In a genome-wide analysis of the transcriptional changes in Arabidopsis thaliana in response to Cu deficiency, about 13% are found to depend on the transcription factor SPL7. These include the genes encoding Cu(II) reductases FRO4 and FRO5, which are shown to act in high-affinity root Cu uptake. Severe physiological Cu deficiency results in a disruption of Fe homeostasis.
Abstract
The transition metal copper (Cu) is essential for all living organisms but is toxic when present in excess. To identify Cu deficiency responses comprehensively, we conducted genome-wide sequencing-based transcript profiling of Arabidopsis thaliana wild-type plants and of a mutant defective in the gene encoding SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 (SPL7), which acts as a transcriptional regulator of Cu deficiency responses. In response to Cu deficiency, FERRIC REDUCTASE OXIDASE5 (FRO5) and FRO4 transcript levels increased strongly, in an SPL7-dependent manner. Biochemical assays and confocal imaging of a Cu-specific fluorophore showed that high-affinity root Cu uptake requires prior FRO5/FRO4-dependent Cu(II)-specific reduction to Cu(I) and SPL7 function. Plant iron (Fe) deficiency markers were activated in Cu-deficient media, in which reduced growth of the spl7 mutant was partially rescued by Fe supplementation. Cultivation in Cu-deficient media caused a defect in root-to-shoot Fe translocation, which was exacerbated in spl7 and associated with a lack of ferroxidase activity. This is consistent with a possible role for a multicopper oxidase in Arabidopsis Fe homeostasis, as previously described in yeast, humans, and green algae. These insights into root Cu uptake and the interaction between Cu and Fe homeostasis will advance plant nutrition, crop breeding, and biogeochemical research.
INTRODUCTION
Cu is an essential micronutrient required by all organisms. During evolution, the element Cu has been selected as a cofactor in the active centers of a diverse group of proteins because of its unique and powerful chemical properties. Cu can adopt both the cuprous (+I) and cupric (+II) oxidation states at physiologically relevant conditions and is thus suitable for catalysis of single-electron transfer reactions. Furthermore, among all essential transition metals, Cu cations undergo the most stable coordinative interactions with donor electron pairs from ligand molecules (Fraústo da Silva and Williams, 2001).
To be able to exploit the potent chemistry of Cu without succumbing to its deleterious interactions, organisms have to regulate Cu uptake, internal movement, and binding partners precisely (Puig and Thiele, 2002; De Freitas et al., 2003). In recent years, the Cu homeostasis network has emerged as a paradigm in transition metal homeostasis, owing to the pioneering work on the roles of metallochaperone proteins in its operation and its surprising conservation among eukaryotes (Burkhead et al., 2009; Robinson and Winge, 2010). Discoveries were facilitated by the comparably sparse use in biology of Cu, which was much less available before the oxygenation of the Earth’s atmosphere (Ridge et al., 2008). The potent and important biochemical functions of Cu in single-electron transfer reactions, Fe homeostasis, energy metabolism, and redox homeostasis are reflected in the dramatic consequences of defects in Cu homeostasis, as known in human Menkes and Wilson diseases (Shim and Harris, 2003).
Cu has also adopted critical functions at the very core of plant-specific biochemistry. A Cu metalloprotein of central importance is plastocyanin, which acts as the soluble electron carrier between the cytochrome b6f complex and photosystem I in the thylakoid lumen (Redinbo et al., 1994). The role of Cu in gas binding by the receptors for the plant hormone ethylene exemplifies an as yet only marginally explored functional implication of Cu homeostasis in plant signaling and developmental regulation (Rodriguez et al., 1999). Finally, the use of Cu in mitochondrial cytochrome c oxidase (Tsukihara et al., 1995) and in the synthesis of extracellular matrix components (Fraústo da Silva and Williams, 2001), which is rather widespread in biology, is also found in plants (Burkhead et al., 2009).
Plants require Cu at minimum concentrations of around 5 μg g−1 leaf dry biomass and encounter Cu toxicity when concentrations exceed ~20 μg g−1 dry biomass (Marschner, 1995; Burkhead et al., 2009). In crops, well-described symptoms of Cu limitation are reduced growth rates due to impaired photosynthesis, leaf bleaching, and reductions in the physical stability of plants, pollen fertility, seed set, and yield, as well as an increased incidence of ergots in wheat (Triticum aestivum; Marschner, 1995; Solberg et al., 1999). Cu deficiency is widespread in agricultural soils high in organic matter content and affects, for example, an estimated 19% of arable land in Europe (Shorrocks and Alloway, 1988). Based on experimental work and the presence of conserved Cu binding motifs, the Arabidopsis thaliana Cu metalloproteome can be estimated to comprise more than 200 proteins, the majority of which are of unknown function (Andreini et al., 2008). Plastocyanin is the most abundant Cu protein in vascular plants and contains more than half of the chloroplast-localized Cu (Marschner, 1995). Cu/Zn superoxide dismutases in the cytosol, peroxisomes, and chloroplasts and cytochrome c oxidase in the mitochondria are also highly abundant. A large class of Cu-dependent proteins are multicopper oxidases (MCOs), many of which are annotated as laccase (LAC)-like and are generally thought to catalyze the radical-mediated oxidative polymerization of phenyl propanoids into lignins in secondary cell wall formation (McCaig et al., 2005; Burkhead et al., 2009).
Arabidopsis is known to respond to Cu deficiency by enhancing the transcription of genes encoding membrane proteins implicated in Cu acquisition and transport, in particular COPPER TRANSPORTER1 (COPT1), COPT2, YELLOW STRIPE-LIKE2 (YSL2), ZINC-REGULATED TRANSPORTER IRON-REGULATED TRANSPORTER PROTEIN2 (ZIP2), and FERRIC REDUCTASE OXIDASE3 (FRO3), as well as the metallochaperone COPPER CHAPERONE (CCH) (Wintz et al., 2003; Mukherjee et al., 2006; Yamasaki et al., 2009). The second type of response has so far uniquely been described under deficiency of Cu and involves a reorganization of metabolism to economize on Cu. According to the current working model, the Cu-dependent proteins of major abundance, Cu/Zn superoxide dismutases CSD1 and CSD2, are replaced by Fe superoxide dismutase isoforms to maintain the function of the Cu protein plastocyanin, more specifically of isoform 1 (PETE1), under Cu deficiency (Wintz et al., 2003; Abdel-Ghany et al., 2005; Puig et al., 2007; Abdel-Ghany, 2009). This fundamental principle of Cu conservation was first described in the green alga Chlamydomonas reinhardtii, in which under Cu deficiency plastocyanin is replaced by the haem-containing cytochrome c6 (Quinn and Merchant, 1995; Kropat et al., 2005). In C. reinhardtii, Cu deficiency–responsive transcriptional activation and the downregulation of the levels of specific proteins are both dependent on the COPPER RESPONSE REGULATOR1 (CRR1). In Cu-deficient cells, CRR1 binds to a Cu response element (CuRE) containing the sequence motif GTAC as an essential core element (Quinn and Merchant, 1995; Kropat et al., 2005; Sommer et al., 2010). The CuRE can be located in the promoter region or further upstream of genes, so that under Cu deficiency, either increased levels of functional or nonfunctional transcripts are synthesized, respectively (Moseley et al., 2002; Merchant et al., 2006). Importantly, CRR1 also regulates the expression of a protease that mediates the degradation of plastocyanin under Cu deficiency, thus effecting Cu economy (Castruita et al., 2011).
In Arabidopsis, Cu economy is achieved through a homologous pathway involving the transcriptional regulator SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 (SPL7) and related CuREs, but the mechanism of action is distinct (Yamasaki et al., 2009; Sommer et al., 2010). In fact, the specificity of the Zn finger–containing DNA binding domain of SPL7 for binding of double-stranded DNA containing a GTAC core motif was shown even before both were implicated in Cu homeostasis (Yamasaki et al., 2004). Cu-deficient plants downregulate the abundance of specific Cu-dependent proteins primarily at the transcript level through an increase in the abundance of the so-called Cu-microRNAs (miRNAs) miR408, miR397, miR398, and miR857, which in turn cause the cleavage of target transcripts (Yamasaki et al., 2007, 2009; Abdel-Ghany and Pilon, 2008). In addition to CSD1 and CSD2, further target transcripts of miR398 are COPPER CHAPERONE FOR SOD (CCS), encoding the cognate metallochaperone (Cohu et al., 2009; Beauclair et al., 2010), and COX-5b, which encodes the plant homolog of the Zn binding subunit of cytochrome c oxidase (Yamasaki et al., 2007). Subsequently, miR408 was shown to target PLANTACYANIN (ARPN), LAC3, LAC12, LAC13, miR397 LAC2, LAC4, LAC17, and miR857 LAC7 (Abdel-Ghany and Pilon, 2008).
C. reinhardtii CRR1 and Arabidopsis SPL7 are members of the squamosa promoter binding protein (SBP) domain family of transcription factors, which is unique to green algae and plants (Klein et al., 1996). Different from the Cu homeostasis network itself, the transcriptional regulators orchestrating it are not conserved across kingdoms (Winge, 1998).
Here, we present a genome-wide analysis of the transcriptional responses of Arabidopsis to Cu deficiency, based on Illumina RNA-Seq technology. In parallel, the dependence of the Arabidopsis Cu deficiency regulon on the transcription factor SPL7 is monitored using a newly identified spl7 allele. We identify 1563 genes that respond to Cu deficiency at the transcript level, among them 883 in roots and 731 in shoots. Of these, transcript levels of the physically linked FRO4 and FRO5 genes are highly responsive to Cu deficiency. The biological significance of this regulation is validated by its dependence on SPL7 and the obliterating impact of FRO4/FRO5 knockdown on root surface Cu(II)-specific reductase activity and Cu uptake rates of root tips. In addition, we diagnose secondary Fe deficiency in Cu-deficient wild-type plants, which is exacerbated in the spl7 mutant. A defect in root-to-shoot Fe movement in Cu-deficient spl7 mutant plants is associated with a loss of ferroxidase activity, suggesting the involvement of a MCO in Fe homeostasis of a vascular plant, by analogy to related pathways in yeast, humans, and green algae. In wild-type plants, this function is prioritized by the Cu homeostasis network.
RESULTS
Transcriptional Cu Deficiency Responses of Arabidopsis as Determined by RNA-Seq
To identify transcriptomic Cu deficiency responses comprehensively and simultaneously assess their dependence on SPL7, two independent T-DNA insertion lines disrupted in the SPL7 gene were isolated in the Columbia-0 (Col-0) background and characterized. In the spl7-1 (Yamasaki et al., 2009) and the newly isolated spl7-2 mutant (see Methods), insertion of T-DNAs in the 5th exon and 8th intron of the SPL7 gene, respectively, result in a >11-fold reduction in full-length SPL7 transcript levels in roots and a >10-fold reduction in shoots when compared with the wild-type or to an spl7-2 mutant complemented with a 5.9-kb genomic fragment containing the SPL7 gene (see Supplemental Figure 1 online). The effects of Cu deficiency on growth of wild-type and spl7 mutant plants were then documented at the seedling stage in the previously used Murashige and Skoog medium (Yamasaki et al., 2009) and a newly designed EDTA-free agar-based solid Hoagland medium and in 6-week-old vegetative plants grown hydroponically (see Supplemental Figure 2 online). In the hydroponic growth system, wild-type (Col-0) and spl7-2 mutant plants were either maintained in control solution or transferred to a Cu-deficient hydroponic solution for the final 3 weeks of cultivation (for details, see Methods; see Supplemental Figures 2A to 2C online). Under all Cu-sufficient conditions, the appearance and biomass production of wild-type plants and the two spl7 mutant lines were similar (see Supplemental Figure 2 online). Under Cu-deficient conditions, typical symptoms of Cu deficiency were observed in the wild type, including chlorosis and strongly reduced shoot and root biomass production (Marschner, 1995). Whereas these symptoms were evident in seedlings maintained in Cu-deficient media from germination on, vegetative plants grown hydroponically appeared healthy and exhibited merely a slight reduction in shoot biomass under the Cu deficiency conditions employed here (see Supplemental Figure 2 online). In all cultivation systems, Cu deficiency symptoms were considerably more severe in spl7 mutants than in wild-type plants or in the complemented spl7-2 mutant (see Supplemental Figure 2 online). SPL7 function is thus of major importance for plant growth under Cu deficiency not only in seedlings (Yamasaki et al., 2009), but also in older, vegetative plants.
The physiological status of hydroponically grown plants was confirmed by measuring the abundance of previously described Cu-responsive transcripts in roots and shoots using real-time RT-PCR (Sancenón et al., 2003; Wintz et al., 2003). Compared with plants grown under control conditions, shoot transcript levels of FSD1 and YSL2 (DiDonato et al., 2004) were upregulated, and those of CCS, CSD1, and CSD2 were downregulated, in response to Cu deficiency in the wild type, but not in the spl7-2 mutant (see Supplemental Figure 3 online). Transcript levels of CSD2 remained slightly downregulated in response to Cu deficiency in the spl7-2 mutant, indicating that the repression of CSD2 transcript abundance is not exclusively SPL7 dependent. YSL2, CCH, CCS, and CSD1 were observed to respond similarly in both shoots and roots. The upregulation of root transcript levels of COPT1, COPT2, ZIP2, and CCH detected in the wild type under Cu deficiency was absent in spl7-2. An SPL7-dependent upregulation of MIR398b and c, but not MIR398a, precursor transcripts levels under Cu deficiency was observed in shoots and to an even higher degree in roots (see Supplemental Figure 4 online). Wild-type plants grown hydroponically under Cu deficiency exhibited the characteristic biochemical Cu deficiency responses of superoxide dismutase activities but were free of any visible pleiotropic symptoms of general stress (see below and Supplemental Figure 2 online). Across all genotypes and treatments, constant transcript levels of ASCORBATE PEROXIDASE1 (APX1) as a known general stress marker gene and of IRON-REGULATED TRANSPORTER3 (IRT3) as a known Zn deficiency marker gene indicated the absence of general stress and Zn deficiency, respectively (see Supplemental Figure 3C online). Taken together, these results indicated that known SPL7-dependent transcriptional Cu deficiency responses reported previously in whole seedlings (Yamasaki et al., 2009) were also observed in roots or shoots of hydroponically grown older, vegetative plants.
The whole-transcriptome analysis protocol from Illumina (RNA-Seq) was subsequently used for a genome-wide analysis of mRNA abundance in roots and shoots of Cu-deficient and wild-type (Col-0) and spl7-2 mutant plants. We obtained a total of 90 million reads (>3 Gb), which were mapped to the Arabidopsis genome and employed for the estimation of transcript abundance and differential expression (for details, see Methods; see Supplemental Data Set 1 online). On this basis, we identified 1563 genes responding to Cu deficiency through an at least twofold upward or downward change in transcript levels with a false discovery rate (FDR) below 1%. Of these, we found transcript levels of 206 genes (13.2%) to be regulated in an SPL7-dependent fashion (see Supplemental Data Set 1 online; see Methods for details on data filtering). For 537 genes in shoots and 398 genes in roots, we observed an at least twofold upward change in transcript levels in response to Cu deficiency in the wild type, and 346 genes in shoots and 333 genes in roots showed an at least twofold downward change (Figure 1A). Overall, 19.7% of the Cu-responsive transcripts in roots and 9.1% in shoots were regulated in an SPL7-dependent fashion. The proportion of SPL7-dependently regulated Cu deficiency–responsive transcripts for which a common regulation was shared between roots and shoots was 4 times higher (8.1%) than among the Cu deficiency–responsive transcripts regulated independently of SPL7 (2.1%) (Figure 1B).
Figure 1.
The Transcriptional Cu Deficiency Response of Arabidopsis and Its Dependence on SPL7.
(A) Genes responding transcriptionally to Cu deficiency in an SPL7-dependent or independent fashion according to RNA-Seq. Bars represent the number of genes for which transcript levels were changed in response to Cu deficiency in the wild type. Arrows indicate up- or downregulation under Cu deficiency. The striped portion of the bars shows the number of genes regulated in an SPL7-dependent manner, and the remainder (black portion) the number regulated independent of SPL7. Transcript abundance was concluded to increase/decrease under Cu deficiency for a gene when arithmetic means of transcript abundances differed by a factor of at least 2 (FDR < 0.01) in 6-week-old wild-type Arabidopsis (Col-0) cultivated in Cu-deficient (–Cu) hydroponic solutions for 3 weeks when compared with control plants cultivated in modified Hoagland solution containing 0.25 μM Cu (+Cu) throughout. Furthermore, changes in transcript levels were concluded to be dependent on SPL7 if, additionally, [log2 FC (wild type –Cu versus spl7 –Cu) ≥ 0.5 log2 FC (wild type –Cu versus wild type +Cu] for log2 FC (wild type –Cu versus wild type +Cu) > 1, and [log2 FC (wild type –Cu versus spl7 –Cu) ≤ 0.5 log2 FC (wild type –Cu versus wild type +Cu)] for log2 FC (wild type –Cu versus wild type +Cu) < -1. Note that 0.1 hpm was assigned to all genes for which no hits were obtained in the respective sample.
(B) Genes responding transcriptionally to Cu deficiency in an SPL7-dependent or -independent fashion in roots and shoots. Arrows indicate up- or downregulation under Cu deficiency. Ovals represent the number of genes for which transcript levels were changed in response to Cu deficiency in shoots (black) or roots (gray) according to RNA-Seq. The overlap between the ovals shows the number of genes regulated in both tissues. Data and data filtering were as described in (A).
(C) Functional categories represented by the Cu deficiency–responsive genes. For each group of genes, identically colored bars indicate the fraction of genes assigned to the MapMan functional categories (BIN) specified. Shown are only those functional categories that were significantly overrepresented in at least one of the groups. Genes in each group are characterized by common regulation at the transcript level as indicated, based on RNA-Seq data (see Supplemental Data Set 1 online). Values for all expressed genes (gray) and values for genes encoding Cu metalloproteins (black) are shown for reference; 15 to 30% of genes are of unknown function. Significant enrichment of a functional category in a group of genes (P < 0.05, Fisher's exact test with Bonferroni corrections) is marked by asterisks. Data and data filtering were as described in (A). Refer to Methods and Supplemental Methods 1 online for detailed information concerning plant cultivation and timing of sample collection. TCA, tricarboxylic acid.
[See online article for color version of this figure.]
Global functional analysis based on MapMan bins suggested that among Cu deficiency–responsive transcripts there was an overrepresentation of biotic stress, cell wall, hormone metabolism, metal homeostasis and transport, photosynthesis, secondary metabolism, signaling, and general membrane transport functions (Figure 1C) (Thimm et al., 2004). Among these functional classes, SPL7-dependent Cu-responsive regulation was prominent in metal homeostasis/transport and general transport. Additionally, genes encoding miRNAs and proteins with redox functions were overrepresented among the transcripts that were both Cu responsive and SPL7 dependent. Despite an overrepresentation of Cu-dependent proteins in mitochondrial electron transport and the tricarboxylic acid cycle, very little Cu-dependent transcriptional regulation occurred within these functional classes. Conversely, there was an overrepresentation of biotic stress, cell wall, hormone metabolism, and photosynthesis-related transcripts among Cu-responsive transcripts, although Cu metalloproteins are not generally overrepresented in these classes.
The RNA-Seq technology not only covers more genes than previous expression profiling platforms, it also generates more information at the single-gene level than most microarray-based methods. The transcript coverage of aligned reads across the genome was visualized based on the existing gene models on a local installation of the UCSC browser (http://genomes.mcdb.ucla.edu/ArthCopper/). For example, it was evident that the abundance of FRO4 transcripts was higher in Cu-deficient than in Cu-sufficient roots (see Supplemental Figure 5A online) and that compared with wild-type plants, SPL7 transcript abundance is strongly reduced in the spl7-2 mutant, with additional evidence for 3′-terminal truncation, as expected (see Supplemental Figures 1 and 5B and Supplemental Data Set 1 online).
Data Validation and Identification of FRO4 and FRO5 Transcripts as Highly Cu Deficiency Responsive in Dependence of SPL7
An unsupervised motif search (Castruita et al., 2011) identified a core GTAC motif with AT-rich flanking nucleotides as highly overrepresented in the proximal regions upstream of genes that transcriptionally responded to Cu deficiency in the wild type but not in the spl7-2 mutant (Figure 1A; see Supplemental Figure 6 online). This was in agreement with the known core motif bound by SPL7 and other SBP-domain proteins (Klein et al., 1996; Birkenbihl et al., 2005; Kropat et al., 2005; Liang et al., 2008; Yamasaki et al., 2009; Sommer et al., 2010).
We chose to focus initially on strongly Cu-responsive transcripts (Tables 1 and 2) that had not previously been identified as regulated in an SPL7-dependent manner. Out of these, transcript levels of the predicted ferric-chelate reductase-encoding gene FRO5 were quantitatively the most Cu responsive (Table 1). Root transcript levels increased from undetectable levels under control conditions to 705 hits per million (hpm) under Cu deficiency. Root transcript levels of a second, highly homologous gene of the same family, FRO4, increased ~50-fold in response to Cu deficiency, again fully dependent on SPL7 (Table 1). Transcript levels of FRO5, but not of FRO4, were also increased in shoots in response to Cu deficiency, but to a much lower level of only 3.5 hpm (Table 2).
Table 1.
Transcripts Highly Responsive to Cu Deficiency in an SPL7-Dependent Fashion in Roots
| AGI ID | Gene | Short Description | hpm (Wild Type +Cu) | hpm (Wild Type –Cu) | hpm (spl7 +Cu) | hpm (spl7 –Cu) | Log2 FC (–Cu versus +Cu) in the Wild Type | Log2 FC (Wild Type versus spl7) in –Cu |
| At5g23990 | FRO5 | Ferric Reductase Oxidase5; ferric-chelate reductase family | 0.1 | 705.0 | 0.4 | 0.1 | 12.8 | 12.8 |
| At1g71200 | bHLH160 | Basic helix-loop-helix family protein number 160; DNA binding/transcription factor | 0.1 | 25.8 | 0.1 | 0.1 | 8.0 | 8.0 |
| At2g47015 | MIR408 | miRNA 408 family | 0.1 | 14.6 | 0.1 | 0.1 | 6.7 | 7.2 |
| At4g25100 | FSD1 | Ferric Superoxide Dismutase1; Fe superoxide dismutase | 18.7 | 1112.1 | 0.1 | 0.4 | 5.9 | 11.3 |
| At5g23980 | FRO4 | Ferric Reductase Oxidase4; ferric-chelate reductase family | 1.9 | 96.9 | 1.4 | 0.8 | 5.7 | 7.0 |
| At2g47010 | Unknown | Unknown | 0.4 | 11.8 | 1.1 | 0.4 | 4.9 | 4.9 |
| At5g14570 | NRT2.7 | High Affinity Nitrate Transporter2.7; nitrate transmembrane transporter family | 0.1 | 2.8 | 0.1 | 0.1 | 4.8 | 4.8 |
| At4g13554 | MIR857a | miRNA 857 family | 0.1 | 2.6 | 0.1 | 0.1 | 4.7 | 4.7 |
| At5g23989 | Unknown | Unknown protein | 0.1 | 2.3 | 0.1 | 0.1 | 4.5 | 4.5 |
| At3g46900 | COPT2 | Cu transporter2; Cu ion transmembrane transporter family | 15.5 | 347.9 | 10.9 | 25.4 | 4.5 | 3.8 |
| At5g27230 | Unknown | Unknown | 0.1 | 2.2 | 0.9 | 0.2 | 4.4 | 3.6 |
| At3g06560 | PAPS3 | Poly(A) polymerase3/polynucleotide adenylyltransferase | 0.1 | 2.1 | 0.1 | 0.3 | 4.4 | 2.7 |
| At4g28790 | bHLH23 | Basic helix-loop-helix family protein number 23; DNA binding/transcription factor | 1.0 | 19.9 | 0.9 | 2.7 | 4.3 | 2.9 |
| At1g32350 | AOX1D | Alternative Oxidase1D; alternative oxidase | 0.1 | 1.9 | 0.1 | 0.2 | 4.2 | 3.4 |
| At5g59520 | ZIP2 | Zn transporter 2 precursor; Zn-regulated transporter, Fe-regulated transporter protein family | 17.0 | 300.0 | 2.3 | 3.5 | 4.1 | 6.4 |
| At2g11280 | Unknown | Unknown | 0.1 | 1.7 | 0.6 | 0.2 | 4.1 | 3.2 |
| At5g39635 | MIR169C | miRNA 169 family | 0.1 | 1.7 | 0.1 | 0.1 | 4.1 | 4.1 |
| At3g57180 | BPG2 | Brassinazole-insensitive Pale Green 2/GTP binding | 0.1 | 2.0 | 0.5 | 0.3 | 3.9 | 2.7 |
| At4g34850 | PKSB/ LAP5 | Polyketide Synthase B/Less Adhesive Pollen 5/chalcone and stilbene synthase family | 0.1 | 1.5 | 0.1 | 0.1 | 3.9 | 3.9 |
| At4g35660 | Unknown | Unknown | 0.1 | 1.3 | 0.1 | 0.1 | 3.7 | 3.7 |
| At4g11730 | AHA12 | Putative plasma membrane proton P3A-ATPase/proton pump | 0.1 | 1.2 | 0.8 | 0.1 | 3.5 | 3.5 |
| At4g13550 | Unknown | Lipase class 3 family protein, triglyceride lipase | 1.6 | 13.7 | 3.1 | 1.6 | 3.1 | 3.1 |
| At1g47395 | Unknown | Unknown | 2.1 | 0.3 | 1.3 | 2.5 | −3.0 | −3.3 |
| At1g72060 | Unknown | Ser-type endopeptidase inhibitor | 3.1 | 0.4 | 3.7 | 8.0 | −3.1 | −4.5 |
| At4g27440 | PORB | Protochlorophyllide Oxidoreductase B; oxidoreductase/protochlorophyllide reductase protein | 1.9 | 0.2 | 1.9 | 1.7 | −3.2 | −3.1 |
| At5g22788 | Unknown | Unknown | 1.9 | 0.2 | 0.3 | 1.7 | −3.3 | −3.0 |
| At3g33076 | Unknown | Gypsy-like retrotransposon | 24.1 | 2.5 | 6.5 | 37.1 | −3.3 | −3.9 |
| At2g28190 | CSD2 | Cu/Zn Superoxide Dismutase 2; Cu/Zn superoxide dismutase in plastid | 150.1 | 15.2 | 138.3 | 158.0 | −3.3 | −3.4 |
| At1g69485 | Unknown | Ribosomal L32p family protein | 1.2 | 0.1 | 0.5 | 1.4 | −3.5 | −3.8 |
| At2g33760 | Unknown | Pentatricopeptide repeat–containing protein | 1.3 | 0.1 | 1.7 | 1.6 | −3.7 | −4.0 |
| At5g01110 | Unknown | Tetratricopeptide repeat-like superfamily protein | 1.4 | 0.1 | 1.1 | 2.2 | −3.8 | −4.5 |
| At1g08830 | CSD1 | Cu/Zn Superoxide Dismutase1; Cu/Zn superoxide dismutase in cytosol | 483.2 | 32.5 | 540.0 | 490.5 | −3.9 | −3.9 |
| At1g64360 | Unknown | Unknown | 1.6 | 0.1 | 0.6 | 1.3 | −4.0 | −3.7 |
| At1g53130 | GRI | Grim Reaper; regulation of cell death by extracellular ROS | 1.6 | 0.1 | 1.4 | 2.0 | −4.0 | −4.3 |
| At4g15660 | ROXY15 | Glutaredoxin family protein | 1.9 | 0.1 | 2.8 | 2.1 | −4.3 | −4.4 |
Genes are shown if root transcript abundances were (1) changed by a factor of at least 8 in 6-week-old wild-type Arabidopsis (Col-0) cultivated in Cu-deficient (−Cu) hydroponic solutions when compared to control plants cultivated in modified Hoagland solutions (+Cu) (FDR < 0.01) and if Cu-dependent differences in transcript levels were dependent on SPL7, as concluded if (2) transcript levels were significantly different in the spl7 mutant compared to the wild type under Cu deficiency [FDR < 0.01 for (wild type −Cu versus spl7 −Cu)] and (3) [log2 FC (wild type −Cu versus spl7 −Cu) ≥ 0.5 log2 FC (wild type −Cu versus wild type +Cu) for positive log2 FC (wild type −Cu versus wild type +Cu), and log2 FC (wild type −Cu versus spl7 −Cu) ≤ 0.5 log2 FC (wild type −Cu versus wild type +Cu) for negative log2 FC (wild type −Cu versus wild type +Cu)]. Arabidopsis wild-type and spl7-2 mutant plants were cultivated in a hydroponic solution containing normal concentrations of 0.25 μM CuSO4 continuously (control) or in a solution lacking added Cu for the final 3 weeks before harvest. Genes are ranked by decreasing log2 fold change in (wild type −Cu versus wild type +Cu). Note that 0.1 hpm was assigned to all genes for which no hits were obtained in the respective sample. Arabidopsis Genome Initiative identifiers (AGI ID) are underlined for genes shared between Tables 1 and 2.
Table 2.
Transcripts Highly Responsive to Cu Deficiency in an SPL7-Dependent Fashion in Shoots
| AGI ID | Gene | Short Description | hpm (Wild Type +Cu) | hpm ( Wild Type –Cu) | hpm (spl7 +Cu) | hpm (spl7 –Cu) | Log2 FC (–Cu versus +Cu) in the Wild Type | Log2 FC (Wild Type versus spl7) in –Cu |
| At2g47015 | MIR408 | miRNA 408 family | 0.1 | 12.8 | 0.1 | 0.1 | 7.0 | 7.0 |
| At4g25100 | FSD1 | Ferric Superoxide Dismutase 1; Fe superoxide dismutase | 13.7 | 1565.5 | 0.8 | 0.8 | 6.8 | 11.0 |
| At5g23990 | FRO5 | Ferric Reductase Oxidase 5; ferric-chelate reductase family | 0.1 | 3.5 | 0.1 | 0.1 | 5.1 | 5.1 |
| At5g24380 | YSL2 | Yellow Stripe Like 2; oligopeptide transporter | 2.7 | 68.2 | 2.2 | 2.2 | 4.6 | 4.9 |
| At3g46900 | COPT2 | Cu Transporter 2; Cu ion transmembrane transporter family | 0.7 | 16.8 | 0.8 | 0.7 | 4.5 | 4.7 |
| At1g71200 | bHLH160 | Basic helix-loop-helix family protein number 160; DNA binding/transcription factor | 0.1 | 2.2 | 0.1 | 0.1 | 4.5 | 4.5 |
| At5g14565 | MIR398C | miRNA 398 family | 0.1 | 1.9 | 0.1 | 0.2 | 4.3 | 3.4 |
| At5g09443 | Unknown | Potential natural antisense transcript | 0.2 | 4.4 | 0.9 | 0.8 | 4.1 | 2.5 |
| At4g28790 | bHLH23 | Basic helix-loop-helix family protein number 23; DNA binding/transcription factor | 0.6 | 8.4 | 0.1 | 0.1 | 3.7 | 6.4 |
| At4g28900 | Unknown | Transposable element gene | 3.9 | 0.4 | 2.9 | 3.5 | −3.2 | −3.1 |
| At5g57560 | TCH4 | Touch 4; hydrolase acting on glycosyl bonds/xyloglucan: xyloglucosyl transferase | 170.3 | 15.6 | 27.5 | 53.9 | −3.4 | −1.8 |
| At1g12520 | CCS | Cu chaperone for superoxide dismutase 1; superoxide dismutase Cu chaperone family | 165.1 | 11.3 | 173.8 | 168.5 | −3.9 | −3.9 |
| At1g08830 | CSD1 | Cu/Zn Superoxide Dismutase 1; Cu/Zn superoxide dismutase in cytosol | 502.9 | 29.4 | 536.9 | 616.0 | −4.1 | −4.4 |
| At2g02850 | ARPN | Plantacyanin; Cu ion binding protein | 22.7 | 1.3 | 17.5 | 22.8 | −4.1 | −4.2 |
| At3g45710 | Unknown | Proton-dependent oligopeptide transporter family | 1.9 | 0.1 | 0.5 | 2.1 | −4.3 | −4.4 |
| At2g29130 | LAC2 | Laccase 2; Cu ion binding/oxidoreductase, MCO | 4.4 | 0.1 | 3.4 | 2.0 | −5.4 | −4.3 |
| At2g28190 | CSD2 | Cu/Zn Superoxide Dismutase 2; Cu/Zn superoxide dismutase in chloroplast | 570.6 | 12.2 | 637.3 | 436.6 | −5.6 | −5.2 |
Genes are shown if shoot steady state transcript levels were (1) higher or lower by a factor of at least 8 in 6-week-old wild-type Arabidopsis (Col-0) cultivated in Cu-deficient (−Cu) hydroponic solutions when compared to control plants cultivated in modified Hoagland solutions (+Cu, control) (FDR < 0.01) and if Cu-dependent differences in transcript levels were dependent on SPL7, as concluded if (2) transcript levels were significantly different in the spl7 mutant compared to the wild type under Cu deficiency [FDR < 0.01 for (wild type −Cu versus spl7 −Cu)] and (3) [log2 FC (wild type −Cu versus spl7 −Cu) ≥ 0.5 log2 FC (wild type −Cu versus wild type +Cu) for positive log2 FC (wild type −Cu versus wild type +Cu), and log2 FC (wild type −Cu versus spl7 −Cu) ≤ 0.5 log2 FC (wild type −Cu versus wild type +Cu) for negative log2 FC (wild type −Cu versus wild type +Cu)]. Arabidopsis wild-type and spl7-2 mutant plants were cultivated in a hydroponic solution containing normal concentrations of 0.25 μM CuSO4 continuously or in a solution lacking added Cu for the final 3 weeks before harvest. Genes are ranked by decreasing log2 fold change in (wild type −Cu versus wild type +Cu). Note that 0.1 hpm was assigned to all genes for which no hits were obtained in the respective sample. Arabidopsis Genome Initiative identifiers (AGI ID) are given in underlined font for genes shared between Tables 1 and 2.
To confirm gene expression differences identified by RNA-Seq, we performed real-time RT-PCR analysis on the RNA from the two replicate experiments subjected to RNA-Seq, as well as from an additional independent experiment. For both FRO4 and FRO5, there was very good quantitative agreement between RNA-Seq and real-time RT-PCR data (Figures 2A and 2B). This was also found for other genes chosen to span the entire range of Cu-dependent transcriptional changes. Transcript levels of NITRATE TRANSPORTER2.7 (NRT2.7), encoding a tonoplast nitrate transporter previously reported to be expressed specifically in seeds (Chopin et al., 2007), were highly upregulated under Cu deficiency in roots, dependent on SPL7 (Table 1, Figure 2A). In addition, root and shoot transcript levels of LAC2, previously proposed to be a target of miR397a (Abdel-Ghany and Pilon, 2008), and shoot transcript levels of ARPN, proposed to be a target of miR408 (Abdel-Ghany and Pilon, 2008), were highly repressed under Cu deficiency dependent on SPL7, consistent with the known SPL7 dependence of both MIR397a and MIR408 (Yamasaki et al., 2009) (Table 2, Figure 2B; see Supplemental Data Set 1 online for root LAC2). Transcript levels of Cox5b-1, which is one of the two Arabidopsis genes encoding the Zn binding subunit 5b of mitochondrial cytochrome c oxidase (Ferguson-Miller and Babcock, 1996), were previously proposed to be targeted by miR398 (Yamasaki et al., 2007) and showed intermediate, SPL7-dependent downregulation under Cu deficiency in both roots and shoots (to between 25 and 30% of Cu-sufficient controls based on RNA-Seq [see Supplemental Data Set 1 online] and to ~25 to 34% based on real-time RT-PCR [Figures 2A and 2B]). Finally, transcript levels of YSL3, reported to contribute to fertility and normal seed development as well as the allocation of metals (Cu and Fe) into various organs of Arabidopsis (Waters et al., 2006; Chu et al., 2010), showed slight upregulation under Cu deficiency in both roots and shoots (~2.5- to 3.5-fold according to RNA-Seq (see Supplemental Data Set 1 online) and ~2.5-fold according to real-time RT-PCR (Figures 2A and 2B), and this was only partially SPL7 dependent.
Figure 2.
Validation by Real-Time RT-PCR of Cu Deficiency–Responsive, SPL7-Dependent Regulation.
(A) and (B) Relative transcript levels determined by real-time RT-PCR of FRO5, FRO4, YSL3, NRT2.7, LAC2, ARPN, and COX1 genes in roots (A) and shoots (B) of 6-week-old wild-type (WT; Col-0) and spl7-2 plants cultivated continuously in a hydroponic solution containing the usual concentrations of 0.25 μM CuSO4 (+Cu, control) or cultivated in a solution lacking added Cu (–Cu) for the final 3 weeks before harvest. Values are arithmetic means ± sd of n = 4 technical replicates from one experiment representative of two independent biological experiments. Data shown are transcript levels relative to EF1α as a constitutively expressed control gene, multiplied by 1000 (see Supplemental Methods 1 online). Tissues from at least five culture vessels containing two plants each were pooled for each genotype and treatment per experiment.
(C) Correlation between RNA-Seq and real-time RT-PCR data. Data points representing transcript level ratios of –Cu versus control treatments are shown in black, and data points representing transcript level ratios of spl7-2 versus the wild type are shown in gray. Data are from two independent biological experiments.
(D) Correlation showing between-experiment reproducibility. Data from a third independent experiment are shown as a function of the arithmetic mean of data from the two experiments analyzed by RNA-Seq. Data points represent ratios of transcript levels of –Cu versus control treatments (black) and spl7-2 versus the wild type (gray) as determined by real-time RT-PCR.
Overall, there was a linear correlation between results from RNA-Seq and real-time RT-PCR performed on the same RNA samples from two independent experiments (Figure 2C; y = 0.9593x + 0.0908 and R2= 0.92 for differences between treatments, and y = 0.9667x + 0.0106 and R2= 0.95 for differences between genotypes). This confirmed the reliability of estimating relative differences in transcript abundance using RNA-Seq and emphasized the large dynamic range of the RNA-Seq technique, in contrast with microarray-based analyses of gene expression (Czechowski et al., 2004). Reproducibility between experiments was confirmed using RNA from a third, independent biological experiment for comparison by real-time RT-PCR (Figure 2D; y = 0.9897x + 0.3492 and R2 = 0.99 for differences between treatments, and y = 1.0014x – 0.1898 and R2= 0.98 for differences between genotypes).
Increase in Root Surface Cu(II) Chelate Reductase Activity in Response to Cu Deficiency Requires FRO4/FRO5 and SPL7
Among the most strongly Cu deficiency–responsive out of all Arabidopsis transcripts were those of FRO4 and FRO5, two genes located in tandem on Arabidopsis chromosome 5 (Tables 1 and 2, Figure 2). These genes encode two functionally uncharacterized putative ferric chelate reductases of the FRO family (Mukherjee et al., 2006) of gp91phox-related haem-containing membrane proteins. Known root Cu uptake proteins of the COPT/Ctr-like family transport Cu in the reduced Cu(I) form (Hassett and Kosman, 1995; Eisses and Kaplan, 2005), and Arabidopsis possesses six genes encoding members of this protein family. However, the most bioavailable and chemically stable oxidation state of Cu in soils is Cu(II), so that a reduction step is required for Cu acquisition via Cu(I) transporters. Nevertheless, no specific Cu(II) reductase has been identified in vascular plants so far (Robinson et al., 1999; Mukherjee et al., 2006; Burkhead et al., 2009).
To assess whether FRO4 and FRO5 might act as Cu(II)-specific reductases, we measured root surface Cu(II) chelate reductase activities in Cu-deficient and Cu-sufficient wild-type and spl7 mutant seedlings cultivated under sterile conditions. Roots of Cu-deficient wild-type seedlings exhibited clearly detectable Cu(II) chelate reductase activity of ~8 μmol Cu(I) g−1 fresh biomass h−1 (Figure 3A). By contrast, activities were below 0.4 μmol Cu(I) g−1 fresh biomass h−1 in wild-type seedlings grown under Cu-sufficient conditions as well as in both Cu-sufficient and Cu-deficient spl7-1 and spl7-2 seedlings. This indicated that under Cu deficiency, root surface Cu(II) chelate reductase activity is strongly upregulated and that this is dependent on SPL7.
Figure 3.
Root Surface Cu(II) Chelate Reductase Activity Does Not Respond to Cu Deficiency in spl7 Mutants and Depends on FRO4 and FRO5.
Root surface Cu(II) chelate reductase activity of wild-type (WT; Col-0) and spl7-2 seedlings (A) and of wild-type (Col gl-1), fro4, amiR-FRO5, and amiR-FRO4FRO5 seedlings (B) grown for 3 weeks on vertical glass plates in an agarose-solidified nutrient medium containing 0.5 μM CuSO4 (+Cu, control) or no added Cu (–Cu). Values are arithmetic means ± sd of n = 3 replicate plates, from each of which 20 seedlings were pooled for analysis. Data are from one experiment representative of a total of four independent experiments. Note that basal activities in Cu-sufficient Col gl-1 seedlings are considerably higher than in Col-0 (see Yi and Guerinot, 1996). Note that data in Figures 3A and 6A are based on plant material grown in a common experiment. Different letters denote statistically significant differences (P < 0.05) between means based on ANOVA (Tukey’s HSD). FW, fresh biomass.
To test our hypothesis that FRO4 and FRO5 contribute to this activity, we isolated a loss-of-function fro4 T-DNA insertion line exhibiting substantially reduced FRO4 transcript levels in comparison to the wild type (see Supplemental Figure 7 online). Moreover, we generated transgenic knockdown lines transformed with artificial microRNA (amiRNA) constructs targeting either FRO5 or both FRO4 and FRO5, in which the levels of intact target transcripts or of target proteins were substantially reduced by posttranscriptional or translational silencing, respectively (see Supplemental Figures 8 and 9 online). In FRO5 and FRO4FRO5 amiRNA lines, which differed from each other by a shift in the amiRNA target site of only 2 nucleotides (see Supplemental Figures 8A and 9A online), FRO5 transcripts were still detectable (see Supplemental Figures 8B and 9B online). This was also observed in double transgenic Arabidopsis FRO5 amiRNA plants additionally carrying a 35S-FRO5-YFP-HA construct, which encodes a chimeric fusion protein of FRO5 with yellow fluorescent protein (YFP) and a human influenza virus hemagglutinin (HA) tag (see Supplemental Figure 8C online). However, FRO5-YFP-HA fusion protein signals were undetectable by either immunoblot or confocal microscopy in double transgenic FRO5 amiRNA lines, whereas the fusion protein was present in single transgenic lines devoid of the FRO5 amiRNA transgene (see Supplemental Figures 8D and 8E online). This indicated that the silencing of FRO5 occurred at the level of translation in FRO5 amiRNA lines and, by similarity, also in the FRO4FRO5 amiRNA lines, in which, different from FRO5, FRO4 transcript levels were strongly reduced (see Supplemental Figure 9B online).
In contrast with the corresponding wild type (Col gl-1), neither the fro4 T-DNA insertion line nor the FRO5 or FRO4FRO5 amiRNA lines showed an increase in root surface Cu(II) chelate reductase activity under Cu deficiency when compared with control conditions (Figure 3B). This is consistent with a strong prediction for secretory pathway targeting of FRO4 and FRO5 (http://aramemnon.uni-koeln.de). Taken together, our results suggest that FRO4 and FRO5 encode root surface Cu(II) chelate reductases that are transcriptionally upregulated dependent on SPL7 in response to Cu deficiency, leading to increased enzyme activity.
Cu Uptake into Roots of Cu-Deficient Plants Is Dependent on FRO4/FRO5 and SPL7
To determine whether SPL7 and FRO4/FRO5 functions are required for root Cu uptake, we used the Cu-specific fluorophore Coppersensor-1 (CS1) as a tool to image intracellular Cu in root tips of 2-week-old seedlings by confocal laser scanning microscopy. Fifteen replicate root tips were imaged per genotype and treatment, applying fixed confocal laser scanning microscopy settings (Figures 4A to 4D; see Supplemental Figures 10 to 12 online), and the average pixel intensity was calculated keeping both the image area and the length of the imaged root tip segment constant in each image (Figures 4E and 4F). CS1 fluorescence signals were clearly visible in root tips of both wild-type (Col-0) and spl7-2 seedlings grown under Cu-sufficient conditions, but barely detectable upon growth under Cu-deficient conditions (Figures 4A and 4E; see Supplemental Figures 10A to 10D online). For a comparative estimate of root Cu uptake rates, roots of wild-type and spl7-2 mutant seedlings, which had been precultivated in Cu-deficient or Cu-sufficient media for 2 weeks, were incubated in a solution containing 10 nM CuSO4 for 10 min prior to staining with CS1. The resulting high intensities of fluorescence signals suggested that root Cu uptake rates were strongly increased in Cu-deficient wild-type seedlings when compared with Cu-sufficient seedlings (Figures 4B and 4E; see Supplemental Figures 10E and 10F online). By contrast, no CS1 fluorescence signal was detectable even after short-term incubation in 10 nM exogenous Cu in root tips of spl7-2 seedlings precultivated under Cu-deficient conditions (Figures 4B and 4E; see Supplemental Figures 10G and 10H online). This suggested that root Cu uptake was strongly impaired in Cu-deficient spl7-2 seedlings under these conditions. This observation confirmed that SPL7 function is required for root Cu uptake by Cu-deficient plants.
Figure 4.
CS1 Visualization of Cu Uptake in Root Tips of Arabidopsis Seedlings.
(A) to (D) Confocal fluorescence microscopy images taken at the central longitudinal plane of root tips stained with the Cu(I)-specific dye CS1 (top panels) and bright-field micrographs showing the outline of the corresponding root tips (bottom panels) for wild-type (WT; Col-0) and spl7-2 mutant seedlings ([A] and [B]) and for wild-type (Col gl-1), fro4, amiR-FRO5, and amiR-FRO4FRO5 seedlings ([C] and [D]). Seedlings cultivated on vertical agarose glass plates containing 0.5 μM CuSO4 (+Cu) or no added Cu (–Cu) for 2 weeks were directly stained with CS1 ([A] and [C]) or incubated in a solution containing 10 nM CuSO4 ([B] and [D]) for 10 min prior to staining with CS1. Photographs are shown from one root tip representative of a total of 15 root tips (see Supplemental Figures 10 to 12 online) from one experiment representative of two independent biological experiments in which seedlings of different genotypes were grown in groups of five individuals per genotype per plate. Bars = 5 μm.
(E) and (F) Relative quantification of Cu(I)-based on fluorescence intensity of the Cu(I)CS1 complex in root tips of 2-week-old wild-type and spl7-2 mutant seedlings (E) and wild-type, fro4, amiR-FRO5, and amiR-FRO4FRO5 seedlings (F) without (Control) and with incubation in 10 nM CuSO4 prior to staining with CS1 (see [A] to [D] above). Values are arithmetic means ± sd of average pixel intensities over the imaged area of n = 3 replicate plates, for each of which data were averaged from five root tips, from one experiment representative of a total of two independent biological experiments. Different letters denote statistically significant differences (P < 0.05) between means based on ANOVA (Tukey’s HSD). A.U., arbitrary units.
To analyze whether the SPL7 dependence of root Cu uptake is attributable to FRO4 and FRO5, root tips of the wild type and fro4 and amiR-FRO5 and amiR-FRO4FRO5 seedlings grown on Cu-sufficient and Cu-deficient media were stained with CS1. Again, a fluorescence signal was present in Cu-sufficient seedlings of the wild type (Col gl-1) and the three transgenic lines, but virtually undetectable upon growth under Cu-deficient conditions (Figures 4C and 4F; see Supplemental Figure 11 online). Upon short-term supply of 10 nM exogenous Cu, the CS1 fluorescence signal was higher in root tips of Cu-deficient wild-type seedlings compared with Cu-sufficient wild-type seedlings (Figures 4D and 4F; see Supplemental Figure 12 online), as observed above (Figure 4B). By contrast, CS1 fluorescence signal intensities indicated that Cu uptake rates were strongly reduced in roots of Cu-deficient fro4 and amiR-FRO5 seedlings and that there was no Cu uptake in roots of Cu-deficient amiR-FRO4FRO5 seedlings (Figures 4D and 4F; see Supplemental Figure 12 online). In agreement with this, upon cultivation in Cu-deficient media, biomass production was reduced to lower levels in amiR-FRO4FRO5 plants than in the wild type (see Supplemental Figures 9C to 9E online). No phenotype was observed on normal fertilized soil. Taken together, these results suggest that the increase in high-affinity root Cu uptake rates under Cu deficiency is dependent on SPL7 and requires FRO4 and FRO5. Consequently, Cu(II) reduction is required for high-affinity Cu uptake into roots.
Impaired Cu Accumulation in spl7-2 Mutant Plants
To test whether plant Cu levels reflect decreased root Cu uptake rates in spl7 mutants when compared with the wild type, metal contents were measured in shoots and roots of 6-week-old hydroponically grown plants by inductively coupled plasma atomic emission spectrometry. In shoots of spl7 plants grown under Cu-sufficient conditions, total Cu content was ~60% of the wild type (Figure 5A; see Supplemental Figure 13A online), although the mutant grew normally (see Supplemental Figures 2A to 2C online). Total Cu content in roots of the spl7 mutants was only slightly lower than in the wild type (Figure 5B). Cu concentrations in spl7 were around 70% of the wild type values of 9.2 mg kg−1 in shoots and 22.5 mg kg−1 in roots, respectively (see Supplemental Figure 13B online).
Figure 5.
Cu Content of Wild-Type and spl7 Mutant Plants.
Total Cu content in shoots (A) and roots (B) of 6-week-old wild-type (WT; Col-0) and spl7 mutant plants upon continuous cultivation in a hydroponic solution containing the usual concentration of 0.25 μM CuSO4 (+Cu, control) or cultivation in a solution lacking added Cu (–Cu) for the final 3 weeks before harvest. Values are arithmetic means ± sd (n = 6 individuals for shoots, n = 3 pools from two individuals for roots). Data are shown from one experiment representative of a total of three independent experiments. Different letters denote statistically significant differences (P < 0.05) between means based on ANOVA (Tukey’s HSD).
Upon growth under Cu-deficient conditions, total Cu contents were drastically reduced in all genotypes to less than a quarter of Cu-sufficient controls (Figure 5). Total Cu contents in roots and shoots of spl7 were around 70% of those in the wild type. Contrastingly, tissue concentrations of Cu in spl7 mutants were ~20 and 50% higher in roots and shoots, respectively, than in wild-type plants grown in Cu-deficient media (see Supplemental Figure 13 online). We interpret this as reflecting the inability of spl7 to economize on Cu because it cannot replace, for example, Cu/Zn superoxide dismutases by Fe superoxide dismutase isoforms under Cu deficiency (see Supplemental Figure 3 online) (Yamasaki et al., 2007). Thus, in Cu-deficient media, spl7 mutants encounter severe growth impairment as a consequence of Cu limitation at higher tissue Cu concentrations than the wild type (see Supplemental Figure 2 online). Taken together, these data support our hypothesis that SPL7 is required for high-affinity root Cu uptake and confirm that Cu is limiting for growth of spl7 mutants when Cu availability is low.
Activation of Fe Deficiency Responses in Roots and a Low Fe Status in Shoots of Severely Cu-Deficient Plants
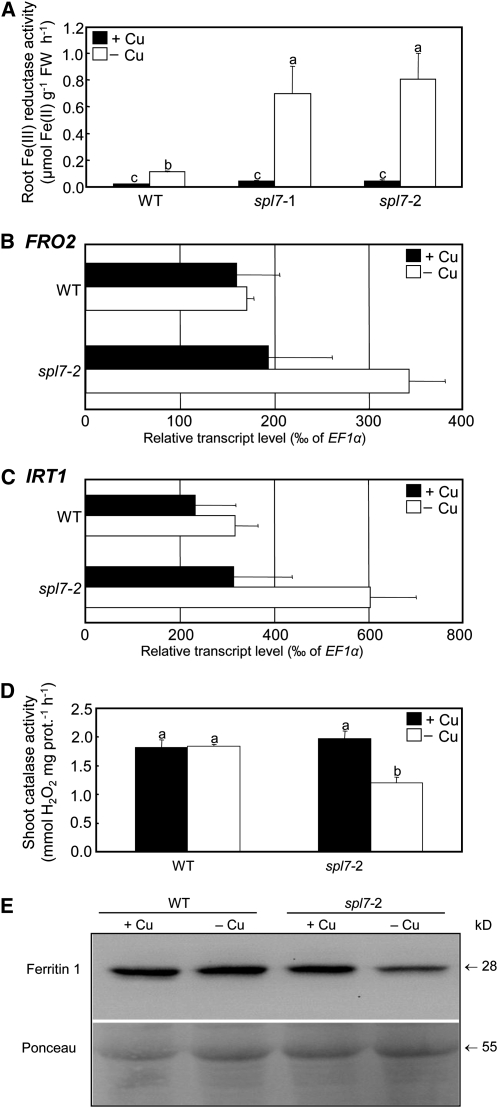
Our RNA-Seq transcriptome analysis suggested that upon growth in Cu-deficient media transcript levels of the characteristic Fe deficiency response genes, IRT1 and FRO2, were slightly, between 1.6- and 2-fold, increased in spl7-2 when compared with the wild type (see Supplemental Data Set 1 online). To test whether even these small changes in gene expression reflected physiological differences between mutant and wild-type plants, we analyzed various Fe deficiency markers in wild-type and spl7-2 plants. Compared with seedlings grown under control conditions, root surface ferric chelate reductase activity was increased in wild-type plants grown on Cu-deficient media and strongly, to approximately fivefold higher levels, in spl7 mutants (Figure 6A). Analysis of transcript levels by real-time RT-PCR quantitatively confirmed increased root FRO2 and IRT1 transcript levels in Cu-deficient spl7-2 when compared with the wild type (Figures 6B and 6C). At least in spl7-2, this was not dependent on a change in root FE-DEFICIENCY INDUCED TRANSCRIPTION FACTOR (FIT) transcript levels, which did not respond to Cu deficiency in the mutant and increased very slightly (1.16-fold, FDR = 0.02) in the wild type (see Supplemental Data Set 1 online) (Colangelo and Guerinot, 2004). Root Fe deficiency responses are known to be systemically controlled through the shoot Fe status (Vert et al., 2003). To test whether the transcriptional activation of Fe deficiency responses in roots was a consequence of a low physiological Fe status in shoots of spl7-2 under Cu deficiency, we measured biochemical markers of shoot Fe status. Indeed, the activity of catalase, a haem-Fe–dependent enzyme as well as Ferritin1 protein levels were decreased in shoots of spl7-2 mutants when compared with the wild type (Figures 6D and 6E). Taken together, these results suggest that Cu deficiency can cause secondary physiological Fe deficiency in plants and that this is particularly severe in spl7 mutants showing defects in high-affinity root Cu uptake and internal Cu economy.
Figure 6.
Activation of Fe Deficiency Markers in Plants Cultivated under Cu-Deficient Conditions.
(A) Root surface ferric chelate reductase activity of wild-type (WT; Col-0) and spl7-2 mutant seedlings grown on vertical agarose glass plates containing 0.5 μM CuSO4 (+Cu, control) or no added Cu (–Cu) for 3 weeks. Each value is the arithmetic mean ± sd of n = 3 replicate pools of 20 seedlings grown per plate from one experiment representative of four independent biological experiments. Note that data in Figures 3A and 6A are based on plant material grown in a common experiment. FW, fresh biomass.
(B) and (C) Real-time RT-PCR analysis of transcript levels of FRO2 (B) and IRT1 (C) in roots of 6-week-old wild-type and spl7-2 plants grown continuously in a hydroponic solution containing 0.25 μM CuSO4 (+Cu, control) or grown in a solution lacking added Cu (–Cu) for the final 3 weeks before harvest. Values are arithmetic means ± sd of relative transcript levels normalized to EF1α, then multiplied by 1000, and were calculated from n = 4 technical replicates from one experiment representative of two independent biological experiments (see Supplemental Methods 1 online).
(D) Specific activities of the haem-dependent enzyme catalase in shoots of wild-type and spl7-2 plants cultivated as described for (B) and (C) above. Values are arithmetic means ± sd of n = 3 replicate measurements from one experiment representative of two independent biological experiments. Different letters denote statistically significant differences (P < 0.05) between means based on ANOVA (Tukey’s HSD).
(E) Abundance of Ferritin1 (FER1) protein in shoots of wild-type and spl7-2 Arabidopsis plants cultivated as described for (B) and (C) above. Soluble protein extracts (30 μg) were separated on a denaturing polyacrylamide gel and transferred to a polyvinylidene fluoride membrane for immunoblot analysis using an anti-FER1 antibody (top panel). Protein loading was visualized on the membrane through Ponceau Red staining prior to immunodetection. Data shown are from one experiment representative of two independent biological experiments.
Low plant Fe status in spl7 grown under Cu deficiency was associated with an ~60% reduction in total shoot Fe content compared with the wild type (Figure 7A). By contrast, total root Fe content was ~30% higher in spl7 than in the wild type grown under the same conditions (Figure 7B). This suggested a strong impairment of root-to-shoot Fe movement in spl7 mutant plants grown under Cu-deficient conditions when compared with spl7 grown on Cu-sufficient media or to the wild type. Cu deficiency–induced root Fe accumulation was also apparent based on root and shoot Fe concentrations (see Supplemental Figure 14 online). Moreover, in shoots of spl7 grown on Cu-deficient media, a physiological Fe deficiency (see Figure 6 above) despite overall Fe concentrations equivalent to those in the wild type further suggested that intercellular Fe distribution in the shoot is also Cu dependent. In comparison to Cu sufficiency, a trend toward reduced root-to-shoot Fe movement under Cu deficiency was also apparent in the wild type (Figure 7; see Supplemental Figure 14 online). Perls’ stain for ferric Fe confirmed Fe accumulation in roots of spl7-2 and, to a lesser extent, of wild-type seedlings grown in Cu-deficient media (Figure 7C). Taken together, these data suggest that Cu deficiency leads to a reduction in root-to-shoot Fe translocation and that this causes Fe deficiency in shoots, which in turn triggers Fe deficiency responses.
Figure 7.
Fe Content and Root Fe Accumulation of Wild-Type and spl7 Mutant Plants.
(A) and (B) Total Fe content in shoots (A) and roots (B) of 6-week-old wild-type (WT; Col-0) and spl7 mutant plants upon continuous cultivation in a hydroponic solution containing a normal concentration of 0.25 μM CuSO4 (+Cu, control) or cultivation in a solution lacking added Cu (–Cu) for the final 3 weeks. Values are arithmetic means ± sd (n = 6 individuals for shoots, n = 3 pools from two individuals for roots). Data are shown from one experiment representative of a total of three independent experiments. Different letters denote statistically significant differences (P < 0.05) between means based on ANOVA (Tukey’s HSD).
(C) Detection of Fe(III) using Perls’ stain in roots of wild-type and spl7-2 plants. Plants were cultivated as described above. Data shown are from one experiment representative of a total of three independent biological experiments. Bars = 1 mm.
Partial Phenotypic Complementation of the Growth Defect of spl7-2 by High Fe Supply
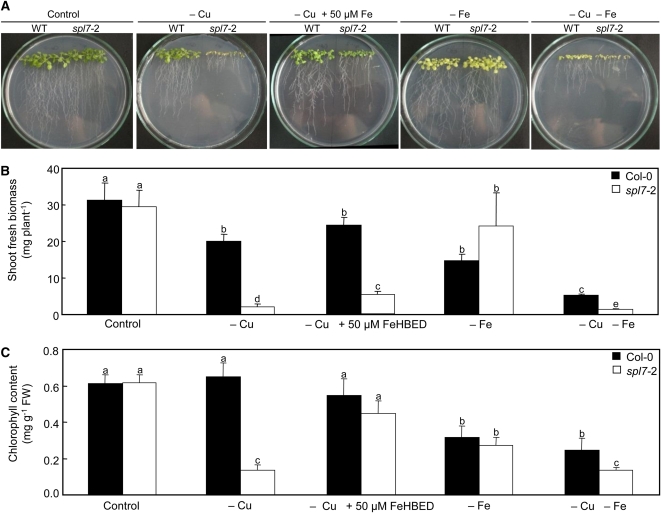
We tested whether the spl7-2 mutant can be phenotypically complemented by adding extra Fe to the growth media (see Supplemental Figure 2 online). As shown earlier, spl7-2 seedlings grew normally on modified Hoagland medium but grew very poorly and were severely chlorotic in modified Hoagland medium lacking added Cu. When 50 μM extra Fe(III) N,N′-bis(2-hydroxybenzyl)-ethylenediamine-N,N′-diacetic acid was added to Cu-deficient medium, growth of the spl7-2 mutant was improved and chlorophyll concentrations were restored to ~85% of wild-type levels (Figure 8). This confirmed that the symptoms in the spl7-2 mutant are not exclusively caused by Cu homeostasis defects but partially reflect secondary Fe deficiency resulting from a defect in long-distance Fe movement in severely Cu-deficient plants.
Figure 8.
Phenotypic Complementation of the Growth Defect of spl7-2 by High Fe Supply.
Photographs (A), shoot fresh biomass (B), and leaf chlorophyll concentrations (C) of 21-d-old wild-type (WT) and spl7-2 mutant seedlings grown on vertical glass plates containing agarose-solidified nutrient medium (0.5 μM CuSO4, control) or the same medium containing no added Cu (–Cu), no added Cu and supplemented with 50 μM Fe(III)HBED (–Cu +50 μM Fe), no added Fe (–Fe), and no added Cu or Fe (–Cu –Fe), respectively. Photographs are shown from one experiment representative of two independent biological experiments. Values in (B) and (C) are arithmetic means ± sd of n = 3 replicate plates, from each of which material was pooled of three to four seedlings grown on a common plate, from one experiment representative of two independent biological experiments. Different letters denote statistically significant differences (P < 0.05) between means based on ANOVA (Tukey’s HSD). FW, fresh biomass.
[See online article for color version of this figure.]
Reduced MCO Activities in Severely Cu-Deficient Arabidopsis
In yeast and humans, Cu deficiency is known to disrupt Fe homeostasis because these organisms depend on Cu-containing enzymes (MCOs) that oxidize Fe(II) to Fe(III) in association with specific transmembrane Fe transport functions (Askwith et al., 1994; Harris et al., 1995; Muckenthaler et al., 2008). In combination with the Fe homeostasis defects in the spl7 mutant, this prompted us to test for the presence and Cu dependence of MCO-related enzyme activities in plants (Figure 9). An in-gel enzyme assay showed that ferroxidase activity is detectable in roots of wild-type Arabidopsis plants and that this activity is severely reduced in spl7-2 plants cultivated under Cu deficiency, coinciding with the occurrence of Fe homeostasis defects (Figure 9A). Arabidopsis ferroxidase activity might be mediated by one or several of the 41 putative MCOs with homology to Saccharomyces cerevisiae Fet3p, C. reinhardtii FOX1, or human caeruloplasmin, encoded in the Arabidopsis genome. Phenoloxidase activity, which is classically attributed to laccases, migrated at approximately the same position as ferroxidase activity in denaturing SDS-PAGE gels and was also strongly reduced in spl7-2 grown under Cu-deficient conditions (Figure 9B). In-gel activity assays also confirmed that superoxide dismutase activities were changed as expected in the different genotypes under the employed conditions (Figure 9C). Taken together, our data suggest that the disruption of a MCO activity acting as ferroxidase in Arabidopsis Fe homeostasis could underlie the defect in long-distance Fe transport observed in severely Cu-deficient spl7-2 mutant plants.
Figure 9.
MCO Activities in Wild-Type and spl7-2 Mutant Plants.
(A) to (C) In-gel detection of ferroxidase (A), phenoloxidase (B), and superoxide dismutase (C) activities in extracts of total soluble protein from roots of 6-week-old wild-type (WT) and spl7-2 plants grown continuously in a hydroponic solution containing 0.25 μM CuSO4 (+Cu, control) or grown in a solution lacking added Cu (–Cu) for the final 3 weeks before harvest. Thirty micrograms of protein was separated on a denaturing ([A] and [B]) or native (C) polyacrylamide gels. Data shown are from one experiment representative of two independent biological experiments.
(D) Protein loading was visualized by Coomassie blue staining.
DISCUSSION
An Important Role for SPL7 in Cu Deficiency Responses of Shoots and Roots of Arabidopsis
RNA-Seq technology was employed to obtain a genome-wide account of transcriptional responses to Cu deficiency in Arabidopsis as well as of the role of SPL7 therein. Expanding on previous work, 206 target genes for SPL7-dependent transcriptional regulation were identified, and our results highlight an important role for SPL7 not only in roots, but also in shoots, of Cu-deficient plants (Tables 1 and 2, Figure 2; see Supplemental Figures 1, 3, and 4 online; compare Yamasaki et al., 2009). There is no doubt that transcriptional Cu deficiency responses exhibit some dynamics over time. This genome-wide approach included only a single time point of Cu deficiency. We observed a substantial degree of qualitative consistency between responses in the different growth systems used in this study and previously (Sancenón et al., 2003; Wintz et al., 2003; Abdel-Ghany and Pilon, 2008; Yamasaki et al., 2009), overall spanning a wide range of plant ages and different extents of Cu deficiency. Therefore, we feel confident that this study captured a large number of transcriptional responses of fundamental and general importance.
SPL7-Dependent Transcriptional Repression Is Prominent in Arabidopsis
Among the SPL7-dependent transcriptional Cu deficiency responses of Arabidopsis, more transcripts were downregulated in abundance than upregulated. Downregulation of the expression of some genes under Cu deficiency is known to operate through the SPL7-mediated activation of transcription of the Cu-miRNAs (see Supplemental Data Set 1 and Supplemental Figure 4 online) (Yamasaki et al., 2009). These miRNAs function in the downregulation of transcript levels of a known set of target genes, which have largely been identified (Yamasaki et al., 2007, 2009; Abdel-Ghany and Pilon, 2008). Consequently, SPL7 either activates the transcription of additional unidentified noncoding repressive transcripts or of downstream transcriptional repressors, or SPL7 itself might alternatively be able to repress transcription under Cu deficiency (Moseley et al., 2002). By contrast, in Cu-deficient C. reinhardtii, most of the significant alterations in mRNA abundance involve increases rather than decreases, and as many as about half of these genes are direct targets of the master regulator of nutritional Cu sensing CRR1 (Castruita et al., 2011).
Additional Pathways of Cu Deficiency–Responsive Transcriptional Regulation Are SPL7 Independent
Genome-wide transcriptomics conducted here suggest that ~87% out of more than 1500 identified Cu deficiency–responsive transcripts are regulated in an SPL7-independent manner (Figure 1; see Supplemental Data Set 1 online). While a small subset of these responses might constitute symptoms of, rather than responses to, Cu deficiency, these results indicate that some transcriptional responses to Cu deficiency are controlled by yet unidentified transcription factors acting in SPL7-independent pathways. These are likely to make major contributions to the noteworthy transcriptional remodeling of secondary metabolism, biotic stress, cell wall, hormone metabolism, photosynthesis, and signaling in response to Cu deficiency (Figure 1). The existence of SPL7-independent pathways of Cu deficiency responsive transcriptional regulation is further supported by an only partial SPL7 dependence of transcriptional regulation of CSD2 and YSL3 (Figure 2; see Supplemental Figure 3 online). In this context, it is worth noting the existence of genes encoding the SPL7-related transcription factors SPL1 and SPL12 in the Arabidopsis genome. Transcript levels of numerous genes related to pathogen responses are remarkably increased under Cu deficiency, mostly independent of SPL7 (see Supplemental Data Set 1 online, shoots). Cu deficiency has profound effects on cell wall composition (Marschner, 1995), and cell wall–derived signals have been implicated as elicitors of plant responses to pathogens (Hématy et al., 2009). The activation of pathogen defense gene expression in Cu-deficient plants requires further investigation.
Metabolic Reorganization under Cu Deficiency Depends Partly on SPL7
A hallmark of Cu deficiency responses in algae and vascular plants appears to be the reorganization of metabolism for the replacement of Cu metalloproteins by functionally equivalent Cu-independent proteins. This is a result of transcriptional regulation dependent on CRR1 in C. reinhardtii and SPL7 in Arabidopsis (Yamasaki et al., 2009; Castruita et al., 2011). Differing overall transcriptional responses suggest fundamental differences between the mechanisms and targets of Cu deficiency–triggered metabolic reorganization in C. reinhardtii and Arabidopsis (Castruita et al., 2011) (see Supplemental Data Set 1 online; Figure 1).
Arabidopsis SPL7 acts as the transcriptional activator of Cu-miRNAs suppressing CSD transcript levels as well as of FSD1 (Yamasaki et al., 2009). Using transcript levels adjusted for Cu binding stoichiometry to approximate Cu quota, our data are consistent with the known major contribution to overall Cu economy through the reduction in CSD1 and CSD2 transcript levels in both roots and shoots, as well as with the SPL7 dependence of this (see Supplemental Table 1 online) (Burkhead et al., 2009). Based on transcript levels, there appears to be only a single additional SPL7-dependently regulated target of metabolic reorganization with major consequences for global Cu use. Transcript levels of UCLACYANIN2 (UCC2) were the highest of all transcripts encoding Cu-dependent proteins in roots and reduced by 50% in Cu-deficient plants. This would reduce the estimated proportion of Cu-containing UCC2 from 24% of the Cu metalloproteome in sufficient plants to only 17% in Cu-deficient plants (see Supplemental Table 1 online). According to miRBase (http://www.mirbase.org), the UCC2 transcript carries a degenerate putative target sequence for miR408 at the beginning of its 3′-untranslated region. By comparison to UCC2, our data suggest that the known Cu deficiency–dependent reduction in LAC transcripts makes much smaller contributions to overall Cu economy in roots, mainly through LAC2, LAC7, and LAC3, and virtually no contribution in shoots. Consequently, the downregulation of LAC expression under Cu deficiency might have a locally confined role in Cu economy or serve a different physiological function. Our finding that in wild-type roots neither ferroxidase nor phenoloxidase activity was strongly reduced under Cu deficiency suggests that the sum of activities of the contributing holoproteins is hardly affected in the wild type but strongly reduced in spl7-2 (Figure 9).
Overall, in Cu-deficient wild-type plants, the estimated consumption of Cu through Cu-dependent proteins was reduced to 69 and 76% of Cu-sufficient control plants in roots and shoots, respectively. By contrast, such a reduction in Cu consumption through transcriptional regulation did not occur in roots of the spl7-2 mutants and was only very slight in spl7-2 shoots. This supports our hypothesis that spl7-2 faces a growth limitation partly because of a misallocation of Cu so that critical Cu-dependent apometalloproteins cannot be supplied with their cofactor and thus remain inactive. Yamasaki et al. (2009) observed similar Cu concentrations in wild-type and spl7-1 mutant seedlings grown under a given Cu supply, which is overall consistent with the data shown in this article (see Supplemental Figure 13 online). Based on this earlier observation, it was proposed that reduced expression of genes encoding Cu uptake systems, such as COPT1 and COPT2, is compensated by alternative Cu uptake transporters in spl7 (Yamasaki et al., 2009). Instead, we propose here that under Cu deficiency conditions, Cu is limiting for biomass production of spl7 mutant plants, and the misallocation of Cu in spl7 results in increased Cu concentrations, whereas total plant Cu content is decreased when compared with the wild type (Figure 5; see Supplemental Figures 2 and 13 and Supplemental Table 1 online). Interestingly, a progressive increase in tissue Cu concentrations was observed under severe Cu limitation in barley (Hordeum vulgare) and is known as the Steenbjerg effect (Steenbjerg, 1951). Thus, for Cu in particular, it can be misleading to estimate root nutrient uptake rates based on measurements of tissue concentrations, which reflect the outcome of numerous growth and nutrient homeostasis processes over the long term.
Taken together, our data are consistent with a paramount role of SPL7 in metabolic reorganization under Cu deficiency, although quantitatively minor aspects of this reorganization are SPL7 independent (see Supplemental Table 1 online). In the future, it will be important to address this at the protein level (Abdel-Ghany, 2009).
SPL7-Dependent Increase in FRO4 and FRO5 Transcript Levels in Response to Cu Deficiency
Both our RNA-Seq and real-time RT-PCR data confirmed all 14 Cu-responsive transcripts that were recently shown to be regulated dependent on SPL7 (Sancenón et al., 2003, 2004; Wintz et al., 2003; Abdel-Ghany et al., 2005; Abdel-Ghany and Pilon, 2008; Yamasaki et al., 2009). However, of the 32 putative direct or indirect SPL7 targets identified recently using microarrays (Yamasaki et al., 2009), only six were found in this study (see Supplemental Data Set 1 online). Moreover, this study identified 186 previously unreported putative SPL7 targets. These discrepancies might be attributable to a difference in plant age and cultivation system between our study and the earlier one. Moreover, the precision and large dynamic range of RNA-Seq allowed us to identify transcripts not known to be Cu regulated or SPL7 dependent before, either because microarrays were insufficiently sensitive or because these genes were not represented, for example, FRO4, FRO5 (cf. Mukherjee et al., 2006), YSL3, NRT2.7, BASIC HELIX-LOOP-HELIX FAMILY PROTEIN23, MIR169C, TOUCH4, ALTERNATIVE OXIDASE 1D (AOX1D), PROTOCHLOROPHYLLIDE OXIDOREDUCTASE B, GRIM REAPER (Tables 1 and 2, Figure 2). These data can serve as a basis for the identification of candidate proteins that may act in the replacement or compensation of reduced Cu-dependent functions under Cu deficiency (see Supplemental Data Set 1 online). Examples are POLYKETIDE SYNTHASE B, so far believed to be pollen specific, in the extracellular matrix (Kim et al., 2010), or AOX1D to accommodate a possible reduction in activity of the Cu enzyme cytochrome c oxidase (Table 1). Under Cu deficiency, the abundance of the NRT2.7 transcript encoding a transporter implicated in the vacuolar storage of nitrate increases in an SPL7-dependent manner in roots (Table 1, Figure 2), constituting a regulatory link between Cu status and nitrogen metabolism that remains to be explained (Chopin et al., 2007). The formation of a Cu-bound intermediate appears to be required in the biosynthetic pathway of the molybdenum cofactor that forms part of the enzyme nitrate reductase (Mendel, 2011).
FRO4 and FRO5, which encode two functionally uncharacterized putative ferric chelate reductases of the FRO family (Mukherjee et al., 2006), were among the most strongly Cu deficiency responsive out of all SPL7-dependently regulated transcripts, predominantly in roots (Table 1, Figure 2). The core binding motif of SBP domain proteins, including C. reinhardtii CRR1 and Arabidopsis SPL7 in vitro, is GTAC (Klein et al., 1996; Birkenbihl et al., 2005; Kropat et al., 2005; Liang et al., 2008; Yamasaki et al., 2009), consistent with the results of motif enrichment analysis within promoter regions of putative SPL7 targets identified in this study (see Supplemental Figure 6 online). A motif search (http://Arabidopsis.med.ohio-state.edu/) identifies five and 11 GTAC motifs within curated promoter sequences of FRO4 and FRO5, respectively (Davuluri et al., 2003), both of which include four [AT]GTAC[AT] motifs (see Supplemental Figure 6 online). Consequently, both genes are likely to be direct SPL7 targets.
Ferric Chelate Reductases FRO4 and FRO5 Mediate Cu(II) Reduction at the Root Surface and Are Required for High-Affinity Root Cu Uptake
Wild-type Arabidopsis plants responded to Cu deficiency by strongly inducing root surface Cu(II) chelate reductase activity, whereas this response was absent in both spl7 mutants and genotypes disrupted in the expression of either FRO4 or FRO5, or both (Figure 3; see Supplemental Figures 7 to 9 online). This is consistent with our hypothesis that these genes encode root surface Cu(II) chelate reductases. There was no correlation between root surface cupric and ferric chelate reductase activities across genotypes and conditions (Figures 3 and 6). Thus, cupric ion reduction is not a side activity of a ferric reductase such as FRO2 (Robinson et al., 1999) but instead involves a Cu(II)-specific enzyme activity that can be estimated to have <1% activity toward Fe(III) as a substrate. Consistent with this, previous experiments addressing Cu accumulation and Cu sensitivity in FRO2-overexpressing plants have not been conclusive regarding an in vivo function for FRO2 in root Cu(II) reduction (Yi and Guerinot, 1996; Connolly et al., 2003). In C. reinhardtii, four candidate reductases have been annotated in the genome (Merchant et al., 2006; Allen et al., 2007), but it is not yet known whether any of these participates in Cu uptake.
Our results indicate that FRO4/FRO5-dependent Cu(II) reduction is required for high-affinity Cu uptake by Cu-deficient root tips of Arabidopsis (Figure 4; see Supplemental Figures 10 to 12 online). This finding implies that at least in root tips of Cu-deficient seedlings, Cu(I) uptake into roots by proteins of the COPT family, such as COPT1, is predominant (Sancenón et al., 2004). In agreement with this, promoter-β-glucuronidase reporter studies conducted by these authors suggested the presence of COPT1 promoter activity in root tips. The major role of reductases in Cu uptake also indicates that the uptake of Cu(II), for example by ZIP2 and ZIP4 as suggested earlier (Wintz et al., 2003; Puig et al., 2007; Burkhead et al., 2009; Yamasaki et al., 2009), plays only a minor, if any, role at the Cu concentrations employed here. This finding is further corroborated by a strongly reduced total Cu content in vegetative spl7-2 mutant plants when compared with the wild type (Figure 5).
The FRO4 and FRO5 genes are located in tandem on chromosome 5, suggesting that they arose from a recent gene duplication event. A phylogenetic analysis of the amino acid sequences of the encoded proteins (Mukherjee et al., 2006) explains the partial functional redundancy between the two genes. The sum of Cu deficiency–inducible root surface Cu(II) chelate reductase activities of genotypes disrupted in either single FRO4 or FRO5 function alone was lower than the activity in the wild type (Figure 3B). An analogous observation was made with respect to initial root Cu uptake rates (Figure 4F). In both the fro4 T-DNA insertion line and the FRO5 amiRNA line, transcripts of the nontarget homolog were detectable (see Supplemental Figures 7B and 8B online). Consequently, it is worth investigating whether FRO4 and FRO5 operate as functionally interdependent heterooligomers.
Interestingly, FRO4 and FRO5 are not only expressed in roots, but also in shoots, yet at a substantially lower level than in roots (Figure 2). Different from roots, where both genes respond to Cu deficiency and are regulated dependently on SPL7, FRO4 expression is constitutive and independent of SPL7 in shoots. Cu was proposed to be transported in the xylem as Cu(II)-nicotianamine complex (Pich and Scholz, 1996; Krämer and Clemens, 2005), and the Cu(II) chelate reductase activities of FRO4 and FRO5 could contribute to reducing Cu(II) to Cu(I) before its cellular uptake in the shoot (Mukherjee et al., 2006; Yamasaki et al., 2009).
A Cu Requirement for Fe Homeostasis in Arabidopsis
The requirement of Cu for root-to-shoot and intercellular mobility of Fe and the occurrence of secondary Fe deficiency in Cu-deficient Arabidopsis are surprisingly reminiscent of the situation in mammals (Figures 6 to 8; see Supplemental Figure 14 online). In humans, the extracellular ferroxidase activities of the MCOs caeruloplasmin and haephestin are required for cellular Fe(II) export via the plasma membrane protein ferroportin 1 (Hellman and Gitlin, 2002). Defects in human MCO activities cause pathological Fe accumulation in cells and organs acting as sources for Fe (re-)distribution, whereas sites of major Fe demand experience Fe shortage. In S. cerevisiae, high-affinity Fe uptake depends on the oxidation of Fe(II) by the MCO Fet3p prior to cellular Fe(III) import through Ftr1p (Askwith et al., 1994). Likewise, in C. reinhardtii, the ferroxidase-like MCO FOX1 contributes to Fe assimilation under Fe deficiency (La Fontaine et al., 2002), but Cu deficiency does not cause secondary Fe deficiency in this organism unlike in humans, yeast, and Arabidopsis.
The lack of ferroxidase activity in protein extracts from roots of spl7-2 grown under Cu deficiency (Figure 9), which coincided with a root-to-shoot Fe translocation defect (Figures 6 and 7), was consistent with the possibility that one or several membrane transport steps in Fe homeostasis, likely in cellular Fe export, depend on MCO-mediated ferroxidase activity (Hoopes and Dean, 2004; McCaig et al., 2005). If the decrease in a ferroxidase activity under Cu deficiency is the cause of the observed impairment in root-to-shoot Fe translocation, then the primary accumulation of Fe will occur in the ferrous oxidation state. This may explain the low intensity of the Perls’ stain that is specific for Fe(III) when compared with total Fe accumulation (Figure 7; see Supplemental Figure 14 online). However, when accumulated in biological tissues, Fe(II) can readily undergo conversion into Fe(III) through the Fe-catalyzed Fenton reaction (Krämer and Clemens, 2005). The characteristic Cu binding motifs associated with MCO functionality are conserved in the Arabidopsis proteins annotated as ascorbate oxidases (three proteins), laccases (17 proteins), and Low Phosphate Root proteins (two proteins), but only poorly in SKU5 Similar proteins (19 proteins). Laccases have been associated with functions in lignin biosynthesis and the oxidative polymerization of flavonoids, although the in vivo functions of most of them remain to be identified (McCaig et al., 2005; Pourcel et al., 2005; Cai et al., 2006; Liang et al., 2006; Berthet et al., 2011). Direct testing of our current working model will require the identification of the genes that account for the Cu requirement in plant Fe homeostasis.
If adequate Cu nutritional status is required to maintain Fe partitioning in Arabidopsis, we would expect the activation of Cu acquisition under Fe deficiency. Indeed, root COPT2 transcript levels were approximately sixfold upregulated in response to Fe deficiency in a partly FIT-dependent manner (Colangelo and Guerinot, 2004).
In summary, comparative RNA-Seq analysis identified novel global transcriptional and physiological responses to Cu deficiency in Arabidopsis, as well as a number of prime candidate genes. We were able to show that SPL7 function is required for high-affinity root Cu uptake involving Cu(II) reduction via FRO4 and FRO5. Furthermore, our results show that severe physiological Cu deficiency results in a disruption of Fe homeostasis. Future work will aim to identify the Cu-dependent proteins in Fe partitioning, determine the functions of major Cu metalloproteins, and confirm major Cu deficiency responses at the protein level.
METHODS
Plant Material
The Arabidopsis thaliana T-DNA insertion lines SALK_093849 (spl7-1, Col-0 background) and SALK_125385 (spl7-2, Col-0 background) were obtained from the Nottingham Arabidopsis Stock Centre, based on http://signal.salk.edu. A fro4 T-DNA insertion line (SAIL_159_H09; Col gl-1 background) was obtained from the ABRC (see Supplemental Methods 1 online for more information concerning the identification of the homozygous plants). For complementation of the spl7-2 mutant, a 5.9-kb genomic region spanning the entire SPL7 locus was PCR-amplified from Col-0. amiRNAs were designed to silence FRO5 and both FRO4 and FRO5, respectively, using the amiRNA designer interface WMD2 (http://wmd2.weigelworld.org) and constructed by overlapping PCR using a template plasmid (pRS300) containing the MIR319a precursor, kindly provided by Detlef Weigel (Schwab et al., 2006; Ossowski et al., 2008). To generate the initial 35S-FRO5-YFP-HA construct, the Gateway cloning system was used (Earley et al., 2006) (see Supplemental Methods 1 online for a detailed description of the generation of transgenic lines).
Growth Conditions and Experimental Treatments
For soil cultivation, plants were grown in plastic trays filled with ready-to-use commercial, prefertilized soil mixture Type Minitray (Terreau Professionell GePAC). After stratification of seeds on moist paper at 4°C in the dark for 4 to 5 d, seeds were transferred onto soil. Germination and cultivation were conducted in a growth chamber at 20°C, 50% relative humidity, in long-day conditions (16 h light, 8 h dark) at a light intensity of 100 to 120 μmol m−2 s−1 provided by fluorescent tubes (L58W/840 and L5W/25; Osram). Hydroponic cultivation was done in short days as described previously (Becher et al., 2004). Cu deficiency treatments were initiated when plants were 3 weeks old and were continued for 3 weeks (see Supplemental Methods 1 online). Sterile plant growth was performed in vertically positioned round glass Petri plates (diameter of 0.15 m) in a climate-controlled growth chamber with a photoperiod of 11 h light/13 h dark at 145 μE m−2 s−1 during the day and day and night temperatures of 20 and 18°C, respectively. Twenty sterilized seeds per plate were plated on control (+Cu; 0.5 μM CuSO4) or Cu-deficient (−Cu; Cu omitted from solution) full-strength nutrient solution (Becher et al., 2004) supplemented with 1% (w/v) Suc and 1% (w/v) agar (Agar Type M; Sigma-Aldrich). After plating the seeds, plates were kept in the dark at 4°C for 1 week. Three-week-old seedlings were harvested for chlorophyll quantification and measurement of root-surface Cu(II) and Fe(III) chelate reductase activities (see Supplemental Methods 1 online).
Determination of Biomass and Elemental Concentrations
After harvest of hydroponically grown plants, fresh biomass of shoot and root material was determined. Subsequently, shoots were rinsed in ultrapure water and blotted dry, and roots from two individuals of the same genotype, treatment, and hydroponic culture vessel were desorbed together in 150 mL of an ice-cold solution of 5 mM CaCl2 and 1 mM MES-KOH, pH 5.7, for 20 min, with a replacement of the solution after 5 min, followed by two washes with ice-cold ultrapure water, each for 3 min. Shoot and root tissues were dried at 60°C for 3 d. Dried shoot and root material was homogenized, weighed, and subsequently digested for the measurement of elemental concentrations by inductively coupled plasma atomic emission spectrometry (IRIS Advantage HX Duo; Thermo-Fisher) as described previously (Becher et al., 2004). Three independent experiments were conducted, each comprising three replicate culture vessels per genotype and treatment.
RNA Extraction and Quantitative Real-Time RT-PCR
Total RNA extraction and quantitative real-time RT-PCR were done essentially as described earlier (Talke et al., 2006), with primer sequences given in Supplemental Table 2 online (see Supplemental Methods 1 and Supplemental Tables 2 and 3 online).
High-Throughput Sequencing of mRNA Using the SOLEXA Technology
Ten micrograms of total RNA per sample was used to generate the cDNA Colony Template Libraries (CTLs) for high-throughput DNA sequencing using SOLEXA technology (Fasteris Genome Analyzer Service). Briefly, poly(A) transcripts were purified, and double-stranded cDNA synthesis was performed using oligo(dT) priming for first-strand synthesis. cDNA was fragmented into 50- to 200-bp fragments through nebulization, followed by end repair, addition of 3′ adenine nucleotides, the ligation of adapters, gel purification to isolate fragments of 150 to 500 bp, and PCR amplification. For quality control analysis, an aliquot of each CTL was cloned into the TOPO plasmid, and 5 to 10 clones were sequenced using capillary sequencing. Subsequently, the CTLs were sequenced on the Illumina Genome Analyzer, generating two to seven million reads of 35 bases in length per sample. In total, 16 lanes (35 cycles each) were used to sequence all the samples (see Supplemental Data Set 1 online), with one lane per sample. Two replicate samples that were generated in independently conducted biological experiments were run for each treatment and genotype. The standard Illumina analysis pipeline was used for collecting raw images and base calling to generate sequence files, which were used as primary data files for further in-house analysis. Raw reads and processed data are available in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE24696.
Data Analysis
Mapping Sequences
Raw sequence files from the Illumina pipeline were used for alignment against the TAIR7 genome release of Arabidopsis using SOAP (Li et al., 2008). An iterative mapping strategy was employed to maximize the number of aligned sequences (González-Ballester et al., 2010). First, the original 35-mers were aligned with a tolerance of up to two mismatches. On average, we found a unique hit for 67.3% of the reads. The remaining reads were trimmed one base at a time (up to 21 nucleotides) from both the 5′ and 3′ ends and mapped again with no mismatches allowed, until a successful hit was found. Reads from both the 3′ end of the transcript [containing a fraction of the poly(A) signal] or from exon-exon junctions, along with reads with more than two sequencing errors are expected to be rescued with this additional alignment round. On average, 5.6 million reads per library were mapped uniquely to the Arabidopsis genome.
Transcript Abundance
The oligo(dT) priming used for cDNA synthesis resulted in a uniform decay of coverage from 3′-end in 5′-direction over the length of each transcript, with 90% of reads mapping within 1050 nucleotides from the 3′-end (see Supplemental Figure 15 online). The transcript level of each gene was calculated as the number of reads mapping to it, normalized to the total number of reads for the respective sample, omitting normalization to the full length of the transcript (as in the standard reads per kilobase of exon model per million normalization for random-primed RNA-Seq libraries). Let Hijk denote the matrix containing the number of alignment hits for each lane I, gene j and hit length k ∈ [21…35]. For each gene j and each lane I, the transcript relative abundance (TRA) is computed after pooling all of the hits overlapping j and normalizing by the number of aligned reads for lane i:
 |
where the “·” symbol indicates sum over the corresponding index and c = 106, so that Tij has units of hits per million. Because of the lack of full-length transcript coverage upon oligo(dT) priming, these values are not expected to be proportional to the number of mRNAs in the cell in a given experiment, especially for transcripts shorter than 1000 nucleotides. However, both the number of alignment hits and the TRAs can be used for a quantitative evaluation of differential expression between two samples (see Figure 2 for a comparison with real-time RT-PCR estimates).
Differential Expression
The genes for which no hits were recorded across all the samples were discarded from the data set. For the genes for which hits were recorded in only a subset of the samples (e.g., low expression or strongly regulated genes), we attempted to account for the missing data. We replaced missing values with a small value of expression (0.1 hits per million). This data set, which we will refer to as “imputed data set,” was used for downstream analyses. Differential expression can be detected by identifying the genes exhibiting significant variation in their TRA estimates. It has been shown (Marioni et al., 2008) that the distribution of total reads aligned per gene can be approximated by a Poisson distribution assuming random sampling (i.e., each read is sampled uniformly and independently from every possible read in the sample). A Poisson distribution is used to model Hij· and compute P values for differential expression using the method originally proposed for SAGE data by Audic and Claverie (1997) on pooled lanes for the same sample. Additionally, posterior probabilities of differential expression ((P) ϵ [0, 1]) can be computed from the P values using a mixture-model approach (Allison et al., 2002). This was implemented by the Cyber-T software (Baldi and Long, 2001) and used to filter genes above a given value of FDR. TRA values presented in this work correspond to the mean across independent repeats i of the experiment a(i)
 . Differential expression between two conditions (a(i),b(i)) for a gene j was considered significant only if
. Differential expression between two conditions (a(i),b(i)) for a gene j was considered significant only if  , and its FDR was lower or equal to 1%. The subset of genes thus determined to be differentially expressed were used for further comparative analyses.
, and its FDR was lower or equal to 1%. The subset of genes thus determined to be differentially expressed were used for further comparative analyses.
Functional Overrepresentation Analysis
The enrichment of the set of target genes in a particular biological function was addressed by means of an overrepresentation analysis based on the MapMan functional ontology (Thimm et al., 2004). MapMan is hierarchical in nature, with 36 primary functional categories called BINs, which further branch into subBINs distributed at several hierarchical levels. To avoid multiple assignments of a BIN/functional category to the same gene due to the hierarchical nature of MapMan, the subBINs were excluded from the overrepresentation analysis. Arabidopsis Genome Initiative codes are used as primary gene identifiers to avoid any further redundancy in assigning functional categories. For a list containing genes of interest, we calculated the percentage of genes in the list that were assigned to a particular BIN and made direct comparisons with the percentages of genes in the same BIN among all expressed genes, which correspond to the imputed data set referred to earlier. To test statistically if the BIN is overrepresented in the gene list compared with the imputed data set, we used Fisher’s exact test for overrepresentation and corrected for multiple testing by the Benjamini-Hochberg method.
In Vivo Imaging of Cu(I) Using the CS1 Dye
Arabidopsis seedlings were grown on vertically positioned glass plates for 2 weeks. Apical root segments of ~10 mm in length were cut off with a razor blade, washed in 10 mM EDTA, pH 7.5, for 5 min, and then washed with ultrapure water three times for 5 min each. Root segments were subsequently incubated with 10 μM CS1 for 30 min in darkness (Zheng et al., 2006). Fluorescence of CS1 was observed using a confocal laser scanning microscope (Leica SP5) with excitation at 543 nm using a Rhodamine filter and emission collected at 555 to 600 nm. Images of all root tips were collected at identical settings for gain, pinhole, and z position. To confirm that differences in fluorescence intensity reflect relative levels of intracellular Cu, root tips were incubated with 100 μM bathocuproine disulfonic acid [BCDS; a Cu(I) chelator] for 10 min and subsequently washed with 10 mM EDTA, pH 7.5, and three times with ultrapure water, prior to staining with 10 μM CS1 for 30 min in darkness. For the approximation of root Cu uptake activities, root tips were incubated with 10 nM CuSO4 for 10 min and subsequently washed with 10 mM EDTA, pH 7.5, and three times with ultrapure water prior to staining with 10 μM CS1 as described above. Working solutions of CS1 in ultrapure water were diluted from a 2 mM stock made up in DMSO and stored at −20°C. The quantification of the CS1 fluorescence signal was done using the ImageJ software as described (Sinclair et al., 2007).
Root Surface Cu(II) Reductase and Fe(III) Chelate Reductase Activities
Root surface activities of Cu(II) and Fe(III) chelate reductases were determined using intact roots of 3-week-old seedlings grown in sterile culture on vertically positioned round glass plates. As described previously (Yi and Guerinot, 1996), the assay solution consisted of 0.1 mM Fe(III)NaEDTA and 0.3 mM FerroZine [3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine-4′,4′′-disulfonic acid] (Sigma-Aldrich) in ultrapure water for measurement of Fe(III) chelate reductase activity or 0.2 mM CuSO4, 0.6 mM Na3citrate, and 0.4 mM BCDS (Sigma-Aldrich) in ultrapure water for measurement of Cu(II) reductase activity, respectively. Roots from 20 seedlings were pooled and submerged in 1 mL of solution for each reductase assay. The assays were conducted in the dark and at room temperature. After 30 min for Fe(III) chelate reductase and 20 min for Cu(II) reductase, the absorbance of assay solutions was measured at 562 nm for the Fe(II)FerroZine complex and at 483 nm for the Cu(I)BCDS complex, respectively. Extinction coefficients were 28.6 mM−1 cm−1 for the Fe(II)FerroZine complex (Gibbs, 1976) and 12.25 mM−1 cm−1 for the Cu(I)BCDS complex (Welch et al., 1993).
Localization of Fe(III) with Perls’ Stain and Measurement of Chlorophyll Concentrations
Roots of plants cultivated hydroponically were used to determine Fe(III) localization as described previously (Green and Rogers, 2004), with slight modifications (see Supplemental Methods 1 online). Concentrations of chlorophyll a/b and total chlorophyll were measured in the shoots of 3-week-old seedlings grown on vertically oriented round glass plates as described (Porra et al., 1989).
Immunoblots and Determination of Enzyme Activities
For immunoblotting, 30 μg of soluble protein extract (see Supplemental Methods 1 online) was separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane with a Bio-Rad transfer system (Mini Trans Blot Cell; Bio-Rad). The membrane was incubated with anti-At-FER1 antibody (dilution 1:1000) kindly provided by F. Gaymard (Arnaud et al., 2007), followed by a horseradish peroxidase–conjugated secondary antibody (dilution 1:10,000) using standard protocols (Sambrook and Russel, 2001). In-gel detection of phenol oxidase and ferroxidase activities was conducted as described (Hoopes and Dean, 2001), with some modifications (see Supplemental Methods 1 online). Electrophoresis and in-gel detection of superoxide dismutase activities were done as previously described (Ghasemi et al., 2009). Catalase activity was determined using a UV spectrophotometric method as described (Ghasemi et al., 2009).
Statistical Analysis
Single comparison of means were performed using Student’s t test, and multiple comparisons were performed by two-way analysis of variance (ANOVA; Tukey’s honestly significant difference [HSD]) using Statgraphics software (version XV.I; Statpoint Technologies) unless indicated otherwise. In silico promoter analysis was conducted as described by Castruita et al. (2011).
Accession Numbers
Raw reads and processed data are available in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE24696. Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At5g18830 (SPL7), At5g23990 (FRO5), At5g23980 (FRO4), At2g03445 (MIR398A), At5g14545 (MIR398B), At3g56240 (CCH), At5g59030 (COPT1), At1g07890 (APX1), At1g60960 (IRT3), At4g19690 (IRT1), At1g01580 (FRO2), At5g53550 (YSL3), and At3g15640 (COX5b-1) (see also Tables 1 and 2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Molecular Characterization of spl7 Mutants and Complementation by Introduction of a Wild-Type SPL7 Transgene.
Supplemental Figure 2. Arabidopsis spl7 Mutants Are Hypersensitive to Cu Deficiency.
Supplemental Figure 3. Validation by Real-Time RT-PCR Analysis of Transcript Levels of Previously Described Cu-Regulated Transcripts.
Supplemental Figure 4. Validation by Real-Time RT-PCR Analysis of Transcript Levels of Precursors of Previously Described Cu-Regulated MicroRNAs.
Supplemental Figure 5. Browser View of RNA-Seq Read Alignments on the FRO4 and SPL7 Loci.
Supplemental Figure 6. Promoter Analysis of Genes Exhibiting SPL7-Dependent Cu Deficiency–Responsive Transcriptional Regulation.
Supplemental Figure 7. Molecular Characterization of fro4 Mutants.
Supplemental Figure 8. Molecular Characterization of amiR-FRO5 Lines.
Supplemental Figure 9. Molecular and Phenotypic Characterization of amiR-FRO4FRO5 Lines.
Supplemental Figure 10. Cu(I) Detection in Root Tips Using Coppersensor-1 in Wild-Type Arabidopsis and the spl7-2 Mutant.
Supplemental Figure 11. Cu(I) Detection in Root Tips Using Coppersensor-1 in Lines with Reduced FRO4 and FRO5 Protein Levels.
Supplemental Figure 12. Cu(I) Detection in Root Tips Using Coppersensor-1 in Lines with Reduced FRO4 and FRO5 Protein Levels upon Short-Term Cu Supply.
Supplemental Figure 13. Cu Concentrations in Wild-Type and spl7 Plants.
Supplemental Figure 14. Fe Concentrations in Wild-Type and spl7 Plants.
Supplemental Figure 15. Average Cumulative Transcript Coverage for Oligo(dT) and Random-Hexamer Primed RNA-Seq Libraries as a Function of the Distance from the 3′-End of Transcripts.
Supplemental Table 1. Transcriptome-Based Approximation of the Effect of Cu Deficiency on Cu Quota within the Cu-Dependent Proteome of Arabidopsis Wild-Type and spl7-2 Mutant Plants.
Supplemental Table 2. Primers Used for Real-Time RT-PCR Analysis.
Supplemental Table 3. Primers Used in the Characterization of fro4 and amiFRO5 Mutant Lines.
Supplemental Methods 1. Details on Methods and Methods Used in the Supplemental Data.
Supplemental Data Set 1. RNA-Seq Analysis of the Transcriptomic Response of Arabidopsis to Cu Deficiency and the Role of the Cu Regulatory Transcription Factor SPL7 Therein.
Supplementary Material
Acknowledgments
We acknowledge postdoctoral fellowships to M.B. from the Alexander von Humboldt Foundation and the Spanish Ministry of Science and Innovation; a Deutsche Forschungsgemeinshaft Heisenberg fellowship and funding from the FRONTIERS program at the University of Heidelberg, Germany, and the European Union InP Public Health Impact of Long-Term, Low-Level Mixed Element Exposure in Susceptible Population Strata (FOOD-CT-2006-016253) to U.K.; a grant from the National Science Foundation (IOS-0919739) to E.L.C.; a postdoctoral fellowship from the Spanish Foundation of Science and Technology (MEC-FECYT) to D.C.; National Institutes of Health Grant GM42143 to S.S.M.; and support from the University of California, Los Angeles–Department of Energy Institute for Genomics and Proteomics under Contract DE-FC02-02ER63421 to M.P. We thank Chris Chang for providing the CS1 (NIH GM79465), Frédéric Gaymard for the antibody against At-FER1, Nicolaus von Wirén for helpful interactions, and Dudley Page for critical reading of the manuscript.
AUTHOR CONTRIBUTIONS
M.B. performed experiments and generated figures (Figures 2 to 9; Supplemental Figures 1C, 1D, 2 to 4, 8E, 9C to 9E, and 10 to 14 online) and, jointly with U.K., coordinated the experimental and bioinformatic work as well as the writing of the article. D.C. instructed V.S., and they jointly analyzed the RNA-Seq data and generated figures and tables (Tables 1 and 2, Figure 1; Supplemental Data Set 1 and Supplemental Figures 5, 6, and 15 online). M.P. supervised all bioinformatic analysis. G.T.W. performed experiments and generated figures (Supplemental Figures 7, 8A to 8D, 9A, and 9B online), and H.Y. for Figure 3B and preparatory experiments. E.L.C. designed experiments for the isolation and characterization of lines with reduced FRO4/FRO5 protein levels. S.C.D. produced and provided the CS1 dye. P.H. directed the isolation and genetic characterization of spl7 mutants, which was experimentally implemented by A.G. (Supplemental Figures 1A, 1B, and 2E online). P.H., S.S.M., and U.K. jointly designed the RNA-Seq study, and U.K. designed the follow-up experiments in consultation with S.S.M., E.L.C., and P.H. M.B. and U.K. wrote the article, U.K. generated Supplemental Table 1 online, and D.C., V.S., and G.T.W. wrote specific methods sections. S.S.M., E.L.C., D.C., P.H., V.S., and M.P. edited the article. All authors commented on the article.
References
- Abdel-Ghany S.E. (2009). Contribution of plastocyanin isoforms to photosynthesis and copper homeostasis in Arabidopsis thaliana grown at different copper regimes. Planta 229: 767–779 [DOI] [PubMed] [Google Scholar]
- Abdel-Ghany S.E., Müller-Moulé P., Niyog K.K., Pilon M., Shikanai T. (2005). Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17: 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Ghany S.E., Pilon M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283: 15932–15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.D., Kropat J., Tottey S., Del Campo J.A., Merchant S.S. (2007). Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 143: 263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison D.B., Gadbury G.L., Heo M.S., Fernández J.R., Lee C.K., Prolla T.A., Weindruch R. (2002). A mixture model approach for the analysis of microarray gene expression data. Comput. Stat. Data Anal. 39: 1–20 [Google Scholar]
- Andreini C., Banci L., Bertini I., Rosato A. (2008). Occurrence of copper proteins through the three domains of life: A bioinformatic approach. J. Proteome Res. 7: 209–216 [DOI] [PubMed] [Google Scholar]
- Arnaud N., Ravet K., Borlotti A., Touraine B., Boucherez J., Fizames C., Briat J.-F., Cellier F., Gaymard F. (2007). The iron-responsive element (IRE)/iron-regulatory protein 1 (IRP1)–cytosolic aconitase iron-regulatory switch does not operate in plants. Biochem. J. 405: 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askwith C., Eide D., Van Ho A., Bernard P.S., Li L., Davis-Kaplan S., Sipe D.M., Kaplan J. (1994). The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76: 403–410 [DOI] [PubMed] [Google Scholar]
- Audic S., Claverie J.M. (1997). The significance of digital gene expression profiles. Genome Res. 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Baldi P., Long A.D. (2001). A Bayesian framework for the analysis of microarray expression data: Regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- Beauclair L., Yu A., Bouché N. (2010). MicroRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 62: 454–462 [DOI] [PubMed] [Google Scholar]
- Becher M., Talke I., Krall L., Krämer U. (2004). Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the znc hyperaccumulator Arabidopsis halleri. Plant J. 37: 251–268 [DOI] [PubMed] [Google Scholar]
- Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cezard L., Le Bris P., Borrega N., Herve J., Blondet E., Balzergue S., Lapierre C., Jouanin L. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl R.P., Jach G., Saedler H., Huijser P. (2005). Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352: 585–596 [DOI] [PubMed] [Google Scholar]
- Burkhead J.L., Reynolds K.A., Abdel-Ghany S.E., Cohu C.M., Pilon M. (2009). Copper homeostasis. New Phytol. 182: 799–816 [DOI] [PubMed] [Google Scholar]
- Cai X., Davis E.J., Ballif J., Liang M., Bushman E., Haroldsen V., Torabinejad J., Wu Y. (2006). Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 57: 2563–2569 [DOI] [PubMed] [Google Scholar]
- Castruita M., Casero D., Karpowicz S.J., Kropat J., Vieler A., Hsieh S.I., Yan W., Cokus S., Loo J.A., Benning C., Pellegrini M., Merchant S.S. (2011). Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin F., Orsel M., Dorbe M.F., Chardon F., Truong H.N., Miller A.J., Krapp A., Daniel-Vedele F. (2007). The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.H., Chiecko J., Punshon T., Lanzirotti A., Lahner B., Salt D.E., Walker E.L. (2010). Successful reproduction requires the function of Arabidopsis YELLOW STRIPE-LIKE1 and YELLOW STRIPE-LIKE3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol. 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohu C.M., Abdel-Ghany S.E., Gogolin Reynolds K.A., Onofrio A.M., Bodecker J.R., Kimbrela J.A., Niyogib K.K., Pilon M. (2009). Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: Regulation and unexpected phenotypes in an Arabidopsis mutant. Mol. Plant 2: 1336–1350 [DOI] [PubMed] [Google Scholar]
- Colangelo E.P., Guerinot M.L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly E.L., Campbell N.H., Grotz N., Prichard C.L., Guerinot M.L. (2003). Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 133: 1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Bari R.P., Stitt M., Scheible W.R., Udvardi M.K. (2004). Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: Unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Davuluri R.V., Sun H., Palaniswamy S.K., Matthews N., Molina C., Kurtz M., Grotewold E. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Freitas J., Wintz H., Kim J.H., Poynton H., Fox T., Vulpe C. (2003). Yeast, a model organism for iron and copper metabolism studies. Biometals 16: 185–197 [DOI] [PubMed] [Google Scholar]
- DiDonato R.J., Jr, Roberts L.A., Sanderson T., Eisley R.B., Walker E.L. (2004). Arabidopsis Yellow Stripe-Like2 (YSL2): A metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J. 39: 403–414 [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Eisses J.F., Kaplan J.H. (2005). The mechanism of copper uptake mediated by human CTR1: A mutational analysis. J. Biol. Chem. 280: 37159–37168 [DOI] [PubMed] [Google Scholar]
- Ferguson-Miller S., Babcock G.T. (1996). Heme/copper terminal oxidases. Chem. Rev. 96: 2889–2907 [DOI] [PubMed] [Google Scholar]
- Fraústo da Silva J.J.R., Williams R.J.P. (2001). The Biological Chemistry of the Elements. (Oxford, UK: Clarendon Press)
- Ghasemi R., Ghaderian M., Krämer U. (2009). Interference of nickel with copper and iron homeostasis contributes to metal toxicity symptoms in the nickel hyperaccumulator plant Alyssum inflatum. New Phytol. 184: 566–580 [DOI] [PubMed] [Google Scholar]
- Gibbs C.R. (1976). Characterization and application of FerroZine iron reagent as a ferrous iron indicator. Anal. Chem. 48: 1197–1201 [Google Scholar]
- González-Ballester D., Casero D., Cokus S., Pellegrini M., Merchant S.S., Grossman A.R. (2010). RNA-Seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L.S., Rogers E.E. (2004). FRD3 controls iron localization in Arabidopsis. Plant Physiol. 136: 2523–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris Z.L., Takahashi Y., Miyajima H., Serizawa M., MacGillivray R.T., Gitlin J.D. (1995). Aceruloplasminemia: Molecular characterization of this disorder of iron metabolism. Proc. Natl. Acad. Sci. USA 92: 2539–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett R., Kosman D.J. (1995). Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J. Biol. Chem. 270: 128–134 [DOI] [PubMed] [Google Scholar]
- Hellman N.E., Gitlin J.D. (2002). Ceruloplasmin metabolism and function. Annu. Rev. Nutr. 22: 439–458 [DOI] [PubMed] [Google Scholar]
- Hématy K., Cherk C., Somerville S. (2009). Host-pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 12: 406–413 [DOI] [PubMed] [Google Scholar]
- Hoopes J.T., Dean J.F. (2004). Ferroxidase activity in a laccase-like multicopper oxidase from Liriodendron tulipifera. Plant Physiol. Biochem. 42: 27–33 [DOI] [PubMed] [Google Scholar]
- Hoopes J.T., Dean J.F.D. (2001). Staining electrophoretic gels for laccase and peroxidase activity using 1,8-diaminonaphtalene. Anal. Biochem. 293: 96–101 [DOI] [PubMed] [Google Scholar]
- Kim S.S., et al. (2010). LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl alpha-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J., Saedler H., Huijser P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250: 7–16 [DOI] [PubMed] [Google Scholar]
- Krämer U., Clemens S. (2005). Functions and homeostasis of zinc, copper, and nickel in plants. In Molecular Biology of Metal Homeostasis and Detoxification, Topics in Current Genetics, M. Tamás and E. Martinoia, eds (Berlin/Heidelberg: Springer-Verlag), pp. 216–271
- Kropat J., Tottey S., Birkenbihl R.P., Depege N., Huijser P., Merchant S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fontaine S., Quinn J.M., Nakamoto S.S., Page M.D., Göhre V., Moseley J.L., Kropat J., Merchant S. (2002). Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot. Cell 1: 736–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Li Y., Kristiansen K., Wang J. (2008). SOAP: Short oligonucleotide alignment program. Bioinformatics 24: 713–714
- Liang M., Davis E., Gardner D., Cai X., Wu Y. (2006). Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224: 1185–1196 [DOI] [PubMed] [Google Scholar]
- Liang X., Nazarenus T.J., Stone J.M. (2008). Identification of a consensus DNA-binding site for the Arabidopsis thaliana SBP domain transcription factor, AtSPL14, and binding kinetics by surface plasmon resonance. Biochemistry 47: 3645–3653 [DOI] [PubMed] [Google Scholar]
- Marioni J.C., Mason C.E., Mane S.M., Stephens M., Gilad Y. (2008). RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. (1995). Mineral Nutrition of Higher Plants. (Boston: Academic Press)
- McCaig B.C., Meagher R.B., Dean J.F. (2005). Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221: 619–636 [DOI] [PubMed] [Google Scholar]
- Mendel R.R. (2011). Cell biology of molybdenum in plants. Plant Cell Rep. 30: 1787–1797 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., Allen M.D., Kropat J., Moseley J.L., Long J.C., Tottey S., Terauchi A.M. (2006). Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763: 578–594 [DOI] [PubMed] [Google Scholar]
- Moseley J.L., Page M.D., Alder N.P., Eriksson M., Quinn J., Soto F., Theg S.M., Hippler M., Merchant S. (2002). Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler M.U., Galy B., Hentze M.W. (2008). Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 28: 197–213 [DOI] [PubMed] [Google Scholar]
- Mukherjee I., Campbell N.H., Ash J.S., Connolly E.L. (2006). Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 223: 1178–1190 [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Pich A., Scholz G. (1996). Translocation of copper and other mcronutrients in tomato plants (Lycopersicom esculetum Mill.): Nicotianamine-stimulated copper transpor. J. Exp. Bot. 47: 41–47 [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Pourcel L., Routaboul J.M., Kerhoas L., Caboche M., Lepiniec L., Debeaujon I. (2005). TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S., Andrés-Colás N., García-Molina A., Peñarrubia L. (2007). Copper and iron homeostasis in Arabidopsis: Responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ. 30: 271–290 [DOI] [PubMed] [Google Scholar]
- Puig S., Thiele D.J. (2002). Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 6: 171–180 [DOI] [PubMed] [Google Scholar]
- Quinn J.M., Merchant S. (1995). Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7: 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redinbo M.R., Yeates T.O., Merchant S. (1994). Plastocyanin: Structural and functional analysis. J. Bioenerg. Biomembr. 26: 49–66 [DOI] [PubMed] [Google Scholar]
- Ridge P.G., Zhang Y., Gladyshev V.N. (2008). Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS ONE 3: e1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N.J., Procter C.M., Connolly E.L., Guerinot M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Robinson N.J., Winge D.R. (2010). Copper metallochaperones. Annu. Rev. Biochem. 79: 537–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F.I., Esch J.J., Hall A.E., Binder B.M., Schaller G.E., Bleecker A.B. (1999). A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283: 996–998 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russel D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press)
- Sancenón V., Puig S., Mateu-Andres I., Dorcey E., Thiele D.J., Peñarrubia L. (2004). The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J. Biol. Chem. 279: 15348–15355 [DOI] [PubMed] [Google Scholar]
- Sancenón V., Puig S., Mira H., Thiele D.J., Peñarrubia L. (2003). Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 51: 577–587 [DOI] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H., Harris Z.L. (2003). Genetic defects in copper metabolism. J. Nutr. 133: 1527S–1531S [DOI] [PubMed] [Google Scholar]
- Shorrocks V.M., Alloway B.J. (1988). Copper in plant, animal and human nutrition. In Copper Development Association TN35 (Potters Bar, UK: Orchard House), pp. 15–17
- Sinclair S.A., Sherson S.M., Jarvis R., Camakaris J., Cobbett C.S. (2007). The use of the zinc-fluorophore, Zinpyr-1, in the study of zinc homeostasis in Arabidopsis roots. New Phytol. 174: 39–45 [DOI] [PubMed] [Google Scholar]
- Solberg E., Evans I., Penny D. (1999). Copper deficiency: Diagnosis and correction. Agri-facts. Soil Fertility/Crop Nutrition. Alberta Agriculture, Food and Rural Development, Agdex 532–3, pp. 1–9
- Sommer F., Kropat J., Malasarna D., Grossoehmec N.E., Chend X., Giedroc D.P., Merchant S. (2010). The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22: 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbjerg F. (1951). Yield curves and chemical plant analyses. Plant Soil 3: 97–109 [Google Scholar]
- Talke I., Hanikenne M., Krämer U. (2006). Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number of genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol. 142: 148–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1995). Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science 269: 1069–1074 [DOI] [PubMed] [Google Scholar]
- Vert G., Briat J.F., Curie C. (2003). Dual regulation of the Arabidopsis high affinity root iron uptake system by local and long-distance signals. Plant Physiol. 132: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters B.M., Chu H.H., Didonato R.J., Roberts L.A., Eisley R.B., Lahner B., Salt D.E., Walker E.L. (2006). Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch R.M., Norvell W.A., Schaefer S.C., Shaff J.E., Kochian L.V. (1993). Induction of iron(III) and copper(II) reduction in pea (Pisum sativum L.) roots by Fe and Cu status: Does the root-cell plasmalemma Fe(III)-chelate reductase perform a general role in regulating cation uptake? Planta 190: 555–561 [Google Scholar]
- Winge D.R. (1998). Copper-regulatory domain involved in gene expression. Prog. Nucleic Acid Res. Mol. Biol. 58: 165–195 [DOI] [PubMed] [Google Scholar]
- Wintz H., Fox T., Wu Y.-Y., Feng V., Chen W., Chang H.-S., Zhu T., Vulpe C. (2003). Expression profiles of A. thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 278: 47644–47653 [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Abdel-Ghany S.E., Cohu C.M., Kobayashi Y., Shikanai T., Pilon M. (2007). Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 282: 16369–16378 [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009). SQUAMOSA Promoter Binding Protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K., et al. (2004). A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 337: 49–63 [DOI] [PubMed] [Google Scholar]
- Yi Y., Guerinot M.L. (1996). Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Zheng L., Miller E.W., Pralle A., Isacoff E.I., Chang C. (2006). A selective turn-on fluorescent sensor for imaging copper in living cells. J. Am. Chem. Soc. 128: 10–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.