This work identifies the SIP2 MAP kinase kinase as an interacting partner of the symbiosis receptor kinase SymRK. It shows that SymRK inhibits the kinase activity of SIP2 and establishes that a MAP kinase cascade plays a role in the signaling network leading to nodulation in legumes.
Abstract
The symbiosis receptor kinase, SymRK, is required for root nodule development. A SymRK-interacting protein (SIP2) was found to form protein complex with SymRK in vitro and in planta. The interaction between SymRK and SIP2 is conserved in legumes. The SIP2 gene was expressed in all Lotus japonicus tissues examined. SIP2 represents a typical plant mitogen-activated protein kinase kinase (MAPKK) and exhibited autophosphorylation and transphosphorylation activities. Recombinant SIP2 protein could phosphorylate casein and the Arabidopsis thaliana MAP kinase MPK6. SymRK and SIP2 could not use one another as a substrate for phosphorylation. Instead, SymRK acted as an inhibitor of SIP2 kinase when MPK6 was used as a substrate, suggesting that SymRK may serve as a negative regulator of the SIP2 signaling pathway. Knockdown expression of SIP2 via RNA interference (RNAi) resulted in drastic reduction of nodules formed in transgenic hairy roots. A significant portion of SIP2 RNAi hairy roots failed to form a nodule. In these roots, the expression levels of SIP2 and three marker genes for infection thread and nodule primordium formation were downregulated drastically, while the expression of two other MAPKK genes were not altered. These observations demonstrate an essential role of SIP2 in the early symbiosis signaling and nodule organogenesis.
INTRODUCTION
The symbiotic interaction between legumes and rhizobia results in the formation of nitrogen-fixing root nodules. Rhizobium cells enter into a symbiosis with the legume host, in which the bacterium gains carbohydrates from the host and exports reduced nitrogen in return (Gresshoff, 2003). The establishment of a symbiosis between rhizobia and legumes is highly specific. The mutual recognition process consists of a series of complex events starting from the perception of legume-derived flavonoids by rhizobia. In response, the rhizobial symbiont begins to produce the lipochitooligosaccharide signal molecules, known as Nod factors (NFs). NFs are chitin (N-acetylglucosamine oligomers) derivatives with N-acetylation at their nonreducing end and other modifications at the reducing end (Spaink et al., 1995). The specific NF structure determines the strict specificity between rhizobium and host legume species. Only successful recognition of an NF by the host elicits the signal transduction cascade that results in both the rhizobial infection process and nodule organogenesis.
At the onset of nodule organogenesis, a nodule primordium is formed from the already differentiated cortical cells. Rhizobia are guided to this primordium in a host-regulated manner (Kouchi et al., 2004; Geurts et al., 2005). After decades of research in two model legume species, Lotus japonicus and Medicago truncatula, more than two dozen legume genes have been implicated in NF perception, symbiotic signal transduction, bacterial infection, nodule organogenesis, and regulation of nitrogen fixation (Kouchi et al., 2010). Most of these genes have been isolated from symbiotic mutants of the model legumes by the conventional mutagenesis approach, which appears to be practically reaching saturation (Kouchi et al., 2010). The exact biochemical functions of these genes and their regulation mechanisms remain ambiguous.
L. japonicus Symbiosis Receptor Kinase (SymRK) and its homologs, Nodulation Receptor Kinase (NORK) from Medicago sativa and Does-not-Make-Infections2 (DMI2) from M. truncatula, have been demonstrated to be required for nodulation in legumes as well as for actinorhiza formation in the trees Casuarina glauca (Gherbi et al., 2008) and Datisca glomerata (Markmann et al., 2008). It encodes a protein with three Leu-rich repeat (LRR) domains in the predicted extracellular region and a protein kinase domain facing the cytoplasm (Endre et al., 2002; Stracke et al., 2002). Genetic dissection indicates that the L. japonicus SymRK mutant is impaired in Ca2+ spiking (Miwa et al., 2006) and is unable to form root nodules or be infected by arbuscular mycorrhizal fungi (Stracke et al., 2002). The three LRR domains of SymRK are believed to be involved in the ligand–protein interaction that leads to activation of the SymRK receptor through phosphorylation (Yoshida and Parniske, 2005). It is paramount to understand the exact biochemical functions and regulation mechanisms of SymRK because it is a receptor kinase that appears to govern the entire symbiotic process (Kouchi et al., 2010). By screening for proteins that interact with the DMI2 kinase, a putative mevalonate synthase (HMGR1) involved in the synthesis of isoprenoid compounds has been identified previously. Its involvement in the rhizobial infection process and nodule organogenesis in M. truncatula has also been shown by knockdown analysis of HMGR1 (Kevei et al., 2007). Recently, we isolated SIP1 (for SymRK-interacting protein 1), a DNA binding protein with an AT-rich domain, through a two-hybrid screening approach using the SymRK kinase domain as bait (Zhu et al., 2008). SIP1 specifically binds to the promoter of NIN in L. japonicus. The ligands of the LRR domain and phosphorylation targets of SymRK protein kinase are still unknown.
A mitogen-activated protein kinase (MAPK) cascade is likely to be a downstream target of the NF signaling pathway. MAPKs are phosphorylated and activated by MAPK kinases (MAPKKs), which themselves are activated by MAPKK kinases (MAPKKKs) (Davis et al., 2000; Chang and Karin, 2001; Jonak et al., 2002). Signaling through MAPK cascades can lead to cellular responses, including cell division, cell differentiation, and physiological adjustments to various stresses (Mishra et al., 2006; Colcombet and Hirt, 2008; Pitzschke et al., 2009). In plants, MAPKs can be organized into a complex network for efficient transmission of specific stimuli (Mishra et al., 2006). Targets of MAPKs can be various transcription factors and protein kinases as well as upstream components of the MAPK cascade, such as MAPKKs, MAPKKKs, or the receptors themselves (Karin, 1998; Whitmarsh and Davis, 1998; Cardinale et al., 2002). The Arabidopsis thaliana genome contains ~110 genes coding for the MAPK cascade components: 20 MAPKs, 10 MAPKKs, and more than 80 MAPKKKs (MAPK Group, 2002; Pitzschke et al., 2009). Large-scale proteomic analysis using protein chips has suggested that a total of 570 protein targets are phosphorylated by 10 MAPKs in Arabidopsis. These substrates are enriched in transcription factors implicated in the regulation of development, defense, and stress responses (Popescu et al., 2009) Phosphorylation can alter transcription factor activity in several ways, including via changes in protein turnover, transcriptional activity, subcellular localization, and ability to interact with other proteins or nucleic acids (Fiil et al., 2009).
There have been only a few MAPK cascade components that have been studied in detail in plants (Pitzschke et al., 2009). Compared with MAPKKKs and MAPKs, MAPKKs serve as the key point in MAPK cascades (Kong et al., 2011). Plant MAPKKs can be divided into four groups based on their sequence similarity (MAPK Group, 2002; Hamel et al., 2006). The Arabidopsis MKK1/MKK2-MPK4/MPK6 cascades have previously been shown to play important roles in the responses to salt and cold stresses and pathogen attack (Teige et al., 2004; Meszáros et al., 2006; Gao et al., 2008; Qiu et al., 2008b). Arabidopsis MKK3 participates in signaling cascades elicited by pathogen infection (Dóczi et al., 2007; Takahashi et al., 2007). The Arabidopsis MKK4/MKK5-MPK3/MPK6 cascades play important roles in the regulation of plant development and biotic stress (Asai et al., 2002; Wang et al., 2007). The Arabidopsis MKK6-MPK4/MPK11 cascades directly regulate cytokinesis and mitosis (Beck et al., 2010, 2011; Takahashi et al., 2010). MKK7 leads to activation of plant basal and systemic-acquired resistance in Arabidopsis (Zhang et al., 2007). The MKK9-MPK3/MPK6 cascades regulate ethylene signaling and camalexin biosynthesis and may also play a role in leaf senescence (Xu et al., 2008; Yoo et al., 2008; Zhou et al., 2009).
In this study, we identified L. japonicus SymRK-interacting protein 2 (SIP2) as an interacting partner of the kinase domain of SymRK in vitro and in planta. SIP2 is a MAPKK and its kinase activity was negatively regulated by SymRK. Our results obtained from SIP2 RNA interference (RNAi) knockdown in plants strongly suggest that SIP2 may participate in the regulation of early symbiotic signal transduction and nodule organogenesis in L. japonicus.
RESULTS
SIP2 Is a MAPKK
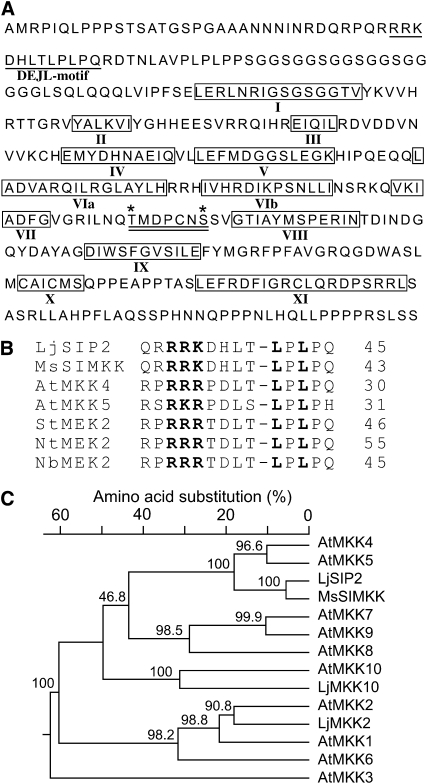
In search of potential interacting proteins of SymRK, we identified SIP2 from a library screen in the yeast two-hybrid system. Protein BLAST analysis revealed that SIP2 belongs to the plant MAPKK family (see Supplemental Figure 1 online). The full-length cDNA of SIP2 contained an open reading frame of 1122 nucleotides encoding a peptide of 373 amino acid residues with the predicated molecular mass of 41 kD. The protein contains typical domain structures of MAPKKs (Figure 1A), including 11 catalytic kinase subdomains (Hanks and Hunter, 1995), two putative phosphorylation sites, and an activation loop (Kiegerl et al., 2000; Cardinale et al., 2002). Plant MAPKKs including SIP2 have an activation loop with a consensus of S/TXXXXXS/T (Figure 1A, double underlined), which is slightly different from the S/TXXXS/T found in the mammalian enzymes (MAPK Group, 2002). The activation loop of MAPKKs is thought to be a putative target of an upstream MAPKKK (Cardinale et al., 2002). As shown in Figure 1B, the N terminus of SIP2 contains a DEJL motif -K/R-K/R-K/R-X (1-5)-L/I-X-L/I, which is known to function as a MAPK docking site in mammals and is conserved in animal and yeast proteins (Bardwell et al., 1996; Jacobs et al., 1999). Based on the phylogenetic tree analysis, SIP2 is closest to At-MKK4 and At-MKK5 from Arabidopsis (Figure 1C) and can be grouped with the Arabidopsis Group C MKKs. Both SymRK and SIP2 are protein kinases. In order to test if the kinase activity is required for their interaction, we generated kinase-negative mutants of both protein kinases by replacing the essential Lys (K) residue with Arg (R) at the ATP binding site (Figure 2A). This Lys-to-Arg substitution (SymRK-PK-KR and SIP2-KR) abolished the kinase activity completely (Figure 3C). However, it did not affect the interaction between SymRK and SIP2. As shown in Figure 2A, the kinase-negative SymRK-PK-KR interacted with SIP2, and vice versa, the kinase-negative SIP2-KR also interacted with SymRK-PK as efficiently as their corresponding wild-type proteins did, as assessed by the quantitative β-galactosidase activity assays (see Supplemental Figure 2 online). This suggests that the kinase activities of both SymRK and SIP2 are not required for the physical interaction of the two proteins.
Figure 1.
SIP2 Is a MAPKK from L. japonicus.
(A) Deduced amino acid sequence of SIP2. The conserved DEJL motif is underlined. The catalytic subdomains I to XI of protein kinases are boxed and indicated by Roman numbers. Two putative phosphorylation sites of Thr and Ser are marked with asterisks. The activation loop, which comprises the two phosphorylation residues and is a putative target of upstream MAPKKKs, is underlined twice.
(B) The highly conserved DEJL motif of L. japonicus SIP2 and Arabidopsis MAPKKs. It has a consensus of K/R-K/R-K/R-X(1-5)-L/I-X-L/I and is designated for its putative role as the docking site for mammalian ERK and JNK MAPKs or LXL. The highly conserved Leu-X-Leu/Ile (LXL) motif provides a hydrophobic site for binding with MAPKs via hydrophobic interaction. The basic residues (RRK) within the DEJL motif interact with acidic residues of MAPKs via salt bridges. The last residue in each protein is indicated by a number.
(C) Phylogenetic tree of L. japonicus SIP2, MAPKK2, and MAPKK10, M. sativa SIMKK, and Arabidopsis MAPKK family members. SIP2 is most closely related to At-MKK4 and At-MKK5. Multiple sequence alignment and neighbor-joining phylogenetic analysis were performed using DNAStar software. Bootstrap values (%) obtained from 1000 trials are given at branch nodes. The alignment used to generate this tree is available as Supplemental Data Set 1 online.
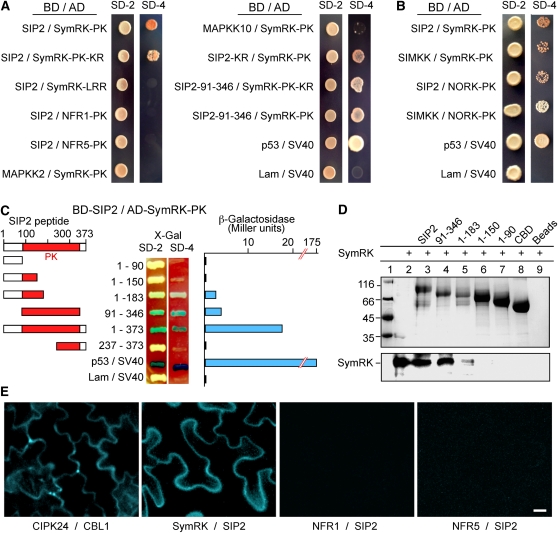
Figure 2.
Interaction of SIP2 with SymRK in Vitro and in Planta.
(A) SIP2 interacts with SymRK in yeast cells. Yeast Y187 cells carrying the Gal4 DNA binding domain (BD) fusion constructs were mated with yeast AH109 cells harboring the Gal4 activation domain (AD) fusion constructs. The diploid cells were selected on SD media lacking Leu and Trp (SD-2), and interaction was assessed according to their ability to grow on selective SD media lacking Leu, Trp, His, and Ade (SD-4) for 5 d. SymRK-LRR, the extracellular LRR region of SymRK; SymRK-PK, the intracellular PK domain of SymRK; NFR1-PK, the intracellular PK domain of NFR1; NFR5-PK, the intracellular PK domain of NFR5; SIP2-KR, a Lys-to-Arg substitution kinase-negative mutant of SIP2; SIP2-91-346, the PK domain of SIP2. Note that SIP2 did not interact with the putative NF receptors (NFR1 and NFR5), and SymRK did not interact with Lj-MAPKK2 and Lj-MKK10. SIP2 did interact with the SymRK ortholog (NORK) from alfalfa, while SymRK also interacted with the SIP2 ortholog (SIMKK) from alfalfa. The interaction between mammalian p53 and SV40 served as a positive control, whereas coexpression of lamin (Lam) and SV40 served as a negative control.
(B) Interactions between SymRK and SIP2 orthologs from Lotus and Medicago. Lotus SymRK interacted with the SIP2 ortholog, SIMKK, from alfalfa, and similarly, Lotus SIP2 interacted with the SymRK ortholog, NORK, from alfalfa. The interaction between alfalfa SIMKK and NORK-PK was relatively weak, and the yeast colonies were selected on SD/-Leu/-Trp/-Ade media, followed by selection on SD/-Leu/-Trp/-Ade/-His.
(C) The PK domain of SIP2 is responsible for interaction with SymRK. SIP2 and its truncated constructs were expressed as fusion proteins with the GAL4 binding domain (BD) in pGBKT7. SymRK-PK was expressed as a recombinant protein fused with the GAL4 activation domain (AD) in pGADT7. Yeast cells containing both plasmids were grown on SD-Leu-Trp medium (SD-2) containing X-Gal (80 mg/L) and assessed for interactions on SD-Leu-Trp-His-Ade medium (SD-4) containing X-Gal. The strength of interaction was evaluated by assaying β-galactosidase activities in soluble extracts prepared from yeast colonies.
(D) In vitro protein pull-down assay for the interaction between SIP2 and SymRK-PK. CBD-tagged SIP2 and four of its truncated products (numbers indicate residues of SIP2) were absorbed to chitin beads and mixed with purified soluble SymRK-PK. After washing, proteins pulled down by the chitin beads were separated via SDS-PAGE and visualized with Coomassie blue dye (top). The same gel was used for immunoblotting with anti-SymRK antibody (bottom). Note that the full-length SIP2 and its PK domain (residues 91 to 346) could pull down SymRK, and the N-terminal half (1 to 183) of SIP2 also could interact with SymRK with a reduced affinity.
(E) BiFC on the interaction between SIP2 and SymRK in planta. N. benthamiana leaves were cotransformed with SCFPC155:SymRK-full and SIP2-full:SCFPN173 (SymRK / SIP2). Leaf epidermal cells were observed via fluorescence microscopy. Because the CFP is split into N- and C-terminal halves and fused with different proteins, no cyan fluorescence would be observed if the fusion proteins did not interact. As a positive control, Arabidopsis CIPK24 (CBL-interacting protein kinase 24) and calcineurin B-like 1 (CBL1) were fused with the C-terminal CFPC155 and N-terminal CFPN173, respectively, and coexpressed. Note that no interactions were observed between NFR1 and SIP2, or NFR5 and SIP2 using similar fusion approach. Bar = 20 μm.
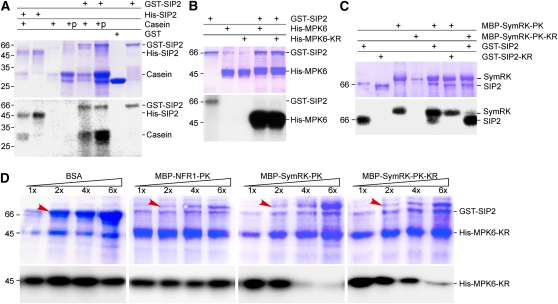
Figure 3.
In Vitro Kinase Activity of SIP2.
(A) Protein kinase activity of SIP2. SIP2 was expressed as a GST fusion protein in vector pGEX-6P1. GST alone served as a control. SIP2 was also expressed as a His tag fusion protein in pET28a. Affinity-purified GST, GST-SIP2, and His-SIP2 were incubated with casein or calf intestinal phosphatase–treated casein in the presence of [γ-32P]ATP. The reaction products were analyzed in SDS-PAGE. The gel was stained with Coomassie blue (top), dried, and subjected to autoradiography (bottom). Molecular mass standard (kD) is shown on left side.
(B) Phosphorylation of MPK6 MAPK by SIP2. Purified GST-SIP2 was incubated with Arabidopsis MPK6 or its kinase-negative mutant, MPK6-KR, in the presence of [γ-32P]ATP. Kinase reaction products were resolved on SDS-PAGE. The gel was stained with Coomassie blue (top) and subjected to autoradiography (bottom).
(C) SIP2 and SymRK did not phosphorylate each other. Because both SIP2 and SymRK could undergo autophosphorylation, their kinase-negative mutants were used as substrates. SIP2 and its kinase-negative mutant were purified on GST beads, whereas SymRK and its kinase-negative mutant were isolated via the MBP tag using chitin beads. In the presence of [γ-32P]ATP, the kinase-negative SymRK-PK-KR was not phosphorylated by SIP2. Similarly, the kinase-negative SIP2-KR was not phosphorylated by SymRK-PK.
(D) SymRK inhibited the phosphorylation of MPK6 by SIP2. The kinase-negative MPK6-KR was purified on Ni beads and used as substrate for SIP2 in the presence of [γ-32P]ATP. An increasing amount of SymRK-PK or its kinase-negative SymRK-PK-KR was added to the reaction mix. Increasing amounts of BSA and purified NFR1-PK were used to test the specificity of inhibition. The base amount (1×) of effector proteins (BSA, NFR1, and SymRK) was 1.0 μg per reaction, and the increasing amounts are indicated as relative quantities (2× to 6×). The bands corresponding to the increased amount of added proteins are labeled by arrows. The reaction products were analyzed on SDS-PAGE gels stained with Coomassie blue (top). Autoradiographs show the corresponding phosphorylated MPK6-KR (bottom).
[See online article for color version of this figure.]
The Interaction between SymRK and SIP2 Is Specific and Occurs in Planta
We asked if SIP2 also interacted with other receptor-like kinases implicated in the NF perception. NFR1 and NFR5 contain a LysM domain in the extracellular domain and have been shown to function as the NF receptors in L. japonicus (Madsen et al., 2003; Radutoiu et al., 2003). Our results show that SIP2 did not interact with the PK domain of NFR1 and NFR5 (Figure 2A). We then asked if SymRK also interacted with other MAPKKs from L. japonicus. To test this, we needed to clone MAPKK cDNAs from L. japonicus. From the available genome sequencing database, we found cDNA sequence information for three potential full-length MAPKKs. We designed three pairs of primers accordingly and performed RT-PCR reactions using total RNA from roots and nodules. We were able to amplify and clone two of them. Based on the homology of the deduced peptide sequences with the Arabidopsis MAPKK family members, we designated them as MAPKK2 and MAKK10 (Figure 1C) and tested them for any potential interaction with SymRK. Our results showed that MAPKK2 did not interact with SymRK-PK at all in yeast (Figure 2A). The interaction between MAPKK10 and SymRK was very weak in the yeast colony growth assay (Figure 2A) and was negligible when assessed by the quantitative β-galactosidase activity measurement (see Supplemental Figure 2 online). We also performed protein gel blot analysis on the yeast cells to assure that all fusion proteins were expressed properly (see Supplemental Figure 3 online). These results indicate that there is a specific interaction between SymRK and SIP2.
To determine if the interaction between SymRK and SIP2 is conserved among legumes, we cloned orthologs of SymRK and SIP2 from alfalfa (Medicago sativa). NORK from alfalfa is the ortholog of Lotus SymRK, whereas SIMKK from alfalfa is the closest homolog of Lotus SIP2 (having 88% similarity at the amino acid level). Our results showed that the protein kinase domain of Ms-NORK (NORK-PK) indeed interacted with Lj-SIP2 and Ms-SIMKK also interacted with SymRK-PK (Figure 2B; see Supplemental Figures 2 and 3 online). Like the partnership in Lotus, Ms-NORK-PK and Ms-SIMKK from alfalfa also interacted with each other, although the interaction was weaker than that between Lj-SymRK and Lj-SIP2 (Figure 2B; see Supplemental Figure 2 online). These results demonstrate that similar interactions between SIP2 and SymRK orthologs may also be conserved in other legumes and may play an essential role in early signaling during nodule organogenesis.
To confirm further the occurrence of this interaction in planta, we coexpressed SymRK and SIP2 in plant cells as recombinant proteins fused with split cyan fluorescent protein (CFP) tags. If the two proteins of interest interacted with each other in plant cells, the interaction would reconstitute the CFP and lead to cyan fluorescence in transgenic cells, a technique known as bimolecular fluorescence complementation (BiFC; Hu et al., 2002; Hu and Kerppola, 2003; Walter et al., 2004; Kim et al., 2007; Waadt et al., 2008). For this, we constructed plasmids that would express a fusion protein of the C-terminal split CFP with SymRK (SCFPC155:SymRK) and a fusion protein between SIP2 and the N-terminal CFP (SIP2:SCFPN173). The constructs were coexpressed in Nicotiana benthamiana epidermis leaves via Agrobacterium tumefaciens–mediated transient transformation. Leaf cells were analyzed 2 to 3 d after Agrobacterium infiltration. Using a confocal laser scanning microscope, we observed strong cyan fluorescent signals in the plasma membrane and the cytoplasm of leaf cells, suggesting that SymRK and SIP2 interact with each other in plant cells (Figure 2E). We also constructed plasmids that would express NFR1 and NFR5 fused with the C-terminal split CFP (SCFPC155:NFR1 and SCFPC155:NFR5). We coexpressed these constructs with the SIP2 plasmid in N. benthamiana leaf cells. The expression of these fusion proteins was confirmed by immunoblot analyses. We did not detect any cyan fluorescence (Figure 2E), suggesting that SIP2 does not interact with NF receptors NFR1 and NFR5 in planta. The expression of the fusion proteins was confirmed by immunoblot analyses using monoclonal antibodies against the hemagglutinin (HA) and FLAG tags present in the fusion proteins (see Supplemental Figure 4 online). These observations demonstrate that SIP2 and SymRK interact with each other in plant cells, and the interaction does not involve NFR1 and NFR5.
The Kinase Domain of SIP2 Is Responsible for Its Interaction with SymRK
In order to characterize the structural requirements of the interaction between SIP2 and SymRK, SIP2 and five truncated derivatives of it were fused to the GAL4 activation domain and analyzed for potential interactions with the SymRK protein kinase domain (SymRK-PK) (Figure 2C). The SIP2 constructs included a full-length SIP2 (residues 1 to 373, SIP21-373), an N terminus with a MAPK docking site (residues 1 to 90, SIP21-90), an N terminus supplemented with catalytic subdomains I and III (residues 1 to 150, SIP21-150), an N terminus supplemented with catalytic subdomains I to V (residues 1 to 183, SIP21-183), a complete kinase domain (residues 91 to 346, SIP291-346), and a C terminus supplemented with catalytic subdomains VIII to XI (residues 237 to 373, SIP2237-373). We observed positive interactions of SymRK-PK with SIP21-373, SIP291-346, and SIP21-183 (Figure 2C). These interactions were further confirmed by the detection of high levels of β-galactosidase activities using yeast cell extracts (Figure 2C). Among the three constructs, the full-length SIP21-373 exhibited the strongest interaction with SymRK-PK. Although SIP291-346 and SIP21-183 had weaker interactions with SymRK-PK in the colony growth assay, the interactions were repeatedly detectable using the β-galactosidase activity assay (Figure 2C). The remaining three truncated SIP2 constructs did not show an interaction with SymRK-PK in either colony growth tests or assays of β-galactosidase activity measurement. The expression of fusion proteins in yeast cells was confirmed by immunoblot analysis (see Supplemental Figure 3 online). These results indicate that SIP2 interacts with SymRK-PK and that the kinase domain in SIP2 is critical for the interaction. The noncatalytic N terminus and catalytic C terminus alone did not interact with SymRK-PK. However, the deletion of the noncatalytic N terminus and C terminus of SIP2 decreased the strength of interaction, whereas a noncatalytic N terminus supplemented with catalytic subdomains I to V promoted the interaction with SymRK. These data suggest that both the N and C termini contribute to the interaction of SIP2 with SymRK. These results are consistent with previous reports showing that SIMKK can interact with SIMK in alfalfa and the N-terminal MAPK docking site of SIMKK contributes, but is not essential for, to the interaction with SIMK (Kiegerl et al., 2000).
We further verified the interactions between SIP2 constructs and SymRK-PK using an in vitro protein–protein pull-down assay. The SIP2 constructs were fused to a chitin binding domain (CBD), which was immobilized to chitin beads. After incubating the beads with purified SymRK-PK, proteins retained on the beads were separated via SDS-PAGE. The SymRK bound to the beads was detected by immunoblotting with anti-SymRK antibody. SIP21-373, SIP291-346, and SIP21-183 could pull down SymRK-PK, and the weakest interaction was with SIP21-183 (Figure 2D). We conclude that SIP2 interacts with SymRK in vitro and the kinase domain of SIP2 is responsible for the interaction with SymRK.
SIP2 Is a Functional Protein Kinase
We expressed recombinant SIP2 proteins tagged with glutathione S-transferase (GST) or poly-His (HIS) in pGEX-6P1 and pET28a vectors. The recombinant proteins were expressed in Escherichia coli cells and purified using appropriate affinity beads. Purified GST-SIP2 and HIS-SIP2 proteins were subjected to in vitro kinase assays using casein as a substrate in the presence of [γ-32P]ATP. Phosphorylated proteins were visualized by autoradiography (Figure 3A). As a MAPKK, SIP2 was able to autophosphorylate itself, suggesting that the tags did not disrupt the kinase activity. In addition to the autophosphorylation band, an additional band corresponding to casein was observed, suggesting that SIP2 can also transphosphorylate a protein substrate. Neither the GST nor the HIS tag appeared to affect the transphosphorylation activity of SIP2.
In a typical MAPK cascade, a MAPKK activates a specific MAPK(s). Because there is more information on the members and their relationships for MAPKs in Arabidopsis than in Lotus, we selected two Arabidopsis MAPK representatives, MPK3 and MPK6, for testing phosphorylation by SIP2. We first tested their potential interaction with SIP2 in the yeast two-hybrid system. Our result showed that SIP2 interacted with At-MPK6 but not At-MPK3 (see Supplemental Figures 2, 3, and 5 online). At-MPK6 appears to be a less specific substrate than other MAPKs and can be used as a substrate for phosphorylation by a wider set of MAPKKs, including MKK2 (Teige et al., 2004), MKK3 (Takahashi et al., 2007), MKK4, MKK5 (Asai et al., 2002), and MKK9 (Yoo et al., 2008). To determine if MPK6 could serve as a substrate of SIP2, we created a kinase-negative form of MPK6 (MPK6-KR) by replacing the absolutely conserved Lys residue with Arg in the ATP binding site (Kiegerl et al., 2000; Zhou et al., 2009). We used both MPK6 and MPK6-KR as substrates for in vitro kinase assays, and our results showed that both of them could be phosphorylated strongly by SIP2 (Figure 3B). Thus, SIP2 represents a typical MAPKK and can use MPK6 as a substrate for phosphorylation.
SymRK Is Not Phosphorylated by SIP2
SymRK is a Ser/Thr protein kinase with both autophosphorylation and transphosphorylation activities (Yoshida and Parniske, 2005). To test if SymRK could be phosphorylated by SIP2, the kinase activity of SymRK had to be abolished. We generated a kinase-negative SymRK SymRK-PK-KR (K622R) mutant through site-directed mutagenesis. GST-tagged SymRK-PK, maltose binding protein (MBP)-tagged SymRK-PK-KR was used as substrate for testing the GST-tagged SIP2 kinase (Figure 3C). The results showed that SIP2 could not phosphorylate SymRK-PK-KR. We further asked if SIP2 could be phosphorylated by SymRK. For testing this, the autophosphorylation activity of SIP2 had to be abolished. We generated a kinase-negative mutant, SIP2-KR (K120R), as a GST-tagged recombinant protein. As shown in Figure 3C, both Lys-to-Arg mutants of SIP2 and SymRK (i.e., SIP2-KR and SymRK-PK-KR) had no kinase activity and exhibited no autophosphorylation (Figure 3C). We used MBP-tagged SymRK as the kinase source. As shown in Figure 3C, SymRK could autophosphorylate itself but failed to phosphorylate the kinase-negative SIP2-KR, suggesting that SymRK is neither a potential phosphorylation target nor a kinase source of SIP2.
SymRK Is an Inhibitor of SIP2 Kinase
We then asked if the interaction between SymRK and SIP2 would affect the kinase activity of SIP2. To test this, we added an increasing amount of either kinase-active (SymRK-PK) or kinase-negative SymRK (SymRK-PK-KR) as an effector to the kinase-active SIP2 assays in the presence of MPK6-KR or casein as a substrate. The results showed that the kinase activity of SIP2 decreased with the increase in SymRK in a dose-dependent manner (Figure 3D; see Supplemental Figure 6 online). Both kinase-active and kinase-negative SymRKs had similar inhibitory effect on SIP2 kinase activity. To eliminate any potential nonspecific effect of proteins in the reaction mix, we added an increasing amount of BSA to the SIP2 kinase assays and did not observe any inhibitory effect of BSA on the SIP2 kinase activity (Figure 3D). We also tested if NFR1 might have any effect on the SIP2 kinase activity. We did not observe any inhibitory effect of NFR1 on the SIP2 kinase activity, suggesting that the inhibitory effect of SymRK is specific. We conclude that SymRK is not a substrate of SIP2 but has a specific inhibitory effect on the kinase activity of SIP2 toward the MPK6 substrate in vitro.
Constitutive Expression of SIP2
Lotus SymRK is constitutively expressed in roots, and its expression level is not affected by treatment with NF or inoculation with Mesorhizobium loti during the first 48 h (Stracke et al., 2002). A different report using real-time PCR suggests that Rhizobium inoculation for 12 d may raise the expression level of SymRK in the infected roots (Radutoiu et al., 2003). In this study, young roots were harvested at different time points after inoculation with M. loti MAFF303099, and the expression levels of SIP2 were measured using real-time PCR. We did not observe significant changes in SIP2 mRNA level in 12 d after inoculation with M. loti MAFF303099 (Figure 4A). We also asked if there would be any tissue-specific expression of the SIP2 gene. Our results showed that SIP2 was expressed in all tissues tested, including roots, stem, leaves, and nodules (Figure 4A). It is likely that SIP2 may have other roles in the development of L. japonicus plants in addition to nodulation.
Figure 4.
Gene Expression and Subcellular Localization of SIP2.
(A) Expression of SIP2 mRNA in L. japonicus. Roots were harvested 1, 2, 4, 6, 8, 10, and 12 d after inoculation with M. loti. Roots treated with water (uninoculated roots) were also harvested at the same time intervals and served as the mock control. Rhizobium-inoculated roots (IR), stems (S), and leaves (L) were harvested 8 d after inoculation, while nodules (N) were collected 42 d after inoculation. Total RNA was isolated and used for real-time PCR to measure the expression levels of the SIP2 mRNA. The ATPase gene (AW719841) was used as an internal control. Error bars represent sd of the experimental values obtained from three technical replicates.
(B) Subcellular localization of SIP2 in L. japonicus hairy roots. SIP2 was expressed as a GFP fusion protein (GFP:SIP2) under the control of the CaMV35S promoter in L. japonicus hairy roots induced by A. rhizogenes LBA1334. GFP alone served as a control. Bars = 20 μm.
(C) Subcellular localization of GFP-tagged SIP2 in onion cells. SIP2 was expressed as a fusion protein with GFP (GFP:SIP2) under the control of the 35S promoter. Plasmid expressing GFP alone served as a control. The plasmids were delivered to the onion epidermal cells via particle bombardment, and fluorescence images were taken using a confocal laser scanning microscope. Onion epidermal cells expressing GFP:SIP2 were treated with 4% NaCl for 5 min to induce plasmolysis before imaging. Green fluorescence (left) and the corresponding bright-field images (right) were taken using a confocal laser scanning microscope. The overlay images (middle) were produced using Photoshop software. Bars = 50 μm.
Subcellular Localization of SIP2 Protein
We constructed a plasmid that would express a green fluorescent protein (GFP)–tagged SIP2 fusion protein under the control of the cauliflower mosaic virus 35S (CaMV35S) promoter. The plasmid was delivered to onion epidermal cells via particle bombardment. GFP alone, which was distributed in the cytoplasm and nucleus, was used as a control. In cells expressing GFP-tagged SIP2, the green fluorescence signal was observed in the plasma membrane and cytoplasm but was excluded from the nucleus (Figure 4C). Similar results were also observed in Lotus hairy roots expressing GFP:SIP2 through Agrobacterium rhizogenes LBA1334-mediated transformation (Figure 4B). In order to ensure a clear identification of the cell wall versus the cytoplasm, we treated onion epidermal cells with 4% NaCl for 5 min to induce plasmolysis right before microscopy observation. The fluorescence signal was localized to the cytoplasm and plasma membrane. We conclude that the SIP2 protein is localized to the plasma membrane and cytoplasm.
Impairment of Nodulation by SIP2 Knockdown Expression via RNAi
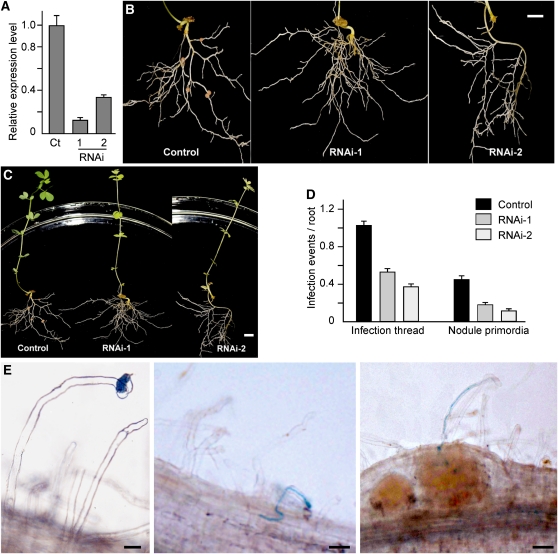
We generated transgenic L. japonicus hairy roots that had a downregulated SIP2 expression level via RNAi. To induce the formation of hairy roots, hypocotyls of L. japonicus seedlings were treated with A. rhizogenes LBA1334 containing appropriate plasmids (see Methods). Transgenic hairy roots were identified and selected based on β-glucuronidase (GUS) staining. The normal root system and the nontransgenic hairy roots were excised from the L. japonicus seedlings. For SIP2 knockdown expression, a fragment of SIP2 cDNA including 111 bp of coding sequence and either 129 bp of 3′-untranslated region (RNAi-1) or 309 bp of 3′-untranslated region (RNAi-2) was selected to generate SIP2 RNAi constructs (see Supplemental Figure 7 online). The two RNAi constructs were used to generate L. japonicus hairy roots via A. rhizogenes LBA1334 infection. The suppression of SIP2 expression was confirmed using real-time PCR amplification of a region in the SIP2 mRNA. Compared with the control hairy roots generated using the empty vector, hairy roots expressing SIP2 RNAi had downregulated SIP2 mRNA levels in ~70% of transgenic hairy roots. The hairy roots were inoculated with M. loti MAFF303099 to induce nodule formation, and their nodulation phenotypes were analyzed 4 weeks after inoculation. The average nodule number per root was 4.4 and 3.8 in SIP2 RNAi-1 and RNAi-2 plants, respectively, which were significantly lower than that of the control hairy roots (8.2; see Supplemental Table 1 online). We observed that 21.4% (22/103) of SIP2-RNAi-1 roots and 37.9% (22/58) of SIP2-RNAi-2 roots did not develop any nodules (Figure 5B). Analysis using real-time RT-PCR revealed that the SIP2 mRNA levels in the RNAi hairy roots was significantly lower than that in the control hairy roots, suggesting a correlation between the observed nodulation phenotypes and the SIP2 expression levels (Figure 5A; see Supplemental Figure 8A online). These data clearly suggest that the knockdown expression of SIP2 via RNAi has a deleterious effect on nodule formation.
Figure 5.
Expression of SIP2-RNAi in L. japonicus Hairy Roots.
(A) Real-time RT-PCR analysis of SIP2 expression levels in the control hairy roots expressing the empty vector (Ct) and representatives of hairy roots expressing the SIP2-RNAi constructs (RNAi-1 and RNAi-2). Error bars represent sd of the experimental values obtained from three technical replicates.
(B) The control hairy roots expressing the empty vector and representative hairy roots expressing the SIP2-RNAi constructs (RNAi-1 and RNAi-2). L. japonicus plants were maintained in a nitrogen fertilizer-free environment and root images were taken 4 weeks after Rhizobium infection. Bar = 1 cm.
(C) Images of whole plants whose roots were shown in (B). Bar = 1 cm.
(D) Numbers of ITs and nodule primordia per root of the control hairy roots and SIP2-RNAi-1 and SIP2-RNAi-2 hairy roots. The data on ITs and nodule primordia were collected from 15 to 20 independent hairy roots in each group of plants. Error bars represent sd of the experimental values.
(E) IT formation in SIP2-RNAi hairy roots. ITs were visualized after staining with X-Gal in hairy roots 8 d after infection with M. loti expressing a lacZ construct. The images show the key steps of Rhizobium infection in hairy roots, including bacterial entry and root hair deformation (left), IT growth (middle), and release of rhizobial cells in developing nodule cells (right). Bars = 20 μm in left and middle panels and 50 μm in the right panel.
Suppression of Infection Thread Formation and Nodule Initiation by SIP2 RNAi
To address the possible involvement of SIP2 in the early infection process, we examined the formation of infection threads (ITs) in the hairy roots expressing SIP2 RNAi. An M. loti strain labeled with lacZ (Tansengco et al., 2003; Kumagai et al., 2006; Kang et al., 2011) was used to infect hairy roots. In SIP2 RNAi hairy roots, the initiation of ITs and IT growth could still be observed (Figure 5E). Some ITs could even reach the nodule primordia, followed by the rhizobial release from ITs into the nodule cells (Figure 5E). Compared with the control hairy roots (~1.0 infection event per root segment), the SIP2 RNAi roots produced a reduced number of ITs, having only 0.55 and 0.38 infection events per root segment in RNAi-1 and RNAi-2 roots, respectively (Figure 5D). The number of ITs that reached to the nodule primordia was also significantly reduced in the SIP2 RNAi-roots (Figure 5D). Thus, IT formation appeared to be impaired by the knockdown expression of SIP2 in RNAi hairy roots.
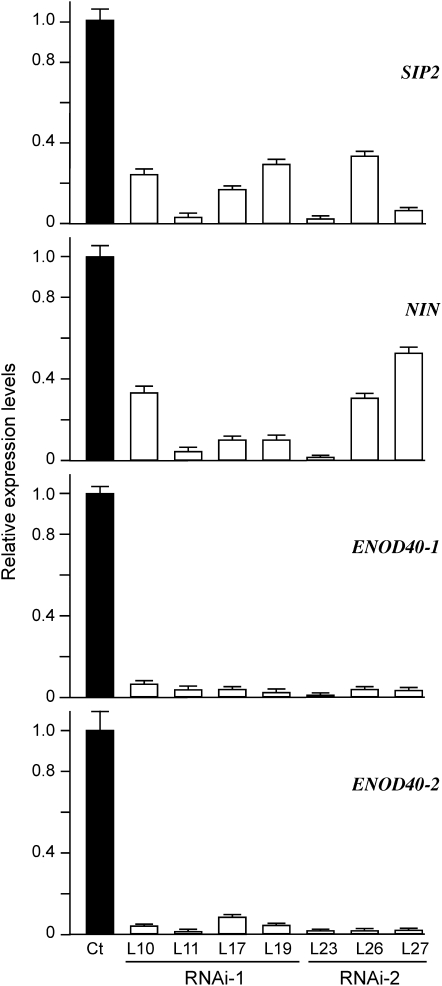
We also analyzed the expression levels of marker genes associated with the early nodule formation in SIP2 RNAi hairy roots. For this analysis, we chose the NIN gene, which is required for IT formation and nodule primordia initiation (Schauser et al., 1999), and ENOD40-1 and ENOD40-2, which are expressed in the early stage of bacterial infection and in mature nodules, respectively (Kumagai et al., 2006). Representative hairy roots expressing SIP2 RNAi-1 and RNAi-2 were selected, and the suppression level of SIP2 expression was also measured via real-time PCR (Figure 6). An average of 80 and 63% downregulation in SIP2 mRNA was observed in SIP2 RNAi-1 and RNAi-2 hairy roots, respectively (Figure 5A). To assure that the SIP2 RNAi did not nonspecifically target other MAPKKs, we measured the expression levels of two other MAPKK genes by real-time PCR. The mRNA levels of both MAPKK2 and MAPKK10 in these independent nodule-deficient hairy roots were not significantly different from the control (see Supplemental Figure 9 online), suggesting that the SIP2 RNAi constructs specifically target SIP2 and do not affect the expression of other MAPKK genes. Correlated to the downregulation of SIP2, the transcript levels of NIN, ENOD40-1, and ENOD40-2 were all lowered compared with those of the control hairy roots (Figure 6). These observations suggest that the downregulated SIP2 expression in the RNAi hairy roots has repressive effects on the expression of other nodulation-related genes, possibly through which IT initiation and nodule formation were impaired.
Figure 6.
Suppression of Marker Genes Related to IT Formation and Nodule Organogenesis.
Total RNA was isolated from the control hairy roots (Ct) expressing the empty vector and representative plants expressing SIP2-RNAi-1 (L10, L11, L17, and L19) and SIP2-RNAi-2 (L23, L26, and L27). Real-time RT-PCR analysis was performed to assess the expression levels of SIP2, NIN, ENOD40-1, and ENOD40-2 genes. Error bars represent sd of the experimental values obtained from three technical replicates.
SymRK participates in the symbiosis signaling pathway, which mediates early root responses to infection by both Rhizobium and arbuscular mycorrhizal fungi (Kistner et al., 2005; Parniske, 2008). We asked if the SymRK-SIP2 interaction played a role in mycorrhization of legume roots. We infected the SIP2-RNAi hairy roots with Glomus intraradices, a common arbuscular mycorrhizal fungus. Two weeks after inoculation, the fungus penetrated into the outer cell layers, colonized the root cortex, and formed arbuscules. There was no observable difference in the efficiency of hyphal and arbuscular colonization between SIP2-RNAi hairy roots and the control hairy roots expressing the empty vector (see Supplemental Figure 10 online). Therefore, we conclude that the SymRK-SIP2 interaction is specific for the root response to Rhizobium infection and is not required for arbuscular mycorrhizal colonization.
In addition to the drastic reduction in IT formation and nodule primordial initiation in the SIP2 RNAi hairy roots, a significant portion of the transgenic hairy roots, 21.4% in SIP2 RNAi-1 and 37.9% of SIP2 RNAi-2, failed to develop any nodule at all. This observation, together with the data on the suppression of the three nodulation marker genes, clearly demonstrates a pivotal role of SIP2 in IT formation and nodule primordium initiation in L. japonicus. Thus, SIP2 may represent a previously undescribed and crucial regulatory pathway required for the establishment of a symbiotic relationship in legumes.
DISCUSSION
SymRK is a member of the LRR RLK family of receptor-like kinases in plants and is required for the recognition of both arbuscular mycorrhizal fungi and nitrogen-fixing rhizobial bacteria. In the Rhizobium-legume symbiosis, SymRK is required for the early signal transduction pathway from the NF perception to the rapid symbiosis-related gene activation (Stracke et al., 2002; Radutoiu et al., 2003; Yoshida and Parniske, 2005). SymRK is essential for nodulation in L. japonicus and occupies an enigmatic position in the NF signaling pathways. However, little is known about how it is linked to downstream responses. In this study, we describe SIP2, a SymRK-interacting protein and provide evidence showing that SIP2 plays an important role in the regulation of early symbiotic signal transduction and nodule organogenesis in L. japonicus. We also showed that the interaction between SymRK and SIP2 is not involved in arbuscular mycorrhizal colonization.
The SIP2 gene was identified in a yeast two-hybrid screening of a cDNA library constructed using mRNAs isolated from L. japonicus roots 2 to 12 d after inoculation with Rhizobium. The SIP2 protein was shown to interact with the PK domain of SymRK. The interaction between SIP2 and SymRK-PK was verified in vitro and in planta (Figures 2A, 2D, and 2E). SymRK is expressed constitutively in the roots of L. japonicus (Stracke et al., 2002), and its ortholog gene of the water-tolerant legume Sesbania rostrata is detected in the leaves, flowers, and uninfected adventitious root primordia (Capoen et al., 2005). Our data on SIP2 gene expression showed that it is expressed in all tissues examined, including stems, leaves, Rhizobium-inoculated roots, uninoculated roots, and nodules in L. japonicus. SymRK is localized in the plasma membrane and IT membranes (Capoen et al., 2005; Limpens et al., 2005). Similarly, SIP2 is also localized at the plasma membrane and cytoplasm (Figure 4B). The overlap in gene expression and subcellular localization suggests that the two proteins could have both temporal and spatial opportunities to interact with each other in planta. We provided further evidence using BiFC to show that the two proteins indeed interact with each other when coexpressed in plant cells (Figure 2E).
Our biochemical data clearly demonstrate that SIP2 is a functional MAPKK (Figure 3A). Several plant MAPKKs have been shown to play important roles in the cellular responses to different biotic and abiotic cues (Kiegerl et al., 2000; Mody et al., 2003; Dóczi et al., 2007; Takahashi et al., 2007). SIMKK, an alfalfa MAPKK, exhibiting 88% similarity with Lotus SIP2 at the amino acid level, has been implicated in the salt-induced activation of SIMK during the response to fungal elicitor (Kiegerl et al., 2000; Cardinale et al., 2002). Arabidopsis MKK4 and MKK5, having 69 and 66% similarity with Lotus SIP2, respectively, are involved in the activation of several downstream MAPKs that modulate the responses to environmental stimuli in plants (Tena et al., 2001; Zhang and Klessig, 2001; Nakagami et al., 2005; Pedley and Martin, 2005), regulate stomatal development (Wang et al., 2007), and control floral organ abscission (Cho et al., 2008). Tobacco (Nicotiana tabacum) MEK2, sharing 67% similarity with Lotus SIP2, functions to activate SIPK and WIPK in the defense response signaling pathway (Yang et al., 2001). MAPK signal cascades are composed of three kinase modules, including MAPKKK, MAPKK, and MAPK, and are linked in various ways to specific upstream receptors and downstream targets. MAPK signaling modules also have overlapping roles in regulating cell division, plant development, hormone biosynthesis and signaling, and responses to abiotic stress and pathogens. So far, only a few MAPK cascade components have been studied in detail because their biological functions are often intertwined, redundant, and pleiotropic (Friedman and Perrimon, 2006). Previous work on tobacco SIPKK has shown that it interacts with SIPK but cannot phosphorylate it in vitro (Liu et al., 2000). By contrast, the recombinant alfalfa PRKK is an inactive MAPKK and requires constitutive activation by MAPKKK for MAPKK activity (Cardinale et al., 2002). The function of the MAPK signaling cascade in the establishment of nitrogen fixation symbiosis in legumes is unclear and deserves further intense investigations. Our data clearly show that SymRK and SIP2 could not phosphorylate one another, despite the strong interaction. Interestingly, SymRK appeared to be a specific inhibitor of SIP2 kinase activity (Figure 3D). The biological importance of this inhibitory effect remains to be explored in future work.
Typically, MAPKK can activate its downstream MAPKs through phosphorylation. The well-characterized MAPKs are represented by Arabidopsis MPK3, MPK4, and MPK6, which are activated by a diversity of stimuli, including abiotic stress, pathogens, and oxidative stress (Petersen et al., 2000; Asai et al., 2002; Wang et al., 2007; Qiu et al., 2008a). It would be interesting to know what MAPKs are activated by the SIP2 MAPKK. To test if SIP2 acted as an MAPKK to target a MAPK in planta, we cloned MPK3 and MPK6 cDNAs from Arabidopsis. SIP2 interacted with MPK6 but not MPK3 in yeast cells (see Supplemental Figures 2, 3, and 5 online). SIP2 phosphorylated MPK6 in vitro (Figure 3B). These data suggest that there may be a SIP2 to MPK6 signaling cascade in L. japonicus. Because SymRK-PK acted as a kinase inhibitor of SIP2 in vitro (Figure 3D), it is likely that SymRK may play a role as a negative regulator of the SIP2-MPK6 signaling cascade in planta. It is not clear how this negative regulation of SIP2 by SymRK fits into the signaling cascade and what are the downstream cellular targets of the SIP2 signaling pathway during nodulation in L. japonicus. Because MAPK cascades are known to have crosstalk between pathways (Cardinale et al., 2002; Jonak et al., 2002; Popescu et al., 2009; Sinha et al., 2011; Tena et al., 2011), how other MAPKKs contribute to early symbiosis signaling also remains to be investigated.
SIP2 appears to play very important roles in nodule organogenesis. In hairy roots expressing SIP2 RNAi, the knockdown expression of SIP2 resulted in a reduced number of ITs, nodule primordia, and nodule number (Figure 5; see Supplemental Table 1 online). Moreover, SIP2 suppression by RNAi also caused a much higher rate of nodule-deficient hairy roots (see Supplemental Figure 8 online) and significant downregulation of the three marker genes related to the IT formation and nodule primordia initiation (Figure 6).
Orthologs of Lotus SymRK from other legumes, including alfalfa (NORK), M. truncatula (DMI2), pea (Pisum sativum; SYM19), S. rostrata (Sr-SymRK), and C. glauca (Cg-SymRK), have been shown to be required for nodule initiation and bacterial internalization in the cortical cells during symbiosome formation (Endre et al., 2002; Stracke et al., 2002; Capoen et al., 2005; Limpens et al., 2005; Oldroyd and Downie, 2006). SymRK is likely active near the junction of fungal and rhizobial signaling cascades (Stracke et al., 2002). SymRK might also play a crucial role at the symbiotic interface in guarding and defense responses for the benefit of microbial accommodation (Holsters, 2008). In plants, MAPK pathways are involved in the regulation of development, growth, programmed cell death, and in responses to various environmental stimuli, including cold, heat, reactive oxygen species, UV, drought, and pathogen attack (Colcombet and Hirt, 2008). Two alfalfa MAPKKs, SIMKK and PRKK, have been shown to activate different sets of MAPKs in response to the same fungal elicitor (Cardinale et al., 2002). Tobacco MEK2 appears to be activated by WIPK and SIPK in response to the various pathogenic signals (Zhang and Liu, 2001). In Arabidopsis, the perception of the bacterial elicitor peptide flg22 by the LRR receptor kinase FLS2 leads to the activation of a MAPK signaling cascade, consisting of MEKK1, MKK4/5, and MPK3/6. In L. japonicus, the defense response induced by flg22 has been shown to result in inhibition of rhizobial infection and delay in nodule organogenesis, suggesting an antagonistic effect of the pathway on the nodule formation in the initial rhizobium–legume interaction. (Lopez-Gomez et al., 2012). We propose that SIP2 may mediate a MAPK signaling cascade in response to the infection of Rhizobium. Different from bacterial pathogens, rhizobia are engaged in a symbiotic relationship with the host plants and are housed in a specific root organ, the nodule. It is unknown if SIP2 also participates in plant responses to bacterial pathogen attacks.
In this report, we described the identification of SIP2 as an interacting partner of SymRK and demonstrated that SIP2 plays an important role in early symbiotic signal transduction, nodule organogenesis, and root development in L. japonicus. As a typical MAPKK, SIP2 is likely to function through MAPK cascades. Thus, this work has extended the MAPK signaling network to the establishment of Rhizobium-legume symbiosis and opened a new avenue for future research in the fields of MAPK signaling and biological nitrogen fixation.
METHODS
Plant Materials and Growth Conditions
Seeds of Lotus japonicus MG-20 were gently rubbed by sandpaper and frozen in liquid nitrogen for 1 min. Surface sterilization of seeds was performed in 70% ethanol for 1 min, followed by immersion in 7% sodium hypochlorite for 10 min. After six washes with sterile water, the seeds were transferred to Murashige and Skoog (MS) solid medium and incubated in dark for 3 d. Seed germination was performed at 22°C in continuous light for 3 d. The seedlings were used for transformation with Agrobacterium rhizogenes LBA1334 or planted in pots with sterile sand supplemented with nitrogen-free Fahraeus medium (Fahraeus, 1957). Seedlings were grown in a growth chamber maintained at 22°C with a 16/8-h day/night cycle. Five-day-old seedlings were inoculated with ~107 colony-forming units/mL of Mesorhizobium loti MAFF303099. Tobacco (Nicotiana benthamiana) plants were grown in a growth chamber at 25°C with a 16/8-h day/night cycle.
Yeast Two-Hybrid Screen
The cDNA encoding the SymRK kinase domain was amplified by PCR and cloned into pGBKT7 vector (Zhu et al., 2008). Screening of interaction clones was performed according to the manufacturer’s instructions (Clontech). A total of 1 × 107 transformants from the cDNA library were screened for growth on the SD/-Leu-Trp-His-Ade media. Positive clones were verified by retransformation of the rescued plasmid into yeast AH109 cells containing the bait plasmid (pGBKT7-SymRK-PK). Plasmid pGBKT7-Lam (Clontech) was used as a negative control. Colonies growing on the SD/-Leu-Trp-His-Ade media were transferred to selective media containing X-Gal (80 mg/mL) or were lifted on filters as described (Ma et al., 2007).
Plasmid Construction for Protein–Protein Interactions in Yeast
The SIP2 cDNA was amplified by PCR using SIP2-yF and SIP2-yR (for primer sequences, see Supplemental Table 2 online) and inserted into EcoRI of pGBKT7 and pGADT7. The SymRK-PK and SymRK-LRR cDNAs were amplified by PCR and inserted into NdeI-EcoRI sites of pGBKT7 and pGADT7. The NFR1-PK and NFR1-PK cDNAs were amplified by PCR and inserted into EcoRI-XhoI of pGADT7. The full-length Lj-MAPKK2 and Lj-MAPKK10 coding regions were inserted into NdeI-EcoRI and EcoRI-PstI of pGBKT7, respectively. The Ms-NORK and Ms-SIMKK coding regions were cloned into NdeI-EcoRI of pGADT7 and NdeI-SalI of pGBKT7, respectively. The At-MPK3 and At-MPK6 coding regions were cloned into EcoRI-SalI and NdeI-SalI of pGBKT7.
β-Galactosidase Assay
Yeast cells grown in liquid selection media were pelleted and washed twice with Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgSO4, pH 7.0). The cells were resuspended in 300 μL of Z-buffer and permeabilized by two freeze-thaw cycles in liquid nitrogen and 37°C. Cell extracts were mixed with 0.7 mL of Z-buffer containing 50 mM β-mercaptoethanol and 160 μL of O-nitrophenyl β-d-galactopyranoside (4 mg/mL in Z-buffer). After incubation at 30°C for 30 min or until the yellow color appeared, the reaction was terminated by the addition of 0.4 mL of 1.0 M Na2CO3. The reaction mixture was centrifuged for 10 min at 12,000g to remove cell debris. β-Galactosidase activity in the supernatant was measured at OD420 and expressed in Miller units (Miller, 1972).
Protein Extraction from Yeast Cells for Immunoblot Analysis
To prepare protein extracts for immunoblotting, fresh diploid colonies on SD selection plates were inoculated into 5 mL liquid SD selection medium and grown overnight at 28°C. The yeast cell pellets were washed with cold sterile water, frozen immediately in liquid nitrogen, and stored at −70°C. The cell pellets were thawed by suspension in 200 μL of lysis buffer (8 M urea, 5% SDS, 40 mM Tris-HCI, pH 6.8, 0.1 mM EDTA, 1% β-mercaptoethanol, 0.4 mg/mL bromphenol blue, and 7% protease inhibitor solution). The suspension was warmed to 60°C and transferred to a 1.5-mL screw-cap microcentrifuge tube containing 80 μL of glass beads. Samples were heated at 70°C for 10 min, followed by vortexing vigorously for 5 min. Cell debris was discarded after centrifugation at 12,000g for 10 min. The supernatant was transferred to a fresh 1.5-mL tube, boiled for 2 min, and loaded onto a 12% SDS-PAGE gel. Proteins were transferred to nitrocellulose membrane and probed with monoclonal antibodies against HA (Sigma-Aldrich) and Myc (Invitrogen). Primary antibodies were detected with a peroxidase -conjugated goat anti-mouse IgG secondary antibody (Abmart), followed by signal detection using Super Signal West Pico Chemiluminescent Substrate (Pierce).
Expression and Purification of Fusion Proteins
The full-length cDNA of SIP2 was amplified by PCR using SIP2-eF and SIP2-eR primers (see Supplemental Table 2 online) with additional restriction enzyme sites and inserted into SpeI-EcoRI of pTYB12 (New England Biolabs) for CBD fusion. The full-length SIP2 and SIP2-K120R coding regions were cloned into EcoRI-NotI of pGEX-6P1 (Amersham Pharmacia Biotech) for GST fusion. The full-length SIP2 coding region was inserted into EcoRI-HindIII of pET-28a (Novagen) for expression of His-tagged recombinant protein. The SymRK and SymRK-PK coding regions were inserted into EcoRI-SalI of pMAL-c2X (New England Biolabs) for MBP fusion. The SymRK-PK and SymRK-PK-K622R were cloned into pGEX-KG vector (Amersham Pharmacia Biotech) for GST fusion. The NFR1-PK coding region was inserted into EcoRI-SalI of pMAL-c2X for MBP fusion.
For protein expression, Escherichia coli BL21-Codon Plus (DE3)-RIL (Stratagene) harboring the plasmids was induced with 0.3 mM isopropyl 1-thio-β-d-galactopyranoside in Luria-Bertani broth for 6 h at 20°C. GST fusion proteins were purified using glutathione Sepharose 4B columns (Sigma-Aldrich). MBP fusion proteins were purified using Amylose resin (New England Biolabs). His-tagged proteins were purified using nickel-agarose beads (Qiagen) under native conditions and eluted with a buffer solution (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, and 200 mM imidazole, pH 8.0). Purified proteins were desalted by Millipore Amicon Ultra-15 and stored at −80°C.
In Vitro Protein–Protein Interaction
To assay the interaction between SymRK-PK and SIP2, CBD-tagged full-length or truncated SIP2 was absorbed on chitin beads, which were then incubated with 5 mg of purified SymRK-PK protein in the interaction buffer (20 mM Tris-HCl, 100 mM KCl, 2 mM MgCl2, and 5% glycerol, pH 8.0) for 2 h on ice with gentle shaking. The chitin beads were washed 10 times with 1.0 mL of TEG buffer (20 mM Tris-HCl, 1 mM EDTA, and 5% glycerol, pH 8.0). Retained proteins were eluted by boiling for 5 min in 3× SDS sample buffer (2% SDS, 29.1 mmol/L Tris, pH 6.8, 10% glycerol, and 0.01% bromphenol blue; Zhang et al., 2003). Samples were analyzed on 12% SDS-PAGE, followed by immunoblotting with anti-Lj-SymRK antibody (Zhu et al., 2008).
In Vitro Phosphorylation Assay
For protein kinase assays, ~5 μg of kinase and 5 μg of substrate protein were incubated in 40 μL of 20 mM HEPES, pH 7.4, 15 mM MgCl2, 5 mM EGTA, 1 mM DTT, 10 μM ATP, and 2 μCi of [γ-32P]ATP at 26°C for 30 min. The reaction was stopped by adding 5× SDS loading buffer and boiling for 5 min. Samples were analyzed directly via 12% SDS-PAGE.
RNA Isolation and Quantitative Real-Time PCR
Total RNA was isolated from stems, leaves, nodules, control roots, and roots inoculated with M. loti MAFF303099, using TRIZOL reagent (Invitrogen). RNA samples were treated with DNase I to remove potential contaminating genomic DNA, followed by extraction with phenol:chloroform. First-strand cDNA was prepared using oligo(dT) primer. Quantitative PCR was performed on a Mini Opticon real-time PCR system (LightCycler 480) using One-Step SYBR PrimeScript RT-PCR kit II (Takara). Cycling conditions were as follows: 95°C for 30s, 95°C for 5s, 60°C for 15s, and 72°C for 12s. The reaction was performed for 40 cycles, followed by a step at 72°C for 5 s. ATP synthase was used as a reference gene (AW719841). Melting curve analysis and sequencing of the amplified products was used to determine the identity of the amplified PCR products. The experiment was performed with three replicates, and the averages of measurements were presented.
Transient Expression in N. benthamiana Leaves and BiFC Experiments
The full-length SymRK cDNA was amplified by PCR using SymRK-bF and SymRK-bR (see Supplemental Table 2 online) and cloned into SpeI-SalI of pSCYCE-R to obtain SymRK:SCFP3Ac. The NFR1 and NFR5 coding regions were cloned into SpeI-XhoI of pSCYCE-R to obtain NFR1:SCFP3Ac and NFR5:SCFP3Ac, respectively. The SIP2 coding region was cloned into SpeI-KpnI of pSCYNE to obtain SIP2:SCFP3AN. These constructs were transferred into Agrobacterium tumefaciens strain GV3101/PMP90 by electroporation. Agrobacterium cells harvested by centrifugation were resuspended in infiltration buffer (10 mM MES, pH 5.7, 10 mM MgCl2, and 150 μM acetosyringone). Agrobacterium cells containing different plasmids were mixed and adjusted to a final OD600 of 0.5. After incubation for 2 to 4 h at room temperature, the Agrobacterium mixture was infiltrated into the top leaves of 6-week-old N. benthamiana plants with a 1- to 2-mL syringe. Fluorescence was observed 2 to 3 d after infiltration using a Zeiss LSM510 laser scanning microscope with CFP (excitation wavelength of 405 nm; emission wavelength of 477 nm) for SCFP3AC/SCFP3AN complexes.
Protein Extraction from Plant Cells for Immunoblot Analysis
Leaf discs were harvested, frozen in liquid nitrogen, and stored at −80°C. Leaf discs were ground in liquid nitrogen and the resulting powders were resuspended in extraction buffer (50 mM Tris-HCl, pH 7.5,150 mM NaCl, 0.1% Nonidet P-40, 4 M urea, and 1 mM phenylmethylsulfonyl fluoride). After centrifugation at 9000g for 30 min at 4°C, the supernatant was mixed with SDS loading buffer. Proteins were separated on 12% SDS-PAGE gels and electroblotted to nitrocellulose membrane (Hybond-C; Amersham) at 25 V for 50 min. After blocking with 5% skim milk, the membrane was incubated with primary antibody overnight and then with secondary antibody for 2 h. Protein bands were detected with Super Signal West Pico Chemiluminescent Substrate (Pierce). The dilutions used in these experiments were 1:1000 for anti-HA antibody (Sigma-Aldrich), 1:1000 for anti-FLAG (Cell Signaling Technology), 1:1000 for anti-actin (Abmart), and 1:5000 for goat anti-mouse horseradish peroxidase–conjugated antibody (Abmart).
Expression of GFP Fusion Protein for Subcellular Localization
For subcellular localization, the SIP2 coding region was cloned into SpeI of pCAMBIA1302 (CAMBIA). The plasmid was used for transient expression in onion (Allium cepa) epidermal cells by particles bombardment using a Biolistic PDS-1000/He particle delivery system (Bio-Rad). After incubation for 24 to 48 h at 25°C in dark, the epidermal cell layers were examined using a confocal laser scanning microscope (Leica). The plasmid was modified by replacing the hygromycin resistance gene with the GUS gene and used for transformation of hairy roots in L. japonicus. A. rhizogenes LBA1334 cells containing the plasmid were used to induce hairy root formation on hypocotyls of L. japonicus seedlings. Transgenic hairy roots, identified based on GUS staining, were examined using a Zeiss LSM510 confocal microscope.
SIP2 RNAi
Two SIP2-RNAi constructs (see Supplemental Figure 7 online) were used in this work. RNAi-1 contained 111 bp of the coding sequence and 129 bp of the 3′ untranslated region, while RNAi-2 encompassed the 111-bp coding sequence and 309-bp 3′ untranslated region. The SIP2 fragments were cloned into pCAMBIA1301-35S-int-T7 in inverse orientations with an Actin intron between them. The RNAi constructs were expressed under the control of the CaMV35S promoter.
Induction and Selection of Transgenic Hairy Roots
A. rhizogenes LBA1334 cells containing the binary vectors for RNAi suppression were used to induce hairy root formation on L. japonicus seedlings as described previously (Kumagai and Kouchi, 2003). Briefly, seedlings were cut at the base of hypocotyls and placed in the A. rhizogenes suspension in a Petri dish for 30 min. The seedlings were transferred onto MS medium agar plates containing 1.5% Suc and cocultivated for 5 d in a growth chamber. The plants were transferred onto fresh MS medium plates containing 300 μg/mL cefotaxime and grown for 10 more days until hairy roots developed from hypocotyls. For selection of transgenic hairy roots, a root tip (2 to 3 mm) was excised for GUS staining overnight at 37°C in a solution containing 100 mM sodium phosphate, pH 7.0, 0.1% Triton X-100, 0.1% N-laurylsarcosine, 10 mM EDTA, 1 mM K3Fe(CN)6, 1 mM K4Fe(CN)6, and 0.5 mg/mL X-Gluc. The remaining portion of the hairy root (20 to 30 mm long) attached to the seedling was labeled. If its tip was GUS-negative, the whole hairy root would be discarded. If the hairy root tip was GUS-positive, the remaining portion of the hairy root would be saved. One to three transgenic hairy roots were preserved for each seedling. Plants harboring transgenic hairy roots were transferred to pots filled with vermiculite and sand (1:1) with half-strength B&D medium and grown in a chamber in a 16/8-h day/night cycle at 22°C. After 5 to 7 d, plants were inoculated with M. loti MAFF303099. Nodule number was scored 4 weeks after Rhizobium inoculation.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HQ910409 (Lj-SIP2), JQ581600 (Lj-MapKK2), and JQ581601 (Lj-MapKK10). Other sequences cited in this article have the following accession numbers: AAB97145 (At-MKK1), BAA28828 (At-MKK2), BAA28829 (At-MKK3), BAA28830 (At-MKK4), BAA28831 (At-MKK5), NP_200469 (At-MKK6), AAF25995 (At-MKK7), AAF30316 (At-MKK8), AAG30984 (At-MKK9), AAF81327 (At-MKK10), BAA04866 (AtMPK3), BAA04869 (AtMPK6), AJ293274 (Ms-SIMKK), and AB264547 (Nt-MEK2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Multiple Sequence Alignment of SIP2-Related MAPKKs.
Supplemental Figure 2. β-Galactosidase Activities of Yeast Cells Used for Protein–Protein Interaction Assays.
Supplemental Figure 3. Immunoblot Analysis of Protein Expression in Yeast Cells.
Supplemental Figure 4. Immunoblot Analysis of Protein Expression in Plant Cells.
Supplemental Figure 5. Interactions between Lotus SIP2 and Arabidopsis Map Kinases.
Supplemental Figure 6. Inhibition of SIP2 Kinase Activity by SymRK.
Supplemental Figure 7. SIP2-RNAi Constructs.
Supplemental Figure 8. Nodulation Phenotypes of SIP2-RNAi Hairy Roots.
Supplemental Figure 9. Expression of Lj-MKK2 and Lj-MKK10 in SIP2-RNAi Hairy Roots.
Supplemental Figure 10. Mycorrhization of Lotus japonicus Hairy Roots Expressing SIP2-RNAi.
Supplemental Table 1. Number of Nodules Formed on the SIP2 RNAi Hairy Roots.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Alignment Used for the Phylogenetic Analysis in Figure 1C.
Supplementary Material
Acknowledgments
We thank Shiping Wang for providing pCAMBIA1301U, Guojiang Wu for M. loti MAFF303099 strain, and Da Luo for pCAMBIA1301-35S-int-T7. We also thank Yao Hang for technical advice on transient expression and microscopy. This work was supported by funds from the National Basic Research Program of China (973 Program Grant 2010CB126502), the National Natural Science Foundation of China (Grant 30870186), the Ministry of Agriculture of China (Grant 2009ZX08009-116B), the State Key Laboratory of Agricultural Microbiology (Grant AMLKF200804), and the Graduate Education Innovation Fund of Huazhong Agricultural University.
AUTHOR CONTRIBUTIONS
T.C., Z.Z., and Z.H. designed this work and wrote the article. T.C. performed most of the experiments, and H.Z., D.K., K.C., C.W., and H.G. provided substantial help in specific experiments.
References
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bardwell L., Cook J.G., Chang E.C., Cairns B.R., Thorner J. (1996). Signaling in the yeast pheromone response pathway: Specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol. 16: 3637–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Komis G., Müller J., Menzel D., Samaj J. (2010). Arabidopsis homologs of nucleus- and phragmoplast-localized kinase 2 and 3 and mitogen-activated protein kinase 4 are essential for microtubule organization. Plant Cell 22: 755–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Komis G., Ziemann A., Menzel D., Samaj J. (2011). Mitogen-activated protein kinase 4 is involved in the regulation of mitotic and cytokinetic microtubule transitions in Arabidopsis thaliana. New Phytol. 189: 1069–1083 [DOI] [PubMed] [Google Scholar]
- Capoen W., Goormachtig S., De Rycke R., Schroeyers K., Holsters M. (2005). SrSymRK, a plant receptor essential for symbiosome formation. Proc. Natl. Acad. Sci. USA 102: 10369–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale F., Meskiene I., Ouaked F., Hirt H. (2002). Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14: 703–711 [PMC free article] [PubMed] [Google Scholar]
- Chang L., Karin M. (2001). Mammalian MAP kinase signalling cascades. Nature 410: 37–40 [DOI] [PubMed] [Google Scholar]
- Cho S.K., Larue C.T., Chevalier D., Wang H., Jinn T.L., Zhang S., Walker J.C. (2008). Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Davis S., Vanhoutte P., Pages C., Caboche J., Laroche S. (2000). The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. J. Neurosci. 20: 4563–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dóczi R., Brader G., Pettkó-Szandtner A., Rajh I., Djamei A., Pitzschke A., Teige M., Hirt H. (2007). The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19: 3266–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Fahraeus G. (1957). The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Fiil B.K., Petersen K., Petersen M., Mundy J. (2009). Gene regulation by MAP kinase cascades. Curr. Opin. Plant Biol. 12: 615–621 [DOI] [PubMed] [Google Scholar]
- Friedman A., Perrimon N. (2006). High-throughput approaches to dissecting MAPK signaling pathways. Methods 40: 262–271 [DOI] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Geurts R., Fedorova E., Bisseling T. (2005). Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8: 346–352 [DOI] [PubMed] [Google Scholar]
- Gherbi H., Markmann K., Svistoonoff S., Estevan J., Autran D., Giczey G., Auguy F., Péret B., Laplaze L., Franche C., Parniske M., Bogusz D. (2008). SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankiabacteria. Proc. Natl. Acad. Sci. USA 105: 4928–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresshoff P.M. (2003). Post-genomic insights into plant nodulation symbioses. Genome Biol. 4: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel L.P., et al. (2006). Ancient signals: Comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11: 192–198 [DOI] [PubMed] [Google Scholar]
- Hanks S.K., Hunter T. (1995). Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 9: 576–596 [PubMed] [Google Scholar]
- Holsters M. (2008). SYMRK, an enigmatic receptor guarding and guiding microbial endosymbioses with plant roots. Proc. Natl. Acad. Sci. USA 105: 4537–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Hu C.D., Kerppola T.K. (2003). Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs D., Glossip D., Xing H., Muslin A.J., Kornfeld K. (1999). Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13: 163–175 [PMC free article] [PubMed] [Google Scholar]
- Jonak C., Okrész L., Bögre L., Hirt H. (2002). Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 5: 415–424 [DOI] [PubMed] [Google Scholar]
- Kang H., Zhu H., Chu X., Yang Z., Yuan S., Yu D., Wang C., Hong Z., Zhang Z. (2011). A novel interaction between CCaMK and a protein containing the Scythe_N ubiquitin-like domain in Lotus japonicus. Plant Physiol. 155: 1312–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. (1998). Mitogen-activated protein kinase cascades as regulators of stress responses. Ann. N. Y. Acad. Sci. 851: 139–146 [DOI] [PubMed] [Google Scholar]
- Kevei Z., et al. (2007). 3-Hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19: 3974–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiegerl S., Cardinale F., Siligan C., Gross A., Baudouin E., Liwosz A., Eklöf S., Till S., Bögre L., Hirt H., Meskiene I. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12: 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.H., Roh H.E., Lee M.N., Hur M.W. (2007). New fast BiFC plasmid assay system for in vivo protein-protein interactions. Cell. Physiol. Biochem. 20: 703–714 [DOI] [PubMed] [Google Scholar]
- Kistner C., Winzer T., Pitzschke A., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Webb K.J., Szczyglowski K., Parniske M. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Sun L., Zhou Y., Zhang M., Liu Y., Pan J., Li D. (2011). ZmMKK4 regulates osmotic stress through reactive oxygen species scavenging in transgenic tobacco. Plant Cell Rep. 30: 2097–2104 [DOI] [PubMed] [Google Scholar]
- Kouchi H., Imaizumi-Anraku H., Hayashi M., Hakoyama T., Nakagawa T., Umehara Y., Suganuma N., Kawaguchi M. (2010). How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H., et al. (2004). Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res. 11: 263–274 [DOI] [PubMed] [Google Scholar]
- Kumagai H., Kinoshita E., Ridge R.W., Kouchi H. (2006). RNAi knock-down of ENOD40s leads to significant suppression of nodule formation in Lotus japonicus. Plant Cell Physiol. 47: 1102–1111 [DOI] [PubMed] [Google Scholar]
- Kumagai H., Kouchi H. (2003). Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol. Plant Microbe Interact. 16: 663–668 [DOI] [PubMed] [Google Scholar]
- Limpens E., Mirabella R., Fedorova E., Franken C., Franssen H., Bisseling T., Geurts R. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 102: 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S., Klessig D.F. (2000). Molecular cloning and characterization of a tobacco MAP kinase kinase that interacts with SIPK. Mol. Plant Microbe Interact. 13: 118–124 [DOI] [PubMed] [Google Scholar]
- Lopez-Gomez M., Sandal N., Stougaard J., Boller T. (2012). Interplay of flg22-induced defence responses and nodulation in Lotus japonicus. J. Exp. Bot. 63: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Hong Z., Zhang Z. (2007). Perinuclear and nuclear envelope localizations of Arabidopsis Ran proteins. Plant Cell Rep. 26: 1373–1382 [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Markmann K., Giczey G., Parniske M. (2008). Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol. 6: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros T., Helfer A., Hatzimasoura E., Magyar Z., Serazetdinova L., Rios G., Bardóczy V., Teige M., Koncz C., Peck S., Bögre L. (2006). The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 48: 485–498 [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1972). Experiments in Molecular Gentics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Mishra N.S., Tuteja R., Tuteja N. (2006). Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 452: 55–68 [DOI] [PubMed] [Google Scholar]
- Miwa H., Sun J., Oldroyd G.E., Downie J.A. (2006). Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant Microbe Interact. 19: 914–923 [DOI] [PubMed] [Google Scholar]
- Mody N., Campbell D.G., Morrice N., Peggie M., Cohen P. (2003). An analysis of the phosphorylation and activation of extracellular-signal-regulated protein kinase 5 (ERK5) by mitogen-activated protein kinase kinase 5 (MKK5) in vitro. Biochem. J. 372: 567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Pitzschke A., Hirt H. (2005). Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2006). Nuclear calcium changes at the core of symbiosis signalling. Curr. Opin. Plant Biol. 9: 351–357 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008). Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Pedley K.F., Martin G.B. (2005). Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8: 541–547 [DOI] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.L., et al. (2008b). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.L., Zhou L., Yun B.W., Nielsen H.B., Fiil B.K., Petersen K., Mackinlay J., Loake G.J., Mundy J., Morris P.C. (2008a). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Sinha A.K., Jaggi M., Raghuram B., Tuteja N. (2011). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 6: 196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink H.P., Wijfjes A.H., Lugtenberg B.J. (1995). Rhizobium NodI and NodJ proteins play a role in the efficiency of secretion of lipochitin oligosaccharides. J. Bacteriol. 177: 6276–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Takahashi F., Yoshida R., Ichimura K., Mizoguchi T., Seo S., Yonezawa M., Maruyama K., Yamaguchi-Shinozaki K., Shinozaki K. (2007). The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19: 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Soyano T., Kosetsu K., Sasabe M., Machida Y. (2010). HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 51: 1766–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansengco M.L., Hayashi M., Kawaguchi M., Imaizumi-Anraku H., Murooka Y. (2003). crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus. Plant Physiol. 131: 1054–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15: 141–152 [DOI] [PubMed] [Google Scholar]
- Tena G., Asai T., Chiu W.L., Sheen J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4: 392–400 [DOI] [PubMed] [Google Scholar]
- Tena G., Boudsocq M., Sheen J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh A.J., Davis R.J. (1998). Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 23: 481–485 [DOI] [PubMed] [Google Scholar]
- Xu J., Li Y., Wang Y., Liu H., Lei L., Yang H., Liu G., Ren D. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283: 26996–27006 [DOI] [PubMed] [Google Scholar]
- Yang K.Y., Liu Y., Zhang S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Parniske M. (2005). Regulation of plant symbiosis receptor kinase through serine and threonine phosphorylation. J. Biol. Chem. 280: 9203–9209 [DOI] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zhang S., Liu Y. (2001). Activation of salicylic acid-induced protein kinase, a mitogen-activated protein kinase, induces multiple defense responses in tobacco. Plant Cell 13: 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Dai Y., Xiong Y., DeFraia C., Li J., Dong X., Mou Z. (2007). Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J. 52: 1066–1079 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Quick M.K., Kanelakis K.C., Gijzen M., Krishna P. (2003). Characterization of a plant homolog of hop, a cochaperone of hsp90. Plant Physiol. 131: 525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Cai Z., Guo Y., Gan S. (2009). An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 150: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Chen T., Zhu M., Fang Q., Kang H., Hong Z., Zhang Z. (2008). A novel ARID DNA-binding protein interacts with SymRK and is expressed during early nodule development in Lotus japonicus. Plant Physiol. 148: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.