Calcium- and calmodulin-dependent protein kinase (CCaMK) is a protein kinase that is crucial for plant-microbe symbioses. This work shows the importance of nuclear localization of CCaMK for its function in plant symbiotic responses. Moreover, activation of CCaMK in the nucleus induces cytological changes similar to those important for fungal infection, without presence of the symbiotic fungi.
Abstract
The common symbiosis pathway is at the core of symbiosis signaling between plants and soil microbes. In this pathway, calcium- and calmodulin-dependent protein kinase (CCaMK) plays a crucial role in integrating the signals both in arbuscular mycorrhizal symbiosis (AMS) and in root nodule symbiosis (RNS). However, the molecular mechanism by which CCaMK coordinates AMS and RNS is largely unknown. Here, we report that the gain-of-function (GOF) variants of CCaMK without the regulatory domains activate both AMS and RNS signaling pathways in the absence of symbiotic partners. This activation requires nuclear localization of CCaMK. Enforced nuclear localization of the GOF-CCaMK variants by fusion with a canonical nuclear localization signal enhances signaling activity of AMS and RNS. The GOF-CCaMK variant triggers formation of a structure similar to the prepenetration apparatus, which guides infection of arbuscular mycorrhizal fungi to host root cells. In addition, the GOF-CCaMK variants without the regulatory domains partly restore AMS but fail to support rhizobial infection in ccamk mutants. These data indicate that AMS, the more ancient type of symbiosis, can be mainly regulated by the kinase activity of CCaMK, whereas RNS, which evolved more recently, requires complex regulation performed by the regulatory domains of CCaMK.
INTRODUCTION
Legume plants benefit from two major mutualistic plant–microbe interactions: arbuscular mycorrhizal symbiosis (AMS) and root nodule symbiosis (RNS). AMS and RNS provide inorganic materials (mainly phosphate and nitrogen sources, respectively) to host plants. In return, the host plant supplies photosynthetic products to the symbionts and an anaerobic environment for RNS. These symbiotic nutrient supplies have contributed to survival and fitness of the host plants on land.
Arbuscular mycorrhizal (AM) fungi absorb inorganic materials and water from the rhizosphere through extraradical hyphae, which attach to and penetrate into the host root (Harrison, 2005). The intraradical hyphae elongate longitudinally in the apoplastic spaces between cortical cells, in which a tree-like symbiotic structure called the arbuscule is surrounded by the plant membrane (Bonfante and Perotto, 1995; Gianinazzi-Pearson, 1996). In the arbuscule, nutrients are exchanged across the fungal and plant membranes by transporters (Harrison and van Buuren, 1995; Bago et al., 2003).
In RNS, the root nodule is a symbiotic secondary organ that develops on the host roots for accommodation of the symbiont. Symbiotic partner rhizobia colonize root hairs and secrete rhizobial symbiosis–signaling molecules called Nod factors (Lerouge et al., 1990; Dénarié and Cullimore, 1993). Cortical cell division is stimulated by recognition of Nod factors, resulting in nodule formation. The host plant takes rhizobia into the nodule cells through an infection thread, a structure that allows bacterial entry into the host roots (Gage, 2004). In the nodules, rhizobia produce nitrogenase to fix (convert) atmospheric nitrogen into ammonia, which provides a usable nitrogen source for the host plants (Udvardi and Day, 1997). The host plant produces the oxygen binding protein leghemoglobin in the infected cells and maintains the anaerobic environment necessary for nitrogenase function.
AMS and RNS share several similarities despite the differences between fungal and bacterial symbionts. These similarities have presumably originated from a common evolutionary origin. RNS, which is the more recent type of symbiosis, has retained some of the same symbiosis factors and mechanisms as the more ancient AMS type of symbiosis (Parniske, 2008; Markmann and Parniske, 2009). Indeed, previous studies have identified symbiosis mutants in legume plants that abolish both AMS and RNS. Several such symbiosis (Sym) genes have been identified through studies of these mutants (Endre et al., 2002; Stracke et al., 2002; Ané et al., 2004; Lévy et al., 2004; Imaizumi-Anraku et al., 2005; Kanamori et al., 2006; Tirichine et al., 2006; Saito et al., 2007; Yano et al., 2008; Groth et al., 2010; Murray et al., 2011). A signaling pathway composed of these symbiosis genes is termed the “common Sym pathway” (Parniske, 2008).
One of the common Sym proteins, calcium- and calmodulin-dependent protein kinase (CCaMK), has an essential role in transduction of both AMS and RNS signals (Lévy et al., 2004; Gleason et al., 2006; Tirichine et al., 2006). CCaMK is composed of a kinase domain, a calmodulin binding domain (CaM BD), and the EF-hand domain (Patil et al., 1995; Takezawa et al., 1996). The regulatory domains, including the CaM BD and the EF hands, are believed to function as sensors for changes in calcium ion (Ca2+) concentration generated by upstream symbiosis signaling. According to the model, the binding of Ca2+ to CCaMK causes a conformational change of the protein and deregulates the kinase activity, which in turn activates a downstream signaling pathway by phosphorylation of substrates (Sathyanarayanan et al., 2000).
Previously, a gain-of-function (GOF) type of CCaMK mutant, spontaneous nodule formation1 (snf1), was isolated in Lotus japonicus (Tirichine et al., 2006). The kinase domain of the mutated CCaMK contains a Thr-to-Ile amino acid substitution at residue 265 (T265I), which is believed to mimic autophosphorylation for deregulation/activation of the CCaMK kinase domain. In snf1 mutants, deregulation of CCaMK appears to activate the RNS signaling pathway, which results in formation of a nodule-like structure called the spontaneous nodule (SPN) in the absence of rhizobia. In addition, another type of GOF-CCaMK has been identified in Medicago truncatula by truncation of the regulatory domains, resulting in similar phenotypes (Gleason et al., 2006). This indicates that the common Sym pathway is sufficient for activation of cortical cell division and consequent nodule formation in RNS (Hayashi et al., 2010; Madsen et al., 2010). However, the functions of GOF-CCaMK in AMS are largely unknown. Since AMS does not trigger obvious structural changes, such as nodule formation, induction of AMS-related genes can be used as a proxy to examine the functions of GOF-CCaMK in AMS.
In L. japonicus, expression of the subtilisin-like Ser protease (subtilase) gene SbtM1 and the phosphate transporter gene PT4 is induced by AM fungal infection (Takeda et al., 2009, 2011). Expression of SbtM1 is exclusively induced in AMS not in RNS, which is under the regulation of the common Sym pathway. RNA interference silencing of SbtM1 reduces hyphal elongation and arbuscule formation in the host roots (Takeda et al., 2009). Lj PT4 is an apparent ortholog of the Mt PT4 gene isolated from M. truncatula (Liu et al., 1998; Harrison et al., 2002; Guether et al., 2009). Mt PT4 is required for AMS, essential for maintenance of arbuscules in the later stages of AMS (Javot et al., 2007). In addition to the changes in gene expression, development of the prepenetration apparatus (PPA), an intracellular preinfection structure produced during AM fungal colonization, has been reported in carrot (Daucus carota) and M. truncatula roots (Genre et al., 2005, 2008). During PPA formation, the epidermal and cortical cells that are proximate to the fungal hyphae show dense endoplasmic reticulum (ER) and migration of the nuclei. Formation of the PPA is likely to facilitate fungal penetration into the host cells by reorganization of cytoplasmic structure. Since the PPA is abolished in the common sym mutants, formation of the PPA requires a trigger through the common Sym pathway (Genre et al., 2005). Despite the importance of CCaMK in AMS, the functions of GOF-CCaMK in induction of AMS-specific genes or formation of the PPA are unclear.
Similar to expression of SbtM1 in AMS, expression of NIN (for Nodule inception) is specifically induced in RNS (Radutoiu et al., 2003). It is expressed in epidermal cells early in the infection stage of RNS and subsequently in dividing cortical cells and mature nodules in the later stages of nodule development (Schauser et al., 1999; Grønlund et al., 2005). In snf1 mutants, NIN expression is detected merely in dividing cortical cells and in SPNs (Tirichine et al., 2006), whereas early spontaneous NIN expression in snf1 mutants is undetectable at epidermal cells. Expression of Early Nodulin11 (ENOD11) in M. truncatula is induced both by RNS and by AMS (Journet et al., 2001). Similar to snf1 in L. japonicus, GOF-CCaMK in M. truncatula constitutively induces ENOD11 expression in the absence of rhizobia (Gleason et al., 2006). In L. japonicus, expression of subtilase genes SbtM4 and SbtS is induced 2 to 4 d after inoculation of either rhizobia or AM fungi (Takeda et al., 2009). Whereas SbtM4 expression is maintained throughout nodule development, SbtS expression is induced only in the early stage of nodule development and is hardly detected in nodules. In snf1 mutants, SbtM4 expression is observed only in the late stage of SPN formation, whereas SbtS is not detected (Takeda et al., 2011). These facts indicate that RNS responses are not fully activated in snf1. This led us to hypothesize that another form of GOF-CCaMK (i.e., truncation of the regulatory domains) may have a different spectrum in symbiosis signaling.
In this study, we examined the functions of GOF-CCaMK both in AMS and in RNS. We constructed a GOF-CCaMK by truncation of the regulatory domains and examined whether downstream gene expression was affected by its nuclear localization. Fusion of GOF-CCaMK with a nuclear localization signal (NLS) enhanced symbiosis gene expression in the absence of the symbionts. Furthermore, a cytoplasmic structure similar to the PPA was induced by nuclear-localized GOF-CCaMK.
RESULTS
Nuclear Localization of Deregulated CCaMK Strongly Induced Both RNS- and AMS-Dependent Genes
To analyze the role of CCaMK in AMS and RNS, we constructed a series of vectors containing the GOF-CCaMK variants expressed under the control of the cauliflower mosaic virus 35S promoter (see Supplemental Figure 1 online). In previous studies, CCaMKTD, which has the amino acid substitution Thr265Asp (T265D) in wild-type CCaMK (CCaMKTT), was shown to trigger SPN formation in L. japonicus (Banba et al., 2008; Hayashi et al., 2010), but regulation of symbiosis gene expression downstream of CCaMKTD was not examined. In addition, the functions of truncated CCaMK lacking the regulatory domains were not tested in L. japonicus. Therefore, we constructed CCaMK314, a C-terminally truncated CCaMK containing amino acids 1 to 314 that corresponded to the DMI3 (CCaMK of M. truncatula) kinase domain used by Gleason et al. (2006). We also tested the T265D substituted form of CCaMK314, designated CCaMK314TD. Analysis in L. japonicus revealed that green fluorescent protein (GFP)–fused CCaMK is localized in the nucleus (Yano et al., 2008), although there is no canonical NLS in CCaMK. However, GFP-fused CCaMK314TD was found both in the cytosol and in the nucleus of leaf epidermal cells of Nicotiana benthamiana (see Supplemental Figures 2A to 2C online), indicating that these proteins had lost the ability to specifically localize to the nucleus. To address whether nuclear activity of CCaMK is essential for symbiosis signaling, we fused an NLS to CCaMK314TD. The GFP-fused CCaMK314TD-NLS protein was predominantly retained in the nucleus in N. benthamiana leaves (see Supplemental Figures 2D to 2F online). We also constructed a CCaMK314TD fusion with a nuclear export signal (NES), and localization of GFP-CCaMK314TD-NES was analyzed in N. benthamiana leaf epidermal cells. Although weak fluorescence of GFP-CCaMK314TD-NES could be seen in the nucleus (see Supplemental Figure 2G online), a significant reduction in nuclear localization of the fusion protein was detected (see Supplemental Figures 2G to 2I online), compared with GFP-CCaMK314TD (see Supplemental Figure 2A online) or GFP-CCaMK314TD-NLS (see Supplemental Figure 2D online).
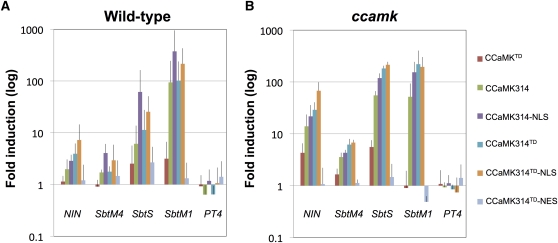
We introduced full-length CCaMKTD and the kinase-only GOF-CCaMK variants (CCaMK314, CCaMK314-NLS, CCaMK314TD, CCaMK314TD-NLS, and CCaMK314TD-NES) into L. japonicus by hairy root transformation and analyzed expression of symbiosis genes. Among the series of GOF-CCaMK transformants, hairy roots carrying CCaMK314, CCaMK314-NLS, CCaMK314TD, and CCaMK314TD-NLS showed induction of RNS-induced genes NIN, SbtM4, and SbtS at 2 weeks after generation of hairy roots (Figure 1A). As we expected, the enforced nuclear localization and the T265D substitution of CCaMK314 enhanced its effect on the induction of the RNS-dependent marker genes (NIN, SbtM4, and SbtS). On the other hand, the nuclear-exported variant (CCaMK314TD-NES) triggered only weak induction of those genes (Figure 1A). These results indicate that nuclear localization of GOF-CCaMK is important for its ability to induce expression of RNS-induced genes. Expression of RNS-induced genes was not statistically significant in hairy roots carrying the full-length CCaMKTD variant (Figure 1A). In snf1 mutants, NIN expression is not observed before cortical cell division (Tirichine et al., 2006). The lack of induction of NIN by full-length CCaMKTD agrees with the results previously obtained in snf1 mutants (Tirichine et al., 2006).
Figure 1.
Symbiosis-Induced Gene Expression in Wild-Type and ccamk Mutant Roots Induced by Transformation with GOF-CCaMK Variants.
Induction of NIN, SbtM4, SbtS, SbtM1, and PT4 in 2-week-old hairy roots carrying each of the GOF-CCaMK variants was analyzed. Fold induction levels in wild-type Gifu (A) and in ccamk (B) were calculated compared with that of transgenic hairy roots carrying a control vector (p35S:GFP) (expression level = 1). Total RNA was extracted from more than 10 roots per experiment. Error bars indicate sd (n = 3 to 6 independent experiments).
CCaMK is known to be a key factor both in AMS and in RNS because deficiency of a CCaMK function abolishes both types of symbioses (Catoira et al., 2000; Senoo et al., 2000). Therefore, we also analyzed expression of two AMS-specific genes in L. japonicus, SbtM1 and PT4. The GOF-CCaMK variants were introduced into wild-type plants to examine AMS-specific gene expression (Figure 1A). The expression analysis revealed that all of the kinase-only GOF-CCaMK variants except for the NES fusion could induce expression of SbtM1 but not that of PT4, whereas full-length CCaMKTD and snf1 could not induce expression of either gene (Figure 1A; Takeda et al., 2011). Because SbtM1 is involved in an early stage of AM fungal colonization (Takeda et al., 2009), these results showed that kinase-only GOF-CCaMK could activate the early signaling pathway in AMS. Failure of induction by full-length CCaMKTD indicates that the regulatory domains play an inhibitory role in the activation of the AMS signaling pathway upstream of the AMS-induced gene SbtM1.
Histochemical Examination of the RNS and AMS Genes Induced by GOF-CCaMK
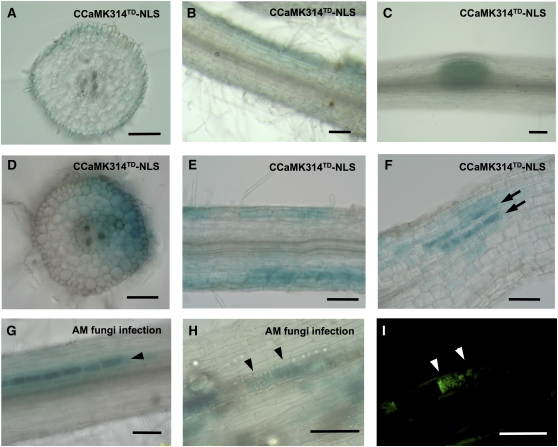
Since some symbiosis genes, such as NIN, express in epidermis as well as in cortex in RNS, a similar regulation mechanism is supposed in AMS. Therefore, we wondered whether our GOF-CCaMK could mimic tissue expression pattern of symbiosis genes. The tissue-specific activation of the RNS and AMS signaling pathways was examined histochemically using transgenic plants carrying β-glucuronidase (GUS) genes driven by the NIN and SbtM1 promoters, respectively. In snf1 mutants, NIN promoter–GUS (NINpro:GUS) expression is limited to dividing cortical cells and SPNs, and no GUS activity is detected in epidermal cells (Tirichine et al., 2006). The series of GOF-CCaMK variants was introduced into the NINpro:GUS plants to examine the activation of the RNS pathway. At 1 week after transfer of the plants to sterilized soil, GUS activity was not detected in hairy roots carrying full-length CCaMKTD (0/22; GUS-positive roots/all transgenic roots examined), in agreement with the results in snf1 mutants. However, hairy roots carrying any of the kinase-only GOF-CCaMK variants (CCaMK314, 12/29; CCaMK314-NLS, 16/20; CCaMK314TD, 24/36; CCaMK314TD-NLS, 37/47; GUS-positive roots/all transgenic roots examined) except for the NES fusion (CCaMK314TD-NES, 0/9; GUS-positive roots/all transgenic roots examined) showed GUS activity. The GUS activity was initially detected in the root epidermal layer (Figures 2A and 2B) and shifted later to dividing cortical cells (Figure 2C). The spatial expression pattern was similar to NIN expression induced upon rhizobial infection (Grønlund et al., 2005). The fact that no induction of GUS activity can be observed in NINpro:GUS roots carrying CCaMK314TD-NES is consistent with the results of the expression analysis by real-time PCR (Figure 1A). The histochemical expression analysis demonstrates that the nuclear-localized kinase-only GOF-CCaMK variants, but not full-length CCaMKTD, activate the RNS signaling pathway in epidermal cells at the time points examined.
Figure 2.
Histochemical Detection of NIN and SbtM1 Induction by GOF-CCaMK.
(A) to (F) CCaMK314TD-NLS was introduced into transgenic plants carrying NINpro:GUS ([A] to [C]) or SbtM1pro:GUS fusions ([D] to [F]) by hairy root transformation. Activity of GUS was detected 1 week ([D] and [E]) or 2 weeks after transferring 10-d-old hairy roots to sterilized soil ([A] to [C] and [F]).
(A) and (B) Cross section (A) and longitudinal section (B) (100-μm thick) of the hairy root carrying CCaMK314TD-NLS showed the NINpro:GUS activity in epidermal cells.
(C) The activity was also detected in dividing cortical cells which would develop into a SPN (whole mount).
(D) to (F) Cross section (D) and longitudinal sections ([E] and [F]) (100-μm thick) of the hairy root containing CCaMK314TD-NLS showed the SbtM1pro:GUS activity in epidermal and cortical cells. Arrows indicate cortical cells with strong GUS activity (F).
(G) to (I) SbtM1 promoter GUS plants without GOF-CCaMK were infected with G. intraradices for 2 weeks and examined for the GUS activity (whole mount). The AM infected roots were stained for the GUS activity ([G] and [H]), and the AM fungi were visualized with WGA Alexa Fluor 488 (I). Arrowheads indicate cortical cells with strong GUS activity ([G] and [H]). The GUS-positive cells in (H) correspond to the arbuscule-containing cells (indicated by white arrowheads in [I]).
Bars = 100 μm.
The series of GOF-CCaMK variants was introduced into SbtM1 promoter–GUS (SbtM1pro:GUS) plants by means of hairy root transformation, and the transgenic roots were grown in the absence of AM fungal infection. The roots showed GUS staining induced by the GOF-CCaMK variants consistent with the real-time PCR analysis of SbtM1 expression (Figures 1A). All of the kinase-only GOF-CCaMK variants activated the AMS signaling pathway since GUS staining was observed in the uninfected roots (CCaMK314, 12/40; CCaMK314-NLS, 30/50; CCaMK314TD, 46/87; CCaMK314TD-NLS, 57/97; GUS-positive roots/all transgenic roots examined) except for the NES fusion (CCaMK314TD-NES, 0/49; GUS-positive roots/all transgenic roots examined). Full-length CCaMKTD (0/75; GUS-positive roots/all transgenic roots examined) did not activate this pathway. The GUS activity was detected both in epidermal and in cortical cells at 1 week after transferring to sterilized soil (Figures 2D and 2E), whereas the GUS staining was confined only to cortical cells in 2-week-old hairy roots (Figure 2F). Interestingly, discrete rows of the GUS-positive cortical cells showed higher GUS activity than that seen in neighboring cells (Figure 2F). This type of GUS staining pattern was similar to that induced by AM fungal infection (Figures 2G to 2I). In SbtM1pro:GUS roots infected with AM fungi, the intense GUS staining corresponded to cells that adjoined elongated fungal hyphae or that contained arbuscules (Figures 2H and 2I; Takeda et al., 2009). These results indicate that the position of the cortical cells where fungal hyphae elongate in between and/or where arbuscules develop is likely to be determined downstream of CCaMK and independent of actual infection by AM fungi.
Activation of SPN Formation by Nuclear-Localized GOF-CCaMK Variants
All of the GOF-CCaMK variants induced SPN formation in transgenic hairy roots, in contrast with wild-type CCaMK (CCaMKTT) (Table 1; see Supplemental Figure 3 online). The SPNs induced by GOF-CCaMK variants were structurally comparable to those in snf1 (Tirichine et al., 2006). The frequency of SPN formation induced by kinase-only GOF-CCaMK was lower than that induced by full-length CCaMKTD (Table 1), despite the fact that hairy roots carrying full-length CCaMKTD showed lower expression of RNS-induced genes than those carrying the nuclear-localized kinase-only GOF-CCaMK variants (Figure 1A). The difference in frequency of SPN formation between full-length CCaMKTD and kinase-only GOF-CCaMK is likely to be caused by the presence of the regulatory domains of CCaMK (i.e., the CaM BD and EF hands). RNS-induced gene expression in cortical cells appears to be necessary and sufficient for SPN formation, indicating that epidermal expression may not correlate with SPN formation. The transgenic roots carrying the NES fusion formed small, distorted SPNs (see Supplemental Figure 3 online), but the frequency was very low (Table 1). This SPN formation might have been induced by residual nuclear localization of CCaMK314TD-NES (see Supplemental Figure 2G online). These results show that the nuclear localization of GOF-CCaMK is required for SPN formation.
Table 1.
SPN Formation and Complementation of ccamk Mutants by GOF-CCaMK
| Spontaneous Nodule Formationa |
Symbiont Entry into ccamk Mutantsa |
|||
| CCaMK Variant | Wild Type | ccamk | Rhizobia | Mycorrhiza |
| Vector control | 0% (0/64) | 0% (0/35) | 0% (0/16) | 0%(0/55) |
| CCaMKTT (wild type) | 0% (0/40) | 0% (0/45) | 86% (32/37) | 79% (27/34) |
| CCaMKTD | 87% (41/47) | 41%(14/34) | 75% (15/20) | 64% (18/28) |
| CCaMK314 | 16% (5/32) | 32%(11/34) | 0% (0/14) | 29% (9/31) |
| CCaMK314-NLS | 39% (9/23) | 26% (6/23) | 0% (0/27) | 17% (5/30) |
| CCaMK314TD | 17% (5/29) | 33% (9/27) | 0% (0/24) | 18% (9/50) |
| CCaMK314TD-NLS | 15% (3/19) | 15% (6/39) | 0% (0/25) | 57% (16/28) |
| CCaMK314TD-NES | 2% (1/48) | 2% (1/53) | 0% (0/35) | 2% (1/43) |
Values in parentheses indicate numbers of SPN formed or complemented roots/total transformed roots.
SPN formation was observed at 4 weeks after transferring to a sterilized soil. Infection of rhizobia or arbuscular mycorrhiza was observed at 4 and 3 weeks after inoculation, respectively.
Both RNS and AMS gene induction as well as SPN formation might be caused by the activity of endogenous CCaMK in wild-type plants. To test this possibility, we introduced the series of GOF-CCaMK variants into ccamk mutant roots and examined the symbiotic responses. In ccamk transgenic hairy roots, nuclear-localized GOF-CCaMK triggered expression of RNS- and AMS-induced genes and SPN formation similar to those in wild-type plants (Figure 1B, Table 1). These results suggest that activation of the RNS/AMS signaling pathway and induction of the downstream symbiosis genes by the nuclear-localized kinase-only GOF-CCaMK variants are independent of endogenous wild-type CCaMK, strengthening the idea that the regulatory domains are not essential for RNS/AMS gene induction and SPN formation. In the expression analysis, the level of RNS/AMS gene induction in ccamk mutants was generally higher than that in wild-type plants, and induction of RNS-induced genes was also observed in hairy roots carrying full-length CCaMKTD (Figure 1B). Relatively higher expression in ccamk mutants could be caused by the difference in basal gene expression in uninfected roots carrying the vector control between ccamk mutants and wild-type plants.
Complementation of Symbiont Infection Defects in ccamk mutants by the GOF-CCaMK Variants
To further examine the functional features of the GOF-CCaMK variants, we attempted to complement the defect in bacterial entry in ccamk mutants by transformation with each of the variants. Among the ccamk mutants transformed with GOF-CCaMK, only hairy roots carrying full-length CCaMKTD recovered the ability to form infection threads and to allow entry of rhizobia into nodules, comparable to that of the wild-type CCaMK (CCaMKTT) (Table 1; see Supplemental Figure 3 online). On the other hand, the nuclear-localized kinase-only GOF-CCaMK variants did not complement the bacterial entry process in ccamk mutants (Table 1; see Supplemental Figure 3 online) even though they induced expression of RNS genes to higher levels than did full-length CCaMKTD (Figure 1B) and caused SPN formation. (We presume that the nodule-like structures observed in hairy roots transformed with the GOF-CCaMK variants are SPNs induced by the variants independent of bacterial infection.) These results suggest that the regulatory domains of CCaMK play a crucial role in infection thread formation and bacterial entry into nodules, as shown by Gleason et al. (2006).
We subsequently examined whether any of the GOF-CCaMK variants could complement the AMS-deficient phenotypes in ccamk mutant roots. In this analysis, the defect in AM fungal colonization in the ccamk mutants was complemented by all of the GOF-CCaMK constructs except for the NES fusion (Table 1; see Supplemental Figure 3 online). Hyphal entry into the host root as well as formation of arbuscules and vesicles was observed in the transformed hairy roots. The very low complementation frequency obtained with the NES fusion (Table 1) was consistent with the result that AMS-induced gene expression was not significant in inoculated hairy roots carrying the NES fusion (see Supplemental Figure 4 online), again suggesting that the nuclear localization of GOF-CCaMK is also important for its signaling activity in AMS. Induction of PT4 expression was also restored by the series of GOF-CCaMK variants, except for the NES fusion, upon infection by AM fungi (see Supplemental Figure 4 online). PT4 expression was not induced in hairy roots carrying GOF-CCaMK in the absence of AM fungi, despite the presence of SbtM1 expression (Figure 1). These results indicate either that infection by AM fungi and activation by CCaMK are both required for PT4 induction or that fungal infection suppresses a mechanism that inhibits PT4 expression downstream of CCaMK.
Remodeling of a Cytoplasmic Structure by GOF-CCaMK Variants
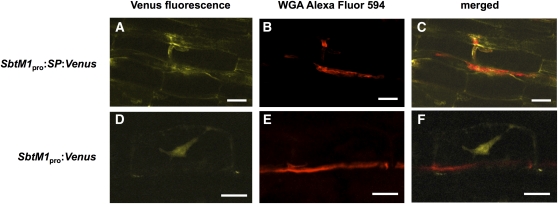
In addition to AMS-induced gene expression, we examined cytological changes induced by the GOF-CCaMK variants at the preinfection stage of AMS. SbtM1 is expressed in and secreted from arbuscule-containing cells (Takeda et al., 2009). In addition, a gene encoding a fluorescent protein (Venus) fused with the signal peptide of SbtM1 under the control of the SbtM1 promoter (SbtM1pro:SP:Venus; Takeda et al., 2009) showed that hyphal penetration into cortical layers induced expression of SbtM1 (Figures 3A to 3C). The result indicates that the cortical cells expressing SbtM1 correspond to cells penetrated by hyphae. To examine the cytoplasmic remodeling in the cells before fungal penetration, the cytoplasm was visualized with Venus fusion protein driven by the SbtM1 promoter (SbtM1pro:Venus; Takeda et al., 2009). PPA formation was expected to appear as a dense cytoplasmic bridge in cortical cells. The SbtM1pro:Venus construct was introduced by means of hairy root transformation, and AM fungi were allowed to infect the roots for 2 weeks. Cortical cells along which the AM hyphae elongated expressed SbtM1pro:Venus, and the fluorescence revealed development of a cytoplasmic bridge before fungal penetration into the cells (Figures 3D to 3F). The cytoplasmic bridges visualized by Venus fluorescence were dense and often showed a particular pattern: Strands of the cytoplasm elongated from the nucleus to the opposite side of the cell membrane (Figure 3D). This polar distribution of the dense cytoplasm was not typically observed in cortical cells without fungal infection, in which normal transvacuolar strands, if any, are very thin and spread randomly from the nucleus (see Supplemental Figures 5A and 5B online). These results demonstrate that PPAs are also formed in L. japonicus, in which expression of SbtM1 is a suitable marker for the PPA-containing cells.
Figure 3.
Induction of a PPA by AM Fungal Infection.
Transgenic hairy roots carrying SbtM1pro:SP:Venus ([A] to [C]) or SbtM1pro:Venus ([D] to [F]) were infected with G. intraradices for 2 weeks. The images of Venus fluorescence ([A] and [D]) and fungal structure visualized by WGA Alexa Fluor 594 ([B] and [E]) are merged ([C] and [F]). Secreted Venus fluorescent protein was detected in the apoplastic space where the AM fungal hypha had penetrated into the host cell (C). A dense cytoplasmic bridge visualized by Venus fluorescence was detected in a cortical cell adjacent to an AM hypha (F). Bars = 25 μm.
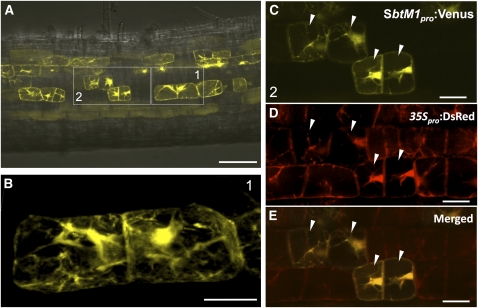
Using this test system, we investigated whether remodeling of a cytoplasmic structure was induced by the kinase-only GOF-CCaMK variants. We examined transgenic roots carrying both CCaMK314TD-NLS and SbtM1pro:Venus in the absence of AM fungal infection. In the transgenic roots, the SbtM1 promoter was activated by CCaMK314TD-NLS, and Venus fluorescence was observed in cortical cells (Figure 4). As we observed in the histochemical expression analysis using the GUS fusion (Figure 2F), various strengths of fluorescence were detected in cortical cells (Figure 4A). The AMS signaling pathway appeared to be activated in the fluorescent cells. Although a majority of the cells showed weak Venus fluorescence in the nucleus and in thin transvacuolar strands, a few numbers of cells exhibiting strong fluorescence (>2% of detectable Venus-expressing cells, n = 122) showed dense cytoplasmic bridges (Figures 4A and 4B) in the cortical layers but not in the epidermal layer. Development of the cytoplasm was especially obvious around the nucleus, and the structures were nearly identical to the PPAs induced by AM fungal infection (Figures 3D to 3F; Genre et al., 2005, 2008). Formation of the PPA-like structure was observed independent of the developmental stage of cortical cells in which SbtM1 expression was highly induced, but the well-developed PPA-like structures tended to be found more often in the young cortical cells (Figures 4A and 4B; see Supplemental Figures 5C and 5D online). The cells with the PPA-like structures were often found to be next each other (Figure 4A) and adjacent to each other (Figure 4B; see Supplemental Figures 5C and 5D online). The other neighboring cells without Venus fluorescence did not show such a dense cytoplasmic bridge (Figures 4C to 4E). Formation of the PPA-like structure was also observed using GFP fused with the ER retention signal (HDEL = HisAspGluLeu) under the control of the SbtM1 promoter (SbtM1pro:GFP-HDEL) (see Supplemental Figure 6 online). The PPAs were induced in GFP-HDEL–expressing cortical cells by AM fungal infection (see Supplemental Figure 6A online). In the PPA-like structure, tubular ER structures induced by CCaMK314TD-NLS were formed in the cytosol (see Supplemental Figure 6B online; Genre et al., 2005, 2008). However, the ER structures were not evident when compared with those induced by AM fungal infection, which indicates that actual infection is necessary for developing the extensive ER structures.
Figure 4.
Induction of a PPA-Like Structure by GOF-CCaMK.
(A) Hairy roots cotransformed with CCaMK314TD-NLS and SbtM1pro:Venus were observed after 3 weeks of growth under aseptic conditions. Laser transmission and Venus fluorescence images were merged.
(B) A stack of images obtained along the z axis of cells corresponding to box 1 region in (A) strongly expressed SbtM1pro:Venus and showed dense cytoplasmic bridges.
(C) to (E) Venus, DsRed, and merged images in the box 2 region in (A).
(C) Cortical cells expressing SbtM1pro:Venus showed the PPA-like structure.
(D) Cytosol was visualized with a transformation marker DsRed under the control of the 35S promoter.
(E) Merged image of Venus (C) and DsRed (D) showed cells (arrowheads) with dense cytoplasmic bridges compared with the neighboring cells.
Bars = 100 μm in (A) and 25 μm in (B) to (E).
It was reported that the PPA-containing cells exhibit enlargement of the nucleus (Genre et al., 2008). To confirm whether the PPA-like structure accompanied enlarged nuclei, the nuclear size was measured in cortical cells (Table 2; see Supplemental Figure 7 online). With AM fungal infection, the PPAs were induced in SbtM1pro:Venus-expressing cells, in which the nucleus was significantly enlarged compared with that in cortical cells of uninfected roots or cortical cells without Venus fluorescence neighboring to the PPA formed cells. Cells with Venus fluorescence induced by CCaMK314TD-NLS also showed statistically significant enlargement of the nucleus, although the degree of enlargement was less than that induced by AM fungal infection. Enlargement of the nucleus was not observed in cortical cells without Venus fluorescence in the same region. Above observations confirm that the PPA-like structure induced by the nuclear-localized kinase-only GOF-CCaMK variants alone appears to have similar identity to the PPA that is formed in response to AM fungi.
Table 2.
Nuclear Size in Cortical Cells
| Treatment | Nuclear Diameter (μm) | Minimum (μm) | Maximum (μm) | n |
| Uninfected root (vector control) | 5.60 (0.79)a | 4.05 | 7.54 | 56 |
| AM infected root (no fluorescence) | 6.28 (1.10)b | 4.38 | 9.24 | 53 |
| AM infected root (Venus fluorescence) | 9.18 (1.58)c | 6.54 | 13.19 | 25 |
| CCaMK314TD-NLS (no fluorescence) | 6.68 (1.43)b | 4.55 | 10.41 | 84 |
| CCaMK314TD-NLS (Venus fluorescence) | 7.85 (1.44)d | 5.64 | 11.12 | 71 |
Values in parentheses indicate standard deviation. Letters indicate statistically significant differences at P < 0.05 (Tukey-Kramer’s multiple comparison).
DISCUSSION
Regulation of CCaMK by Ca2+ Signals
In this study, we investigated the functions of CCaMK in symbiosis signaling by conferring a GOF to the protein (a summary of the results is shown in Table 3). The nuclear-localized kinase-only GOF-CCaMK variants, in contrast with full-length CCaMKTD, triggered strong RNS-induced gene expression in epidermal cells of L. japonicus (Figures 1 and 2). In M. truncatula, there is no substantial difference in spontaneous activation of one of the early nodulins ENOD11 expression between the kinase-only and the full-length GOF-CCaMK variants (Gleason et al., 2006). The available information on tissue-specific expression of NIN led us to hypothesize that epidermal expression of NIN by kinase-only GOF-CCaMK would be sufficient for infection thread formation and bacterial entry into the host, leading to complementation of ccamk mutants. However, our analysis revealed that epidermal activation of the RNS signaling pathway by the nuclear-localized kinase-only GOF-CCaMK variants was not sufficient for complementation of the infection defect in ccamk mutants (Table 1). This result indicates importance of the regulatory domains at the C terminus of CCaMK for bacterial entry (see Supplemental Figure 8 online). It has been suggested that different Nod factor molecular structures are required between bacterial entry and cortical cell division, resulting in bacterial accommodation (Ardourel et al., 1994). Our analyses showed that CCaMK integrates both events. According to our model (see Supplemental Figure 8 online), three functions of CCaMK (nuclear localization, kinase activity, and regulation by the C-terminal domains) regulate symbiosis signaling for expression of symbiosis-induced genes, cortical cell division/SPN formation, and bacterial entry. Importantly, whereas the regulatory domains play both a negative role in kinase activity and a positive role in bacterial entry in RNS, they are less important in AMS, in which the regulatory domains are not strictly required for successful AMS. AMS is an evolutionarily older type of symbiosis than RNS (Parniske, 2008), so that the signaling pathway in AMS is expected to be simpler than that in RNS. This may explain why the nuclear-localized kinase-only CCaMK variants are able to complement the defective AMS phenotype in the ccamk mutant but are unable to complement part of the defective RNS phenotype in the same mutant. Activation of the AMS signaling pathway is likely to be one of the fundamental aspects of the CCaMK functions in the symbioses.
Table 3.
Symbiotic Functions of the CCaMK Variants in RNS and AMS
| GOF (without Symbionts) |
Complementation |
|||||||
| Gene Induction |
||||||||
| Structural Change |
RNS (NIN) |
AMS |
Symbiont Entry |
|||||
| CCaMK Variants | SPN | PPA | Epi | Cor | SbtM1 | PT4 | RNS | AMS |
| CCaMKTT (wild type) | − | − | − | − | − | − | + | + |
| CCaMKTD | + | − | − | + | − | − | + | + |
| CCaMK314TD-NLS | + | + | + | + | + | − | − | + |
| CCaMK314TD-NES | − | − | − | − | + | − | − | − |
Cor, expression in cortical cells; Epi, expression in epidermal cells; +, detected, −, not or scarcely detected.
The deregulation of symbiosis signaling by the truncated CCaMK protein emphasizes the negative regulatory role of the C-terminal domains, that is, the CaM BD and the three EF hands (Gleason et al., 2006). The C-terminal domains have been reported to possess an autoinhibitory activity through their ability to change conformation (Sathyanarayanan et al., 2000; Harper et al., 2004). In the inactive form, the folded C-terminal region covers the kinase domain. The physical barrier created by the C-terminal region inhibits the activation of the kinase domain. Activation of the symbiosis signaling pathway leads to unfolding of the protein, which enables the kinase domain either to make contact with an upstream activator or to be self-activated through autophosphorylation, or a combination of both. The presence of the CaM BD and EF hands in the C terminus of CCaMK indicates that Ca2+ is a trigger for the conformational change of CCaMK. Alteration of Ca2+ concentration is sensed by the EF hands or the CaM BD through calmodulin, or both. Thus, Ca2+ binding affects the conformation of CCaMK. In RNS, Ca2+ influx and Ca2+ spiking are known to occur early in the symbiotic response. The Ca2+ influx, which is a transient flux of Ca2+ into a root hair cell, is observed several minutes after application of Nod factors (Cárdenas et al., 1999; Felle et al., 1999). Periodic Ca2+ oscillation known as Ca2+ spiking is observed in the root hair ~10 min after treatment with Nod factor (Ehrhardt et al., 1996). The largest amplitude of Ca2+ spiking is associated with the nucleus, in which a characteristic fluctuation of Ca2+ is thought to encode a symbiosis signal (Sieberer et al., 2009). Furthermore, Ca2+ spiking is also observed in AMS, indicating that Ca2+ spiking is a signaling factor in the common Sym pathway (Kosuta et al., 2008; Chabaud et al., 2011). These facts suggest that Ca2+ spiking is a candidate for an activator of CCaMK. The importance of the nuclear localization of CCaMK demonstrated in this work further supports the relationship of CCaMK with Ca2+ spiking. Although it has not yet been proven by direct evidence, it is worth considering the possibility that CCaMK is a decoder of the Ca2+ spiking signal.
Importance of Nuclear Localization of CCaMK for Downstream Signaling
The exclusion of GOF-CCaMK from the nucleus abolished or reduced its signaling activity, which indicates that the nuclear localization of CCaMK is essential for the proper activation of the signaling pathway both in AMS and in RNS. Activation of downstream signaling events such as that of transcription factors is likely to occur in the nucleus. It is still unknown whether the C terminus of CCaMK contains a functional NLS or interacts with a protein containing an NLS. CYCLOPS, a common Sym protein and a CCaMK interactor, is predicted to possess two NLS motifs and is localized in the nucleus (Yano et al., 2008). It is therefore possible that the association with CYCLOPS may enhance the nuclear localization of CCaMK. In addition, kinase-only CCaMK fails to interact with CYCLOPS (Yano et al., 2008). This is in agreement with the observation that epidermal expression of NIN, which is normally triggered by rhizobial infection, seems severely compromised in cyclops mutants (Yano et al., 2008). However, because ccamk mutant roots transformed with the nuclear-localized kinase-only GOF-CCaMK variants by NLS fusion still show defects in infection similar to those seen in cyclops mutants, it is unlikely that the function of CYCLOPS is solely to confer nuclear localization to CCaMK.
The proper CCaMK function requires its nuclear localization, indicating that the target protein is probably also nuclear localized. CYCLOPS is a target of phosphorylation by CCaMK (Yano et al., 2008); however, its loss-of-function phenotype differs from that of CCaMK. In cyclops, infection by rhizobia induces formation of nodule primordia in the cortex and colonization of rhizobia on root hairs. On the other hand, ccamk mutants show no distinct symbiotic phenotypes in RNS other than root hair deformation and Ca2+ spiking. These facts indicate that CYCLOPS functions as a secondary target of CCaMK. Initial phosphorylation targets of CCaMK are likely to be symbiosis-related transcriptional regulators, such as NSP1, NSP2, or NIN (Schauser et al., 1999; Hirsch et al., 2009). Since the loss-of-function phenotypes of these three regulators are partially similar to those of CCaMK (i.e., no cortical cell division upon infection), it is plausible that these regulators are directly or indirectly phosphorylated by CCaMK and then activate transcription of a number of symbiosis-induced genes. The AMS-specific pathway may be regulated by a similar mechanism, although no such AMS-specific transcriptional regulators have yet been identified from genetic analyses.
AMS Signaling and Downstream Events Regulated by CCaMK
The interaction of L. japonicus and Glomus intraradices shows Arum-type hyphal elongation, in which intercellular hyphae develop to form intracellular arbuscules, as occurs in M. truncatula (Dickson, 2004; Genre et al., 2005, 2008). AM hyphae elongate longitudinally between cortical cells and often penetrate into the cell when they cross the cell transversely or form an arbuscule. Our results revealed that PPA formation coincided with SbtM1 expression, suggesting that PPA formation would be observed not only in cortical layers where fungal penetration occurs, but also in arbuscule-containing cells where SbtM1 is strongly induced (Figure 3). The system for fungal penetration into the host cell may be shared by the processes of hyphal elongation and arbuscule formation, as indicated by Genre et al. (2008). Our experimental method was unable to show whether AM fungi could enter the cells containing the PPA-like structures that had been induced by kinase-only GOF-CCaMK. However, Genre et al. (2005) have shown that PPA formation precedes fungal invasion into the host cell, which requires the common Sym genes. Our results have confirmed that the PPA forms before fungal penetration, which is likely to be regulated by CCaMK in the common Sym pathway. The PPA-like structures we observed here are confirmed by two characters: cytoplasmic remodeling and nuclear enlargement (Figure 4, Table 2; see Supplemental Figure 6 online). The PPAs induced by AM fungal infection have three characteristics: cytoplasmic remodeling that is visualized by cytoskeletons or ER, nuclear migration, and nuclear enlargement (Genre et al., 2005, 2008). Nuclear migration is triggered by AM fungal infection even in common Sym mutants (Genre et al., 2005), so that we regard two characteristics (i.e., cytoplasmic remodeling and nuclear enlargement) as being sufficient for the identity of the PPAs, which are also observed in the PPA-like structures. The PPA-like structures were not formed at the epidermal layer despite of SbtM1 expression in this layer. This can be due to the fact that in L. japonicus, the majority of AM fungal invasion in the epidermal layer is apoplastic and requires epidermal opening (Demchenko et al., 2004; Parniske, 2004; Kistner et al., 2005). The identity and/or responses of the epidermal layer to AM fungi may differ between L. japonicus and M. truncatula.
We found that expression of L. japonicus PT4, an AMS-specific gene, responded differently to the GOF-CCaMK variants and AM fungal infection. The lack of PT4 induction by the GOF-CCaMK variants in the absence of fungal infection indicates that one or more other factors, possibly triggered by the presence of AM fungi, are required (see Supplemental Figure 8 online). In addition, the regulatory domains, which are required for bacterial entry in RNS, are not required for induction of PT4 expression. These facts indicate that other signals may be produced after fungal entry that additionally activate the kinase domain of CCaMK, which is not conferred by the GOF mutation. Alternatively, it is possible that a completely different pathway exists for signaling of symbiosis responses downstream of fungal entry. One candidate for the triggering factor is lysophosphatidylcholine, identified by Drissner et al. (2007) as an inducer of the phosphate transporter in maize (Zea mays). After PPA formation and fungal entry, this factor may induce late symbiotic responses, including PT4 expression. Another, but not exclusive, idea is that supply of phosphorus by AM fungi directly induces PT4 expression. In RNS, symbiosis-induced genes have been categorized as early-nodulin and late-nodulin genes (Mylona et al., 1995). AMS-induced responses can also be categorized as early and late; for example, SbtM1 and PT4 can be considered as early and late AMS-induced genes, respectively.
Our analyses of the GOF-CCaMK variants revealed that alterations in the regulatory domains and nuclear localization resulted in different degrees of activation of CCaMK and, as a consequence, different symbiotic responses. This feature of CCaMK suggests the possibility that CCaMK may function in sorting AMS and RNS signals within the common Sym pathway. The detailed regulation mechanism of the regulatory domains of CCaMK is currently under investigation. Such analysis will provide further insights into the mechanisms of CCaMK signaling and possibly serve to identify upstream activators and downstream signaling factors.
METHODS
Plant Growth
Seeds of Lotus japonicus accession Gifu B-129, ccamk-3 (Tirichine et al., 2006), NINpro:GUS (Radutoiu et al., 2003), or SbtM1pro:GUS (Takeda et al., 2009) transgenic plants were scarified, germinated on 0.8% agar plates, and sterilized with 0.1% sodium hypochlorite. Seedlings were grown in a growth chamber (24°C, 16 h light/8 h dark). For SPN formation, the transgenic plants carrying GOF-CCaMK were grown in sterilized expanded vermiculite (Nittai) supplied with water for 4 weeks. The ccamk-3 plants transformed with GOF-CCaMK variants were inoculated with Mesorhizobium loti carrying DsRED (Maekawa et al., 2009) and then grown under the same conditions as above. For induction of the PPA-like structure, plants transformed with the GOF-CCaMK variants were grown in sterilized 1:1 (v/v) mixture of vermiculite and commercial soil for turf grass (Sunbellex) according to Banba et al. (2008) and supplied with water. The AM fungus Glomus intraradices (DAOM197198; PremierTech) was propagated in a chive (Allium schoenoprasum) nurse pot system (Demchenko et al., 2004) made using the same soil as induction of the PPA-like structure. The pot was used for AM fungi inoculation after removal of the chive plants.
Construction of Binary Vectors
The kinase-only GOF-CCaMK variants were amplified by PCR (forward primer, 5′-CACCATGGGATATGATCAAACCAGAA-3′; reverse primer, 5′-TTAAATCTCAGGGTCCATTTGCT-3′) from CCaMKTT or CCaMKTD constructs (Banba et al., 2008). NLS (ProLysLysLysArgLysValGluAsp) and NES (AsnGluLeuAlaLeuLysLeuAlaGlyLeuAspIleAsnLysThr) (Wen et al., 1995; Shen et al., 2007) were C-terminally joined to the coding sequence for the truncated CCaMK protein by PCR (NLS coding sequence, 5′-CCTAAGAAGAAGAGAAAGGTTGAGGAT-3′; NES coding sequence, 5′-AACGAGCTTGCTTTGAAATTGGCAGGATTAGATATTAACAAGACG-3′). The amplified fragments were cloned into pENTR D/TOPO (pENTR D/TOPO cloning kit; Invitrogen). The resulting entry clones were converted with the destination vector p35S-GW-GFP (Yano et al. 2008) or p35S-GW-DsRed (K. Yano and M. Hayashi, unpublished data) by the LR reaction (Gateway LR clonase II Enzyme Mix; Invitrogen). The destination vector without the Gateway cassette (p35S-GFP or p35S-DsRed) was used for the vector control.
Plant Transformation
Agrobacterium tumefaciens AGL1 was used for the tobacco (Nicotiana benthamiana) infiltration transformation. The infiltration was performed as described by Yano et al. (2008). Transgenic hairy roots were induced by Agrobacterium rhizogenes LBA1334 (Offringa et al., 1986) following the method described by Díaz et al. (2005).
Expression Analysis
Total RNA was extracted from roots using the RNeasy plant mini kit (Qiagen). Reverse transcription (RT) and real-time PCR were performed using the QuantiTect reverse transcription kit (Qiagen) and Light Cycler FastStart DNA MasterPLUS SYBR Green I (Roche) with Light Cycler (Roche) according to the manufacturer’s instructions. cDNA was synthesized from 100 ng total RNA in a 20-μL RT reaction mixture. The RT product (0.8 μL) was added to a total of 20 μL real-time PCR reaction mixture. Real-time PCR primer sets and thermal cycler conditions were as described by Takeda et al. (2009). Ubiquitin was used as a reference gene (Takeda et al., 2005). The relative expression level of each gene was calculated with the delta cycle threshold method (Takeda et al., 2005) and normalized to the transcription level of ubiquitin. Three to six biologically independent experiments were performed to calculate the mean and sd values.
Histochemistry and Microscopy Observation
Transgenic roots carrying NINpro:GUS or SbtM1pro:GUS were treated with GUS staining buffer (0.5 mg/mL X-Gluc, 100 mM phosphate buffer, pH 7.0, 100 mM EDTA, 0.5 mM K4[Fe(CN)6], 0.5 mM K3[Fe(CN)6], and 0.1% Triton X-100) at 37°C for 1 to 6 h.
Wheat germ agglutinin (WGA) Alexa Fluor 488 or 594 (Invitrogen) was used for visualization of fungal cell walls (Harrison et al., 2002). The transgenic roots infected with AM fungi were boiled with 2% KOH at 95°C for 20 min and washed three times with PBS. Infected roots were then stained with WGA Alexa Fluor (1 μg/mL final concentration) and kept at room temperature for more than 1 h. For the staining of living cells, transgenic roots carrying the Venus fusion constructs were embedded in 5% low-melting agarose (SeaPlaque Agarose; CAMBREX) without fixing, kept at 30°C during embedding, and sectioned with a VT1200S vibrating-blade microtome (Leica). The root sections (100 to 120 μm) were stained with 1 μg/mL WGA Alexa Fluor 594 or 488 in PBS for more than 1 h at room temperature. Nuclear staining in root cells was performed with 4′,6-diamidino-2-phenylindole (Dojindo). Roots were soaked in 10 μg/mL 4′,6-diamidino-2-phenylindole in half-strength PBS and vacuumed for 30 min. The roots were incubated for 3 h at room temperature and washed three times with half-strength PBS.
Bright-field and fluorescence microscopy were performed with a stereomicroscope (SZX12; Olympus) or an inverted microscope (BZ-9000; Keyence) equipped with ×10 dry (numerical aperture 0.45) and ×20 dry (numerical aperture 0.75) objectives. Venus fluorescence was detected with a confocal microscope (TCS SP5; Leica, Nikon A1) equipped with ×20 or ×40 dry objective lenses. Images were acquired and analyzed using the LAS-AF software package (Leica), NIS elements (Nikon), and Adobe Photoshop (Adobe Systems).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AJ239041.1 (NIN), AP009542 (SBtM4), AP006864 (SbtS), AP009544 (SbtM1), AP010874 (PT4), and DQ249171.1 (Ubiquitin).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Wild-Type CCaMK and GOF-CCaMK Variants Used in This Study.
Supplemental Figure 2. Localization of GOF-CCaMK.
Supplemental Figure 3. SPN Formation and Complementation of ccamk Mutants by GOF-CCaMK.
Supplemental Figure 4. Activation of AMS-Induced Genes by GOF-CCaMK in ccamk Mutants.
Supplemental Figure 5. Transvacuolar Strands and PPA-Like Structures Induced by GOF-CCaMK.
Supplemental Figure 6. PPA-Like Structures Visualized by ER-Retained GFP.
Supplemental Figure 7. Histograms of Nuclear Sizes in Cortical Cells.
Supplemental Figure 8. Model of CCaMK-Regulated AMS and RNS Signaling.
Supplementary Material
Acknowledgments
Seeds of SbtM1pro:GUS plants and binary vectors SbtM1pro:Venus and SbtM1pro:SP:Venus were kindly provided by Martin Parniske. We thank Jens Stougaard for providing seeds of NINpro:GUS plants. We also thank Haruko Imaizumi-Anraku and Masayoshi Kawaguchi for experimental supports. A plant binary vector p35S-GW-DsRed was constructed and kindly provided by Koji Yano. Some of the confocal images were acquired at Spectrography and Bioimaging Facility, National Institute for Basic Biology Core Research Facilities. N.T. was supported by a Research Fellow of the Japan Society for the Promotion of Science and Japan Ecology Foundation. M.H. was supported by grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project Grant PMI-0001) and from the Japan Society for the Promotion of Science (Funding Program for Next Generation World-Leading Researchers).
AUTHOR CONTRIBUTIONS
N.T., T.M., and M.H. conceived and designed the experiments. N.T. and T.M. performed the experiments. N.T., T.M., and M.H. analyzed the data. N.T. and M.H. wrote the article with the assistance of T.M.
References
- Ané J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Ardourel M., Demont N., Debellé F., Maillet F., de Billy F., Promé J.C., Dénarié J., Truchet G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B., Pfeffer P.E., Abubaker J., Jun J., Allen J.W., Brouillette J., Douds D.D., Lammers P.J., Shachar-Hill Y. (2003). Carbon export from arbuscular mycorrhizal roots involves the translocation of carbohydrate as well as lipid. Plant Physiol. 131: 1496–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banba M., Gutjahr C., Miyao A., Hirochika H., Paszkowski U., Kouchi H., Imaizumi-Anraku H. (2008). Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol. 49: 1659–1671 [DOI] [PubMed] [Google Scholar]
- Bonfante P., Perotto S. (1995). Strategies of arbuscular mycorrhizal fungi when infecting host plants. New Phytol. 130: 3–21 [Google Scholar]
- Cárdenas L., Feijó J.A., Kunkel J.G., Sánchez F., Holdaway-Clarke T., Hepler P.K., Quinto C. (1999). Rhizobium nod factors induce increases in intracellular free calcium and extracellular calcium influxes in bean root hairs. Plant J. 19: 347–352 [DOI] [PubMed] [Google Scholar]
- Catoira R., Galera C., de Billy F., Penmetsa R.V., Journet E.P., Maillet F., Rosenberg C., Cook D., Gough C., Dénarié J. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M., Genre A., Sieberer B.J., Faccio A., Fournier J., Novero M., Barker D.G., Bonfante P. (2011). Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol. 189: 347–355 [DOI] [PubMed] [Google Scholar]
- Demchenko K., Winzer T., Stougaard J., Parniske M., Pawlowski K. (2004). Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytol. 163: 381–392 [DOI] [PubMed] [Google Scholar]
- Dénarié J., Cullimore J. (1993). Lipo-oligosaccharide nodulation factors: A minireview new class of signaling molecules mediating recognition and morphogenesis. Cell 74: 951–954 [DOI] [PubMed] [Google Scholar]
- Díaz C.L., Grønlund M., Schlaman H.R.M., Spaink H.P. (2005). Induction of hairy roots for symbiotic gene expression studies. In Lotus japonicus Handbook, A.J. Marquez, ed (Dordrecht, The Netherlands: Springer), pp. 261–277
- Dickson S. (2004). The Arum&Paris continuum of mycorrhizal symbioses. New Phytol. 163: 187–200 [DOI] [PubMed] [Google Scholar]
- Drissner D., Kunze G., Callewaert N., Gehrig P., Tamasloukht M., Boller T., Felix G., Amrhein N., Bucher M. (2007). Lyso-phosphatidylcholine is a signal in the arbuscular mycorrhizal symbiosis. Science 318: 265–268 [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W., Wais R., Long S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Felle H.H., Kondorosi E., Kondorosi A., Schultze M. (1999). Elevation of the cytosolic free [Ca2+] is indispensable for the transduction of the Nod factor signal in alfalfa. Plant Physiol. 121: 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage D.J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68: 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Faccio A., Barker D.G., Bonfante P. (2008). Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Timmers T., Bonfante P., Barker D.G. (2005). Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V. (1996). Plant cell responses to arbuscular mycorrhizal fungi: Getting to the roots of the symbiosis. Plant Cell 8: 1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B.W., Oldroyd G.E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Grønlund M., Roussis A., Flemetakis E., Quaedvlieg N.E., Schlaman H.R., Umehara Y., Katinakis P., Stougaard J., Spaink H.P. (2005). Analysis of promoter activity of the early nodulin Enod40 in Lotus japonicus. Mol. Plant Microbe Interact. 18: 414–427 [DOI] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Draxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L., Parniske M. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M., Balestrini R., Hannah M., He J., Udvardi M.K., Bonfante P. (2009). Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 182: 200–212 [DOI] [PubMed] [Google Scholar]
- Harper J.F., Breton G., Harmon A. (2004). Decoding Ca(2+) signals through plant protein kinases. Annu. Rev. Plant Biol. 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Harrison M.J. (2005). Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 59: 19–42 [DOI] [PubMed] [Google Scholar]
- Harrison M.J., Dewbre G.R., Liu J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.J., van Buuren M.L. (1995). A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature 378: 626–629 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Banba M., Shimoda Y., Kouchi H., Hayashi M., Imaizumi-Anraku H. (2010). A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J. 63: 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433: 527–531 [DOI] [PubMed] [Google Scholar]
- Javot H., Penmetsa R.V., Terzaghi N., Cook D.R., Harrison M.J. (2007). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet E.P., El-Gachtouli N., Vernoud V., de Billy F., Pichon M., Dedieu A., Arnould C., Morandi D., Barker D.G., Gianinazzi-Pearson V. (2001). Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Kanamori N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C., Winzer T., Pitzschke A., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Webb K.J., Szczyglowski K., Parniske M. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R.J., Downie J.A., Oldroyd G.E. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J.C., Dénarié J. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Lévy J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Liu H., Trieu A.T., Blaylock L.A., Harrison M.J. (1998). Cloning and characterization of two phosphate transporters from Medicago truncatula roots: Regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol. Plant Microbe Interact. 11: 14–22 [DOI] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Maekawa-Yoshikawa M., Takeda N., Imaizumi-Anraku H., Murooka Y., Hayashi M. (2009). Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 58: 183–194 [DOI] [PubMed] [Google Scholar]
- Markmann K., Parniske M. (2009). Evolution of root endosymbiosis with bacteria: How novel are nodules? Trends Plant Sci. 14: 77–86 [DOI] [PubMed] [Google Scholar]
- Murray J.D., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65: 244–252 [DOI] [PubMed] [Google Scholar]
- Mylona P., Pawlowski K., Bisseling T. (1995). Symbiotic nitrogen fixation. Plant Cell 7: 869–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offringa I.A., Melchers L.S., Regensburg-Tuink A.J., Costantino P., Schilperoort R.A., Hooykaas P.J. (1986). Complementation of Agrobacterium tumefaciens tumor-inducing aux mutants by genes from the T(R)-region of the Ri plasmid of Agrobacterium rhizogenes. Proc. Natl. Acad. Sci. USA 83: 6935–6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M. (2004). Molecular genetics of the arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 7: 414–421 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008). Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nature Rev. Microbiol. 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Patil S., Takezawa D., Poovaiah B.W. (1995). Chimeric plant calcium/calmodulin-dependent protein kinase gene with a neural visinin-like calcium-binding domain. Proc. Natl. Acad. Sci. USA 92: 4897–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Saito K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan P.V., Cremo C.R., Poovaiah B.W. (2000). Plant chimeric Ca2+/calmodulin-dependent protein kinase. Role of the neural visinin-like domain in regulating autophosphorylation and calmodulin affinity. J. Biol. Chem. 275: 30417–30422 [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Senoo K., Solaiman M.Z., Kawaguchi M., Imaizumi-Anraku H., Akao S., Tanaka A., Obata H. (2000). Isolation of two different phenotypes of mycorrhizal mutants in the model legume plant Lotus japonicus after EMS-treatment. Plant Cell Physiol. 41: 726–732 [DOI] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Sieberer B.J., Chabaud M., Timmers A.C., Monin A., Fournier J., Barker D.G. (2009). A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 151: 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Takeda N., Haage K., Sato S., Tabata S., Parniske M. (2011). Activation of a Lotus japonicus subtilase gene during arbuscular mycorrhiza is dependent on the common symbiosis genes and two cis-active promoter regions. Mol. Plant Microbe Interact. 24: 662–670 [DOI] [PubMed] [Google Scholar]
- Takeda N., Okamoto S., Hayashi M., Murooka Y. (2005). Expression of LjENOD40 genes in response to symbiotic and non-symbiotic signals: LjENOD40-1 and LjENOD40-2 are differentially regulated in Lotus japonicus. Plant Cell Physiol. 46: 1291–1298 [DOI] [PubMed] [Google Scholar]
- Takeda N., Sato S., Asamizu E., Tabata S., Parniske M. (2009). Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J. 58: 766–777 [DOI] [PubMed] [Google Scholar]
- Takezawa D., Ramachandiran S., Paranjape V., Poovaiah B.W. (1996). Dual regulation of a chimeric plant serine/threonine kinase by calcium and calcium/calmodulin. J. Biol. Chem. 271: 8126–8132 [DOI] [PubMed] [Google Scholar]
- Tirichine L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Udvardi M.K., Day D.A. (1997). Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 493–523 [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth J.L., Tsien R.Y., Taylor S.S. (1995). Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473 [DOI] [PubMed] [Google Scholar]
- Yano K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.