This work examines the molecular basis of cell death–associated root colonization of Arabidopsis by the mutualistic fungus P. indica. The data support the existence of a previously unknown pathway in plants, in which ER stress induces a vacuole-mediated cell death dependent on VPE/caspase 1–like activities.
Abstract
In Arabidopsis thaliana roots, the mutualistic fungus Piriformospora indica initially colonizes living cells, which die as the colonization proceeds. We aimed to clarify the molecular basis of this colonization-associated cell death. Our cytological analyses revealed endoplasmic reticulum (ER) swelling and vacuolar collapse in invaded cells, indicative of ER stress and cell death during root colonization. Consistent with this, P. indica–colonized plants were hypersensitive to the ER stress inducer tunicamycin. By clear contrast, ER stress sensors bZIP60 and bZIP28 as well as canonical markers for the ER stress response pathway, termed the unfolded protein response (UPR), were suppressed at the same time. Arabidopsis mutants compromised in caspase 1–like activity, mediated by cell death–regulating vacuolar processing enzymes (VPEs), showed reduced colonization and decreased cell death incidence. We propose a previously unreported microbial invasion strategy during which P. indica induces ER stress but inhibits the adaptive UPR. This disturbance results in a VPE/caspase 1–like-mediated cell death, which is required for the establishment of the symbiosis. Our results suggest the presence of an at least partially conserved ER stress–induced caspase-dependent cell death pathway in plants as has been reported for metazoans.
INTRODUCTION
The endoplasmic reticulum (ER) plays a vital role in the processing of glycoproteins destined for secretion. These secretory proteins pass the ER to obtain their proper three-dimensional structures, which is a prerequisite for their functionality. The ER makes essential contributions to the protein composition of the plasma membrane, extracellular matrix (apoplast), and vacuoles. Because of these functions, it is intimately involved in plant developmental processes (Padmanabhan and Dinesh-Kumar, 2010). The ER machinery recognizes defects in nascent proteins, which results in improper processing and their degradation, as demonstrated for the plasma membrane–localized brassinosteroid receptor BRI1 (Jin et al., 2007). In Arabidopsis thaliana, accurate protein processing in the ER is termed ER quality control (ER-QC) and regulated by at least three systems: the STROMAL-DERIVED FACTOR2 (SDF2)–HEAT SHOCK PROTEIN40/ER LUMEN-LOCALIZED DnaJ PROTEIN3b (ERdj3b)–LUMINAL BINDING PROTEIN (BIP) complex, the CALRETICULIN/CALNEXIN (CRT/CNX) cycle, and the protein disulfide isomerase (PDI) system (Anelli and Sitia, 2008). After cotranslational translocation into the ER, for which the SEC61 translocon is required, nascent proteins are N-glycosylated by the oligosaccharyl transferase complex (OST). This OST complex consists of various subunits (e.g., DEFENDER AGAINST APOPTOTIC DEATH1 [DAD1]; Kelleher and Gilmore, 2006). After N-glycosylation, nascent proteins bind to the SDF2-ERdj3b-BIP complex. Thereafter, the coordinated action of CRTs and CNXs is required to mediate protein folding. For certain glycoproteins, PDIs perform intramolecular disulfide isomerization that is eventually required for correct folding. Misfolded proteins leave the ER for degradation by cytosolic proteasomes (Anelli and Sitia, 2008; Vitale and Boston, 2008). The ER-QC machinery is apparently highly conserved among eukaryotes, including plants (Liu and Howell, 2010a). The ER workload and, thus, ER-QC activities vary depending on the developmental stage, the type of tissue, and the occurrence of external stresses. In case ER-QC does not meet the demand of protein processing, the enrichment of misfolded proteins in the ER triggers ER stress. In mammals, ER stress is sensed by the ER membrane–localized receptors INOSITOL REQUIRING PROTEIN1, ACTIVATING TRANSCRIPTION FACTOR6 (ATF6), and PROTEIN KINASE RNA-LIKE ER KINASE, which activate the unfolded protein response (UPR) (Schröder, 2006). The UPR functions as an adaptive process of cells to enhance quality control and to relieve ER stress (Malhotra and Kaufman, 2007). In mammals, the UPR consists of induction of ER chaperones, elevated ER-associated degradation, and attenuated translation of secreted proteins (Malhotra and Kaufman, 2007). By contrast, prolonged ER stress or malfunctional UPR results in proapoptotic signaling and programmed cell death (PCD), which is mediated by the same set of ER stress sensors that are activating the UPR. ER stress–induced apoptosis relies on the activation of a set of cell death–associated Cys proteases, the so-called caspases (Szegezdi et al., 2006; Rasheva and Domingos, 2009). By contrast, ER-QC, ER stress sensing/signaling, and ER stress–induced cell death are less well understood in plants, although molecular studies indicate that ER stress sensing/signaling and the UPR are conserved in plants (Kamauchi et al., 2005; Vitale and Boston, 2008). Recent studies identified the transcription factors BASIC REGION/LEU ZIPPER MOTIF (bZIP) bZIP28 and bZIP60 as ER membrane–localized ER stress sensors that are involved in the induction of UPR (Liu et al., 2007a; Iwata et al., 2008). Both proteins are similar to the mammalian bZIP transcription factor ATF6 (Urade, 2009). Moreover, the ER is known to participate in plant PCD pathways as it modulates drought stress–induced PCD (Duan et al., 2010), and ER dysfunction initiates PCD (Malerba et al., 2004; Watanabe and Lam, 2006). Whereas a regulatory role of the ER in plant cell death initiation has been demonstrated (Watanabe and Lam, 2009), the molecular basis of PCD initiation and execution in response to ER stress has remained elusive.
Plants sense microbial invaders via plasma membrane–localized pattern recognition receptors (PRRs). PRRs specifically recognize microbial molecules known as microbe-associated molecular patterns (MAMPs). In plants, the PRRs EF-TU RECEPTOR (EFR), FLAGELLIN-SENSITIVE2 (FLS2), and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) recognize the MAMPs bacterial flagellin, elongation factor TU, and fungal chitin, respectively, and induce a highly effective immune response (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006; Petutschnig et al., 2010). The ER is essential for the function of the plant immune system. A considerable number of antimicrobial proteins traverse the ER to their final destination, such as the apoplast, vacuole, or plasma membrane (Jelitto-Van Dooren et al., 1999; Wang et al., 2005; Lipka et al., 2007; Kwon et al., 2008). Disturbance of ER-QC results in improper processing of immunity-related proteins and impairs plant immunity, as recently reported for EFR (Nekrasov et al., 2009; Saijo et al., 2009). Mutants lacking components of the ER-QC were specifically impaired in EFR processing and its delivery to the plasma membrane. Consequently, plants were more susceptible to bacterial pathogens. Interestingly, the induction of genes encoding ER-QC proteins accompanies pathogen recognition, thereby priming the ER for higher processing capacities for defense components (Wang et al., 2005). Mutants lacking SEC61α (a subunit of ER membrane translocon), DAD1 (a subunit of the OST complex), or BIP2 (a chaperone of the SDF2-ERdj3b-BIP complex) exhibit reduced immune activation by the salicylic acid analog benzothiadiazole (Wang et al., 2005).
In this study, we investigated the colonization strategy of the mutualistic fungus Piriformospora indica in Arabidopsis roots. P. indica was discovered in the 1990s and thereafter found to confer various beneficial effects to host plants, such as growth promotion in different plants, seed yield increase, abiotic stress tolerance, and biotic stress resistance (Verma et al., 1998; Varma et al., 1999; Waller et al., 2005). The fungus has obviously evolved efficient invasion strategies as it colonizes a broad array of plants, and nonhosts have not been identified (Qiang et al., 2011). Recent studies indicated that its colonization success relies on the broad suppression of root immunity (Schäfer et al., 2009; Jacobs et al., 2011). Elucidation of the P. indica genome sequence and the fungal transcriptome during colonization of living and dead barley (Hordeum vulgare) roots identified traits commonly associated with biotrophy and saprotrophy (Zuccaro et al., 2011). Consistent with this, our recent studies revealed a biphasic colonization strategy in Arabidopsis roots (Jacobs et al., 2011). P. indica initially colonizes living cells hallmarked by plasma membrane invagination and by the intactness of all cell organelles. This biotrophic phase is observed until 3 d after inoculation (DAI) and is followed by a cell death–associated colonization phase (>3 DAI), which is consistent with previous reports on interactions of P. indica with barley (Deshmukh et al., 2006). In barley, the latter phase involved transcriptional regulation of the ER membrane–localized cell death inhibitor BAX INHIBITOR-1 (BI-1), raising the possibility that the fungus recruits a host cell death program to establish a successful interaction. P. indica shows a patchy colonization pattern and preferentially colonizes the root maturation zone, while it is hardly detectable at the root meristematic and elongation zones. In addition, the fungus is obviously unable to pass the root endodermis and enter the central cylinder (Deshmukh et al., 2006; Jacobs et al., 2011). In this study, we analyzed the molecular basis of those processes involved in the initiation and execution of cell death during P. indica colonization. Based on cytological and molecular studies, we suggest that P. indica successfully colonizes Arabidopsis roots by triggering ER stress and simultaneously suppressing the adaptive UPR. Furthermore, we hypothesize that the inability of colonized root cells to relieve ER stress results in the induction of a vacuolar processing enzyme (VPE)/caspase 1–like activity-dependent vacuolar cell death program. We propose a model in which fungal colonization success is dependent on ER stress–induced cell death.
RESULTS
P. indica Impairs the Integrity of the ER, Thereby Enhancing Mutualistic Root Colonization
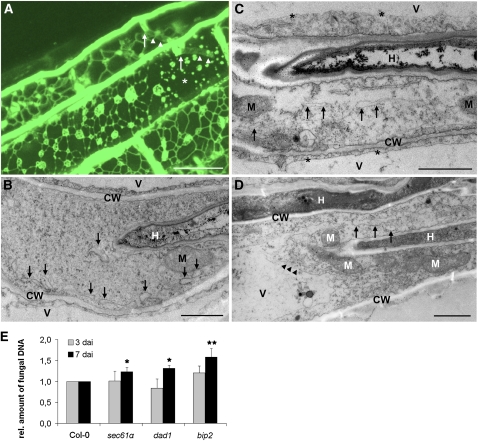
By analyzing P. indica colonization of roots of Arabidopsis line green fluorescent protein (GFP)-tmKKXX, in which the ER is GFP tagged (Benghezal et al., 2000), we observed a loss of the continuous ER network and the local appearance of globular ER structures around intracellular hyphae during biotrophic root colonization, suggesting an disintegration of the ER (see asterisk in Figure 1A). In comparison, surrounding cells showed continuous ER networks, reflecting the structural intactness of the ER (Figure 1A). To assess subcellular changes occurring during fungal colonization of Arabidopsis roots, we performed transmission electron microscopy. Transmission electron microscopy studies confirmed that ER swelling accompanies biotrophic root colonization (Figure 1B). Since other organelles were ultrastructurally unaltered and we did not observe lysis of the cytoplasm, these colonized cells were considered alive. As colonization proceeded (>3 DAI), we observed ER swelling that was followed by tonoplast rupture and lysis of the cytoplasm (Figures 1C and 1D), which are hallmarks of PCD (van Doorn et al., 2011). Interestingly, even at this stage, plastids and mitochondria remained ultrastructurally unaltered (Figures 1C and 1D). These observations prompted us to hypothesize that P. indica disturbs ER function and that the resulting ER stress might induce a vacuole-mediated cell death. ER swelling thus represents an ultrastructural hallmark for the termination of biotrophic and the initiation of cell death–associated colonization by P. indica.
Figure 1.
P. indica Impairs ER Integrity and Induces Vacuole Collapse in Colonized Root Cells.
(A) Confocal microscopy of P. indica–colonized GFP-tmKKXX (ER marker)–expressing Arabidopsis roots at 3 DAI. The fungus penetrated (arrows) two cells and intracellular hyphae are visible (arrowheads). The ER of the upper colonized cell is still intact, while ER disintegration is associated with colonization of the lower cell (asterisk). Note the ER of surrounding, noncolonized cells is intact. P. indica was stained with chitin-specific WGA-AF488. Intracellular hyphae are faintly stained due to limited dye diffusion. Bar = 20 μm.
(B) Micrograph of a biotrophic colonization phase. Intracellular hypha invaginated the host plasma membrane. The ER of this cell is partially swollen (arrows). CW, cell wall; H, hypha; M, mitochondria; V, vacuole. Bar = 2 μm.
(C) ER swelling and lysis of the cytoplasm during cell death–associated root colonization. Arrows indicate ER swelling in a cell harboring an intracellular hypha (H). Cell lysis is restricted to the colonized cell, while noncolonized neighboring cells are unaffected, as indicated by the intact tonoplasts (asterisks). CW, cell wall; H, hypha; M, mitochondria; V, vacuole. Bar = 2 μm.
(D) ER swelling and vacuolar collapse during cell death–associated colonization. Intracellularly colonized cell shows ER swelling (arrows), vacuolar collapse, as indicated by tonoplast rupture (arrowheads), and lysis of the cytoplasm. The micrograph shows intracellular (top H) and intercellular (bottom H) hyphae. CW, cell wall; H, hyphae; M, mitochondria; V, vacuole. Bar = 2 μm.
(E) ER dysfunction improves cell death–associated colonization of Arabidopsis roots by P. indica. Arabidopsis Col-0 and the mutants sec61α, dad1, and bip2, which are impaired in ER function, were inoculated with P. indica and fungal biomass was determined at biotrophic (3 DAI) and cell death–associated colonization stages (7 DAI) by qRT-PCR. Fungal colonization levels in all mutants were normalized with wild-type Col-0 colonization (set to 1). Results shown are means ± se of three independent experiments. For each experiment, ~200 plants were analyzed per line at each time point. Asterisks indicate significant differences in the colonization of mutants compared with Col-0 at 7 DAI at P < 0.05 (*) or P < 0.01 (**) as analyzed by two-way analysis of variance (ANOVA).
[See online article for color version of this figure.]
Next, we examined to what extent the disturbed ER function affects colonization success. To this end, the Arabidopsis mutants bip2 (deficient in the chaperone BIP2), dad1 (deficient in a subunit of the oligosaccharyl transferase DAD1), and sec61α (deficient in a component of the SEC61 translocon complex), all of which lack components of the ER-QC, were selected and analyzed for altered fungal colonization at 3 DAI (biotrophic phase) and 7 DAI (cell death phase). Quantitative real-time PCR (qRT-PCR)–based quantification of fungal biomass showed enhanced fungal colonization rates at 7 DAI in all mutants compared with wild-type Columbia-0 (Col-0) (Figure 1E), suggesting that impaired ER-QC supports fungal development during cell death–associated colonization. Alternatively, malfunctional immunity in these ER-QC mutants might explain the altered colonization. Therefore, we examined their responsiveness to MAMPs as well as their colonization by two pathogens following two different lifestyles, the biotrophic powdery mildew fungus Erysiphe cruciferarum and necrotrophic Botrytis cinerea. In the first assay, we analyzed the flg22-induced seedling growth inhibition in wild-type Col-0 and the ER-QC mutants. flg22 treatment reduced the biomass of wild-type Col-0 and all mutant plants, as opposed to the flg22-insensitive fls2c mutant (see Supplemental Figure 1A online). Second, we analyzed the occurrence of flg22- or chitin-induced oxidative burst in roots of the ER-QC mutants. Consistently, we observed transient root oxidative bursts upon treatment with flg22 or chitin in bip2, dad1, and sec61α mutants like in wild-type Col-0, but not in flg22- and chitin-insensitive mutants fls2c and cerk1-2, respectively (see Supplemental Figures 1B and 1C online), indicating intactness of MAMP-triggered immunity in the mutants. In addition, bip2, dad1, and sec61α showed no significant alterations in colonization by biotrophic E. cruciferarum and necrotrophic B. cinerea compared with wild-type Col-0 (see Supplemental Figures 2A and 2B online).
P. indica–Colonized Plants Are Hypersensitive to ER Stress but Disturbed in the UPR
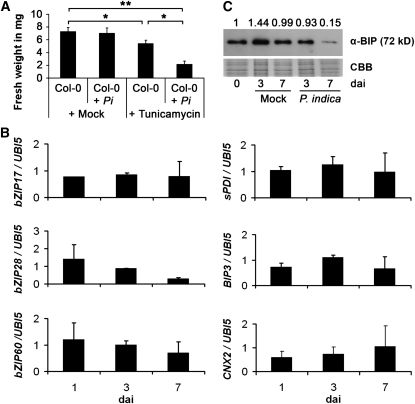
Since analysis of fungal growth in roots indicated improved colonization of mutants lacking crucial components of the ER-QC, we investigated whether P. indica affected tolerance of colonized plants to ER stress. We applied the ER stress inducer tunicamycin (TM), which specifically blocks UDP-N-ACETYLGLUCOSAMINE:DOLICHOL PHOSPHATE N-ACETYGLUCOSAMINE-1-P TRANSFERASE and thereby inhibits protein N-glycosylation in the ER (Pattison and Amtmann, 2009). P. indica–colonized (3 DAI, biotrophic stage) and noncolonized (mock-treated) Col-0 plants were treated with TM or DMSO (control). Plant fresh weights were determined at 7 d after treatment (DAT). In noncolonized plants, TM treatment resulted in an ~20% reduction in fresh weight compared with DMSO-treated plants. In P. indica–colonized plants, TM significantly reduced plant biomass by ~60% compared with untreated controls (Figure 2A). This supports the view that P. indica disturbs ER function already during biotrophic colonization and further suggests that P. indica–colonized plants are hypersensitive to ER stress.
Figure 2.
P. indica–Colonized Plants Are Hypersensitive to ER Stress, but ER Stress Signaling Is Not Activated in Colonized Roots.
(A) Arabidopsis Col-0 plants colonized by P. indica (Pi) are more sensitive to the ER stress inducer TM than are noncolonized plants, as evidenced by a reduced biomass. Plant biomass was determined at 7 DAT. Data presented show means of three independent experiments ± se. For each experiment, 10 plants were analyzed per treatment. Asterisks indicate significance at P < 0.05 (*) and P < 0.01 (**) as analyzed by two-way ANOVA.
(B) Expression of ER stress sensors (bZIP17, bZIP28, and bZIP60) and markers for the UPR (sPDI, BIP3, and CNX2) was measured by qRT-PCR. Arabidopsis plants were inoculated with P. indica or mock treated. Data shown represent fold changes of genes and display the ratio of candidate gene expression to housekeeping gene UBIQUITIN5 in colonized roots relative to mock-treated roots. Data presented show means of three independent experiments ± se.
(C) BIP protein accumulation is reduced during P. indica colonization. Arabidopsis roots were inoculated with P. indica or mock treated and harvested for protein extraction. The staining with Coomassie blue (CBB) indicates equal loading of all samples. Numbers on top of the immunoblot indicate relative BIP protein band intensities (0 DAI was set to 1) as determined by ImageJ.
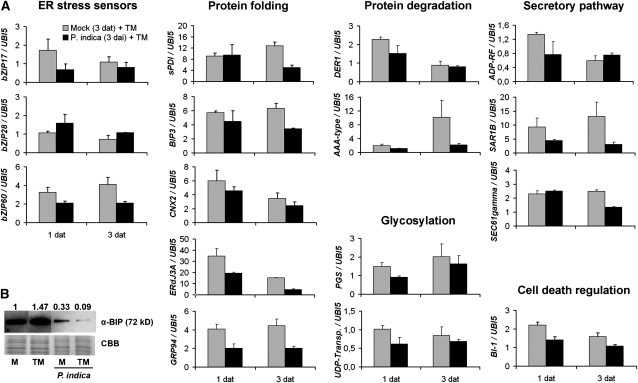
Next, we examined whether colonization-associated ER stress resulted in both activation of ER stress sensing and subsequent UPR. P. indica predominantly colonizes the maturation zone of roots, while the fungus is hardly detectable in the juvenile root sections, the elongation and meristematic zones. Thus, to avoid dilution effects of noncolonized root tissues, we harvested the maturation zone of P. indica–colonized and noncolonized roots at 1, 3, and 7 DAI and monitored gene expression levels of putative ER stress sensors (bZIP17, bZIP28, and bZIP60) and markers of the UPR (sPDI, BIP3, and CNX2) by qRT-PCR. Unexpectedly, none of the tested genes was expressed at higher levels during colonization (Figure 2B). Instead, expression of bZIP28, BIP3, and CNX2 was suppressed in colonized roots at some of the investigated time points (Figure 2B). Consistent with this, BIP protein levels were reduced in colonized roots at 3 and 7 DAI by 35 and 85%, respectively, compared with noncolonized roots (Figure 2C). To elucidate whether the selected ER stress markers were induced by ER stress in roots and whether P. indica might suppress ER stress signaling, we treated noncolonized and P. indica–colonized roots (3 DAI, biotrophic stage) with TM or DMSO (control). The root samples were harvested at 1 and 3 d after TM treatment, and the expression levels of ER stress sensors and UPR markers were analyzed by qRT-PCR. TM treatment induced the expression of all these genes (except bZIP28) in noncolonized roots (Figure 3A, gray columns). By contrast, induction of bZIP17, bZIP60, BIP3, and sPDI expression was much weaker in P. indica–colonized roots, while CNX2 and BI-1 showed only slightly reduced expression levels (Figure 3A). These data indicated impaired ER stress signaling at the transcript level.
Figure 3.
P. indica Colonization Results in the Suppression of the UPR.
(A) Arabidopsis roots were inoculated with P. indica or mock treated. Inoculated and mock-treated plants were treated with TM (5 μg mL−1) or DMSO (control). Root samples from different treatments (Pi + TM, Pi + DMSO; mock + TM, mock + DMSO) were harvested at 1 and 3 DAT. Data shown represent fold changes of genes and display the ratio of candidate gene expression to housekeeping gene UBIQUITIN5 using the ΔΔCT method (Schmittgen and Livak, 2008). ΔΔCT values obtained from Pi + TM samples were divided by ΔΔCT values of Pi + DMSO to obtain the displayed fold changes. Similarly, ΔΔCT values of samples mock + TM were divided by ΔΔCT values of mock + DMSO. Fold changes >1 or <1 indicate induction or suppression of genes, respectively. Data are means of three independent experiments ± se.
(B) BIP protein accumulation after TM treatment and P. indica inoculation. Samples were run on the same blot, but the lanes were arranged for presentation. Arabidopsis roots were inoculated with P. indica or mock treated prior to treatment with TM or DMSO (control) and harvested 2 d later. The staining with Coomassie blue (CBB) indicates equal loading of all samples. Numbers on top of the immunoblots represent relative BIP protein band intensities (mock treatment was set to 1) as determined by ImageJ. M, mock.
In addition to sPDI, BIP3, and CNX2, gene products of ERDj3A and GRP94 participate in ER-localized protein folding and are induced during ER stress (Iwata et al., 2008). Notably, TM-induced expression of ERDj3A and GRP94 was also suppressed in P. indica–colonized roots (Figure 3A). We further tested the expression of ER stress–induced genes, whose products function in protein degradation (Derlin-like 1 and AAA-type ATPase), glycosylation (putative galactinol synthase and UDP-galactose/UDP-glucose transporter [UDP-transp.]), and the secretory pathway (ADP-ribosylation factor [ADP-RF], SAR1B, and SEC61γ). These genes are induced in leaves by TM (Urade, 2007; Iwata et al., 2008). Except for UDP-transp., TM induced the expression of these genes in roots at 1 and/or 3 DAT. By clear contrast, expression of all genes (except CNX2) was suppressed in P. indica–colonized roots at 1 and/or 3 d after TM treatment (Figure 3A). We further examined whether root colonization by P. indica also affected BIP accumulation in response to TM. For this, P. indica–colonized (3 DAI) and noncolonized roots were treated with TM or DMSO (control), and roots were harvested 2 d later. BIP accumulated in response to TM treatment in noncolonized roots (increase of 47%) (Figure 3B). By contrast, BIP protein synthesis was suppressed in P. indica–colonized mock (decrease of 67%) and TM-treated roots (decrease of 95%). Taken together, the analyses suggested that the fungus disturbed ER stress signaling as well as UPR-associated processes such as protein folding, glycosylation, protein degradation, and secretion.
Vacuole-Mediated Cell Death Is Downstream of ER Stress Induction and Affects Mutualistic Root Colonization
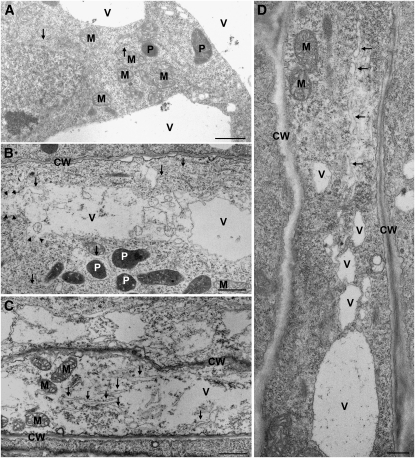
Our electron microscopy studies indicated that colonization-associated ER swelling preceded vacuolar collapse during cell death–associated colonization (Figure 1). Therefore, we wanted to know which proteins were essential for vacuole collapse following ER stress. VACUOLAR PROCESSING ENZYMES (VPEs) mediate vacuolar collapse and execution of virus-induced cell death (hypersensitive response) in tobacco (Nicotiana tabacum; Hatsugai et al., 2004). VPEs form a small gene family consisting of four members (αVPE, βVPE, γVPE, and δVPE) (Hatsugai et al., 2006). All VPEs but δVPE were expressed in roots as indicated by RT-PCR (see Supplemental Figure 3 online). In a first assay, we treated roots of wild-type Col-0 and the quadruple vpe-null mutant, which is deficient in the four VPEs, with TM or DMSO (control) and examined root cells for ultrastructural changes (Figure 4). In addition, we included dad1, in which the impairment in ER function might result in TM hypersensitivity. We did not observe any changes in wild-type Col-0 or mutant root cells after mock treatment or at 1 DAT with TM (Figure 4A; data not shown). At 3 DAT, we observed ER swelling, vacuolar collapse, and lysis of the cytoplasm in root cells of wild-type Col-0 and dad1 (Figures 4B and 4C). dad1 showed the most severe effects, as lysis of the cytoplasm was most pronounced and detected in all cells at 3 DAT. By contrast, vacuolar collapse was not detected in root cells of vpe-null, although ER swelling occurred (Figure 4D). These results indicated that the genetic impairment of ER homeostasis (in dad1) accelerated the ER stress–induced cell death. Moreover, VPEs are required for ER stress–induced collapse of the vacuole. Notably, ultrastructural changes, associated with ER stress–induced vacuolar cell death after TM treatment, were highly similar to those observed in root cells during P. indica colonization (Figure 1). In both cases, ER stress seems to lie upstream of vacuolar collapse, and ER swelling most likely is not the consequence of cell death.
Figure 4.
Execution of TM-Induced Vacuolar Cell Death Depends on VPEs.
(A) Root cell of mock-treated Col-0 does not show any ultrastructural changes at 3 DAT. Bar = 1 μm.
(B) Root cell of Col-0 treated with TM (5 μg mL−1) shows ER swelling (arrows) and vacuolar collapse as indicated by the lack of a tonoplast or tonoplast rupture (arrowheads) at 3 DAT. Bar = 1 μm.
(C) Root cell of dad1 treated with TM shows severe ER swelling (arrows) and cytoplasmic lysis. Vacuoles are completely collapsed, as indicated by the absence of the tonoplast. Bar = 1 μm.
(D) Root cell of Col-0 mutant vpe-null treated with TM shows ER swelling (arrows), while the vacuole is completely intact at 3 DAT. Bar = 1 μm.
CW, cell wall; M, mitochondria; P, plastid; V, vacuole.
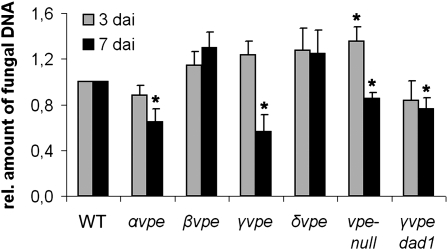
The results prompted us to test whether vacuolar collapse is essential for root colonization. We quantified P. indica colonization of αvpe, βvpe, γvpe, δvpe, and vpe-null roots at 3 (biotrophic phase) and 7 DAI (cell death phase) by qRT-PCR. vpe-null mutant showed higher fungal colonization at 3 DAI, consistent with earlier studies demonstrating an immune-related function of VPEs (Hatsugai et al., 2004; Rojo et al., 2004). We observed significantly reduced colonization of αvpe, γvpe, and vpe-null mutants at 7 DAI, while βvpe and δvpe mutants showed little if any enhancement in colonization (Figure 5), indicating that VPE-related activities contribute to cell death–associated colonization. To confirm that VPE-related activities act downstream of P. indica–induced ER stress, we generated γvpe dad1 double mutants and quantified P. indica colonization by qRT-PCR. γvpe dad1 displayed reduced colonization at 7 DAI, which was highly similar to the colonization phenotype of γvpe (Figure 5). Together, these data suggest that VPEs participate in the execution of ER stress–induced cell death and that γVPE plays a critical role in cell death–associated root colonization by P. indica.
Figure 5.
Colonization of Arabidopsis Roots by P. indica Is Dependent on VPEs.
Arabidopsis wild type (WT) and mutants αvpe, βvpe, γvpe, δvpe, and vpe-null, as well as double mutant γvpe dad1 were inoculated with P. indica. Fungal biomass was determined at biotrophic (3 DAI) and cell death–associated colonization stages (7 DAI) by qRT-PCR. Fungal colonization levels in all mutants were normalized with wild-type colonization (set to 1). Results shown are means ± se of three independent experiments. For each experiment, around 200 plants were analyzed per line at each time point. Asterisks indicate significant differences in the colonization of mutants compared with Col-0 at 3 or 7 DAI at P < 0.05 (*) as analyzed by two-way ANOVA.
VPE- and Caspase 1–Like Activities Are Enhanced in TM-Treated and P. indica–Colonized Roots
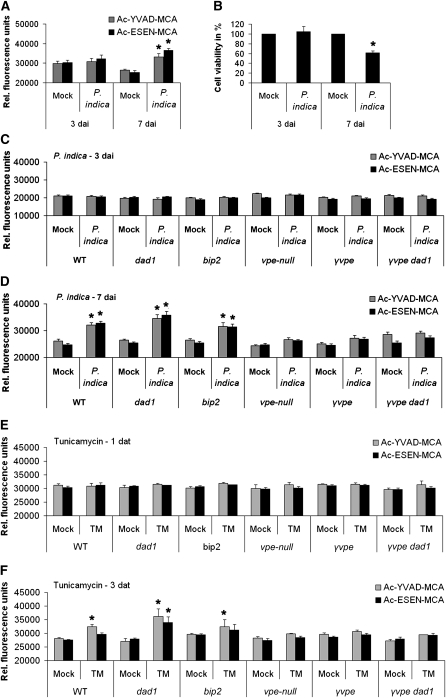
In addition to VPE activity, VPEs have caspase 1–like protease activity. It is suggested that both enzyme activities are required for vacuole-mediated plant cell death execution (Hatsugai et al., 2004; Kuroyanagi et al., 2005), although the enzyme targets are unknown. To examine the occurrence of these protease activities during root colonization, we measured VPE- and caspase 1–like activities in wild-type Col-0 roots during biotrophic (3 DAI) and cell death–associated colonization (7 DAI). To this end, we set up an assay to measure VPE- and caspase 1–like activities in root extracts from P. indica–colonized and noncolonized roots. Upon addition of either 1 mM VPE specific substrate Ac-ESEN-MCA or caspase 1–specific substrate Ac-YVAD-MCA to root extracts, VPE-mediated cleavage of MCA was spectrometrically determined. P. indica itself was unable to cleave these substrates (data not shown). The analyses did not reveal P. indica–dependent changes in enzyme activities at 3 DAI (biotrophic phase) but significantly enhanced VPE- and caspase 1–like activities at 7 DAI (cell death phase) with P. indica (Figure 6A). To relate the enhanced enzyme activities to colonization-associated cell death, we performed a fluorescein diacetate (FDA)-based cell viability assay. Esterases cleave off fluorescein in living cells, and the degree of cleavage can be quantified spectrometrically. We found a strong correlation between the length of analyzed root segments and the measured absolute fluorescence indicating FDA cleavage (see Supplemental Figure 4 online). The FDA assay revealed unaltered cell viability at 3 DAI but significantly reduced cell viability at 7 DAI with P. indica (Figure 6B). Thus, the experiments indicate a clear correlation of enhanced enzyme activities with the occurrence of enhanced cell death at 7 DAI.
Figure 6.
VPEs Mediate VPE and Caspase 1–Like Activities in Roots during Cell Death–Associated Colonization by P. indica or after TM Treatment.
(A) VPE- and caspase 1–like activities during biotrophic (3 DAI) and cell death–associated colonization (7 DAI) of roots by P. indica. For the assay, VPE substrate Ac-ESEN-MCA or caspase 1–like substrate Ac-YVAD-MCA was added to the root extracts for spectrophotometric determination of VPE and caspase 1–like activities, respectively. The values are given as relative fluorescence units (RFU). Data displayed are means (± se) of eight independent measurements per treatment of four independent biological experiments.
(B) FDA-based cell viability assay indicative of cell death in wild type roots at 3 and 7 days after P. indica inoculation or mock-treatment. Root segments were stained with FDA, and fluorescence intensities were spectrophotometrically determined. The values are given as relative fluorescence units relative to mock-treated roots (set to 100%). Data displayed are means (±se) of eight independent measurements per treatment of four independent biological experiments. Asterisks indicate significant differences in enzyme activities (A) and cell viability (B) in mock-treated compared with P. indica–colonized roots at P < 0.05 (*) analyzed by two-way ANOVA.
(C) and (D) VPE- and caspase 1–like activities during root colonization of various mutants by P. indica. The experiments were performed as described in (A). Root samples were harvested at 3 (C) and 7 DAI (D). Data displayed are means with (±se) of four independent measurements per treatment of four biological experiments. Asterisks indicate significant differences in respective enzyme activities between P. indica–colonized and mock-treated roots at P < 0.05 (*) as analyzed by two-way ANOVA.
(E) and (F) VPE- and caspase 1–like activities in roots of various mutants after TM or mock treatment. The experiments were performed as described in (A). Root samples were harvested at 1 (E) and 3 DAT (F). Data displayed are means (±se) of four independent measurements per treatment of four biological experiments. Asterisks indicate significant difference in respective enzyme activities between TM- and mock-treated roots at P < 0.05 (*) as analyzed by two-way ANOVA. WT, wild type.
We next examined whether the altered P. indica colonization of the ER-QC mutants bip2 and dad1 as well as γvpe, vpe-null, and γvpe dad1 mutants during biotrophic or cell death–associated interaction stages (Figures 1C, 1D, and 5) was associated with altered VPE- and caspase 1–like activities (Figures 6C and 6D). In a complementary experiment, we determined whether TM-induced cell death observed in our cytological studies (Figure 4) correlated with changed VPE- and caspase 1–like activities (Figures 6E and 6F). Since colonization data indicated a central role of γVPE during cell death–associated colonization, we performed both sets of experiments with γvpe and used vpe-null as control. To unravel whether improved colonization of ER-QC mutants (Figure 1E) and the hypersensitivity of dad1 to TM (Figure 4C) were linked to altered VPE and caspase 1–like activities, we examined bip2 besides the dad1 mutant. Finally, we included the double mutant γvpe dad1 in this enzyme activity studies because of its reduced colonization phenotype (Figure 5). We did not detect altered VPE and caspase 1–like activities in all mutants at 3 DAI (Figure 6C). However, we detected changes in VPE and caspase 1–like activities in a mutant-specific manner at 7 DAI. Enzyme activities were elevated in wild-type, dad1, and bip2 roots at a similar level at 7 DAI. By contrast, neither VPE nor caspase 1–like activities were observed in vpe-null and in γvpe at 7 DAI (Figure 6D). These findings suggested that the P. indica–induced activation of both enzyme activities relied on γVPE. Interestingly, elevated VPE and caspase 1–like activities in P. indica–colonized dad1 roots at 7 DAI was not detected in colonized γvpe dad1 roots at 7 DAI. In fact, enzyme activities in γvpe dad1 roots were highly similar to those in γvpe mutants (Figure 6D). TM treatment of all mutant roots resulted in changes of VPE and caspase 1–like activities that were similar to our analyses with P. indica–colonized roots. We did not observe altered enzyme activities at 1 DAT (Figure 6E). Caspase 1–like activity increased in dad1, bip2, and wild-type roots, while VPE activities were enhanced in dad1 roots at 3 d after TM treatment (Figure 6F). TM-induced VPE and caspase 1–like activities were not detectable in vpe-null and γvpe. Again, γvpe dad1 did not show TM-induced elevation of enzyme activities as detected in dad1 at 3 DAT, and the enzyme activity phenotypes strongly resembled those of γvpe. In summary, the results of the enzyme activity assays were consistent with the colonization and TM-induced cell death phenotypes (Figures 1E, 4, and 5).
ER Dysfunction Enhances P. indica– and TM-Induced Cell Death in a VPE-Dependent Manner
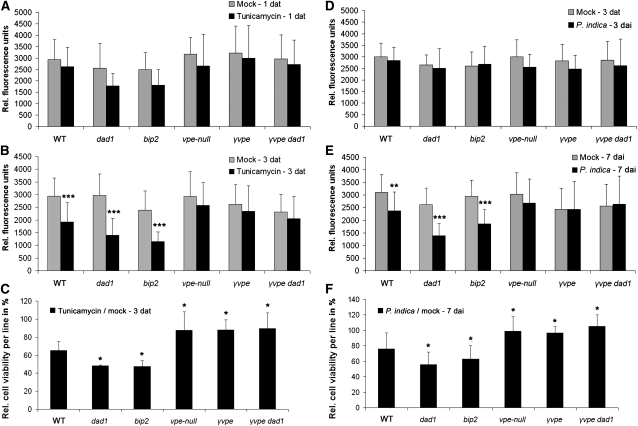
Finally, we were interested to know whether the variation in VPE and caspase 1–like enzyme activities in the various mutants after TM treatment or during cell death–associated root colonization (Figures 6D and 6F) was associated with an altered occurrence of cell death. In addition, these analyses should reveal whether P. indica and TM-induced ER stress initiated cell death and whether this was a VPE/caspase 1–like-mediated cell death. Therefore, we applied the FDA-based cell viability assay as described above. First, we treated dad1, bip2, vpe-null, γvpe, and γvpe dad1 mutants and the respective wild type with TM and stained root segments with FDA at 1 and 3 DAT. Whereas all mutants exhibited unaltered cell viability compared with the wild type at 1 DAT (Figure 7A), wild-type dad1 and bip2 mutants displayed a reduced cell viability at 3 DAT, which was not observed in vpe-null, γvpe, and γvpe dad1 mutants (Figures 7B and 7C). Notably, we detected higher cell death ratios in dad1 and bip2 as well as higher cell viability ratios in all vpe mutants compared with the wild type at 3 DAT (Figure 7C). We next determined FDA cleavage in the same mutants during biotrophic (3 DAI) and cell death–associated (7 DAI) colonization by P. indica. Consistent with our enzyme assays, none of the mutants showed an altered viability phenotype at 3 DAI (Figure 7D). Similar to the results obtained in the TM assay, the wild type, dad1, and bip2 exhibited more cell death at 7 DAI with P. indica. Again, cell viability was unaltered in vpe-null, γvpe, and γvpe dad1 mutants upon P. indica colonization at 7 DAI (Figures 7E and 7F). Unaltered cell viability in γvpe dad1 was in clear contrast with the reduced cell viability in dad1. Again, cell death ratios were enhanced in dad1 and bip2, while cell viability ratios were enhanced in all vpe mutants compared with the wild type at 7 DAI (Figure 7F). These results were consistent with the enzyme activity assays in that enhanced cell death in wild-type, dad1, and bip2 roots coincided with an enhanced caspase 1–like activity at 3 DAT with TM and enhanced VPE and caspase 1–like activities at 7 DAI with P. indica in these mutants. Accordingly, at those time points (1 DAT [TM] and 3 DAI [P. indica]) and in those mutants (vpe-null, γvpe, and γvpe dad1) where we did not detect altered enzyme activities, we did not observe reduced cell viability. Together, these data confirm enhanced ER stress (TM)- and P. indica–induced cell death in mutants impaired in ER-QC. Moreover, our data identified γVPE as a key factor in the execution of ER stress–induced cell death triggered by TM or by fungal colonization in the mutualistic interaction of P. indica and Arabidopsis.
Figure 7.
VPEs Are Required for Cell Death Execution during Colonization by P. indica or after TM Treatment.
(A) and (B) FDA-based assay indicative of cell death in dad1, bip2, γvpe, vpe-null, and γvpe dad1 roots compared with the wild type (WT) at 1 (A) and 3 (B) d after TM or mock treatment.
(C) Relative cell viability in wild-type and mutant lines at 3 d after TM treatment. Relative fluorescence values (shown in [B]) of TM-treated roots were divided by the respective values of mock-treated roots.
(D) and (E) FDA-based assay indicative of cell death in dad1, bip2, γvpe, vpe-null, and γvpe dad1 roots compared with the wild type at 3 (D) and 7 (E) d after P. indica inoculation (DAI) or mock treatment.
(F) Relative cell viability in wild-type and mutant lines at 7 DAI. Relative fluorescence values (shown in [E]) of inoculated roots were divided by the respective values of mock-treated roots.
Data displayed are means (±sd) of eight independent measurements per treatment of at least three biological experiment. Asterisks indicate significant differences in relative fluorescence values ([B] and [E]) or cell viability ([C] and [F]) in mutants compared with the wild type at 3 DAT (TM; [B] and [C]) or at 7 DAI (P. indica; [E] and [F]) at P < 0.05, P < 0.01 (**), and P < 0.001 (***) as analyzed by two-way ANOVA.
DISCUSSION
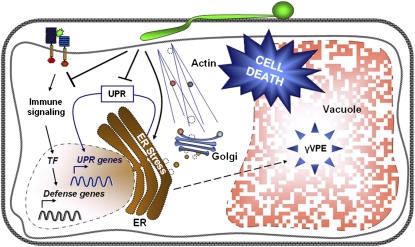
In this study, we examine the molecular basis of cell death–associated root colonization of Arabidopsis by the mutualistic fungus P. indica. Our study suggests that P. indica suppresses ER stress signaling as initial step, which eventually results in a vacuole-mediated cell death that is dependent on VPE/caspase 1–like activities (Figure 8). Cell death–dependent colonization and mutualism is counterintuitive, but, notably, P. indica only colonizes parts of the root maturation zone and does not enter the root vasculature (Deshmukh et al., 2006; Jacobs et al., 2011). Therefore, P. indica colonization does not impair root function and development. It also is not known at which interaction stage P. indica deploys its beneficial potential to plants. Our cytological data suggest that, after cell death–associated colonization, P. indica continues its biphasic lifestyle in neighboring cells. Fungal sporulation can be observed from 7 DAI onwards (Jacobs et al., 2011).
Figure 8.
Summary of Molecular Events Associated with Cell Death–Dependent Colonization of Arabidopsis Roots by P. indica.
We formerly demonstrated suppression of immune signaling by P. indica downstream of its recognition by the root surveillance system (Jacobs et al., 2011). Based on our data, P. indica induces ER stress but simultaneously suppresses the UPR in colonized cells. This fungal strategy results in ER swelling and the subsequent activation of a γVPE/caspase 1–like-mediated cell death program, which is preceded by vacuolar collapse. Impaired ER integrity is thought to disturb vesicular protein secretion. TF, transcription factors (e.g., WRKYs).
[See online article for color version of this figure.]
ER stress occurrence during mutualistic colonization is evident from cytological and pharmacological analyses, as colonized cells showed ER disintegration (Figures 1A to 1D), and P. indica–colonized plants were hypersensitive to the ER stress inducer TM (Figure 2A). Importantly, fungal colonization did not result in the induction of ER stress signaling, known as the UPR. The UPR usually encompasses translational attenuation, induction of ER chaperones (e.g., BIPs), and elevated degradation of misfolded proteins. By these means, eukaryotic cells aim to relieve ER stress that occurs under abiotic and biotic stress conditions as well as at certain developmental stages (Malhotra and Kaufman, 2007; Vitale and Boston, 2008). Microarray studies with Arabidopsis plants exposed to TM revealed the induction of UPR genes involved in protein processing within the ER, protein degradation and protein trafficking (Kamauchi et al., 2005; Urade, 2007; Iwata et al., 2008). In our study, a randomly selected subset of these genes was induced by TM in roots (Figure 3A). By clear contrast, P. indica suppressed TM-induced ER stress already during biotrophic colonization (3 DAI), as indicated by the reduced expression of all tested TM-responsive UPR components (Figure 3A) and the decreased BIP protein accumulation (Figure 3B). bZIP17, bZIP28, and bZIP60 transcription factors participate in ER stress signaling (Liu et al., 2007a, 2007b, 2008; Iwata et al., 2008; Liu and Howell, 2010a). Although bZIP17 and bZIP60 were induced by TM in roots (Figure 3A), none of these genes were induced during P. indica colonization (Figure 2B). This suggests that the fungus apparently inhibits the initiation of ER stress signaling. Consistent with this, TM treatment could not induce the expression of bZIP60- and bZIP28-regulated UPR genes (Iwata et al., 2008; Liu and Howell, 2010b) in P. indica–colonized roots (Figure 3A).
Most probably, fungal suppression of the UPR (Figures 3A and 3B) is required for cell death initiation. Failed ER stress adaptation or severe ER stress can result in the activation of cell death (Szegezdi et al., 2006). In mammals, the principles of ER stress–induced proapoptotic signaling have been intensively studied (Szegezdi et al., 2006; Rasheva and Domingos, 2009). The same plasma membrane–localized ER stress sensors that induce the UPR also initiate apoptotic signaling under severe ER stress by activating the bZIP transcription factor ATF4, the c-Jun N-terminal kinase pathway, and a caspase cascade. Central to this proapoptotic state is the activation of Bcl2-ASSOCIATED X PROTEIN (BAX) and B-CELL LYMPHOMA 2 INTERACTING MEDIATOR OF CELL DEATH (BIM), which contribute to the execution of apoptosis by enhancing Ca2+ release from ER and mitochondria. In addition, BAX and BIM mediate cytochrome c release from mitochondria, thereby activating the apoptosome (Szegezdi et al., 2006). Several studies suggest a conservation of ER stress signaling in plants and mammals. BI-1 is a negative regulator of cell death in mammals that antagonizes BAX-induced lethality (Xu and Reed, 1998). Although BAX homologs are not present in plants, barley BI-1 and other plant BI-1 proteins suppress BAX-induced cell death in planta (Hückelhoven, 2004; Eichmann et al., 2006; Watanabe and Lam, 2009). BI-1 is thought to control Ca2+ release from the ER under stress conditions in plants and mammals (Chae et al., 2004; Watanabe and Lam, 2009). Arabidopsis plants overexpressing BI-1 exhibit enhanced TM tolerance, which indicates its negative regulatory function in ER-PCD (Watanabe and Lam, 2008). In barley, P. indica suppresses BI-1 transcription and BI-1 overexpression resulted in reduced P. indica colonization (Deshmukh et al., 2006). Consistent with this, we observed a suppression of BI-1 by P. indica after TM application (Figure 3A). However, 3-week-old Arabidopsis mutants lacking BI-1 (atbi1-2) (Watanabe and Lam, 2006) did not show an altered P. indica colonization at 3 and 7 DAI (data not shown) using the qRT-PCR–based fungal colonization assay (see Methods).
Similar to mammalian ER-PCD (Szegezdi et al., 2006), we present several lines of evidence that ER stress is an initiator of PCD in Arabidopsis roots and that this PCD is dependent on VPEs. Application of TM to Col-0 resulted in ER swelling followed by vacuolar collapse and lysis of the cytoplasm (Figure 4B). Whereas TM triggered ER swelling in vpe-null mutants, vacuolar collapse and lysis of the cytoplasm did not occur (Figure 4D). Consistent with this, TM treatment did not induce VPE and caspase 1–like activities in vpe-null and in γvpe at 3 DAT. This coincided with the inability of TM to induce cell death in both mutants (Figures 6F, 7B, and 7C). By contrast, TM hypersensitivity of dad1 was associated with an increased cell death occurrence (Figures 4C, 7B, and 7C). Caspase 1–like activity might be most important to transduce cell death after TM treatment. Despite the lack of VPE activity, cell death occurrence was similar in bip2 compared with dad1 (Figures 6F, 7B, and 7C). Notably, the γvpe dad1 double mutant behaved almost identically to γvpe in terms of enzyme activities and cell viability at 3 DAT (Figures 6F, 7B, and 7C). Therefore, TM-induced ER-PCD most probably strongly relies on the activity of γVPE for the execution of cell death even in those cases where ER-QC is impaired, as indicated by the double mutant γvpe dad1.
Interestingly, P. indica activated an ER-PCD that was similar to TM-induced ER-PCD. P. indica–induced ER-PCD was associated with enhanced VPE/caspase 1–like activities and reduced cell viability at 7 DAI (Figures 6A, 6B, and 7E). Enzyme activities and cell death occurrence were unaltered in vpe-null, γvpe, and γvpe dad1 at 7 DAI (Figures 6D, 7E, and 7F), and this was linked to a reduced colonization of these mutants (Figure 5). Moreover, P. indica colonization resulted in enhanced VPE and caspase 1–like activities and cell death occurrence in roots of dad1 and bip2 mutants at 7 DAI, and this was associated with improved colonization (Figures 1E). This means that the fungus depends on this ER-PCD for successful colonization of Arabidopsis roots. Apparently, γVPE mediates most of the P. indica–induced VPE and caspase 1–like activities (Figures 6D, 7E, and 7F), while participation of αVPE and βVPE in ER-PCD remains elusive. Based on the colonization experiments (Figure 5), αVPE might function in the same signaling pathway as γVPE. It is tempting to speculate that βvpe might antagonize cell death signaling since it showed a tendency to higher colonization by P. indica. In contrast with previous studies (Kinoshita et al., 1995; Gruis et al., 2004), our PCR analyses indicate expression of all VPEs but δVPE in roots (see Supplemental Figure 3 online). The reduction in VPE and caspase 1–like activities in roots lacking γVPE further confirmed its presence in roots. Since we did not observe enhanced VPE and caspase 1–like activities or the occurrence of cell death in wild-type or vpe mutants at 3 DAI (biotrophic stage) (Figures 6C and 7D), the enhanced colonization of γvpe at 3 DAI (Figure 5) might indicate an additional immunity-related function of γVPE. Although γvpe dad1 and γvpe showed a similar degree of P. indica colonization at 7 DAI (Figure 5), γvpe dad1 displayed a colonization phenotype comparable to dad1 at 3 DAI. One might argue that ER stress induction may not be upstream of γVPE-mediated PCD. However, we monitored virtually identical VPE/caspase 1–like activities and cell viability in γvpe and γvpe dad1 at 7 DAI (P. indica) and at 3 DAT (TM) (Figures 6D, 6F, 7B, and 7E). Therefore, we speculate that, during biotrophic root colonization (3 DAI), dad1 might counteract a yet unknown process in γvpe that is unlinked to ER-PCD.
Impairment of ER integrity might not only serve to induce PCD. The ER plays a crucial role in plant innate immunity by processing antimicrobial proteins (Wang et al., 2005), whose delivery to the site of microbial attack relies on vesicle-mediated transport processes (Lipka et al., 2005; Hückelhoven, 2007). In addition, specific components of the ER machinery were identified to mediate processing of the PRR EFR (Nekrasov et al., 2009; Saijo et al., 2009), which activates innate immunity after recognition of the bacterial MAMP elongation factor TU (Zipfel et al., 2006). Recently, we found that P. indica suppresses early root immune signaling and that this is essential for biotrophic root colonization (Jacobs et al., 2011). In this study, we recorded a disturbance in ER homeostasis as early as 3 DAI (Figures 1A, 1B, 3A, and 3B). Although improved root colonization of sec61α, dad1, and bip2 mutants might reflect disturbance in immune signaling, several indications contradict this assumption: (1) The improved colonization of these mutants occurred only during cell death–dependent but not biotrophic colonization. By contrast, colonization of cerk1 mutants lacking a functional chitin receptor showed improved colonization at 3 DAI (Jacobs et al., 2011). (2) flg22- and chitin-triggered root oxidative burst and flg22-induced seedling growth inhibition, representing early and late MAMP responses of plants, are not impaired in these mutants (see Supplemental Figure 1 online). (3) All three mutants show an unaltered susceptibility to the biotrophic powdery mildew fungus E. cruciferarum and the necrotrophic fungus B. cinerea (see Supplemental Figure 2 online). Notably, these mutants are impaired in systemic immune signaling induced by the salicylic acid analog benzothiadiazole (Wang et al., 2005). This indicates an impairment of these mutants in a distinct immune pathway rather than a general defect in various immune pathways. (4) The γvpe dad1 double mutant showed a colonization phenotype similar to γvpe at 7 DAI (Figure 5). Therefore, the improved root colonization of dad1 (Figure 1E) rather indicates an altered ER stress–induced cell death threshold as supported by our enzyme and cell viability assays (Figures 6 and 7).
Our data support the existence of a previously unknown pathway in plants, in which ER stress induces a vacuolar cell death dependent on VPE/caspase 1–like activities, which occurs after TM treatment and during microbial colonization. It further indicates a sophisticated strategy for the mutualistic fungus P. indica to colonize Arabidopsis roots successfully. It will be interesting to see in future studies to what extent P. indica–triggered ER dysfunction in roots impairs generation and secretion of MTI components (e.g., antimicrobial proteins and PRRs). Furthermore, it is tempting to speculate that impaired ER integrity also affects the function of the ER in vacuole loading with antimicrobial proteins. This would explain why VPE-mediated vacuolar collapse supports rather than stops fungal growth as was reported for bacterial pathogens (Hatsugai et al., 2009). We speculate that the impairment of ER function is a more common strategy of microbes to colonize (and kill) eukaryotic host cells.
METHODS
Plant, Fungal Material, Plant Inoculation, and TM Treatment
bip2, dad1, and sec61α mutants were provided by X. Dong (Wang et al., 2005); αvpe, βvpe, γvpe, δvpe, and vpe-null were provided by I. Hara-Nishimura (Kuroyanagi et al., 2005); the cerk1-2 mutant was provided by V. Lipka (Petutschnig et al., 2010); the fls2c mutant was provided by C. Zipfel (Zipfel et al., 2004); the atbi1-2 mutant was provided by E. Lam (Watanabe and Lam, 2006); and GFP-tmKKXX was provided by D.A. Jones (Benghezal et al., 2000). γvpe dad1 mutant was generated within this study by crossing dad1 and γvpe. The isolate of Piriformospora indica DSM11827 was obtained from the German collection of microorganisms and cell cultures in Braunschweig, Germany. For inoculation, all Arabidopsis thaliana seeds were sterilized in 3% sodium hypochlorite and grown in squared Petri dishes on half-strength Murashige and Skoog (MS) under 8 h light (180 μmol m−2 s−1 photon flux density)/16 h night, 22°C/18°C, and 60% relative humidity. If not stated otherwise, 3-week-old plant roots were inoculated with P. indica chlamydospores at a concentration of 500,000 spores mL−1 or treated with TM (5 μg mL−1). One milliliter of spore suspension was used per squared Petri dish containing ~40 plants. For all gene expression and protein analyses, we removed the elongation and meristematic zones and only harvested the maturation zones of roots. For TM assays, control treatment was performed by applying DMSO. For Erysiphe cruciferarum inoculation (see Supplemental Figure 2A online), Arabidopsis plants were grown at 22°C and 65% relative humidity in a 10 h photoperiod with 120 μmol m−2 s−1 light in a 2:1 soil sand mixture (Frühstofer Erde, Type P; Quarzsand, granulation: 0.1 to 0.5 mm; Sakret Trockenbaustoffe Europa). E. cruciferarum was grown on Col-0, to maintain constant aggressiveness, and on susceptible phytoalexin deficient 4 (pad4) mutants (provided by the European Arabidopsis Stock Centre; Zhou et al., 1998), for strong conidia production. For Botrytis cinerea inoculation (see Supplemental Figure 2B online), Arabidopsis plants were placed under an inoculation box covered with a polyamide net (0.2 mm2). Conidia were brushed off from pad4 plants through the net until a density of three to four conidia mm−2 was reached. Inoculated plants were kept in a growth chamber under the above conditions. Leaves were photographed 11 d after inoculation. B. cinerea strain B05.10 was grown on HA agar (1% malt extract, 0.4% Glc, 0.4% yeast extract, and 1.5% agar, pH 5.5) as described previously (Doehlemann et al., 2006). Detached rosette leaves from 6- to 7-week-old soil grown Arabidopsis plants were placed in Petri dishes containing 1% agar and inoculated with 5 μL spore suspension (2 × 105 spores mL−1 in 12 g L−1 potato dextrose broth; Duchefa Biochemie). To maintain high humidity, Petri dishes were covered with a transparent lid. Digital photographs were taken at different time points and analyzed using ImageJ to measure lesion size (Abramoff et al., 2004).
Quantification of Fungal Colonization by qRT-PCR
Genomic DNA was isolated from 100 mg root material with the Plant DNeasy kit (Qiagen). In qRT-PCR analysis, 40 ng of genomic DNA served as template. The amplification reaction was performed using 10 μL of SYBR green JumpStart Taq ReadyMix (Sigma-Aldrich) and 350 nM of oligonucleotides specifically amplifying P. indica internal transcribed spacer (Pi ITS) and Arabidopsis ubiquitin 5 (At Ubi5) (see Supplemental Table 1 online). The standard quantification program from the 7500 fast thermal cycler (Applied Biosystems) was applied. To determine the relative amount of P. indica in plant roots, the 2−Δcycle threshold (CT) method (Schmittgen and Livak, 2008) was used. Therefore, the raw CT of P. indica ITS were subtracted from those obtained for At UBI5. Thereafter, the 2−ΔCT of wild types were divided by 2−ΔCT of mutants. In all cases, colonization of the respective mutant is given relative to wild-type colonization (set to one).
Gene Expression Analysis
For qRT-PCR–based gene expression studies, 3-week-old plants were inoculated with P. indica or mock treated and harvested at 1, 3, and 7 DAI. For TM treatment, inoculated or mock-treated plants were treated with TM (5 μg mL−1) or DMSO (control) at 3 DAI. Roots were harvested at 12, 24, 48, and 72 h after TM treatment. For RT-PCR–based studies, roots of 3- and 5-week-old plants were harvested (see Supplemental Figure 3 online). In all cases, RNA was extracted from homogenized root material using TRIzol (Invitrogen). For cDNA synthesis, 500 ng of RNA was DNase-I digested and transcribed into cDNA using a qScript cDNA synthesis kit (Quanta Biosciences). For qRT-PCR analysis, 10 ng of cDNA was used as template for determining the amplification of candidate genes. As described above the 2−ΔCT method was applied to evaluate the level of gene expression. For RT-PCR, 10 ng of cDNA served as template. Primers are used as listed in Supplemental Table 1 online.
Protein Extraction and Immunodetection of BIP
Roots were inoculated with P. indica or mock treated and harvested at 1, 3, and 7 DAI. For TM assays, P. indica or mock-treated roots were treated with TM 5 μg mL−1 or DMSO (control) at 3 DAI and harvested at 2 DAT. Total protein was extracted with a buffer containing 250 mM Suc, 50 mM HEPES-KOH, 5% glycerin, 1 mM Na2MoO4 × 2H2O, 25 mM NaF, and 10 mM EDTA. Subsequently, 20 μg of each protein sample was separated by SDS-PAGE and transferred to Roti-polyvinylidene fluoride membrane (Roth). The proteins were probed with an Arabidopsis anti-BIP antibody (Santa Cruz Biotechnology), followed by an anti-rabbit IgG-alkaline phosphate antibody (Sigma-Aldrich). The anti-BIP antibody was raised against the conserved C-terminal region of BIP and most probably detects BIP and BIP2. The gels were stained in a solution containing 20% (v/v) Coomassie Brilliant Blue R 250 (Roth) and 20% methanol and were later destained with a solution of 40% methanol/10% glacial acid/50% water (v/v/v). Protein bands were quantified using ImageJ.
Cell Death Assay
Root segments (1.5 cm) of the maturation zone from 2-week-old plants inoculated with P. indica or treated with TM (control DMSO) were transferred to half-strength MS containing FDA. After 10 min of incubation, root segments were washed five times and the fluorescence intensities were measured at 535 nm after excitation at 485 nm using a fluorescence microplate reader (TECAN infinite 200). See Supplemental Figure 4 online for further details on the linearity in the quantification of cell death using FDA.
Caspase and VPE Activity Assays
Three-week-old roots were inoculated with P. indica or mock treated, and the roots were harvested at 3 and 7 DAI. For TM assays, roots were treated with TM 5 μg mL−1 or DMSO and harvested at 1 and 3 DAT. A buffer containing 100 mM sodium acetate, pH 5.5, 100 mM NaCl, 1 mM EDTA, and 1mM phenylmethylsulfonyl fluoride was applied to get root extracts. To measure caspase 1–like and VPE activity, 1 mM fluorogenic caspase 1 substrate (Ac-YVAD-MCA) and VPE substrate (Ac-ESEN-MCA) (Peptide Institute) were added to the root extracts. Fluorescence intensities were measured at 465 nm after excitation at 360 nm using a fluorescence microplate reader (TECAN infinite 200).
Cytological Analyses
Root samples were either fixed or directly stained with chitin-specific WGA-AF488 (Molecular Probes) as described (Deshmukh et al., 2006). Confocal images were recorded on a TCS SP2 microscope (Leica). WGA-AF488 and GFP were excited with a 488-nm laser line and detected at 505 to 540 nm. For ultrastructural studies, roots were embedded as described (Zechmann et al., 2007), and ultrathin sections (80 nm) were investigated after staining with uranyl acetate and lead citrate with a Philips CM10 transmission electron microscope.
Growth Retardation Assays
For the growth retardation assay with TM, Arabidopsis seedlings were grown on half-strength MS medium containing 1% Suc for 14 d. Plants were inoculated with P. indica or mock treated and transferred to liquid half-strength MS medium containing 1% Suc and TM (25 ng mL−1) or DMSO (control). Seedling fresh weight was determined 7 d after TM/DMSO treatment. For the growth retardation assay with flg22, plants were grown on half-strength MS medium containing 1% Suc for 14 d and thereafter transferred to liquid half-strength MS medium containing 1% Suc and 10 μM flg22. Plant fresh weight was determined 10 d after treatment. flg22 peptide sequence was used as described (Gómez-Gómez et al., 1999).
MAMP-Induced Root Oxidative Burst
Two-week-old plant roots were treated with either 1 μM flg22 or 1 μM N-acetylchitooctaose (see Supplemental Figure 1 online). For determination of the oxidative burst, roots were cut in 1-cm-long pieces (10 mg per assay) and subjected to a luminol-based assay as described (Gómez-Gómez et al., 1999).
Statistical Analyses
Results are expressed as means ± sd or ± se as indicated in the figure legends and represent at least three similar experiments. Data analysis was performed by R software (www.R-project.org) applying variance analysis using the lm procedure. Data were analyzed as a randomized block design with experiment as blocking factor and the factors treatment and type and their interactions. Comparisons of means were performed by linear contrasts using the function estimable from the package gmodels. P ≤ 0.05 was considered significant.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis: UBI5 (AT3G62250 and AEE80327.1), AAA-TYPE ATPase (AT5G40010 and AED94501.1), ADP-RF (AT1G70490 and AAG40377.1), BI-1 (AT5G47120 and AED95473.1), BIP1 (AT5G28540 and AED93812.1), BIP2 (AT5G42020 and AAP37765.1), BIP3 (AT1G09080 and AEE28393.1), bZIP17 (AT2G40950 and AEC09905.1), bZIP28 (AT3G10800 and AEE74956.1), bZIP60 (AT1G42990 and AEE31935.1), CERK1 (AT3G21630 and AEE76532.1), CNX2 (AT5G07340 and AAQ56828.1), DERLIN-LIKE1 (AT4G21810 and AEE84506.1), DAD1 (AT1G32210 and ABD38892.1), ERdJ3A (AT3G08970 and AEE74703.1), FLS2 (AT5G46330 and AED95370.1), GRP94 (AT4G24190 and AEE84862.1), PUTATIVE GALACTINOL SYNTHASE (AT2G47180 and AEC10811.1), SEC61α (AT2G34250 and BAH19870.1), SEC61γ (AT5G50460 and AED95948.1), SAR1B (AT1G09180 and AEE28409.1), sPDI (AT1G77510 and AEE35987.1), UDP-TRANSPORTER (AT2G02810 and AAW28558.1), αVPE (AT2G25940 and AEC07775.1), βVPE (AT1G62710 and AEE33996.1) γVPE (AT4G32940 and AEE86150.1), and δVPE (AT3G20210 and AEE76348.1); P. indica: ITS (AF019636).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MAMP-Induced Responses in Seedlings of sec61α, dad1, and bip2 Mutants.
Supplemental Figure 2. bip2, dad1, and sec61α Showed an Unaltered Susceptibility to Biotrophic E. cruciferarum and Necrotrophic B. cinerea.
Supplemental Figure 3. Expression of VPE Genes in Roots of Arabidopsis Seedlings.
Supplemental Figure 4. Correlation between Esterase-Mediated Cleavage of FDA in Vivo and the Length of Analyzed Root Segments.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank the European Arabidopsis Stock Centre, Xinnian Dong, David A. Jones, Ikuko Hara-Nishimura, Eric Lam, Volker Lipka, and Cyril Zipfel for providing seeds. We also thank Ralph Hückelhoven for critical reading of the manuscript, Rebekka Fensch for technical assistance, Jörn Pons-Kühnemann for the statistical analyses, and Ruth Eichmann for experiments with E. cruciferarum and critical comments on the manuscript. The work was funded by Grant DFG-SPP1212 to K.-H.K. and P.S. and by the Austrian Science Fund (FWF P20619-B16) to B.Z.
AUTHOR CONTRIBUTIONS
P.S. designed the research. X.Q., B.Z., and M.U.R. performed research. X.Q., B.Z., and P.S. analyzed data. X.Q., K.-H.K., and P.S. wrote the article with help of all coauthors.
References
- Abramoff M.D., Magalhaes P.J., Ram S.J. (2004). Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Anelli T., Sitia R. (2008). Protein quality control in the early secretory pathway. EMBO J. 27: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M., Wasteneys G.O., Jones D.A. (2000). The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. Plant Cell 12: 1179–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae H.J., et al. (2004). BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol. Cell 15: 355–366 [DOI] [PubMed] [Google Scholar]
- Deshmukh S., Hückelhoven R., Schäfer P., Imani J., Sharma M., Weiss M., Waller F., Kogel K.H. (2006). The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc. Natl. Acad. Sci. USA 103: 18450–18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Berndt P., Hahn M. (2006). Different signalling pathways involving a Galpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol. Microbiol. 59: 821–835 [DOI] [PubMed] [Google Scholar]
- Duan Y., Zhang W., Li B., Wang Y., Li K., Sodmergen , Han C., Zhang Y., Li X. (2010). An endoplasmic reticulum response pathway mediates programmed cell death of root tip induced by water stress in Arabidopsis. New Phytol. 186: 681–695 [DOI] [PubMed] [Google Scholar]
- Eichmann R., Dechert C., Kogel K.H., Hückelhoven R. (2006). Transient over-expression of barley BAX Inhibitor-1 weakens oxidative defence and MLA12-mediated resistance to Blumeria graminis f.sp. hordei. Mol. Plant Pathol. 7: 543–552 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Felix G., Boller T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Gruis D., Schulze J., Jung R. (2004). Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 16: 270–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N., Iwasaki S., Tamura K., Kondo M., Fuji K., Ogasawara K., Nishimura M., Hara-Nishimura I. (2009). A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 23: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N., Kuroyanagi M., Nishimura M., Hara-Nishimura I. (2006). A cellular suicide strategy of plants: Vacuole-mediated cell death. Apoptosis 11: 905–911 [DOI] [PubMed] [Google Scholar]
- Hatsugai N., Kuroyanagi M., Yamada K., Meshi T., Tsuda S., Kondo M., Nishimura M., Hara-Nishimura I. (2004). A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R. (2004). BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis 9: 299–307 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R. (2007). Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45: 101–127 [DOI] [PubMed] [Google Scholar]
- Iwata Y., Fedoroff N.V., Koizumi N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S., Zechmann B., Molitor A., Trujillo M., Petutschnig E., Lipka V., Kogel K.H., Schäfer P. (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 156: 726–740 Erratum. Plant Physiol. 157: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelitto-Van Dooren E.P., Vidal S., Denecke J. (1999). Anticipating endoplasmic reticulum stress. A novel early response before pathogenesis-related gene induction. Plant Cell 11: 1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Yan Z., Nam K.H., Li J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamauchi S., Nakatani H., Nakano C., Urade R. (2005). Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 272: 3461–3476 [DOI] [PubMed] [Google Scholar]
- Kelleher D.J., Gilmore R. (2006). An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16: 47R–62R [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Nishimura M., Hara-Nishimura I. (1995). The sequence and expression of the gamma-VPE gene, one member of a family of three genes for vacuolar processing enzymes in Arabidopsis thaliana. Plant Cell Physiol. 36: 1555–1562 [PubMed] [Google Scholar]
- Kuroyanagi M., Yamada K., Hatsugai N., Kondo M., Nishimura M., Hara-Nishimura I. (2005). Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J. Biol. Chem. 280: 32914–32920 [DOI] [PubMed] [Google Scholar]
- Kwon C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Lipka V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Lipka V., Kwon C., Panstruga R. (2007). SNARE-ware: The role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 23: 147–174 [DOI] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010a). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010b). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. (2007a). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. (2007b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51: 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Howell S.H. (2008). Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 31: 1735–1743 [DOI] [PubMed] [Google Scholar]
- Malerba M., Cerana R., Crosti P. (2004). Comparison between the effects of fusicoccin, Tunicamycin, and Brefeldin A on programmed cell death of cultured sycamore (Acer pseudoplatanus L.) cells. Protoplasma 224: 61–70 [DOI] [PubMed] [Google Scholar]
- Malhotra J.D., Kaufman R.J. (2007). The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18: 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan M.S., Dinesh-Kumar S.P. (2010). All hands on deck—The role of chloroplasts, endoplasmic reticulum, and the nucleus in driving plant innate immunity. Mol. Plant Microbe Interact. 23: 1368–1380 [DOI] [PubMed] [Google Scholar]
- Pattison R.J., Amtmann A. (2009). N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 14: 92–99 [DOI] [PubMed] [Google Scholar]
- Petutschnig E.K., Jones A.M., Serazetdinova L., Lipka U., Lipka V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285: 28902–28911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang X., Weiss M., Kogel K.H., Schäfer P. (November 24, 2011). Piriformospora indica - A mutualistic basidiomycete with an exceptionally large plant host range. Mol. Plant Pathol. http://dx.doi.org/10.1111/J.1364-3703.2011.00764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheva V.I., Domingos P.M. (2009). Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis 14: 996–1007 [DOI] [PubMed] [Google Scholar]
- Rojo E., Martín R., Carter C., Zouhar J., Pan S., Plotnikova J., Jin H., Paneque M., Sánchez-Serrano J.J., Baker B., Ausubel F.M., Raikhel N.V. (2004). VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr. Biol. 14: 1897–1906 [DOI] [PubMed] [Google Scholar]
- Saijo Y., Tintor N., Lu X., Rauf P., Pajerowska-Mukhtar K., Häweker H., Dong X., Robatzek S., Schulze-Lefert P. (2009). Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 28: 3439–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer P., Pfiffi S., Voll L.M., Zajic D., Chandler P.M., Waller F., Scholz U., Pons-Kühnemann J., Sonnewald S., Sonnewald U., Kogel K.H. (2009). Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica. Plant J. 59: 461–474 [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schröder M. (2006). The unfolded protein response. Mol. Biotechnol. 34: 279–290 [DOI] [PubMed] [Google Scholar]
- Szegezdi E., Logue S.E., Gorman A.M., Samali A. (2006). Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7: 880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urade R. (2007). Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 274: 1152–1171 [DOI] [PubMed] [Google Scholar]
- Urade R. (2009). The endoplasmic reticulum stress signaling pathways in plants. Biofactors 35: 326–331 [DOI] [PubMed] [Google Scholar]
- van Doorn W.G., et al. (2011). Morphological classification of plant cell deaths. Cell Death Differ. 18: 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A., Savita Verma, Sudha , Sahay N., Butehorn B., Franken P. (1999). Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl. Environ. Microbiol. 65: 2741–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Varma A., Rexer K., Hassel A., Kost G., Sarbhoy A., Bisen P., Bütehorn B., Franken P. (1998). Piriformospora indica, gen. et sp. nov., a new root-colonizing fungus. Mycologia 90: 896–903 [Google Scholar]
- Vitale A., Boston R.S. (2008). Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic 9: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Waller F., Achatz B., Baltruschat H., Fodor J., Becker K., Fischer M., Heier T., Hückelhoven R., Neumann C., von Wettstein D., Franken P., Kogel K.H. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. USA 102: 13386–13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Weaver N.D., Kesarwani M., Dong X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2006). Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 45: 884–894 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2008). BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem. 283: 3200–3210 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2009). Bax inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int. J. Mol. Sci. 10: 3149–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Reed J.C. (1998). Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol. Cell 1: 337–346 [DOI] [PubMed] [Google Scholar]
- Zechmann B., Müller M., Zellnig G. (2007). Membrane associated qualitative differences in cell ultrastructure of chemically and high pressure cryofixed plant cells. J. Struct. Biol. 158: 370–377 [DOI] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Tsui F., Klessig D.F., Glazebrook J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
- Zuccaro A., et al. (2011). Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog. 7: e1002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.