This study demonstrates that sequence variation in the ligand-interacting degron modulates the stability of JAZ8 and, as a consequence, the extent to which JAZ8 represses jasmonate-responsive gene expression. This study also shows that JAZ8 is an EAR motif–containing protein that represses gene expression by direct recruitment of the corepressor TOPLESS to cognate transcription factors.
Abstract
The lipid-derived hormone jasmonoyl-l-Ile (JA-Ile) initiates large-scale changes in gene expression by stabilizing the interaction of JASMONATE ZIM domain (JAZ) repressors with the F-box protein CORONATINE INSENSITIVE1 (COI1), which results in JAZ degradation by the ubiquitin-proteasome pathway. Recent structural studies show that the JAZ1 degradation signal (degron) includes a short conserved LPIAR motif that seals JA-Ile in its binding pocket at the COI1-JAZ interface. Here, we show that Arabidopsis thaliana JAZ8 lacks this motif and thus is unable to associate strongly with COI1 in the presence of JA-Ile. As a consequence, JAZ8 is stabilized against jasmonate (JA)-mediated degradation and, when ectopically expressed in Arabidopsis, represses JA-regulated growth and defense responses. These findings indicate that sequence variation in a hypervariable region of the degron affects JAZ stability and JA-regulated physiological responses. We also show that JAZ8-mediated repression depends on an LxLxL-type EAR (for ERF-associated amphiphilic repression) motif at the JAZ8 N terminus that binds the corepressor TOPLESS and represses transcriptional activation. JAZ8-mediated repression does not require the ZIM domain, which, in other JAZ proteins, recruits TOPLESS through the EAR motif–containing adaptor protein NINJA. These findings show that EAR repression domains in a subgroup of JAZ proteins repress gene expression through direct recruitment of corepressors to cognate transcription factors.
INTRODUCTION
The lipid-derived hormone jasmonoyl-l-Ile (JA-Ile) and structurally related jasmonates (collectively referred to as JAs) play an essential role in controlling plant growth, development, and responses to environmental stress. Among the major functions ascribed to JAs are activation of defense responses to insect attack and pathogen infection, reproductive development, and growth inhibition (Glazebrook, 2005; Wasternack, 2007; Howe and Jander, 2008; Browse, 2009). These general roles for the hormone suggest that JA signaling evolved as a mechanism to optimize plant fitness in rapidly changing terrestrial environments. This hypothesis is supported by emerging mechanistic evidence for extensive crosstalk between JA and other hormones that mediate developmental plasticity (Dombrecht et al., 2007; Moreno et al., 2009; Pieterse et al., 2009; Hou et al., 2010; Robson et al., 2010; Ballaré, 2011; Kazan and Manners, 2011; Zhu et al., 2011). A greater understanding of the molecular mechanism of JA signaling is therefore expected to impact broad areas of plant biology (Howe, 2010; Kazan and Manners, 2012).
The discovery of JASMONATE ZIM-domain (JAZ) proteins spurred remarkable progress in understanding how JAs regulate large-scale changes in gene expression (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Pauwels and Goossens, 2011). JAZs belong to the plant-specific TIFY family of transcriptional regulators that are defined by the presence of a TIF[F/Y]XG motif within a larger (~28 amino acids) conserved region known as the ZIM (or TIFY) domain (Vanholme et al., 2007; Chung et al., 2009; Bai et al., 2011). JAZs are distinguished from other TIFY proteins by the presence of a multifunctional C-terminal region known as the Jas motif. Both the ZIM domain and Jas motif are required for JAZ-mediated repression of JA responses in cells containing low JA-Ile levels. The Jas motif interacts with members of the basic helix-loop-helix (bHLH) (e.g., MYC2) and R2R3 MYB family of transcription factors that promote the expression of JA response genes (Chini et al., 2007; Melotto et al., 2008; Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011; Qi et al., 2011; Song et al., 2011). The ZIM domain and its associated TIFY motif mediate JAZ interaction with an adaptor protein called NOVEL INTERACTOR OF JAZ (NINJA), which functions to recruit the transcription corepressor TOPLESS (TPL) and TPL-related proteins (TPRs) (Pauwels et al., 2010; Pauwels and Goossens, 2011). As is the case for other proteins that repress transcription through TPL and associated chromatin remodeling enzymes (Ohta et al., 2001; Krogan and Long, 2009; Kagale and Rozwadowski, 2011), NINJA contains an EAR (for ERF-associated amphiphilic repression) motif that is necessary and sufficient for TPL interaction (Pauwels et al., 2010). The ZIM domain also promotes homo- and heteromeric interactions between JAZ proteins, which in Arabidopsis thaliana are encoded by 12 genes (JAZ1 to JAZ12) (Chini et al., 2009; Chung and Howe, 2009; Chung et al., 2009). Alternative splicing of JAZ genes expands the repertoire and potential combinatorial diversity of JAZ–JAZ interactions (Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010). A current challenge is to determine the contribution of individual JAZ isoforms to the regulation of JA signaling.
JAZ-mediated repression is relieved in response to stimuli that activate the production of JA-Ile and subsequent degradation of JAZ proteins via the ubiquitin/26S proteasome pathway. JA-Ile initiates the signaling cascade by promoting interaction of JAZ with the F-box protein CORONATINE INSENSITIVE1 (COI1). As the specificity determinant of the SCF (for SKP1-CUL1-F-box)–type ubiquitin ligase SCFCOI1, COI1 binding to many JAZ proteins is stimulated directly by JA-Ile (Xie et al., 1998; Thines et al., 2007; Katsir et al., 2008b; Melotto et al., 2008; Fonseca et al., 2009; Yan et al., 2009). Recent structural studies revealed that the N-terminal 20 amino acids of the Jas motif adopt a bipartite structure consisting of a loop region that interacts with both COI1 and JA-Ile and an α-helix that contacts the surface of COI1 adjacent to the ligand binding pocket (Sheard et al., 2010). Point mutations affecting conserved basic amino acids in the loop region of JAZ1 and JAZ9 abrogate COI1 binding and enhance the repression activity of ectopically expressed JAZ1 (Melotto et al., 2008). High-affinity ligand binding to the COI1-JAZ1 coreceptor complex also requires an inositol pentakisphosphate (IP5) cofactor that interacts with COI1 and JAZ near the hormone binding pocket (Sheard et al., 2010).
Initial insight into the identity of JAZ proteins as repressors came from the observation that truncated JAZs lacking the Jas motif are resistant to JA-mediated degradation and, when expressed in Arabidopsis, confer dominant insensitivity to exogenous JA (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). A role for JAZ10 as a negative regulator is supported by the phenotype of jaz10 loss-of-function mutants, which exhibit increased sensitivity to JA (Yan et al., 2007; Demianski et al., 2012). The repressive function of JAZ10 appears to result from the action of alternative splice variants that lack either a portion of the Jas motif (JAZ10.3) or the entire Jas motif (JAZ10.4) (Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010). These naturally occurring truncated JAZs have reduced capacity to interact with COI1 and thus are stabilized against hormone-dependent degradation (Chung and Howe, 2009). Ectopic expression of JAZ10.3 and JAZ10.4, which retain the ability to interact with MYC2, results in reduced sensitivity to JA. Transgenic lines overexpressing a JAZ10 genomic clone also show reduced sensitivity to JA, presumably as a consequence of overproduction of JAZ10.3 and JAZ10.4 (Chung et al., 2010).
Although it is established that dominant repression by JAZ10 splice variants and artificially truncated JAZs results from decreased association with COI1, a role for full-length (i.e., Jas motif–containing) JAZs as transcriptional repressors remains to be shown. For example, JA-insensitive phenotypes have not been observed in transgenic lines of Arabidopsis that overexpress full-length JAZ1, JAZ2, JAZ3, or JAZ10 (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010). Other than jaz10, the only jaz mutant reported to exhibit enhanced sensitivity to JA is an RNA interference line of JAZ1 (Grunewald et al., 2009). However, this phenotype was not observed in a T-DNA insertion mutant (jaz1-1) that was characterized as a likely null mutant (Demianski et al., 2012). The lack of obvious JA-hypersensitive phenotypes among most jaz mutants reported to date may result from functional redundancy between JAZ proteins. Determining the role of specific JAZ isoforms in transcriptional repression is needed to further understand how JAs control myriad responses during the plant life cycle.
Here, we establish a role for full-length JAZ8 as a transcriptional repressor and describe two novel features of the protein that are critical for repression activity. First, we show that JAZ8 lacks the canonical degron that promotes JA-Ile–dependent interaction of other JAZ proteins with COI1. Second, we demonstrate that an LxLxL-type EAR motif near the N terminus of JAZ8 mediates transcriptional repression. Significantly, JAZ8-mediated repression does not require the ZIM domain, which in other JAZs recruits TPL through the EAR motif of NINJA. Based on these findings, we propose that JAZ8 and other EAR motif–containing JAZ proteins directly recruit TPL/TPRs to transcription factors as a mechanism for repressing JA responses.
RESULTS
JAZ8 Weakly Associates with COI1 in the Presence of Known Receptor Ligands
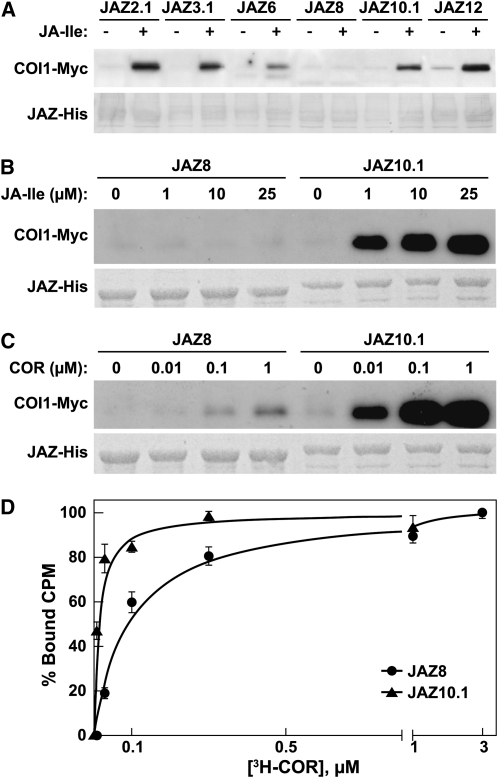
We used in vitro pull-down assays to investigate the relationship between sequence diversity in the Jas motif and hormone-dependent binding of full-length JAZ proteins to COI1. A phylogenetic tree constructed from the 27–amino acid Jas motif of all 12 Arabidopsis JAZs divided the family into six groups: JAZ1/2, JAZ3/4/9, JAZ5/6, JAZ7/8, JAZ10.1, and JAZ11/12 (see Supplemental Figure 1 and Supplemental Data Set 1 online). Representative full-length proteins from each group were expressed in Escherichia coli as maltose binding protein (MBP)-JAZ-6xHis fusions and tested for their ability to interact with COI1 in the presence or absence of the active form of the hormone, (3R,7S)-JA-Ile. At a concentration of 1 μM, JA-Ile stimulated binding of COI1 to JAZ2.1, JAZ3.1, JAZ6, JAZ10.1, and JAZ12, but not JAZ8 (Figure 1A). Higher concentrations of JA-Ile increased the amount of COI1 binding to JAZ10.1 but failed to promote interaction with JAZ8 (Figure 1B). The phytotoxin coronatine, which is a potent agonist of the COI1-JAZ receptor system (Katsir et al., 2008b; Melotto et al., 2008; Yan et al., 2009; Sheard et al., 2010), stimulated low-level, dose-dependent binding of COI1 to JAZ8 (Figure 1C). Saturation binding assays confirmed that 3H-coronatine binds specifically to COI1-JAZ8 receptor complexes, albeit with much lower affinity (91.4 nM) than that observed for COI1-JAZ10.1 complexes (7.0 nM) (Figure 1D).
Figure 1.
JAZ Proteins Differentially Interact with COI1 in the Presence of JA-Ile.
(A) JA-Ile differentially promotes COI1 interaction with various JAZ proteins. Pull-down assays were performed using crude leaf extracts from 35S:COI1-Myc transgenic plants and purified recombinant JAZ-His proteins. Reactions were supplemented (+) with 1 μM (3R,7S)-JA-Ile or an equivalent volume of assay buffer (−). Protein bound to JAZ-His was separated by SDS-PAGE and analyzed by immunoblotting with anti-Myc antibody for the presence of COI1-Myc. The blotted membrane was stained with Coomassie blue to visualize the amount of JAZ-His loaded.
(B) JAZ8 does not associate with COI1 in the presence of JA-Ile. Pull-down assays with purified JAZ8-His and JAZ10.1-His were performed as described in (A). Reaction mixtures were supplemented with the indicated concentration of (3R,7S)-JA-Ile.
(C) JAZ8 weakly associates with COI1 in the presence of coronatine. Pull-down assays were performed as described in (A). Reactions were supplemented with the indicated concentration of coronatine (COR).
(D) Saturation binding of 3H-labeled coronatine to COI1-JAZ8 (circle) and COI1-JAZ10.1 (triangle) complexes. Data show the mean ± se of two replicates performed in duplicate. CPM, counts per minute.
JAZ8 Attenuates JA Responses and Is Resistant to JA-Mediated Degradation in Vivo
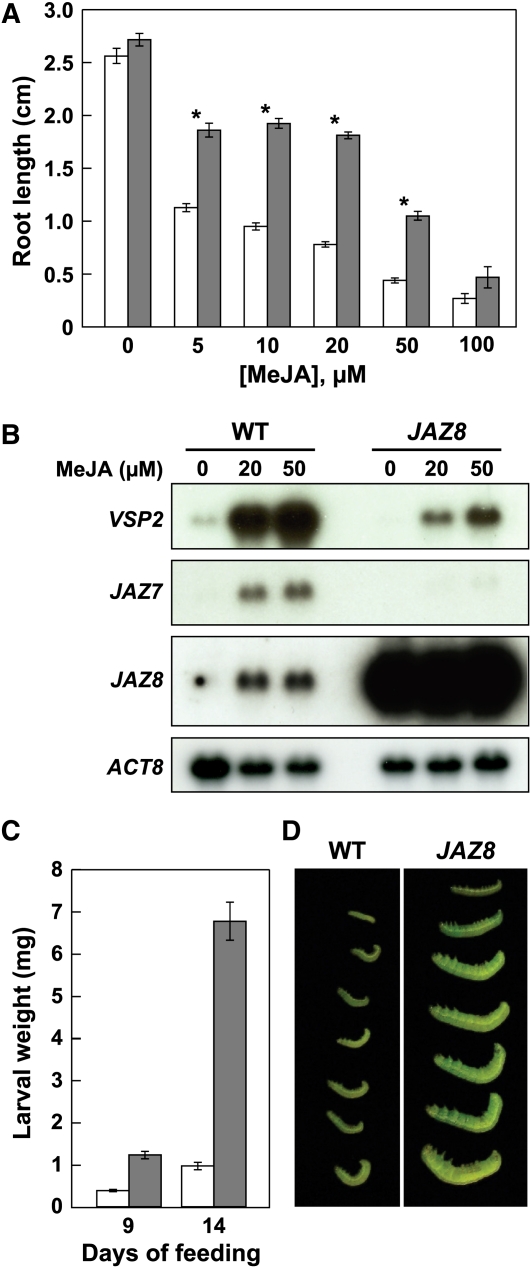
Truncated JAZ proteins that fail to interact with COI1 in the presence of JA-Ile can repress JA signaling when overexpressed in Arabidopsis (Thines et al., 2007; Melotto et al., 2008; Chung et al., 2010). To determine whether full-length JAZ8 exerts dominant repression in vivo, we constructed transgenic lines of Arabidopsis that express the JAZ8 cDNA under the control of the cauliflower mosaic virus (CaMV) 35S promoter. T1 plants (24 independent lines) selected for the presence of the 35S:JAZ8 transgene did not exhibit obvious defects in flower development or fertility. Subsequently, homozygous lines (T3 generation) that were selected for high expression of the transgene (see Methods) were tested in root growth inhibition assays. All 35S:JAZ8 lines (n = 5) selected in this manner were insensitive to JA-mediated root growth inhibition and were significantly more insensitive to JA than the myc2 mutant, jin1-7 (see Supplemental Figure 2 online) (Lorenzo et al., 2004). Dose–response experiments performed with a representative 35S:JAZ8 homozygous line (#24) showed that JAZ8 overexpression confers insensitivity to a broad range of concentrations of exogenous methyl-JA (MeJA) but does not affect root growth in seedlings grown on medium lacking the compound (Figure 2A). Consistent with this finding, RNA gel blot analysis demonstrated that 35S:JAZ8 seedlings are impaired in the expression of the JA-responsive genes VSP2 and JAZ7 (Figure 2B). To test the effect of JAZ8 overexpression on JA-mediated defense responses, we compared the weight gain of Spodoptera exigua larvae grown on adult wild-type and 35S:JAZ8 plants. Insects reared on 35S:JAZ8 lines for either 9 or 14 d were significantly heavier (P < 0.0001) than insects grown on the wild type (Figures 2C and 2D). The increased mass of caterpillars grown on 35S:JAZ8 plants was associated with increased consumption of leaf tissue (see Supplemental Figure 3 online). In summary, these results show that overexpression of full-length JAZ8 impairs JA-mediated root growth inhibition and defense responses, as well as JA-dependent gene expression.
Figure 2.
Overexpression of JAZ8 Results in Decreased Sensitivity to JA.
(A) Root growth inhibition assay of wild-type (open bars) and 35S:JAZ8 (closed bars) seedlings grown on MS media containing the indicated concentration of MeJA. Root length measurements were made 6 d after seed germination. Data show the mean ± se (n > 20 seedlings per genotype for each concentration of MeJA except 100 μM, in which n = 6). Asterisks denote statistically significant differences (P < 0.05, Student’s t test) between the two plant genotypes at the indicated concentration of MeJA.
(B) JA-responsive gene expression in wild-type (WT) and 35S:JAZ8 (JAZ8) seedlings. Seedlings were grown in liquid MS medium for 9 d and then treated for 2 h with the indicated concentration of MeJA or a mock control (0). Total RNA isolated from the treated seedlings was subjected to RNA gel blot analysis with the indicated probes. A JAZ8 probe was used to verify overexpression of JAZ8 in the transgenic line, and an ACTIN8 (ACT8) probe was used as a loading control.
(C) Weight of S. exigua larvae reared on wild-type (open bars) or 35S:JAZ8 (closed bars) plants for 9 or 14 d. Data show the mean ± se (n = 56 to 65 larvae per plant genotype). The experiment was repeated four times with similar results. Data from a representative experiment are shown.
(D) Photograph of representative S. exigua larvae recovered from wild-type and 35S:JAZ8 (JAZ8) plants after 14 d of feeding.
[See online article for color version of this figure.]
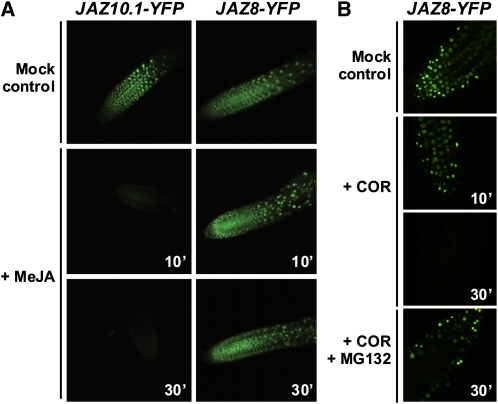
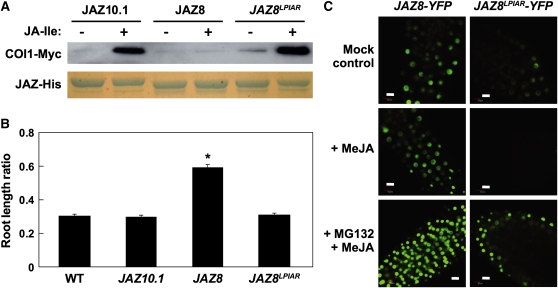
To determine whether the JA-insensitive phenotype of 35S:JAZ8 plants is associated with increased JAZ8 stability in vivo, we used confocal laser scanning microscopy to compare JA-mediated changes in fluorescence of JAZ8-yellow fluorescent protein (YFP) and JAZ10.1-YFP fusion proteins that were stably expressed in Arabidopsis. Root growth inhibition assays showed that 35S:JAZ8-YFP seedlings, similar to 35S:JAZ8 lines, exhibit reduced sensitivity to exogenous MeJA (see Supplemental Figure 4A online). In the absence of JA treatment, a nuclear-localized YFP signal was observed in both 35S:JAZ10.1-YFP and 35S:JAZ8-YFP seedlings (Figure 3A). Within 10 min of treatment with 50 μM MeJA, this signal was largely eliminated in 35S:JAZ10.1-YFP seedlings, as previously reported (Chung and Howe, 2009). The nuclear-localized YFP signal in roots of 35S:JAZ8-YFP seedlings persisted up to 30 min after MeJA treatment. Consistent with the ability of coronatine to associate with COI1-JAZ8 complexes in vitro (Figure 1), exogenous coronatine stimulated JAZ8-YFP turnover in vivo (Figure 3B). This effect was partially inhibited by pretreatment of seedlings with the 26S proteasome inhibitor MG132. These findings show that JAZ8 is more resistant than JAZ10.1 to JA-mediated degradation in vivo but can be degraded in response to ligands (i.e., coronatine) that promote COI1-JAZ8 association.
Figure 3.
JAZ8 Is Resistant to JA-Mediated Degradation in Vivo.
(A) Differential stability of JAZ8 and JAZ10.1 in the presence of JA. Transgenic seedlings (7 d old) expressing JAZ8-YFP or JAZ10.1-YFP fusion proteins were treated either with water (mock) or with 50 μM MeJA for the indicated amount of time prior to imaging of roots by confocal fluorescence microscopy. Image settings used for seedlings of the same genotype at different time points were identical.
(B) Coronatine stimulates JAZ8 turnover in a 26S proteasome-dependent manner. Transgenic seedlings (4 d old) expressing JAZ8-YFP were pretreated with water or the 26S proteasome inhibitor MG132 (100 μM) for 80 min, at which time seedlings were treated with coronatine (1 μM) for an additional 10 or 30 min. Seedlings treated with both coronatine and MG132 were imaged at the 30 min time point. YFP signal in root tissue was visualized by confocal microscopy. Microscope settings were identical for all images.
[See online article for color version of this figure.]
JAZ8 Lacks a Canonical JA-Ile Degron
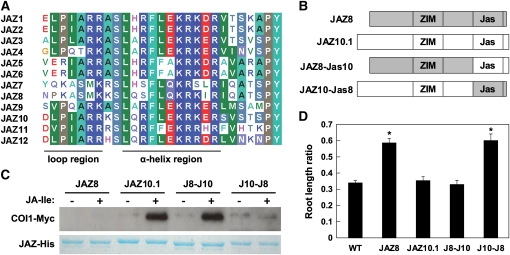
We performed domain swap experiments to test whether the observed functional differences between JAZ8 and JAZ10.1 can be attributed to sequence variation in the Jas motif (Figure 4A). For this purpose, we generated constructs encoding chimeric proteins in which the Jas motif and remaining C-terminal amino acids (two and five amino acids for JAZ8 and JAZ10.1, respectively) of JAZ8 and JAZ10.1 were reciprocally exchanged as shown schematically in Figure 4B. In vitro pull-down assays performed with the resulting fusion proteins showed that the Jas motif of JAZ10.1 confers on JAZ8 the ability to interact with COI1 in the presence of 2.5 μM JA-Ile (Figure 4C). Moreover, binding of COI1 to the JAZ10-Jas8 chimera, in which the C-terminal region of JAZ10.1 is replaced by the Jas motif from JAZ8, was not stimulated by JA-Ile. To determine whether these changes in hormone-dependent COI1 interaction impact JA-regulated physiological responses, we constitutively expressed the chimeric proteins in Arabidopsis. Seedlings that express the JAZ8-Jas10 chimera were fully sensitive to JA. Conversely, lines expressing JAZ10-Jas8 exhibited a JA-insensitive root growth phenotype that was indistinguishable from that of 35S:JAZ8 plants (Figure 4D). We conclude that the Jas motif of JAZ8 lacks sequence determinants that are required for JA-Ile–dependent binding to COI1.
Figure 4.
A Jas Domain Swap Converts JAZ8 to a COI1-Interacting Protein That Does Not Exert Dominant Repression of JA Responses.
(A) Amino acid alignment of the Jas motif in 12 Arabidopsis full-length JAZ proteins. Structural features within the Jas motif that physically associate with COI1 and JA-Ile (Sheard et al., 2010) are indicated.
(B) Schematic diagram of chimeric proteins constructed by swapping of the Jas motif. Sequence regions derived from JAZ8 and JAZ10.1 are shown in gray and white, respectively.
(C) Pull-down assays performed with parental (JAZ8 and JAZ10.1) and chimeric (JAZ10-Jas8, J10-J8; JAZ8-Jas10, J8-J10) proteins in the presence (+) or absence (−) of 2.5 μM JA-Ile.
(D) JA-mediated root growth inhibition in wild-type (WT) and transgenic seedlings (35S:JAZ8, JAZ8; 35S:JAZ10.1, JAZ10.1; 35S:JAZ8-Jas10, J8-J10; and 35S:JAZ10-Jas8, J10-J8). Seedlings were grown for 6 d on MS medium supplemented or not supplemented with 20 μM MeJA. The root length ratio was calculated by dividing the average the root length of seedlings grown on MeJA-containing medium by the average root length of seedlings of the same genotype grown in the absence of MeJA. Data points show the mean ± se (n = 18 to 24 seedlings per data point). Asterisks denote significant differences (P < 0.05, Student’s t test) in comparisons between the indicated transgenic line and the wild type .
[See online article for color version of this figure.]
The COI1-interacting degron of JAZ1 includes a six–amino acid (LPIARR) loop region that encloses JA-Ile in its binding pocket (Sheard et al., 2010). This sequence is largely conserved in other JAZs (e.g., JAZ10.1) that strongly interact with COI1 in the presence of JA-Ile (Figure 4A). The sequence (PKASMK) of the corresponding region in JAZ8 differs markedly from the canonical LPIARR motif. To determine whether this variation is responsible for the weak COI1-JAZ8 association observed in vitro (Figure 1), we constructed a modified version (JAZ8LPIAR) of JAZ8 in which five amino acids (PKASM) in the loop region were replaced with the canonical LPIAR. In vitro pull-down assays showed that this substitution is sufficient to confer JA-Ile–dependent interaction of JAZ8 with CO11 (Figure 5A). Consistent with this finding, the PKASM→LPIAR substitution completely suppressed the JA-insensitive root growth phenotype of 35S:JAZ8 seedlings (Figure 5B). The ability of JAZ8 to interact with MYC2 (Chini et al., 2009) raised the possibility that the PKASM→LPIAR substitution impairs this interaction, which could account for the JA-sensitive phenotype of 35S:JAZ8LPIAR seedlings. Yeast two-hybrid assays showed that JAZ8LPIAR, like JAZ8, interacts with MYC2, thus excluding this possibility (see Supplemental Figure 5 online). Fluorescence microscopy showed that a JAZ8LPIAR-YFP fusion protein accumulates in the nucleus (Figure 5C), further indicating that the substitution does not impair expression or localization of the protein. Imaging of 35S:JAZ8-YFP and 35S:JAZ8LPIAR-YFP seedlings treated with MeJA (for 15 min) demonstrated that JAZ8LPIAR-YFP is more sensitive than JAZ8-YFP to JA-mediated degradation. The ability of MG132 to block JA-mediated turnover of JAZ8LPIAR-YFP showed that degradation of JAZ8LPIAR is dependent on the 26S proteasome. We conclude that the noncanonical degron sequence (PKASM) in JAZ8 does not effectively interact with COI1 in a JA-Ile–dependent manner and that this sequence is important for JAZ8’s function as a dominant repressor of JA signaling.
Figure 5.
The Canonical LPIAR Degron Promotes JAZ8 Binding to COI1, Destabilizes JAZ8, and Restores JA Responsiveness.
(A) JAZ8LPIAR interacts with COI1 in a JA-Ile–dependent manner. Pull-down reactions were performed in the presence (+) or absence (−) of 2.5 μM JA-Ile as described in the legend for Figure 1A. Protein bound to the indicted JAZ-His fusion protein was separated by SDS-PAGE and analyzed by immunoblotting with anti-Myc antibody for the presence of COI1-Myc.
(B) Transgenic lines overexpressing JAZ8LPIAR are sensitive to JA. Root growth assays were performed with wild-type (WT), 35S:JAZ10 (JAZ10.1), 35S:JAZ8 (JAZ8), and 35S:JAZ8LPIAR (JAZ8LPIAR) seedlings. Data show the mean ± se for each genotype (n = 20). Asterisk denotes significant differences (P < 0.05, Student’s t test) in comparisons between the indicated transgenic line and the wild type.
(C) JAZ8LPIAR-YFP is degraded in vivo in response to JA treatment. Six-day-old transgenic seedlings expressing JAZ8-YFP (left) or JAZ8LPIAR-YFP (right) were pretreated with water or the 26S proteasome inhibitor MG132 (100 μM) for 75 min, at which time seedlings were treated with either MeJA (+MeJA, 50 μM) or water (mock control) for an additional 15 min. YFP signal in root tissue was visualized by fluorescence microscopy. Microscope settings were identical for all images. Bars = 50 μm.
[See online article for color version of this figure.]
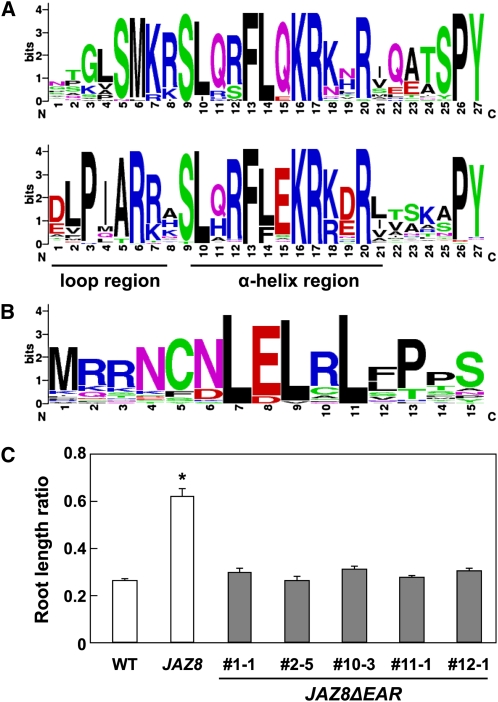
These results led us to investigate whether JAZ proteins in other plant species have a JAZ8-like Jas motif. Phylogenetic analyses indicate that angiosperm JAZs cluster into five groups, including those represented by JAZ7/8 (group IV) and JAZ10 (group III) (Bai et al., 2011). Alignment of the predicted Jas motif in group IV proteins from diverse plant species showed that Ser and Met residues within the PKASM motif are highly conserved in other members of this group, whereas the PKA sequence is more variable (Figure 6A). Comparison of this motif with a consensus Jas motif constructed from canonical JAZ10-like proteins showed that these two groups of JAZs differ markedly in the N-terminal degron sequence but not in other regions of the Jas motif (Figure 6A).
Figure 6.
The EAR Motif of JAZ8 Is Required for Repression of JA Responses.
(A) Consensus sequence of the Jas motif in JAZ8-like (top) and JAZ10-like (bottom) proteins from various plant species (see Supplemental Table 2 online).
(B) Consensus sequence of the N-terminal 15 amino acids from JAZ8-like proteins from diverse plant species (see Supplemental Table 2 online).
(C) Comparison of JA-mediated root growth inhibition in 35S:JAZ8 and 35S:JAZ8ΔEAR seedlings. Wild-type (WT), 35S:JAZ8 (line #24), and 35S:JAZ8ΔEAR (five independent homozygous lines) were grown for 8 d on medium supplemented or not supplemented with 20 μM MeJA. Root length ratios were calculated as described in the legend of Figure 4D. Data show the mean ± se for each genotype (n > 14 seedlings per genotype). Asterisks denote a significant difference (P < 0.05, Student’s t test) in comparisons between transgenic and wild-type seedlings. The mean root length ratio calculated for seedlings (n = 92) from all five independent 35S:JAZ8ΔEAR lines was not significantly different (P value = 0.068, Student’s t test) from that of the wild type.
[See online article for color version of this figure.]
An EAR Motif in JAZ8 Mediates Transcriptional Repression
Previous genome-wide sequence searches identified an LxLxL-type EAR motif in Arabidopsis JAZ8 and its close relative, JAZ7 (Kagale et al., 2010). This sequence (LELRL) is located close to the N terminus of JAZ8 and is highly conserved among group IV JAZs from diverse plant species (Figure 6B) (Bai et al., 2011). To investigate the functional relevance of the motif in JAZ8-mediated repression of JA responses, we generated transgenic lines that overexpress a mutant version (JAZ8ΔEAR) of JAZ8 in which the LELRL sequence was deleted. Root growth assays performed with multiple homozygous 35S:JAZ8ΔEAR lines showed that deletion of the EAR motif suppresses the JA-insensitive phenotype conferred by JAZ8 overexpression (Figure 6C). Similar results were obtained with lines expressing YFP-tagged JAZ8 proteins in which the LELRL motif was deleted (JAZ8ΔEAR-YFP) or substituted with Ala residues (JAZ8LELRL→5xA-YFP) (see Supplemental Figure 4B online). Fluorescence microscopy showed that these JAZ8-YFP derivatives are expressed in these lines and, like JAZ8-YFP, accumulate in the nucleus (see Supplemental Figure 6 online). These results support the hypothesis that the conserved LELRL motif performs a critical role in JAZ8-mediated inhibition of JA responses.
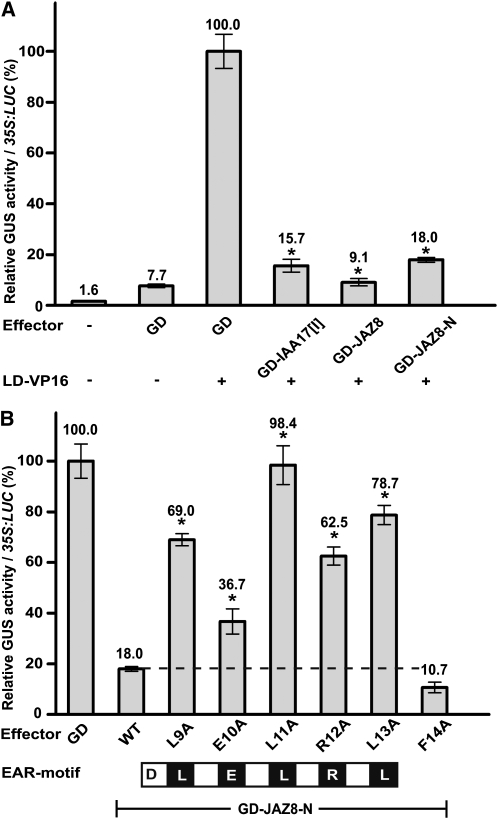
We next used a heterologous carrot (Daucus carota) protoplast transfection system (Tiwari et al., 2004) to test directly whether JAZ8 functions as a transcriptional repressor. This assay relied on the use of a β-glucuronidase (GUS) reporter gene [35S(-46)LexA(2x)-Gal4(2x):GUS] that is driven by a 35S minimal promoter harboring LexA and Gal4 DNA binding sites. The reporter was cotransfected with two effector constructs: one encoding a fusion protein (LD-VP16) consisting of the LexA DNA binding domain (LD) fused to the VP16 activation domain and a second effector encoding the Gal4 DNA binding domain (GD) fused to either full-length JAZ8 (GD-JAZ8) or the N-terminal region of JAZ8 (GD-JAZ8-N). Cotransfection of the GUS reporter with LD-VP16 and a construct encoding only GD resulted in strong activation (~13-fold) of the reporter in comparison to the GD construct alone (Figure 7A). The EAR motif repression domain of INDOLE-3-ACETIC ACID INDUCIBLE17 (IAA17) was used as a positive control for transcriptional repression; cotransfection of this effector (GD-IAA17[I]) resulted in strong reduction in reporter gene expression, as previously reported (Tiwari et al., 2004). GD-JAZ8 repressed LD-VP16–mediated transcription of the reporter to a level similar to that obtained with GD-IAA17[I]. Cotransfection with GD-JAZ-N, which encodes the EAR motif–containing N-terminal 44 amino acids of JAZ8, also resulted in strong repression (Figure 7A). These findings show that JAZ8 directly represses a transcriptional activator and that the repression activity resides within the N-terminal 44 amino acids of JAZ8.
Figure 7.
The EAR Motif Is Required for Transcriptional Repression by JAZ8.
(A) Carrot protoplasts were transfected with the GUS reporter [35S(-46)LexA(2x)-Gal4(2x):GUS] alone or in combination with the indicated effector and LD-VP16 constructs as described in the text. GUS activities are expressed relative to the sample in which LD-VP16 was cotransfected with the GD effector (Gal4 DNA binding domain alone; set to 100%). GUS activities were normalized by cotransfection with a 35S:LUC construct. Asterisks denote statistically significant differences (P < 0.01, Student’s t test) in comparisons to the LD-VP16 + GD activation control. Data show the mean ± sd of three replicate assays.
(B) Carrot protoplasts were cotransfected with the GUS reporter gene described in (A), LD-VP16, and the indicated effector construct. L9A, E10A, L11A, R12A, and L13A correspond to single Ala substitution mutations in the EAR motif of JAZ8-N (black boxes). The F14A mutation is located C-terminal to the EAR motif and serves as a control. GUS activities are expressed as described in (A). Asterisks denote statistically significant differences (P < 0.01, Student’s t test) in comparisons to the unmodified GD-JAZ8-N construct. Data points show the mean ± sd of three replicate assays. WT, wild type.
Ala-scanning mutagenesis was used to determine whether the LELRL motif of JAZ8 is required for transcriptional repression. Individual Ala substitutions within the LELRL motif resulted in partial or complete loss of repression by GD-JAZ-N in the protoplast transfection assay (Figure 7B). The effect of mutating any one of the three Leu residues was stronger than that of mutating the intervening hydrophilic residues. The Leu11→Ala substitution completely impaired GD-JAZ8-N repression activity, suggesting that this residue is important for recruitment of corepressors. Similar findings were reported for the characterization of EAR motifs in SUPERMAN and IAA17 repressors (Hiratsu et al., 2003; Tiwari et al., 2004).
Mechanism of Repression by JAZ8
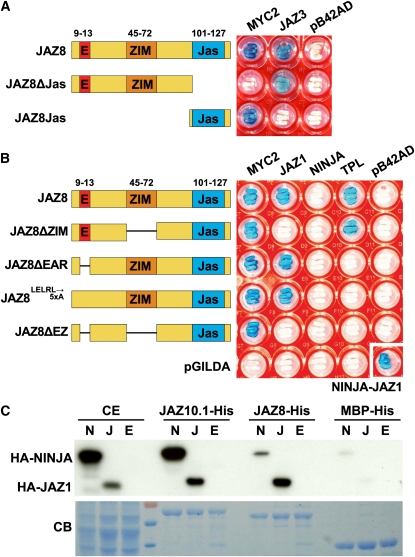
We used the yeast two-hybrid (Y2H) system to investigate how interaction of JAZ8 with other components of the JA signaling module may contribute to the mechanism of JAZ8-mediated repression. We first investigated the interaction of JAZ8 with the bHLH transcription factor MYC2, which plays a major role in controlling JA-dependent root growth inhibition (Lorenzo et al., 2004; Fernández-Calvo et al., 2011). Full-length JAZ8 interacted with MYC2 (Figure 8A), as previously reported (Chini et al., 2009). A JAZ8ΔJas construct lacking the Jas motif did not bind MYC2 but maintained the ability to heterodimerize with JAZs. Moreover, a 36–amino acid C-terminal fragment containing the Jas motif was sufficient for binding of MYC2.
Figure 8.
Protein–Protein Interaction Domains in JAZ8.
(A) Y2H assay of JAZ8 and JAZ8 deletion derivatives with MYC2 and JAZ3. Yeast strains expressing both the bait (JAZ8) and prey (MYC2 or JAZ3) proteins were plated on media containing X-Gal. LacZ-mediated blue-color formation is indicative of protein–protein interaction. JAZ8 proteins were coexpressed with an empty prey vector (pB42AD) as a negative control. Photographic images of yeast cells were taken after 48 h of incubation at 30°C.
(B) Y2H analysis of JAZ8 deletion proteins with MYC2, JAZ1, NINJA, and TPL. Yeast strains expressing both the bait (JAZ8 or JAZ8 deletion) and prey (MYC2, JAZ1, NINJA, or TPL) proteins were tested as described in (A). Empty bait (pGILDA) and prey (pB42AD) vectors were used as negative controls. As a positive control for NINJA interaction, yeast cells were cotransformed with pB42AD-NINJA and pGILDA-JAZ1 (bottom right).
(C) JAZ8 interacts weakly with NINJA in vitro. Purified JAZ-His proteins (fused to MBP) were incubated with crude extract from yeast cells expressing HA-tagged derivatives of NINJA (N), JAZ1 (J), or an empty vector control (E). MBP-His was used as a control for specificity. Purified protein complexes were separated by SDS-PAGE and probed with an anti-HA antibody for the presence of HA-tagged NINJA (HA-NINJA) or JAZ1 (HA-JAZ1). The bottom panel shows a Coomassie blue (CB)–stained gel to visualize the amount of protein loaded. CE, crude yeast extracts that were used as an input control. Protein molecular weight markers were run in the fourth lane from the left.
Consistent with previous Y2H studies (Pauwels et al., 2010; Arabidopsis Interactome Mapping Consortium, 2011), full-length JAZ8 interacted with TPL but not with NINJA (Figure 8B). Deletion of the ZIM domain abolished JAZ8 heterodimerization with JAZ1 and all other JAZs tested (see Supplemental Figure 7 online) but did not affect the interaction with MYC2 or TPL. Removal of the LELRL motif (JAZ8ΔEAR construct), or substitution of this sequence with five Ala residues (JAZ8LELRL→5xA), eliminated the interaction with TPL but did not affect binding to JAZ1 or MYC2 (Figure 8B). A construct (JAZ8ΔEZ) that lacks both the EAR and ZIM domains interacted with MYC2 but not with TPL or JAZ1. These collective results show that the EAR, ZIM, and Jas motifs mediate JAZ8 interaction with TPL, other JAZ proteins, and MYC2, respectively.
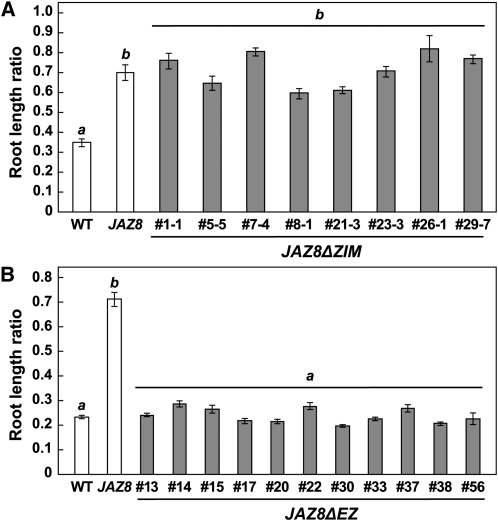
Pauwels et al. (2010) showed that although JAZ8 does not interact with NINJA in the Y2H system, the two proteins do interact in pull-down assays performed with green fluorescent protein–tagged NINJA expressed in planta. We used a yeast-expressed, hemagglutinin (HA)-tagged NINJA derivative to reexamine this question. Whereas JAZ8-His and JAZ10.1-His recovered comparable amounts of HA-JAZ1, the amount of HA-NINJA recovered by JAZ8-His was much less than that associated with JAZ10.1-His (Figure 8C). The amount of HA-NINJA bound by JAZ8-His, however, was consistently greater than that recovered by MBP-His alone. This finding, together with the results of Pauwels et al. (2010), raised the possibility that JAZ8-mediated repression may depend in part on recruitment of TPL via JAZ8’s ZIM domain. To test this hypothesis, we constructed transgenic lines (35S:JAZ8ΔZIM) that overexpress JAZ8ΔZIM. Multiple independent T1 lines showing high transgene expression were selected for use in root growth inhibition assays. The mean root length ratio (0.72 ± 0.01; n = 127) exhibited by seedlings from eight independent homozygous 35S:JAZ8ΔZIM lines was not significantly different from that of 35S:JAZ8 seedlings (0.70 ± 0.04; n = 23) (Figure 9A). As an additional control, we constructed transgenic lines that express the JAZ8ΔEZ construct (Figure 8B) in which both the EAR and ZIM motifs of JAZ8 are deleted. Eleven independent lines that express the 35S:JAZ8ΔEZ transgene to high levels were selected for root growth inhibition assays. The mean root length ratio of seedlings from all lines was not different (P value = 0.62, Student’s t test) from that of wild-type seedlings (Figure 9B), indicating that overexpression of JAZ8ΔEZ does not significantly repress JA signaling. These results show that the ZIM domain is not required for repression of JA-mediated root growth inhibition by ectopically expressed JAZ8.
Figure 9.
The ZIM Domain Is Not Required for JAZ8-Mediated Repression of JA-Induced Root Growth Inhibition.
(A) 35S:JAZ8ΔZIM plants are insensitive to JA-mediated root growth inhibition. Assays were performed with wild-type (WT), 35S:JAZ8 (JAZ8; line #24), and eight independent 35S:JAZ8ΔZIM (JAZ8ΔZIM; gray bars) lines that are homozygous for the transgene. Root length ratios were calculated as described in the legend of Figure 4D. Data show the mean ± se for each genotype (n = 12 to 24 seedlings per genotype). The mean root length ratio of seedlings (n = 127) from all eight 35S:JAZ8ΔZIM lines was not significantly different than that of 35S:JAZ8 seedlings (P value = 0.69, Student’s t test). Means with different italicized letters are significantly different at P < 0.001.
(B) JA-mediated root growth phenotype of 35S:JAZ8ΔEZ lines expressing a JAZ8 derivative lacking both the EAR and ZIM motifs. Assays were performed as described in (A) and included 11 independent 35S:JAZ8ΔEZ T2 lines (JAZ8ΔEZ; gray bars). Data show the mean ± se for each genotype (n = 13 to 22 seedlings per genotype). The mean root length ratio of seedlings (n = 212) from all 11 35S:JAZ8ΔEZ lines was not significantly different than that of wild-type seedlings (P value = 0.62, Student’s t test). Means with different italicized letters are significantly different at P < 0.001.
DISCUSSION
Sequence Diversity in the JAZ Degron Modulates JA Responsiveness
Our results provide new insight into sequence determinants within the Jas motif that promote hormone-dependent interaction of JAZ proteins with COI1. Recent structural studies showed that the JAZ1 degron is located in the N-terminal region of the Jas motif and includes a hexapeptide motif (LPIARR) that seals JA-Ile at the COI1-JAZ1 interface (Sheard et al., 2010). The C-terminal end (ARR) of the motif makes direct contact with JA-Ile, whereas N-terminal residues (LPI) clamp down on the COI1 surface to trap the hormone in place. These structural studies are supported by genetic evidence showing that mutations in the C-terminal basic residues (RR) result in dominant repression of JA responses by JAZ1 (Melotto et al., 2008). Here, we show that the LPIARR sequence is largely conserved in JAZ proteins (including JAZ2, JAZ3, JAZ9, JAZ10.1, and JAZ12) that strongly interact with COI1 in the presence of JA-Ile. This finding is consistent with the rapid JA-mediated turnover of this group of JAZs (Chini et al., 2007; Thines et al., 2007; Chung and Howe, 2009; Grunewald et al., 2009; Pauwels et al., 2010).
The importance of the LPIARR motif as a JAZ degradation signal is highlighted by the finding that JAZ8, which lacks this motif, does not readily associate with COI1 in the presence of JA-Ile. The weak COI1–JAZ8 interaction was associated with increased stability of JAZ8 in JA-treated cells and the ability of JAZ8 to repress JA responses in 35S:JAZ8 plants. Domain swap and site-directed mutagenesis experiments demonstrated that these unique features of JAZ8 are attributed to sequence variation in the degron region; replacement of PKASM in the Jas motif of JAZ8 with LPIAR conferred JA-Ile–dependent interaction with COI1 and decreased the stability of JAZ8. The PKASM→LPIAR substitution also suppressed the JA-insensitive phenotype of 35S:JAZ8 plants, thus establishing a direct link between sequence variation in the ligand-contacting region of the degron and JA signal output. These findings show that the LPIARR motif functions in vivo as a critical part of the JAZ degron, thus confirming and extending our previous structural studies (Sheard et al., 2010).
The central (SLX2FX2KRX2R) and C-terminal (X5PY) regions of the Jas motif are highly conserved among all JAZs, whereas the N-terminal amino acids corresponding to the degron loop are much more variable (Figure 6A) (Chung et al., 2009; Sheard et al., 2010). Our work on JAZ8 indicates that sequence variation in the N terminus reflects the existence of functionally distinct JAZ subgroups that differ in their stability. These subgroups include the canonical LPIAR(R/K)-containing JAZs (e.g., JAZ1) that interact robustly with COI1 in the presence of JA-Ile and X3SMK-containing JAZs, such as JAZ8, that do not strongly associate with COI1 under these conditions. The degron loop region of JAZ5 and JAZ6 (Figure 4A) is unusual in that it contains the canonical C-terminal residues (IARR) that contact JA-Ile (Sheard et al., 2010) but lacks the N-terminal residues (LP) that clamp the hormone in the binding pocket. Interestingly, this sequence variation correlates with the reduced capacity of JAZ6 to recover COI1 in pull-down assays (Figure 1A). Differential association of JAZ isoforms with COI1, mediated by sequence variation in the degron, may enable plants to perceive and respond appropriately to a wide dynamic range of JA-Ile. Our results support a scenario in which the intracellular level of JA-Ile dictates which JAZs in a given cell associate with COI1. It is likely, for example, that some JAZs are recruited to COI1 in response to low JA-Ile concentrations, whereas other JAZs associate with COI1 only in the presence of high JA-Ile levels. Sequence variation in the auxin degron has also been shown to contribute to the diversification of Aux/IAA proteins (Dreher et al., 2006; Sato and Yamamoto, 2008), further highlighting the conserved nature of the COI1-JAZ and TIR1-Aux/IAA receptor systems (Katsir et al., 2008a).
The mechanisms by which JAZ8 and other stable JAZs are removed from cells remain unknown. One hypothesis is that stable JAZs are eliminated via COI1-independent proteolytic pathways, perhaps involving other F-box proteins. However, the ability of coronatine to destabilize JAZ8 in vivo, together with the inhibitory effect of MG132 on this process, suggests that JAZ8 can be degraded by a pathway involving COI1 and the 26S proteasome. Given that coronatine functions as a potent agonist of the JA-Ile receptor, it is possible that JAZ8 is slowly degraded by a COI1-dependent route in the presence of very high intracellular levels of JA-Ile. Our results also raise the possibility that recruitment of JAZ8 to COI1 is mediated by a small molecule other than (3R,7S)-JA-Ile. The ability of JAZ8 to associate with COI1 in the presence of coronatine demonstrates that JAZ8 is capable of interacting with COI1 in a ligand-dependent manner. Although we cannot exclude the possibility that this interaction is mediated by a cryptic COI1 binding site located outside the Jas motif, the most straightforward interpretation is that the noncanonical loop region of JAZ8 is a weak substrate for coronatine-triggered COI1 association. The putative degron loop of JAZ8 contains a KK motif (Figure 4A) that may partially fulfill the function of the critical dibasic motif (R205R206) in JAZ1, which interacts directly with the carboxyl group of the ligand at the bottom of the binding pocket (Sheard et al., 2010). Among the small molecules that could potentially promote COI1-JAZ8 association are metabolic precursors or derivatives of JA-Ile. Several studies have provided evidence that the JA-Ile precursors 12-oxo-phytodienoic acid and jasmonic acid elicit JA-related responses without their prior conversion to JA-Ile (Hopke et al., 1994; Blechert et al., 1999; Miersch et al., 1999; Stintzi et al., 2001; Ribot et al., 2008; Wang et al., 2008). To date, however, the only naturally occurring JA derivatives known to promote COI1-JAZ binding are JA-Ile, structurally related JA–amino acid conjugates (e.g., JA-Leu), and 12-hydroxy-JA-Ile (Thines et al., 2007; Katsir et al., 2008b; Melotto et al., 2008; Chini et al., 2009; Koo et al., 2011).
The expression pattern of JAZ8 in wild-type plants provides insight into the physiological role of JAZ8 as a repressor of JA responses. The most striking aspect of JAZ8 expression is its rapid and strong induction in response to exogenous JA and other stress-related cues that activate the pathway (Figure 2B; Thines et al., 2007; Chini et al., 2007; Chung et al., 2008). In mature Arabidopsis leaves, for example, JAZ8 and its paralog, JAZ7, are highly expressed in response to mechanical wounding and infection with coronatine-producing strains of Pseudomonas syringae (Chung et al., 2008; Koo et al., 2009; Demianski et al., 2012). The enhanced expression of JAZ8 in wounded leaves of a cyp94b3 mutant that hyperaccumulates JA-Ile further suggests that expression of JAZ8 is promoted by high endogenous levels of JA-Ile (Koo et al., 2011). Given that the repressive activity of JAZ8 is dominant and persists in the presence of JA-Ile, stress-induced expression of JAZ8 is expected to result in attenuation of JA responses, consistent with the JA-insensitive phenotype exhibited by 35S:JAZ8 plants. We therefore propose that JAZ8, together with alternative splice variants of JAZ10 (Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010), is a component of a JA-triggered negative feedback loop to prevent runaway activation of JA responses that could lead to inhibition of plant growth and reduced fitness (Zhang and Turner, 2008).
Repression of JA Responses by EAR Motif–Containing JAZ Proteins
The recent identification of NINJA provided an important advance in understanding how JAZ proteins repress JA-dependent transcriptional responses (Pauwels et al., 2010). NINJA negatively regulates the expression of JA response genes by bridging the ZIM domain of target JAZs to members of the TPL family of corepressors. As is the case for other TPL-interacting proteins (Szemenyei et al., 2008; Krogan and Long, 2009), NINJA contains an EAR motif that functions to recruit TPL. Our results indicate that JAZ8 employs a distinct and more direct mechanism to repress JA-dependent transcription; through its N-terminal EAR and C-terminal Jas motifs, we propose that JAZ8 provides a direct link between TPL and cognate transcription factors. Three independent lines of evidence demonstrate a key role for the EAR motif in JAZ8-mediated repression. First, we show that JAZ8 interacts with TPL in yeast and that a canonical LxLxL-type EAR motif at the N terminus of JAZ8 is required for this interaction. These findings are in agreement with a recent large-scale Y2H study in which TPL was shown to interact with JAZ8 (Arabidopsis Interactome Mapping Consortium, 2011). Second, we demonstrate that the EAR motif is required for JAZ8-mediated repression of JA-dependent root growth inhibition in 35S:JAZ8 plants. Finally, we show that JAZ8 represses VP16-mediated transcriptional activation in a protoplast transfection assay and, moreover, that the EAR motif is important for the mechanism of repression.
A key role for the EAR motif in JAZ8-mediated repression is supported by our finding that 35S:JAZ8 and 35S:JAZ8ΔZIM lines exhibit a similar level of insensitivity to JA. This finding shows that the ZIM domain is not strictly required for repression by ectopically expressed JAZ8, thus suggesting that JAZ8 action under these conditions depends neither on NINJA nor on interaction with other JAZs. Such a ZIM/TIFY-independent mechanism of repression differs from that of JAZ10.4, whose function as a dominant repressor is blocked by point mutations within the ZIM domain (i.e., TIFY motif) that disrupt JAZ10.4 homo- and heterodimerization (Chung and Howe, 2009). In this context, JAZ8ΔZIM appears to define the minimal domain architecture of a JAZ repressor, which in this case consists only of an EAR repression domain and a transcription factor binding Jas motif. Given the ability of JAZ8 to interact weakly with NINJA in pull-down assays (Pauwels et al., 2010; this study), we cannot exclude the hypothesis that JAZ8 is capable of repressing gene expression through both a NINJA-independent pathway involving direct binding of TPL/TPR to JAZ8’s EAR motif as well as a NINJA-dependent pathway in which the ZIM domain of JAZ8 recruits TPL/TPR via NINJA. However, our collective results favor the hypothesis that JAZ8 represses gene expression mainly via a direct, NINJA-independent mechanism.
The bHLH transcription factor MYC2 plays a major role in JA-mediated root growth inhibition (Fernández-Calvo et al., 2011). We found that the JA-insensitive root growth phenotype of JAZ8-overexpressing plants is stronger than that of a myc2 mutant (jin1-7). This finding, together with the fact that JAZ8 physically associates with MYC2 (Cheng et al., 2011; Fernández-Calvo et al., 2011; this study), suggests that JAZ8 targets MYC2 for repression. JAZ8 also interacts with the MYC2-related proteins MYC3 and MYC4, which serve a minor but significant role in activating JA responses in the root (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011), as well as MYB-type transcription factors (MYB21 and MYB24) implicated in JA-regulated male fertility (Song et al., 2011). In contrast with the male-sterile phenotype of transgenic lines that overexpress various truncated JAZ proteins (Thines et al., 2007; Melotto et al., 2008; Chung and Howe, 2009), we did not observe reproductive defects associated with overexpression of JAZ8. Identification of transcription factors and physiological processes that are regulated by endogenous JAZ8 is an important question for future studies.
Genome-wide sequence surveys showed that JAZ5, JAZ6, and JAZ7 also contain predicted EAR motifs (Kagale et al., 2010). Moreover, proteome-wide mapping of the Arabidopsis interactome identified JAZ5 and JAZ6 as TPL binding proteins in yeast (Arabidopsis Interactome Mapping Consortium, 2011; Causier et al., 2012). It thus seems likely that repression of JA-responsive genes through direct JAZ-TPL coupling is not unique to JAZ8. Several reported features of JAZ7 are reminiscent of JAZ8, including the N-terminal location of the EAR motif and the ability to interact with MYC-related transcription factors but not NINJA (Pauwels et al., 2010; Cheng et al., 2011; Fernández-Calvo et al., 2011). JAZ5 and JAZ6 both interact with NINJA but are distinct in that they each contain two predicted EAR motifs in the central and C-terminal regions of the protein (Kagale et al., 2010; Pauwels et al., 2010; Fernández-Calvo et al., 2011). These observations support our general conclusion that different subgroups of JAZ employ distinct domain architectures to recruit corepressors to cognate transcription factors. Future work aimed at delineating the mechanisms and evolutionary origins of NINJA-dependent and NINJA-independent JAZ repression pathways should help to unravel the complexities of JA biology.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 was used as the wild-type parent for all experiments. Plants were grown in soil under controlled conditions and were transformed with Agrobacterium tumefaciens as previously described (Chung et al., 2008; Chung and Howe, 2009). 35S:JAZ10.1 and 35S:JAZ10.1-YFP transgenic lines were described previously (Chung and Howe, 2009). The jin1-7 mutant (SALK_040500) was previously described (Lorenzo et al., 2004). Lines that express transgenes to high levels were identified as follows. Seedlings (T1 generation) of transformed lines were screened on Murashige and Skoog (MS) agar medium containing Suc (0.8%), kanamycin (50 μg/mL), and vancomycin (100 μg/mL). For each construct, 32 independent T1 plants were transferred to soil for evaluation of transgene expression level by RNA gel blot analysis. RNA isolated from untreated leaf tissue was hybridized with a JAZ8 cDNA as a probe as previously described (Chung et al., 2008). Selected high-expressing lines (typically five per construct) were further propagated for identification of homozygous T3 lines. Lines expressing JAZ8-YFP fusion proteins were screened at the seedling stage by fluorescence microscopy (Axio Scope; Carl Zeiss).
Plant Treatments
The effect of exogenous JA on root growth inhibition was determined as described previously (Chung and Howe, 2009), with minor modifications. Surface-sterilized and stratified seeds were sown on square Petri plates containing MS agar medium (0.8% agar; Caisson Labs) supplemented with 0.8% Suc (w/v) and the indicated concentration of MeJA (Sigma-Aldrich). Wild-type and transgenic lines were grown on the same plate to control for plate-to-plate variation. Petri plates were incubated vertically in a growth chamber maintained at 21°C and continuous light, unless otherwise indicated. Following 8 d of growth, the length of primary roots was measured using Image J software (http://rsbweb.nih.gov/ij/index.html). Root length ratios (percentage of inhibition by MeJA) were calculated by dividing the average the root length of seedlings grown on MeJA-containing medium by the average root length of seedlings of the same genotype grown in the absence of MeJA.
Spodoptera exigua eggs were obtained from Benzon Research and hatched at 30°C. Newly hatched larvae were transferred to fully expanded rosette leaves of 5-week-old wild-type and 35S:JAZ8 transgenic (line 24) plants. Eight larvae were reared on each of 12 plants per genotype. Insect-challenged and unchallenged control plants were maintained under standard growth conditions (Chung et al., 2008). Larval weights were measured at various times after challenge, and weighed larvae were returned to their plant of origin. The experiment was independently repeated four times with similar results. RNA gel blot analysis of RNA isolated from liquid-grown seedlings was performed as previously described (Chung et al., 2008).
Transgene Constructs and Site-Directed Mutagenesis
A complete list of constructs and primers used to generate transgenic lines is provided in Supplemental Table 1 online. Three PCR reactions were used to generate constructs encoding the JAZ8-JAZ10 chimeric proteins. In the case of the JAZ8-Jas10 chimera, one reaction used the JAZ8 cDNA as a template with primer set JAZ8-XbaI-FP and JAZ8-Jas10-RP to amplify a portion (amino acids 1 to 100) of the JAZ8 coding sequence that lacks the Jas motif. A second PCR reaction used the JAZ10 cDNA as a template with a primer set (JAZ8-Jas10-FP and JAZ10-KpnI-RP) to amplify the Jas motif–containing region of JAZ10 (amino acids 166 to 205). The PCR products from these two reactions contain complementary sequences (built into the primers) that allow the products to anneal as template in a third PCR reaction with primer set JAZ8-XbaI-FP and JAZ10-KpnI-RP. The resulting chimeric product was cloned into pGEM-T Easy (Promega) for sequencing and subcloning into the final destination vector. A similar approach was used to construct the JAZ10-Jas8 chimera using the primer sets listed in Supplemental Table 1 online.
The JAZ8ΔZIM construct was generated using three PCR reactions. The first PCR reaction used JAZ8 cDNA as a template with a primer set (JAZ8-NcoI-FP and JAZ8-deltaZIM-RP) to amplify the N-terminal region (amino acids 1 to 44) of JAZ8. A second PCR was used to amplify the C-terminal region of JAZ8 (amino acids 72 to 131) with primers JAZ8-ΔZIM-FP and JAZ8-XhoI-RP. The two PCR products contain complementary sequences built into the primers (JAZ8-ΔZIM-FP and JAZ8-ΔZIM-RP). In a third PCR reaction (primers JAZ8-NcoI-FP and JAZ8-XhoI-RP), the two PCR products were annealed as a template to generate JAZ8ΔZIM (i.e., JAZ8 lacking amino acids 45 to 71). The resulting product was cloned into pGEM-T Easy for sequencing and subcloning into Y2H and overexpression vectors. The JAZ8ΔEAR construct was generated in two PCR reactions. The first reaction used the JAZ8 cDNA as a template with a primer set (JAZ8-deltaEAR-FP and JAZ8-pENTR-RP) that amplifies a portion of the JAZ8 coding region lacking the EAR motif. This PCR product was then used as template in the second PCR (primers JAZ8-pENTR-FP and JAZ8-pENTR-RP) to generate the JAZ8ΔEAR product, which was subsequently cloned into pENTR/D-TOPO (Invitrogen) and pGEM-T Easy for sequencing and subcloning.
Site-directed mutagenesis of the Jas and EAR motif of JAZ8 was performed with Pfu Turbo DNA polymerase (Stratagene) as previously described (Chung and Howe, 2009). PCR reactions were performed with JAZ8 cDNA in the vector pGEM-T Easy. PCR-amplified products were treated with restriction enzyme DpnI to remove the parental plasmid. The presence of the desired mutation was verified by DNA sequencing.
In Vivo Degradation of JAZ-YFP Fusion Proteins
Transgenic seedlings expressing JAZ-YFP fusion proteins were grown on MS plates (containing kanamycin) and then transferred to water (0.6 mL) on 48-well microtiter plates. Seedlings were incubated in the presence or absence of MeJA or coronatine at room temperature on an orbital shaker (70 rpm). YFP fluorescence was analyzed on an Olympus Fluoview confocal microscope with imaging software provided by the manufacturer. In some experiments, seedlings were pretreated with water or 100 μM MG132 (Sigma-Aldrich) for ~75 min prior to the addition of MeJA. Images shown in a single panel were taken at the same exposure time and microscope parameters, unless otherwise indicated in the figure legend.
Protein–Protein Interaction Assays
JAZ cDNAs were cloned into pRMG-nMAL to produce plasmids that encode MBP and hexa-histidine (His6)–tagged fusion proteins, referred to as JAZ-His (Thines et al., 2007). Primers used for amplifying JAZ cDNAs are listed in Supplemental Table 1 online. JAZ-His fusion proteins were expressed in Escherichia coli and purified by Ni-affinity chromatography as previously described (Katsir et al., 2008b; Chung et al., 2010). Leaf extracts prepared from a transgenic line of Arabidopsis that expresses Myc-tagged Arabidopsis COI1 were used as the source of COI1 (Melotto et al., 2008; Chung et al., 2010). In vitro COI1-JAZ pull-down assays were performed with chemically synthesized (3R,7S)-JA-Ile or coronatine as previously described (Chung et al., 2010). Purified JAZ-COI1 complexes were separated by SDS-PAGE on a 10% acrylamide gel, transferred to polyvinylidene fluoride membrane, and probed with an anti-c-Myc antibody (Roche). In vitro JAZ8-NINJA and JAZ8-JAZ1 interaction assays were performed with purified JAZ-His protein (25 μg) and crude extract (600 μg) from yeast cells expressing N-terminal HA-tagged derivatives of NINJA or JAZ1, which were cloned into the yeast expression vector pB42AD (Clontech) using primers listed in Supplemental Table 1 online. JAZ-His complexes were recovered by Ni-affinity chromatography, and protein–protein interactions were detected with an anti-HA antibody (Covance), as described above.
Y2H assays were performed with the Matchmaker LexA system (Clontech) as previously described (Melotto et al., 2008; Chung and Howe, 2009). JAZ8 and JAZ8-deletion constructs were subcloned into the pGILDA bait vector to generate translational fusions with the LexA DNA binding domain. Full-length cDNAs encoding MYC2, other JAZs, NINJA, and TPL were subcloned into the pB42AD prey vector to generate fusions with B42 activation domain (AD). Bait and prey vectors were cotransformed into yeast (Saccharomyces cerevisiae) strain EGY48 using the frozen-EZ yeast transformation II kit (Zymo Research). Transformants were selected for colorimetric detection (β-galactosidase) of protein–protein interaction as described by Chung and Howe (2009). Photographic images of Y2H plates were taken after 48 h of incubation at 30°C.
Coronatine Binding Assays
Full-length Arabidopsis COI1 and ASK1 were coexpressed as a glutathione S-transferase fusion protein and an untagged protein, respectively, in Hi5 suspension insect cells. The COI1-ASK1 complex was isolated from the soluble cell lysate by glutathione affinity chromatography. Full-length JAZ substrate proteins were expressed as 6xHis-fusion proteins in E. coli and purified on nickel-nitrilotriacetic acid resin with subsequent dialysis into 20 mM Tris-HCl, pH 8.0, 200 mM NaCl, and 10% glycerol. Radioligand binding was assayed with purified COI1-ASK1 complex (2 μg) and JAZ proteins at a 1:3 molar ratio. Reactions were prepared in 100 μL final volume and in a binding buffer containing 20 mM Tris-HCl, 200 mM NaCl, and 10% glycerol. Saturation binding experiments were conducted with serial dilutions of 3H-coronatine in binding buffer. Nonspecific binding was determined in the presence of 300 μM coronatine (Sigma-Aldrich). Specific binding was calculated by subtracting nonspecific binding from total binding. Following incubation with mixing at 4°C, all samples were collected with a cell harvester (Brandel) on polyethyleneimine-treated (Sigma-Aldrich) filters. Samples were incubated in liquid scintillation fluid for >1 h before counting with a Packard Tri-Carb 2200 CA liquid scintillation analyzer (Packard Instrument Company). Saturation binding experiments were analyzed by nonlinear regression using GraphPad Prism version 5.00 for MacOSX. 3H-coronatine was synthesized commercially as previously described (Katsir et al., 2008b).
Carrot Protoplasts Transactivation Assay
The effector constructs 35S:GD (GD: Gal4 DNA binding domain), 35S:GD-IAA17[I] (IAA17/AXR3 domain I, amino acids 1 to 29), and 35S:LD-VP16 (LD: LexA DNA binding domain; VP16: Herpes simplex virus VP16 activation domain) were all driven by the CaMV 35S promoter and have been described previously (Tiwari et al., 2004). The 35S:GD vector was converted into a Gateway destination version (pGD-RfA) by inserting the reading frame A cassette (RfA; Invitrogen) downstream of the sequence encoding the Gal4 DNA binding domain. JAZ8 (amino acids 1 to 131) and JAZ8-N (N terminus; amino acids 1 to 44) coding regions were amplified by RT-PCR using RNA from Arabidopsis seedlings as template and the primer pairs listed in Supplemental Table 1 online. cDNAs were cloned into pDONR/Zeo via Gateway BP cloning reactions (Invitrogen). GD-JAZ8 and GD-JAZ8-N effector constructs were generated by recombination between pDNOR clones described above and pGD-RfA via Gateway LR cloning reactions. The reporter constructs 35S:LexA(2x)-Gal4(2x):GUS, which is driven by CaMV 35S-46 minimal promoter containing LexA and Gal4 DNA binding sites, and 35S:LUC have been described previously (Tiwari et al., 2004). To generate JAZ8 effector constructs encoding mutated proteins (L9A, E10A, L11A, R12A, L13A, and F14A), the JAZ8-N coding region was mutagenized with a Quickchange II site-directed mutagenesis kit (Stratagene) and a complementary pair of primers (see Supplemental Table 1 online) containing the relevant mutation. All mutations were confirmed by DNA sequencing. Reporter and effector plasmids were isolated from the E. coli strain ER2925 (New England Biolabs) using Wizard midiprep DNA purification kit (Promega).
Isolation of protoplasts from carrot (Daucus carota) suspension culture cells, transfections, luciferase (LUC), and GUS assays have been described (Niu et al., 2011). GUS activities for each measurement were normalized by cotransfection with a 35S:LUC reporter gene (Liu et al., 1994). GUS activity measurements were performed with 4-methylumbelliferyl-β-glucuronide (Sigma-Aldrich) as a substrate and excitation and emission wavelengths of 365 and 455 nm, respectively. LUC activity was determined with a Luciferase Assay System (Promega) using an emission wavelength of 550 nm and a photomultiplier gain of 775 V. Fluorescence and luminescence were measured with a 96-well luminescence spectrometer LS-50B (Perkin-Elmer). Three transfections were performed for each construct and the measurements were averaged. All transfections were repeated two or three times using independent protoplast preparations. Results from representative experiments are reported. Significant differences were determined by Student’s t test using Graphpad Prism version 4.00 for Windows software (GraphPad Software). Differences between mean values are reported as significant at P < 0.01.
Phylogenetic Analysis
JAZ8-related sequences were identified using BLAST (BLASTP and TBLASTN) and the Phytozome database (http://www.phytozome.net/) with JAZ8 as the query sequence. Of the resulting 43 sequences identified from 18 monocot and dicot species, 33 proteins contained a complete ZIM domain and Jas motif and were used to generate the Weblogo (Crooks et al., 2004) shown in Figure 6. Manual annotation identified 30 JAZ8-like proteins that contain a canonical an EAR motif at the N terminus, and these sequences were used to generate the Weblogo shown in Figure 6. A similar approach was used to identify 94 JAZ10-like proteins for construction of the Weblogo. Phylogenetic trees were generated with the neighbor-joining method using MEGA54 software (http://www.megasoftware.net/).
Accession Numbers
Arabidopsis Genome Initiative numbers described in this article are listed below: ACT8 (At1g49240), COI1 (At2g39940), IAA17 (At1g04250), JAZ1 (At1g19180), JAZ2 (At1g74950), JAZ3 (At3g17880), JAZ4 (At1g48500), JAZ5 (At1g17380), JAZ6 (At1g72450), JAZ7 (At2g34600), JAZ8 (At1g30135), JAZ9 (At1g70700), JAZ10.1 (At5g13220.1), JAZ11 (At3g43440), JAZ12 (At5g20900), MYC2 (At1g32640), NINJA (At4g28910), TPL (At1g15750), and VSP2 (At5g24770).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Tree Constructed from the Jas Motif of Arabidopsis JAZ Proteins.
Supplemental Figure 2. JA-Mediated Root Growth Inhibition in Independent 35S:JAZ8 Lines.
Supplemental Figure 3. 35S:JAZ8 Plants Are Compromised in Resistance to Herbivory by S. exigua Larvae.
Supplemental Figure 4. JA-Mediated Root Growth Inhibition in 35S:JAZ8-YFP Lines.
Supplemental Figure 5. Substitution of PKASM to LPIAR does Not Affect the Ability of JAZ8 to Interact with MYC2, JAZ1, or TPL.
Supplemental Figure 6. Nuclear Localization of JAZ8-YFP Fusion Proteins.
Supplemental Figure 7. Yeast Two-hybrid Analysis of JAZ8 and JAZ8ΔZIM Interactions with Other Arabidopsis JAZ proteins.
Supplemental Table 1. List of Oligonucleotide Primers Used in This Study.
Supplemental Table 2. List of Plant Species and Genes Used for Construction of JAZ Consensus Sequences Shown in Figure 6.
Supplemental Data Set 1. Sequence Alignment for Phylogenetic Tree Shown in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Chad Seippel and Li Deng (Michigan State University) for technical assistance throughout the project. We thank Kyaw Aung and Jiangping Hu (Michigan State University) for providing Gateway-compatible Y2H vectors and Rob Larkin (Michigan State University) for providing seed for the jin1-7 mutant. We also thank Tom Guilfoyle and Gretchen Hagen (University of Missouri) for carrot cell cultures and plasmid constructs used in transfection assays. Shiv Tiwari (Mendel Biotechnology) provided assistance with protoplasts isolation and transfection, and Yajie Niu (Washington State University) provided the 35S:GD vector. This research was supported by the National Institutes of Health (Grant T32 GM07270 to L.B.S., R01 CA107134 to N.Z., and R01 GM57795 to G.A.H.), the National Science Foundation (0929100 to N.Z.), the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (Grant DE-FG02-99ER20323 to J.B. and DE-FG02-91ER20021 to G.A.H.), the Agricultural Research Center at Washington State University, and the Howard Hughes Medical Institute (N.Z.). We dedicate this work to the memory of Laura B. Sheard, who was an extraordinarily talented and energetic young scientist. As a cherished colleague, she made key discoveries that paved the way for this study. We unfortunately lost her to a car accident while this manuscript was under review.
AUTHOR CONTRIBUTIONS
C.S. and G.A.H. designed the project. C.S., P.F., C.L.D., T.F.C., L.B.S., J.E.M., and L.K. performed experiments, and all authors contributed to analysis and interpretation of results. C.S. and G.A.H. wrote the article with contributions and edits from P.F., L.B.S., N.Z., and J.B.
References
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Meng Y., Huang D., Qi Y., Chen M. (2011). Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98: 128–136 [DOI] [PubMed] [Google Scholar]
- Ballaré C.L. (2011). Jasmonate-induced defenses: A tale of intelligence, collaborators and rascals. Trends Plant Sci. 16: 249–257 [DOI] [PubMed] [Google Scholar]
- Blechert S., Bockelmann C., Fusslein M., Von Schrader T., Stelmach B., Niesel U., Weiler E.W. (1999). Structure-activity analyses reveal the existence of two separate groups of active octadecanoids in elicitation of the tendril-coiling response of Bryonia dioica Jacq. Planta 207: 470–479 [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438 [DOI] [PMC free article] [PubMed]
- Cheng Z., Sun L., Qi T., Zhang B., Peng W., Liu Y., Xie D. (2011). The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4: 279–288 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Cooke T.F., Depew C.L., Patel L.C., Ogawa N., Kobayashi Y., Howe G.A. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Koo A.J.K., Gao X., Jayanty S., Thines B., Jones A.D., Howe G.A. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Niu Y., Browse J., Howe G.A. (2009). Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry 70: 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demianski A.J., Chung K.M., Kunkel B.N. (2012). Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. 13: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K.A., Brown J., Saw R.E., Callis J. (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Grunewald W., Vanholme B., Pauwels L., Plovie E., Inzé D., Gheysen G., Goossens A. (2009). Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 10: 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hopke J., Donath J., Blechert S., Boland W. (1994). Herbivore-induced volatiles: The emission of acyclic homoterpenes from leaves of Phaseolus lunatus and Zea mays can be triggered by a β-glucosidase and jasmonic acid. FEBS Lett. 352: 146–150 [DOI] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Howe G.A. (2010). Ubiquitin ligase-coupled receptors extend their reach to jasmonate. Plant Physiol. 154: 471–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Kagale S., Links M.G., Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Rozwadowski K. (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Chung H.S., Koo A.J., Howe G.A. (2008a). Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008b). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2011). The interplay between light and jasmonate signalling during defence and development. J. Exp. Bot. 62: 4087–4100 [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17: 22–31 [DOI] [PubMed] [Google Scholar]
- Koo A.J., Cooke T.F., Howe G.A. (2011). Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. USA 108: 9298–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A.J.K., Gao X.L., Jones A.D., Howe G.A. (2009). A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59: 974–986 [DOI] [PubMed] [Google Scholar]
- Krogan N.T., Long J.A. (2009). Why so repressed? Turning off transcription during plant growth and development. Curr. Opin. Plant Biol. 12: 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.B., Ulmasov T., Shi X., Hagen G., Guilfoyle T.J. (1994). Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M., Mecey C., Niu Y., Chung H.S., Katsir L., Yao J., Zeng W., Thines B., Staswick P., Browse J., Howe G.A., He S.Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch O., Kramell R., Parthier B., Wasternack C. (1999). Structure-activity relations of substituted, deleted or stereospecifically altered jasmonic acid in gene expression of barley leaves. Phytochemistry 50: 353–361 [Google Scholar]
- Moreno J.E., Tao Y., Chory J., Ballaré C.L. (2009). Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc. Natl. Acad. Sci. USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y.J., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Goossens A. (2011). The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M.J., Leon-Reyes A., Van der Ent S., Van Wees S.C.M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot C., Zimmerli C., Farmer E.E., Reymond P., Poirier Y. (2008). Induction of the Arabidopsis PHO1;H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1-dependent pathway. Plant Physiol. 147: 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Okamoto H., Patrick E., Harris S.R., Wasternack C., Brearley C., Turner J.G. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Yamamoto K.T. (2008). Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant. 133: 397–405 [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Weber H., Reymond P., Browse J., Farmer E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen G., Guilfoyle T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme B., Grunewald W., Bateman A., Kohchi T., Gheysen G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12: 239–244 [DOI] [PubMed] [Google Scholar]
- Wang L., Allmann S., Wu J., Baldwin I.T. (2008). Comparisons of LIPOXYGENASE3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol. 146: 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yan J.B., Zhang C., Gu M., Bai Z.Y., Zhang W.G., Qi T.C., Cheng Z.W., Peng W., Luo H.B., Nan F.J., Wang Z., Xie D.X. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.