Abstract
TDP-43 is linked to neurodegenerative diseases including frontotemporal dementia and amyotrophic lateral sclerosis. Mostly localized in the nucleus, TDP-43 acts in conjunction with other ribonucleoproteins as a splicing co-factor. Several RNA targets of TDP-43 have been identified so far, but its role(s) in pathogenesis remains unclear. Using Affymetrix exon arrays, we have screened for the first time for splicing events upon TDP-43 knockdown. We found alternative splicing of the ribosomal S6 kinase 1 (S6K1) Aly/REF-like target (SKAR) upon TDP-43 knockdown in non-neuronal and neuronal cell lines. Alternative SKAR splicing depended on the first RNA recognition motif (RRM1) of TDP-43 and on 5′-GA-3’ and 5′-UG-3′ repeats within the SKAR pre-mRNA. SKAR is a component of the exon junction complex, which recruits S6K1, thereby facilitating the pioneer round of translation and promoting cell growth. Indeed, we found that expression of the alternatively spliced SKAR enhanced S6K1-dependent signaling pathways and the translational yield of a splice-dependent reporter. Consistent with this, TDP-43 knockdown also increased translational yield and significantly increased cell size. This indicates a novel mechanism of deregulated translational control upon TDP-43 deficiency, which might contribute to pathogenesis of the protein aggregation diseases frontotemporal dementia and amyotrophic lateral sclerosis.
INTRODUCTION
TDP-43 [transactivation response (TAR) DNA binding protein of 43 kDa] is neuropathologically as well as genetically linked to frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) (1–4). Besides hyperphosphorylation, fragmentation and aggregation of TDP-43 in neurodegenerative disease, nuclear depletion of TDP-43 is a hallmark of affected neurons (1). Thus, in addition to a toxic gain of misfunction, loss of (nuclear) TDP-43 function may contribute to disease pathogenesis.
TDP-43 is a RNA binding protein (RBP) involved in various aspects of RNA metabolism (5,6). TDP-43 mediates transcriptional repression (7,8) and acts on mRNA stability (9,10) and miRNA processing (11). Pertaining to alternative splicing, TDP-43 mediates exon skipping of cystic fibrosis transmembrane conductance regulator (CFTR) exon 9 (12) and apolipoprotein A-II exon 3 (13) as well as exon inclusion of survival of motor neuron exon 7 (14). Other reported and validated TDP-43 target RNAs include cyclin-dependent kinase 6 (15), splicing component of 35 kDa (SC35) (16) and histone deacetylase 6 (HDAC6) (17–21). In addition, recent screenings have identified many other novel target RNAs by use of RNA sequencing after crosslinking and immunoprecipitation (CLIP) with TDP-43 antibodies (20,21); however, functional investigation is largely missing so far.
To expand the knowledge about TDP-43 splice targets, we have used Affymetrix exon arrays to identify alternatively spliced transcripts upon TDP-43 knockdown. Thereby, we discovered exon 3 inclusion of S6 kinase 1 (S6K1) Aly/REF-like target (SKAR, also known as POLDIP3 or PDIP46) to be highly dependent on TDP-43, but not on FUS/TLS, another RNA-binding protein involved in FTD/ALS (22–25). RNAi-mediated silencing of TDP-43 in non-neuronal and neuronal cell lines significantly reduced the main SKAR α isoform, containing all nine exons, and concomitantly increased the SKAR β isoform lacking exon 3. Retransfection experiments showed only subtle defects of the C-terminal glycine-rich domain (GRD) as well as of disease-associated TDP-43 point mutations, but highlighted the involvement of the RNA recognition motif (RRM) 1. TDP-43 specifically bound to the proximal intronic region downstream of exon 3 within the SKAR pre-mRNA. Mutagenesis of either a 5′-GA-3′ repeat or the consensus TDP-43 binding motif 5′-UGUGUGU-3′ (26) within this region largely abolished the binding of TDP-43 to the SKAR pre-mRNA and significantly reduced the splicing of SKAR minigene constructs that were generated as splicing reporters. Because SKAR, itself an RRM-containing protein, is a component of the exon junction complex (EJC) (27), we assessed the effects on S6K1-dependent pioneer round of translation and cell growth. We found that the alternative SKAR β isoform is significantly more active than SKAR α. Furthermore, TDP-43 siRNA increased S6K1-dependent signaling and translational yield as well as cell size. Thus, loss of TDP-43 and resulting alternative splicing of SKAR increases splicing-dependent global translation and may thereby contribute to disease pathogenesis by disturbing cellular protein homeostasis.
METHODS
cDNA constructs
Wild-type and mutant Flag-TDP-43 constructs have been described previously (17). SKAR α and β cDNA were PCR amplified from scrambled and siRNATDP-43 treated HEK293E cells, respectively, and were subcloned into pCMV-Myc (Clontech) via BglII/NotI. Intron containing SKAR DNA for in vitro transcription/UV-crosslinking experiments was PCR amplified from human genomic DNA. Different parts of intron containing SKAR DNA (exons 2–3, exons 2–4 and parts 1–11) were subcloned into pGEM-T-Easy (Promega) under control of the T7 promoter. SKAR part 3/4/5 was subloned via NdeI into a pTB splicing reporter construct that has been obtained from Emanuele Buratti, ICGEB, Trieste, Italy (28). Mutant SKAR DNAs were generated by site-directed mutagenesis. Primer sequences can be found in Supplementary Table S2.
Firefly luciferase expression construct (pGL2) has been obtained from Promega. Renilla luciferase expression constructs with or without intron were obtained from Melissa Moore, University of Massachusetts, USA (29). All constructs were sequence verified using BigDye Terminator v.3.1 and an ABI 3100 Genetic Analyzer (Applied Biosystems).
Cell culture and siRNA silencing
HEK293E cells (Invitrogen) were cultured in Dulbecco's modified Eagle medium (DMEM), SH-SY5Y cells (ATCC) in DMEM-F12 (both Biochrom AG), both supplemented with 10% fetal calf serum (PAA Laboratories) at 37°C under humidified 5% CO2/air. Stably silenced TDP-43 HEK293E and SH-SY5Y cells have been described previously (17). DNA transfections were performed with FuGene6 (Roche) or Lipofectamine 2000 (Invitrogen) according to manufacturer's instruction. siRNA transfections were performed using HiPerfect (Qiagen) with 5–20 nmol siRNA. Scrambled siRNA (All Stars Negative Control siRNA) and all other siRNAs were purchased from Qiagen. Target sequences of used siRNAs were as follows (5′→3′): siRNATDP-43A: CACAATAGCAATAGACAGTTA; siRNATDP-43B: CACACTACAATTGATATCAAA; siRNATDP-43C: AAGAAACAATCAAGGTAGTAA; siRNATDP-43D: AAGAATCAGGGTGGATTTGGT; siRNAFUSA: CTGGGTGAGAATGTTACAATT; siRNAFUSB: ACAGCCCATGATTAATTTGTA; siRNAFUSC: ACAGGATAATTCAGACAACAA; siRNAFUSD: AAGATCAATCCTCCATGAGTA; siRNAFUSE: AACTATGTAATTGTAACTATA; siRNASKAR: CCGCTGGGAAGAGTCTATCTA.
Microarray profiling and FIRMA analysis
For expression profiling using the Affymetrix Human Exon 1.0-ST Gene Chip®, 1 µg of total RNA was amplified using the WT Sense Target Labeling kit (Affymetrix). The 5.5 µg fragmented and end labeled cDNA was hybridized to the Human Exon 1.0-ST Gene Chip®. After hybridization the array was washed and stained in a Fluidics Station 450 (Affymetrix) with the recommended washing procedure. Biotinylated cRNA/cDNA bound to target molecules was detected with streptavidin-coupled phycoerithrin, biotinylated anti-streptavidin IgG antibodies and again streptavidin-coupled phycoerithrin according to the protocol. Arrays were scanned using the GCS3000 Gene Chip scanner (Affymetrix) and GCOS 1.4 software. Scanned images were subjected to visual inspection to control for hybridization artifacts and proper grid alignment and analyzed with Expression Console 1.0 (Affymetrix) to generate report files for quality control.
To detect transcripts with significant changes in their splicing profiles the approach finding isoforms using robust multichip analysis (FIRMA) was employed (30). For the overall workflow, we used a custom build CDF file with Ensembl transcript annotations, applied background correction on the individual exon arrays and then applied the FIRMA method. Finally, plots were created to visualize the results on a gene level.
The custom-build CDF annotation file was obtained from the aroma.affymetrix website (http://www.aroma-project.org/) which, in addition to the standard CDF file, contains relationships between probe sets and exons as well as exons and transcripts. These relationships are needed for the statistical analysis on the exon and transcript level. Before applying FIRMA a background correction was employed to each exon array individually. This was done with the robust multichip analysis (RMA)-convolution method implemented in the aroma.affymetrix package (31). This method is based on the quantile normalization protocol described in detail by Bengsston et al. (32).
After normalization the FIRMA based summarization was used to calculate a splicing score for each exon. FIRMA calculates this score for each sample individually. Based on these individual scores, a t-test was used to calculate a P-value for each exon. The P-value was further corrected for multiple testing according to Benjamini and Hochberg (33). To obtain fold-changes, the mean of the treatment and control FIRMA scores were calculated and subsequently the control was subtracted from the treatment (subtraction was used because of the logarithmic scale of the FIRMA score). To reduce the occurrence of false positives, we filtered all probe selection regions containing less than four probes. Often an additional gene expression filter is applied. FIRMA, however, implicitly favors genes with high expression values, thus such a filter was not needed.
The plots were created with GenomeGraphs (34) that allows to overlay annotation and expression data. The transcript coordinates were obtained with the R-package biomaRt (35). The plots show all isoforms reported in Ensembl, the normalized log-intensities for each exon array, the RMA-based intensities and the FIRMA scores and fold-changes.
RT–PCR
Cellular RNA was extracted using the RNeasy Mini Kit (Qiagen) following manufacturer's instruction. For semi-quantitative RT–PCR experiments, 600 ng total RNA were reverse transcribed using Transcriptor High Fidelity cDNA Synthesis Kit (Roche) and anchored oligo-dT primer. RT reaction (2 µl) was used as template for transcript amplification in a 25 µl reaction with 5 µl 5× GoTaq Buffer, 0.1 µl GoTaq Polymerase (Promega) and 2 µM primer. Amplified PCR products were resolved by electrophoresis using 2% agarose gels and stained with ethidium bromide.
For qRT–PCR, 1000 ng total RNA were reverse transcribed using the same conditions as above. The 1/10 dilutions were used in triplicates with 0.2 µM primer and 5 µl LightCycler 480 SYBR Green I Master in a 10 µl reaction and qPCR was executed in a 384-well block on a LightCycler 480 system (Roche). Absolute transcript levels for TDP-43, total SKAR, SKAR α, SKAR β, Renilla and firefly luciferase, and PBGD were obtained by second derivative method. Relative transcript levels were calculated as a ratio to PBGD and normalized to the relative expression level of the mock-transfected control. For expression of transcript levels as a ratio of SKAR isoforms, calibration curves with DNA templates (pCMV-Myc-SKAR α and SKAR β, respectively) were generated. Relative transcript levels were then recalculated into absolute copy numbers and expressed as a ratio to total SKAR copies. Primer sequences can be found in Supplementary Table S2.
Western blotting
Cells were harvested and lysed in lysis buffer [50 mM Tris (pH 7.4), 50 mM NaCl, 1% NP-40, 0.1% deoxycholate and 0.1% SDS, 1× Complete proteinase inhibitor, 1× PhosSTOP phosphatase inhibitor (both Roche)]. Protein concentration was determined by use of bicinchoninic acid (BCA Protein Assay; Pierce Biotechnology). Protein was subjected to SDS–PAGE using 10% polyacrylamide gels or 4–12% Bis–Tris NuPAGE gradient gels (Invitrogen) and transferred onto nitrocellulose. Membranes were incubated with primary antibodies overnight at 4°C followed by HRP-conjugated secondary antibodies (1:15 000; Jackson ImmunoResearch Laboratories). Bands were visualized with ImmobilonWestern Chemiluminescent HRP Substrate (Millipore) on Hyperfilm ECL high performance chemiluminescence (GE Healthcare). For densitometric analysis Image J software version 1.43 u was used.
Antibodies
Following antibodies were used for western blot analysis: rabbit anti-TDP-43 (1:2000; ProteinTech Group), mouse anti-TDP-43 (1:2000, Abnova, clone 2E2-D3), mouse anti-Flag (1:10 000–1:100 000; Sigma, clone M2), mouse anti-FUS (1:2000; Santa Cruz, clone 4H11), mouse anti-GAPDH (1:35 000; Biodesign International), mouse anti-Myc (1:5000; Roche, clone 9E10), rabbit anti-SKAR α/β, rabbit anti-SKAR α, mouse anti-S6 (clone 54D2), rabbit anti-p70 S6 kinase, rabbit anti-phosphoS6 (Ser235/236), rabbit anti-phospho p70 S6 Kinase (Thr389), rabbit anti-phospho-Akt substrate (RXRXXpS/T) (all used 1:1000; Cell Signaling Technologies).
RNA crosslinking
RNA crosslinking experiments were carried out as described recently (17) with slight modifications. Briefly, pGEM-T-Easy containing different parts of SKAR under control of the T7 promoter were linearized with PvuI and NdeI and gel purified. Briefly, 100 ng DNA was then in vitro transcribed and biotinylated using biotin-16-UTP (Biotin RNA Labeling Mix) and T7 RNA Polymerase (both Roche) according to manufacturer's instructions. DNase digest was performed by adding 2 µl of RNase-free DNase (Promega) for 15 min at 37°C and reaction stopped by addition of 1 µl of 0.5 M EDTA.
The 5 pmol biotin-RNA was incubated with 250 µl RNA-X-Link buffer [20 mM HEPES (pH 7.5), 5 mM MgCl2, 50 mM KCl, 150 mM NaCl, 0.5 mM EGTA, 0.5 mM dithiothreitol, 10% glycerol] and 500 µg HEK293E lysate for 20 min at 30°C. HEK293E lysates were generated from HEK293E cells transfected with Flag-TDP-43 for 48 h and lysed in RNA binding buffer + 1% Triton X-100. UV irradiation was performed on ice for 10 min with 0.120 J cm−2 in a Bio-Link BLX device (Vilber-Lourmat). After brief RNase A digest (10 µg ml−1, Sigma) for 15 min streptavidin-beads (Sigma) were added and precipitations carried out overnight at 4°C. Beads were washed 5× in RNA binding buffer + 1% Triton X-100 and elution performed at 95°C with 1× Lämmli buffer. Eluate and input were separated by 10% SDS–PAGE, blotted onto nitrocellulose and probed with antibodies.
Luciferase assay
Translational yield of spliced versus unspliced RNAs was analyzed as described previously (27). In brief, HEK293E cells were double transfected either with intron-containing Renilla luciferase plus firefly control or with intron-less Renilla luciferase plus firefly control in six-well format. At 8-h posttransfection, cells were split into 24-wells and transfected another 16 h later with DNA or another 16 and 36 h later with siRNA. Nearly 48 h after DNA transfection or 24 h after last siRNA transfection luciferase activity was measured in a Mithras LB940 dual-channel luminometer (Berthold Technologies) using the Dual Luciferase Reporter Assay system (Promega). Of each sample, the ratio of Renilla to firefly luminescence was calculated resulting in transfection efficiency controlled values for intron-containing Renilla and intron-less Renilla, respectively. Translational efficiency of spliced versus unspliced RNAs was expressed as a ratio of intron-containing to intron-less transfection-controlled Renilla activities, which were normalized to the control samples (cells treated with scrambled siRNA). The concomitant Renilla and firefly mRNA amount was calculated accordingly.
Total protein measurements
HEK293E cells were transiently transfected with siRNA against TDP-43 or treated with control siRNA on 3 consecutive days. Twenty-four hours after the last transfection cells were detached from the plate and counted three times and 2.5 × 106 cells were harvested and lysed in 100 µl RIPA buffer. Protein concentration was determined in triplicates using BCA assay.
Flow cytometry
HEK293E cells were transiently transfected with siRNA against TDP-43 or treated with control siRNA on three consecutive days. Twenty-four hours after the last transfection cells were detached from the plate and washed twice in PBS, resuspended in PBS and kept on ice until measurement. Mean forward scatter values of 50 000 cells per condition in five independent experiments were recorded using a CyAn flow cytometer (DakoCytomation).
Statistical analysis
Statistical analysis was performed with paired, two-sided Student's t-test. Error bars indicate SEM.
RESULTS
Identification of SKAR as a TDP-43 responsive splice target
To obtain a full survey of TDP-43 splice targets, we performed for the first time a human genome-wide screen using Affymetrix exon arrays. Human embryonic kidney HEK293E cells were transiently silenced by siRNATDP-43 treatment, achieving strong TDP-43 knockdown (17). Total RNA was prepared from four independent siRNATDP-43 and four scrambled control siRNA treatments, and hybridizations of Human exon arrays were performed. The raw data was analyzed by FIRMA, which introduces a score for the likeliness of alternative exon usage, the FIRMA score (30), and further adjusted the primary P-value for multiple testing according to Benjamini and Hochberg to correct for the occurrence of false positives. When the cut-off was set to P < 0.1, only 13 probe sets passed these stringent criteria (Supplementary Table S1). By far, the greatest significance was observed for POLDIP3/SKAR that was selected for in-depth validation.
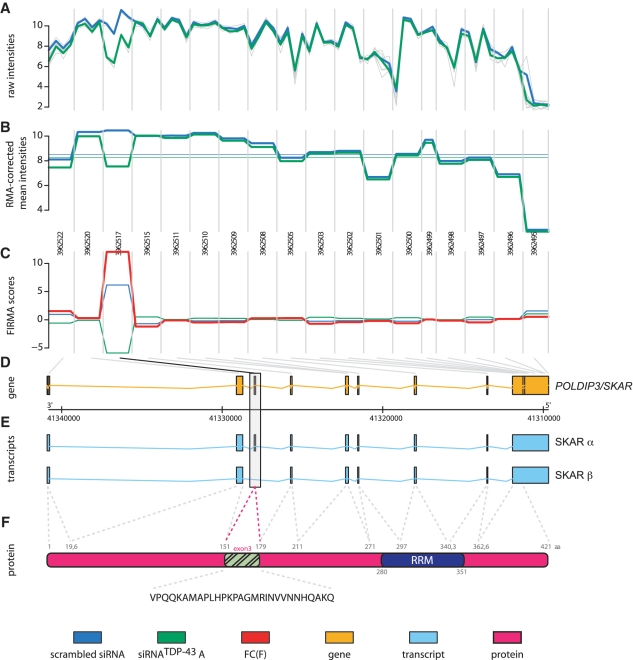
The observed splicing alteration of the SKAR transcript obtained by microarray profiling (raw data Figure 1A, normalized mean values Figure 1B) was traced to a specific hybridization change in exon 3 (FIRMA score Figure 1C, genomic representation of POLDIP3/SKAR Figure 1D). Interestingly, we found two major SKAR transcripts annotated in the Ensembl database (Figure 1E). The SKAR α transcript contains all nine exons, whereas the SKAR β transcript lacks exactly exon 3. Exclusion of the 87 base pair bearing exon 3 results in a 29 amino acids in-frame deletion in the SKAR β protein (Figure 1F). To confirm the identity of SKAR transcripts observed upon control or TDP-43 siRNA treatment, we cloned both variants by RT–PCR. In fact, sequencing of the subcloned variants from control or TDP-43 silenced cells, revealed in conformity with database entries the SKAR α and β isoforms, respectively. Thus, TDP-43 silencing led to an alternative splicing of SKAR, a shift from the SKAR α to the SKAR β transcript.
Figure 1.
GenomeGraph of SKAR as a splice target of TDP-43. HEK293E cells were transfected with control siRNA (scrambled) or treated with siRNA against TDP-43 (siRNATDP-43). Four biological replicates of each group were hybridized on a Human Exon 1.0-ST Gene Chip. Intensity values of microarray hybridizations, single values (gray), mean group intensities of scrambled siRNA (blue) and siRNATDP-43 (green), are shown as normalized background-corrected logarithmic intensities (A) and RMA corrected probe-level data (B). Vertical lines separate the 18 individual probe sets covering the POLDIP3/SKAR gene. (C) Depicted are the mean group values of the FIRMA score. The fold change of the FIRMA score (FC(F)) is shown in red. (D) Genomic representation of the POLDIP3/SKAR gene in orange. Gray lines at the top of this panel indicate localization of the individual probe sets within the genomic coordinates. (E) The two Ensembl annotated alternative splice isoforms SKAR α and SKAR β are depicted in blue. SKAR exon 3 is highlighted by a box. (F) The SKAR α protein isoform is shown in pink, the RRM domain is shown in dark blue. Highlighted in green is the exon 3 derived part. At the bottom the amino acid sequence of exon 3 is given.
Validation of TDP-43-dependent alternative SKAR splicing
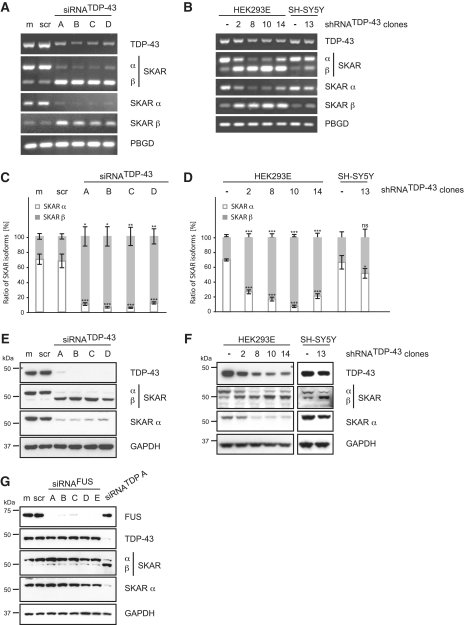
Alternative splicing of SKAR upon knockdown of TDP-43 was validated in non-neuronal HEK293E cells as well as neuronal SH-SY5Y cells (Figure 2). In HEK293E cells, four different siRNAs were used to rule out off-target artifacts. Stable silencing of HEK293E and SH-SY5Y cells by integration of lentiviral delivered shRNATDP-43 was further used to circumvent effects due to general cell stress by transient siRNA transfections. RT–PCR confirmed alternative splicing of SKAR upon TDP-43 knockdown in non-neuronal HEK293E cells as well as neuronal SH-SY5Y cells. SKAR α is the predominant isoform (∼70%) while SKAR β is a minor isoform (∼30%) under basal conditions (Figure 2A–D). Control siRNA treatment did not affect SKAR splicing, while all four siRNATDP-43 treatments led to a dramatic switch from SKAR α to SKAR β expression, now accounting for >90% of the total SKAR transcripts (semi-quantitative RT–PCR Figure 2A, normalized qRT–PCR Figure 2C, for original qRT–PCR data see Supplementary Figure S1A). As for transient silencing, stable knockdown of TDP-43 led to a conspicuous shift of the predominant SKAR α to the SKAR β isoform in several shRNATDP-43 HEK293E cell clones on mRNA level (semi-quantitative RT–PCR Figure 2B, normalized qRT–PCR Figure 2D, for original qRT–PCR data see Supplementary Figure S1B). Importantly, shift from the predominant SKAR α expression to the SKAR β isoform was not only observed in stably silenced TDP-43 non-neuronal HEK293E cells, but also in human neuroblastoma SH-SY5Y cells (Figure 2B and D, for original qRT–PCR data see Supplementary Figure S1B). Consistently, a robust switch from the larger SKAR α isoform to the smaller SKAR β isoform was detected upon TDP-43 knockdown also on protein level by western blot analysis (Figure 2E and F). The observed molecular weight difference between SKAR α and β isoforms is in agreement with sequencing data from subcloned transcripts and the lack of 29 amino acids in isoform β. Moreover, while both isoforms are detected with a total SKAR antibody (CST #3794), isoform β is not recognized by an antibody that has been produced with a synthetic peptide corresponding to human SKAR α (CST #3235).
Figure 2.
Validation of SKAR alternative splicing upon transient silencing of TDP-43. TDP-43 was either silenced transiently by siRNA treatment (A, C, E and G) or stably by use of lentiviral particles encoding for a TDP-43-specific shRNA followed by the selection of single cell clones (B, D and F). For transient silencing, HEK293E cells were either mock treated (m) or transiently transfected with scrambled control siRNA (scr), with one of four different TDP-43-specific siRNAs (siRNATDP-43 A-D) or with one of five specific siRNAs against FUS (siRNAFUS A-E), as indicated. (A–D) Total RNA was extracted and analyzed by RT–PCR. (A and B) Semi-quantitative RT–PCR was performed with primer pairs specific for TDP-43, SKAR (ex2–ex4), SKAR α (ex2|3–ex4) and SKAR β (ex2|4–ex4). (C and D) Real-time PCR was performed with primer pairs against SKAR α (ex2|3–ex4) (white bars), SKAR β (ex2|4–ex4) (gray bars) and total SKAR (ex5|6–ex7). PBGD was used as a housekeeping gene. Resulting relative SKARα/PBGD, SKARβ/PBGD and total SKAR/PBGD ratios were recalculated into absolute copy values and normalized to total SKAR values. Shown are the mean values of five independent experiments ± SEM. *P < 0.05; **P < 0.005; ***P < 0.0005; ns = not significant. Original qRT–PCR data is presented in Supplementary Figure S1A and S1B, respectively. (E–G) Protein was extracted, electrophoresed and resulting western blots probed with antibodies specific for TDP-43, SKAR (both isoforms) and SKAR α. GAPDH was used as a loading control. FUS silencing efficiency was controlled by use of an anti-FUS antibody. Note, that, depending on the primer pair and antibody used, SKAR RNA and protein isoforms, respectively, are visualized as two bands with different molecular weights. The upper band represents SKAR α, the lower corresponds to SKAR β, as indicated.
Importantly and consistent with our present findings, exon 3 skipping of POLDIP3/SKAR had also been found to occur in TDP-43 depleted adult mouse brain samples (20). Thus, TDP-43 knockdown leads to alternative splicing of SKAR in non-neuronal as well as neuronal cells. Noteworthy, SKAR exon 3 skipping is specifically mediated by TDP-43 depletion since knockdown of FUS, the second RBP involved in ALS and FTD (22–24), did not affect SKAR splicing (Figure 2G). Thus, silencing of TDP-43 in non-neuronal and neuronal cells specifically causes alternative splicing of SKAR, shifting from the predominant SKAR α to the SKAR β isoform, both on RNA and protein level.
Restoration of SKAR splicing by TDP-43 wt re-transfection
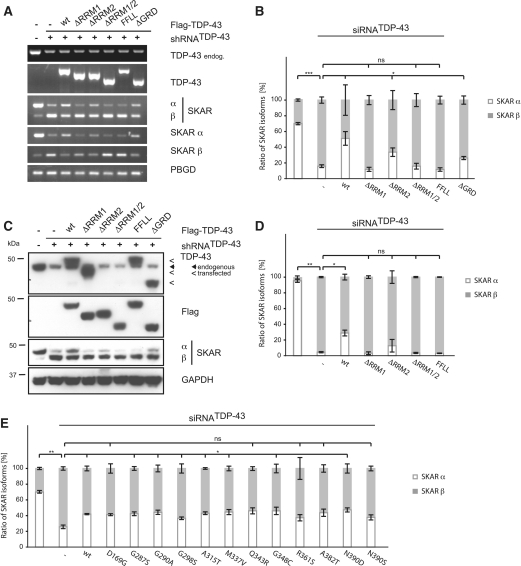
To prove that alternative splicing of SKAR is a direct effect of TDP-43, we performed rescue experiments with wild-type (wt) and mutant TDP-43 (Figure 3). Retransfection of TDP-43 wt significantly restored the expression of SKAR α RNA levels and also reversed, at least partially, the upregulation of SKAR β RNA levels (semi-quantitative RT–PCR Figure 3A, normalized qRT–PCR Figure 3B, for original qRT data see Supplementary Figure S1C). In contrast, mutant TDP-43 lacking RRM1, but not RRM2, failed to restore SKAR α RNA levels. Consistently, retransfection with a double-point mutant F147L/F149L (FFLL) that disrupts the RNA binding potential of RRM1 (12,36) prevented the rescue of SKAR α levels. The effects of the respective TDP-43 mutants were also validated on protein level by western blot analysis (Figure 3C and for densitometric quantification Figure 3D). Hence, the alternative splicing of SKAR strictly depends on the presence and functionality of the TDP-43 RRM1.
Figure 3.
SKAR alternative splicing is dependent on RRM1 of TDP-43. (A) Stably silenced HEK293E cells (shRNATDP-43) or transiently silenced HEK293 cells (siRNATDP-43) were transiently transfected with either control vector (−) or Flag-TDP-43 variants (wt, ΔRRM1, ΔRRM2, ΔRRM1/2, FFLL and ΔGRD or disease-associated mutations, as indicated). Parental HEK293E cells or cells treated with a scrambled siRNA (−) were used as an internal control. (A) Total RNA was extracted and subjected to semi-quantitative RT–PCR using primer pairs amplifying total TDP-43, endogenous TDP-43, total SKAR (ex2–ex4), SKAR α (ex2|3–ex4), SKAR β (ex2|4–ex4) and PBGD as a housekeeping gene. (B and E) RNA was extracted and real-time PCR performed with primer pairs against SKAR α (ex2|3–ex4) (white bars), SKAR β (ex2|4–ex4) (gray bars) and total SKAR (ex5|6–ex7). PBGD was used as a housekeeping gene. Resulting relative SKAR α/PBGD, SKAR β/PBGD and total SKAR/PBGD ratios were re-calculated into absolute copy values and normalized to total SKAR values. Original qRT data is presented in Supplementary Figure S1C and S1D, respectively. *P < 0.05; **P < 0.005; ***P < 0.0005; ns = not significant. (C and D) Protein was extracted, electrophoresed and resulting western blots probed with anti-TDP-43, anti-Flag and anti-SKAR antibodies. GAPDH was used as a loading control. (D) Shown are the mean values ± SEM of densitometric analysis of three independent experiments. *P < 0.05; **P < 0.005; ns = not significant.
We also tested the importance of the C-terminal GRD, in which most disease-associated mutations are located. Deletion of the GRD did not impair the rescue of TDP-43 overtly (Figure 3A–C). However, a tendency of less rescue activity could be seen by qRT–PCR experiments and western blots (Figure 3B and C). When testing discrete pathogenic point-mutations, only some mutations (D169G, Q343R, R361S, A382T and N390S) showed a tendency to restore exon 3 inclusion less efficiently than TDP-43 wt (Figure 3E). Thus, the impact of of disease-associated TDP-43 mutations and the GRD in general for SKAR splicing remains unclear. We suggest that TDP-43 splicing activity toward SKAR specifically depends on direct RRM1 binding to the SKAR pre-mRNA (see below) and potential co-factors believed to interact with the TDP-43 GRD (37) are less important for this particular splice target of TDP-43.
A repeat containing RNA stretch 3′ of exon 3 is necessary for TDP-43 and SKAR splicing
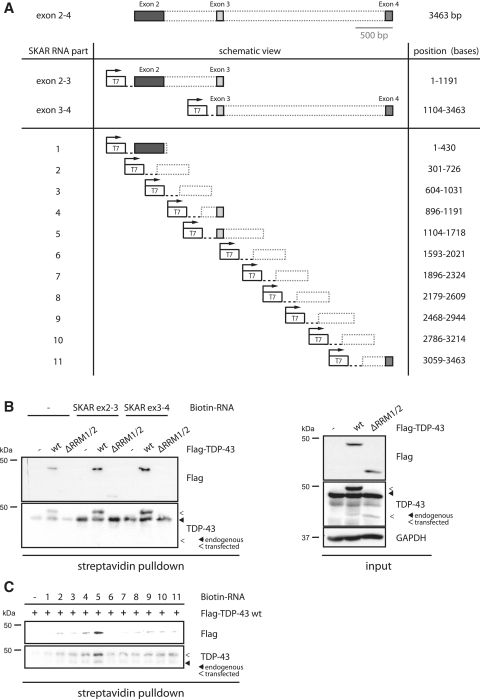
To investigate further, whether TDP-43 mediates exon 3 inclusion by direct binding of the SKAR pre-mRNA, we performed RNA crosslinking experiments. Therefore, we used different parts of the genomic SKAR DNA (Figure 4A) for in vitro transcription/biotinylation. SKAR DNA containing either exons 2–3 or exons 3–4 of the SKAR pre-mRNA was in vitro transcribed/biotinylated, mixed together with lysates from HEK293E cells that were transfected with Flag-TDP-43 wt or ΔRRM1/2, UV-crosslinked and pulled down using streptavidin-coupled agarose. Eluates were separated by SDS–PAGE and western blot probed with TDP-43 and Flag antibodies to detect RNA-bound endogenous and/or transfected TDP-43 protein (note: the ΔRRM1/2 deletion mutant could only detected by the Flag antibody, since the epitope of the TDP-43 antibody is largely deleted in this mutant). Endogenous TDP-43 wt as well as transfected TDP-43 wt did bind to SKAR pre-mRNA (Figure 4B). In contrast, the RRM-lacking mutant ΔRRM1/2, which fails to restore SKAR α levels (Figure 3A–D), was not pulled down efficiently by streptavidin-coupled agarose (Figure 4B), demonstrating the specificity of this assay. Interestingly, SKAR exons 2–3 and exons 3–4 fragments were bound by TDP-43 wt to equal extent, suggesting that TDP-43 has either multiple binding sites or binds to exon 3 itself.
Figure 4.
TDP-43 binds directly to SKAR pre-mRNA. (A) Schematic representation of constructs used for RNA crosslinking assays. (B) SKAR DNAs comprising either exons 2–3 or exons 3–4 including intronic regions were in vitro transcribed/biotinylated and mixed with lysates from untransfected HEK293E cells or HEK293E cells transiently transfected with Flag-TDP-43 wt or ΔRRM1/2. No RNA was added to control samples. Samples were UV crosslinked and precipitated with streptavidin-agarose. Western blots of streptavidin precipitates (left panel) were probed with anti-TDP-43 and anti-Flag to visualize co-precipitated endogenous and exogenous TDP-43. Biotinylated SKAR RNAs pulled down transfected as well as endogenous TDP-43 wt but not ΔRRM1/2. Protein inputs (right panel) of HEK293E lysates confirmed equal transfection efficiencies. Note, that the used TDP-43 antibody does not recognize the TDP-43 ΔRRM1/2 mutant as efficiently as TDP-43 wt because of partial deletion of the epitope. (C) Shorter SKAR fragments, as indicated in (A), were mixed to HEK293E lysates transfected with Flag TDP-43 wt. Anti-Flag and anti-TDP-43 probings of streptavidin pulldowns suggest part-5 to contain the preferential binding site within SKAR mRNA. Endogenous and transfected TDP-43 are indicated by closed and open arrowheads, respectively.
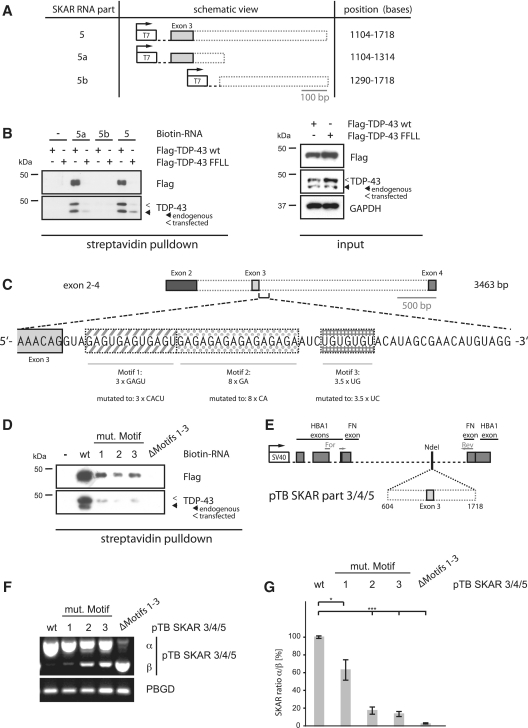
To resolve the binding site more exactly, we went on to generate different, but overlapping, constructs (Figure 4A, bottom) that were subsequently used for in vitro transcription/biotinylation. UV-crosslinking experiments showed that Flag-TDP-43 wt binds strongly to bases 1104–1718 (part-5) within the SKAR exons 2–4 pre-mRNA (Figure 4C), which corresponds to exon 3 plus approximately 500 bases downstream of exon 3. Consistent with the previous experiment using the larger parts exons 2–3 and exons 3–4, TDP-43 bound also to the exon 3 containing overlapping part-4 (bases 896–1191) although to a much lesser extent. Together these data show that TDP-43 binds directly to SKAR pre-mRNA within exon 3 and more prominently to the downstream sequence of exon 3. This binding site within the SKAR pre-mRNA is in accordance with a recently published study that analyzed TDP-43-bound RNAs by CLIP (21). In addition, this binding site is in conformity with the hypothesis of position-dependent regulation of alternative splicing that appears to be a rather general phenomenon: RBPs bind to the 5′ region of an exon or to the exon itself if they mediate skipping whereas binding to the 3′ region often mediates exon inclusion (38), which appears to be the case here.
To narrow down the binding sequence, we generated DNAs containing either only the 5′ (part-5a) or the 3′ (part-5b) region of the preferential bound SKAR pre-mRNA part-5 (Figure 5A). UV-crosslinking experiments using TDP-43 wt and the RNA-binding deficient mutant FFLL showed that transfected TDP-43 wt but not FFLL protein bound to SKAR pre-mRNA part-5 (Figure 5B). Equal binding efficiency was observed for part-5a, which contained the exon 3 plus the first 100 bases downstream. In contrast, TDP-43 did not bind to part-5b, which contained the 3′ region of part-5, indicating that TDP-43 binds the proximal intronic region downstream of SKAR exon 3.
Figure 5.
A repeat containing RNA stretch 3′ of exon 3 is necessary for TDP-43 and SKAR splicing. (A) Schematic representation of constructs used for refined RNA crosslinking assays. (B) Indicated fragments of SKAR DNA were in vitro transcribed/biotinylated and mixed with lysates form HEK293E cells transiently transfected with Flag-TDP-43 wt or FFLL. No RNA was added to control samples. Samples were UV crosslinked and precipitated with streptavidin-agarose. Western blots of streptavidin precipitates (left panel) were probed with anti-TDP-43 and anti-Flag to visualize co-precipitated endogenous and exogenous TDP-43. Biotinylated SKAR RNAs pulled down transfected as well as endogenous TDP-43 wt but not FFLL. Protein inputs (right panel) of HEK293E lysates confirmed even transfection efficiencies. (C) Schematic representation of the three repeat motifs and mutagenized variants within the SKAR pre-RNA 3′ of exon 3. (D) Non-mutated or mutagenized variants of SKAR DNA part-5 were in vitro transcribed/biotinylated and mixed with lysates form HEK293E cells transiently transfected with Flag-TDP-43 wt. No RNA was added to control samples. Samples were UV-crosslinked and precipitated with streptavidin-agarose. Western blots of streptavidin precipitates were probed with anti-TDP-43 and anti-Flag to visualize coprecipitated endogenous and exogenous TDP-43. (E) Schematic representation of the used SKAR minigene construct pTB SKAR part-3/4/5. Primer annealing sites are indicated by arrows. (F and G) HEK293E cells were transfected with pTB SKAR part-3/4/5 variants, as indicated. RNA was extracted and used for RT–PCR using primers for pTB and PBGD as a housekeeping gene. (F) Representative RT–PCR is shown. (G) Shown are the results (mean values ± SEM) of densitometric analysis of seven independent experiments calculated as the ratio of SKAR α to SKAR β. *P < 0.05; ***P < 0.0005.
A closer look at this sequence revealed three neighboring repeats: motif A (containing 3× GAGU), motif B (containing 8× GA) and motif C, the latter representing a short variant of the consensus binding motif (GU)n of TDP-43 (26) 5′-UGUGUGU-3′ (Figure 5C). To analyze the binding of TDP-43 to these repeat motifs, we mutagenized each individually (by changing guanines to cytosines) or deleted them altogether (Figure 5C) and performed RNA crosslinking experiments. Mutagenesis of all three individual motifs led to a strong decrease in the binding affinity toward TDP-43 compared to non-mutated RNA (Figure 5D), suggesting that all three motifs contribute to TDP-43 binding in part. A complete loss of binding could only be achieved by complete deletion of all three motifs (Figure 5D).
We next tested the functional consequences upon mutagenesis of the RNA repeat motifs by generating SKAR splicing reporter constructs. A region of the SKAR pre-mRNA (exon 3 plus 500 bp up- and downstream, corresponding to SKAR parts 3 + 4 + 5) was cloned into an established splicing reporter vector (pTB) with an SV40 promoter and flanking human α-globin and fibronectin exons (28) (Figure 5E). Under basal conditions, expression of this vector led to the appearance of a single RNA product, whereas TDP-43 knockdown led to dramatic and significant switch to an alternatively spliced product, corresponding to the isoforms SKAR α and SKAR β, respectively (Supplementary Figure S2A, for densitometric quantification see Supplementary Figure S2B). Compared to the non-mutagenized splicing reporter, expression of mutagenized SKAR RNA motifs that we showed to be involved in TDP-43 binding (see above) led to the appearance of the alternatively spliced SKAR β isoform even under non-silenced conditions (Figure 5F and G), although to varying extent. Mutagenesis of motif A had the least effect, whereas mutagenesis of motif B and C similarly decreased the splicing efficiency to <20%. In consistence with the RNA-binding experiments, the deletion of all three motifs had the strongest effect onto the splicing efficiency of this SKAR splicing reporter. Thus, deletion of the TDP-43 binding sites leads to exactly the same exon 3 skipping effect as does silencing of TDP-43 for the SKAR wt pre-mRNA.
SKAR β is more active than SKAR α
Rather little is known about the functions of SKAR. It was originally found as a protein interacting with DNA polymerase II (39). More recently, SKAR was identified as a protein interacting with S6K1 and to increase cell size in an S6K1-dependent fashion (40). Moreover, SKAR can recruit S6K1 to the EJC, a complex that forms upon splicing of intron-containing pre-mRNAs at the exon junction. Thereby, SKAR facilitates the EJC-dependent pioneer round of translation (27). Nevertheless, isoform β was not investigated. Bioinformatic analyses of SKAR α and β variants could not reveal any obvious difference between both isoforms. Moreover, exon 3 does not contain a known protein motif or sites that are predicted to be post-translationally modified (Figure 1F).
To detect functional divergences of both SKAR isoforms, we first tested the possibility of different cellular localization of the nuclear cytoplasmic shuttling protein SKAR (27). We could not find any sign for altered subcellular localization (Supplementary Figure S3A) or subnuclear distribution (Supplementary Figure S3B). However, we were restricted to the analysis of overexpressed SKAR α and SKAR β, respectively, because of the low sensitivity of the SKAR antibody toward endogenous protein levels in immunofluorescence stainings.
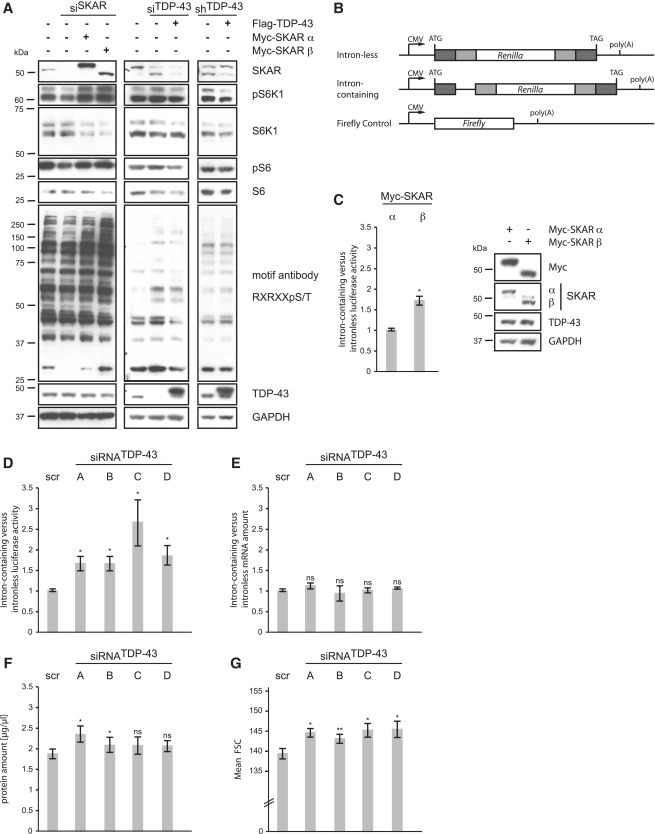
Since SKAR enhances the activity of S6K1 and downstream S6K1-dependent phosphorylation events (27), we tested the activity of both SKAR isoforms on the phosphorylation state of S6K1 itself and of its downstream targets. Therefore, we treated cells with SKAR-specific siRNA and re-expressed Myc-tagged SKAR α or SKAR β cDNAs. Western blotting using phospho-S6K1, phospho-S6 and the S6K1 phospho-motif RXRXXpS/T antibodies showed, in agreement with a previous study (27), that cells treated with SKAR siRNA had lower phosphorylation levels compared to non-silenced cells. As expected, SKAR retransfections restored S6K1 phosphorylations. Interestingly, cells-expressing SKAR β had higher phosphorylation levels compared to SKAR α transfected cells (Figure 6A, left panel). This indicates that SKAR β is more active than the SKAR α isoform. Although the effects of the TDP-43 silencing were less prominent compared to direct effect of SKAR silencing, we found that TDP-43 silenced cells had higher phosphorylation levels of S6K1 and of S6K1-dependent targets than unsilenced cells (Figure 6A, middle panel) or cells that have been additionally retransfected with Flag-TDP-43 wt and therefore express less of the SKAR β isoform, which we suggest to be more active (Figure 6A, middle and right panel).
Figure 6.
SKAR β is more active than SKAR α and leads to enhanced translation and increased cell size. (A) HEK293E cells were treated with control siRNA or transfected with siRNA against SKAR or TDP-43 as indicated. Stably silenced siRNATDP-43 and transiently transfected HEK293E cells were transfected with either control vector (−) or plasmids encoding for Myc-SKAR α, Myc-SKAR β or Flag-TDP-43 wt, as indicated. Cells were serum starved for 16 h. After 6 h serum-stimulation cells were harvested, protein extracted and electrophoresed. Resulting western blots were probed with anti-SKAR, anti-phospho S6K1 (Thr389), anti-S6K1, anti-phospho S6 (Ser235/236), anti-S6, anti-phospho Akt substrate (RXRXXS/T) and anti-TDP-43 antibodies. GAPDH was used as a loading control. Transfection of SKAR β or depletion of TDP-43 results in overall stronger phospho-signal compared to SKAR α. (B) Schematic representation of luciferase constructs used for analysis of translation. (C–G) HEK293E cells were transfected with either Myc-SKAR α or Myc-SKAR β (C) or with control siRNA (scr) and individual siRNATDP-43 A–D, as indicated (D–G). (C–E) Before DNA/siRNA transfection, cells were transfected with firefly control vector plus either intron-containing or intron-less Renilla luciferase constructs. (C and D) Luciferase activity was measured and normalized to control treated HEK293E cells. Shown are the mean values ± SEM of five independent experiments. *P < 0.05. Western blotting confirmed equal expression of Myc-SKAR α and Myc-SKAR β (C, right panel). (E) qRT–PCR confirmed equal RNA levels of Renilla and firefly luciferase in non-silenced and silenced HEK293E cells. (F) Cells were counted and equal numbers of cells was collected. Protein amount was determined using BCA protein assay. Shown are the mean values ± SEM of five independent experiments. *P < 0.05. (G) Cell size was analyzed by flow cytometry, monitoring the forward scatter parameter. Shown are the mean values ± SEM of five independent experiments. *P < 0.05; **P < 0.005.
We went on to investigate the efficiency of the pioneer round of translation, which is restricted to intron-containing pre-mRNAs. Therefore, we measured the luciferase activities of established intron-containing versus intron-less Renilla luciferase constructs (29). A firefly luciferase was used to control for transfection efficiencies (Figure 6B). Compared to SKAR α, cells overexpressing SKAR β showed a significantly higher ratio of intron-containing to intron-less luciferase activity (Figure 6C, left panel), although both isoforms were expressed equally (Figure 6C, right panel). Thus, SKAR β more actively promotes splice-dependent translational yield than SKAR α. Consistently, TDP-43 depleted cells, which express more SKAR β than SKAR α, showed a significantly higher ratio of intron-containing to intron-less luciferase activity compared to non-silenced cells (Figure 6D). The luciferase mRNA levels were not affected by the transfections (Figure 6E), ruling out potential effects on transcription and/or reporter mRNA stability. These results suggest that the pioneer round of translation is more efficiently engaged in TDP-43 silenced cells. In line with this, TDP-43-depleted HEK293E cells also showed a small but clear tendency toward a higher total protein amount, as measured by BCA assay (Figure 6F) and significantly increased cell size compared to non-silenced cells, as measured by flow cytometry (Figure 6G and for stably silenced cells see Supplementary Figure S4). Together these data show that SKAR β is more active than SKAR α and that TDP-43 silencing increases cell size and enhances the splicing-dependent pioneer round of translation. We speculate that disturbed translation as mediated by TDP-43 deficiency may contribute to proteotoxicity in human neurodegenerative diseases.
DISCUSSION
RNA metabolism is linked to neurodegenerative diseases, particularly to FTD and ALS, in a conspicuous manner. The RBPs TDP-43 and FUS/TLS, which compose each specific subsets of tau-negative, ubiquitin-positive aggregates in FTD/ALS are both linked to transcription, mRNA splicing and miRNA processing (5). In addition, other disease-related genes (elongator protein 3, senataxin and angiogenin) have been functionally connected to RNA processing (41–43). Thus, RNA mismetabolism appears as an emerging pathway in FTD/ALS. The vital importance of TDP-43 is further underlined by the embryonic lethality of homozygous knockout animals (36,44–46), which is besides neurological and muscular defects one of the frequently observed phenotypes upon knockout of RBPs (47).
To screen for novel splice targets of TDP-43, we have performed microarray expression profiling using exon array technologies. Currently identified disease-associated genes were neither quantitatively (17) nor qualitatively (this study) altered upon TDP-43 knockdown in HEK293E cells. However, we have discovered a significant shift from the SKAR α to the SKAR β isoform upon depletion of TDP-43. This regulation was further validated in human neuronal cells and has been confirmed by others in mouse adult brain tissue upon TDP-43 depletion (20), emphasizing that this effect is evolutionary conserved and relevant for the nervous system. Using RNA crosslinking assays, we have further shown that TDP-43 binds directly to the SKAR pre-mRNA. For this, the RRM1 is absolutely required, since deletion or mutagenesis of this domain rendered TDP-43 is incapable of binding to the SKAR pre-mRNA and also to restore exon 3 inclusion. In contrast to RRM1, RRM2 of TDP-43 seems dispensible for SKAR exon 3 inclusion, which is in accordance with previous publications on other TDP-43-bound RNAs (12). Similar to reports on CFTR splicing and HDAC6 regulation (17,48), disease-associated TDP-43 point mutations, which mostly cluster within the GRD, did not overtly affect the splicing activity toward SKAR pre-mRNA. Thus, these tested pathogenic mutations do not represent loss-of-function mutations, at least in regard to the until now tested RNA substrates. Surprisingly, deletion of the whole GRD only partially reduces SKAR splice activity of TDP-43. It appears that this particular splicing event depends more on direct TDP-43 binding than potential GRD-interacting co-factors (37).
At the RNA level, our mapping experiments show that TDP-43 binds to a proximal intronic region downstream of the alternatively spliced SKAR exon 3. The identified TDP-43 binding region within the SKAR pre-mRNA is consistent with the reported HITS-CLIP crosslink cluster (21). We detected three repeat motifs in this region, and found that each of the repeats contribute to some TDP-43 binding, but only the deletion of all three motifs abolished TDP-43 binding. Importantly, this was functionally mirrored in a SKAR exon 3 splice reporter assay we generated. Deletion of the TDP-43 binding sites had the same exon 3 skipping effect as TDP-43 silencing for the cell endogenous SKAR pre-mRNA. In conclusion, TDP-43 binds to a defined region just downstream of the alternatively spliced SKAR exon 3, and mediates SKAR exon 3 inclusion.
The functions of SKAR are not very well understood. However, it is known that SKAR positively affects cap-dependent protein translation (27). In addition, SKAR mediates S6K1-dependent effects on cell size (40), probably as a result of enhanced protein translation. However, SKAR α and β isoforms have not been investigated in comparison so far. While bioinformatic analyses have not revealed protein motifs or sites for post-translational modifications in or around exon 3, it may well be that the sequence encoded by exon 3 regulates the activity of SKAR. This may be due to altered protein–protein interactions and/or structural changes caused by either lack of a inhibitory function encoded by exon 3 itself, or by creation of a novel stimulatory function by fusion of exons 2 and 4. Indeed, when comparing the activities of both SKAR isoforms, we found that reported functions of SKAR were enhanced by expression of the SKAR β isoform. This included increased phosphorylation levels of S6K1 downstream targets and S6K1 itself. Thus, SKAR β renders S6K1 more active than SKAR α. Consistently, cells with reduced TDP-43 also showed enhanced phosphorylation levels of S6K1 and of downstream targets. Functionally, this correlated with enhanced protein translation of intron-containing RNAs, and with increased total protein content and cell size.
In summary, we have shown that TDP-43 regulates the alternative splicing of SKAR. TDP-43 knockdown almost completely shifts the expression from the predominant SKAR α to the β isoform, which activates S6K1 more efficiently and increases global protein translation. We have recently shown that TDP-43 knockdown impairs the turnover of toxic proteins by HDAC6 downregulation. In line with this, enhanced S6K1-dependent protein synthesis might be another mechanism by which TDP-43 depletion may disturb cellular protein homeostasis. Thus, TDP-43 appears to be generally involved in protein metabolism by opposite regulation of anabolic and catabolic cellular functions that may eventually contribute to disease pathogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–4.
FUNDING
German Competence Network ‘Degenerative Dementias’ (01GI0705), German Research Foundation (DFG grant KA 1675/3-1), German Center for Neurodegenerative Diseases and the Hertie Foundation. Funding for open access charge: German Research Foundation (DFG grant KA 1675/3-1)
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Emanuele Buratti and Melissa Moore for providing the pTB and Renilla luciferase constructs, respectively, Wolfdieter Springer for correction and commenting the manuscript and the Microarray Facility Tuebingen for execution of the microarray profiling. F.C.F. designed the experiments. F.C.F. and S.S.W. conducted the experiments. J.S. and A.Z. performed the FIRMA analysis. F.C.F. and P.J.K. wrote the manuscript. P.J.K. is the principal investigator.
REFERENCES
- 1.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 2.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 3.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiesel FC, Kahle PJ. TDP-43 and FUS/TLS: cellular functions and implications for neurodegeneration. FEBS J. 2011;278:3550–3568. doi: 10.1111/j.1742-4658.2011.08258.x. [DOI] [PubMed] [Google Scholar]
- 7.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abhyankar MM, Urekar C, Reddi PP. A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues: role for TDP-43 in insulator function. J. Biol. Chem. 2007;282:36143–36154. doi: 10.1074/jbc.M705811200. [DOI] [PubMed] [Google Scholar]
- 9.Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell. Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Ayala YM, De Conti L, Avendano-Vazquez SE, Dhir A, Romano M, D'Ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buratti E, De Conti L, Stuani C, Romano M, Baralle M, Baralle F. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010;277:2268–2281. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- 12.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 13.Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bose JK, Wang IF, Hung L, Tarn WY, Shen CK. TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J. Biol. Chem. 2008;283:28852–28859. doi: 10.1074/jbc.M805376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayala YM, Misteli T, Baralle FE. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc. Natl Acad. Sci. USA. 2008;105:3785–3789. doi: 10.1073/pnas.0800546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dreumont N, Hardy S, Behm-Ansmant I, Kister L, Branlant C, Stevenin J, Bourgeois CF. Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acids Res. 2010;38:1353–1366. doi: 10.1093/nar/gkp1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiesel FC, Voigt A, Weber SS, Van den Haute C, Waldenmaier A, Gorner K, Walter M, Anderson ML, Kern JV, Rasse TM, et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 2010;29:209–221. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SH, Shanware NP, Bowler MJ, Tibbetts RS. Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J. Biol. Chem. 2010;285:34097–34105. doi: 10.1074/jbc.M110.154831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang PM, Ling J, Jeong YH, Price DL, Aja SM, Wong PC. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc. Natl Acad. Sci USA. 2010;107:16320–16324. doi: 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 23.Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 26.Ayala YM, Pantano S, D'Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE. Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J. Mol. Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 27.Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 28.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purdom E, Simpson KM, Robinson MD, Conboy JG, Lapuk AV, Speed TP. FIRMA: a method for detection of alternative splicing from exon array data. Bioinformatics. 2008;24:1707–1714. doi: 10.1093/bioinformatics/btn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bengtsson H, Simpson K, Bullard J, Hansen K. 2008 aroma.affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. Tech Report 745, Department of Statistics, University of California, Berkeley. [Google Scholar]
- 32.Bengtsson H, Irizarry R, Carvalho B, Speed TP. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics. 2008;24:759–767. doi: 10.1093/bioinformatics/btn016. [DOI] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 34.Durinck S, Bullard J, Spellman PT, Dudoit S. GenomeGraphs: integrated genomic data visualization with R. BMC Bioinform. 2009;10:2. doi: 10.1186/1471-2105-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 36.Voigt A, Herholz D, Fiesel FC, Kaur K, Muller D, Karsten P, Weber SS, Kahle PJ, Marquardt T, Schulz JB. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS One. 2010;5:e12247. doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 38.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L, Rodriguez-Belmonte EM, Mazloum N, Xie B, Lee MY. Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen. J. Biol. Chem. 2003;278:10041–10047. doi: 10.1074/jbc.M208694200. [DOI] [PubMed] [Google Scholar]
- 40.Richardson CJ, Broenstrup M, Fingar DC, Julich K, Ballif BA, Gygi S, Blenis J. SKAR is a specific target of S6 kinase 1 in cell growth control. Curr. Biol. 2004;14:1540–1549. doi: 10.1016/j.cub.2004.08.061. [DOI] [PubMed] [Google Scholar]
- 41.Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am. J. Hum. Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suraweera A, Lim Y, Woods R, Birrell GW, Nasim T, Becherel OJ, Lavin MF. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum. Mol. Genet. 2009;18:3384–3396. doi: 10.1093/hmg/ddp278. [DOI] [PubMed] [Google Scholar]
- 43.Svejstrup JQ. Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 2007;19:331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Kraemer BC, Schuck T, Wheeler JM, Robinson LC, Trojanowski JQ, Lee VM, Schellenberg GD. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta. Neuropathol. 2010;119:409–419. doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sephton CF, Good SK, Atkin S, Dewey CM, Mayer P, III, Herz J, Yu G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu LS, Cheng WC, Hou SC, Yan YT, Jiang ST, Shen CK. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 47.Gabut M, Chaudhry S, Blencowe BJ. SnapShot: the splicing regulatory machinery. Cell. 2008;133:192 e191. doi: 10.1016/j.cell.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 48.D'Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, Baralle FE. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.