The sepals and petals of prs wox1 were much narrower than those of the wild type, prs, or wox1 (see Supplemental Figures 2A and 2B online). The number of petal epidermal cells per 100 μm of width in prs wox1 was similar to that of the wild type (see Supplemental Figure 2C online), and the calculated number of petal epidermal cells in the petal blade of prs wox1 (~24.2 cells) was considerably fewer compared with the wild type (~79.1 cells). These results indicate that PRS and WOX1 are redundantly required for cell proliferation in the blade outgrowth of leaves and petals.
Figure 1.
Function of PRS and WOX1 in the Outgrowth of the Leaf Blade.
(A) Images of the eighth leaf of the different genotypes.
(B) The lengths and widths of the first two leaves and the eighth leaves. White, the wild type; yellow, prs-2; cyan, wox1-101; magenta, prs wox1.
(C) The number of palisade cells per 1000-μm width in the first two leaves and the eighth leaves. White, the wild type; magenta, prs wox1. Data in (B) and (C) are represented as the means ± sd. ***P < 0.001 by Tukey’s honestly significant different (HSD) test. The numbers in the bars indicate the number of samples.
(D) to (F) P3 leaf primordia (D), P5 leaf primordia (E), and P8 leaf primordia (F) in cross section. Right drawings show illustrations of cell alignment ([E] and [F]). Yellow, epidermal layer; orange, subepidermal layer; magenta, third layer from surface.
(G) RNA ISH of P3 or P4 leaf primordia cross sections with H4 antisense probe. The number of H4-positive cells per 1000-μm2 area in the whole leaf primordia is indicated below.
(H) The number of H4-positive cells per 1000-μm2 area in cross sections of leaf primordia divided into five parts in width. The photo at right shows an example of the divided primordium. M, the SAM. Data in (G) and (H) are represented as the means ± se. In total, 41 sections from six wild-type plants and 50 sections from seven prs-2 wox1-101 plants were assessed. ***P < 0.001 by Wilcoxon rank-sum test.
(I) RNA ISH of wild-type shoot apex in cross section with WOX1 probe.
(J) Comparison of the expression domain of WOX1 with that of PRS in cross sections of P3 and P5-P6 leaf primordia.
The “M” in (H) and (I) indicates SAM; asterisks in (J), edges of leaf primordia; ladders mark the cell layers of the blade region. Bars = 5 mm in (A), 50 μm in (D), (E), (I), and (J), and 100 μm in (F).
Next, to reveal the spatial-temporal action of PRS and WOX1 in leaf development, we compared cross sections of prs wox1 leaf primordia with those of the wild type at various developmental stages. Plastochron 3 (P3) leaf primordia (the third-youngest leaf primordia) of the wild type appeared triangle or half-circle shaped in the cross section (Figure 1D). At the same stage, prs wox1 leaf primordia were triangle shaped, resembling the wild type (Figure 1D). P4-P5 leaf primordia of the wild type formed a six-cell-layered blade consisting of the epidermal, subepidermal, and two inner cell layers (Figure 1E). At the same stage, prs wox1 leaf primordia were narrower and thicker than those of the wild type, and a six-cell-layered structure was not found (Figure 1E). At this stage, the cell sizes in prs wox1 were not smaller than those in the wild type (Figure 1E), indicating that the number of cells in the leaf was reduced. P6-P8 leaf primordia of the wild type possessed a six-cell-layered blade structure, except around the precursor cells of vascular tissues (Figure 1F). At the same stage, prs wox1 leaf primordia were much narrower and formed a thicker margin compared with those of the wild type (Figure 1F). These results indicate that PRS and WOX1 are required to promote the formation of the leaf blade between the P3 and P4 stages and to keep the leaf margin thin.
To elucidate whether PRS and WOX1 affect cell proliferation in P3-P4 leaf primordia, we examined cell cycle activity by performing an in situ hybridization (ISH) targeting HISTONE H4 (Gaudin et al., 2000). H4-positive cells in prs wox1 leaf primordia (5.64 ± 0.21 per 10−3 mm2 area) were significantly fewer than those in wild-type leaf primordia (4.26 ± 0.16 per 10−3 mm2 area; P < 0.001 by Wilcoxon rank-sum test; Figure 1G), indicating that the studied mutations of PRS and WOX1 lead to reduction of cell division activity. Next, we examined whether the reduced cell division activity is specific to the lateral region. In wild-type leaf primordia, the frequency of H4-positive cells was higher in the lateral region (“a” in Figure 1H) compared with the medial region (“b” and “c” in Figure 1H). This finding is consistent with the expression pattern of the cell cycle marker pCYCB1;1:β-glucuronidase (GUS) (Donnelly et al., 1999). By contrast, in prs wox1, the frequency of H4-positive cells in the lateral region decreased to the same level as in the medial region (Figure 1H). These results suggest that PRS and WOX1 are required for the high frequency of cell division in the lateral region that promotes blade outgrowth.
Next, we investigated whether WOX1 and PRS expression patterns overlap. No WOX1 signal was detected in the SAM and P1-P2 leaf primordia (Figure 1I), whereas PRS was expressed in the margin cells of the leaf primordia beginning at the P1 stage (see Supplemental Figure 3 online; Matsumoto and Okada, 2001). Different levels of the expression of the two genes at these stages indicate that PRS is important for organ initiation compared with WOX1, consistent with the report that PRS functions in the recruitment of lateral-organ founder cells from the SAM (Nardmann et al., 2004). In P3 leaf primordia, WOX1 expression was detected in a few cells located in an area spanning the leaf edge and approximately three cell layers deep (Figures 1I and 1J), similar to PRS expression (Figure 1J). After the P4 stage, strong WOX1 expression was detected in the two middle mesophyll layers of the six-cell-layered blade region and in the margin cells (Figures 1I and 1J). We also found PRS expression in the two middle mesophyll layers of the blade in P4-P6 leaf primordia (Figure 1J; see Supplemental Figure 3 online), similar to WOX1. After the P7 stage, WOX1 expression was detected in the two middle mesophyll layers but was relatively weak in the abaxial-oriented middle mesophyll layer (Figure 1I), whereas PRS expression was very weak and was either only detected in the margin cells or not detected at all (see Supplemental Figure 3 online). These results indicate that the expression of PRS and WOX1 overlap in the margin cells and in the two middle mesophyll layers of the six-cell-layered blade region of P3-P6 leaf primordia. The margin is located at the boundary of the adaxial and abaxial surfaces, and the two middle mesophyll layers are located exactly between the adaxial and abaxial sides of the leaf blade. Thus, we designated the margin and two middle mesophyll layers as the middle domain of the leaf. Our findings suggest that PRS and WOX1 redundantly promote blade outgrowth from the middle domain during the P3-P6 stages.
Ectopic Expression of WOX1 in the Abaxial Region Induces Outgrowth toward the Abaxial Side
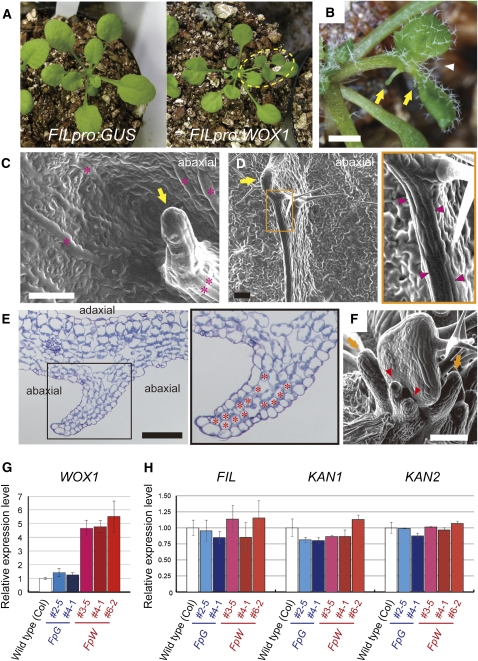
It has been reported that overexpression of PRS induces adventitious outgrowth (Matsumoto and Okada, 2001) and that WOX1 overexpression affects meristem activity (Zhang et al., 2011). To elucidate whether WOX1 is sufficient to induce outgrowth in leaves, we constructed the FILpro:WOX1 line in which WOX1 is ectopically expressed in the abaxial region of the leaf primordia, but not in the SAM. The 13 independent transformants that we obtained frequently formed aberrant leaves with leaflet-like structures and/or deeper serrations at the margin (Figures 2A and 2B). These leaves had filamentous protrusions (Figures 2B and 2C) and/or thin wall-like ridges (Figures 2D and 2E) on the abaxial side. The abaxial outgrowths and leaflet-like growth of the margin were also observed in leaf primordia (Figure 2F). We found that WOX1 mRNA levels in FILpro:WOX1 plants with aberrant leaves increased to approximately fourfold or fivefold of those of the wild-type or FILpro:GUS control plants (Figure 2G). These results indicate that the ectopic expression of WOX1 induces outgrowth toward the exterior of its expression domain in leaf primordia.
Figure 2.
Phenotypes of the FILpro:WOX1 Plants.
(A) Live image of rosette leaves. The dashed oval denotes a leaf with leaflet-like structures.
(B) to (F) The phenotypes of the FILpro:WOX1 plants.
(B) A bifurcated leaf (arrowhead) with abaxial protrusions (arrows).
(C) An abaxial protrusion (arrow) and long, rectangular cells (asterisks) on the abaxial side of a mature leaf.
(D) An abaxial ridge with a hydathode-like structure (arrow) and long, rectangular cells (arrowheads) on the abaxial side of a leaf.
(E) Cross section of an abaxial ridge consisting of compactly packed mesophyll cells (asterisks).
(F) Adventitious outgrowths on the abaxial sides (arrowheads) and around the margins (arrows) of leaf primordia.
(G) The relative level of WOX1 expression.
(H) The relative expression levels of the abaxial-specific genes. FpG, FILpro:GUS; FpW, FILpro:WOX1. Data are represented as the means ± sd in (G) and (H).
Bars = 1 mm in (B) and 100 μm in (C) to (F).
The expression levels of abaxial-specific genes (FIL, KAN1, and KAN2) in the FILpro:WOX1 lines were comparable to those in the wild-type or FILpro:GUS lines (Figure 2H), suggesting that WOX1 induces outgrowth without changing the expression of these three abaxial-specific genes.
We also analyzed the phenotypes of the FILpro:PRS plants. None of 70 T1 plants displayed abnormal true leaves (see Supplemental Figure 4A online) despite the increased expression of PRS (see Supplemental Figure 4B online). The mild phenotype of FILpro:PRS compared with the phenotype of 35Spro:PRS (Matsumoto and Okada, 2001) suggests that the expression level of PRS in FILpro:PRS is not sufficient to promote ectopic outgrowth.
WOX Genes Determine Marginal Cell Fates inside of Their Expression Domain
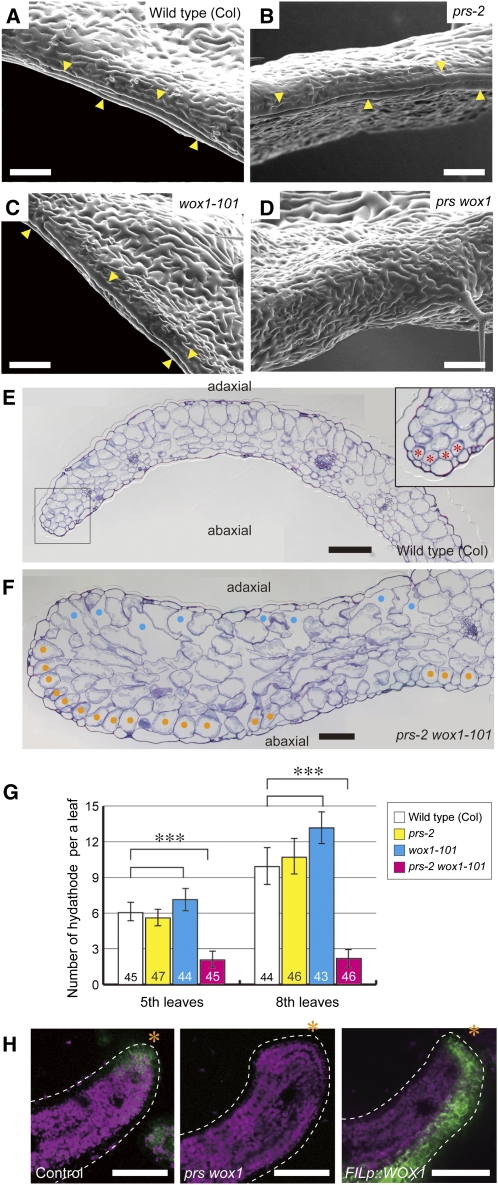
In prs single mutants, the margin-specific structures of the sepals are deformed, but the margin-specific tissues of the leaves, with the exception of the stipules, are normal (Matsumoto and Okada, 2001; Nardmann et al., 2004). To reveal whether WOX1 acts redundantly with PRS in the development of the leaf margin, we analyzed the phenotypes of the leaf tissues of prs wox1 and FILpro:WOX1. The long, rectangular margin cells, which were observed along the leaf margin in wild-type plants (Figure 3A), developed normally in prs and wox1 leaves (Figures 3B and 3C) (Matsumoto and Okada, 2001) but were absent from the margins of the prs wox1 leaves (Figure 3D). The small and compactly packed cells, which were observed at the edges of the mesophyll of wild-type leaves (Figure 3E), were also absent from the margins of prs wox1 leaves (Figure 3F). Furthermore, the number of hydathodes, which formed at the tip of the serration in the wild-type plants, was drastically decreased in prs wox1 compared with the wild type, prs, and wox1 (Figure 3G). This result indicates that PRS and WOX1 are redundantly required for the formation of margin-specific tissues.
Figure 3.
Functions of PRS and WOX1 in the Margin Development of Leaf Primordia.
(A) to (D) Scanning electron micrographs of the leaf margins of the genotypes as indicated. Arrowheads indicate long, rectangular margin cells.
(E) and (F) The leaf margin of mature leaves in cross section. Asterisks, margin-specific compactly packed mesophyll cells; cyan dots, adaxial air spaces; orange dots, abaxial palisade-like cells.
(G) The number of hydathodes. Data are represented as the means ± sd. ***P < 0.001 by Tukey’s HSD test. Numbers in the bars indicate the number of samples.
(H) Fluorescence of KLUpro:vYFPer in cross sections of the leaf blades. Green, YFP fluorescence; magenta, chlorophyll autofluorescence; asterisks, leaf edge.
Bars = 100 μm in (A) to (F) and 50 μm in (H).
FILpro:WOX1 plants possessed long, rectangular cells, similar to wild-type margin cells, and hydathode-like structures on their abaxial surfaces (Figure 2C) and/or in their abaxial protrusions (Figure 2C) and ridges (Figure 2D); compactly packed cells, similar to marginal mesophyll cells, were observed in the abaxial ridges (Figure 2E). The abaxial ridges consisted of margin-like mesophyll cells and spongy cells with air spaces (Figure 2E). These results indicate that WOX1 induces the formation of margin-specific tissues without an ectopic adaxial/abaxial boundary.
Next, to understand better the roles of PRS and WOX1 in margin development, we examined expression patterns in the KLUH (KLU)pro:vYFPer line (vYFPer is an endoplasmic reticulum–localized VENUS–YFP) (Adamski et al., 2009) as a margin marker. The leaf primordia of KLUpro:vYFPer plants showed YFP fluorescence at the blade margins (Figure 3H; see Supplemental Figure 5A online) and leaf bases (see Supplemental Figures 5A and 5B online). The KLUpro:vYFPer signal was absent from the margin of the leaf blade in the prs wox1 background (Figure 3H; see Supplemental Figure 5A online) but persisted in the leaf base (see Supplemental Figures 5A and 5B online). In FILpro:WOX1, the YFP signal was observed in the abaxial region of leaf primordia (Figure 3H; see Supplemental Figure 5A online), including the abaxial outgrowths (Supplemental Figures 5A and 5C online), in addition to the leaf base and margin (Figure 3H; see Supplemental Figures 5B and 5C online). These results indicate that PRS and WOX1 induce the development of margin-specific characteristics.
PRS and WOX1 Are Required for the Maintenance of Adaxial/Abaxial Patterning in the Lateral Regions of Leaves
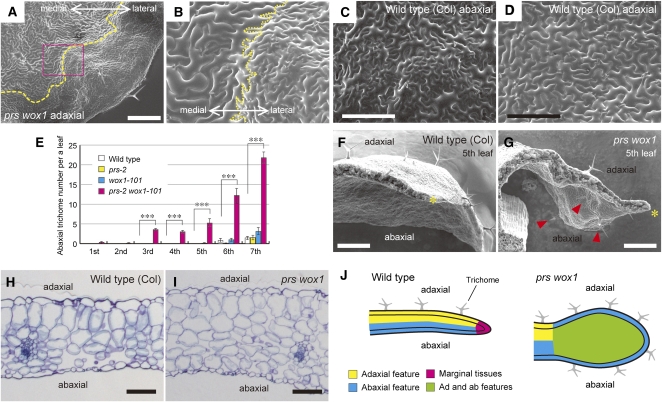
As previously reported (Vandenbussche et al., 2009), large, irregular cells with air spaces, resembling abaxial side–specific spongy cells in wild-type leaves (Figure 3E), were observed on both the adaxial and abaxial sides around the margins of prs wox1 leaves (Figure 3F). In our experiments, we found oblong cells, resembling adaxial side–specific palisade cells in wild-type leaves (Figure 3E), in the subepidermal layers of the margins of the prs wox1 leaves (Figure 3F). Moreover, the adaxial surfaces of the lateral regions of prs wox1 leaves were covered with jigsaw-shaped cells of various sizes with high densities of stomata (Figures 4A and 4B; see Supplemental Figure 6 online), which were restricted to the abaxial leaf surfaces in wild-type leaves (Figure 4C). In the double mutant, the adaxial surface of the medial region was covered with jigsaw-shaped and uniform-sized cells with a low density of stomata (Figures 4A and 4B; see Supplemental Figure 6 online), which were found on the adaxial leaf surface of wild-type leaves (Figure 4D). These two types of pavement cells were in direct contact to each other in prs wox1 and were not separated by long, rectangular margin cells (Figure 4B; see Supplemental Figure 6 online). However, trichomes, which formed only on the adaxial side in the first to fifth rosette leaves in the wild type (Figures 4E and 4F), were observed on the abaxial side neighboring the margin in addition to the adaxial side in prs wox1 (Figures 4E and 4G). The distribution of adaxial and abaxial side–specific tissues was normal in the medial region, even in prs wox1 (Figures 4H and 4I). In summary, in prs wox1, margin-specific tissues are lost, and adaxial and abaxial-like cell types coexist in the region neighboring the margin (Figure 4J). These results indicate that PRS and WOX1 are required for the normal patterning of adaxial and abaxial side–specific tissues in the lateral region.
Figure 4.
Adaxial/Abaxial Patterning of Tissue Differentiation in the Wild-Type and prs wox1 Leaves.
(A) and (B) Scanning electron micrographs of the adaxial surface of prs wox1. Yellow dashed lines, the boundary between the adaxial-type epidermis and abaxial-type epidermis.
(C) and (D) Scanning electron micrographs of the abaxial (C) and adaxial (D) surfaces of the wild-type leaves.
(E) The number of abaxial trichomes. Data are represented as the means ± sd. ***P < 0.001 by Tukey’s HSD test. In total, 11 wild-type and prs-2 and 9 wox1-101 and prs-2 wox1-101 samples were assessed.
(F) and (G) Scanning electron micrographs of the wild-type (F) and prs wox1 leaves (G) in cross section. Asterisks, leaf edges; arrowheads, abaxial trichomes.
(H) and (I) The medial region of the eighth leaves in cross section in the wild type (H) and prs wox1 (I).
(J) Schematic view of leaf margin phenotypes.
Bars = 500 μm in (A), (F), and (G), 200 μm in (C) and (D), and 100 μm in (H) and (I).
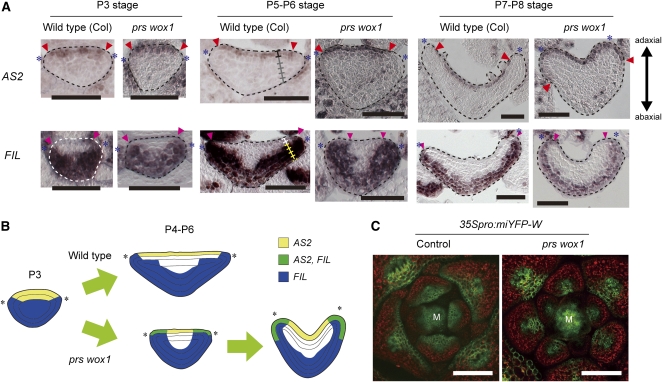
Next, we analyzed whether PRS and WOX1 affect the expression of the adaxial- and abaxial-specific genes. AS2 expression in wild-type leaf primordia was detected in adaxial epidermal cells and rarely in adaxial subepidermal cells but was not detected at the margin and on the abaxial side (Figure 5A). The AS2 expression pattern in prs wox1 was indistinguishable from that of the wild type in P3 leaf primordia, but ectopic expression was detected in the margin cells of P4-P6 leaf primordia and in abaxial epidermal cells of the lateral leaf regions after the P7 stage (Figure 5A). This strongly suggests that PRS and WOX1 maintain a restricted pattern of AS2 expression during blade outgrowth.
Figure 5.
Expression Patterns of AS2, FIL, and 35Spro:miYFP-W in the Wild-Type and prs wox1 Leaf Primordia.
(A) RNA ISH in cross sections of leaf primordia with AS2 and FIL probes. Asterisks, edges of leaf primordia; ladders, cell layers along the adaxial-abaxial axis; arrowheads, edges of the region where genes are expressed.
(B) Schematic view of expression patterns of AS2 and FIL.
(C) YFP fluorescence of 35Spro:miYFP-W in cross sections of shoot apices. M, SAM; green, YFP fluorescence; red, chlorophyll autofluorescence.
Bars = 50 μm in (A) and 100 μm in (C).
The abaxial-specific gene FIL is expressed in P3-P6 leaf primordia of the wild type in the broad region of the abaxial side, including the margin and four cell layers of the six-cell-layered blade on the abaxial side (Figure 5A). After the P7 stage, FIL expression was restricted to three cell layers of the abaxial side (Figure 5A). The FIL expression in the prs wox1 leaf primordia was indistinguishable from that in the wild type at the P3 stage, but unlike the wild type, it was detected in a broader region of P4-P6 leaf primordia, including the lateral region of the adaxial side (Figure 5A). After the P7 stage, FIL expression was restricted to three cell layers of the blade, resembling the wild-type expression pattern, but it was also detected in the adaxial epidermis neighboring the margin (Figure 5A). The expression patterns of these genes indicate that the expression of FIL and AS2 overlap in the cells that are located in the lateral region of prs wox1 leaf primordia (Figure 5B), correlating with the coexistence of adaxial and abaxial side–specific tissues in those cells. The gradually expanding patterns of AS2 and FIL in prs wox1 suggest that PRS and WOX1 are required to maintain adaxial/abaxial expression patterns in the lateral region during blade outgrowth.
The HD-ZIPIII family genes are adaxially expressed in leaf primordia (McConnell et al., 2001). The microRNA miR165/166 is reported to repress the expression of the HD-ZIPIII family genes (Mallory et al., 2004). To investigate whether mutations of the WOX genes affect the pattern of HD-ZIPIII expression via the regulation of miR165/166, we analyzed the expression pattern of the 35Spro:miYFP-W line, in which miR165/166 function can be monitored by a reduction of YFP fluorescence (Toyokura et al., 2011). YFP fluorescence of 35Spro:miYFP-W in the wild type was detected only in the two cell layers of the adaxial side of the leaf primordia (Figure 5C). In prs wox1, YFP fluorescence was further limited to the medial region of the adaxial side (Figure 5C). This indicates that PRS and WOX1 control the expression of the HD-ZIPIII genes through the negative regulation of miR165/166 in the lateral region, suggesting that PRS and WOX1 act in the maintenance of adaxial/abaxial expression patterns during leaf blade outgrowth.
FIL and Its Homologs Upregulate WOX1 Expression
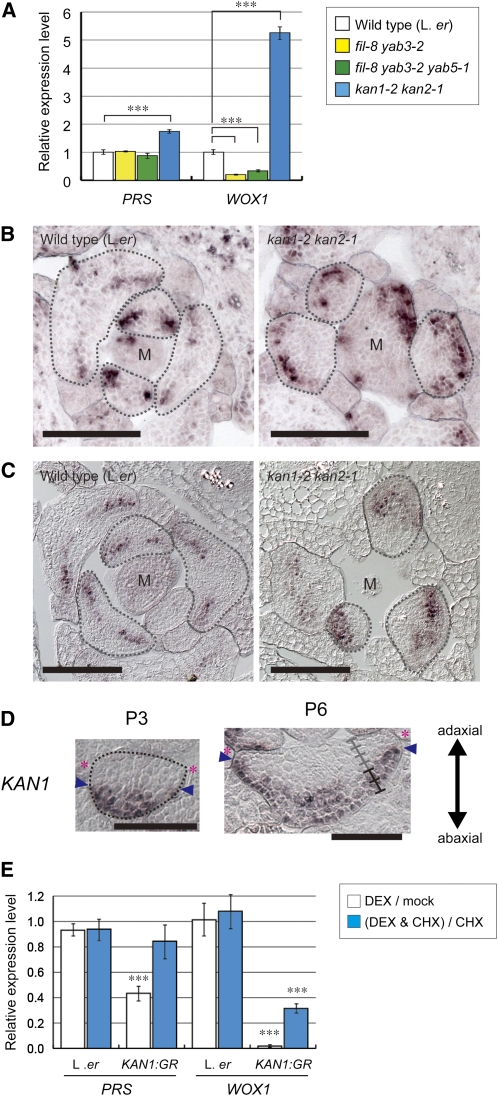
It has been reported that in fil yab3 and fil yab3 yab5, the leaves are narrow and that some types of margin-specific tissues are absent (Eshed et al., 2004; Stahle et al., 2009; Sarojam et al., 2010), similar to prs wox1, but FIL expression did not decrease in prs wox1 (Figure 5A; Vandenbussche et al., 2009) and did not increase in FILpro:WOX1 (Figure 2H). This strongly suggests that PRS and WOX1 do not upregulate FIL expression. To reveal whether FIL/YAB genes upregulate the expression of PRS and WOX1, we examined the expression levels of PRS and WOX1 in fil yab3 and fil yab3 yab5. The expression level of WOX1 in these mutants was approximately half of that in the wild type, whereas the expression level of PRS in the mutants was the same as in the wild type (Figure 6A). These results indicate that FIL and its homologs upregulate WOX1 expression but do not affect the expression level of PRS.
Figure 6.
Control of PRS and WOX1 Expression by the FIL/YAB and KAN Genes.
(A) The relative expression levels of PRS and WOX1. Data are represented as the means ± sd. ***P < 0.001 by paired Student’s t test.
(B) and (C) RNA ISH of cross sections of shoot apices with PRS (B) and WOX1 (C) probes. M, SAM.
(D) RNA ISH of cross sections of leaf primordia with KAN1 probe.
(E) The relative levels of expression of PRS and WOX1 with or without DEX and with or without CHX in each line as indicated. Data are represented as the means ± sd. Asterisks indicate a significant difference between expression levels with and without DEX (***P < 0.001 by Student’s t test).
Bars = 100 μm in (B) and (C) and 50 μm in (D).
KAN Genes Repress the Expression of PRS and WOX1 in the Abaxial Domain
Similar to FILpro:WOX1 transgenic plants, the kan1 kan2 double mutant has been reported to have adventitious outgrowths and ectopic margin-like structures on the abaxial sides of leaves (Eshed et al., 2001, 2004). However, it has been reported that KAN1 expression is not significantly altered in prs wox1 (Vandenbussche et al., 2009), and we found that the expression of KAN1 and KAN2 did not decrease in FILpro:WOX1 (Figure 2H), suggesting that PRS and WOX1 do not repress KAN expression. Therefore, we investigated whether the expression of PRS and/or WOX1 is affected in kan1 kan2. The expression levels of PRS and WOX1 in kan1 kan2 were approximately twofold and fivefold of those of the wild type, respectively (Figure 6A). ISH analyses revealed the expanded accumulation of PRS and WOX1 transcripts in the abaxial region of the kan1 kan2 leaf primordia (Figures 6B and 6C). These results indicate that the KAN genes repress the expression of PRS and WOX1 in the abaxial region of the leaf primordia.
In P3 leaf primordia, KAN1 expression was restricted to a narrow region on the abaxial side and was absent from the cells neighboring the leaf edge (Figure 6D). After the P4 stage, KAN1 signal was detected in two cell layers of the six-cell-layered region on the abaxial side (Figure 6D). These data indicate that KAN1 expression does not overlap with that of PRS and WOX1 (Figure 1J) but does overlap with the domains of the ectopic expression of PRS and WOX1 in the kan1 kan2 double mutant (Figures 6B and 6C), suggesting that KAN1 represses PRS and WOX1 expression in a cell-autonomous manner.
To investigate whether the KAN1 protein directly or indirectly represses PRS and WOX1, we analyzed the effect of dexamethasone (DEX)–inducible expression of a 35Spro:KAN1:glucocorticoid receptor (GR) line, the hormone binding domain of the GR fused to the C terminus of KAN1 under the control of the 35S promoter (Hawker and Bowman, 2004), on the expression of PRS and WOX1. Exposure to DEX for 6 h drastically reduced the amounts of both PRS and WOX1 transcripts (Figure 6E). Exposure to DEX in the presence of cycloheximide (CHX; an inhibitor of protein biosynthesis) led to a substantial reduction in the expression levels of WOX1, although the repression was stronger in the absence of CHX. In contrast with WOX1, following exposure to DEX with CHX, PRS expression levels were only slightly reduced (Figure 6E). These data indicate that the KAN1 protein directly and indirectly represses WOX1 expression and acts indirectly on PRS.
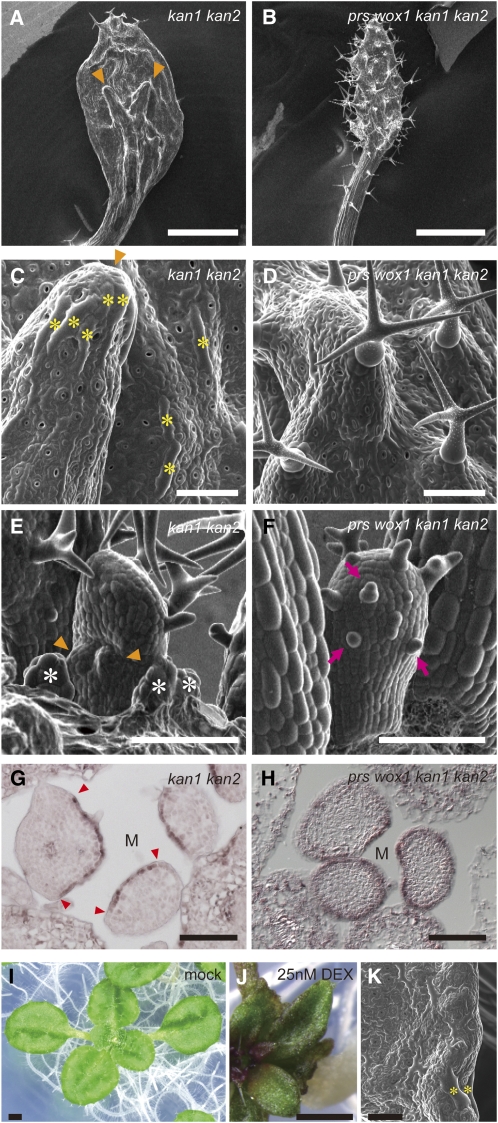
PRS and WOX1 Are Responsible for Outgrowth and Margin Development on the Abaxial Side and Suppress Adaxialization in kan1 kan2 Leaves
To investigate whether the ectopic expression of PRS and WOX1 is responsible for outgrowths and marginal tissue-like cells on the abaxial sides of kan1 kan2 leaves, we compared the leaf phenotypes of kan1 kan2 with those of the prs wox1 kan1 kan2 quadruple mutant. In comparison to kan1 kan2 (Figure 7A), the leaves of prs wox1 kan1 kan2 were narrower and club shaped (Figure 7B). The adventitious outgrowths with a hydathode-like structure, which were observed on the abaxial side of the kan1 kan2 leaves (Figures 7A, 7C, and 7E), were not found in prs wox1 kan1 kan2 (Figures 7B, 7D and 7F). Many stipules and long rectangular cells similar to margin cells formed on the abaxial side in kan1 kan2 (Figures 7C and 7E) but not in prs wox1 kan1 kan2 (Figures 7D and 7F). This finding suggests that the adventitious outgrowths and ectopic formation of marginal tissue-like cells, including long, rectangular cells, hydathode-like structures, and stipules, on the abaxial side of kan1 kan2 are due to the actions of PRS and WOX1. In a complementary experiment, we found that KAN overexpression by DEX treatment, which triggers the downregulation of the expression of PRS and WOX1, led to inhibition of blade outgrowth (Figure 7J; compare with a mock-treated plant in Figure 7I) and loss of long, rectangular margin cells (Figure 7K). These results suggest that the KAN genes determine the direction of blade outgrowth and limit margin development by repressing the expression of PRS and WOX1.
Figure 7.
Genetic Interaction between two WOX Genes and KAN Genes.
(A) to (F) Scanning electron micrographs of the abaxial surfaces of leaves ([A] to [D]) and leaf primordia ([E] and [F]) for indicated genotypes. Arrowheads in (A), (C), and (E), abaxial protrusions; asterisks in (C), long, rectangular cells similar to margin cells; asterisks in (E), stipules; arrows in (F), abaxial trichomes.
(G) and (H) RNA ISH of cross sections of shoot apex with AS2 probe. Arrowheads in (G), edges of AS2 expression domain; M, position above the SAM.
(I) to (K) Phenotype of 35Spro:KAN1:GR plants without (I) and with ([J] and [K]) DEX. Asterisks indicate elongated cells located at the margin.
Bars = 1 mm in (A), (B), (I), and (J) and 100 μm in (C) to (H) and (K).
The leaves of prs wox1 kan1 kan2 also showed severe adaxialization. Few trichomes formed on the abaxial side in the prs wox1 and kan1 kan2 leaves (Figures 4E, 4G, 7A, 7C, and 7E), whereas many trichomes formed on both the adaxial and abaxial sides of the prs wox1 kan1 kan2 leaves (Figures 7B, 7D, and 7F). Consistent with the distribution of the trichomes, AS2 expression was detected only on the adaxial side in the kan1 kan2 leaf primordia (Figure 7G) but in the entire region of the epidermis in the prs wox1 kan1 kan2 leaf primordia (Figure 7H). The phenotype of prs wox1 kan1 kan2 indicates that PRS and WOX1 are involved in adaxial/abaxial patterning in the entire leaf primordium in coordination with the KAN genes.
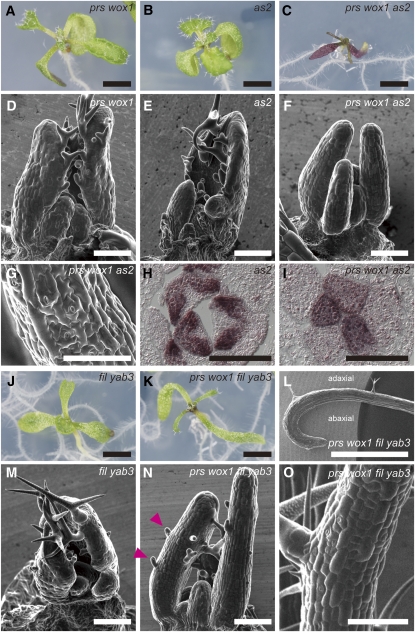
PRS and WOX1 Coordinate Adaxial/Abaxial Patterning in the Entire Leaf Primordium together with AS2 and FIL
Next, we examined whether PRS and WOX1 genetically interact with other adaxial- and abaxial-specific genes. Whereas the prs wox1 and as2-1 plants did not form filamentous leaves in the vegetative phase (Figures 8A, 8B, 8D, and 8E; Iwakawa et al., 2002), the cotyledons were very narrow, and the leaves were severely radialized and had no blades in prs wox1 as2-1 (Figures 8C and 8F). The surfaces of the filamentous organs in prs wox1 as2-1 were covered with short, rectangular cells, small pavement cells, and a high density of stomata (Figure 8G). The first to fifth or sixth filamentous organs formed no trichomes on the surface (Figure 8F; see Supplemental Figure 7 online). The phenotypes of the stomata and trichomes corresponded to those on the abaxial surfaces of the wild-type leaves. In addition, the expression of FIL was restricted to the abaxial region of the as2-1 leaf primordia (Figure 8H) but was detected in the entire region of the prs wox1 as2-1 leaf primordia (Figure 8I), indicating that the filamentous leaves of prs wox1 as2 are severely abaxialized.
Figure 8.
Phenotypes of prs wox1 as2 and prs wox1 fil yab3.
(A) to (I) Phenotypes of prs-2 wox1-101 ([A] and [D]), as2-1 ([B] and [E]), and prs wox1 as2 ([C], [F], and [G]).
(A) to (C) Live images of 8-d-old plants with genotypes as indicated.
(D) to (G) Scanning electron micrographs of leaf primordia ([D] to [F]) and developed filamentous leaves (G).
(H) and (I) FIL expression patterns in cross sections of shoot apex.
(J) to (O) Phenotypes of fil-1 yab3-2 ([J] and [M]) and prs wox1 fil yab3 ([K], [L], [N], and [O]).
(J) and (K) Live images of eight-d-old mutants.
(L) to (O) Scanning electron micrographs of leaves ([L] and [O]) and leaf primordia ([M] and [N]). The first leaf (L) and the third or later leaves ([M] to [O]). Arrowheads in (N), abaxial trichomes.
Bars = 2 mm in (A) to (C) and (J) to (L) and 100 μm in (D) to (I) and (M) to (O).
Severely defective adaxial/abaxial patterning was also observed in prs wox1 fil yab3. The cotyledons of prs wox1 fil yab3 were much narrower than those of prs wox1 and fil-1 yab3-2 (Figures 8A, 8J, and 8K). Although the first two leaves of prs wox1 fil yab3 exhibited slight differences between the adaxial and abaxial sides in the distribution of trichomes, blade outgrowth was severely inhibited (Figures 8K and 8L) compared with prs wox1 and fil yab3 (Figures 8A and 8J), suggesting that the FIL/YAB genes act in blade outgrowth in parallel with PRS and WOX1. Whereas prs wox1 and fil yab3 did not form any filamentous organs during the vegetative phase (Figures 8A, 8D, 8J, and 8M), the third and later leaves of prs wox1 fil yab3 were severely radialized (Figure 8N). The surfaces of these filamentous leaves were covered with short, rectangular cells and many trichomes on both sides (Figures 8N and 8O), indicating that they were severely adaxialized.
In summary, the observed phenotypes suggest that PRS and WOX1 regulate adaxial/abaxial patterning in the leaf primordia in concert with AS2 and the FIL/YAB genes in addition to the KAN genes.
DISCUSSION
We demonstrated that PRS and WOX1, which are expressed in the domain between the adaxial and abaxial sides of the leaf primordia, promote leaf blade outgrowth and margin development and that the expression of PRS and WOX1 is repressed by the abaxial-specific KAN genes. In addition, we have shown that these two WOX genes cooperate in adaxial/abaxial patterning with the adaxial- and abaxial-specific genes. Based on these findings, we propose a new regulatory leaf domain, the middle domain, that is distinct from the adaxial and abaxial domains and plays a key role in the early development of leaves via the functions of PRS and WOX1.
Roles of PRS and WOX1 in Blade Outgrowth
Here, we report that PRS and WOX1 promote cell proliferation to induce blade outgrowth. Although the expression of the two WOX genes is restricted to the margin and two middle mesophyll layers, their loss-of-function mutations affected the frequency of actively dividing cells in the broader lateral region of P3-P4 leaf primordia and led to fewer epidermal and subepidermal cells on both leaf sides in addition to those of the middle mesophyll cells. These data suggest that PRS and WOX1 promote blade outgrowth through the coordinated regulation of proliferation of WOX-expressing cells and surrounding cells.
The coordinated control of cell proliferation implies that PRS and WOX1 modulate factors regulating cell division in a non-cell-autonomous manner. The expression patterns of the KLUpro:vYFPer marker in prs wox1 and FILpro:WOX1 indicate that PRS and WOX1 positively regulate the promoter activity of the KLU gene. KLU encodes the cytochrome P450 CYP78A5 monooxygenase and is reported to prevent the arrest of cell proliferation in a non-cell-autonomous manner (Anastasiou et al., 2007), suggesting that KLU mediates the function of the two WOX genes in blade outgrowth. In addition, the WOX1 orthologous genes M. truncatula STF and tobacco LAM1 have been shown to affect auxin levels (Tadege et al., 2011). The bladeless phenotype of the tobacco lam1 mutant was partially rescued by the coapplication of auxin and cytokinin (Tadege et al., 2011). Auxin and cytokinin are known to induce the cell cycle by upregulating the expression of cell cycle components and repressing inhibitors of cell division (Hartig and Beck, 2006). Thus, the WOX genes may affect the cell cycle via auxin and cytokinin. The YUCCA genes, which encode flavin monooxygenase-like enzymes, have been shown to be involved in auxin biosynthesis and to regulate blade outgrowth and margin development in the context of adaxial/abaxial patterning (Wang et al., 2011) similar to PRS and WOX1, which supports the existence of a relationship between auxin and the WOX genes. Furthermore, WOX1 is also reported to affect the shikimate pathway, sugar metabolism (Tadege et al., 2011), and polyamine homeostasis (Zhang et al., 2011). Taken together, the WOX genes appear to organize leaf blade outgrowth through the regulation of various signaling and metabolic pathways.
During leaf development, the cells of the lateral region actively divide at the P3 stage, after which the cells of the blade region proliferate predominantly in the lateral direction, forming a flat and broad shape. The expression of PRS and WOX1, which is required for lateral-specific blade outgrowth, is restricted to the middle domain during leaf development, and the ectopic expression of PRS and/or WOX1 in the abaxial domain of the leaf primordia in FILpro:WOX1 and kan1 kan2 induces outgrowth toward the abaxial side. Furthermore, outgrowths toward the abaxial side do not form in prs wox1 kan1 kan2 leaf primordia. These results suggest that PRS and WOX1 organize blade outgrowth toward the exterior in their expression domain. Therefore, the restriction of PRS and WOX1 expression patterns to the middle domain would be important to induce lateral-specific outgrowth of the leaf blade.
WOX Genes Lead to Blade Outgrowth Downstream of Adaxial/Abaxial Polarity
It was previously hypothesized that adaxial/abaxial polarity is responsible for blade outgrowth and margin development around the adaxial/abaxial boundary (Waites and Hudson, 1995; Timmermans et al., 1998). However, factors promoting these processes have not yet been identified. In this study, the abaxial-specific KAN genes restricted the transcription of PRS and WOX1 to the middle domain. As shown above, this restriction is important for lateral-specific blade outgrowth and margin-specific development. Furthermore, ectopic expression of WOX1 in the abaxial region leads to abaxial outgrowths without forming an ectopic adaxial/abaxial boundary. Hence, we propose that PRS and WOX1 are the factors that induce blade outgrowth and margin development downstream of the adaxial/abaxial polarity.
FIL, KAN1, and their homologs are reported to act in the development of abaxial side–specific tissues and to be expressed on the abaxial sides of leaf primordia (Sawa et al., 1999; Siegfried et al., 1999; Eshed et al., 2001, 2004; Kerstetter et al., 2001). However, based on the phenotypes of their mutants (Siegfried et al., 1999; Eshed et al., 2001, 2004; Sarojam et al., 2010), these two gene families have opposite effects on both leaf blade outgrowth and margin development; the FIL/YAB genes promote these processes, while the KAN genes inhibit them. We have shown that FIL is more broadly expressed than KAN1 during the P3-P6 stages (Figure 9A) and that the expression domains of PRS and WOX1 are present in that of FIL but do not overlap with that of KAN1 (Figure 9A). Furthermore, the FIL/YAB genes positively regulate WOX1 expression, and the KAN genes negatively regulate the expression of PRS and WOX1, suggesting that the middle domain–specific expression of the WOX genes is largely dependent on the differences in the expression patterns and on the functions of the abaxial side–specific genes, FIL/YAB and KAN. However, because mutations of FIL/YAB genes do not downregulate PRS expression and the FIL expression domain does not completely encompass the WOX1 expression domain after the P7 stage, other adaxial- and/or abaxial-specific genes must contribute to the restrictions of the PRS and WOX1 expression domains.
Figure 9.
Schematic View of the Gene Expression Patterns and a Model of Early Leaf Development.
(A) Schematic view of gene expression patterns in cross sections of P3 and P4-P6 leaf primordia.
(B) Schematic view of the adaxial/middle/abaxial patterns in cross sections of leaf primordia that were observed in this study.
(C) A model of early development of the leaf. A network among adaxial, middle, and abaxial domain–specific genes (below) regulates the establishment and maintenance of the three-domain structure. Lp, leaf primordia.
The Middle Domain Is Distinct from the Adaxial and Abaxial Domains
Based on the expression patterns of AS2, HD-ZIPIII, and FIL, leaf primordia are divided into two domains: the adaxial and abaxial domains (Figure 9A; Toyokura et al., 2011). In this study, we found that the expression domain of FIL includes those of PRS, WOX1, and KAN1 during the P3-P6 stages (Figure 9A). Thus, we conclude that the FIL expression domain is divided into two domains: the middle domain, in which PRS and WOX1 are expressed, and a narrower definition of the abaxial domain, in which KAN1 is expressed. In addition, similar to KAN1, the expression of the ARF3/ETT and ARF4 genes, which are additional abaxial side–specific genes, are absent from the middle domain and restricted to the narrow region of the abaxial side (Chitwood et al., 2009), supporting our conclusion.
Our mutant analyses demonstrated that the middle domain–specific WOX genes are involved in adaxial/abaxial patterning. In Figure 9B, we show a schematic view of the adaxial/abaxial patterns in the mutants that were observed in this study. First, loss of the middle domain–specific WOX genes led to the coexistence of adaxial and abaxial side–specific tissues and the coexpression of AS2 and FIL in the cells neighboring the leaf margin. Second, prs wox1 kan1 kan2 and prs wox1 fil yab3 mutants showed severely adaxialized phenotypes, whereas prs wox1 as2 showed a severely abaxialized phenotype. Third, the ectopic expression of PRS and/or WOX1 on the abaxial side in FILpro:WOX1 or kan1 kan2 led to a mosaic of marginal and abaxial features. Based on these results, we propose that the middle domain is as a new regulatory entity distinct from the adaxial and abaxial domains and is established through the mutual interaction among adaxial, middle, and abaxial domain–specific genes. Further analyses of mutant phenotypes, for example, of the gynoecium or other floral organs, will address whether the middle domain concept in adaxial/abaxial patterning is applicable to a wider range of organs.
Thus, we present a model describing early leaf development in Figure 9C. First, the leaf primordium acquires adaxial/abaxial polarity, which presumably depends on information from the SAM as previously suggested (Bowman et al., 2002; Reinhardt et al., 2005). Second, according to the adaxial/abaxial polarity, the adaxial, middle, and abaxial domain–specific genes are expressed. Next, the adaxial, middle, and abaxial domains are established by a regulatory network among the adaxial, middle, and abaxial domain–specific genes. The mutations of PRS and WOX1 induce the expansion of the AS2 expression domain to the abaxial side of leaf primordia, implying that AS2 may be a direct or indirect target of PRS and WOX1 that is excluded from the middle domain. It has also been reported that mutations of PRS and WOX1 affect the expression of the abaxial-specific ARF4 gene (Vandenbussche et al., 2009). Identification of adaxial and abaxial domain–specific genes regulated by PRS and WOX1 is now in progress. Third, the middle domain–specific WOX genes induce outgrowth and the formation of the six-cell-layered leaf blade. The FIL/YAB genes may promote blade outgrowth in the middle domain together with PRS and WOX1 based on the phenotype of prs wox1 fil yab3. The AINTEGUMENTA (ANT) gene, which encodes an AP2-type transcription factor, is reported to act in blade outgrowth in combination with the FIL/YAB genes and probably in the context of adaxial/abaxial patterning (Nole-Wilson and Krizek, 2006). Thus, the functional relationship between ANT and the middle domain–specific WOX genes is an intriguing problem for future research. Fourth, a regulatory network among adaxial, middle, and abaxial domain–specific genes maintains the three-domain structure of the leaf blade during blade expansion. Finally, domain-specific tissues form along the lines of the three-domain structure. This model provides an in-depth understanding of blade outgrowth that is regulated by adaxial/abaxial polarity and also yields novel insight into patterning along the adaxial-abaxial axis.
Since blade development and adaxial/abaxial patterning of leaves are common among flowering plants, the three-domain structure is expected to play a key role in leaf development in other species. An advanced understanding of the regulatory network for the establishment and maintenance of the three-domain structure in various plant species will unveil the evolutionary processes that led to the flat and broad leaf blade morphology in flowering plants.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana accessions Columbia (Col) and Landsberg erecta were used as the wild types. prs-2 (SALK_108644; Col back ground) was identified from SALK T-DNA populations and carries a T-DNA insertion in the homeodomain (see Supplemental Figure 1A online). wox1-101 (KE1895; Col background) was identified from Kazusa T-DNA insertion populations (Kato et al., 2007) and carries a T-DNA insertion in the second intron of the WOX1 gene (see Supplemental Figure 1B online), resulting in an undetectable level of WOX1 expression as shown by RT-PCR (see Supplemental Figure 1C and Supplemental Methods 1 online). Of the mutants and transgenic lines that were used in this study, 35Spro:miYFP-W (Toyokura et al., 2011), fil-1 (Sawa et al., 1999), fil-8, yab3-2 (Kumaran et al., 2002), yab5-1 (Sarojam et al., 2010), as2-1 (Semiarti et al., 2001), kan1-2 (Eshed et al., 1999), kan2-1 (Eshed et al., 2001), KLUpro:vYFPer (Adamski et al., 2009), and 35Spro:KAN1:GR (Hawker and Bowman, 2004) have been previously described.
For the construction of FILpro:WOX1 and FILpro:PRS, a full-length cDNA fragment of WOX1 and of PRS was amplified by PCR and subcloned into pDONR221 (Invitrogen). Primer sequences for the amplification of the cDNA fragments were as follows: attB1F-WOX1f, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGTGGACGATGGGTTACAAC-3′; attB2R-WOX1r, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTAGTTCTTCAATGGCAGAAA-3′; attB1F-PRSf, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGAGTCCTGTGGCTTCAACG-3′; and attB2R-PRSr, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTAAAGTTTGGTACTGTCTTG-3′.
The cDNA fragments were transferred to the binary vector pGWB-NB1-FILp. The pGWB-NB1-FILp vector is derived from the pGWB-NB1 vector and contains 6011 bp of 5′ upstream sequence of the FIL gene that is inserted into the HindIII site that is located in front of a Gateway cassette. The 6011-bp promoter of FIL was previously described (Watanabe and Okada, 2003). The pGWB-NB1 vector is derived from pGWB401 (Nakagawa et al., 2007) and contains the basta resistance gene. For the construction of FILpro:GUS, the GUS gene was subcloned into pDONR/Zeo (Invitrogen) and transferred into pGWB-NB1-FILp. The three constructs were introduced into wild-type plants (Col) by vacuum infiltration using the Agrobacterium tumefaciens strain ASE. Transgenic plants were screened for BASTA resistance. For analyzing phenotypes of FILpro:WOX1, out of the 67 independent transformants, 13 lines showing abnormal phenotypes in their true leaves were used.
To obtain growing plants, the seeds were sterilized and plated on soil or solid medium containing 0.5× Murashige and Skoog (MS) salts, pH 5.7, 1% Suc, and 0.75% or 1.2% agar; 0.75% agar medium was used only for screening the transgenic plants. Before germination, the seeds were placed at 4°C in the dark for 3 to 4 d. Plants were grown under continuous white fluorescent light at 22°C. To analyze the phenotypes of the DEX-treated plants, the 35Spro:KAN1:GR plants were grown on solid medium with or without 25 nM DEX. For transient exposure to DEX, the 35Spro:KAN1:GR plants were grown on solid medium without DEX for 8 d after germination, transferred to liquid medium containing 0.5× MS salts and 1% Suc with or without 10 μM DEX and with or without 10 μM CHX, and cultured for 6 h.
RNA ISH
Seedlings were fixed and embedded in paraffin according to the previously described microwave protocol (Takahashi et al., 2010) to reduce mRNA degradation. The protocol was modified for ISH as follows: 4% paraformaldehyde in PBS was used for fixation, and Histo-Clear (National Diagnostics) was used instead of n-butyl alcohol. Hybridization was performed as previously described (Ueda et al., 2004) on 8-μm paraffin sections. The FIL (Sawa et al., 1999) and PRS (Matsumoto and Okada, 2001) probes were previously described. The H4 (Dinneny et al., 2004), AS2 (Iwakawa et al., 2002), KAN1 (Kerstetter et al., 2001), and WOX1 (Vandenbussche et al., 2009) probes were generated with reference to previous studies. Probes were labeled using a DIG RNA labeling kit (Roche Applied Science).
Measurement of the Relative Levels of Gene Expression
Total RNA was extracted from the aerial parts of 8-d-old seedlings using the Plant RNeasy mini kit (Qiagen). cDNA was synthesized using the QuantiTect reverse transcription kit (Qiagen). Real-time quantitative PCR was performed on a Rotor-Gene Q (Qiagen) using the QuantiTect SYBR Green PCR kit (Qiagen), and data analysis was conducted using the Rotor-Gene 6000 series software 1.7 (Qiagen). Experiments were performed in experimental triplicate (Figures 2G and 2H; see Supplemental Figure 4B online) or biological duplicate and experimental triplicate (Figures 6A and 6E). The gene expression was normalized against β-TUBULIN-2. Primer sequences for the quantitative RT-PCR analyses of FIL and β-TUBULIN-2 were previously described (Leibfried et al., 2005; Ueno et al., 2007), and those for PRS, WOX1, KAN1, and KAN2 were as follows: PRSf01, 5′-TGTCCTTTGATTGCTGCTCTC-3′; PRSr03, 5′-TCTTCAGCTCCACTTTTGGTGCAG-3′; WOX1f01, 5′-CTGGATATGTTCGGTCGGATG-3′; WOX1r01, 5′-CTCCACCCGTATATTCGCTG-3′; rt-KAN1-F, 5′-ATGTCTATGGAAGGTGTTTTTCTAGAG-3′; rt-KAN1-R, 5′-AGAAGATTCATTGTGATGG-3′; rt-KAN2-F, 5′-ATGGAGCTGTTTCCTGCTCAGC-3′; and rt-KAN2-R, 5′-TGTTCTTGAATCAAGAGCTC-3′.
Microscopy, Histology, and Measurement
For scanning electron microscopy, the seedlings and leaves were frozen in liquid nitrogen and observed with a XL30 scanning electron microscope (FEI).
The fluorescence image was captured by the LSM510 equipped with a META device (Carl Zeiss) and a Leica MZ FLIII microscope equipped with a GFP2 filter and DC500 camera. The fluorescence of marker lines were analyzed in cross section as previously described (Watanabe and Okada, 2003).
For the histological analyses, the leaves were fixed in FAA (45% ethanol, 2.5% acetic acid, and 2.5% formaldehyde) and dehydrated in a graded ethanol series. Infiltration and embedding in Technovit 7100 (Heraus Kulzer) were performed according to the manufacturer’s instructions. Cross sections (3 or 5 μm) were cut with a microtome. Sections were stained with toluidine blue.
For analyzing cell division, the number of H4-positive cells was counted in cross sections of P3-P4 leaf primordia that were hybridized with the HISTONE H4 antisense probe as described above. The area of each cross section was divided into five parts in width, the area was measured using ImageJ v1.41o (National Institutes of Health), and the number of H4-positive cells was counted.
The number of hydathodes in the mature leaves that were cleared and mounted with a clearing solution (chloral hydrate:glycerol:water, 8:1:2) was counted using the Axioplan2 microscope (Carl Zeiss). The number of trichomes on the abaxial sides of mature leaves was counted using the SZX16 microscope (Olympus). The widths of the leaves and petals were measured using ImageJ v1.41o (National Institutes of Health) based on live images. The widths of cells from the leaves and petals were measured using the ImageJ software based on the differential interference contrast images (the first two leaves and petals) or Technovit sections (the eighth leaves). Statistical analyses were performed using the R package (http://www.R-project.org).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: PRS, At2g28610; WOX1, At3g18010; KAN1, At5g16560; KAN2, At1g32240; AS2, At1g65620; FIL, At2g45190; YAB3, At4g00180; and YAB5, At2g26580.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Representations of prs-2 and wox1-101.
Supplemental Figure 2. Phenotypes of Sepals and Petals of prs wox1.
Supplemental Figure 3. RNA in Situ Hybridization of Cross Sections of Shoot Apex with PRS Probes.
Supplemental Figure 4. Phenotypes of the FILpro:PRS Plants.
Supplemental Figure 5. The Pattern of KLUpro:vYFPer.
Supplemental Figure 6. Leaf Phenotype of prs wox1.
Supplemental Figure 7. Phenotype of prs wox1 as2.
Supplemental Methods 1. Expression Analysis of WOX1 in the Wild Type and wox1-101.
Supplementary Material
Acknowledgments
We thank ABRC, Satoshi Tabata (Kazusa), John L. Bowman (Monash University), and Michael Lenhard (Universität Potsdam) for the seeds and Tsuyoshi Nakagawa (Shimane University) and Koichi Toyokura (National Institute for Basic Biology [NIBB]) for the vectors. We thank Kiyoshi Tatematsu (NIBB) and members of Kiyotaka Okada’s lab (NIBB) for their technical advice and helpful discussions and Toshiharu Shikanai (Kyoto University), Taisuke Nishimura (Nagoya University), Mitsuyasu Hasebe (NIBB), Takahiro Yamaguchi (University of Tokyo), Hiroyuki Hirano (University of Tokyo), and Colin Kawaguchi (NIBB) for their helpful discussions. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry for Education, Culture, Sports, Science, and Technology of Japan (No. 19060004) and a Grant-in-Aid for Creative Scientific Research from the Japan Society for the Promotion of Science (No. 19GS0315) to K.O. and by the Arabidopsis Functional Genomics Network, the European Research Area Plant Genomics, and Sonderforschungsbereich 592 grants from the Deutsche Forschungsgemeinschaft to T.L. M.N. was supported by a fellowship from the Japan Society for the Promotion of Science (No. 20-2203).
AUTHOR CONTRIBUTIONS
M.N., N.M., R.T., T.L. and K.O. designed the research. M.N., N.M., and E.R. performed the research and analyzed the data. M.N. and K.O. coordinated the research and wrote the article.
References
- Adamski N.M., Anastasiou E., Eriksson S., O’Neill C.M., Lenhard M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 106: 20115–20120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou E., Kenz S., Gerstung M., MacLean D., Timmer J., Fleck C., Lenhard M. (2007). Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13: 843–856 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Eshed Y., Baum S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18: 134–141 [DOI] [PubMed] [Google Scholar]
- Chitwood D.H., Guo M., Nogueira F.T.S., Timmermans M.C.P. (2007). Establishing leaf polarity: The role of small RNAs and positional signals in the shoot apex. Development 134: 813–823 [DOI] [PubMed] [Google Scholar]
- Chitwood D.H., Nogueira F.T.S., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C.P. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler N.G., Tsukaya H. (2001). Leaf morphogenesis in dicotyledons: Current issues. Int. J. Plant Sci. 162: 459–464 [Google Scholar]
- Dinneny J.R., Yadegari R., Fischer R.L., Yanofsky M.F., Weigel D. (2004). The role of JAGGED in shaping lateral organs. Development 131: 1101–1110 [DOI] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Bowman J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Perea J.V., Bowman J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Izhaki A., Baum S.F., Floyd S.K., Bowman J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Gaudin V., Lunness P.A., Fobert P.R., Towers M., Riou-Khamlichi C., Murray J.A., Coen E., Doonan J.H. (2000). The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol. 122: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A., Gross-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hagemann W., Gleissberg S. (1996). Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199: 121–152 [Google Scholar]
- Hartig K., Beck E. (2006). Crosstalk between auxin, cytokinins, and sugars in the plant cell cycle. Plant Biol. (Stuttg.) 8: 389–396 [DOI] [PubMed] [Google Scholar]
- Hawker N.P., Bowman J.L. (2004). Roles for Class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., Machida Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Kato T., Tabata S., Sato S. (2007). Expression analysis of gene trap lines and mapping of donor loci for Dissociation transposition in Arabidopsis. Plant Biotechnol. 24: 467–479 [Google Scholar]
- Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kumaran M.K., Bowman J.L., Sundaresan V. (2002). YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14: 2761–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. (2004). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N., Okada K. (2001). A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 15: 3355–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- McHale N.A. (1993). LAM-1 and FAT genes control development of the leaf blade in Nicotiana sylvestris. Plant Cell 5: 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nardmann J., Ji J., Werr W., Scanlon M.J. (2004). The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131: 2827–2839 [DOI] [PubMed] [Google Scholar]
- Nole-Wilson S., Krizek B.A. (2006). AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol. 141: 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R., Sussex I. (1985). The developmental morphology and growth dynamics of the tobacco leaf. Planta 165: 158–169 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Frenz M., Mandel T., Kuhlemeier C. (2005). Microsurgical and laser ablation analysis of leaf positioning and dorsoventral patterning in tomato. Development 132: 15–26 [DOI] [PubMed] [Google Scholar]
- Sarojam R., Sappl P.G., Goldshmidt A., Efroni I., Floyd S.K., Eshed Y., Bowman J.L. (2010). Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22: 2113–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Watanabe K., Goto K., Liu Y.G., Shibata D., Kanaya E., Morita E.H., Okada K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon M.J., Schneeberger R.G., Freeling M. (1996). The maize mutant narrow sheath fails to establish leaf margin identity in a meristematic domain. Development 122: 1683–1691 [DOI] [PubMed] [Google Scholar]
- Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Siegfried K.R., Eshed Y., Baum S.F., Otsuga D., Drews G.N., Bowman J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128 [DOI] [PubMed] [Google Scholar]
- Stahle M.I., Kuehlich J., Staron L., von Arnim A.G., Golz J.F. (2009). YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21: 3105–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., et al. (2011). STENOFOLIA regulates blade outgrowth and leaf vascular patterning in Medicago truncatula and Nicotiana sylvestris. Plant Cell 23: 2125–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Kamakura H., Sato Y., Shiono K., Abiko T., Tsutsumi N., Nagamura Y., Nishizawa N.K., Nakazono M. (2010). A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection. J. Plant Res. 123: 807–813 [DOI] [PubMed] [Google Scholar]
- Timmermans M.C., Schultes N.P., Jankovsky J.P., Nelson T. (1998). Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development 125: 2813–2823 [DOI] [PubMed] [Google Scholar]
- Toyokura K., Watanabe K., Oiwaka A., Kusano M., Tameshige T., Tatematsu K., Matsumoto N., Tsugeki R., Saito K., Okada K. (2011). Succinic semialdehyde dehydrogenase is involved in the robust patterning of Arabidopsis leaves along the adaxial-abaxial axis. Plant Cell Physiol. 52: 1340–1353 [DOI] [PubMed] [Google Scholar]
- Ueda M., Matsui K., Ishiguro S., Sano R., Wada T., Paponov I., Palme K., Okada K. (2004). The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development 131: 2101–2111 [DOI] [PubMed] [Google Scholar]
- Ueno Y., Ishikawa T., Watanabe K., Terakura S., Iwakawa H., Okada K., Machida C., Machida Y. (2007). Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19: 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Horstman A., Zethof J., Koes R., Rijpkema A., Gerats T. (2009). Differential recruitment of WOX transcription factors for lateral development and organ fusion in petunia and Arabidopsis Plant Cell 21: 2269–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143 [Google Scholar]
- Wang W., Xu B., Wang H., Li J., Huang H., Xu L. (2011). YUCCA genes are expressed in response to leaf adaxial-abaxial juxtaposition and are required for leaf margin development. Plant Physiol. 157: 1805–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Okada K. (2003). Two discrete cis elements control the abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell 15: 2592–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wu R., Qin G., Chen Z., Gu H., Qu L.J. (2011). Over-expression of WOX1 leads to defects in meristem development and polyamine homeostasis in Arabidopsis. J. Integr. Plant Biol. 53: 493–506 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.