This article describes novel ways that algae may adjust metabolite trafficking when specific branches of fermentation metabolism are blocked. This rerouting of metabolites allows for continued glycolytic energy production under anoxic conditions, which is critical for the cell’s survival. Mechanisms associated with this reengineering of metabolism are almost completely unexplored.
Abstract
Chlamydomonas reinhardtii, a unicellular green alga, often experiences hypoxic/anoxic soil conditions that activate fermentation metabolism. We isolated three Chlamydomonas mutants disrupted for the pyruvate formate lyase (PFL1) gene; the encoded PFL1 protein catalyzes a major fermentative pathway in wild-type Chlamydomonas cells. When the pfl1 mutants were subjected to dark fermentative conditions, they displayed an increased flux of pyruvate to lactate, elevated pyruvate decarboxylation, ethanol accumulation, diminished pyruvate oxidation by pyruvate ferredoxin oxidoreductase, and lowered H2 production. The pfl1-1 mutant also accumulated high intracellular levels of lactate, succinate, alanine, malate, and fumarate. To further probe the system, we generated a double mutant (pfl1-1 adh1) that is unable to synthesize both formate and ethanol. This strain, like the pfl1 mutants, secreted lactate, but it also exhibited a significant increase in the levels of extracellular glycerol, acetate, and intracellular reduced sugars and a decrease in dark, fermentative H2 production. Whereas wild-type Chlamydomonas fermentation primarily produces formate and ethanol, the double mutant reroutes glycolytic carbon to lactate and glycerol. Although the metabolic adjustments observed in the mutants facilitate NADH reoxidation and sustained glycolysis under dark, anoxic conditions, the observed changes could not have been predicted given our current knowledge of the regulation of fermentation metabolism.
INTRODUCTION
Several species of water-oxidizing, photosynthetic algae can catabolize endogenous carbohydrates or secondary metabolites using diverse fermentative pathways that allow them to generate the ATP necessary to drive metabolic and energy-requiring processes during anoxia (Gfeller and Gibbs, 1984; Kreuzberg, 1984; Gfeller and Gibbs, 1985; Gibbs et al., 1986; Ohta et al., 1987). Fermentative metabolism in the model green alga, Chlamydomonas reinhardtii (Chlamydomonas throughout), has been characterized to some extent, and these investigations demonstrate the critical nature of fermentation for survival during the evening and early morning, when diminished levels of photosynthetic activity and high levels of microbial respiration deplete the local environment of O2 (Grossman et al., 2007; Mus et al., 2007; Dubini et al., 2009; Grossman et al., 2011). The fermentation of stored organic compounds in phototrophic microorganisms represents a significant part of their overall energy budget, because many of these ecologically important organisms spend much of their lifecycle under light-limited, hypoxic/anoxic conditions. Additionally, the secretion of energy- and carbon-rich fermentation products by primary producers is likely to shape the composition and density of the microbial consortia that become established in the surrounding aquatic and soil ecosystems.
The primary fermentation pathways that are used during anoxia vary among different algal species (Ohta et al., 1987; Meuser et al., 2009). Metabolic flexibility, even within a single organism, is exemplified by the Chlamydomonas mutant in the iron hydrogenase protein assembly factor hydEF-1, which under anoxic conditions does not synthesize H2 and instead seems to activate reverse tricarboxylic acid (rTCA) reactions and excretes succinate (Dubini et al., 2009). This pathway is not highly active in wild-type strains under similar conditions. Although it is clear that algae have the capacity to implement a diverse set of fermentative pathways when O2 in the environment decreases, little is known regarding the selective pressures that have shaped specific fermentation networks over evolutionary time or how the various branches of fermentation metabolism are regulated by transcriptional, translational, and posttranslational processes.
Chlamydomonas has multiple pathways for the conversion of pyruvate to acetyl-CoA (Wagner et al., 1992; Hemschemeier and Happe, 2005; Atteia et al., 2006; Grossman et al., 2007). Three enzymes involved in these pathways are pyruvate formate lyase (PFL1), pyruvate-ferredoxin-oxidoreductase (PFR1, often referred to as PFOR), and the pyruvate dehydrogenase (PDH) complex. As PDH generates NADH, it is presumed that PFL1 and PFR1 are the favored anoxic pathways, with PFL1 representing the dominant pathway in Chlamydomonas. During anoxia, the acetyl-CoA generated in the PFL1 and PFR1 reactions would be metabolized to either (1) ethanol, via the acetaldehyde/alcohol dehydrogenase (ADH1) pathway, which reoxidizes two NADH-reducing equivalents to maintain redox balance and sustain glycolysis, or (2) acetate, via the phosphotransacetylase/acetate kinase pathway, which generates ATP but does not eliminate reductant (Hemschemeier and Happe, 2005; Atteia et al., 2006).
Formate was demonstrated to be the dominant, secreted organic acid synthesized by dark, anoxic Chlamydomonas cells when maintained at near-neutral pH (Kreuzberg, 1984; Gibbs et al., 1986). Formate synthesis is predominantly a consequence of PFL1 activity, and the administration of an inhibitor of PFL1 activity (hypophosphite) to Chlamydomonas cells severely diminishes formate accumulation and elicits increased secretion of lactate, which is typically only a minor fermentative product in wild-type cells (Kreuzberg, 1984; Hemschemeier and Happe, 2005; Hemschemeier et al., 2008). The PFL1 reaction uses a free-radical mechanism that catalyzes the homolytic cleavage of pyruvate into formate and acetyl-CoA. This reaction is dependent on a radical S-adenosylmethionine–dependent activating enzyme, designated PFL-AE (Atteia et al., 2006; Hemschemeier et al., 2008). PFL-AE is typically present in an inactive form in aerobic cells and is allosterically activated by pyruvate.
Previous studies of Chlamydomonas have suggested that both PFL1 and PFL-AE transcripts and protein are present under aerobic conditions and that the pathway for formate production is poised to be rapidly activated as O2 levels decrease and pyruvate levels increase (Atteia et al., 2006); however, a more recent study indicates an absence of detectable PFL1 transcript under oxic conditions (Philipps et al., 2011). PFL1 of Chlamydomonas has been localized to both chloroplasts and mitochondria (Atteia et al., 2006).
From both regulatory and biotechnological perspectives, it is important to determine how a strain null for PFL1 synthesis modifies electron flux through other pathways of the fermentative network, and how it affects the production of acetyl-CoA, CO2, and reduced ferredoxin. Reduced ferredoxin can be reoxidized by the activity of hydrogenases, which reduce protons to generate H2, a potentially valuable renewable bioenergy carrier. To gain fundamental insights into how anoxic metabolism is regulated and to extend our understanding of metabolism in strains null for PFL1, we isolated several pfl1 insertional mutants that have either greatly reduced expression or no expression of PFL1 and characterized the mutants for accumulation of both internal and external metabolites as the cultures become anoxic. Similar to the data reported for a previously described pfl1 mutant, we observe increases in the production of lactate, ethanol, and CO2, and a decrease in acetate (Philipps et al., 2011). There are some differences in our study relative to those of Philipps et al. (2011), and we have performed many additional key analyses with the pfl1 mutants that have not been reported. In contrast with the knockout strain described by Philipps et al. (2011), we observed an attenuation of fermentative H2 production, but it should be noted that H2 is a minor product relative to the other metabolites and that its levels are easily influenced by algal culturing conditions. However, our findings on H2 production are also supported by both a change in the level of the hydrogenase protein and mRNA; the HYDA1 transcript is significantly lower in the pfl1-1 mutant strain, as is the HYDA protein level. Additionally, we also analyzed internal metabolite concentrations in the pfl1-1 mutant, which has revealed informative changes in the levels of several internal metabolites in response to PFL1 disruption. Finally, we extend our analysis beyond the level of a single gene disruption and have isolated and characterized a double mutant (pfl1-1 adh1) that simultaneously blocks both formate and ethanol production, two of the most dominant products secreted by wild-type cells. This strain compensates by synthesizing higher levels of lactate and glycerol, enabling the oxidation of NADH in the absence of a pathway that generates ethanol; this strain has a completely reconfigured fermentative metabolism relative to wild-type cells. This article represents a continuation of our systematic characterization of fermentative pathways in Chlamydomonas, fills existing knowledge gaps, and highlights our limited understanding of how algal metabolic networks are regulated when specific branches of fermentation metabolism are blocked.
RESULTS
Mutant Generation, Molecular and Biochemical Characterization
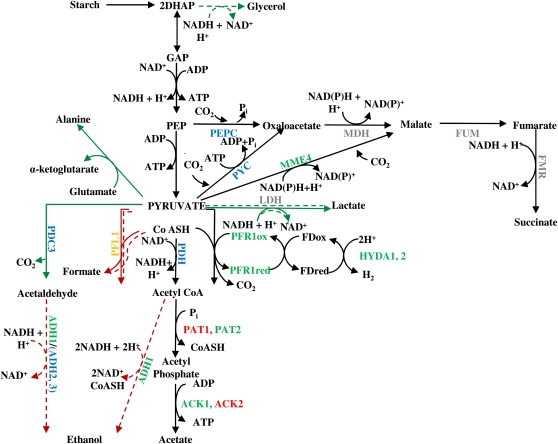
Dark fermentation in Chlamydomonas elicits the concurrent activities of PFL1 and PFR1 (Mus et al., 2007), both of which convert pyruvate to acetyl-CoA, as depicted in Figure 1. PFL1 activity results in the production of formate and acetyl-CoA, whereas PFR1 activity generates CO2 and acetyl-CoA with the concomitant reduction of ferredoxin (Müller, 1993, 1998). The reduced ferredoxin can be used as a substrate by hydrogenases, which catalyze the synthesis of H2. Thus, perturbation of pyruvate metabolism through genetically engineered disruption of the PFL1 pathway represents a strategy that could potentially increase the yield of dark, fermentative H2 production in green algae.
Figure 1.
Anaerobic Metabolic Pathways in Chlamydomonas.
Starch accumulates in the chloroplast during growth in the light and is degraded in the dark. In the absence of the complete TCA cycle (during anoxia), acetyl-CoA produced either via the PFL1 or PFR1 pathways enters the acetate-generating phosphate acetyltransferase-acetate kinase (PAT1/PAT2-ACK2/ACK1) or the bifunctional ADH1 pathways. PFR1 has a proposed role in dark, fermentative H2 production, because it reduces ferredoxin (FD), which in turn provides reductant to the hydrogenase (HYDA1 and possibly HYDA2), which catalyses H2 production. Pyruvate can also be decarboxylated to acetaldehyde by PDC3. The acetaldehyde is subsequently reduced to ethanol, possibly by ADH1 or possibly a second alcohol dehydrogenase (however, this study and Magneschi et al. [2012] suggest that ADH1 is the major enzyme involved in ethanol formation under dark, anoxic conditions). NADH might also be reoxidized by a putative LDH. The pathways on the upper right are proposed to generate succinate and reoxidize NADH in the hydEF-1 mutant (Dubini et al., 2009). In wild-type cells, glycolysis produces two molecules of DHAP/glyceraldehyde 3-phosphate (GAP), which are converted into two molecules of pyruvate; two ATPs are generated in the conversion of each DHAP to pyruvate. When both formate and ethanol production are blocked, the DHAP is converted into glycerol and pyruvate in a ratio of 1:1. Proteins encoded in the Chlamydomonas genome that are associated with the pathways depicted are given above. Red arrows indicate suppressed or downregulated pathways, whereas green arrows indicate the rerouting of electrons in the mutant strains (unbroken arrows for the single mutant pfl1-1, dashed arrows for the double mutant pfl1-1 adh1). Known or putative cellular localizations of the depicted proteins are color coded green for the chloroplast, red for the mitochondrion, orange for dual chloroplast/mitochondrion localization, blue for cytoplasm, and gray for unknown. FMR, fumarate reductase; FUM, fumarase; MDH, malate dehydrogenase; MME4, malic enzyme; PEPC, phosphoenolpyruvate carboxylase; PYC, pyruvate carboxylase; PYK, pyruvate kinase.
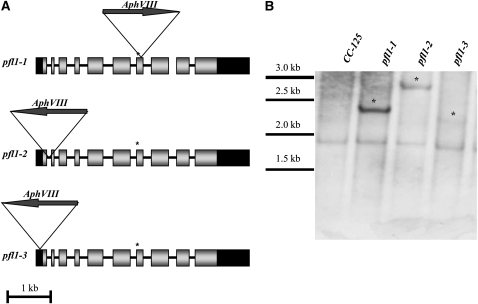
Chlamydomonas contains a single PFL1 gene, and the encoded protein can be targeted to both the chloroplast and mitochondrion. Using the paromomycin marker gene, we generated a library of random insertional mutants in the nuclear genome of Chlamydomonas strain D66 and then screened the library by PCR to identify strains in which PFL1 was interrupted by the marker gene (Figure 2). This screen was performed with a library of >50,000 insertional mutants. To generate the mutants, cells were transformed with a 1.7-kb PCR fragment containing the AphVIII marker gene, which confers resistance to the antibiotic paromomycin, and transformants were plated on selective medium (5 μg mL−1 paromomycin). Genomic DNA isolated from the transformants was screened by PCR using target-gene–specific primers (e.g., primers that anneal to PFL1) and a primer specific to the inserted DNA cassette; the primers that were used for the screen are presented in Supplemental Table 1 online. A more thorough description of this screen has recently been reported (Pootakham et al., 2010; Gonzalez-Ballester et al., 2011). From this screen, we identified the pfl1-1, pfl1-2, and pfl1-3 strains, each having a different disruption of the PFL1 gene. The pfl1-1 transformant was backcrossed four times into strain CC-125 to generate a near-homogeneous genetic background. The other two strains were analyzed in the original D66 genetic background and in the second backcross. Figure 2A shows the positions of the AphVIII gene in each of the strains disrupted for PFL1. In the pfl1-1 mutant, the insertion occurs in the seventh exon of the gene, which is immediately downstream of the sequence encoding the active site. This insertion prevents translation of a full-length product by introducing seven amino acids downstream of the catalytic site followed by a stop codon. The resulting protein is truncated by 318 amino acids (852 amino acids for the full-length protein and 534 amino acids for the truncated protein; see Supplemental Figure 1 online for alignment of the wild type and truncated PFL1). A DNA gel blot with genomic DNA from pfl1-1 indicates that the strain harbors a single copy of the AphVIII gene inserted into PFL1 (Figure 2B); the other pfl1 alleles also seem to have a single insertion in the genome (Figure 2B). These results were confirmed by genetic crosses of the mutant with the wild-type strain, which yielded a 1:1 segregation of paromomycin-resistant to paromomycin-sensitive progeny; all of the progeny tested containing the AphVIII gene showed a similar mutant phenotype (no formate was detected, see below). In pfl1-2 and pfl1-3, the insertion is in the first intron and 5′ untranslated region, respectively, and in both strains, AphVIII integrated in an inverted orientation with respect to PFL1 (Figure 2A). In pfl1-2, the insertion in the intron could potentially be spliced out of the transcript to generate the native transcript or, if the intron is not spliced out of the transcript, could result in the synthesis of a truncated protein as a consequence of an in-frame stop codon after the translation of 145 instead of 852 amino acids (see Supplemental Figure 1 online). In pfl1-3, the insertion resulted in deletion of the first few amino acids, but another in-frame ATG is just upstream of the insertion site. Most of the coding sequence is not perturbed in this strain, and the final protein is predicted to be shorter than the native protein by 10 amino acids (see Supplemental Figure 1 online).
Figure 2.
Mutant Strains.
(A) Marker gene insertion in PFL1 is in the seventh exon, immediately downstream of the sequence encoding the active site in the pfl1-1 allele, whereas it is in the first intron in pfl1-2 and the 5′ untranslated region in pfl1-3. An asterisk designates the position of the active site and black rectangles, gray rectangles, and black bars represent untranslated regions, exons, and introns, respectively.
(B) DNA gel blot analysis of the wild type and the different pfl1 mutants. Genomic DNA was digested with PvuII, and the fragments were separated by agarose gel electrophoresis, transferred to a nitrocellulose membrane, and hybridized with the 1.7-kb PSAD-AphVIII fragment. Asterisks designate the newly introduced PSAD-AphVIII gene sequence. Size standards (in kilobases) are shown to the left.
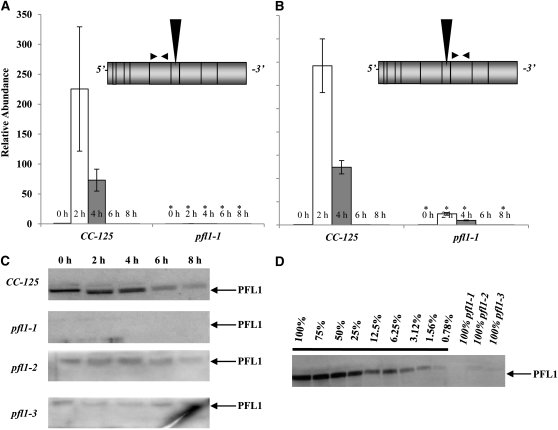
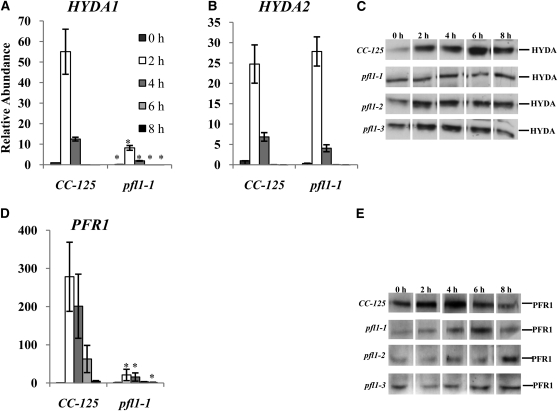
Under anoxic conditions, the PFL1 transcript accumulates to high levels in wild-type parental cells, but shows little accumulation in the pfl1-1 mutant (Figures 3A and 3B). In fact, essentially no transcript was detected by quantitative RT-PCR (qRT-PCR) when the primers used for the reaction annealed 5′ of the insertion site (Figure 3A). The low level of transcript detected 3′ of the insertion site (Figure 3B) might represent read-through from the PSAD promoter, which drives expression of the AphVIII gene. These results suggest that insertion of the marker gene causes transcript instability. Furthermore, we detected no PFL1 protein in the pfl1-1 mutant under either oxic or anoxic conditions, even though wild-type cells have high levels of the protein even prior to the imposition of anoxic conditions (Figure 3C). The strains harboring the pfl1-2 and pfl1-3 alleles accumulate both the PFL1 protein and the PFL1 transcript, but the levels of both are significantly reduced relative to those of wild-type cells (Figure 3C; see Supplemental Figure 2 online). To quantify relative protein levels in pfl1-2 and pfl1-3, an immunoblot of a serial dilution of total protein from wild-type cells after 2 h of dark anaerobic induction was performed and compared with immunoblots for PFL1 in the various mutant strains (Figure 3D). Based on this analysis, <2% of the PFL1 protein accumulated in pfl1-2 and pfl1-3 relative to wild-type cells (Figure 3D). Additionally a double mutant was generated by crossing a single adh1 mutant (Magneschi et al., 2012) with the pfl1-1 strain; this pfl1-1 adh1 mutant produced neither ethanol nor formate (discussed below). In a parallel study, we demonstrated that the adh1 mutant does not synthesize ethanol, but instead synthesizes high levels of glycerol under anoxic conditions; glycerol synthesis would allow for reoxidation of NADH as cultures become hypoxic/anoxic (Magneschi et al., 2012).
Figure 3.
Molecular and Biochemical Characterization of the pfl1-1 Mutant.
(A) and (B) Relative PFL1 transcript abundance in the parental strain (CC-125, left in [A] and [B]) and the pfl1-1 mutant strain (pfl1-1, right in [A] and [B]) under dark, anoxic conditions as determined by qRT-PCR. Expression was monitored for transcripts with sequences upstream (A, Top) and downstream (B, Top) of the insertion site. Total RNA was extracted, and equal amounts of RNA were used for the qRT-PCR analyses. Changes in transcript levels following exposure of the cells to dark, anaerobic conditions (2, 4, 6, and 8 h) are presented as the n-fold change relative to wild-type RNA detected at time 0 h (just prior to transfer). The results show the mean and sd (error bars) for data from three biological replicates, each with two technical replicates. The representation of PFL1 shown in each of the panels is marked for the position of the insert (vertical arrow) and the primers used in the qRT-PCR (paired horizontal arrows). Data were analyzed using a Student’s t test. Asterisk represents transcript levels that are significantly different (P < 0.05) from CC-125 at the respective time points.
(C) The level of PFL1 after exposure of the wild-type strain (CC-125) and pfl1-1, pfl1-2, and pfl1-3 mutants to dark, anoxic conditions (2, 4, 6, and 8 h).
(D) Relative amount of PFL1 in the pfl1 knockdown strains (pfl1-1, pfl1-2, and pfl1-3) relative to the wild-type strain. Serial dilutions of total protein in wild-type cells adapted to 2 h of dark anoxia were used to establish the relative amount of PFL1 in the pfl1-2 and pfl1-3 mutants. The 100% corresponds to 65 μg of total protein in the wild type and mutant strains.
Metabolite Analyses, H2 and CO2 Production
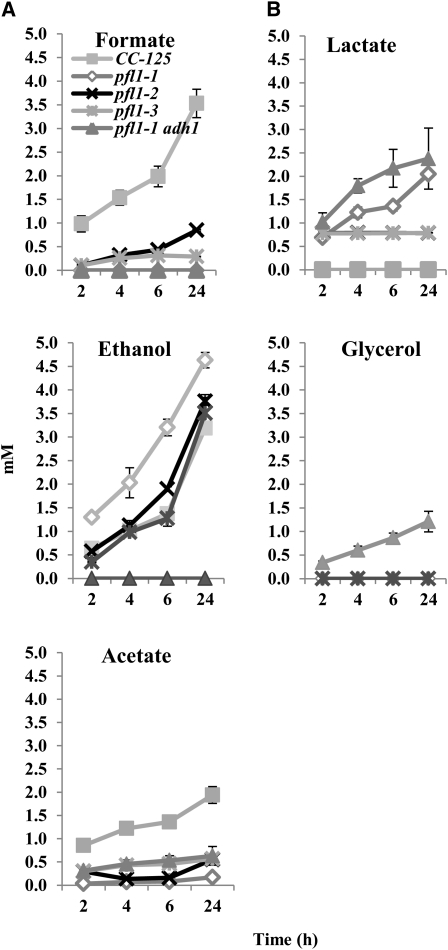
The pfl1-1 mutant lacks pyruvate formate lyase activity based on the inability of the strain to make the PFL1 protein (Figure 3C) and to secrete formate after acclimation of the cells to dark, anoxic conditions (Figure 4A). By contrast, pfl1-2 and pfl1-3, which have highly reduced PFL1 levels, still accumulate formate, although the levels of accumulation in these strains are markedly reduced relative to the wild-type strain CC-125 (a reduction of ~75% at 2 h, with somewhat lesser reductions at 4, 6, and 24 h of dark anoxia induction), as shown in Figure 4A. This likely reflects the accumulation of some active enzyme, albeit at attenuated levels relative to the wild-type strains. Furthermore, all pfl1 mutant strains secrete high levels of lactate, which is not detected in wild-type cultures under the conditions used (Figure 4B). The pfl1 mutants also accumulate ethanol at levels equivalent to or higher than those measured in the wild-type strains (Figure 4A). Furthermore, during dark, anoxic treatment, the acetate concentration in the medium was attenuated for all pfl1 mutants (Figure 4A). By contrast, in wild-type cells, the concentrations of acetate in the medium was generally threefold to fivefold higher than in the mutant strains (Figure 4A). Finally, we constructed a strain containing the pfl1-1 lesion plus a lesion in ADH1 (Magneschi et al., 2012). Although this strain makes neither ethanol nor formate, it excretes lactate, acetate, and glycerol (Figures 4A and 4B); the latter is synthesized by the reduction of dihydroxyacetone phosphate (DHAP) (Figure 1). Glycerol production seems to be the primary mode of NADH reoxidation in the double mutant, which would sustain glycolysis in the absence of ethanol synthesis. This finding, along with the findings of Magneschi et al. (2012), suggest that the production of ethanol as a consequence of PFL1, PFR1, or pyruvate decarboxylase (PDC3) activities (see below) specifically requires ADH1 and that the other Fe-ADH homologs in the Chlamydomonas genome (Joint Genome Institute [JGI] v4.0 Protein ID number 516421 for ADH2 and JGI v4.0 Protein ID number 516422 for ADH3), which should be active in the double mutant strain, cannot replace ADH1, because no ethanol is produced in either the adh1 single mutant (Magneschi et al., 2012) or the pfl1-1 adh1 double mutant.
Figure 4.
External Metabolite Levels in the Wild Type and Mutant Strains during Acclimation of the Cells to Dark Anaerobiosis.
Concentrations of fermentation products secreted by the wild-type strain (CC-125), the different pfl1 mutant strains, and the pfl1-1 adh1 mutant after exposure of the cultures to dark, anoxic conditions for the indicated times. Cells were purged for 30 min with argon in the dark and then maintained in the dark for up to 24 h. Samples were taken at 2, 4, 6, and 24 h; each of the samples was centrifuged and filtered, and the medium was analyzed by HPLC for formate, ethanol, and acetate (A) and for lactate and glycerol (B). Data are from two independent experiments, each with three technical replicates. Errors bars represent sd.
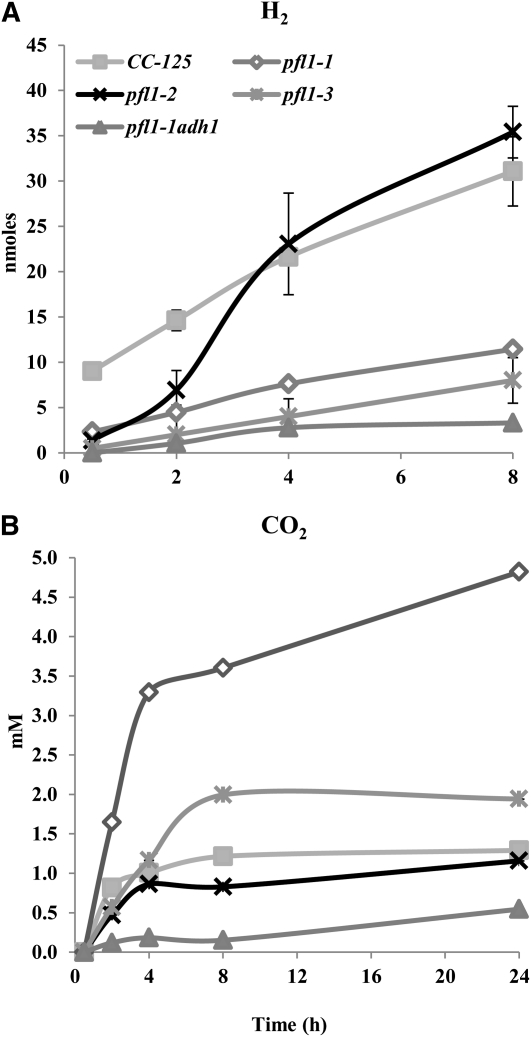
Interestingly, although the pfl1-1 and pfl1-3 mutants could not make (pfl1-1) or produced a small amount of (pfl1-3) formate, and although the level of excreted ethanol was high, these mutants exhibited a significant increase in CO2 production and a decrease in the level of H2 synthesized relative to wild-type cells (Figure 5). The increase in CO2 in the mutant is likely not a result of PFR1 activity, because the mutant strain produces less H2. Hence, other reactions, such as that of PDC3 or the anoxic operation of the pyruvate dehydrogenase complex (PDH1), may be elevated in the mutant (see below). Although the reduced H2 production in the mutant contrasts with the observations reported by Philipps et al. (2011), a comparison between these results should be viewed with caution, because H2 is a minor fermentative product, and its levels are highly sensitive to culturing/induction conditions. qRT-PCR and immunoblot results show that the pfl1-1 mutant exhibits a significant decrease in both hydrogenase transcript and protein levels relative to wild-type cells (Figures 6A to 6C), which is consistent with the results reported by Philipps et al. (2011). Although the antibodies used in this work recognize both the HYDA1 and HYDA2 hydrogenases, transcript analyses suggest that expression from HYDA1 may be more significantly affected than that of HYDA2 (compare Figures 6A and 6B). Moreover, the in vitro hydrogenase activity in the mutant is depressed by 30% when compared with the enzymatic activity in wild-type cells (Table 1). Finally, a significant reduction of both the PFR1 transcript and protein in the pfl1-1 mutant relative to wild-type cells (Figures 6D and 6E) again suggests that there is reduced flux of pyruvate through the pathway, leading to H2 production in the mutant strain exposed to anoxic conditions. An even stronger effect on dark H2 production is observed in the double mutant pfl1-1 adh1 (Figure 5A) with respect to the wild-type strain and the single pfl1 mutant strains. Moreover, the inability of the pfl1-1 adh1 mutant to accumulate ethanol is mirrored by an extreme decrease, relative to pfl1-1 and wild-type CC-125 strains, in the amount of CO2 produced (Figure 5B). These results reinforce our hypothesis that the only active pathway for ethanol production proceeds via ADH1, and that when the ADH1 gene is inactivated, the acetyl-CoA or acetaldehyde cannot be reduced to ethanol (Magneschi et al., 2012).
Figure 5.
H2 and CO2 Accumulation in the Designated pfl1 and pfl1-1 adh1 Mutants during Acclimation to Dark Anoxia.
H2 (A) and CO2 (B) accumulation in the wild-type strain (CC-125), the pfl1 mutant strains, and the pfl1-1 adh1 mutant were measured during acclimation to dark anoxia. For H2 measurements, samples were taken at 0.5, 2, 4, and 8 h after the imposition of anoxic conditions, whereas for CO2, samples were taken up to 24 h after the imposition of anoxic conditions. Both gases were quantified by GC.
Figure 6.
Effect of Dark, Anaerobic Conditions on Levels of Transcripts and Proteins Encoding Enzymes Critical for H2 Production.
Relative HYDA1 (A), HYDA2 (B), and PFR1 (D) transcript abundances in the wild-type CC-125 strain and the pfl1-1 mutant under dark, anoxic conditions, as determined by qRT-PCR. Total RNA was extracted, and equal amounts of RNA were used for the qRT-PCR analyses. Changes in transcript levels after exposure of the cells to dark, anaerobic conditions (2, 4, 6, and 8 h) are presented as an n-fold change relative to RNA from time 0 h (just prior to transfer to anoxic conditions). The results show the mean and sd (error bars) for data from three biological replicates, each with two technical replicates of qRT-PCR experiments. Data were analyzed using a Student’s t test. Asterisk represents transcript levels that are significantly different (P < 0.05) from CC-125 at the respective time points. Levels of the HYDA (C) and PFR1 (E) proteins after exposure of wild-type CC-125 strain and various pfl1 mutants to dark, anaerobic conditions (2, 4, 6, and 8 h) were examined by immunoblot analyses.
Table 1.
In Vitro Hydrogenase Activity in the Mutant (pfl1-1) and the Wild Type (CC-125)
| Hydrogenase Activity (nmol H2 min−1mL−1) |
|||
| Time (h) | CC-125a | pfl1-1a | Activity in Mutant (%) |
| 0.5 | 15.6 ± 1.4 | 4.5 ± 0.6 | 28.7 |
| 2 | 24.7 ± 3.7 | 7.9 ± 2.0 | 32.2 |
| 4 | 27.3 ± 1.0 | 9.2 ± 1.3 | 33.8 |
| 8 | 24.6 ± 2.7 | 7.6 ± 1.3 | 31.0 |
The results show the mean and sd (error bars).
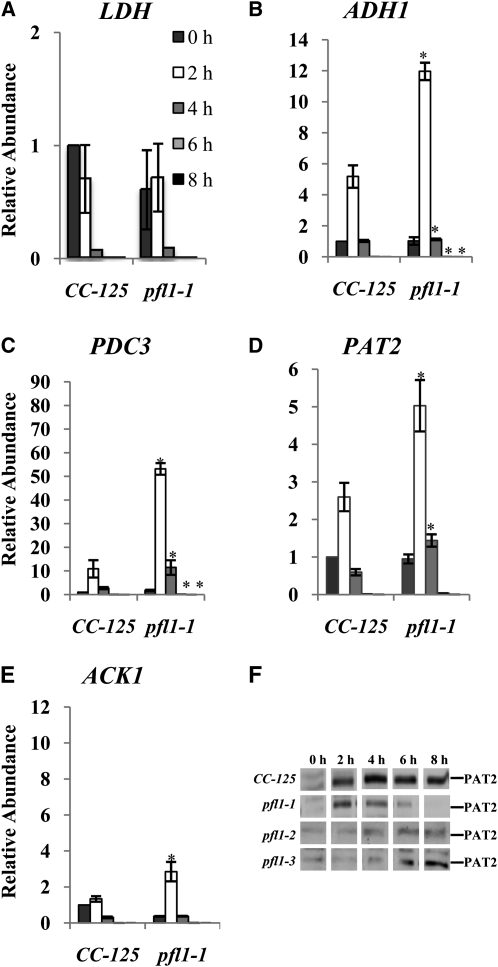
The levels of many transcripts encoding proteins associated with fermentation metabolism are different in the pfl1-1 mutant relative to wild-type cells. The levels of lactate dehydrogenase (LDH) transcripts remain approximately the same in the pfl1-1 mutant and the wild-type cells (Figure 7A), although there is a marked increase in the excretion of lactate by the mutant. This result suggests that LDH can effectively compete for its substrate, pyruvate, in the absence of PFL1, and/or that it is more active in the mutant strain. A biochemical study by Husic and Tolbert (1985) showed the constitutive presence of the LDH protein in Chlamydomonas cells. Significantly, transcripts encoding ADH1 and PDC3 strongly increased (Figures 7B and 7C), whereas those encoding PAT2 and ACK1 increased slightly (Figures 7D and 7E) in pfl1-1 relative to wild-type cells during anoxia. Marked increases in ADH1 and PDC3 may be responsible for both high levels of CO2 and ethanol produced in the pfl1-1 mutant during anoxia. The slight increase in the PAT2 transcript observed in the pfl1-1 mutant does not increase the level of PAT2 protein; indeed, the level of this protein decreases significantly in the mutant strains (Figure 7F), which is consistent with reduced acetate levels.
Figure 7.
Effect of Dark, Anaerobic Conditions on Levels of Transcripts and Proteins Encoding Enzymes Critical for Fermentation Metabolism.
Relative LDH (A), ADH1 (B), PDC3 (C), PAT2 (D), and ACK1 (E) transcript abundances in the wild-type CC-125 and the pfl1-1 mutant strain under dark, anoxic conditions, as determined by qRT-PCR. Total RNA was extracted, and equal amounts of RNA were used for the qRT-PCR analyses. Changes in transcript levels after exposure of the cells to dark, anaerobic conditions (2, 4, 6, and 8 h) are presented as an n-fold change relative to RNA from time 0 h (just prior to transfer). The results show the mean and sd (error bars) for data from three biological replicates, each with two technical replicates of qRT-PCR experiments. Data were analyzed using a Student’s t test. Asterisk represents transcript levels that are significantly different (P < 0.05) from CC-125 at the respective time points. Levels of the PAT2 protein (F) after exposure of wild-type CC-125 strain and various pfl1 mutants to dark, anaerobic conditions (2, 4, 6, and 8 h) were examined by immunoblot analyses.
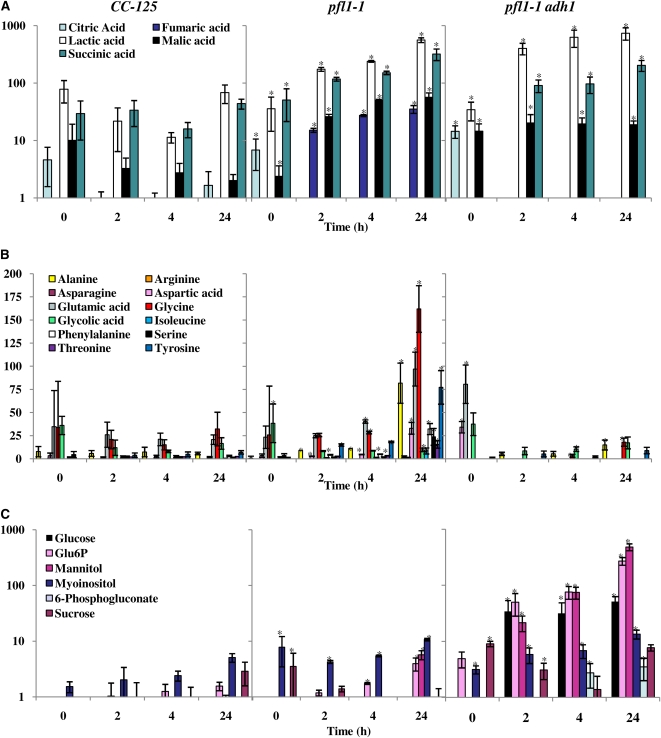
Internal metabolites, analyzed for pfl1-1 (crossed into the CC-125 genetic background) and CC-125 strains, showed that lactic acid and the tricarboxylic acid (TCA) cycle intermediates malate, succinate, and fumarate are elevated in the mutant relative to wild-type cells after exposure to anoxic conditions. Essentially no increase in these metabolites is observed in wild-type cells after the imposition of anaerobiosis (Figure 8A). Additionally, amino acids accumulated to higher levels in the mutant than in the wild-type cells during dark anoxia (Figure 8B). The pfl1-1 adh1 double mutant shows a decrease in the levels of amino acids (Figure 8B) and an increase in the levels of sugars and sugar phosphates relative to the pfl1-1 single mutant (Figure 8C). Among these metabolites, the accumulation of Glc 6-phosphate and 6-phosphogluconate suggests a backup of Glc derivatives from starch catabolism and partial activation of the oxidative pentose phosphate pathway. Overall, these results suggest that the pfl1-1 and pfl1-1 adh1 mutants catabolize internal carbon reserves in a significantly altered manner relative to wild-type cells (Figure 8B).
Figure 8.
Internal Metabolite Levels in the Wild-Type Strain and the pfl1-1 and pfl1-1 adh1 Mutants during Acclimation to Dark Anaerobiosis.
Groups of TCA cycle metabolites (A), amino acids (B), and various sugars (C) as determined by GC-MS measurements. Analyses were performed on 12 replicate samples (three independent biological with four technical replicates each) obtained from CC-125 ([A] to [C], left) pfl1-1 ([A] to [C], center), and pfl1-1 adh1 ([A] to [C], right). Cultures were placed under dark, anoxic conditions for 2, 4, and 24 h. Metabolites were identified based on their retention index as well as by spectral comparisons with prerun standards. Values were obtained from at least nine samples (nine out of 12 samples are shown). Each value represents the relative abundances for individual compounds obtained by normalization of the individual peak areas to that of the internal compound, ribitol. In (A) and (C), the data are shown as log of the relative abundances. Data were analyzed using a Student’s t test. Asterisk represents metabolite levels that are significantly different (P < 0.05) from CC-125 at the respective time points.
In sum, it seems that a loss of PFL1 activity results in reduced PFR1 and hydrogenase activity and that pyruvate catabolism occurs primarily through the action of LDH and PDC3. An increase in these activities would explain the high extracellular and intracellular lactate and ethanol levels, as well as the increase in CO2 production. The acetyl-CoA and acetaldehyde formed in the pfl1-1 strain is primarily converted to ethanol through ADH1, with some acetyl-CoA being converted to acetate via the coupled PAT2/ACK1 reactions. The former reactions seem to specifically use ADH1, as implied by the phenotype of the pfl1-1 adh1 double mutant, whereas the latter reaction seems to be low in the pfl1-1 mutant strain. In addition, there seems to be elevated rTCA activity based on the finding that intracellular levels of malate, fumarate, and succinate increase in the pfl1-1 strain; this is similar to the phenotype of the hydEF-1 mutant (Dubini et al., 2009). There is also a substantial change in the pools of both amino acids and sugars in the pfl1-1 and pfl1-1 adh1 mutant strains. Some of these changes might also be important for balancing redox conditions of the cell and recycling NADH/NAD+, whereas others may control the accumulation of potentially toxic intracellular levels of pyruvate (see below).
DISCUSSION
Characterization of the Chlamydomonas nuclear genome has provided insights into the evolution of green algae and plants and has facilitated identification of numerous polypeptides that function in key metabolic pathways. Both molecular and physiological studies have demonstrated that Chlamydomonas exhibits marked metabolic flexibility, accommodating extreme fluctuations in response to both nutrient availability and environmental O2 levels (Yildiz et al., 1996; Wykoff et al., 1998; Melis et al., 2000; Moseley et al., 2002; Pollock et al., 2003; Grossman et al., 2007; Mus et al., 2007; Dubini et al., 2009; Grossman et al., 2011; Philipps et al., 2011). Although typically grown in the laboratory under conditions optimized for rapid cell proliferation, algae in their natural habitat frequently experience suboptimal growth conditions that could reflect an inadequate supply of nutrients (e.g., N, P, S, and Fe) and/or an inability to rapidly produce fixed carbon backbones (Yildiz et al., 1994; Yildiz et al., 1996; Davies et al., 1999; Shimogawara et al., 1999; Moseley et al., 2009; Pootakham et al., 2010; Gonzalez-Ballester et al., 2011). Furthermore, many compounds that accumulate in aquatic and soil environments (through both anthropogenic and biogeochemical activities) may inhibit specific metabolic pathways, making it advantageous for organisms to maintain metabolic flexibility. Extensive studies have been conducted to elucidate the responses of Chlamydomonas to Fe, Cu, N, S, and P limitation, as well as O2 depletion (Hill et al., 1996; Wykoff et al., 1998; Melis et al., 2000; Moseley et al., 2000; Pollock et al., 2003; Posewitz et al., 2004; Bölling and Fiehn, 2005; Moseley et al., 2006; Naumann et al., 2007). It is essential that we begin to understand the ways in which algal metabolism “adjusts” to environmental challenges if we are to develop an informed picture of (1) the ways in which algae compete and survive in nutrient-limited environments; (2) metabolite exchange and symbiotic/parasitic relationships with other microbiota in natural microbial communities; (3) the role of unicellular algae in global C, N, and P cycling; and (4) potential strategies for engineering phototrophic microorganisms to synthesize molecules of pharmaceutical and nutraceutical value, as well as for the production of energy-rich metabolites for possible future biofuel production.
Anoxic and microoxic conditions are frequently experienced by organisms that inhabit the soil, where there can be high respiratory O2 consumption and low rates of O2 exchange. Fermentative metabolism by Chlamydomonas and other green algae represents a significant ecological component of the soil environment that would enrich that environment through the production of organic acids, alcohols, and H2. Previously, we examined the consequences of anoxia on transcript levels and metabolite secretion in wild-type cells and characterized a mutant defective for HYDEF that is unable to synthesize active hydrogenases (Mus et al., 2007; Dubini et al., 2009). Intriguingly, the hydEF-1 mutant exhibited activation of rTCA reactions that resulted in the production and secretion of succinate. This result was unexpected, because rTCA metabolites are usually only secreted in trace quantities in wild-type strains experiencing anoxic conditions. These findings raise important questions concerning the biotic and abiotic interactions that led to the evolution of the fermentative network and the regulatory mechanisms that integrate the activities among the branches that comprise this network. These studies also demonstrate the ability of this alga to adjust electron and carbon flow through the various branches of fermentation metabolism.
In this study, the conversion of pyruvate to acetyl-CoA by PFL1 was completely eliminated in the pfl1-1 null mutant, as indicated by the absence of formate secretion, and was strongly inhibited in the two other strains with lesions in pfl1. We expected that the major competing reaction driven by PFR1, an enzyme that converts pyruvate to acetyl-CoA, CO2, and reduced ferredoxin, would increase (Figure 1). The reduced ferredoxin generated in this reaction could funnel electrons into hydrogenases, resulting in increased H2 generation. However, rather than observing increased PFR1/hydrogenase activity in the mutants, in vitro hydrogenase activity (Table 1), HYDA1 and PFR1 transcript levels, and both PFR1 and HYDA protein abundances decreased (Figure 6). These observations are counter to our original hypothesis and indicate that metabolic flux through PFR1/HYDA is attenuated in pfl1 mutants. This reduction of PFR1 and HYDA1 transcripts was unexpected and implies that PFL1 activity not only influences metabolic pathway fluxes, but may also affect transcriptional and/or posttranslational processes. Attenuated levels of HYDA1 transcript and protein were also observed in the pfl1 mutant that was characterized by Philipps et al. (2011). These observations necessitate a reexamination of the relationship between PFL1 and PFR1; rather than simply competing for substrate, these two enzymes may be operating in a coordinated fashion to optimally satisfy global metabolic demands during anoxia in Chlamydomonas, perhaps even through their association in a macromolecular complex. Interestingly, the level of the PDC3 transcript was markedly elevated in the pfl1-1 mutant (up to sevenfold) (Figure 7C), which may reflect increased PDC3 and activity. Activation of the PDC3 pathway would provide a mechanism to compensate for reduced PFL1 activity in the pfl1 mutants. This compensatory mechanism could also explain the attenuated PFR1 activity associated with the mutant. An increase in lactate secretion was also observed in our pfl1 mutants, and this was previously observed in wild-type cells treated with the PFL1 inhibitor hypophosphite (Kreuzberg, 1984) as well as in the pfl1 mutant described by Philipps et al. (2011). Hence, when PFL1 activity is eliminated or reduced, the PDC3 and LDH pathways seem to be activated, resulting in rapid metabolism of the pyruvate generated by glycolysis as well as diminished PFR1 activity.
In wild-type cells, ethanol is proposed to be formed from acetyl-CoA and the alcohol/acetaldehyde dehydrogenase (ADH1) pathway. In the pfl1-1 mutant, no acetyl-CoA from the PFL1 reaction is synthesized, and PFR1 transcript and protein abundances are significantly diminished (Figure 3). Hence, both of the major pathways associated with anoxic acetyl-CoA synthesis are compromised in the pfl1-1 mutant, which would severely limit ethanol production from acetyl-CoA. Therefore, it is surprising that there was generally a reproducible increase in ethanol production in the pfl1-1 mutant relative to wild-type cells (Figure 4A), a result consistent with an independent pfl1 mutant study (Philipps et al., 2011). As discussed above, the catalytic activity of PDC3, a predicted pyruvate decarboxylase that converts pyruvate to acetaldehyde and CO2, may coordinate with ADH1 activity, allowing for efficient ethanol production in this mutant strain. The finding that PDC3 transcripts become elevated in the pfl1-1 mutant relative to the wild-type strains (Figure 7C) combined with the metabolite profiles observed in the mutant (Figure 4) support the hypothesis that the PDC3/ADH1 pathway is activated in pfl1 mutants, and that this activation is responsible for most of the ethanol produced in those strains. We favor this hypothesis for several reasons. First, a significant increase in CO2 synthesis (approximately threefold to fourfold) in pfl1-1 relative to wild-type cells (Figure 5B) implies increased pyruvate decarboxylation. Because PFR1 transcript and protein abundances are diminished and H2 production remains low in pfl1-1, it is unlikely that the increased levels of CO2 are a consequence of PFR1 activity, leaving PDC3 as the most likely fermentative enzyme responsible for pyruvate decarboxylation in the mutant. Second, acetate secretion is severely attenuated in pfl1 mutants (Figure 4A), indicating that availability of acetyl-CoA as substrate for acetate synthesis may also be diminished. Finally, ethanol production increases in pfl1 mutants. Although two fermentative pathways for ethanol production have been posited (Figure 1; PFL1-PFR1/ADH1 and PDC3/ADH1), the acetyl-CoA pathway to ethanol requires the oxidation of two NADHs and must be coupled to metabolic pathways that allow either one-half of the 3C metabolic flux to be processed in pathways that do not oxidize NADH (because only one NADH is produced per 3C synthesized in glycolysis) or that generate an additional NADH per 3C in a reaction outside of glycolysis. This redox balance is achieved in wild-type cells by coupling PFL1-PFR1/ADH1 ethanol production to acetate synthesis (Figure 1). The advantage for wild-type cells using the PFL1-PFR1/ADH1-PAT-ACK fermentative pathways is the generation of an additional ATP for each 6C metabolized (through the conversion of acetyl-CoA to acetate). The significant decrease in acetate secretion by pfl1 mutants suggests that these strains activate an alternative pathway to direct pyruvate away from acetyl-CoA and toward the production of ethanol via the PDC3/ADH1 pathway. This requires only a single NADH oxidation, which would exactly balance the generation of reductant in glycolysis. In sum, our currently favored hypothesis is that the PDC3/ADH1 pathway is activated in pfl1 mutants, which is consistent with the observed increases in CO2 and ethanol, as well as the decrease in acetate accumulation. Activation of the PDC3/ADH1 pathway, combined with increased LDH activity, would effectively alleviate the build up of pyruvate that results from the block at PFL1 and attenuated PFR1 activity and would also allow for redox balance, because the generation of NADH by glycolysis would match its use in the LDH and PDC3/ADH1 pathways (both pathways oxidize a single NADH molecule).
Although this is our favored, redox-balanced model for explaining the observed metabolic profiles in pfl1 mutants, two competing models should also be considered. First, an increased pyruvate flux through PFR1 would generate reduced ferredoxin and increased acetyl-CoA and would account for the observed increase in CO2 levels (Figure 5B). The reduced ferredoxin would then have to be used to reduce NAD+ to NADH, either directly or through NAD(P)H synthesis that is catalyzed by a transhydrogenase. This would provide both of the NADH equivalents (one from glycolysis and one from reduced ferredoxin-PFR1) that could be used to reduce acetyl-CoA to ethanol via ADH1. Because two NADH molecules would be produced per acetyl-CoA (one in glycolysis and one by PFR1-ferredoxin-NADH oxidoreductase), all of the acetyl-CoA that is generated would have to be metabolized to ethanol to retain redox balance, which would explain the reduction in acetate levels. We do not favor this model based on the reduction of PFR1 transcript and protein levels in pfl1 mutants. Second, PDH activity could decarboxylate pyruvate, leading to an increase in CO2 levels and concomitant NADH production. Again, this would yield two NADH molecules per acetyl-CoA (one from glycolysis and one from PDH), providing the two NADH molecules necessary to catalyze the conversion of acetyl-CoA to ethanol, and again all acetyl-CoA would have to be metabolized to ethanol to retain redox balance. We do not favor this model, because we assume that PDH is coordinated with oxic metabolism and respiratory electron transport that occurs in mitochondria. However, further experimentation is required to unambiguously determine whether the PDC3/ADH1 model is correct or whether one or both of the alternative pathways are responsible for the observed increases in ethanol and CO2 as well as the diminished levels of acetate in pfl1 mutants.
The Chlamydomonas genome encodes two putative Fe-type alcohol dehydrogenases (JGI v4.0 Protein ID number 516421 for ADH2 and JGI v4.0 Protein ID number 516422 for ADH3) in addition to the bifunctional ADH1. However, the results that we present here suggest that all ethanol production that occurs during fermentation metabolism is coupled to ADH1, because the pfl1-1 adh1 double mutant can no longer synthesize ethanol, and ADH1 is upregulated in the pfl1-1 mutant. Furthermore, results presented here and information from the characterization of the adh1 single mutant (Magneschi et al., 2012) suggest that the conversion of either acetaldehyde (from PDC3 activity) or acetyl-CoA (from PFL1 or PFR1 activity) to ethanol requires ADH1; the single mutant is unable to make any ethanol under dark, anoxic conditions.
Interestingly, both the adh1 single mutant (Magneschi et al., 2012) and the pfl1-1 adh1 double mutant (Figure 4) produce glycerol, which limits the production of pyruvate and oxidizes one NADH [DHAP to glycerol], allowing for the regeneration of NAD+ and sustained glycolytic activity. Glycolysis produces two 3C molecules per Glc at the level of DHAP/glyceraldehyde phosphate (GAP). As shown in Figure 1, the first 3C DHAP is converted to glycerol, oxidizing one NADH in the process, prior to the production of NADH by the GAP dehydrogenase reaction. The second 3C GAP is metabolized to pyruvate, during which one NADH is produced. Therefore, glycerol production allows for one-half of the glycolytic flux to proceed to pyruvate while maintaining redox balance (NADH generated when GAP is converted to pyruvate is consumed in the conversion of DHAP to glycerol). This allows pyruvate to be converted to acetate, producing one ATP without oxidizing any NADH, and explains the increased acetate levels observed in the pfl1-1 adh1 mutant relative to the pfl1 mutant. Glycerol production replaces ADH1 in terms of maintaining cellular redox balance in the double mutant. Lactate production (redox neutral with respect to glycolysis) increases slightly in the pfl1-1 adh1 double mutant above that of the single pfl1 mutants. Therefore, the double mutant responds to the metabolic network perturbations by synthesizing lactate (observed in the pfl1 single mutant), as well as glycerol and acetate (observed in the adh1 single mutant). This combination of reactions reduces the flow of carbon to pyruvate, increases ATP production through the synthesis and excretion of acetate, and sustains redox balance, which in turn allows for continued glycolytic activity. It is remarkable that Chlamydomonas can readily adjust its fermentative metabolism to attain both redox balance and efficient energy production even when the two catabolic pathways that dominate fermenting wild-type cells are disrupted. In essence, introduction of the pfl1-1 adh1 lesions into wild-type Chlamydomonas cells converts it from a formate/acetate/ethanol to a glycerol/lactate/acetate fermentor.
Additionally, mannitol and other sugars are elevated in the double mutant (Figure 8), indicating further adjustments of metabolite fluxes that may be involved in facilitating the regeneration of NAD+ and NAD(P)+ under anoxic conditions. Under anaerobic conditions, yeast cells can use Fru, Xyl, and many other sugars as electron acceptors. The reduction of Fru to mannitol requires reducing equivalents (Baek et al., 2003) and is mediated by an NAD(P)H-dependent mannitol dehydrogenase. The use of reductant by NAD(P)H-dependent enzymes during anoxia suggests that enzymes such as transhydrogenases, which catalyze the efficient interconversion between NAD(P)H and NADH, may also be critical under anaerobic conditions. Our previous work demonstrated a marked increase in the accumulation of Chlamydomonas transhydrogenase mRNA when the cells were experiencing hypoxic/anoxic conditions (Mus et al., 2007).
Although no significant succinate secretion was observed in the pfl1-1 mutant, intracellular levels of malate, fumarate, and succinate all increased under anoxic conditions (Figure 8). This is consistent with activation of rTCA reactions, which we previously observed in the Chlamydomonas hydEF-1 mutant (Dubini et al., 2009). It is not clear why the cells prefer to synthesize and secrete lactate and accumulate intracellular rTCA intermediates when PFL1-dependent formate and acetyl-CoA production are eliminated, or why succinate accumulation and secretion is preferred when hydrogenase activity is abolished. However, it is likely that differential transcriptional and posttranslational processes affect fermentation metabolism, and these processes are almost certainly regulated by both intracellular redox and/or the levels of specific metabolites. In sum, elimination of PFL1 abolishes formate secretion and activates alternative fermentative branches that lead to increases in the intracellular levels of lactate, Ala, succinate, malate, and fumarate and the extracellular accumulation of lactate as well as increases in CO2 and ethanol production. These alternative fermentation reactions allow for pyruvate metabolism and the reoxidation of the NADH produced during glycolysis in the absence of PFL1 activity, which would help sustain ATP production as Chlamydomonas cells become microoxic/anoxic.
The effect of the pfl1-1 lesion on hydrogenase expression is intriguing. Although H2-photoproduction rates are similar to those observed in the control strains (see Supplemental Figure 3 online), this is likely a consequence of the production of sufficient hydrogenase to oxidize all available reduced ferredoxin that is generated by photosynthetic electron transport under the experimental conditions used. Attenuation of HYDA1 but not HYDA2 transcript abundance suggests that regulatory processes elicited in the pfl1-1 mutant primarily affect HYDA1 transcription/transcript abundance and that the two Chlamydomonas hydrogenases are differentially regulated. HYDA1 transcripts were recently shown to be modulated by O2 concentrations, whereas HYDA2 mRNA levels were suggested to be mostly affected by the circadian clock (Whitney et al., 2011). Little is presently known regarding factors that regulate hydrogenase and the transcription of genes encoding enzymes involved in fermentation processes; however, it is clear that anoxic conditions elicit substantial transcriptional regulation of fermentative pathways (Mus et al., 2007). Interestingly, LDH transcript levels do not increase in response to anoxia and are similar in wild-type cells and the pfl1-1 mutant, suggesting that the activity of the LDH enzyme is either translationally or posttranslationally regulated and/or that there is greater availability of its substrates, pyruvate and/or NADH, for the LDH reaction in the absence of PFL1 (for which pyruvate is also a substrate). Prior research indicated that the LDH enzyme is constitutively present (Husic and Tolbert, 1985), which is consistent with our transcript data; however, little is known about its regulation in Chlamydomonas.
Although some of our observations are similar to those of another study of a knockout strain of PFL1 in Chlamydomonas (Philipps et al., 2011), there are also some striking differences for which we have additional supporting data. Differences were particularly noted with respect to dark, fermentative H2 production. In the study of Philipps et al. (2011), the pfl1 knockout strain showed an increase in fermentative H2 production. In our study, we show that the pfl1-1 mutant has lower hydrogenase transcript, protein, and activity and synthesizes less dark, fermentative H2 than wild-type cells. Furthermore, we show that inactivation of PFL1 caused accumulation of intracellular metabolites that were not observed at significant levels in wild-type cells experiencing anoxia. The intracellular levels of lactate, Ala, succinate, malate, and fumarate were all markedly increased, with the accumulation of lactate being the most pronounced. These results suggest that an inability to metabolize pyruvate by PFL1 elicits dramatic metabolic adjustments of pyruvate metabolism. From our analyses, the pfl1-1 mutant seems to generate more lactate (similar to what was observed by Philipps et al. [2011]) and has elevated ethanol production, but exhibits decreased levels of acetyl-CoA, CO2, and H2 through the PFR1 pathway (at odds with what was observed by Philipps et al. [2011]) as well as a change in the activity of the rTCA reactions based on accumulation of specific intracellular metabolites (intracellular metabolites were not specifically measured by Philipps et al. [2011]). Furthermore, there is an elevated intracellular level of Ala, which likely reflects increased Ala transaminase activity. This enzyme catalyzes the production of Ala from pyruvate and Glu (without reoxidation of NADH). Increased Ala synthesis may reflect the need for the cells to maintain low intracellular levels of pyruvate, which would help sustain glycolytic activity and prevent the accumulation of pyruvate to concentrations that would interfere with the regulation of fermentative metabolism and ATP synthesis (Gupta et al., 2009; Zabalza et al., 2009). Recently, Ala biosynthesis was shown to be critical for the survival of Lotus japonicus during waterlogging (Rocha et al., 2010).
In summary, fermentative metabolism in Chlamydomonas readily adapts to the loss of PFL1 activity by synthesizing a variety of other metabolites that would contribute to the reoxidation of NADH and allow for sustained glycolytic production of ATP during anaerobiosis. Additionally, activation of an alternative ethanol-production pathway, probably involving PDC3, is highly likely. This hypothesis is consistent with metabolite data generated from anaerobically acclimated Chlamydomonas cells treated with the PFL1 inhibitor hypophosphite (Kreuzberg, 1984). It will be important to generate a pdc3 mutant to test this hypothesis. Elevated activity of pathways in the pfl1-1 mutant that do not seem to be active in wild-type cells further illustrates the metabolic flexibility of Chlamydomonas, an alga that seems to be well adapted to withstand many environmental challenges, including anoxia. Our inability to predict the consequences of lesions that block specific branches of fermentation metabolism highlights our lack of understanding of metabolism and its regulation in photosynthetic organisms. The availability of the Chlamydomonas genome sequence, high-throughput “omics”-based approaches, and mutants disrupted in many cellular processes will provide the foundation to further elucidate the extent of metabolic flexibility in this alga, the mechanisms that underlie this unexpected capacity to remodel metabolism, and how these capabilities reflect both the life style and interactions among organisms in the soil.
METHODS
Strains and Growth Conditions
The Chlamydomonas reinhardtii strains used in these experiments were CC-125 (nit2−, mt+); D66 (CC-4425; nit2−cw15, mt+) (Pollock et al., 2003); pfl1-1, a mutant originally identified in the D66 (Pollock et al., 2003) genetic background, but backcrossed four times with CC-125; and two other pfl1 insertion mutants, pfl1-2 and pfl1-3; the lesions in the latter two strains were analyzed in the D66 genetic background and in the second backcross. Additionally, an adh1 mutant (Magneschi et al., 2012) was isolated and backcrossed to the pfl1-1 strain to generate the pfl1-1 adh1 double mutant. All of the mutants were identified by a PCR-based screen that was previously described (Pootakham et al., 2010; Gonzalez-Ballester et al., 2011). Both the parental and mutant cells were grown in Tris-acetate-phosphate medium (pH 7.2) at 25°C, 80 μmol photon m−2 s−1 continuous photosynthetically active radiation (cool, white fluorescence lights) at the surface of 1-L Roux culture bottles, stirred with a magnetic stir bar, and vigorously bubbled with air enriched with 3% CO2. The densities of the cell cultures ranged from 2 to 4 × 106 cell/mL.
DNA Gel Blot Analyses
Genomic DNA was isolated from 50-mL liquid cultures of Chlamydomonas CC-125 and the three pfl1 mutant alleles using a standard phenol-chloroform extraction protocol (Sambrook et al., 1989). Ten micrograms of genomic DNA was digested for 2 h with 10 U of PvuII (New England Biolabs), and the DNA fragments were separated by agarose (0.8%) gel electrophoresis and then blotted overnight in 20× SSC onto nylon membranes (Bio-Rad). Transferred DNA was cross-linked to the membrane by UV light illumination. An alkaline phosphatase-labeled DNA probe was synthesized by chemically cross-linking a thermostable alkaline phosphatase to the 1.7-kb PSAD-AphVIII PCR fragment synthesized from plasmid DNA using the primers RB1 5′-ATGGGGCGGTATCGGAGGAAAAG-3′ and RB2 5′- TACCGGCTGTTGGACGAGTTCTTCTG-3′. Probe synthesis and hybridizations were performed using an Amersham AlkPhos Direct Labeling and Detection System Kit following the manufacturer’s protocol (Amersham Biosciences). The cross-linked, membrane-bound genomic DNA was hybridized overnight with the alkaline phosphatase-linked, 1.7-kb PSAD-AphVIII PCR product.
Anaerobic Induction of Liquid Cell Suspensions
Chlamydomonas cultures were grown on Tris-acetate-phosphate medium to 16 to 24 μg mL−1 total chlorophyll and centrifuged at 2500g for 1 min to pellet the cells. Pelleted cells were resuspended in one-tenth volume of anaerobic induction buffer, which contained 50 mM potassium phosphate (pH 7.0) and 3 mM MgCl2 (Ghirardi et al., 1997). The cells were transferred to a sealed anaerobic vial, flushed with argon for 30 min, and then incubated anaerobically in the dark at room temperature. Samples were collected at different time points over a 24-h period.
Extraction of RNA
Total RNA was isolated from frozen cell pellets using a standard phenol-chloroform extraction protocol (Sambrook et al., 1989). The RNA was precipitated overnight in a 1:1 vol/vol with 8 M lithium chloride at 4°C to eliminate most of the DNA in the preparation. To completely free the sample of genomic DNA, ~40 μg of the RNA was treated with 5 U of RNase-free DNase I (Qiagen) for 1 h at room temperature. A Qiagen RNeasy MinElute Kit (Qiagen) was used to purify the DNase-treated total RNA and to remove degraded DNA, tRNA, 5.5 rRNA, DNase, contaminating proteins, and potential inhibitors of the reverse transcriptase reaction. The A260 of the eluted RNA was measured, and its integrity checked on a formaldehyde gel.
Reverse Transcription and qRT-PCR
The abundance of specific transcripts in total mRNA from each sample was quantified by qRT-PCR using the Engine Opticon system (Bio-Rad). First-strand cDNA synthesis was primed from purified, total RNA template using specific primers for each of the Chlamydomonas genes of interest (shown as the reverse primers in Supplemental Table 1 online). The reverse transcription reaction was performed as described previously (Mus et al., 2007; Dubini et al., 2009). Amplifications were performed using the following specific cycling parameters: an initial single step at 95°C for 10 min (denaturation), 40 cycles of (1) 95°C for 30 s (denaturation), (2) 60°C for 45 s (annealing), and (3) 72°C for 30 s (elongation), and (4) the amplification products were quantified by measurements of SYBR green fluorescence after holding the reaction at 80°C for 10 s (this step was incorporated into the protocol to avoid background signals resulting from primer dimer formation). A final elongation step was performed at 72°C for 10 min. Melt curves (65 to 100°C, heating rate of 0.2°C s−1 with continuous fluorescence measurements) were evaluated for all PCRs to ensure that single DNA species were amplified. Both the absolute (Steunou et al., 2006) and relative levels of each specific RNA (normalized to the T0 sample corresponding to the oxic conditions) were determined. All reactions were performed in triplicate with at least two biological replicates.
Extracellular Metabolite Analysis
Organic acid analysis was performed by liquid chromatography using a Hewlett Packard Series 1200 HPLC. Dark-adapted cells were collected at various time points after the imposition of anoxic conditions (Figure 4) and were centrifuged (10,000g) for 1 min. The supernatant was transferred to a new vial, and the samples were frozen in liquid N2 for subsequent analysis. Samples were thawed, centrifuged, and filtered prior to injection of 100 μL of the supernatant onto an Aminex HPX-87H (300 × 7.8 mm) ion exchange column. The separation of metabolites by the column was performed at 45°C with 8 mM filtered sulfuric acid as the mobile phase and a flow rate of 0.6 mL min−1. Retention peaks for the various organic acids were recorded using Agilent ChemStation software, and quantifications were performed by comparisons with the absorption of known amounts of a standard for each of the organic acids. Ethanol was detected using the refracted index detector attached to the HPLC.
Harvesting Cultures and Sample Preparation for Intracellular Gas Chromatography Coupled to Mass Spectrometry Metabolite Analysis
Cultures were exposed to anaerobic conditions, with sample collection at 2, 4, and 24 h. At each collection time, 1.0 mL of culture was transferred with an argon-purged syringe into 3.0 mL of 70% methanol solution prechilled at −80°C and centrifuged at 10,000g for 1 min, and the cell pellet was immediately frozen in liquid nitrogen. The only exception was the initial aerobic time point (t0), for which 2.0 mL of the culture was quenched with 6.0 mL of chilled 70% methanol. In all, 12 samples (three independent biological replicates, each with four technical replicates) were harvested at each of time point, frozen, and stored at −80°C until analysis. To analyze samples, the cell pellets were thawed on ice, resuspended in 1 mL of chilled (−80°C) methanol:chloroform:water in the ratio 10:3:1 (vol:vol:vol) containing 4 μg of ribitol (Sigma-Aldrich) as an internal standard, and then sonicated (three times for 30 s each, with cooling intervals of 30 s on dry ice) followed by centrifugation (10,000g for 2 min) to disrupt the cells and remove cell debris. The supernatant was dried under a N2 atmosphere, and the dried extract was resuspended in 40 μL of methoxyamine hydrochloride (20 mg mL−1 in pyridine), incubated at 37°C for 90 min, and then derivatized at 55°C for 30 min by the addition of 60 μL of N-methyl-N-(trimethylsilyl) trifluoroacetamide containing 1% trimethylchlorosilane (Thermo Fisher Scientific).
Gas Chromatography Coupled to Mass Spectrometry Analytical Procedures
Metabolites were detected using gas chromatography coupled to mass spectrometry (GC-MS). One microliter of derivatized sample (see above) was injected using a splitless injection technique into a GC-MS instrument. This instrument consisted of a 7683 series autosampler, a 7890A GC system, and a 5975C inert XL mass selective detector (MSD) with a Triple-Axis Detector (Agilent Technologies). The inlet temperature was set at 225°C. The compounds were separated using a 30-m DB-35MS column (Agilent Technologies) with a 0.25-mm inner diameter and 0.25-μm film thickness, which is comprised of 35% phenyl and 65% dimethyl arylene siloxane. Helium was used as the carrier gas at a flow rate of 1 mL/min. GC parameters included 50°C isothermal heating for 2 min followed by a 5°C/min increase to 150°C, a hold for 2 min at 150°C, and a second temperature ramping phase of 7°C/min to 320°C, followed by a final hold for 2 min at 320°C. The MSD transfer line and the MS quadrupole were maintained at 280°C and 150°C, respectively, whereas the MS source temperature was 230°C. Compounds were detected using the scan mode with a mass detection range of 40 to 500 atomic mass units.
GC-MS Metabolite Identification and Quantification
Chromatograms were analyzed using MSD Enhanced ChemStation data analysis software (Agilent Technologies). The identity of each metabolite was based on its retention index and spectral comparisons to prerun standards. Unique m/z ions (target ions) were selected for each compound, which were used to manually quantify the compounds based on their peak areas. A minimum of nine (out of 12) samples were used for calculation purposes for each time point. The peak areas were normalized to that of the internal standard, ribitol, followed by multiplication by a constant value (10,000 in this case); hence, reported values are in relative arbitrary units.
CO2 and H2 Measurement
CO2 levels were below detection limits in the serum vial headspace of anaerobically acclimated cells. Therefore, after anaerobic induction, 1 mL of anoxic cells was transferred in a gas-tight syringe to a sealed vial into which 1 mL of 1 M HCl was added. The acidified cell suspension was shaken vigorously to liberate CO2, which was quantified by GC (Hewlett Packard 5890 series II) using a Supelco column (80/100 PORAPAK N 6 ft × 1/8 inch × 2.1 mm) coupled to a thermal conductivity detector.
Fermentative H2 production was measured in 400 μL of head-space gas that was withdrawn from sealed anaerobic vials and analyzed by GC (Agilent Technologies 7890 GC system) using a Supelco column (60/80 mole sieve 5A 6 ft × 1/8 inch) coupled to a thermal conductivity detector. To measure the H2 photoevolution, 250 μL of cells were placed in a Clark-electrode assay chamber as described previously (Posewitz et al., 2004; Mus et al., 2007).
Protein Isolation, SDS-PAGE, and Immunoblot Analysis
Frozen cells were thawed and resuspended in 50 mM Tris buffer (pH 8.0) containing 10 mM EDTA and 2% SDS. For protein concentration determination, a BCA Assay Reagent Kit (Pierce) was used according to the manufacturer’s instructions. For protein gels, cells were solubilized in 2% SDS and 1 mM β-mercaptoethanol and then boiled for 5 min. Proteins were first separated by SDS-PAGE in a 10% polyacrylamide gel and then transferred to polyvinylidene difluoride membranes by a semidry transfer method. The membranes were blocked with a 5% suspension of powdered milk in Tris-buffered saline with 0.1% Tween-20 prior to an incubation of 1 h in the presence of primary antibodies. The dilutions of the primary antibodies used were 1:30,000 for α-PFL1, 1:5,000 for α-HYD, 1:1,000 for α-PFR1, and 1:2,000 for α-PAT2. A 1:10,000 dilution of horseradish peroxidase-conjugated anti-rabbit IgG (Promega) was used as a secondary antibody. The peroxidase activity was detected by an enhanced chemiluminescence assay (Amersham Biosciences). Proteins were quantified by comparison of the band intensities using ImageJ (http://rsbweb.nih.gov/ij/).
Antibodies
Antibodies were produced by Agrisera Antibodies (http://www.agrisera.com/en/info/home.html) against the synthesized peptides, EWLSHENRFQILERK and RSGRNYARDTIDRIF of the Chlamydomonas PFR1 and PAT2, respectively. Synthesized peptides were conjugated to Keyhole Limpet Hemocyanin (KLH) carrier protein via a Cys that was added to the N-terminal of the peptide. Hydrogenase levels were evaluated using commercial antibodies that recognize both HYDA1 and HYDA2 (Agrisera; art number AS09 514). PFL1 antibody was kindly provided by Ariane Atteia at the Laboratoire de Bioénergétique et Ingénierie des Protéines, Institut de Microbiologie de la Mediterranéè, Marseille, France.
Accession Numbers
Sequence data from this article can be found in the GenBank databases under the following accession numbers: PFL1, AJ620191; YP001729802; AAS06904; EFD85284; and YP961832.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignments of Chlamydomonas PFL1 with Known and Predicted PFL1 Enzymes as well as Those Predicted to Be Synthesized in the Mutant Strains.
Supplemental Figure 2. Relative PFL1 Transcript Abundances in the Wild-Type Strain D66, pfl1-2, and pfl1-3 under Dark Anoxic Conditions.
Supplemental Figure 3. Photobiological H2 Production during Acclimation to Dark Anoxia.
Supplemental Table 1. Primers Used for the Genetic Screening and qRT-PCR.
Supplementary Material
Acknowledgments
This work was supported by the Office of Biological and Environmental Research, Genomes to Life Program, Office of Science, U.S. Department of Energy (grants to A.R.G., M.C.P., and M.S.), the National Renewable Energy Laboratory pension program (M.S.), the National Science Foundation Grant MCB-0235878 and U.S. Department of Energy Grant DE-FG02-07ER64427 (A.R.G.), and the Air Force Office of Scientific Research Grant FA9550-05-1-0365 (M.C.P.). L.M. was supported by Scuola Superiore Sant’Anna and Regione Toscana Programma Operativo Regionale Obiettivo 2 Fondo Sociale Europeo. The work at the National Renewable Energy Laboratory was performed under U.S. Department of Energy contract number DE-AC36-08GO28308. The costs of publication for this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
AUTHOR CONTRIBUTIONS
C.C., M.S., M.C.P., and A.R.G. designed the research; C.C., V.S., A.D., W.Y., F.M., and L.M. performed research; C.C., V.S., and A.D. analyzed the data; and M.S., M.C.P., and A.R.G. wrote the article.
References
- Atteia A., van Lis R., Gelius-Dietrich G., Adrait A., Garin J., Joyard J., Rolland N., Martin W. (2006). Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J. Biol. Chem. 281: 9909–9918 [DOI] [PubMed] [Google Scholar]
- Baek H., Song K.H., Park S.M., Kim S.Y., Hyun H.H. (2003). Role of glucose in the bioconversion of fructose into mannitol by Candida magnoliae. Biotechnol. Lett. 25: 761–765 [DOI] [PubMed] [Google Scholar]
- Bölling C., Fiehn O. (2005). Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation. Plant Physiol. 139: 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.P., Yildiz F.H., Grossman A.R. (1999). Sac3, an Snf1-like serine/threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell 11: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubini A., Mus F., Seibert M., Grossman A.R., Posewitz M.C. (2009). Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J. Biol. Chem. 284: 7201–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller R.P., Gibbs M. (1984). Fermentative metabolism of Chlamydomonas reinhardtii: I. Analysis of fermentative products from starch in dark and light. Plant Physiol. 75: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller R.P., Gibbs M. (1985). Fermentative metabolism of Chlamydomonas reinhardtii: II. Role of plastoquinone. Plant Physiol. 77: 509–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi M.L., Togasaki R.K., Seibert M. (1997). Oxygen sensitivity of algal H2- production. Appl. Biochem. Biotechnol. 63-65: 141–151 [DOI] [PubMed] [Google Scholar]
- Gibbs M., Gfeller R.P., Chen C. (1986). Fermentative metabolism of Chlamydomonas reinhardii: III. Photoassimilation of acetate. Plant Physiol. 82: 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ballester D., Pootakham W., Mus F., Yang W., Catalanotti C., Magneschi L., de Montaigu A., Higuera J.J., Prior M., Galván A., Fernandez E., Grossman A.R. (2011). Reverse genetics in Chlamydomonas: A platform for isolating insertional mutants. Plant Methods 7: 24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A.R., Catalanotti C., Yang W., Dubini A., Magneschi L., Subramanian V., Posewitz M.C., Seibert M. (2011). Multiple facets of anoxic metabolism and hydrogen production in the unicellular green alga Chlamydomonas reinhardtii. New Phytol. 190: 279–288 [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Croft M., Gladyshev V.N., Merchant S.S., Posewitz M.C., Prochnik S., Spalding M.H. (2007). Novel metabolism in Chlamydomonas through the lens of genomics. Curr. Opin. Plant Biol. 10: 190–198 [DOI] [PubMed] [Google Scholar]
- Gupta K.J., Zabalza A., van Dongen J.T. (2009). Regulation of respiration when the oxygen availability changes. Physiol. Plant. 137: 383–391 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A., Happe T. (2005). The exceptional photofermentative hydrogen metabolism of the green alga Chlamydomonas reinhardtii. Biochem. Soc. Trans. 33: 39–41 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A., Jacobs J., Happe T. (2008). Biochemical and physiological characterization of the pyruvate formate-lyase Pfl1 of Chlamydomonas reinhardtii, a typically bacterial enzyme in a eukaryotic alga. Eukaryot. Cell 7: 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K.L., Hassett R., Kosman D., Merchant S. (1996). Regulated copper uptake in Chlamydomonas reinhardtii in response to copper availability. Plant Physiol. 112: 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husic D.W., Tolbert N.E. (1985). Anaerobic formation of d-lactate and partial purification and characterization of a pyruvate reductase from Chlamydomonas reinhardtii. Plant Physiol. 78: 277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzberg K. (1984). Starch fermentation via formate producing pathway in Chlamydomonas reinhardtii, Chlorogonium elongatum and Chlorella fusca. Physiol. Plant. 61: 87–94 [Google Scholar]
- Magneschi L., Catalanotti C., Subramanian V., Dubini A., Yang W., Posewitz M.C., Seibert M., Perata P., Grossman A.R. (January 23, 2012). A mutant in the ADH1 gene of Chlamydomonas reinhardtii elicits metabolic restructuring during anaerobiosis. Plant Physiol. http://dx.doi.org/10.1104/pp.111.191569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A., Zhang L., Forestier M., Ghirardi M.L., Seibert M. (2000). Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuser J.E., Ananyev G., Wittig L.E., Kosourov S., Ghirardi M.L., Seibert M., Dismukes G.C., Posewitz M.C. (2009). Phenotypic diversity of hydrogen production in chlorophycean algae reflects distinct anaerobic metabolisms. J. Biotechnol. 142: 21–30 [DOI] [PubMed] [Google Scholar]
- Moseley J.L., Allinger T., Herzog S., Hoerth P., Wehinger E., Merchant S., Hippler M. (2002). Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21: 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.L., Chang C.W., Grossman A.R. (2006). Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot. Cell 5: 26–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.L., Gonzalez-Ballester D., Pootakham W., Bailey S., Grossman A.R. (2009). Genetic interactions between regulators of Chlamydomonas phosphorus and sulfur deprivation responses. Genetics 181: 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J., Quinn J., Eriksson M., Merchant S. (2000). The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19: 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. (1993). The hydrogenosome. J. Gen. Microbiol. 139: 2879–2889 [DOI] [PubMed] [Google Scholar]
- Müller M. (1998). Enzymes and compartmentation of core energy metabolism of anaerobic protist—a special case in eukaryotic evolution. Evolutionary Relationships among Protozoa, Coombs G.H., Vickerman K., Sleigh M.A., Warren A., (Dordrecht, The Netherlands: Kluwer; ), pp. 109–131 [Google Scholar]
- Mus F., Dubini A., Seibert M., Posewitz M.C., Grossman A.R. (2007). Anaerobic acclimation in Chlamydomonas reinhardtii: Anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282: 25475–25486 [DOI] [PubMed] [Google Scholar]
- Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007). Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7: 3964–3979 [DOI] [PubMed] [Google Scholar]
- Ohta S., Miyamoto K., Miura Y. (1987). Hydrogen evolution as a consumption mode of reducing equivalents in green algal fermentation. Plant Physiol. 83: 1022–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps G., Krawietz D., Hemschemeier A., Happe T. (2011). A pyruvate formate lyase-deficient Chlamydomonas reinhardtii strain provides evidence for a link between fermentation and hydrogen production in green algae. Plant J. 66: 330–340 [DOI] [PubMed] [Google Scholar]
- Pollock S.V., Colombo S.L., Prout D.L., Jr, Godfrey A.C., Moroney J.V. (2003). Rubisco activase is required for optimal photosynthesis in the green alga Chlamydomonas reinhardtii in a low-CO(2) atmosphere. Plant Physiol. 133: 1854–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W., Gonzalez-Ballester D., Grossman A.R. (2010). Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 153: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posewitz M.C., King P.W., Smolinski S.L., Zhang L., Seibert M., Ghirardi M.L. (2004). Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J. Biol. Chem. 279: 25711–25720 [DOI] [PubMed] [Google Scholar]
- Rocha M., Licausi F., Araújo W.L., Nunes-Nesi A., Sodek L., Fernie A.R., van Dongen J.T. (2010). Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 152: 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual (New York: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Shimogawara K., Wykoff D.D., Usuda H., Grossman A.R. (1999). Chlamydomonas reinhardtii mutants abnormal in their responses to phosphorus deprivation. Plant Physiol. 120: 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steunou A.S., Bhaya D., Bateson M.M., Melendrez M.C., Ward D.M., Brecht E., Peters J.W., Kühl M., Grossman A.R. (2006). In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl. Acad. Sci. USA 103: 2398–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A.F., Frey M., Neugebauer F.A., Schäfer W., Knappe J. (1992). The free radical in pyruvate formate-lyase is located on glycine-734. Proc. Natl. Acad. Sci. USA 89: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney L.A., Loreti E., Alpi A., Perata P. (2011). Alcohol dehydrogenase and hydrogenase transcript fluctuations during a day-night cycle in Chlamydomonas reinhardtii: The role of anoxia. New Phytol. 190: 488–498 [DOI] [PubMed] [Google Scholar]
- Wykoff D.D., Davies J.P., Melis A., Grossman A.R. (1998). The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz F.H., Davies J.P., Grossman A.R. (1994). Characterization of sulfate transport in Chlamydomonas reinhardtii during sulfur-limited and sulfur-sufficient growth. Plant Physiol. 104: 981–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz F.H., Davies J.P., Grossman A.R. (1996). Sulfur availability and the SAC1 gene control adenosine triphosphate sulfurylase gene expression in Chlamydomonas reinhardtii. Plant Physiol. 112: 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalza A., van Dongen J.T., Froehlich A., Oliver S.N., Faix B., Gupta K.J., Schmälzlin E., Igal M., Orcaray L., Royuela M., Geigenberger P. (2009). Regulation of respiration and fermentation to control the plant internal oxygen concentration. Plant Physiol. 149: 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.