This work finds that the photosystem II (PSII) assembly factor PratA from the cyanobacterium Synechocystis is involved in efficient delivery of manganese to PSII and is organized in distinct structures connecting plasma and thylakoid membranes. It proposes that initial steps of PSII assembly, including its preloading with manganese, take place at these PratA-dependent biogenesis centers.
Abstract
In the cyanobacterium Synechocystis sp PCC 6803, early steps in thylakoid membrane (TM) biogenesis are considered to take place in specialized membrane fractions resembling an interface between the plasma membrane (PM) and TM. This region (the PratA-defined membrane) is defined by the presence of the photosystem II (PSII) assembly factor PratA (for processing-associated TPR protein) and the precursor of the D1 protein (pD1). Here, we show that PratA is a Mn2+ binding protein that contains a high affinity Mn2+ binding site (Kd = 73 μM) and that PratA is required for efficient delivery of Mn2+ to PSII in vivo, as Mn2+ transport is retarded in pratA−. Furthermore, ultrastructural analyses of pratA− depict changes in membrane organization in comparison to the wild type, especially a semicircle-shaped structure, which appears to connect PM and TM, is lacking in pratA−. Immunogold labeling located PratA and pD1 to these distinct regions at the cell periphery. Thus, PratA is necessary for efficient delivery of Mn2+ to PSII, leading to Mn2+ preloading of PSII in the periplasm. We propose an extended model for the spatial organization of Mn2+ transport to PSII, which is suggested to take place concomitantly with early steps of PSII assembly in biogenesis centers at the cell periphery.
INTRODUCTION
Oxygenic photosynthesis supplies the energy for production of most of the biomass on earth. The underlying light-driven photosynthetic electron transport is mediated by multiprotein/pigment complexes (i.e., photosystem II [PSII], the cytochrome b6f complex, and photosystem I), which reside within the thylakoid membrane (TM) system of cyanobacteria, algae, and plants. Electron flow is initiated at PSII, which serves as a light-driven water-plastoquinone oxidoreductase producing oxygen as a by-product of electron extraction from water. The structure of cyanobacterial PSII has been resolved at high resolution and is known to comprise 17 transmembrane protein subunits, three peripheral proteins, 35 chlorophylls, and several additional cofactors, including the catalytic machinery required for water splitting (Ferreira et al., 2004; Yano et al., 2006; Kern et al., 2007; Guskov et al., 2009; Umena et al., 2011). This machinery, the water-oxidizing complex (WOC), is localized on the lumenal side of PSII and contains one calcium atom and four atoms of the transition metal Mn, which are complexed by the D1 and CP43 subunits of PSII (Ferreira et al., 2004; Barber, 2008; Umena et al., 2011). The five metal atoms were found to be linked by five oxygen atoms, and four additional water molecules bind to the Mn4Ca cluster. It is proposed that some of these serve as substrates for the generation of dioxygen (Umena et al., 2011).
The question of how Mn is transported to PSII and assembled into the Mn4Ca cluster has been the subject of intensive research. In the cyanobacterium Synechocystis sp PCC 6803 (Synechocystis 6803), earlier studies have shed much light on the uptake of Mn (as Mn2+; Bartsevich and Pakrasi, 1995, 1996) and assembly/photoactivation of the Mn cluster (Cheniae and Martin, 1971; Tamura and Cheniae, 1987; Zaltsman et al., 1997; Hwang and Burnap, 2005). However, the mechanisms and components involved in transport of Mn to PSII have remained elusive. Two distinct systems for cellular Mn uptake have been described, the so-called MntABC transporter, and a second pathway whose components are yet unidentified (Bartsevich and Pakrasi, 1995, 1996). Moreover, Synechocystis 6803 can store Mn efficiently in the periplasmic space of the cell (Keren et al., 2002). Accumulation occurs rapidly upon transfer of cells to Mn-containing medium and is regulated by the rate of photosynthetic electron transport in an unknown manner (Bartsevich and Pakrasi, 1995, 1996; Keren et al., 2002). This pool may function as a reservoir to keep levels of intracellular Mn constant; however, it is still unknown how Mn is transported from this pool to PSII. Only one periplasmic Mn binding protein (MncA) has been described to date (Tottey et al., 2008), but its precise function remains to be elucidated. In vascular plants, the extrinsic PSII proteins PsbP and PsbO have been reported to bind Mn (Abramowicz and Dismukes, 1984; Bondarava et al., 2007), but these proteins help to stabilize the Mn cluster and do not transport Mn to the WOC (Roose et al., 2007).
Compared with the detailed structural picture of functional PSII, less information is available on its biogenesis. Nevertheless, it is well established that early steps of assembly of PSII subunits include the formation of distinct transient precomplexes (Nixon et al., 2010). Assembly starts with the reaction center proteins D2 and D1, which, together with the PsbE, PsbF, and PsbI subunits, constitute the first detectable intermediate, the so-called RC complex. Upon successive attachment of the inner antennae proteins CP47 and CP43, RC47 and RCC1 core complexes are formed, respectively. At the RCC1 stage, the Mn cluster is photoactivated and thus represents the first assembly intermediate capable of oxygen evolution (Cheniae and Martin, 1971; Becker et al., 2011). Finally, PSII undergoes dimerization and higher order organization within the TM (Mullineaux, 2008).
Several factors have been identified that participate in PSII biogenesis in Synechocystis 6803, including YCF48, a homolog of HCF136 from Arabidopsis thaliana (Plücken et al., 2002; Nickelsen et al., 2007; Komenda et al., 2008; Mulo et al., 2008; Nixon et al., 2010). YCF48 assists in early PSII assembly steps, as it was found to interact directly with the pD1 protein (precursor of D1) and was suggested to be involved in stabilizing newly synthesized pD1 (Komenda et al., 2008). Another factor with a function during early steps of cyanobacterial PSII biogenesis is represented by PratA (for processing-associated TPR protein), a member of the tetratricopeptide repeat (TPR) protein family (Klinkert et al., 2004; Schottkowski et al., 2009a). These proteins contain repetitive motifs of 34 amino acids and are known to mediate protein–protein interactions (Blatch and Lässle, 1999; Main et al., 2005). Consistent with this, PratA was shown to bind directly to the C terminus of D1 and affects its processing by the endoprotease CtpA (Klinkert et al., 2004; Schottkowski et al., 2009a). PratA occurs in two forms: (1) attached to the membrane via its interaction with D1, and (2) as a soluble complex of ~200 kD in the periplasm. Interestingly, although no obvious PratA homologs can be found in vascular plants or green algae (Klinkert et al., 2004), recently, two proteins from Arabidopsis and Chlamydomonas reinhardtii have been characterized that possess similar properties as they were both found to interact directly with D1 and belong to the family of TPR proteins (Peng et al., 2006; Park et al., 2007). This raises the possibility that, despite the apparent lack of sequence similarity to PratA, the role of PratA in PSII biogenesis is conserved in plants and algae.
This periplasmic localization of PratA raises the question on the spatial organization of the PSII assembly process. In cyanobacteria, TMs represent an internal membrane system that is distinct from the cellular envelope formed by the outer membrane and the plasma membrane (PM) enclosing the periplasmic space. Whereas this overall organization is well established, it is unclear how and where the biogenesis of the TM system takes place (Zak et al., 2001; Liberton and Pakrasi, 2008; Mullineaux, 2008; Nixon et al., 2010). Especially, the question of whether direct connections exist between the PM and the TM has been controversial for several years (Liberton et al., 2006; van de Meene et al., 2006; Nickelsen et al., 2011). Earlier membrane fractionation studies in Synechocystis 6803 led to the proposal that assembly of photosynthetic complexes (especially PSII) is initiated at PMs; subsequently, precomplexes are transported to TMs via vesicles or transient fusion of PMs and TMs (Zak et al., 2001; Schneider et al., 2007; Nickelsen et al., 2011). Furthermore, an intermediate membrane subfraction was described and characterized, which is defined by the presence of PratA and was therefore named PDM (for PratA-defined membrane; Schottkowski et al., 2009a; Rengstl et al., 2011). It was speculated that PDMs might resemble PM/TM convergence sites and that they could additionally be identical to previously described thylakoid centers (Hinterstoisser et al., 1993; van de Meene et al., 2006; Nickelsen et al., 2011). Moreover, PDMs have been shown to accumulate the chlorophyll a precursor molecule chlorophyllide a as well as several other PSII assembly factors in a PratA-dependent manner, suggesting that PDMs harbor a network for TM biogenesis where initial steps of PSII assembly take place (Rengstl et al., 2011).
Using Synechocystis 6803 as a model system, here, we focus on two questions: How is Mn delivered to PSII, and where does TM biogenesis originate? Our results demonstrate that both aspects are tightly coupled to the function and localization of PratA. We found that PratA participates in preloading D1 with Mn2+ already during initial PSII assembly steps. Moreover, both markers for initial PSII biogenesis (i.e., PratA and pD1) could be localized to distinct clusters where TMs converge. Taken together, our data allow the proposal of a model on the spatiotemporal organization of TM biogenesis.
RESULTS
The D1 Interaction Partner PratA Influences Mn Homeostasis
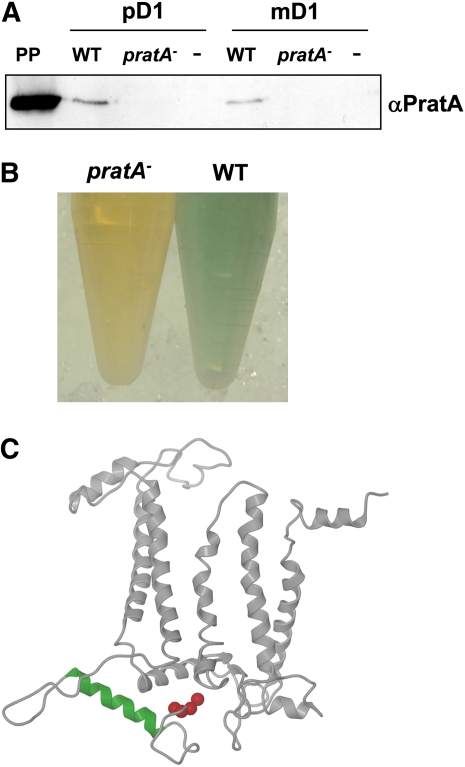
A variety of factors have been characterized in recent years that are required for regulation of PSII assembly (Mulo et al., 2008; Nixon et al., 2010). One of these proteins is PratA, which was previously found to interact directly with the core reaction center protein D1 that plays a major role in complexing the Mn cluster required for oxidation of water (Schottkowski et al., 2009a). This direct binding between PratA and D1 had been shown by yeast two-hybrid and glutathione S-transferase (GST) pull-down experiments, revealing that recombinant PratA (rPratA) binds to the soluble parts of both mature (mD1) and precursor D1 (pD1) C-terminal regions (Schottkowski et al., 2009a). To substantiate further the PratA–D1 interaction, we performed pull-down experiments using heterologously expressed mD1 and pD1 C termini coupled to GST-agarose as applied before (Schottkowski et al., 2009a), but incubated these with isolated periplasmic proteins from both the wild type and a pratA− mutant (Figure 1A). With this approach, native PratA was successfully pulled down from wild-type periplasm by both D1 versions, whereas no PratA signal was detected upon incubation with the pratA− periplasm or when an empty column without bound mD1/pD1 C termini was used (Figure 1A). This verifies the interaction between PratA and D1 in a more physiological system (using native PratA) than applied before.
Figure 1.
Native PratA Binds to the Mature D1 C Terminus Near the Mn Cluster.
(A) Pull-down experiment of GST-mD1 (mature D1) and GST-pD1 (precursor of D1) bound to GST-agarose and incubation with isolated periplasm from Synechocystis wild-type (WT) and pratA− cells. Bound proteins were eluted (see Methods) and subjected to immunoblotting with αPratA. A negative control (−) without mD1/pD1 proteins bound to GST-agarose is included. The first lane includes 10 μg isolated periplasm (PP) without further treatment.
(B) Periplasm samples from Synechocystis wild-type and pratA− cells.
(C) The binding site for PratA on D1 (amino acids 314 to 328; green helix; Schottkowski et al., 2009a) lies close to the residues that form the Mn cluster (His-332, Glu-333, His-337, Asp-342, and Ala-344; red balls; Ferreira et al., 2004; Barber, 2008). The three-dimensional structure of D1 was visualized with Pymol (http://pymol.sourceforge.net, version 0.99, based on the Protein Data Bank file 3BZ1).
Interestingly, after isolation and concentration of periplasm, the color of wild-type periplasm was found to be greenish, whereas the color of the pratA− periplasm appeared yellow (Figure 1B). Since putative pigments could not be extracted by organic solvents, we speculated that the alteration in color might be due to differences in transition metal composition. We were especially interested in Mn because the binding site for PratA on D1 is located in close proximity to those amino acid residues of D1 that are involved in complexing the Mn cluster (Ferreira et al., 2004; Barber, 2008; Schottkowski et al., 2009a; Figure 1C). Measurements of the amounts of Mn and Fe by atomic absorption spectrometry revealed that the level of Mn in pratA− periplasm was reduced by almost two-thirds to 36 ± 7% (sd) relative to the wild type, whereas the Fe concentration was unaltered (99 ± 18%; sd). Hence, the loss of PratA affects the periplasmic concentration of Mn.
PratA Is a Mn Binding Protein
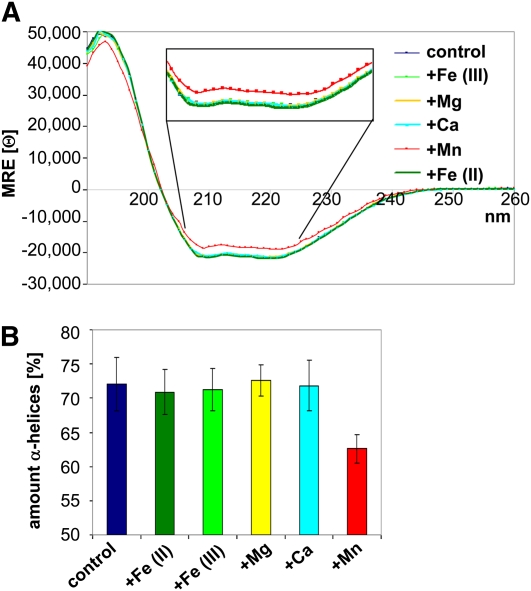
We next tested whether rPratA itself has the capacity to bind Mn ions (see Supplemental Figure 1 online). We first performed circular dichroism (CD) measurements of PratA in the absence or presence of either 1 mM Mn2+, 1 mM Fe3+, 1 mM Fe2+, 1mM Mg2+, or 1 mM Ca2+. The CD data obtained for rPratA alone indicated a structure consisting of 72% α-helices, 5% β-sheets, and 23% turns/coils (Figures 2A and 2B). The high amount of α-helices reflects the nine helix-turn-helix-folded TPR motifs present in PratA that constitute the majority of this protein (Klinkert et al., 2004; Main et al., 2005). When rPratA was incubated with 1 mM Mn2+ prior to the measurement, the α-helical fraction fell from 72% (without Mn2+) to 62%, and a concomitant increase of turns/coils (from 23 to ~30%) was observed (Figures 2A and 2B). This could also be judged by the mean residue ellipticity at 222 nm (Θ222) that was altered to Θ222 = −18762.0 in contrast with Θ222 = −20955.4 without Mn2+. These data indicate a conformational change of rPratA in the presence of Mn2+ and suggest that Mn2+ might be directly bound by rPratA. By contrast, no obvious changes in secondary structure were observed upon incubation with 1 mM Fe3+, 1 mM Fe2+, 1mM Mg2+, or 1 mM Ca2+ (Figures 2A and 2B).
Figure 2.
PratA Undergoes Changes in Secondary Structure upon Incubation with Mn.
(A) CD spectroscopy of recombinant PratA in the absence (dark-blue curve) and presence of 1 mM MnCl2 (red), FeCl3 (light green), MgCl2 (yellow), CaCl2 (light blue), or FeCl2 (dark green). MRE [Θ] represents mean residue ellipticity in degrees cm2 dmol−1 residue−1. The figure shows one representative graph of three independent experiments.
(B) Quantification of the α-helical content from the CD data using the CDSSTR program (protein reference set 7) obtained from the DichroWeb server (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml). Values shown are means (±sd) of three independent experiments.
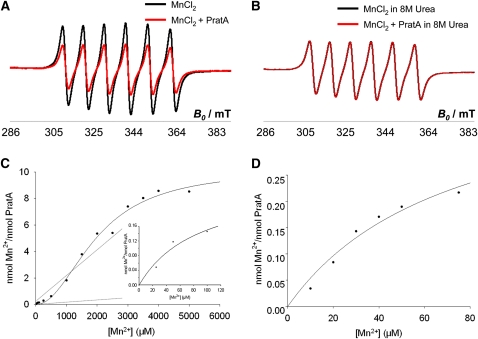
To test directly whether rPratA binds Mn2+, electron paramagnetic resonance (EPR) measurements were conducted. The room temperature (298.15K) spectra of 500 μM Mn2+ recorded in the presence or absence of 100 μM rPratA showed a six-line hyperfine pattern typical of Mn-(H2O)62+ species. In the presence of rPratA, a significantly smaller EPR signal amplitude was observed in comparison to a rPratA-free sample containing identical Mn2+ concentration in the same buffer (Figure 3A). The reduction of the signal amplitude of the Mn2+ spectrum suggests decreased amounts of free Mn2+ in solution due to Mn2+ binding to the protein (Reed and Cohn, 1970; Reed and Markham, 1984; Sen et al., 2006; Hayden and Hendrich, 2010). This effect was abolished upon denaturation of rPratA by 8M urea, indicating specific binding of Mn2+, as folding of the protein is crucial for Mn2+ interaction (Figure 3B). To determine binding stoichiometry and affinities of rPratA, Mn2+ titration experiments were performed and the fraction of bound Mn2+ was calculated from the amplitude of the lowest field transition and plotted against total Mn2+ concentrations (Figure 3C). A maximum of eight Mn2+ ions were found to bind per rPratA molecule. The entire titration curve was shown to exhibit a sigmoid shape indicative of binding of Mn2+ to multiple sites. The data suggest the existence of a high-affinity Mn2+ binding site (Kd1 ~90 μM; Figure 3C, detail) and multiple low-affinity sites (Kd2 > 1 mM; Figure 3C, overview). Because Kd1 could not be precisely determined by the applied EPR method due to technical resolution limitations, we further analyzed it by filter binding assays. In this approach, rPratA was incubated with 10 to 75 μM 54Mn2+, the amount of 54Mn2+ bound to rPratA was counted, and the number of 54Mn2+ per rPratA was calculated from the values obtained from 54Mn2+ in the absence of protein. This allowed us to determine Kd1 to 73 ± 31 μM (Figure 3D).
Figure 3.
PratA Is a Mn Binding Protein and Contains High- and Low-Affinity Binding Sites.
(A) and (B) EPR analyses of 500 μM MnCl2 ± 100 μM PratA in 50 mM Tris, pH 8, and 150 mM NaCl (A) and 500 μM MnCl2 ± 100 μM PratA in Tris/NaCl + 8 M urea (B).
(C) EPR experiment for determination of stoichiometry and binding constant of PratA-Mn. The data show two different binding modes of Mn2+ to PratA, one high-affinity (detail), and several low-affinity binding sites (large graph). The data were fitted using SigmaPlot 11 software.
(D) Filter binding assay for exact determination of the high-affinity binding site. Recombinant PratA was incubated with 10 to 75 μM 54Mn2+, the amount of Mn2+ bound to PratA was measured, and the number of Mn2+ per PratA was calculated from the values obtained from 54Mn2+ without addition of protein.
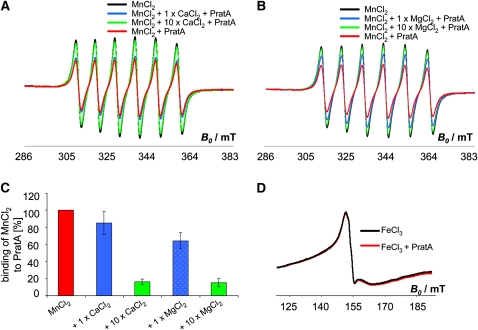
To analyze the specificity of Mn2+ binding, we performed competition experiments, in which 500 μM CaCl2 (equimolar amount) or 5 mM CaCl2 (10-fold excess) were incubated together with 500 μM MnCl2 and 100 μM rPratA prior to EPR analysis. The results suggest that using equimolar amounts, only a minor fraction of Mn2+ could be replaced by Ca2+, since 85.2% of Mn2+ remained bound to PratA (Figures 4A and 4C; fraction of bound Mn2+ was calculated from the amplitude relative to that of total Mn2+ bound without competitor). Considering that a maximum of eight Mn2+ ions were found to bind per rPratA, seven Mn2+ molecules remain attached to rPratA under these conditions. Even upon incubation with 10-fold excess of Ca2+, 16.2% of Mn2+ stayed associated with rPratA, which roughly corresponds to one out of the total eight Mn2+ ions bound (Figures 4A and 4C). Similar effects were observed in competition experiments with MgCl2, although the competition effect was slightly more pronounced in this case (Figures 4B and 4C). Additionally, the same competition experiment was performed using 5 mM MnCl2 (instead of 500 μM) and 5 mM CaCl2 or MgCl2 or 50 mM CaCl2 or MgCl2, respectively. Comparable results were obtained with this approach, as nine Mn2+ ions were found to be attached to one PratA and 1 Mn2+ ion remained bound to PratA upon competition with 10-fold excess of Ca2+ or Mg2+ (percentage of Mn2+ that stays attached: 1× CaCl2, 82.3%; 10× CaCl2, 6.7%; 1× MgCl2, 78.3%; 10× MgCl2, 16.6%; see Supplemental Figure 2 online). Based on these data, it is proposed that rPratA specifically binds Mn2+ with a higher affinity than Ca2+ or Mg2+ and that even with 10-fold excess of Ca2+ or Mg2+, the high-affinity binding site seems to be specific for Mn2+, whereas the residual seven or eight Mn2+ ions are loosely attached to the low-affinity binding sites and, hence, can easily be substituted. Furthermore, in good agreement with the CD spectroscopic data, no evidence for Fe3+ binding to rPratA was obtained in low-temperature (104K) EPR spectra (Figure 4D). To exclude other possible reasons for the observed decay of the EPR signal amplitude (e.g., oxidation of paramagnetic Mn2+; Reaney et al., 2002), EPR measurements were performed on the same Mn2+/rPratA sample before and after the addition of 100 mM CaCl2. To exclude dilution effects caused by addition of CaCl2 to the Mn2+/PratA sample, in parallel an equal amount of buffer was subjected to a second Mn2+/PratA sample used for normalization purposes afterwards. Whereas in the presence of 100 μM rPratA, the signal amplitude decayed to 78.4%, the subsequent addition of CaCl2 returned the signal to 91.4% compared with the signal intensity of 5 mM MnCl2 free in solution (using the amplitude of the first peak of the EPR spectrum for calculation; see Supplemental Figure 3A online). Although the amplitude was not completely restored by the addition of CaCl2, which is likely due to the higher affinity of rPratA for Mn2+ than for Ca2+ (see above), the result suggests that the loss of EPR signal intensity in the presence of rPratA is due to Mn2+ binding rather than to changes in the oxidation state of Mn2+. In the latter case, the presence of CaCl2 would not lead to an EPR signal recovery. As an additional control to determine whether the protein specifically determines the amount of Mn2+ binding, we performed the PratA/Mn2+ binding experiment using 100 μM Mn2+ and varying concentrations of PratA (100, 50, and 25 μM). The results clearly show a linear increase of the amount of PratA-bound Mn2+ dependent on the protein concentration applied (29, 15, and 7 μM Mn2+ attached to PratA, respectively), demonstrating that in this range, the amount of bound Mn2+ is directly proportional to the PratA concentration applied (see Supplemental Figures 3B and 3C online).
Figure 4.
Specificity of Mn2+ Binding to PratA.
(A) and (B) Competition experiment using EPR analysis of 500 μM MnCl2 ± 100 μM PratA in Tris/NaCl + 500 μM (1×) or 5 mM (10×) CaCl2 (A) and 500 μM MnCl2 ± 100 μM PratA in Tris/NaCl + 500 μM (1×) or 5 mM (10×) MgCl2 (B).
(C) Quantification of MnCl2 bound to PratA obtained via EPR spectroscopy. The fraction of bound Mn2+ was calculated from the amplitudes relative to total Mn2+ bound without competitor (=100%; red bar).
(D) EPR analysis of 1 mM FeCl3 before (black curve) and after (red curve) the addition of 100 μM PratA.
To analyze whether Mn2+ binding can also be observed for native PratA, we used a Mn2+-loaded nitrilotriacetic acid column and isolated periplasmic Mn2+ binding proteins via affinity chromatography. Indeed, PratA could be extracted from periplasm by this method, but it did not bind to a Ni2+-containing column or a column not preloaded with metal ions, indicating that PratA isolated from periplasm can specifically bind Mn2+ (Figure 5). Quantification of the eluted PratA in relation to the amount of total periplasmic PratA used for the experiment revealed that 3.4% of PratA was actually precipitated by our approach. Thus, it is likely that the majority of periplasmic PratA might either be already bound to Mn2+ or is involved in complex formation with additional proteins and is thus not accessible for the pull-down assay. Nevertheless, the control samples clearly show the specificity of PratA binding to the Mn2+ column. Taken together, these results indicate that Mn2+ is directly and specifically bound by both recombinant and native PratA.
Figure 5.
Mn2+ Can Be Bound by Native PratA.
Metal-ion chromatography of periplasm from Synechocystis 6803 wild-type cells. Bound proteins were eluted (see Methods) and subjected to immunoblotting with αPratA. E(−), 50% of eluate from column not preloaded with cations (25 μL); E(Mn), 50% of eluate from Mn2+ column (25 μL); E(Ni), 50% of eluate from Ni2+ column (25 μL); FT, 10% of flow-through (20 μL) from Mn2+ column; L, load (periplasm, 10 μg protein); W, 10% of final wash (20 μL) from Mn2+ column.
PratA Functions in Transport of Mn2+ to D1 in Vivo
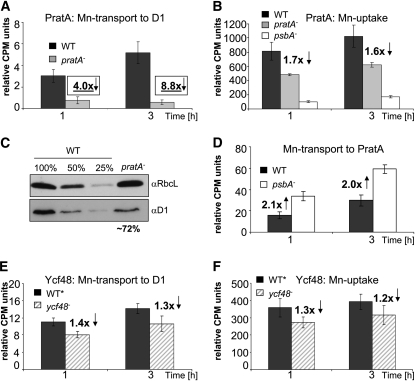
The data obtained so far raised the possibility that PratA might donate Mn2+ to the D1 protein of PSII in vivo. To investigate this hypothesis, we pulse-labeled wild-type and pratA− cells with 54Mn2+ for 1 or 3 h. Subsequently, PSII assembly intermediates were isolated from solubilized protein extracts by immunoprecipitation using an αD1 antibody. Analysis of precipitated 54Mn2+ should then reflect the amount of Mn2+ transported to and incorporated into de novo–synthesized PSII. When protein extracts from a psbA− mutant (TD41; Nixon et al., 1992) were used as a negative control, only minute amounts of background radioactivity were detected in D1-specific precipitates, which were then subtracted from measured values in wild-type and pratA− samples. Intriguingly, rates of incorporation of Mn2+ into PSII were clearly affected in pratA− cells. After 1 h of incubation with 54Mn2+, amounts of bound Mn2+ were reduced 4.0-fold compared with the wild type, and after 3 h, the effect was even more pronounced, resulting in an 8.8-fold reduction of precipitated radioactivity from the pratA− material (Figure 6A). To exclude the possibility that this reduction was solely due to lower cellular Mn2+ uptake rates in pratA−in parallel, levels of radioactivity in whole cells were assessed after incubation with 54Mn2+. After 3 h, the level of 54Mn2+ in the psbA− strain was 6.0-fold less than in the wild type (Figure 6B). This is consistent with earlier reports showing that cellular uptake of Mn2+ depends on photosynthetic activity (Bartsevich and Pakrasi, 1996; Keren et al., 2002). In pratA− cells, 54Mn2+ levels decreased only 1.6-fold, which is likely to be a secondary effect due to lower photosynthesis rates in pratA− (Bartsevich and Pakrasi, 1996; Klinkert et al., 2004; Figure 6B). To address this point further, we quantified the amount of D1 in pratA− using immunodetection with a αD1 antibody in total protein extracts from wild-type and pratA− cells. Previous analyses had revealed a severe reduction of D1 in pratA− in the absence of precise protein quantifications (Klinkert et al., 2004). Here, three independent protein extractions from pratA− were analyzed together with dilution series of wild-type proteins, and the D1 level in pratA- was calculated to be decreased to only 72 ± 8% (= 1.4-fold reduction) of the wild-type level under the growth conditions applied (Figure 6C). Hence, this moderate reduction of D1 amount and, thus, of cellular Mn2+ uptake cannot explain the drastically diminished Mn2+ incorporation into PSII in pratA− cells (8.8-fold reduction compared with the wild type), strongly suggesting that PratA is directly involved in delivery of Mn2+ to PSII. Additionally, we investigated the amount of 54Mn2+ taken up by PratA in the wild type compared with psbA−proposing that if PratA functions in transport of Mn2+ to D1, the metal ion should accumulate bound to PratA when lacking the target. For this purpose, wild-type, pratA−and psbA− cells were incubated with 54Mn2+ as described before; however, after subsequent protein extraction, immunoprecipitation was performed using a αPratA instead of αD1 antibody. After 3 h of incubation with 54Mn2+, indeed, an increase (twofold) of PratA-bound 54Mn2+ was observed in psbA− compared with the wild type (Figure 6D; the background values obtained for pratA− were subtracted from wild-type and psbA− values). This supports the hypothesis of PratA functioning as a Mn2+ transport protein to D1. If and how PratA also exchanges Mn2+ with the periplasmic Mn storage system remain to be determined (Keren et al., 2002).
Figure 6.
PratA Influences Mn Uptake and Transport to PSII in Vivo.
(A) Amounts of Mn delivered to D1 in wild-type and pratA− cells upon incubation of the cells with 54Mn2+ for 1 and 3 h, extraction of proteins, and immunoprecipitation with αD1. Radioactivity levels (in counts per minute [CPM]) measured in samples from psbA− cells (background) were subtracted from values for the wild type (WT) and pratA−.
(B) Uptake of 54Mn2+ into wild-type, pratA−and psbA− cells. For this, cells were used directly for measurement of 54Mn2+ with a scintillation counter.
(C) Immunoblot using αD1 of total protein extracts from the wild type and pratA−. For 100% of the wild type and pratA−10 μg proteins were loaded and the amount of D1 in pratA− was quantified (from three independent experiments) using Aida software (version 3.52.046). The same blot was probed with αRbcL as loading control.
(D) Amount of 54Mn2+ taken up by PratA in wild-type and psbA− cells, measured after incubation with 54Mn2+ for 1 and 3 h and subsequent protein extraction and immunoprecipitation with αPratA. Counts per minute of samples from pratA− cells (background) were subtracted from values for the wild type and psbA−.
(E) and (F) Transport (E) and uptake (F) of 54Mn2+ in wild-type* (Komenda et al., 2008) and ycf48− cells.
In (A) to (F), the levels of radioactivity detected after 1 and 3 h of incubation are expressed relative to the value measured immediately after addition of 54Mn2+ (0 h). Values shown are means (±sd) of four independent experiments.
As PratA is involved in maturation/assembly of D1 (Klinkert et al., 2004; Schottkowski et al., 2009a), we further aimed to test whether other PSII assembly mutants show similar effects in Mn2+ incorporation into PSII. To this end, we analyzed Mn2+ transport and uptake into ycf48−, a mutant deficient in YCF48, a factor that had been shown to be involved in early PSII assembly steps and that, like PratA, is a direct D1 interaction partner (Komenda et al., 2008). However, we detected only a minor reduction in both transport and uptake of Mn2+ (1.3- and 1.2-fold decrease after 3 h incubation with 54Mn2+, respectively) in ycf48−, indicating a specific role of PratA for Mn2+ delivery to PSII (Figures 6E and 6F).
PratA-Dependent Formation of Semicircle-Shaped Structures at the Cell Periphery
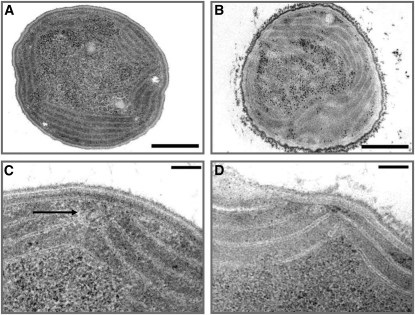
The spatial organization of TM biogenesis has been a matter of debate for several years. To gain further insights into the organization of Mn delivery to PSII, we analyzed the ultrastructure of Synechocystis 6803 wild-type and pratA− cells using transmission electron microscopy. Overview images of whole cells depicted clear changes in the overall membrane appearance between the wild type and pratA− (Figures 7A and 7B). In pratA−TMs appeared less compressed and the outer membrane and PM were less smooth and organized. A closer look at TM convergence sites at the periphery of cells using higher magnifications (110,000- to 140,000-fold) revealed structures of ~60 nm in diameter that are filled by a granular matrix coated with dense material in both wild-type and pratA− sections (Figures 7C and 7D). Most likely these structures represent cross sections of previously described so-called thylakoid centers (van de Meene et al., 2006). Interestingly, in wild-type cells, in some cases membranous semicircle-like structures surrounding thylakoid centers were observed which appeared to contact especially TMs and PMs (Figure 7C). At least eight of these semicircles located between the arcuated and arranged TM layers were found in 360 wild-type cells analyzed. It has to be considered that this structure was described two-dimensionally, meaning that in general only one section was analyzed per cell. Hence, not all of those regions could be detected. The relatively low number indicates that the semicircle-like structures are either located at a central place in the cell and that each cell contains only a few of them or that they are dynamic and form only transiently. However, the semicircles were not detected in any of 1006 analyzed pratA− cells (Figure 7D). To test further the error probability (i.e., to disclaim the null hypothesis “no difference between observed structures in wild-type and pratA− cells”), a Pearson’s χ2 test was performed (in dependence of n = 1 degrees of freedom; Zöfel, 1988). Comparing the expected ratio of 0.022 (wild-type cells, 8/360; expected for pratA− cells, 22.35/1006) of structures per cell with the observed ratio of those structures examined in pratA− cells (pratA− cells, 0/1006), we found a highly significant difference to that for the wild type (χ2 22.35, P value <0.001), indicating a very low error probability. Taken together, our data suggest that the semicircle-shaped structures are drastically reduced, if not lacking, in pratA−indicating that they form in a PratA-dependent manner.
Figure 7.
Ultrastructural Analyses of Wild-Type and pratA− Cells.
Electron microscopy pictures of a typical wild-type ([A] and [C]) and pratA− ([B] and [D]) Synechocystis cell. (A) and (B) show an overview with a magnification of 11,000-fold and (C) and (D) a detailed picture of the PM/TM interface (magnification 110,000-fold). Ultrathin sections (30 to 60 nm) of the cryofixed samples were stained with osmium tetroxide and poststained using lead citrate. The arrow in (C) marks the PratA-dependent semicircular structure. Bars = 500 nm (overview) and 100 nm (details), respectively.
PratA and pD1 Localize to Distinct Structures at the Cell Periphery
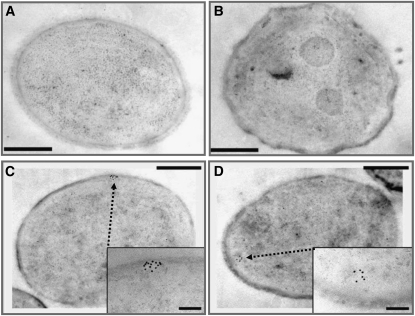
As a next step, we localized PratA within the cyanobacterial cell by performing immunogold labeling experiments using a specific αPratA antibody. No or only few randomly localized signals were detected upon incubation of Synechocystis 6803 wild-type sections without the primary antibody (followed by treatment with the gold-labeled anti-rabbit IgG only; Figure 8A, Table 1) or when sections of pratA− cells were treated with αPratA (Figure 8B, Table 1). However, upon incubation of wild-type sections with αPratA antibody, PratA could be localized to distinct clusters of ~100 nm in diameter at the cell periphery (Figures 8C and 8D). These clusters (defined by more than five gold particles) were detected in 79 out of 520 cells (=15.2%; Table 1), but they were almost completely missing in pratA− sections. Thus, we conclude that they resemble PDMs, and we named these structures “biogenesis centers.”
Figure 8.
PratA Localizes to Distinct Structures at the Cell Periphery.
Ultrathin sections of Synechocystis cells were incubated with αPratA (1:25, rabbit) prior to incubation with gold-conjugated goat anti-rabbit IgG.
(A) Negative control of the wild type without addition of αPratA.
(B) Synechocystis pratA− cell incubated with αPratA.
(C) and (D) Immunogold-labeled sections showing the PratA localization in distinct clusters (marked by arrows). Samples were analyzed without further contrast. Bars = 500 nm (overview) and 100 nm (details), respectively.
Table 1.
Summary of Immunogold Labeling Experiments
| Antibody | Strain | Total No. of Cells Analyzed | No. of Cells with Cluster of Gold Particles (n > 5) | Relative No. of Cluster-Containing Cells (%) |
| – | Wild type | 40 | 0 | 0.0 |
| αPratA | Wild type | 520 | 79 | 15.2 |
| αPratA | pratA− | 254 | 1 | 0.4 |
| αpD1 | Wild type | 50 | 10 | 20.0 |
| αpD1 | TD41 (psbA−) | 214 | 0 | 0.0 |
| αpD1 | pratA− | 235 | 24 | 10.2 |
–, No primary antibody was applied in the control.
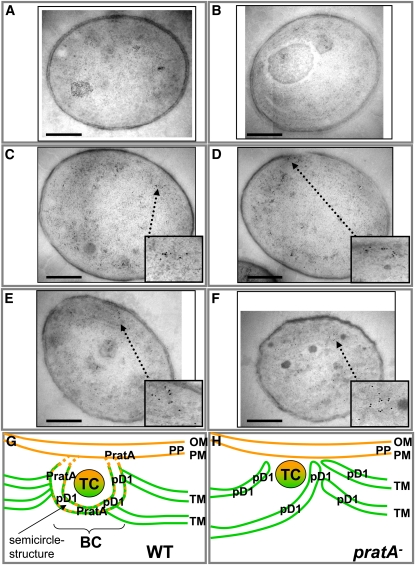
To substantiate this hypothesis further, we performed additional immunogold labeling experiments using an antibody against the pD1 precursor protein, which represents the PratA interaction partner and serves as a second marker for initial PSII assembly steps. Whereas the negative controls showed only few signals that were equally distributed throughout the cells (Figures 9A and 9B, Table 1), pD1 was found to localize to distinct clusters in the wild type, similar to PratA. Although a colocalization of PratA and pD1 on the same sample could not be achieved due to the fact that both antibodies require the same secondary antibody (anti-rabbit), the similar sizes of the clusters (diameter of ~100 nm) and the localization at the convergence sites of the cells in the wild type suggest that both proteins accumulate at the same structures (Figures 9C and 9D, Table 1). Interestingly, in pratA−pD1 was still detected in clusters at the cell periphery, but their number was found to be approximately twofold diminished (Figures 9E and 9F, Table 1). A thorough analysis of the pD1 clusters in the wild type compared with pratA− using as parameters (1) the area of the cluster, (2), the cluster perimeter, (3) the length of the minor and major axes of a suggested ellipse, and (4) an estimation value for the circularity or noncircularity (values between 1 = circle and 0 = other structures) revealed no obvious differences (see Supplemental Figure 4 online). The parameter of circularity is a calculated mean value that represents the Gaussian distribution of circularity of clusters in a selection of cells. If this value comes close to 1, this means that the clusters adopt on average a more circle-like structure. By contrast, a value of nearly 0 indicates random cluster shapes. For the cluster, two different definitions were used (i.e., an accumulation of n ≥ 5 or n ≥ 7 gold particles, respectively; n ≥ 7, n = 11 for anti pD1-immunolabeled structures in wild-type cells, n = 12 for those in pratA− cells), both revealing no obvious changes in pratA− compared with the wild type. Additionally, two ways of analysis were applied, one connecting the particles forming a single cluster and one where the overall shape of the particles building each cluster was plotted by hand. Again, both variations did not lead to the recognition of alterations in pratA− compared with the wild type. However, in pratA−the gold particles adjacent to the clusters often appeared to form streak-like structures. This observation, together with the reduced overall amount of pD1 organized in clusters (20.0% in wild-type cells and 10.2% in pratA− cells; Table 1) corresponds to the loss of semicircle-like structures observed in the ultrastructure of pratA− cells (Figures 7C and 7D), supporting the idea that both PratA and pD1 are colocalizing in these semicircles in the wild type and that this spatial organization is lost in the absence of PratA (Figures 9G and 9H).
Figure 9.
Localization of pD1.
Ultrathin sections of Synechocystis cells were incubated with αpD1 (1:50, rabbit) prior to incubation with gold-conjugated goat anti-rabbit IgG.
(A) Control of the wild type without αpD1.
(B) Synechocystis psbA− cell incubated with αpD1.
(C) to (F) Immunogold-labeled electron microscopy pictures depicting the pD1 localization in the wild type ([C] and [D]) and pratA− ([E] and [F]). Samples were analyzed without further contrast. Bars = 500 nm.
(G) and (H) Schematic model depicting the structure of TM convergence sites and the localization of PratA and pD1 in the wild type and pratA−. BC, biogenesis center; OM, outer membrane; PP, periplasm; TC, thylakoid center; WT, wild type. For further details, see text.
[See online article for color version of this figure.]
DISCUSSION
PratA Is a Mn2+ Binding Protein
In this study, we assigned a yet unidentified function to the periplasmic TPR protein PratA from Synechocystis 6803. The data strongly suggest that it works as a Mn2+ binding and transport protein that delivers Mn2+ ions directly and efficiently to PSII. Our conclusions are based on the findings that (1) PratA inactivation affects intercellular Mn levels, (2) recombinant as well as native PratA specifically bind Mn2+ ions, and (3) Mn2+ incorporation into PSII is reduced in a pratA− mutant.
EPR measurements and filter binding assays revealed at least one high-affinity Mn2+ binding site within PratA with a binding constant of Kd1 = 73 μM. Based on titration and competition experiments with Ca2+ and Mg2+ ions, we postulate a stoichiometry of 1 Mn2+:1 PratA at this site (Figures 3 and 4). The observed Kd1 value is in the range of affinities of other Mn2+ binding proteins, such as the Mn-dependent catalase MnCat (Kd = 40 μM) or the oxidative stress protecting protein SsDPS (Kd = 48 μM) (Meier et al., 1996; Crowley et al., 2000; Pierce et al., 2003; Hayden and Hendrich, 2010). Nevertheless, much higher binding constants for Mn2+ to protein have been reported, for instance, in case of MnSOD and PsbP proteins (Mizuno et al., 2004; Bondarava et al., 2007). It has to be taken into account, however, that metal binding to these proteins is irreversible, in contrast with PratA, for which we postulate a function in Mn2+ transport and, thus, transient Mn2+ binding. Consequently, the observed binding constant of PratA is in good agreement with its proposed function. We further postulate that the high-affinity Mn2+-specific binding site cannot be occupied by Ca2+, Mg2+, or Fe3+, which provides an explanation why changes in the secondary structure of PratA using CD spectroscopy have only been observed in the presence of Mn2+ ions (Figure 2). In addition, low-affinity Mn2+ binding sites were determined (Kd2 > 1 mM), at which Mn2+ is loosely and probably unspecifically attached to the surface of PratA. This is suggested by the above-mentioned competition experiments, which showed that the weakly attached Mn2+ ions could be replaced by Ca2+ or Mg2+, whereas the tightly bound Mn2+ remained associated with PratA, even in the presence of 10-fold excess of Ca2+ or Mg2+. Thus, it remains questionable whether Mn2+ is bound to the low-affinity binding sites in vivo under physiological conditions. Interestingly, PratA is rich in Asp and Glu residues and has a pI of ~5.1. As it is known that Mn2+ prefers such ligands, it seems likely that these residues are involved in loose attachment of Mn2+ to the surface of PratA. A similar effect has been described for PsbP, for which a stoichiometry of 10 Mn2+:1 PsbP has been found; however, further analyses revealed two different binding modes as well, as most Mn2+ ions were also merely bound with a low affinity to the Asp/Glu-containing surface of PsbP (Bondarava et al., 2007). The exact localization of the high-affinity Mn2+ binding site for PratA including the amino acids involved has to be determined in future work. In addition, the relation of PratA to another recently described periplasmic Mn binding protein, named MncA (Tottey et al., 2008), remains to be further investigated. This cupin-folded protein is the only other Mn binding protein localized in the periplasm described so far. However, the function of this protein remains elusive, and a potential connection to PSII assembly has not been analyzed yet. Taken into account that MncA was suggested to be the principal periplasmic Mn binding protein (Tottey et al., 2008), and knowing that Synechocystis 6803 is able to store Mn very efficiently in high concentrations in the periplasm in a yet unknown manner (Bartsevich and Pakrasi, 1996; Keren et al., 2002), it can be hypothesized that MncA might rather function in Mn storage than in delivery of Mn to PSII.
Delivery of Mn2+ to PSII
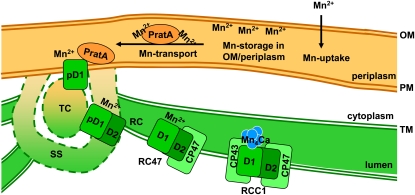
The radioactive pulse labeling experiments (Figure 6) revealed that the kinetics of Mn2+ incorporation into PSII are affected in a pratA− mutant background. Furthermore, the parallel analysis of the PSII assembly mutant ycf48− verified that inefficient Mn2+ transport to PSII in pratA− is not a secondary effect of distorted PSII assembly, but PratA specific. Hence, the question arises at which stage of PSII assembly Mn2+ is delivered to PSII by PratA. Earlier studies revealed that PratA associates only with very early PSII assembly intermediates, probably already with newly membrane inserted pD1 protein (Schottkowski et al., 2009a; Rengstl et al., 2011). Furthermore, a pratA− mutant showed defects in C-terminal processing of D1 (Klinkert et al., 2004), which is required for assembly of the Mn4Ca cluster in the WOC. Processing occurs at the level of RC complex formation of PSII; thus, this might represent the intermediate PSII assembly stage at which Mn2+ is delivered by PratA, resulting in a preloading of D1 with Mn2+. After the transfer, PratA leaves the RC complex, and the inner antennae subunits CP47 and CP43 attach to this intermediate in a stepwise fashion to form RCC1 complexes (Rengstl et al., 2011). At this stage, all ligands for complexing the four Mn ions are available and photoactivation of the WOC takes place (Figure 10; Becker et al., 2011).
Figure 10.
Schematic Model of Mn Delivery to PSII.
Mn is first taken up into the periplasm (as Mn2+). Subsequently, it can be stored in the periplasm or is transported to PSII via the assistance of PratA. It is then bound by D1, which is thus preloaded with Mn2+ to assemble a functional Mn cluster (depicted in blue) during later steps of PSII biogenesis. OM, outer membrane; RC, reaction complex; SS, semicircular structure; TC, thylakoid center. Since the precise architecture of the biogenesis centers (= TC + SS) remains to be resolved at higher resolution, it is depicted with broken lines. For further details, see text.
[See online article for color version of this figure.]
In pratA−, the Mn2+ transport rate to PSII was found to be strongly reduced; nevertheless, Mn2+ was still incorporated by PSII, allowing photoautotrophic growth of mutant cells although with reduced rates compared with the wild type (Klinkert et al., 2004). Therefore, complementing Mn2+ delivery systems to PSII apart from the periplasmic PratA-assisted transport have to exist; alternatively, Mn2+ might reach D1 without the coupling to transport proteins. However, PratA seems to be responsible for a direct Mn2+ transport to PSII, and it drastically increases the transport efficiency. To date, at least two different systems for cellular Mn uptake have been described (i.e., an ABC transporter [Mnt system] and a yet unidentified system) (Bartsevich and Pakrasi, 1995,1996). The presence of a Mn transporter system in the PM suggests that Mn is also supplied from the cytoplasmic compartment of the cell in a yet unknown way. PratA-mediated capture of Mn2+ by D1 already at the periplasm would, however, be an elegant way to bypass the need for active transport of Mn2+ across the PM to the thylakoids via the Mnt system. This is likely to be crucial when the cell faces a high demand for Mn, for instance, during/after cell division when substantial amounts of new TMs have to be synthesized. Moreover, periplasmic Mn2+ loading of PSII would prevent nonspecific oxidation of Mn2+ during subsequent transfer processes. The exact mechanism of Mn2+ transfer from PratA to D1 and the possible involvement of additional factors required for this process have to be determined structurally and biochemically in future studies.
Spatial Organization of PSII Assembly
PratA defines the PDM system, an intermediate membrane fraction proposed to be the site for initial steps of PSII biogenesis, which harbors especially the pD1 precursor protein as well as several PSII assembly factors and the chlorophyll precursor chlorophyllide a (Schottkowski et al., 2009a; Nickelsen et al., 2011; Rengstl et al., 2011). Our data suggest that the first steps in Mn incorporation into PSII (i.e., binding to D1) also occur in PDMs, whereas further assembly of RC47 and RCC1 complexes and photoactivation of the Mn cluster take place in the TM (Cheniae and Martin, 1971; Hwang and Burnap, 2005). Based on several facts it has been postulated that PDMs connect the PM and the TM system (Nickelsen et al., 2011) and that they function in transport of precomplexes from PM to TM: (1) D1 and D2 are not exclusively present in TMs, but also in PM/PDM fractions of Synechocystis 6803 (Smith and Howe, 1993; Zak et al., 2001; Keren et al., 2005; Rengstl et al., 2011); (2) CtpA, the protease required for cleavage of the C-terminal extension of D1, is exclusively found in the PM/PDM (Zak et al., 2001); (3) PratA is localized in the periplasm and in the PDMs; (4) pD1 accumulates in the PDMs upon knockout of PratA (Schottkowski et al., 2009a). However, the existence of structures connecting PM and TM being the site of early PSII assembly processes has been discussed controversially (Liberton et al., 2006; Liberton and Pakrasi, 2008; Mullineaux, 2008; Nixon et al., 2010; Nickelsen et al., 2011). Here, we present experimental evidence from ultrastructural analyses that show the existence of PratA-dependent semicircular structures of ~100 nm in diameter surrounding thylakoid centers at the cellular periphery, which indeed appear to connect TMs and PM (Figure 7) and which we called biogenesis centers (= thylakoid center + semicircular structure; Figure 9G). Although these structures have to be analyzed three-dimensionally in more detail in further studies, it can be hypothesized that the observed semicircles surround the rod-like thylakoid centers detected before (van de Meene et al., 2006). In none of the >1000 pratA− cells analyzed have these semicircle-shaped structures been found, suggesting that although we cannot exclude that certain prestructures already form PratA independently, PratA seems to be required for either the stability of these structures or its fixation at the cell periphery. Furthermore, both PratA and pD1 signals in immunogold labeling experiments form clusters of similar size close to the PM. The localization of these two markers for initial PSII biogenesis at structures apparently connecting PMs and TMs provides a solution to the long-standing, controversial discussion whether or not the PM is involved in TM biogenesis in cyanobacteria. This is in line with membrane fractionation experiments in Synechocystis 6803 that also indicated a crucial role of PratA for membrane organization (Rengstl et al., 2011). Due to technical limitations in staining of images in gold-labeling assays, we were not able to localize precisely the clusters to the semicircular structures. However, their common localization and size as well as their absence in the pratA− mutant strongly suggest that these structures at the cell periphery represent biogenesis centers formed by PDMs. Further high-resolution imaging techniques will be required to resolve the structure of the biogenesis centers in more detail and three-dimensionally. Interestingly, similar biogenesis centers have additionally been detected in C. reinhardtii (Uniacke and Zerges, 2007). In this case, the biogenesis regions seem to consist of membranes surrounding the pyrenoid structure of the chloroplast. Thus, it can be postulated that the general mechanisms of TM biogenesis are evolutionary conserved from cyanobacteria to algae and possibly also to plants. Taken together, our data allow the proposal of a coherent model for the spatiotemporal organization of TM biogenesis in cyanobacteria, which includes not only biogenesis centers as PM/TM connecting sites responsible for early PSII assembly processes, but which additionally involves Mn2+ preloading of PSII from the periplasm (Figure 10).
A Function of PratA during Repair of PSII?
Our results provide evidence for a function of PratA in direct and efficient delivery of Mn2+ to D1 during PSII de novo assembly. It remains unknown if and how PratA functions in PSII repair, as a role in this process has not been investigated yet. Although light-induced damage to PSII (photoinhibition) has been studied in great detail in vitro (Adir et al., 2003; Edelman and Mattoo, 2008; Nixon et al., 2010), the in vivo mechanism of PSII photodamage is controversial (Vass and Cser, 2009). Furthermore, it is unclear whether PSII repair in cyanobacteria is spatially separated from PSII de novo assembly like in chloroplasts of green algae (Zak et al., 2001; Komenda et al., 2006; Nixon et al., 2010; Uniacke and Zerges, 2007). Concerning Mn, it has even been suggested that the Mn cluster itself is the initial site of damage (Hakala et al., 2005; Ohnishi et al., 2005), and it can be speculated that Mn is merely recycled during PSII repair; however, direct evidence for this is missing. Hence, future studies have to analyze further PSII repair in general and a potential photodamage-related function of PratA in particular.
Evolutionary Aspects of PratA Function
PratA is a cyanobacterial protein for which no homologs exist in eukaryotes (Klinkert et al., 2004). However, Lpa1 (for Low PSII Accumulation I) from Arabidopsis has recently been shown to possess similar properties: It interacts directly with D1 and belongs to the TPR protein family along with its homolog from C. reinhardtii named REP27 (for repair-aberrant mutant 27; Peng et al., 2006; Park et al., 2007). Lpa1 contains two TPR motifs near its N terminus, which are arranged like the first two of the nine TPR domains in PratA, but it shows no additional sequence similarities to PratA (see Supplemental Figure 5A online). Interestingly, initial EPR measurements suggest that Lpa1, similar to PratA, has Mn2+ binding activity, suggesting that the participation of TPR proteins in Mn2+ transport to PSII by direct interaction with D1 might be evolutionary conserved from cyanobacteria to vascular plants (see Supplemental Figure 5B online).
Recently, membrane domains with specialized functions have been localized even in the ancient cyanobacterium Gloeobacter violoceus, the only known organism performing oxygenic photosynthesis without internal TMs (Rexroth et al., 2011). The cytoplasmic membrane of G. violoceus was found to contain a “green fraction” and an “orange fraction,” which, regarding protein and lipid content, seem to resemble the TM and PM of other cyanobacteria, respectively. This idea is supported by the observation that components of the photosynthetic and respiratory electron transfer chain are highly enriched in the green fraction (Rexroth et al., 2011). Interestingly, a PratA homologous protein was detected exclusively in the orange fraction, which allows the proposal that this specialized membrane region might be the evolutionary origin of the later-developed biogenesis centers. Taken together, we hypothesize that PratA-related proteins function in early PSII assembly steps, including its preloading with Mn2+ in all photosynthetic organisms from G. violoceus to vascular plants.
METHODS
Strains, Growth Conditions, and Sequence Analysis
Synechocystis sp PCC 6803 (Synechocystis 6803) wild-type and mutant strains (pratA−, psbA−, and ycf48−, including the respective wild type used; Komenda et al., 2008) were grown on solid or in liquid BG11 medium (Rippka et al., 1979) at 23°C at a continuous photon irradiance of 30 μE m−2 s−1. The mutant strains pratA− and ycf48− and the psbA− deletion strain TD41 were previously constructed as described (Nixon et al., 1992; Klinkert et al., 2004; Komenda et al., 2008). For sequence analysis of PratA (slr2048) and Lpa1 (At1g02910), the full-length protein sequences were aligned using ClustalX2 (see Supplemental Figure 5A online).
Isolation of Periplasm and Atomic Absorption Spectrometry
Periplasm was isolated from 9 liters of Synechocystis wild-type and pratA− cultures (Fulda et al., 2000) and concentrated to 6 mL by ultrafiltration (Millipore). Fe and Mn levels were measured with a multielement hollow-cathode lamp (slit width, 0.2 nm; lamp current, Fe, 5 mA; Mn, 4 mA) on a Varian AA240 atomic absorption spectrometer at 248.3 and 279.5 nm, respectively. All measurements were performed in an air (13.5 L/min)-acetylene (2 L/min) flame. Single element solutions (Roth) were used for calibration. Experiments were performed in triplicate, and results are expressed in nanograms of metal per milligram of chlorophyll.
Overexpression and Purification of Proteins
For heterologous expression of N-terminal GST fusion proteins, GST-Lpa1, GST-PratA, GST-mD1, and GST-pD1 (Schottkowski et al., 2009a) were used. For Lpa1, the coding region, excluding the transit sequence (Peng et al., 2006), was cloned into the SalI-BamHI sites of pGEX-4T-1 (GE Healthcare) using the primers Lpa_for (5′-GATCGGATCCATGGATGCTCTTGTTCAGTTTG-3−) and Lpa_rev (5′-GATCGTCGACGTCTTTCTAACTTGCTGAGAA-3′). Overexpression (in Escherichia coli BL21 [DE3] cells) was performed at 12°C overnight. Proteins were purified via GST-agarose (Biontex) under native conditions in the presence of 5 mM EDTA and, if required, recovered from the matrix by removing the GST tag by incubation with 1 unit of thrombin (GE Healthcare) per 100 μg of fusion protein for 6 h at room temperature (RT). Thrombin was removed by benzamidine-sepharose (GE Healthcare). Freshly prepared proteins (see Supplemental Figure 1 online) were used for CD or EPR analysis after dialysis against 10 mM Tris/H2SO4, pH 8.0, or 50 mM Tris/HCl, pH 8.0, and 150 mM NaCl, respectively.
CD Measurements
CD experiments with PratA were performed at RT using a Jasco J-810 spectropolarimeter flushed with nitrogen (Stengel et al., 2008). Spectra were collected from 260 to 190 nm using a cylindrical quartz cell (path length of 1 mm). Each spectrum was the average of three scans taken at a scan rate of 50 nm/min with a spectral bandwidth of 1 nm. For the final representation, the baseline was subtracted from the spectrum. The protein concentration varied from 0.080 to 0.122 mg/mL. Where indicated, 1 mM MnCl2, FeCl3, FeCl2, MgCl2, or CaCl2 was added and incubated with the protein for 15 min at RT prior to the measurement. Experiments were performed in triplicate. Data analysis was performed using CDSSTR (reference set 7) from the DichroWeb server (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml), and spectra were plotted after subtraction of the baseline.
EPR Analysis
Continuous-wave EPR spectra were recorded on a Miniscope MS200 X-band spectrometer (microwave frequency ≈ 9.4 GHz) equipped with a rectangular TE102 resonator (Magnettech). The temperature was adjusted with a TC H02 control unit (Magnettech). Measurements were performed using 298.15K, 3.162 mW (microwave power), 100 kHz (modulation frequency), and 0.5 mT (modulation amplitude) for Mn(II); and 104K, 15.85 mW, 100 kHz, and 0.5 mT for Fe(III). Prior to the measurements, 500 μM MnCl2 (or 5 mM MnCl2 where indicated) or 1 mM FeCl3 were incubated for 5 min at RT with PratA (100 μM if not indicated differently). For the competition experiments, 500 μM (1×) or 5 mM (10×) CaCl2 or MgCl2 was incubated together with 500 μM MnCl2 and PratA before analysis. For Mn titration experiments, 100 μM PratA was titrated with 25 to 5000 μM MnCl2, and continuous-wave EPR spectra were recorded after incubation for 5 min. The amount of protein-bound Mn2+ was calculated from the signal amplitude of the lowest-field transition. Thereby, the peak-to-peak height of the lowest-field line of a Mn2+ standard was plotted against Mn2+ concentration to obtain a standard curve. The concentration of the free Mn2+ in solutions containing rPratA was calculated by comparison to the standard curve, and the amount bound was determined by difference. The stoichiometry of bound Mn2+ per rPratA molecule was then obtained by dividing the amount of protein-bound Mn2+ by the concentration of rPratA in solution. Data were analyzed by plotting according to Scatchard using SigmaPlot 11 with the Enzyme Kinetics module 1.3.
Filter Binding Assays
For determination of the PratA-Mn binding properties, recombinant PratA (100 μM; in 50 mM Tris/HCl, pH 8.0, and 150 mM NaCl) was incubated with 10 to 75 μM of 54Mn2+ (as MnCl2; Hartmann Analytics) for 15 min at RT, loaded on Amicon Ultra-0.5 10 kD columns (Millipore), concentrated, and washed once with Tris/NaCl. Recovered samples were suspended in scintillation mixture (Optiphase Supermix; Perkin-Elmer), and 54Mn2+ was measured with a LS 6500 multipurpose scintillation counter (Beckman Coulter). For calculation of the amount of bound Mn2+ per PratA, the obtained counts per minute of the control (rPratA without added 54Mn2+; background) was subtracted from the measured counts per minute of samples that had been incubated with 10 to 75 μM of 54Mn2+ and calculated relative to the counts per minute detected for total 54Mn2+ used for the experiment. From these values, the concentration of 54Mn2+ that was bound by the protein was obtained, and using the known concentration of rPratA (100 μM), the amount of bound Mn2+ per PratA was calculated. Data were analyzed by plotting according to Scatchard using SigmaPlot 11 with the Enzyme Kinetics module 1.3.
Affinity Chromatography Using Metal-Loaded Columns
His-Bind Fractogel Matrix (Novagen; 100 μL) was incubated with 100 mM MnCl2, NiSO4, or buffer as control (50 mM Tris, pH 8.0, and 200 mM NaCl) for 1 h at RT and washed five times with Tris/NaCl. The beads were subsequently incubated (2 h, RT) with periplasm (200 μg protein) isolated from wild-type Synechocystis (Fulda et al., 2000) and washed five times with Tris/NaCl. Proteins were eluted by incubation with 50 μL 2× Roti-Load 1 (Roth) for 5 min at 95°C. Samples of load, flow-through, wash, and eluate were analyzed by immunoblotting using a αPratA antibody (Klinkert et al., 2004). Quantification of signals was performed using AIDA software (version 3.52.046).
GST Pull-Down Assays
GST fusion proteins (mD1 and pD1; 100 μg each) were bound to GST-agarose (Biontex) for 2 h at RT. The matrix was then incubated for 2 h at RT with periplasm (300 μg protein) from Synechocystis wild-type and pratA− cultures (Fulda et al., 2000). As control, wild-type periplasm was incubated with an equivalent amount of GST-agarose matrix without bound mD1/pD1 protein. The beads were washed five times with 50 mM Tris, pH 8.0, and 150 mM NaCl, proteins were eluted by incubation with 50 μL 2× Roti-Load 1 (Roth) for 5 min at 95°C, and samples were analyzed by immunoblotting using αPratA.
Mn Uptake Assay and Immunoprecipitation with αD1
Synechocystis wild-type and mutant strains (pratA−psbA− TD41, and ycf48− with the respective wild type; Komenda et al., 2008) were grown in 50 mL liquid BG11 for 5 to 6 d. Chlorophyll concentrations were measured after extraction with 100% methanol and calculated from the absorbance values at 666 and 720 nm (Wellburn and Lichtenthaler, 1984). Cells were resuspended in BG11 (chlorophyll concentration 5 μg/mL), and 54Mn2+ was added for 1 and 3 h at 1 μCi/mL. As control, one sample was analyzed immediately after addition of 54Mn2+ (0 h), and values obtained after incubation for 1 and 3 h were calculated relative to it. Samples (10 μg chlorophyll) were washed twice in TMK buffer (10 mM Tris/HCl, pH 6.8, 10 mM MgCl2, and 20 mM KCl). For analysis of Mn uptake, cells were used directly for measurement of 54Mn2+ with a scintillation counter. To monitor the delivery of Mn to PS II (Mn transport), cells were resuspended in 200 μL TMK buffer and broken by shaking with glass beads (0.1 mm diameter) on a vortex for 2 × 1 min separated by 1 min on ice. The suspension was centrifuged for 1 min at 6000g, and the supernatant was solubilized with dodecylmaltoside (1.5%, 10 min on ice). After centrifugation for 10 min at 20,000g, the supernatant was diluted 1:10 with TMK buffer and supplied with 5 μL primary antiserum (αD1; Schottkowski et al., 2009a). After incubation for 2 h at RT, 25 μL Protein A-Sepharose (Roche) was added, and the suspension was incubated overnight at 4°C. The beads were washed five times in TMK buffer and analyzed with a scintillation counter (Beckman Coulter). For normalization purposes, radioactivity levels (in cpm) measured in samples from psbA− cells (background) were subtracted from values for the wild type and pratA−. Experiments were performed four times. Protein extracts for determination of D1 content in wild-type and pratA− cells were prepared according to Schottkowski et al. (2009b). Isolated proteins were separated by SDS-PAGE and probed with the indicated antibodies. Quantification of signals was performed using AIDA software (version 3.52.046), and the levels in the pratA- strain were compared with those in the wild-type strain (set to 100%). Means (±sd) were calculated from three independently inoculated cultures.
Transmission Electron Microscopy and Immunogold Labeling
Synechocystis cells (the wild type, pratA−, and psbB−) were harvested and 20 μL of the moist cell pellet was placed into a HPF aluminum platelet (Leica, BAL-TEC, HPM 100 carriers type A and B; 100 μM in depth), high-pressure frozen (Leica EM HPM100) at 2100 bar, and stored in liquid nitrogen. No cryoprotectants were used. For analyzing the ultrastructure, cryofixed samples were freeze-substituted (Leica EM AFS2) at −85°C for 72 h in acetone anhydrous with 1% glutaraldehyde and 1% tannic acid according to van de Meene et al. (2006) and washed three times in acetone anhydrous (−85°C, 15 min each). Infiltration of cells was achieved using acetone including 1% osmium tetroxide for 3 h at −85°C, followed by incubation for 20 h at −20°C, 3 h at 4°C, and 1 h at RT. Cells were washed four times in acetone and infiltrated with Spurr’s resin (Spurr, 1969). For immunodetection experiments, cryofixed samples were freeze-substituted in acetone containing 0.5% uranyl acetate (Pfeiffer and Krupinska, 2005) for 20 h at −90°C. Dehydration was performed by two infiltration steps with acetone (8 h at −70°C and 8 h at −50°C). Afterwards, cells were infiltrated with HM-20 (Lowicryl HM-20; Polysciences) by washing 30 min with acetone at −50°C, infiltrated with 30% HM-20/70% acetone overnight, followed by 70% HM-20/30% acetone for 2 h, finally three times in 100% HM-20 for 2 h at −50°C. Polymerization was performed under UV light at −50°C for 48 h and at RT for 24 h. Ultrathin sections (30 to 60 nm) were cut with a diamond knife (Ultramicrotome Leica EM UC6). For ultrastructural analysis, they were poststained with lead citrate (Reynolds, 1963). For immunogold labeling, the sections were collected on coated Ni grids, incubated twice with glycin (50 mM in PBS, pH 7.4), once in blocking buffer (FCS 10% inactivated at 56°C for 30 min, 0.01% gelatin, 0.05% Tween 20 in PBS, pH 7.4) for 5 min, and twice in blocking buffer for 60 min. Sections were incubated for 18 h at 4°C with αPratA 1:25 (rabbit) or αpD1 1:50 (rabbit). The sections were washed six times for 5 min and incubated with gold-conjugated goat anti-rabbit IgG (BBInternational; 5 nm, 1:50) for 90 min at RT before washing three times in blocking buffer, twice with 50 mM glycin in PBS, pH 7.4, twice with PBS, pH 7.4, and twice with double-distilled water. Samples were analyzed without further contrast. Micrographs were taken at 80 kV on a Fei Morgagni 268 electron microscope; ×1800 and 11,000-fold (overviews), 44,000-fold and 110,000-fold (details). As negative control, Synechocystis wild-type sections were incubated without the primary antibody, followed by treatment with the gold-labeled anti-rabbit IgG. The cluster analysis (determination of cluster area, perimeter, major axis, minor axis, and circularity) was performed with ImageJ (http://rsb.info.nih.gov/ij/; see Supplemental Figure 4 online).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: slr2048 (PratA), slr2034 (Ycf48), and At1g02910 (Lpa1). The three-dimensional structure of D1 is derived from the Protein Data Bank file 3BZ1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Purification of Recombinant PratA.
Supplemental Figure 2. Competition of Mn2+ Binding to PratA Using 5 mM MnCl2.
Supplemental Figure 3. Recovery of Mn2+ Signal and Dependency of Mn2+ Binding on PratA Concentration.
Supplemental Figure 4. Determination of Parameters Used for Analysis of Gold Clusters in Immunogold Labeling Experiments with αpD1.
Supplemental Figure 5. Evolutionary Investigation of PratA Function.
Supplementary Material
Acknowledgments
We thank Christian Schwarz for generation of the three-dimensional illustration of D1, Norio Murata for providing the pD1 antibody, and Gunnar Jeschke and Jürgen Soll for helpful discussions and suggestions. This work is supported by grants from the Deutsche Forschungsgemeinshaft (SFB TR1 TPC10 to J.N.; Ju333/4-3 to H.J.) and by the Bayerische Gleichstellungsförderung Scholarship of Ludwig-Maximilians-Universität Munich (A.S.).
AUTHOR CONTRIBUTIONS
A.S. and J.N. designed the research. A.S., I.L.G., D.H., and B.R. performed research. A.S., I.L.G., H.J., and J.N. analyzed data. A.S. and J.N. wrote the article.
References
- Abramowicz D.A., Dismukes G.C. (1984). Manganese proteins isolated from spinach thylakoid membranes and their role in O2 evolution. II. A binuclear manganese-containing 34 kilodalton protein, a probable component of the water dehydrogenase enzyme. Biochim. Biophys. Acta 765: 318–328 [DOI] [PubMed] [Google Scholar]
- Adir N., Zer H., Shochat S., Ohad I. (2003). Photoinhibition - A historical perspective. Photosynth. Res. 76: 343–370 [DOI] [PubMed] [Google Scholar]
- Barber J. (2008). Crystal structure of the oxygen-evolving complex of photosystem II. Inorg. Chem. 47: 1700–1710 [DOI] [PubMed] [Google Scholar]
- Bartsevich V.V., Pakrasi H.B. (1995). Molecular identification of an ABC transporter complex for manganese: Analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 14: 1845–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsevich V.V., Pakrasi H.B. (1996). Manganese transport in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 271: 26057–26061 [DOI] [PubMed] [Google Scholar]
- Becker K., Cormann K.U., Nowaczyk M.M. (2011). Assembly of the water-oxidizing complex in photosystem II. J. Photochem. Photobiol. B 104: 204–211 [DOI] [PubMed] [Google Scholar]
- Blatch G.L., Lässle M. (1999). The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 21: 932–939 [DOI] [PubMed] [Google Scholar]
- Bondarava N., Un S., Krieger-Liszkay A. (2007). Manganese binding to the 23 kDa extrinsic protein of photosystem II. Biochim. Biophys. Acta 1767: 583–588 [DOI] [PubMed] [Google Scholar]
- Cheniae G.M., Martin I.F. (1971). Photoactivation of the manganese catalyst of O 2 evolution. I. Biochemical and kinetic aspects. Biochim. Biophys. Acta 253: 167–181 [DOI] [PubMed] [Google Scholar]
- Crowley J.D., Traynor D.A., Weatherburn D.C. (2000). Enzymes and proteins containing manganese: An overview. In Metal Ions in Biological Systems, Sigel A., Sigel H., (New York: Marcel Dekker, Inc.), pp. 211–261 [PubMed] [Google Scholar]
- Edelman M., Mattoo A.K. (2008). D1-protein dynamics in photosystem II: The lingering enigma. Photosynth. Res. 98: 609–620 [DOI] [PubMed] [Google Scholar]
- Ferreira K.N., Iverson T.M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838 [DOI] [PubMed] [Google Scholar]
- Fulda S., Huang F., Nilsson F., Hagemann M., Norling B. (2000). Proteomics of Synechocystis sp. strain PCC 6803. Identification of periplasmic proteins in cells grown at low and high salt concentrations. Eur. J. Biochem. 267: 5900–5907 [DOI] [PubMed] [Google Scholar]
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009). Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16: 334–342 [DOI] [PubMed] [Google Scholar]
- Hakala M., Tuominen I., Keränen M., Tyystjärvi T., Tyystjärvi E. (2005). Evidence for the role of the oxygen-evolving manganese complex in photoinhibition of photosystem II. Biochim. Biophys. Acta 1706: 68–80 [DOI] [PubMed] [Google Scholar]
- Hayden J.A., Hendrich M.P. (2010). EPR spectroscopy and catalase activity of manganese-bound DNA-binding protein from nutrient starved cells. J. Biol. Inorg. Chem. 15: 729–736 [DOI] [PubMed] [Google Scholar]
- Hinterstoisser B., Chichna M., Kuntner O., Peschek G.A. (1993). Cooperation of plasma and thylakoid membranes for the biosynthesis of chlorophyll in cyanobacteria: The role of the thylakoid centers. J. Plant Physiol. 142: 407–413 [Google Scholar]
- Hwang H.J., Burnap R.L. (2005). Multiflash experiments reveal a new kinetic phase of photosystem II manganese cluster assembly in Synechocystis sp. PCC6803 in vivo. Biochemistry 44: 9766–9774 [DOI] [PubMed] [Google Scholar]
- Keren N., Kidd M.J., Penner-Hahn J.E., Pakrasi H.B. (2002). A light-dependent mechanism for massive accumulation of manganese in the photosynthetic bacterium Synechocystis sp. PCC 6803. Biochemistry 41: 15085–15092 [DOI] [PubMed] [Google Scholar]
- Keren N., Liberton M., Pakrasi H.B. (2005). Photochemical competence of assembled photosystem II core complex in cyanobacterial plasma membrane. J. Biol. Chem. 280: 6548–6553 [DOI] [PubMed] [Google Scholar]
- Kern J., Biesiadka J., Loll B., Saenger W., Zouni A. (2007). Structure of the Mn4-Ca cluster as derived from X-ray diffraction. Photosynth. Res. 92: 389–405 [DOI] [PubMed] [Google Scholar]
- Klinkert B., Ossenbühl F., Sikorski M., Berry S., Eichacker L., Nickelsen J. (2004). PratA, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 279: 44639–44644 [DOI] [PubMed] [Google Scholar]
- Komenda J., Barker M., Kuviková S., de Vries R., Mullineaux C.W., Tichy M., Nixon P.J. (2006). The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 281: 1145–1151 [DOI] [PubMed] [Google Scholar]
- Komenda J., Nickelsen J., Tichý M., Prásil O., Eichacker L.A., Nixon P.J. (2008). The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 283: 22390–22399 [DOI] [PubMed] [Google Scholar]
- Liberton M., Howard Berg R., Heuser J., Roth R., Pakrasi H.B. (2006). Ultrastructure of the membrane systems in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. Protoplasma 227: 129–138 [DOI] [PubMed] [Google Scholar]
- Liberton M., Pakrasi H.B. (2008). Membrane systems in cyanobacteria. In The Cyanobacteria, A. Herrero and E. Flores, eds (Norfolk, UK: Caister Academic Press), pp. 271–288 [Google Scholar]
- Main E.R., Stott K., Jackson S.E., Regan L. (2005). Local and long-range stability in tandemly arrayed tetratricopeptide repeats. Proc. Natl. Acad. Sci. USA 102: 5721–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A.E., Whittaker M.M., Whittaker J.W. (1996). EPR polarization studies on Mn catalase from Lactobacillus plantarum. Biochemistry 35: 348–360 [DOI] [PubMed] [Google Scholar]
- Mizuno K., Whittaker M.M., Bächinger H.P., Whittaker J.W. (2004). Calorimetric studies on the tight binding metal interactions of Escherichia coli manganese superoxide dismutase. J. Biol. Chem. 279: 27339–27344 [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W. (2008). Biogenesis and dynamics of thylakoid membranes and the photosynthetic apparatus. In The Cyanobacteria, A. Herrero and E. Flores, eds (Norfolk, UK: Caister Academic Press), pp. 289–304 [Google Scholar]
- Mulo P., Sirpiö S., Suorsa M., Aro E.M. (2008). Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth. Res. 98: 489–501 [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Nowaczyk M.M., Klinkert B. (2007). Assembly factors of the photosynthetic machinery in cyanobacteria. Prog. Bot. 68: 57–79 [Google Scholar]
- Nickelsen J., Rengstl B., Stengel A., Schottkowski M., Soll J., Ankele E. (2011). Biogenesis of the cyanobacterial thylakoid membrane system—An update. FEMS Microbiol. Lett. 315: 1–5 [DOI] [PubMed] [Google Scholar]
- Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. (Lond.) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon P.J., Trost J.T., Diner B.A. (1992). Role of the carboxy terminus of polypeptide D1 in the assembly of a functional water-oxidizing manganese cluster in photosystem II of the cyanobacterium Synechocystis sp. PCC 6803: Assembly requires a free carboxyl group at C-terminal position 344. Biochemistry 31: 10859–10871 [DOI] [PubMed] [Google Scholar]
- Ohnishi N., Allakhverdiev S.I., Takahashi S., Higashi S., Watanabe M., Nishiyama Y., Murata N. (2005). Two-step mechanism of photodamage to photosystem II: Step 1 occurs at the oxygen-evolving complex and step 2 occurs at the photochemical reaction center. Biochemistry 44: 8494–8499 [DOI] [PubMed] [Google Scholar]
- Park S., Khamai P., Garcia-Cerdan J.G., Melis A. (2007). REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast-localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiol. 143: 1547–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer S., Krupinska K. (2005). New insights in thylakoid membrane organization. Plant Cell Physiol. 46: 1443–1451 [DOI] [PubMed] [Google Scholar]
- Pierce B.S., Elgren T.E., Hendrich M.P. (2003). Mechanistic implications for the formation of the diiron cluster in ribonucleotide reductase provided by quantitative EPR spectroscopy. J. Am. Chem. Soc. 125: 8748–8759 [DOI] [PubMed] [Google Scholar]
- Plücken H., Müller B., Grohmann D., Westhoff P., Eichacker L.A. (2002). The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 532: 85–90 [DOI] [PubMed] [Google Scholar]
- Reaney S.H., Kwik-Uribe C.L., Smith D.R. (2002). Manganese oxidation state and its implications for toxicity. Chem. Res. Toxicol. 15: 1119–1126 [DOI] [PubMed] [Google Scholar]
- Reed G.H., Cohn M. (1970). Electron paramagnetic resonance spectra of manganese (II)-protein complexes. Manganese (II)-concanavalin A. J. Biol. Chem. 245: 662–664 [PubMed] [Google Scholar]
- Reed G.H., Markham G.D. (1984). EPR of Mn(II) complexes with enzymes and other proteins. In Biological Magnetic Resonance, L.J. Berliner and J. Reuben, eds (New York: Plenum Press), pp. 73–142 [Google Scholar]
- Rengstl B., Oster U., Stengel A., Nickelsen J. (2011). An intermediate membrane subfraction in cyanobacteria is involved in an assembly network for photosystem II biogenesis. J. Biol. Chem. 286: 21944–21951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexroth S., Mullineaux C.W., Ellinger D., Sendtko E., Rögner M., Koenig F. (2011). The plasma membrane of the cyanobacterium Gloeobacter violaceus contains segregated bioenergetic domains. Plant Cell 23: 2379–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds E.S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17: 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J.B., Herdmann M., Stanier R. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111: 1–61 [Google Scholar]
- Roose J.L., Wegener K.M., Pakrasi H.B. (2007). The extrinsic proteins of photosystem II. Photosynth. Res. 92: 369–387 [DOI] [PubMed] [Google Scholar]
- Schneider D., Fuhrmann E., Scholz I., Hess W.R., Graumann P.L. (2007). Fluorescence staining of live cyanobacterial cells suggest non-stringent chromosome segregation and absence of a connection between cytoplasmic and thylakoid membranes. BMC Cell Biol. 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottkowski M., Gkalympoudis S., Tzekova N., Stelljes C., Schünemann D., Ankele E., Nickelsen J. (2009a). Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 284: 1813–1819 [DOI] [PubMed] [Google Scholar]
- Schottkowski M., Ratke J., Oster U., Nowaczyk M., Nickelsen J. (2009b). Pitt, a novel tetratricopeptide repeat protein involved in light-dependent chlorophyll biosynthesis and thylakoid membrane biogenesis in Synechocystis sp. PCC 6803. Mol. Plant 2: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Sen K.I., Sienkiewicz A., Love J.F., vanderSpek J.C., Fajer P.G., Logan T.M. (2006). Mn(II) binding by the anthracis repressor from Bacillus anthracis. Biochemistry 45: 4295–4303 [DOI] [PubMed] [Google Scholar]
- Smith D., Howe C.J. (1993). The distribution of photosystem-I and photosystem-II polypeptides between the cytoplasmic and thylakoid membranes of cyanobacteria. FEMS Microbiol. Lett. 110: 341–347 [Google Scholar]
- Spurr A.R. (1969). A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Stengel A., Benz P., Balsera M., Soll J., Bölter B. (2008). TIC62 redox-regulated translocon composition and dynamics. J. Biol. Chem. 283: 6656–6667 [DOI] [PubMed] [Google Scholar]
- Tamura N., Cheniae G. (1987). Photoactivation of the water-oxidizing complex in photosystem-ii membranes depleted of mn and extrinsic proteins. 1. Biochemical and kinetic characterization. Biochim. Biophys. Acta 890: 179–194 [Google Scholar]
- Tottey S., Waldron K.J., Firbank S.J., Reale B., Bessant C., Sato K., Cheek T.R., Gray J., Banfield M.J., Dennison C., Robinson N.J. (2008). Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature 455: 1138–1142 [DOI] [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J.R., Kamiya N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Uniacke J., Zerges W. (2007). Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell 19: 3640–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Meene A.M.L., Hohmann-Marriott M.F., Vermaas W.F.J., Roberson R.W. (2006). The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184: 259–270 [DOI] [PubMed] [Google Scholar]
- Vass I., Cser K. (2009). Janus-faced charge recombinations in photosystem II photoinhibition. Trends Plant Sci. 14: 200–205 [DOI] [PubMed] [Google Scholar]
- Wellburn A.R., Lichtenthaler H.K. (1984). Formulae and programme to determine total carotenoids and chlorophyll a and b of leaf extracts in different solvents. In Advances in Photosynthesis Research, Vol. II, C. Sybesma, ed (The Hague, Lancaster, Boston: Martinus Nijhoff/Dr. W. Junk Publishers), pp. 9–12 [Google Scholar]
- Yano J., Kern J., Sauer K., Latimer M.J., Pushkar Y., Biesiadka J., Loll B., Saenger W., Messinger J., Zouni A., Yachandra V.K. (2006). Where water is oxidized to dioxygen: structure of the photosynthetic Mn4Ca cluster. Science 314: 821–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak E., Norling B., Maitra R., Huang F., Andersson B., Pakrasi H.B. (2001). The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc. Natl. Acad. Sci. USA 98: 13443–13448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman L., Ananyev G.M., Bruntrager E., Dismukes G.C. (1997). Quantitative kinetic model for photoassembly of the photosynthetic water oxidase from its inorganic constituents: requirements for manganese and calcium in the kinetically resolved steps. Biochemistry 36: 8914–8922 [DOI] [PubMed] [Google Scholar]
- Zöfel P. (1988). Statistik in der Praxis. (Stuttgart, Germany: Gustav Fischer Verlag). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.