This work demonstrates that plant microsporocytes have specific meiosis initiation machinery by characterizing the rice CC-type glutaredoxin MICROSPORELESS1 (MIL1). The mil1 mutant shows defects both in the meiotic entry of sporogenous cell progenies and in the differentiation of surrounding somatic cell layers.
Abstract
The switch from mitosis to meiosis is one of the most pivotal events in eukaryotes undergoing sexual reproduction. However, the mechanisms orchestrating meiosis initiation remain elusive, particularly in plants. Flowering plants are heterosporous, with male and female spore genesis adopting different developmental courses. We show here that plant pollen mother cells contain a specific meiosis initiation machinery through characterization of a rice (Oryza sativa) gene, MICROSPORELESS1 (MIL1). The mil1 mutant does not produce microspores in anthers but has the normal female fertility. Detailed molecular and cytological investigations demonstrate that mil1 anthers are defective in the meiotic entry of sporogenous cell progenies and in the differentiation of surrounding somatic cell layers, resulting in locules filled with somatic cells instead of microspores. Furthermore, analysis of mil1 msp1 double mutants reveals that due to the absence of MIL1, the cells in their anther locule center do not activate meiotic cell cycle either, generating a similar anther phenotype to mil1. MIL1 encodes a plant-specific CC-type glutaredoxin, which could interact with TGA transcription factors. These results suggest meiotic entry in microsporocytes is directed by an anther-specific mechanism, which requires MIL1 activity, and redox regulation might play important roles in this process.
INTRODUCTION
Sexual reproduction is the predominant mode of reproduction in nature. Successful completion of sexual reproduction hinges on activating the meiotic cell cycle to form haploid gametes at appropriate time and places. Commitment to the meiotic cell cycle in premeiotic cells involves an exquisitely choreographed signal cascade, which varies among species (Pawlowski et al., 2007). In fission yeast (Schizosaccharomyces pombe) and budding yeast (Saccharomyces cerevisiae), though the switch from mitotic cell cycle to meiosis in both species is subject to the nutritional control, they hold significantly different nutritional signals and meiosis-triggering signaling networks. In budding yeast, the lack of at least one essential growth nutrient, the absence of Glc, and the presence of a nonfermentable carbon source in its nutritional environment combined trigger the meiotic cycle; while in fission yeast, the environmental signals required for the transition to meiosis are nitrogen starvation and cellular stress (Honigberg and Purnapatre, 2003; Harigaya and Yamamoto, 2007). In multicellular eukaryotes, spore mother cells are surrounded by somatic nursing cells; thus, meiosis initiation must integrate developmental cues that might be species distinctive. Meiosis in mouse (Mus musculus) is induced by retinoic acid, a derivative of vitamin A, and there are different timing and signal regulations of meiosis entry in male and female germ cells (Bowles et al., 2006; Koubova et al., 2006; Wolgemuth, 2006; Bowles and Koopman, 2007).
In most animals, the germ line cells are predetermined during early embryonic development and remain as a distinct population throughout their lives. On the contrary, flowering plants have their cells programmed to undergo meiosis established from somatic cells much later during development (Walbot and Evans, 2003). The establishment of reproductive cell fate in plants involves exquisitely timed developmental events, including cell division, cell-type differentiation, as well as cell–cell communication, suggesting that plants possess a unique way to recruit cells into the meiotic pathway, in which they have to response to both environmental and developmental signals (Bhatt et al., 2001). Despite the widespread sexual reproduction in angiosperms, and important applications that can be derived from an enhanced understanding of this process, very few genes have been reported to be involved in meiosis initiation in plants. Maize (Zea mays) has played a pioneering role in this field with the ameiotic1 (am1) mutant characterized decades ago (Staiger and Cande, 1992; Golubovskaya et al., 1993, 1997; Pawlowski et al., 2009). In am1, meiosis in the microsporocyte is blocked and switched to mitosis, and the responsible gene has been manifested lately to be likely required for the initiation of meiosis. In addition, AM1 is also required for the progression through the early stages of meiotic prophase I in maize, as it regulates the leptotene–zygotene transition. The homolog of AM1 in Arabidopsis thaliana, SWITCH1 (SWI1), shares functions on the initiation of meiosis with AM1. The female meiocytes of swi1 divide in a mitosis-like pattern (Siddiqi et al., 2000; Mercier et al., 2001, 2003; Agashe et al., 2002; Ravi et al., 2008). However, its rice (Oryza sativa) homolog has not been manifested to be involved in meiosis entry. Microsporocytes in the rice am1 mutant enter meiosis successfully, although they would stall at leptotene. Rice AM1 shares functions with maize AM1 in the transition through a leptotene-zygotene checkpoint (Che et al., 2011). The functional divergences of these three homologous genes point to the diversity of the meiosis initiation mechanisms in the plant kingdom. In addition, a recent report shows that the rice MEL2 gene plays important roles in meiotic entry. It is essential for premeiotic G1/S phase transition of the male and female sporocytes as well as synchrony of male meiosis (Nonomura et al., 2011).
Angiosperms are heterosporous plants in which two kinds of spore mother cells (microsporocytes in the anther and megasporocytes in the ovule) are developed, and they employ distinctive developmental programs between micro- and megaspore genesis. In the male reproductive organ anthers, the microsporocytes are usually enveloped by four layers of somatic cells (Wilson and Zhang, 2009; Zhang and Wilson, 2009). The reproductive cell progenitors (sporogenous cells) originate from pluripotent precursors, archesporial cells, which produce both sporogenous cells and the three layers of subepidermal parietal cells. Sporogenous cells undergo several rounds of mitosis to develop into microsporocytes competent for meiosis. The transformation from asynchronous mitosis to synchronous meiotic cycle in sporogenous cells might involve multiple developmental events, in the context of an elaborately arranged microsporangium. In light of the significant differences in micro- and megaspore genesis, it will be fascinating to determine whether male sporocytes have their specific mechanism in triggering meiotic switch. In addition, male sterility plays an integral role in hybrid seed production, particularly in rice. Therefore, the investigation of plant male sporocytes development is of a great theoretical and practical value.
Glutaredoxins are small oxidoreductases using glutathione as a cofactor. They mediate reversible reduction of intracellular disulfide bonds, resulting in posttranslational modifications of target proteins, probably in response to an altered cellular redox state. In land plants, glutaredoxin isoforms can be classified into three distinct subgroups according to the residues of their active-site motifs, which are composed of four amino acids (CxxC or CxxS). The CC-type glutaredoxins, which contain a Cys in the second position of the active-site motif that leads to the designation of this subclass, are restricted to land plants (Rouhier et al., 2008). Limited knowledge has been reported about functions of CC-type glutaredoxins, beyond the areas of floral organ development, microsporogenesis, and pathogen responses (Xing et al., 2005; Ndamukong et al., 2007; Xing and Zachgo, 2008; Li et al., 2009). However, the number of CC-type glutaredoxins expands apparently from lower plants, such as bryophytes, to higher plants, such as Arabidopsis, rice, and poplar (Populus trichocarpa), concomitantly to the development of more complex plant structure, suggesting that this type of glutaredoxin might play as yet uncharacterized but indispensable roles in the survival and adaption of higher plants (Xing et al., 2006).
To elucidate the mechanisms regulating the transition to meiosis in plant anthers, we isolated a rice mutant, microsporeless1 (mil1), which did not produce microspores but had normal female fertility. In mil1 anthers, meiosis was replaced by continued mitosis, resulting in the mature anther locules filled with somatic cells. The MIL1 gene encodes a meiocyte centromere-localized CC-type glutaredoxin, which could interact with TGA transcription factors. Our data indicated that anthers might have their specific mechanism to induce meiosis, which necessitates MIL1’s function.
RESULTS
mil1 Anthers Have Defects in Microspore Genesis
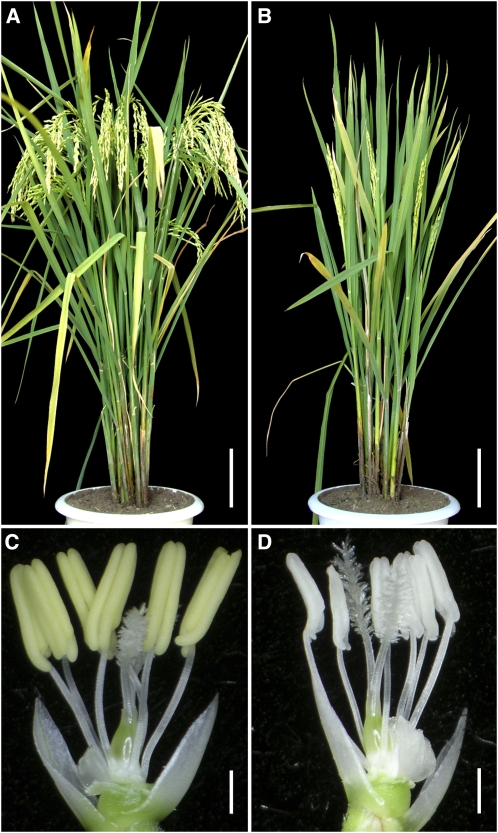
Our initial phenotypic analysis of the homozygous mil1 plants suggested its complete male sterility (Figures 1A and 1B). Careful examination revealed that mil1 anthers were white, lacked dehiscence, and were apparently devoid of pollen grains compared with wild-type anthers (Figures 1C and 1D). Except for the male sterility, no additional obvious mutational phenotypes were observed in the mil1 mutant. When mil1 mutant plants were pollinated with pollen from wild-type plants, the seeds developed normally. The absence of pollen in mil1 mutant flowers combined with the fact that mil1 mutation did not affect fertility in megasporocytes indicated that MIL1 function is required for male fertility.
Figure 1.
Phenotypic Analysis of the Wild Type and mil1.
(A) A wild-type plant after heading.
(B) A mil1 plant after heading.
(C) Wild-type anthers.
(D) Sterile mil1 anthers.
Bars = 10 cm in (A) and (B) and 1 mm in (C) and (D).
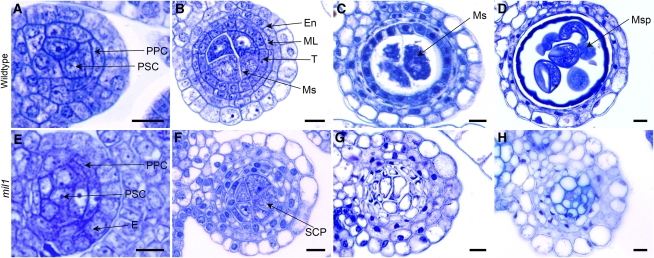
To determine the point at which the mutant phenotype was first detectable and determine what cell types were affected by the mutation, we observed transverse sections of wild-type and mil1 anthers. Young wild-type anthers generated primary sporogenous cells and primary parietal cells from the archesporial initials underneath the epidermis (Figure 2A). At the subsequent development stage, in wild-type microsporangia, the primary sporogenous cells underwent several rounds of mitotic divisions to produce microsoprocytes, which were much larger than the surrounding nonreproductive cells and occupied the center of the locules, with two to four sporocytes transected per locule. The primary parietal cells divided periclinally and anticlinally into two sets of secondary parietal cells. The outer set of secondary parietal cells divided further and differentiated into endothecium, and the secondary parietal cells in the inner set underwent one more periclinal division to form the middle layer and the tapetal layer adjacent to the microsoprocytes. The tapetal cells were clearly recognizable, close to rectangular in shape compared with the thin middle layer (Figure 2B). The middle layer degraded during meiosis stage and was observed only as crushed remnants between the tapetal and endothecial layers, while the tapetum started to degenerate after microspores released from the tetrads and before the commencement of pollen mitosis (Figures 2C and 2D).
Figure 2.
Toluidine Blue–Stained Transverse Sections of Anthers in the Wild Type and mil1.
(A) to (D) The wild type.
(E) to (H) mil1.
(A) and (E) Anthers with primary parietal cells and primary sporogenous cells.
(B) Wild-type anthers with microsporocytes and four parietal layers.
(C) Wild-type anthers at meiotic stage exhibiting crushed middle layer.
(D) Wild-type anthers filled with microspores, with middle layer and tapetum degraded.
(F) mil1 anthers with SCPs in the center. They generally have four parietal layers but show extra layers in some regions.
(G) mil1 SCPs increase in number and decrease in size as anthers develop. No cell degeneration is observed in wall cells.
(H) mil1 anthers are composed of somatic cells.
En, endothecium; ML, middle layer; Ms, microsporocyte; Msp, microspore; PPC, primary parietal cell; PSC, primary sporogenous cell; T, tapetum. Bars = 10 μm.
[See online article for color version of this figure.]
mil1 anthers completed the biological events similar to the wild type at an early developmental stage. There was obvious differentiation of parietal cells and sporogenous cells due to the asymmetrical division of archesporial cells (Figure 2E). At the comparable developmental stage, prospective microsporocytes appeared in the center of mil1 anthers, and these cells were also much larger than the surrounding somatic cells, with two to four cells per transverse locule section (Figure 2F). Nevertheless, unlike wild-type microsporocytes, which would develop into microspores (Figures 2C and 2D), these progenies of mil1 sporogenous cells divided into many smaller cells, so that finally the mature mil1 anthers were composed of highly vacuolated and tightly arrayed cells instead of pollen grains (Figures 2G and 2H). The differentiation of endothecium and epidermis was initially superficially normal, but they displayed quite different appearances from the wild-type ones at later development stages. On the other hand, the inner two somatic cell layers were in disarray. In most of the anthers, these two layers could be clearly discerned, but in some regions, the cells were arranged so irregularly that it could hardly be determined whether there were two or three cell layers. Besides, these cell layers were represented by undifferentiated cells with characteristics of neither the middle layer nor the tapetal layer (Figure 2F). These inner indeterminate cell layers never degenerated; instead, these cells enlarged and vacuolated and finally developed into the tightly packed cells that occupied the anther locules, having a similar fate as the prospective microsporocytes. In addition, the cells at the central part of the locules had thickened their cell walls (Figures 2G and 2H). Therefore, in mil1 anthers, both the sporocyte and the surrounding cell development were defective, and all cells developed into somatic cells.
Progenies of the mil1 Sporogenous Cells Have Defects in Meiotic Entry
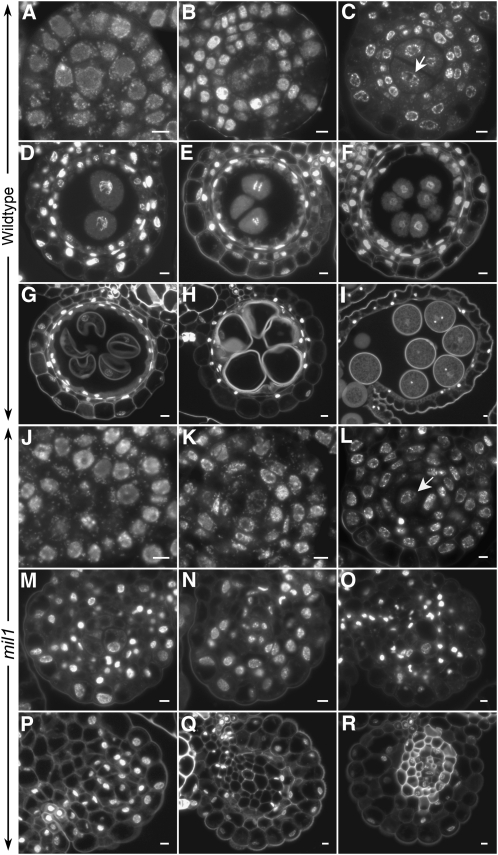
To investigate what happened to the mil1 microsporocytes, we then obtained detailed knowledge of the mutant phenotype by in situ observations of chromosome behavior of anther cells at different developmental stages using transverse anther sections stained by 4′,6-diamidino-2-phenylindole (DAPI). The spikelet length was used as a surrogate for the reproductive stage progression to compare the phenotypes between the wild type and the mutant. Archesporial cell division and differentiation appeared normal in mil1 anthers (Figures 3A and 3J). Subsequently, from primary parietal cells, three inner initial cell layers were formed. At these stages, chromosomes in all the cells, no matter somatic cells or the sporogenous cells in the center, had similar morphology (Figures 3B and 3K). After establishing three subepidermal parietal cell layers, mil1 anthers showed obvious patterns that deviated from the wild-type ones. In the wild-type anthers at this developmental stage, the microsporocyte nuclei were at premeiotic interphase, distinguishing them from those of the surrounding somatic cells by the presence of more extended tenuous chromosomes (Figure 3C). On the entry into meiotic cycle, the microsporocyte nuclei exhibited rather distinct appearance, with their chromosomes highly contracted and with brighter DAPI staining (Figures 3D and 3E). Whereas in mil1 microsporangia, the chromosomes of the central microsporocytes displayed patterns typical of the surrounding somatic cells, and no meiotic chromosome behavior was detected (Figures 3L and 3M). Since no meiosis was observed in mil1 microsporocytes, they were called sporogenous cell progenies (SCPs) instead of microsporocytes in the following text. In the subsequent stage, wild-type pollen mother cells generated tetrads of haploid microspores, which subsequently proceeded through two rounds of mitosis to develop into mature pollens. Concomitantly, the inner parietal layers degenerated as pollen maturing (Figures 3F to 3I). In mil1 microsporangiums, no cell degeneration was observed. The central SCPs increased in number and decreased in size as the anther developed. Gradually, the mil1 SCPs lost their featured morphology and became indistinguishable from the surrounding somatic cells. The nuclei of the mil1 anther cells condensed and finally disappeared, and cells at the locule center had walls thickened (Figures 3M to 3R).
Figure 3.
DAPI-Stained Transverse Sections of Anthers in Both the Wild Type and mil1.
(A) to (I) The wild type.
(J) to (R) mil1.
(A) and (J) Anthers with sporogenous cells and primary parietal cells.
(B) and (K) Anthers with two secondary parietal cells.
(C) and (L) Anthers with four parietal cells established. Wild-type microsporocytes (arrow in [C]) are in premeiotic interphase, while mil1 SCPs (arrow in [L]) behave like somatic cells.
(D) and (E) Wild-type anthers with microsporocytes in meiosis. The middle layer is crushed.
(F) Wild-type anthers with tetrads formed.
(G) to (I) Wild-type anthers showing developing microspores. The tapetum is degenerated.
(M) to (R) mil1 SCPs show no sign of meiosis, and together with anther wall cells they develop into somatic cells.
Bars = 5 μm.
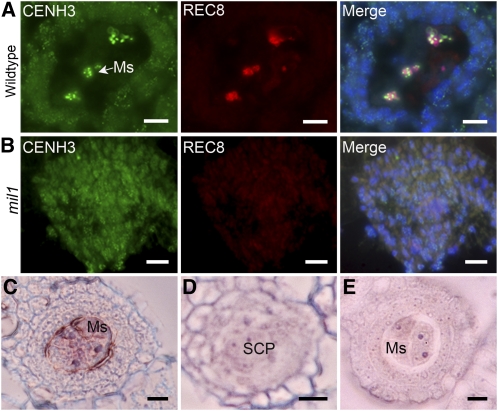
Chromosome morphology in the mil1 mutant indicated that the mil1 SCPs did not undergo meiosis. Therefore, we examined the distribution patterns of a meiotic-specific protein REC8 in the wild-type and mil1 microsporangium through fluorescence immunolocalization with correspondent polyclonal antibodies. REC8 is attached to the meiotic chromosomes at early prophase I and is a valuable marker for identifying meiotic cells (Wang et al., 2009; Shao et al., 2011). In addition, we applied CENH3 as a positive control, which is a histone H3 variant specific to centromere histones in rice (Nagaki et al., 2004). The rice REC8 protein was discretely distributed along chromosomes during early prophase I in the wild-type microsporocytes. By contrast, REC8 foci were not detected in mil1 SCPs, while CENH3 signals were observed in both wild-type and mil1 cells with distinct staining patterns. CENH3 staining signals in wild-type microsporocytes were much brighter than those in the surrounding somatic cells, although we cannot yet provide an explanation for this difference. CENH3 labeling was of a similar intensity in all mil1 anther cells, just as those in the wild-type somatic cells (Figures 4A and 4B). Thus, it appears that the mil1 SCPs behave like somatic cells.
Figure 4.
Detection of Meiosis Using Meiosis-Specific Markers in Both the Wild Type and mil1.
(A) and B) Anther transverse sections immunostained with antibodies against CENH3 (green) and REC8 (red). Images in the left and middle columns are merged in the right column.
(A) The wild type. Microsporocytes show strong immunosignals for both proteins.
(B) mil1. SCPs exhibit weak CENH3 staining signals and are totally absent of REC8 staining signals just like somatic cells.
(C) and (D) In situ hybridization for REC8 mRNA in anthers using an antisense probe.
(C) REC8 is expressed preferentially in the wild-type microsporocytes.
(D) Expression of REC8 is negligible in mil1 SCPs.
(E) Wild-type anthers hybridized with the REC8 sense probe as a control.
Ms, microsporocyte. Bars = 10 μm.
We further confirmed the character of mil1 SCPs by testing REC8 expression using RNA in situ hybridization. As shown in Figure 4C, the REC8 transcripts were specifically expressed in microsporocytes of the wild-type anther, but REC8 signal was negligible above the background level in mil1 SCPs (Figures 4D and 4E). We then investigated expression of several rice meiotic-related genes, including MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1) (Nonomura et al., 2007), MEL2 (Nonomura et al., 2011), MER3 (Wang et al., 2009), HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 (PAIR1) (Nonomura et al., 2004), AM1 (Che et al., 2011), REC8 (Shao et al., 2011), SOLO DANCERS (SDS) (Chang et al., 2009), SPO11-1 (Yu et al., 2010), and ZEP1 (Wang et al., 2010), in both wild-type panicles undergoing meiosis and mil1 panicles at the similar length. Using RT-PCR and quantitative RT-PCR (qRT-PCR), we found that the tested genes all had their transcripts significantly decreased in mil1 panicles, suggesting MIL1 precedes these genes in action (see Supplemental Figures 1A and 1B online). Collectively, these results indicate the mil1 SCPs have defects in activating the meiotic cell cycle.
MIL1 Encodes a Glutaredoxin
Genetic analysis revealed that a single recessive gene controlled the mil1 phenotype. To unravel the molecular basis of MIL1 function on male reproduction, we applied map-based cloning to get the MIL1 gene. The gene was located to a narrow region ~13 kb between two markers S8 and S9 on chromosome 7 (see Supplemental Figure 2A online). According to the information from the National Center for Biotechnology Information, there are four putative genes in this region and through sequencing three of them were found to have the same sequences in mil1 and the wild type. In the remaining gene, we found that there was a sequence inserted between nucleotides 225 and 226 of the open reading frame (ORF); thus, we regarded this gene as the candidate gene and designated it as MIL1 (see Supplemental Figure 2B online). We were unable to obtain the full-length sequence of the insert, but through PCR, we recovered the sequence information of both ends of the insert, which showed high similarity to the sequences of a rice CACTA transposon in the locus LOC_Os08g27980.
Complementation experiments further confirmed MIL1 as the candidate gene. A plasmid containing the entire ORF, 4-kb upstream region and 1.3-kb downstream region, rescued the mil1 mutant, in contrast with the control vector carrying the truncated ORF (see Supplemental Figure 2C online). We examined the expression of MIL1 in mil1 and found that in contrast with the substantial expression in wild-type meiotic panicles, MIL1 had no detectable transcripts in mil1 panicles at a comparable developmental stage, indicating that mil1 is a null mutant of MIL1 (see Supplemental Figure 3 online).
Since there is no full-length cDNA for MIL1 in the rice cDNA databases, we performed 3′ rapid amplification of cDNA ends (3′RACE) and 5′RACE and obtained a 624-bp product composed of a 384-bp ORF with no introns, a 197-bp 3′ noncoding region, and a 59-bp 5′ noncoding region. The MIL1 protein contains 127 amino acids (see Supplemental Figure 2D online). Homology searches revealed that MIL1 is a glutaredoxin belonging to the plant-specific CC type, with CCMC composing the active site. There are 21 CC-type glutaredoxins in Arabidopsis and 17 in rice (Xing et al., 2006). Protein sequence analysis of these CC-type glutaredoxins from Arabidopsis and rice revealed that MIL1 occupied a relatively isolated position on the phylogenetic tree, so that it was difficult to decide which glutaredoxin is the closest homolog of MIL1 in Arabidopsis (see Supplemental Figure 4 online).
MIL1 Is Preferentially Expressed in Sporogenous and Inner Parietal Cells
MIL1 requirement for the reproductive and surrounding cell development was confirmed by its temporal and spatial expression patterns. qRT-PCR was applied to examine MIL1 transcripts in different tissues from the wild-type plant. As expected, there was specific accumulation of MIL1 mRNAs in young panicles (length of 1 to 10 cm), and the most abundant transcripts were detected in panicles at meiosis stage (length of 5 cm). No product could be found in other tissues, including mature panicles, leaves, roots, leaf sheaths, and internodes (see Supplemental Figure 5 online).
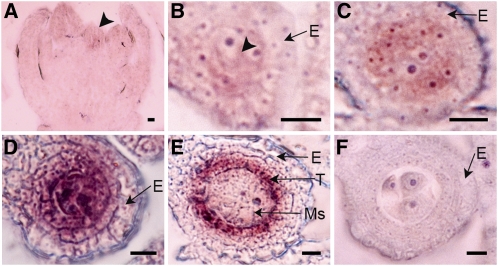
RNA in situ hybridization was performed to localize the MIL1 transcripts in panicles in detail. Its transcripts were first detected in the inner floral meristem where stamen primordia would initiate (Figure 5A). Later, the expression was detectable in young stamens. Transverse sections showed that the transcripts were localized in the four corners of young anthers where archesporial cells were located (Figure 5B). Afterwards, high levels of MIL1 transcripts preferentially accumulated in both parietal cells and sporogenous cells and later persisted into microsporocytes and inner subepidermal wall cells (Figures 5C and 5D). At meiotic stages, MIL1 was expressed mainly in the somatic cell layers encasing the microsporocytes, especially in the tapetum, with weak expression in microsporocytes, whereas hardly any signals were detectable in the outer two layers (Figure 5E). The signals were not visualized in anthers probed with a sense strand MIL1 RNA as a negative control (Figure 5F; see Supplemental Figures 6A to 6C online).
Figure 5.
In Situ Localization of MIL1 mRNA.
(A) to (E) Wild-type anthers hybridized with MIL1 antisense probe.
(A) MIL1 transcripts are first detected in the stamen primordia, as indicated by the arrowhead.
(B) Transverse sections show that MIL1 is transcribed early in archesporial cells, as indicated by the arrowhead.
(C) MIL1 is transcribed later in parietal and sporogenous cells.
(D) Dramatically enhanced MIL1 expression in inner parietal cells and microsporocytes.
(E) MIL1 expression is stronger in tapetum and weaker in meiocytes.
(F) Control hybridization with MIL1 sense probe.
E, epidermis; Ms, microsporocyte; T, tapetum. Bars = 10 μm.
mil1 msp1-5 SCPs Have Defects in Meiotic Entry
Because mil1 anthers exhibited apparent abnormalities both in SCPs and somatic cells, we wondered which cell type exhibited the primary defects. To address this question, we tried to assess the effect of MIL1’s inactivation on microsporogenesis in the absence of defective somatic cell layers. In rice, the MULTIPLE SPOROCYTE1 (MSP1) gene is critical for initiating anther wall formation (Nonomura et al., 2003). In msp1-1 anthers, no tapetal layer is formed, and often the middle layer is absent; meanwhile, an extra number of male sporocytes are generated. We isolated an allelic mutant of msp1-1, named msp1-5, which had a retrotransposon Tos 17 insertion in the ORF of the MSP1 gene (see Supplemental Figure 7 online) and exhibited the same phenotype as the characterized msp1-1 mutant (Figures 6A to 6D).
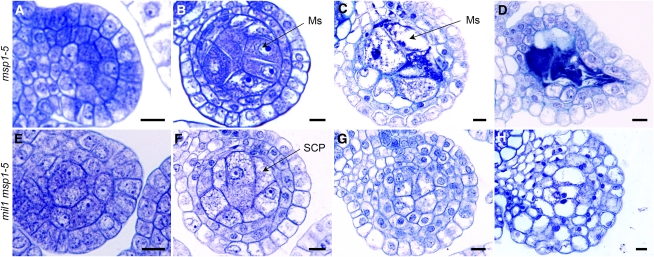
Figure 6.
Toluidine Blue–Stained Transverse Sections of Anthers in Both msp1-5 and the mil1 msp1-5 Double Mutant.
(A) to (D) msp1-5.
(E) to (H) mil1 msp1-5.
(A) and (E) msp1-5 and mil1 msp1-5 anthers develop normally at early developmental stage, with primary parietal cells and primary sporogenous cells.
(B) and (F) msp1-5 and mil1 msp1-5 both have excess microsporocytes (or SCPs) and almost two parietal layers.
(C) and (D) msp1-5 anthers have degraded microsporocytes, but intact parietal cells remained.
(G) and (H) mil1 msp1-5 SCPs behave like mil1 SCPs, increasing in number and decreasing in size as anthers develop, finally developing into somatic cells.
Ms, microsporocyte. Bars = 10 μm.
[See online article for color version of this figure.]
Double mutants of mil1 and msp1-5 were produced and investigated. Transverse section analysis illustrated that anther development at early stages in the mil1 msp1-5 double mutant were quite similar to that in msp1-5. The differentiation of primary parietal cells and primary sporogenous cells were normal, but the parietal cell layer was not able to divide periclinally further (Figures 6A and 6E). At the stage when four wall layers were formed in wild-type anthers, the msp1-5 and mil1 msp1-5 microsporangia had only two obvious layers of wall cells, though additional wall layers could be detected in some region, whereas excess number of microsporocytes were readily observed (Figures 6B and 6F). After this stage, mil1 msp1-5 presented a phenotype quite different from that of msp1-5. The msp1-5 microsporocytes swelled and disintegrated, leaving empty and compressed locules with perfect epidermal and endothecial layers (Figures 6C and 6D). By contrast, the mil1 msp1-5 microsporocytes continued cell division, and finally the microsporangium took the morphology quite similar to that of mil1 (Figures 6G and 6H).
Because mil1 msp1-5 bypassed the meiocyte degradation occurring in msp1-5 anthers, we observed the chromosome morphology to learn what brought about the differences in msp1-5 and mil1 msp1-5 anthers. msp1-5 microsporocytes were detected to enter meiosis but they failed to complete successfully; finally, these undivided microsporocytes degenerated (Figures 7A and 7B). By contrast, the mil1 msp1-5 microsporocytes did not initiate meiosis nor degrade; instead, they divided into tightly arranged somatic cells (Figures 7C and 7D). Since the mil1 msp1-5 microsporocytes did not undergo meiosis either, they were also called SCPs. In addition, we checked MIL1’s acting region in msp1-5 microsporangiums and found that MIL1 transcripts were only detected in the msp1-5 microsporocytes where REC8 also had specific distribution (Figures 7E and 7F). Therefore, eliminating MIL1 from msp1-5 microsporocytes blocked meiosis initiation. In other words, meiotic entry defects in mil1 msp1-5 were derived from the MIL1 inactivation in SCPs.
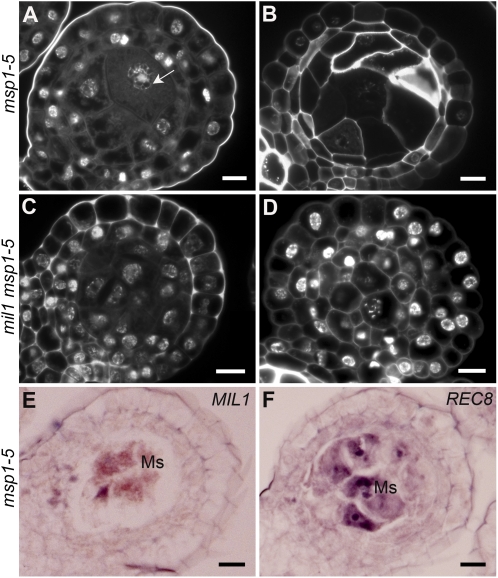
Figure 7.
Characterization of Anther Cell Development in Both msp1-5 and mil1 msp1-5.
(A) to (D) DAPI-stained transverse sections of anthers.
(A) Microsporocytes enter meiosis in msp1-5, as the arrow indicates.
(B) Microsporocytes abort later in msp1-5.
(C) mil1 msp1-5 SCPs show no signs of meiotic cell cycle activation.
(D) mil1 msp1-5 SCPs finally develop into somatic cells.
(E) msp1-5 anthers probed with MIL1 antisense probe.
(F) msp1-5 anthers probed with REC8 antisense probe.
Ms, microsporocyte. Bars = 10 μm.
[See online article for color version of this figure.]
MIL1 Is Localized in Meiocyte Nuclei
Localization studies were further performed on MIL1 to understand its mechanism of action. The intracellular distribution of MIL1 protein in microsporangium was investigated by fluorescence immunolocalization on transverse anther sections using a polyclonal antibody against recombinant MIL1. The results revealed that in wild-type anther, the protein had a predominant distribution in both tapetal cells and meiocytes, displaying strong parallels with the transcripts distribution. Furthermore, a significantly strong MIL1 labeling was found on the meiocyte nuclei region (Figure 8A). Consequently, to determine more detailed distribution of MIL1 in meiotic nuclei, dual immunolocalization was performed on wild-type anther squashes using anti-MIL1 antibody in combination with anti-REC8 antibody. We found MIL1 appeared as numerous discrete foci in premeiotic interphase nuclei. Some of the fluorescence foci were much larger than others and colocalized with REC8-labeled axes, whereas other smaller foci were not axis associated and showed scattering in the nuclei and even the cytoplasm (Figure 8B). At pachytene stage, MIL1 was mainly located in the centromeric region of each chromosome, although some weaker immunostaining signals could be detected in both other chromosome regions and the cytoplasm.
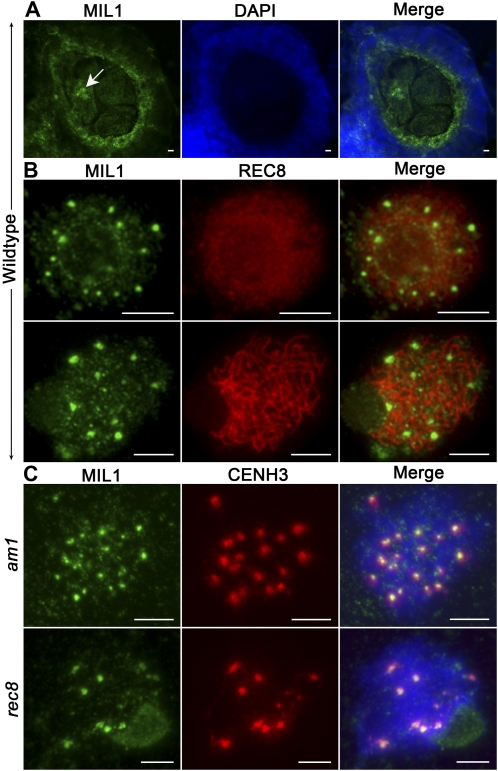
Figure 8.
Immunofluorescent Localization of MIL1.
(A) Immunolocalization of MIL1 (green) on wild-type rice anther sections counterstained with DAPI (blue). MIL1 shows a strong labeling on the microsporocyte nuclei region (arrow).
(B) Dual immunolocalization of MIL1 (green) and REC8 (red) in wild-type rice on chromosome spreads at premeiotic interphase (top part) and pachytene (bottom part).
(C) Dual immunolocalization of MIL1 (green) and CENH3 (red) on chromosome spreads of am1 and rec8 microsporocytes.
Bars = 5 μm.
Rice proteins AM1 and REC8 are localized to meiotic chromosomes quite early at meiotic prophase I (Wang et al., 2009; Che et al., 2011; Shao et al., 2011). To define the relationship between these two proteins and MIL1, MIL1 immunolocalization was investigated in anther squashes from the rice am1 mutant and rec8 mutant, in combination with CENH3. MIL1 appeared normally as discrete foci in meiotic nuclei of these two mutants, just like that in the wild type (Figure 8C). This provides further evidence to support the proposal that MIL1 works prior to those two proteins. Besides, the larger MIL1 foci showed evident colocalization with CENH3 foci, indicating that MIL1 localizes mainly on the centromeres of meiotic chromosomes.
MIL1 Might Cooperate with TGA Transcription Factors
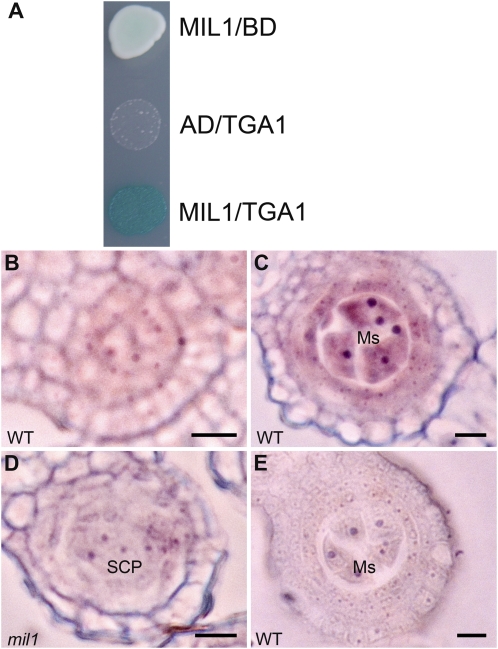
Several Arabidopsis glutaredoxins have been confirmed to act by posttranslationally modifying TGA transcription factors (Ndamukong et al., 2007; Xing and Zachgo, 2008; Murmu et al., 2010). Therefore, we wondered whether there were rice TGAs that might interact with MIL1. We searched the rice genome using Arabidopsis TGA sequences as the template and identified 14 rice TGA genes (see Supplemental Figure 8 online). Among these genes, LOC_Os05g37170 exhibited preferential panicle expression according to its EST profile; we consequently named the gene TGA1 and took it for further investigation. The TGA1 and MIL1 interaction was tested in a yeast two-hybrid analysis, and an obvious physical interaction was detected (Figure 9A).
Figure 9.
TGA1’s Expression Pattern and Its Relationship with MIL1.
(A) Interaction of MIL1 and TGA1 in yeast grown on medium with X-gal, Gal, raffinose, and without Ura, Leu, Trp, and His. AD, pB42AD; BD, pLexA; MIL1, pLexA-MIL1; TGA1, pB42AD-TGA1.
(B) and (C) Wild-type (WT) anthers hybridized with TGA1 antisense probe. TGA1 is transcribed weakly in primary parietal and sporogenous cells (B), and later more strongly in microsporocytes and inner parietal cells (C).
(D) TGA1 hardly expresses in mil1 anthers.
(E) Wild-type anthers hybridized with TGA1 sense probe.
Ms, microsporocyte. Bars = 10 μm.
To gain insights into the relationship between TGA1 and MIL1, the expression of TGA1 was analyzed based on in situ RNA. TGA1 showed weak expression at early anther development stage, mainly active in sporogenous cells and parietal cells beneath epidermis (Figure 9B). The transcriptional activity was dramatically enhanced as microsporangium expanded, with products confined to microsporocytes and nurse cells, which overlapped with the MIL1 expression region (Figure 9C). Hardly any TGA1 signal above background was detected in mil1 anthers (Figures 9D and 9E). qRT-PCR using cDNA from meiotic panicles (length 5 cm) also showed that TGA1 had much fewer transcripts in mil1 compared with that in the wild type (see Supplemental Figure 9 online). Collectively, these data indicated that there might be interactions between MIL1 and TGA transcription factors in vivo. As to TGA1’s lower transcription level in mil1 anthers, considering that glutaredoxins usually regulate target protein’s activity at a posttranslational level, we proposed that it might result from the anther cell differentiation failure, although we could not rule out the possibility that MIL1 might also exert effect on TGA1’s transcription.
DISCUSSION
In this study, we demonstrate that MIL1 is essential for the microsporogenesis. The most conspicuous phenotype of mil1 is the microsporangiums eventually filled with tightly packed somatic cells instead of microspores. Nonetheless, mil1 microsporangiums at early development stage generate primary sporogenous cells normally, which produce progeny cells distinct from the surrounding somatic cells. Thus, what happens in mil1 SCPs preventing the generation of microspores? We perform an extensive characterization of mil1 SCPs. Morphological, cytological, and molecular investigations show that no sign of meiosis could be observed in these SCPs, on the ground of the observations that chromosomes in mil1 SCPs (1) exhibit the morphology of somatic cell chromosome, (2) have CENH3 distribution in a somatic cell chromosome pattern, and (3) lack meiotic specific REC8 proteins deposition. Besides, mil1 SCPs display obvious diminution of REC8 transcripts and substantial transcription decline of meiosis genes. Overall, the data aforementioned demonstrate that meiosis never happens in mil1 SCPs. In light of the increasing cell number in mil1 locule centers, we deduce that, like am1 mutants (Palmer, 1971; Staiger and Cande, 1992; Golubovskaya et al., 1993, 1997; Pawlowski et al., 2009), meiosis might be replaced with a mitotic division in mil1 SCPs. Thus, the MIL1 gene may exert an impact on meiosis initiation in microsporocytes, which, on mutation, will cause microsporocytes to continue the mitotic way other than initiate the meiotic program. MIL1’s influence occurs prior to the action of most of the genes involved in early meiotic processes, including the rice AM1 gene.
In view of the fact that loss of MIL1 results in development aberrations both in SCPs and somatic nursing cells, this gene might exert its impact on meiosis activation directly or indirectly. In the latter case, the primary phenotype of mil1 would be development defects in nursing cells and the meiosis bypass phenotype as the consequence of this initial defect. Alternatively, meiosis entry failure could be contributable chiefly to the defects in SCP development other than the compromise of inner parietal cell functions. Our phenotypic analyses of mil1 msp1-5 are certainly consistent with this proposition. mil1 msp1-5 and msp1-5 share quite similar phenotypes at the anther cell differentiation stages except that mil1 msp1-5 SCPs do not make commitment to meiosis, which leads to distinctive anther phenotypes at maturity. MIL1 distributed exclusively in msp1-5 microsporocytes; therefore, it is reasonable to deduce that the only reason for the meiosis entry failure in mil1 msp1-5 is the MIL1 absence in its SCPs. To summarize, we conclude that MIL1 plays a direct role in triggering meiosis commencement, as we depict in the diagram in Supplemental Figure 10 online. This contention is given added credence by our observations that MIL1 appears mainly on meiocyte chromosome centromeres, and MIL1 transcripts distribute abundantly in microsporocytes when transition to meiosis is taking place.
In mil1 microsporangium, initial differentiation of the archesporial cells appears normal and two secondary parietal layers are formed. The inner secondary parietal layer divides into two cell layers, but these two layers fail to differentiate into the middle and tapetal layers, resulting in two somatic cell layers represented by undifferentiated cells that occupy the position where the middle and tapetal layers should be. In some regions of the mil1 parietal tissues, more than four cell layers could be observed, which might result from the periclinal division of the inner two cell layers. We propose that due to the undifferentiating state of these two layers, their cells might take on further periclinal division in addition to the anticlinal division that usually happens in the middle and tapetal layers. This suggests that MIL1 might regulate the differentiation of tapetal and middle layer, or more exactly, the differentiation of the inner secondary parietal cells. Similarly, MIL1’s impact on nursing cell development could also be direct or indirect. Concerning the prominent distribution of MIL1 transcripts and proteins in the inner subepidermal parietal cells, we are in favor of MIL1’s direct roles in nursing cell establishment. Besides, mil1 endothecium and epidermis also display quite different appearances from the wild-type ones at later development stages. In view that MIL1 shows little accumulation of transcripts in the outer two layers, we propose that these abnormalities are secondary effects to the defects in SCPs and inner parietal cells.
Therefore, we propose that MIL1 not only regulates meiosis initiation in microsporocytes, but also controls differentiation of the inner secondary parietal cells into middle and tapetal layers. The dual role of MIL1 in microsporogenesis may reflect the proposition that functional microspore formation requires the integrity of reproductive and nonreproductive tissues and successful communication between both tissue types (Ma, 2005).
Studies in yeast and mammals indicate that the cell cycle switch from mitotic to meiotic is controlled by a large set of genes that ensure the activation of meiosis is coordinated with environmental or developmental conditions (Honigberg and Purnapatre, 2003; Marston and Amon, 2004; Baltus et al., 2006; Bowles et al., 2006; Koubova et al., 2006; Bowles and Koopman, 2007; Harigaya and Yamamoto, 2007; Pawlowski et al., 2007). Owing to their sedentary lifestyle, plants must be capable of cooperating with a variety of factors in their environments. To complete meiosis and sexual life cycles successfully, plants have to integrate environmental and developmental cues and activate the meiotic cell cycle at the correct time and place. Consequently, making the commitment to enter meiosis in such a complicated developmental context might also involve an intricate network of genes.
In heterosporous plants, such as gymnosperms, angiosperms, and some ferns, a large number of microsporocytes are generated in anthers; however, the ovule produces one sporocyte. Furthermore, in many angiosperms, including Arabidopsis and rice, meiosis in the male reproductive structure is not synchronized with meiosis in the female (Armstrong and Jones, 2001; Itoh et al., 2005). Therefore, it is reasonable to infer that the mechanism(s) triggering male meiosis might differ from those initiating female meiosis. On the other hand, it has been manifested that the micro- and macrosporocytes share common components in the meiotic process (Ma, 2005). Thereby, the plant mitosis/meiosis switch mechanism might involve both unisexual components, like MIL1, and bisexual players, such as MEL2 in rice and AM1 in maize (Pawlowski et al., 2009; Nonomura et al., 2011). Although it is not yet known how these players in this machinery are integrated to coordinately orchestrate this critical part of the life cycle of flowering plants, we learn from this study that MIL1 works prior to MEL2. Furthermore, with continuing effort to unravel the mystery of reproductive development in plants, it will soon be possible to build a genetic network of these regulators and to advance our understanding of this mechanism.
Glutathione has been manifested to be involved in cell proceeding and differentiation as well as cell signaling, whereas glutaredoxins serve functions in a broad array of cellular processes and perform versatile roles in plant development (Rouhier et al., 2008). It has always been supposed that signal transduction pathways contribute to the early anther cell differentiation, but what kinds of signal molecules are involved have not been identified (Sun et al., 2007). MIL1’s glutaredoxin property, combined with the fact that redox modifications play significant roles in cell signaling, suggests MIL1 might influence male meiosis initiation via participation in an anther-specific meiosis initiation signaling pathway. The interaction between MIL1 and TGA1 further supports this hypothesis. According to the model proposed by Murmu et al. (2010), glutaredoxins will modulate TGA transcription factor activity in response to redox changes triggered by developmental signals. Further analysis of MIL1 function will contribute to an enhanced understanding of the plant-specific mechanisms that commit specific regulatory/developmental genes to meiosis and elucidate the roles of glutaredoxin and redox regulation involved in this process. Moreover, the study of MIL1-like proteins in species other than rice will hint at the possible roles redox regulation may play in reproductive cell development in other species.
METHODS
Plant Materials
The rice (Oryza sativa) mil1 mutant, msp1-5 mutant, and the wild type (O. sativa cv Zhongxian 3037) were used in this study. Rice mil1 is a spontaneous mutant of Zhongxian 3037 (indica). The F2 mapping population was constructed from crosses between mil1 and Zhonghua 11 (japonica). msp1-5 is a spontaneous mutant of Nipponbare (japonica) obtained from rice tissue culture. To generate the mil1 msp1-5 double mutant, msp1-5 (+/) heterozygotes were crossed with mil1 homozygotes using the latter as the female parent, and F2 plants were phenotypically screened and genotyped.
Map-Based Cloning and Complementation Tests
Marker details used for fine mapping mil1 are provided in Supplemental Table 1 online. The MIL1 locus was located to a narrow, ~13-kb region between the two sequence tagged site markers S8 and S9. An insertion was identified in the LOC_Os07g05630 gene. We created the complementation construct pCMIL by inserting a 5.5-kb genomic DNA fragment containing the entire MIL1 ORF and the 3870-bp upstream and 1306-bp downstream regions to the binary vector pCAMBIA1300 (CAMBIA). The pCMILC control vector containing only a partial upstream region (−3870 to −493 bp) was also constructed. These two plasmids were introduced into calli derived from seeds of plants heterozygous for the MIL1 locus using an Agrobacterium tumefaciens–mediated method as previously described (Lee et al., 1999).
Antibody Production
The recombinant MIL1 protein was obtained by cloning the MIL1 ORF into the expression vector pMAL-c5X (New England Biolabs), which was transformed into Escherichia coli strain BL21 (DE3) (Ding Guo). The MIL1-MBP fusion protein was purified using the pMAL protein fusion and purification system following the instruction manual (New England Biolabs). The primer sequences are listed in Supplemental Table 2 online. The polyclonal antibody was raised in mouse (Mus musculus). MIL1 recombinant proteins were injected into a mouse every 2 weeks. Immune sera were extracted 2 months after the first injection.
Phenotypic Characterization
Histological studies were performed as previously described with minor modifications (Wang et al., 2009; Che et al., 2011). Wild-type, mil1, and msp1-5 spikelets representing various developmental stages were fixed in FAA (5% formaldehyde, 5% glacial acetic acid, and 63% ethanol) or Carnoy’s solution (ethanol:glacial acetic acid, 3:1) overnight at 4°C. Following dehydration in a graded ethanol series, the samples were embedded in Technovit 7100 resin (Hereaus Kulzer). Four-micrometer transverse sections were cut using a Leica microtome, stained with 0.25% toluidine blue or DAPI, and observed. For immunofluorescence on anther sections, spikelets were embedded in optimal cutting temperature compound (Sakura Finetek) after fixation, and 15-μm transverse sections were cut using a Leica CM1850 cryostat at −20°C. Immunofluorescence on anther squashes was performed as described by Wang et al. (2009).
Gene Expression Analysis and RACE
Total RNA was extracted from various tissues using TRIpure reagent (BioTeke). For RT-PCR, 25 ng of cDNA was applied using corresponding gene-specific primer pairs. Ubiquitin was amplified as the control. qRT-PCR was performed on a cycler apparatus (Bio-Rad) using the SYBR Green PCR Master Mix (Tiangen) according to the manufacturer's instructions. Amplification was conducted in 96-well optical reaction plates with the following protocol: 94°C for 4 min, 40 cycles of 94°C for 15 s, 58°C for 15 s, and 72°C for 15 s. Expression levels of target genes were quantified using the Bio-Rad CFX96 real-time PCR detection system (Bio-Rad) by a relative quantitation method (δδ cycle threshold). The statistical significance was analyzed by Student’s t test. Data were presented as mean values of at least two biological repeats with se. Ubiquitin was chosen as a control to normalize all data. The MIL1, REC8, and TGA1 sense and antisense probes were synthesized with T7 RNA polymerase using the digoxigenin RNA labeling kit (Roche). Tissue fixation and RNA in situ hybridization were performed as previously described (Hong et al., 2010). RACE-PCR was conducted with the RACE cDNA amplification kit (Clontech) according to the manufacturer's instructions. Primer sequences are listed in Supplemental Table 2 online.
Yeast Two-Hybrid Analysis
The yeast two-hybrid assay was performed using a MATCHMAKER LexA two-hybrid system (Clontech). The MIL1 and TGA1 ORF was fused in frame to pLexA and pB42AD, respectively, to construct pLexA-MIL1 and pB42AD-TGA1. Both constructs were used to transform EGY48 containing p8op-lacZ. Transformants were selected on selective medium plates at 30°C for 3 d. The activation ability was assayed on Gal/Raf/(Ura-/His-/Leu-/Trp-)/X-gal plates to test LacZ reporter gene activation for 3 d. The primer sequences are listed in Supplemental Table 2 online.
Phylogenetic Analysis
The amino acid sequences of rice TGA proteins were retrieved through a database search using the amino acid sequences of Arabidopsis thaliana TGAs, TGA9 (At1g08320) and TGA10 (At5g06839). The multiple sequence alignments were performed by Clustal_X 1.83 (Thompson et al., 1997) with the following parameters: weight matrix, Gonnet; gap opening penalty, 10.0; and gap extension penalty, 0.10. Phylogenetic trees were constructed with the aligned CC-type glutaredoxin protein sequences and TGA protein sequences from Arabidopsis and rice (see Supplemental Data Sets 1 and 2 online) using MEGA 3.1 software (Kumar et al., 2004) based on the neighbor-joining method with parameters of Poisson correction model, pairwise deletion, and bootstrap (1000 replicates; random seed).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL and/or Oryzabase databases under the following accession numbers: CENH3, AY438639; MEL1, AB292928; MEL2, AB522964; MER3, FJ008126; MIL1, JN417767; MSP1, AB103395; AM1, AK101644; REC8, AY371049; SDS, AK074008; SPO11-1, AB219537; TGA1, AK109520; PAIR1, AB158462; and ZEP1, GU479042.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Meiosis-Related Genes Are Downregulated in mil1 Anthers.
Supplemental Figure 2. Isolation of the MIL1 Gene and Molecular Characterization.
Supplemental Figure 3. RT-PCR Analysis Shows No Detectable MIL1 Transcripts in mil1 Panicles at Meiotic Stages, with UBQ Used as a Control.
Supplemental Figure 4. Phylogeny of the CC-Type Glutaredoxins.
Supplemental Figure 5. qRT-PCR Analysis of MIL1 Expression.
Supplemental Figure 6. Control Hybridization of MIL1 RNA in Situ Analysis.
Supplemental Figure 7. Structure of the msp1-5 Allele.
Supplemental Figure 8. Phylogenetic Tree of the Arabidopsis and Rice TGA Proteins.
Supplemental Figure 9. qRT-PCR Analysis Shows TGA1 Having a Much Lower Expression Level in mil1 Panicles at Meiotic Stages, with UBQ Used as a Control.
Supplemental Figure 10. Diagram for MSP1 and MIL1 Function in Male Reproductive Cell Development.
Supplemental Table 1. List of the PCR-Based Molecular Markers Developed in This Study.
Supplemental Table 2. The Primers Used for MIL1 Molecular Characterization.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 4.
Supplemental Data Set 2. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 8.
Supplementary Material
Acknowledgments
We thank Shunong Bai (Peking University, China) for valuable suggestions. This work was supported by grants from the Ministry of Sciences and Technology of China (2012AA100101, 2011CB944602, and 2011CB100200) and the National Natural Science Foundation of China (30921061).
AUTHOR CONTRIBUTIONS
L.H. and Z.C. designed the research and wrote the article. L.H., D.T., and K.Z. performed the research and analyzed the data. K.W. and M.L. provided the technical assistance.
References
- Agashe B., Prasad C.K., Siddiqi I. (2002). Identification and analysis of DYAD: A gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129: 3935–3943 [DOI] [PubMed] [Google Scholar]
- Armstrong S., Jones G. (2001). Female meiosis in wild-type Arabidopsis thaliana and in two meiotic mutants. Sex. Plant Reprod. 13: 177–183 [Google Scholar]
- Baltus A.E., Menke D.B., Hu Y.C., Goodheart M.L., Carpenter A.E., de Rooij D.G., Page D.C. (2006). In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat. Genet. 38: 1430–1434 [DOI] [PubMed] [Google Scholar]
- Bhatt A.M., Canales C., Dickinson H.G. (2001). Plant meiosis: The means to 1N. Trends Plant Sci. 6: 114–121 [DOI] [PubMed] [Google Scholar]
- Bowles J., Knight D., Smith C., Wilhelm D., Richman J., Mamiya S., Yashiro K., Chawengsaksophak K., Wilson M.J., Rossant J., Hamada H., Koopman P. (2006). Retinoid signaling determines germ cell fate in mice. Science 312: 596–600 [DOI] [PubMed] [Google Scholar]
- Bowles J., Koopman P. (2007). Retinoic acid, meiosis and germ cell fate in mammals. Development 134: 3401–3411 [DOI] [PubMed] [Google Scholar]
- Chang L., Ma H., Xue H.W. (2009). Functional conservation of the meiotic genes SDS and RCK in male meiosis in the monocot rice. Cell Res. 19: 768–782 [DOI] [PubMed] [Google Scholar]
- Che L.X., Tang D., Wang K.J., Wang M., Zhu K.M., Yu H.X., Gu M.H., Cheng Z.K. (2011). OsAM1 is required for leptotene-zygotene transition in rice. Cell Res. 21: 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya I., Avalkina N., Sheridan W.F. (1997). New insights into the role of the maize ameiotic1 locus. Genetics 147: 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya I., Grebennikova Z.K., Avalkina N.A., Sheridan W.F. (1993). The role of the ameiotic1 gene in the initiation of meiosis and in subsequent meiotic events in maize. Genetics 135: 1151–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y., Yamamoto M. (2007). Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res. 15: 523–537 [DOI] [PubMed] [Google Scholar]
- Hong L.L., Qian Q., Zhu K.M., Tang D., Huang Z.J., Gao L., Li M., Gu M.H., Cheng Z.K. (2010). ELE restrains empty glumes from developing into lemmas. J. Genet. Genomics 37: 101–115 [DOI] [PubMed] [Google Scholar]
- Honigberg S.M., Purnapatre K. (2003). Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J. Cell Sci. 116: 2137–2147 [DOI] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Koubova J., Menke D.B., Zhou Q., Capel B., Griswold M.D., Page D.C. (2006). Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. USA 103: 2474–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lee S., Jeon J.S., Jung K.-H., An G. (1999). Binary vectors for efficient transformation of rice. J. Plant Biol. 42: 310–316 [Google Scholar]
- Li S., Lauri A., Ziemann M., Busch A., Bhave M., Zachgo S. (2009). Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21: 429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Marston A.L., Amon A. (2004). Meiosis: cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 5: 983–997 [DOI] [PubMed] [Google Scholar]
- Mercier R., Armstrong S.J., Horlow C., Jackson N.P., Makaroff C.A., Vezon D., Pelletier G., Jones G.H., Franklin F.C.H. (2003). The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130: 3309–3318 [DOI] [PubMed] [Google Scholar]
- Mercier R., Vezon D., Bullier E., Motamayor J.C., Sellier A., Lefèvre F., Pelletier G., Horlow C. (2001). SWITCH1 (SWI1): A novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev. 15: 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmu J., Bush M.J., DeLong C., Li S., Xu M., Khan M., Malcolmson C., Fobert P.R., Zachgo S., Hepworth S.R. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 154: 1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K., Cheng Z.K., Ouyang S., Talbert P.B., Kim M., Jones K.M., Henikoff S., Buell C.R., Jiang J.M. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36: 138–145 [DOI] [PubMed] [Google Scholar]
- Ndamukong I., Abdallat A.A., Thurow C., Fode B., Zander M., Weigel R., Gatz C. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50: 128–139 [DOI] [PubMed] [Google Scholar]
- Nonomura K.I., Eiguchi M., Nakano M., Takashima K., Komeda N., Fukuchi S., Miyazaki S., Miyao A., Hirochika H., Kurata N. (2011). A novel RNA-recognition-motif protein is required for premeiotic G1/S-phase transition in rice (Oryza sativa L.). PLoS Genet. 7: e1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K.I., Miyoshi K., Eiguchi M., Suzuki T., Miyao A., Hirochika H., Kurata N. (2003). The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K.I., Morohoshi A., Nakano M., Eiguchi M., Miyao A., Hirochika H., Kurata N. (2007). A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K.I., Nakano M., Fukuda T., Eiguchi M., Miyao A., Hirochika H., Kurata N. (2004). The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16: 1008–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R.G. (1971). Cytological studies of ameiotic and normal maize with reference to premeiotic pairing. Chromosoma 35: 233–246 [Google Scholar]
- Pawlowski W.P., Sheehan M.J., Ronceret A. (2007). In the beginning: the initiation of meiosis. Bioessays 29: 511–514 [DOI] [PubMed] [Google Scholar]
- Pawlowski W.P., Wang C.J.R., Golubovskaya I.N., Szymaniak J.M., Shi L., Hamant O., Zhu T., Harper L., Sheridan W.F., Cande W.Z. (2009). Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proc. Natl. Acad. Sci. USA 106: 3603–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M., Marimuthu M.P.A., Siddiqi I. (2008). Gamete formation without meiosis in Arabidopsis. Nature 451: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Rouhier N., Lemaire S.D., Jacquot J.P. (2008). The role of glutathione in photosynthetic organisms: Emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59: 143–166 [DOI] [PubMed] [Google Scholar]
- Shao T., Tang D., Wang K., Wang M., Che L., Qin B., Yu H., Li M., Gu M., Cheng Z. (2011). OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol. 156: 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi I., Ganesh G., Grossniklaus U., Subbiah V. (2000). The dyad gene is required for progression through female meiosis in Arabidopsis. Development 127: 197–207 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Cande W.Z. (1992). Ameiotic, a gene that controls meiotic chromosome and cytoskeletal behavior in maize. Dev. Biol. 154: 226–230 [DOI] [PubMed] [Google Scholar]
- Sun Y.J., Hord C.L.H., Chen C.B., Ma H. (2007). Regulation of Arabidopsis early anther development by putative cell-cell signaling molecules and transcriptional regulators. J. Integr. Plant Biol. 49: 60–68 [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V., Evans M.M.S. (2003). Unique features of the plant life cycle and their consequences. Nat. Rev. Genet. 4: 369–379 [DOI] [PubMed] [Google Scholar]
- Wang K., Tang D., Wang M., Lu J., Yu H., Liu J., Qian B., Gong Z., Wang X., Chen J., Gu M., Cheng Z. (2009). MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J. Cell Sci. 122: 2055–2063 [DOI] [PubMed] [Google Scholar]
- Wang M., Wang K.J., Tang D., Wei C.X., Li M., Shen Y., Chi Z.C., Gu M.H., Cheng Z.K. (2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Z.A., Zhang D.B. (2009). From Arabidopsis to rice: Pathways in pollen development. J. Exp. Bot. 60: 1479–1492 [DOI] [PubMed] [Google Scholar]
- Wolgemuth D.J. (2006). Making the commitment to meiosis. Nat Genet 38: 1362–1363 [DOI] [PubMed] [Google Scholar]
- Xing S.P., Lauri A., Zachgo S. (2006). Redox regulation and flower development: A novel function for glutaredoxins. Plant Biol (Stuttg) 8: 547–555 [DOI] [PubMed] [Google Scholar]
- Xing S.P., Rosso M.G., Zachgo S. (2005). ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132: 1555–1565 [DOI] [PubMed] [Google Scholar]
- Xing S.P., Zachgo S. (2008). ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 53: 790–801 [DOI] [PubMed] [Google Scholar]
- Yu H.X., Wang M., Tang D., Wang K.J., Chen F.L., Gong Z.Y., Gu M.H., Cheng Z.K. (2010). OsSPO11-1 is essential for both homologous chromosome pairing and crossover formation in rice. Chromosoma 119: 625–636 [DOI] [PubMed] [Google Scholar]
- Zhang D.B., Wilson Z.A. (2009). Stamen specification and anther development in rice. Chin. Sci. Bull. 54: 2342–2353 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.