Abstract
Exposure to aristolochic acid (AA), a component of Aristolochia plants used in herbal remedies, is associated with chronic kidney disease and urothelial carcinomas of the upper urinary tract. Following metabolic activation, AA reacts with dA and dG residues in DNA to form aristolactam (AL)-DNA adducts. These mutagenic lesions generate a unique TP53 mutation spectrum, dominated by A : T to T : A transversions with mutations at dA residues located almost exclusively on the non-transcribed strand. We determined the level of AL-dA adducts in human fibroblasts treated with AA to determine if this marked strand bias could be accounted for by selective resistance to global-genome nucleotide excision repair (GG-NER). AL-dA adduct levels were elevated in cells deficient in GG-NER and transcription-coupled NER, but not in XPC cell lines lacking GG-NER only. In vitro, plasmids containing a single AL-dA adduct were resistant to the early recognition and incision steps of NER. Additionally, the NER damage sensor, XPC-RAD23B, failed to specifically bind to AL-DNA adducts. However, placing AL-dA in mismatched sequences promotes XPC-RAD23B binding and renders this adduct susceptible to NER, suggesting that specific structural features of this adduct prevent processing by NER. We conclude that AL-dA adducts are not recognized by GG-NER, explaining their high mutagenicity and persistence in target tissues.
INTRODUCTION
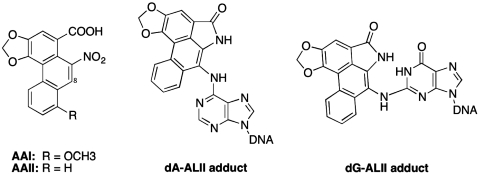
Aristolochia herbals have been used for centuries for medicinal purposes; only recently have their serious toxic effects been recognized (1,2). Aristolochic acid (AA) is a collective term used to describe the complex mixture of structurally related nitrophenanthrene carboxylic acids produced by various Aristolochia species (3). The principal constituents of this mixture are AA-I and AA–II, which differ only in the presence or absence of an O-methoxy group at C-8 (Figure 1). Both AA-I and AA-II react with DNA to form adducts—we used AA-II in all experiments reported here.
Figure 1.
Structures of AA and dA-AL and dG-AL adducts. The DNA adducts are formed by cellular reduction of the nitro group, hydroxy lactone formation and activation to react with the exocyclic amine groups of dA and dG.
In the early 1990s, more than 100 Belgian women developed chronic kidney disease and urothelial carcinomas after ingesting a slimming regimen that contained Aristolochia fangchi (4). Aristolactam (AL)-DNA adducts were found in the renal and urothelial tissues of these patients, confirming prior exposure to AA (5,6). The syndrome, initially called Chinese herbs nephropathy (CHN), later was renamed aristolochic acid nephropathy (AAN). AAN is characterized by tubulointerstitial nephritis with urothelial carcinomas of the upper urinary tract developing in 50% of the cases (7). Many cases of AAN have subsequently been reported worldwide (1,2).
Aristolochic acids are activated by cellular nitroreductases (8), forming reactive intermediates that bind covalently to DNA. Via this mechanism, AAI and AAII produce the following DNA adducts: 7-(deoxyadenosin-N6-yl)aristolactam I (dA-ALI), 7-(deoxyguanosin-N2-yl)aristolactam I (dG-ALI), 7-(deoxyadenosin-N6-yl)aristolactam II (dA-ALII) and 7-(deoxyguanosin-N2-yl)aristolactam II (dG-ALII) (Figure 1) (9). Both dG and dA adducts block DNA replication and exhibit miscoding properties (10–12). dA-AL adducts are significantly more mutagenic and/or persistent than dG-AL adducts, reflected in the observation that chronic exposure of humans to AA leads predominantly to A : T to T : A transversions (13,14). A remarkable feature of dA-AL adducts is their persistence in target tissues of rodents and humans (6,13,15–17), suggesting that these adducts are at least partially resistant to repair.
In humans, nucleotide excision repair (NER) is the primary pathway responsible for the removal of bulky DNA lesions, including those formed by AA (18). There are two subpathways of NER, transcription-coupled repair (TC-NER) and global-genome repair GG-NER) that differ with respect to the manner in which the substrate lesions are recognized. Following lesion recognition, these two subpathways converge to a common set of steps to complete the repair process, including DNA opening, strand incisions by XPF and XPG, and filling of the gap.
TC-NER has been shown to operate specifically on lesions in the transcribed strand of expressed genes and is initiated when RNA polymerase is stalled at the lesion site (19). In contrast, GG-NER monitors the entire genome and is initiated by the damage sensor XPC-RAD23B (20,21), sometimes in conjunction with an ubiquitin–ligase complex containing the UV-damage binding protein subunit DDB2 (22). In the GG-NER pathway, there is a strong correlation between the degree of thermodynamic destabilization induced by the presence of a DNA lesion and the efficiency of NER (23,24). In line with this observation, XPC-RAD23B recognizes lesions indirectly by binding to the undamaged strand opposite the lesion, which is rendered more accessible by destabilization of duplex DNA (25–27). No such correlation has been established for TC-NER, which does not employ XPC-RAD23B, and it is believed that most bulky lesions will effectively block transcription by RNA polymerase II (28–31) leading to the initiation of repair.
Recently, AA was shown to be the etiologic agent responsible for Balkan endemic nephropathy (BEN) (13,17). This environmental disease is characterized by renal tubulointerstitial fibrosis associated with a high incidence of upper urinary tract carcinomas (32). Analysis of the mutational profile of the p53 tumor suppressor gene in BEN patients revealed a unique pattern of A : T→T : A transversions characteristic of exposure to AA (14). Importantly, mutated adenosines were located exclusively on the non-transcribed strand, suggesting that dA-AL adducts on either strand are likely to be refractory to GG-NER, while adducts on the transcribed strand are repaired by TC-NER. A similar strand bias was observed in studies of Hupki mice exposed to AAI (33). Both the persistence of dA-AL adducts and the marked strand bias for A→T mutations suggest that dA-AL adducts are selectively repaired by TC-NER.
To explore the involvement of TC-NER and GG-NER in repair of dA-AL DNA adducts, we investigated the cytotoxicity and genotoxicity of ALII in cell lines with defects in one or both of these repair pathways. Additionally, we investigated NER susceptibility and XPC-RAD23B binding affinity for dA-ALII adducts in vitro. Taken together, these studies demonstrate that dA-AL-DNA adducts fail to bind XPC-RAD23B and, therefore, are refractory to repair by GG-NER, while being processed efficiently by TC-NER.
MATERIALS AND METHODS
Chemicals
γ-32P-ATP (6000 Ci/mmol) and α-32P-dCTP were purchased from PerkinElmer (Boston, USA). Cis-diamminedichloroplatinum(II), cisplatin, obtained from Sigma Co. (St. Louis, MO, USA), was dissolved, at 10 mM, in DMSO and stored at −80°C. Aristolochic acid II with a purity of >97%, was dissolved, at 25 mM, in DMSO and stored at −20°C. The concentration of AAII was established by UV absorption at 250 nm (34). Enzymes used for 32P-post-labeling analysis were obtained from Worthington (Newark, NJ, USA) and Sigma. dG-ALII, dA-ALII, N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF), and N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AF), containing oligonucleotides were synthesized as described (12,35). All other oligonucleotides were purchased from Integrated DNA Technologies (IDT).
Cell lines
Table 1 lists the cell lines used for the cytotoxicity and genotoxicity assays. XP cell lines were obtained from the Coriell repository. CS1AN/pc3.1, CS1AN/p3.1-CSBwt and CS1ANE7/p3.1-CSBE646Q (CSB deficient and CSB corrected cell lines, CSB, CSB+, CSBmut, respectively) were kindly provided by Vilhelm Bohr (National Institute of Aging, Baltimore, MD, USA) (36). All cell lines were SV-40 transformed.
Table 1.
Cell lines used in the study
| Cell line | Phenotype | Name | Expected GGR status | Expected TCR status |
|---|---|---|---|---|
| GM00637 | Wild-type | Control | + | + |
| CS1AN/p3.1 | CSB | CSB | + | − |
| CS1AN/p3.1-CSBwt | CSB corrected | CSB+ | + | + |
| CS1ANE7/p3.1-CSBE646Q | CSB mutant corrected | CSBmut | + | − |
| GM16094 | CSA | CSA | + | − |
| GM15983 | XPC | XPC | − | + |
| GM04312 | XPA | XPA | − | − |
| GM15876 | XPA corrected | XPA+ | + | + |
| GM14930 | XPG | XPG | − | − |
| GM08207 | XPD | XPD | − | − |
| GM15877 | XPD corrected | XPD+ | + | + |
Cell culture and chemical exposures
Cells were grown in 75 cm2 flasks containing Dulbecco's modified minimum essential medium (high glucose DMEM) supplemented with fetal bovine serum (10%), penicillin (100 U/ml) and streptomycin (100 mg/ml). The medium for CS1AN and XPD complemented cell lines contained geneticin (0.4 mg/ml). Cultures were maintained at standard conditions in a humidified atmosphere containing 5% CO2 at 37°C. Cultures were split in a 1 : 5 ratio every third to fourth day. Before treatment, cells were passed in a 1 : 6 ratio and 0.5 ml culture maintained on 24-well plates for cytotoxicity assays. Also, 5 ml of the culture was grown to confluence on 60-mm plates. Following exposure to drug, DNA was isolated for adduct analysis. Before treatment with the DNA-damaging agents, cultures were washed thoroughly with Dulbecco's phosphate buffered saline (PBS) without CaCl2 and MgCl2. Then 5–200 µM AAII or 1–10 µM cisplatin, diluted in DMEM without supplements, were added to the cells as follows and incubated under standard conditions: CSB+ and XPD+ cell lines were exposed in the presence of geneticin (0.4 mg/ml) to maintain CSB and XPD complementation. Cells were treated with AAII or cisplatin for 48 h for adduct analyses and with AAII for 72 h for cytotoxicity studies. To establish the time course for adduct formation, wild-type and selected cell lines were exposed to 100 and 10 µM AAII, for up to 72 h, respectively.
Cell viability assay
To measure cell viability, ATP content was assayed using a FLASC kit from Sigma. After exposure to AAII, cells in 24-well plates were washed with PBS, and 400 µl lysis buffer (FL-SAR) was added to each well with the following incubation at room temperature for 10 min. Luciferase (FL-AAM) was diluted 20-fold in the FL-AAB buffer and the luminescence of 5 µl sample in 100 µl enzyme was measured on a TD-20/20 ‘Luminometer’ (Turner Designs). The toxicity for each chemical dose was defined as the ratio of ATP in treated cells to ATP in the untreated control. Treatments were conducted in six different wells for each exposure. Ratio data were analyzed using Sigma Plot v8.0 (SPSS Inc.).
32P-post-labeling adduct analysis with gel electrophoresis
DNA adduct levels were measured as described previously (37). A total of 15 fmol of each of the following 24-mer oligonucleotides were used as standards. This corresponds to 1 adduct/106 nt in 5 µg DNA.
5′-TCT TCT TCT GTG CXC TCT TCT TCT-3′ X = dA-ALII
5′-TCT TCT TCT GTX CAC TCT TCT TCT-3′ X = dG-ALII
Following exposure to AAII, cells were washed with phosphate buffered saline, collected by centrifugation and stored at −80°C for DNA adduct analysis. Briefly, DNA was isolated using the DNeasy Blood and Tissue Qiagen kit (Cat. N 69506, QIAGEN Sciences, MD, USA). DNA (5–20 µg) was digested at 37°C overnight with phosphodiesterase and micrococcal nuclease. Following further digestion with nuclease-P1 for 1 h at 37°C: AL-DNA adducts were extracted with butanol as described (37), then treated with 20 µCi γ-32P-ATP and 10 U of OptiKinase (Affymetrix Inc.). After evaporating the solution to dryness, 5 µl of formamide-EDTA buffer with xylencyanol and bromophenol blue was added. Samples were loaded on 30% non-denaturating acrylamide gel prerun for 40 min at 1600 V. The running buffer consisted of 200 mM boric acid, 2.5 mM EDTA, 100 mM Tris–base, pH 7.0. The gel was allowed to run for 4 h at 1800 V and then visualized by phosphorimaging. Image QuaNT v5.2 (Molecular Dynamics) was used to estimate the amount of adducts present. The dependence of adduct levels on AAII dose was analyzed using Sigma Plot v8.0 (SPSS Inc.).
XPC-RAD23B purification and HeLa whole-cell extract preparation
Polyhistidine-tagged RAD23B was expressed in E. coli BL21(DE3)LysS using the expression vector pET-24d and purified on nickel beads (Qiagen) as described (38). Polyhistidine-tagged XPC was expressed in Sf9 cells using the expression vector pFastBac1. Cells were lysed as described and the third supernatant fraction S3 was combined with partially purified RAD23B (25). The correctly folded heterodimer was further purified through nickel beads (Qiagen), gel filteration (Pharmacia) and heparin (Amersham) columns. HeLa whole-cell extracts were prepared as described (39).
Preparation of oligonucleotides
The 44-mer oligonucleotides containing dA-ALII were prepared as described (12), purified by HPLC and characterized by ESI-MS using a Micromass Platform LC/MS. 24-mer and 44-mer oligonucleotides containing a single, dG-AAF/AF lesion were prepared and purified as described (35).
Sequences of oligonucleotides containing dA-ALII and dG-AAF/AF were as follows (5′–3′) with the modified residue indicated in bold: C209-ALII: d(AGACAGCCCTAGTACGATGACA(ALII)GAAACACTGCGTGC ATGGATCC); C209-ALII with mismatch: d(AGACAGCCCTAGTACGATG ACA(ALII)CAAACACTGCGTGCATGGATCC); SI6-ALII: d(AGACAGCCCTAGT ACTCTCCTA(ALII)GGTTGGCTCGGTGCATGG ATC); SI6-ALII with mismatch: d(AGACAGCCCTAGTACTCTCCCA(ALII)CGTTGGCTCGCGTGCATGGATC); 24-mer NarI-AAF: d(CTATTACCGGCG(AAF)CCACATGTCAGC); 24-mer NarI-AF: d(CTATTACCGGCG(AF)CCACATGTCAGC); 44-mer NarI-AAF: d(CCCT AGCTAGAGCTACGTAGCTATTACCGGCG(AAF)CCACATGTC AGC); 44-mer NarI-AF: d(CCCTAGCTAGAGCTACGTAGCTATTACCGGCG(AF)CCACATGTC AGC).
Preparation of plasmids for in vitro NER assay
The following oligonucleotides (5′–3′) were cloned into pBluescript II SK+ using two BbsI restriction sites. Primer 1 for C209: d(CCCTAGTACGATGACAGAAACACTGC); primer 2 for C209: d(GCACGCAGTGTTTCTGTCATCGTACT); primer 3 for C209 with mismatch: d(CCCTAGTACGATGAGGGAAACACTGC); primer 4 for C209 with mismatch: d(GCACGCAGTGTTTCCCTCATCGTACT); primer 5 for SI6: d(CCCTAGTACTCTCCTAGGTTGGCTCGC); primer 6 for SI6: d(GCACGCGAGCCAACCTAGGAGAGTACT); primer 7 for SI6 with mismatch: d(CCCTAGTACTCTCCGGGGTTGGCTCGC); primer 8 for SI6 with mismatch: d(GCACGCGAGCCAACCCCGGAGAGTACT). Single-stranded circular DNA was generated from the resulting plasmids as described (35) and used in primer extension reaction to generate the lesion-containing double-stranded plasmids as follows: 100 pmol of the 44-mer oligonucleotides was 5′-phosphorylated by incubation with 20 U of T4 PNK enzyme and 2 mM of ATP for 2 h. After annealing with 31 pmol of single-stranded modified pBluescript II SK+ (35), further incubation with dNTPs, T4 DNA polymerase and T4 DNA ligase yielded covalently closed circular DNA containing a single ALII adduct (39). The closed circular DNA was purified by cesium chloride/ethidium bromide density gradient centrifugation followed by consecutive butanol extractions to remove the ethidium bromide and concentrated on a Centricon YM-30 (Millipore). The plasmid was further purified by sucrose gradient centrifugation (39). Fractions containing the closed circular plasmid were pooled and concentrated on a Centricon YM-30 (Millipore). The purified plasmids containing single dA-ALII and dG-AAF/AF lesions were aliquoted and stored at −80°C. The NER dual incision assay was performed using an established protocol (39). HeLa cell extracts (2 μl, 21 mg/ml) or XP-F cell extract (3 μl, 12.5 mg/ml), 2 μl of 5× repair buffer (200 mM Hepes-KOH, 25 mM MgCl2, 110 mM phosphocreatine (di-Tris salt, Sigma), 10 mM ATP, 2.5 mM DTT and 1.8 mg/ml BSA, adjusted to pH 7.8), 0.2 μl 2.5 mg/ml creatine phosphokinase (rabbit muscle CPK, Sigma) and either purified ERCC1-XPF protein (0.5 µl, 0.18 mg/ml) or 400 mM NaCl (0.5 µl, final NaCl concentration was 70 mM) in a total volume of 10 μl were pre-incubated at 30°C for 10 min. dA-AL or dG-AAF/AF-containing plasmid DNA (1 µl, 50 ng/µl) was added and the mixture incubated at 30°C for 45 min. Samples were placed on ice, 0.5 μl of 1 μM of a 3′-phophorylated complementary oligonucleotide was added and the mixtures heated at 95°C for 5 min. The samples were allowed to cool at room temperature for 15 min to allow the DNA to anneal. One microliter of a sequenase/[α-32P]-dCTP mix (0.25 U of sequenase and 2.5 μCi of [α-32P]-dCTP per reaction) was added, the mixture was incubated at 37°C for 3 min, and 1.2 μl of dNTP mix (100 μM of each dATP, dTTP, dGTP, 50 μM dCTP) was added and the mixture incubated for another 12 min. The reactions were stopped by adding 12 μl of loading dye (80% formamide, 10 mM EDTA) and heating at 95°C for 5 min. The samples were analyzed on a 14% sequencing gel (0.5× TBE) at 45 V for 2.5 h. Reaction products were visualized using a PhosphorImager (Typhoon 9400, Amersham Biosciences).
Sequences (5′–3′) of complementary strands (CS) used in in vitro NER assay were as follows: CS 1 for NarI-AAF/AF-containing plasmids: d(GGGGAGTGTTTCTGTCATCGTACTAGGGCTGT); CS 2 for a C209-containing plasmid: d(GGGGAGTGTTTCT GTCATCGTACTAGGGCTGT); CS 3 for a C209 with mismatch-containing plasmid: d(GGGGAGTGTTTGTGTCATCGTACTAGGGCTGT); CS 4 for a SI6-containing plasmid: d(GGGGAGCCAACCTAGGAGAGTACTAGGGCTGT); CS 5 for a SI6 with mismatch-containing plasmid: d(GGGGAGCCAACGTGGGAGAGTACTAGGGC).
Electrophoretic mobility shift assays of XPC-RAD23B binding to dA-AL and dG-AAF/AF-containing oligonucleotides
dA-AL, dG-AAF or dG-AF-containing 44-mer oligonucleotides (4 nM) were annealed to a 5′ Cy5 labeled complementary strand (1.3 nM) in the presence of 10 mM Tris–HCl, pH 8, containing 6.6 mM NaCl and 0.66 mM MgCl. The resulting duplexes were incubated with 4 nM non-modified duplex 44-mer competitor and increasing amounts of XPC-RAD23B (0–125 nM), in 25 mM Tris–HCl, pH 7.5, 40 mM NaCl, 0.1 mg/ml BSA and 10% glycerol at 25°C for 30 min in a reaction volume of 15 μl. Reaction mixtures were loaded onto a pre-equilibrated native 5% polyacrylamide gel with 0.5× TBE buffer and run at 4°C for 50 min at 20 mA. Gels were scanned using a Typhoon 9400 imager. The intensities of the bands for free 44-mer oligonucleotides and the complex formed with XPC-RAD23B were determined by Image Quant TL program from Amersham Biosciences. Each reaction was performed a minimum of three times to determine the binding constant.
The equilibrium dissociation constants (Kd) for the XPC-RAD23B bound to the 44-mer duplexes containing ALII or AAF/AF were determined using the Hill equation y = [L]n/(Kd+[L]n), (Y: percentage of protein–DNA complex and L: concentration (nM) of XPC-RAD23B (protein), n: Hill coefficient, describing cooperativity). Kd values were determined using the SigmaPlot program 10.0 by fitting the data to sigmoid curves. The sequences (5′–3′) of the oligonucleotides used in the binding assays were as follows.
C209: d(AGACAGCCCTAGTACGATGACAGAAACACTGCGTGCATGGATCC); C209-ALII: d(AGACAGCCCTAGTACGATGACA(ALII)GAAACACTGCGTGCAT GGATCC); Complementary strand (CS) for C209 and C209-ALII: d(Cy5-GGATCCATGCACGCAGTG TT TCTGTCATCGTACTAGGGCTGTCT); C209 with mismatch: d(AGACAGCCCTAGTACGATGACACAAACACTGCGTGCATGGATCC) C209-ALII with mismatch: d(AGACAGCCCTAGTACGATGACA(ALII)CAAACAC TGCGTGCATGGATCC); CS for C209-ALII and C209 with mismatch: d(Cy5-GGATCCATGCACGCAGTGTTTCCCTCATCGTACTAGGGCTGTCT); SI6: d(AGA CAGCCCTAGTACTCTCCTAGGTTGGCTCGGTGCATGGATC); SI6-ALII: d(AGA CAGCCCTAGTACTCTCCTA(ALII)GGTTGGCTCGGTGCATGGATC); CS for SI6 and SI6-ALII: d(Cy5-GATCCATGCACGCGAGCCAACCTAGGAGAGTACTAGGGC TGTCT); SI6 with mismatch: d(AGACAGCCCTAGTACTCTCCCACGTTGGCTC GCGTGCATGGATC); SI6-ALII with mismatch: d(AGACAGCCCTAGTACTCTC CCA(ALII)CGTTGGCTCGCGT GCATGGATC); CS for SI6 and SI6-ALII with mismatch: d(Cy5-GATCCATGCACGCGAGCCAACCCCGGAGAGTACTAGGGCTG TCT).
RESULTS
Cell lines with defects in both GG-NER and TC-NER are sensitive to AAII
We used a series of fibroblast cell lines with mutations in selected components of TCR-NER and/or GGR-NER to investigate the relative roles of these repair processes in the genotoxic and the cytotoxic response to AAII (Table 1). Cell lines deficient in one of the genes of a pathway responsible for AL-DNA adduct repair are expected to be more sensitive to AA exposure. Cells lacking functional CSA or CSB are deficient in TC-NER, those lacking XPC are deficient in GG-NER, whereas cells deficient in XPA, XPD or XPG have deficiencies in the steps common to both pathways. The following corrected human fibroblast cell lines were used: CSB+, CSBmut (E646Q an ATPase-deficient mutant of CSB), XPA+ and XPD+. Cells were treated with different doses of AAII and their viability was measured by an ATP luciferase-dependent proliferation assay.
Cisplatin was used as a positive control, as it has been shown that cells with a defect in TC-NER (CSA-, XPD-, XPG- and XPA-deficient human fibroblasts) are hypersensitive to this drug in an MTT assay (40). Results obtained with the ATP assay were fully consistent with these observations (Supplementary Figure S1).
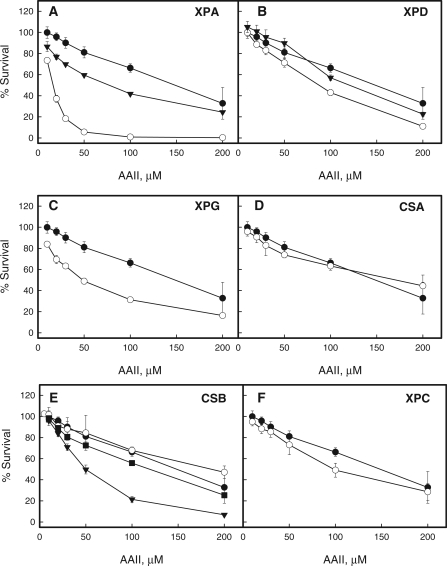
Among GGR/TCR-deficient cell lines, XPA was most sensitive to AAII. XPA complementation rescued partially the phenotype when compared to the control (Figure 2A). XPG and XPD cells were more affected than the control, with XPG being more sensitive than XPD (Figure 2B and C). The sensitivity of the XPD+ cell line was similar to that of normal human fibroblasts (Figure 2B). The XPA and XPG cell lines exhibit unscheduled DNA synthesis (UDS) level less than 2% of normal (41) while XPD cells retain approximately 5% of UDS (42). Thus, variations in the level of UDS between different cell lines may account for observed differences in sensitivity to AAII.
Figure 2.
AAII cytotoxicity study. (A–F) XP-, CS-deficient and control normal human fibroblasts cell lines were treated with 5–200 µM AAII for 72 h with the following ATP level measurements. Filled circles indicate the control normal human fibroblasts cell line, open circles—the deficient cell lines corresponding to the panel names, filled triangles—complemented cell lines if available. In the case of CSB-deficient cell lines, the complementation of the deficient cell line was achieved through pc3.1 plasmid vector harboring the cDNA of wild-type or ATPase mutant ERCC6 gene. On the F, filled squaresindicate the CSB-deficient cell line complemented with ATPase mutant gene. To correct the abnormal phenotype XPA-deficient cells are transfected with the full-length cDNA of the XPAC gene. The XPD cell line is transfected with p2E-ER2, a complementary DNA expression construct of the ERCC2 (XPD) gene, to correct the abnormal phenotype. The point for each AAII dose corresponds to the ATP ratio between treated and untreated cells and presents mean ± standard deviation for at least six measurements in two independent experiments.
Cells with deficiencies in either GG-NER or TC-NER were not hypersensitive to AAII. CSA, CSB and XPC showed no or marginal hypersensitivity to treatment with AAII compared to normal fibroblasts (Figure 2D, E and F). Intriguingly, complementation of CSB cells with wild-type protein, but not with the E646Q mutant with a deficiency in ATPase activity (36), result in increased cytotoxicity, suggesting that overexpression of active CSB adversely affects cellular response to AAII (Figure 2E).
Our studies therefore show that deficiencies limited to either the TC-NER (CSA, CSB) or GG-NER (XPC) pathway did not result in AAII hypersensitivity, while cells lacking components involved in both pathways (XPA, XPD or XPG) displayed increased cytotoxicity, suggesting that, in vivo, AL-DNA adducts are repaired by NER.
Defects in TC-NER, but not GG-NER lead to increased levels of ALII-DNA adducts
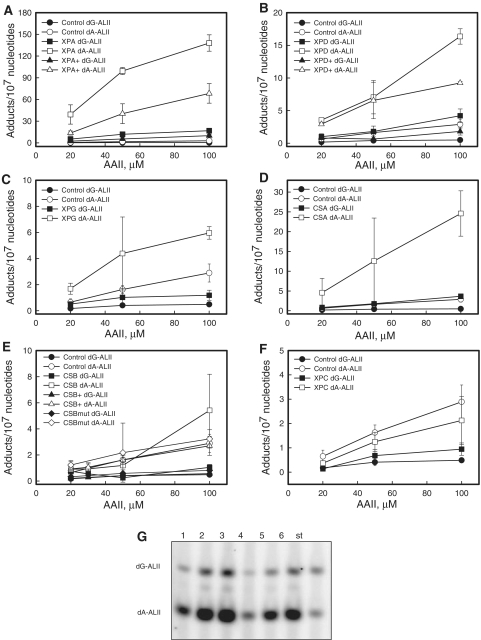
To establish the genotoxicity of AAII and to correlate cytotoxicity with AII-DNA adduct levels, cell lines were treated with different doses of AAII for 48 h and the levels of adducts were determined. The time of exposure was chosen so as to avoid cell death and hence the depletion of cells containing adducts from the plates. Following exposure, the cells were washed, collected and stored at −80°C. DNA was digested to single nucleotides, labeled with γ-[32P]-ATP and analyzed by gel electrophoresis. This post-labeling approach allows for the determination of adduct levels in many samples on a single gel (37). Initially, we performed a time course study with 10 and 100 µM AAII for selected cell lines to establish the linearity of adduct formation (Supplementary Figure S2). To quantify adducts, synthetic oligonucleotides with single dG-ALII or dA-ALII adduct were used as standards. Following mock treatment, no bands corresponding to the dA and dG adduct were found (data not shown). Figure 3G shows a fragment of the polyacrylamide gel used for the adduct analysis.
Figure 3.
AAII genotoxicity study. XP-, CS-deficient, normal fibroblast and complemented cell lines if available were treated with 20, 50, 100 µM AAII for 48 h. DNA was isolated and analyzed for adduct levels with 32P-post-labeling assay. (A–F) – The dependence of dG- and dA-ALII adduct levels from the treatment AAII dose. Control indicates the control human fibroblasts cell line. XPA-, XPD-, XPG-, CSA-, CSB-, CSBmut-, XPC-deficient human fibroblasts cell lines. XPA+, XPD+, CSB+ are corresponding complemented cells. All treatments for every cell line were done in triplicate and the results are presented as means ± standard deviations. (G) The fragment of a 30% polyacrylamide gel after 32P-labeling of DNA adduct nucleosides. 1–3, XPA-deficient cell line treated with 20, 50, 100 µM AAII for 48 h; 4–6, XPA complemented cell line treated the same way; st, The standard mixture of 24-mer oligonucleotides containing 15 fmol of each of the single dG-ALII or dA-ALII, the upper and lower bands, respectively. Each standard band corresponds to 1 adduct/106 nucleotides for 5 µg DNA. For each DNA digestion, at least three standard mixtures were used.
We detected 5- to 10-fold higher levels of dA-AL adducts, compared to dG-AL, in all cell lines after treatment with AAII over a dose range of 20–100 uM (Figure 3). Measurement of repair is complicated by intracellular accumulation of the chemical and differing growth rates of the cell lines employed. After removal of AAII from the medium, washing and continued growth in AA-free medium, cells continued to divide, though at reduced rates. After incubation for 80 h, cells contained significant levels (>70% of initial adduct levels) of both dG-AL and dA-AL adducts (data not shown). These results are consistent with measurements of adduct levels in primary human urothelial cell cultures (Sidorenko, unpublished data).
The levels of dA-AL and dG-AL adducts in cells with deficiencies in different NER genes decreased in the following order: XPA > XPD ≈ CSA > XPG >CSB ≈ CSB+ ≈ CSBmut ≈ XPC ≈ WT control (Figure 3). CSB deficiency led to higher adduct levels when compared to the control cell line only at the highest AAII dose (Figure 3E). Complementation of XPA and XPD deficiency led to significant decreases in the levels of dG-AL and dA-AL (Figure 3A and B), while adduct levels in XPC-deficient cells were comparable to control values, suggesting that GG-NER plays only a minor role in adduct repair (Figure 3G). However, after exposure to 100 µM AAII, we did observe a small, but significant, difference in the levels of dG-ALII, but not dA-ALII, adduct levels between XPC cells and the control, suggesting that dG-ALII adducts are repaired by both TC-NER and GG-NER. The lack of difference at lower doses might reflect the limit of detection for the post-labeling assay where the signal cannot be reliably separated from the noise. These in vivo studies suggest that TC-NER is the main pathway for repair of dA-ALII adducts, as suggested by the TP53 mutation spectrum of patients with upper urothelial cancers caused by dietary exposure to AA (14).
dA-ALII adducts are refractory to repair by GG-NER in vitro
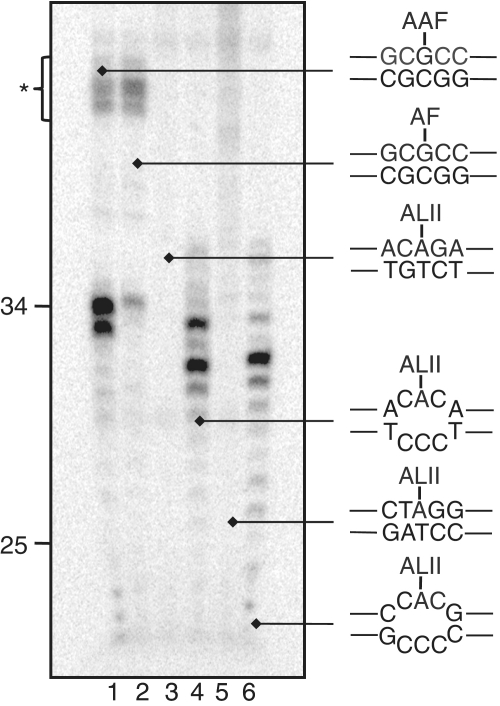
We next sought to investigate whether dA-ALII adducts are repaired by GG-NER in vitro and, if they proved refractory to NER, to develop an understanding of why they are not repaired. We decided to investigate NER of dA-ALII in the context of hot spots for mutagenesis in the p53 gene of patients with upper urothelial cancer as likely sites with inefficient GG-NER (14). Oligonucleotides containing dA-ALII at codon 209 (ACAGA) or at a splice site in intron 6 (CTAGG) (Table 2) were synthesized (12) and used to prepare plasmids containing single site-specifically placed adducts (35). These plasmids were incubated with an NER-proficient HeLa whole-cell extract and the products, expected to be ∼25–30 nt in length, were detected after annealing to complementary oligonucleotide with an overhang of four Gs and a ‘fill-in’ labeling reaction with 32P-dCTP (35,39). We compared repair of dA-ALII to that of dG-AAF, an excellent NER substrate, and dG-AF, an inefficient NER substrate. Consistent with our previous observations (JEY and ODS, manuscript in preparation), dG-AAF was processed efficiently in the extracts, yielding the characteristic multi band pattern of excised fragments, while the dG-AF substrate was repaired with much lower efficiency (Figure 4, lanes 1 and 2). In contrast, no product was observed for the two dA-ALII containing plasmids (C209-ALII and SI6-ALII) (Figure 4, lanes 3 and 5), indicating that dA-ALII adducts were not repaired by GG-NER under these conditions.
Table 2.
Oligonucleotide sequences used in this study
| Name | Sequence |
|---|---|
| AAF | 5′-GGCG(AAF)CC- |
| AF | 5′-GGCG(AF)CC- |
| C209 | 5′-ACAGA- |
| C209-ALII | 5′-ACA(AL)GA- |
| C209 with mismatch | 5′-ACACA− |
| -TCCCT-5′ | |
| C209-ALII with mismatch | 5′-ACA(AL)CA- |
| -TC C CT-5′ | |
| SI6 | 5′-CTAGG- |
| SI6-ALII | 5′-CTA(AL)GG- |
| SI6 with mismatch | 5′-CCACG- |
| -GCCCC-5′ | |
| SI6-ALII with mismatch | 5′-CCA(AL)CG- |
| -GC C CC-5′ |
AAF (dG-AAF), N-(deoxyguanosin-8-yl)-2-acetylaminofluorene; C209, codon 209; SI6, splice site, intron 6; AF (dG-AF), N-(deoxyguanosin-8-yl)-2-aminofluorene; ALII (dA-ALII), 7-(deoxyadenosin-N6-yl) aristolactam II (R = H). The modified residue is indicated in bold.
Figure 4.
NER dual incision activity on dG-AAF, dG-AF and dA-ALII containing plasmids. Plasmids containing site-specific dG-AAF (lane 1), dG-AF (lane 2) or dA-ALII (lanes 3–6) residues were incubated with a HeLa cell extract. Excision products containing were detected by annealing to complementary oligonucleotides with a 4G overhang, which served as a template for end-labeling with [α-32P] dCTP and sequenase. The reaction products were resolved on a 14% denaturing polyacrylamide gel. A low molecular weight ladder from NER was used as a size marker and the position of 25 and 34 nt bands are indicated. Asterisks denotes non-specific, NER-independent bands.
We then sought to understand whether the absence of NER was due to a lack of thermodynamic destabilization induced by dA-ALII lesions. Previous studies have shown that certain lesions, such as UV-induced cyclopyrimidine dimers, are inefficiently repaired by NER and that the NER activity can be dramatically increased by placing the lesion in the context of a mismatch (25). To test if this was also the case for dA-ALII, we placed dA-ALII in the C209 and SI6 sequences in the context of a CCC mismatch and measured whether this additional distortion reflected NER activity. This was indeed the case (Figure 4, lanes 4 and 6) and the repair efficiency of the dA-ALII adducts in the context of a mismatch was similar to that of dG-AAF.
As is often observed during NER in vitro assays, the pattern of the bands of the excised fragments and non-specific bands was different for the dG-AAF and dA-ALII adducts and we wished to confirm that the activities observed were indeed due to GG-NER. We therefore performed additional NER assays using extracts from cell lines derived from xeroderma group F patients with a mutation in the XPF protein. We have shown previously that these extracts are devoid of NER activity and that activity can be restored by adding recombinant purified ERCC1-XPF (43,44). No NER-specific bands were observed for any of the substrates upon incubation with the XP-F extract alone (Supplementary Figure S3, odd lanes), but addition of ERCC1-XPF restored robust NER activity to the reactions with the dG-AAF plasmid (Supplementary Figure S3, lane 2) and with dA-ALII plasmids, if dA-ALII was located in a mismatch (Supplementary Figure S3, lanes 8 and 12). These studies show that dA-ALII adducts are specifically repaired by NER only if they are placed in the context of a destabilized duplex.
dA-ALII adducts are poorly recognized by XPC-RAD23B in duplex DNA
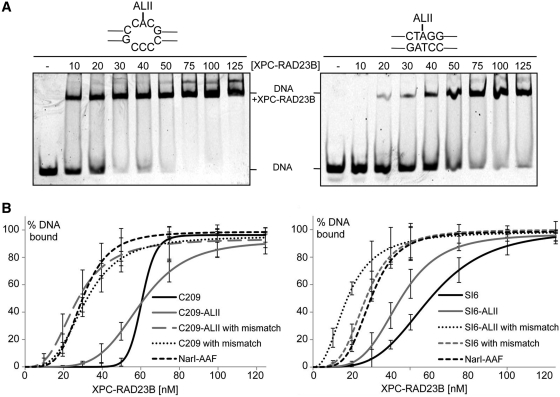
XPC-RAD23B is responsible for recognition of DNA lesions in GG-NER and for the subsequent recruitment of downstream NER factors (18). It is generally believed that the thermodynamic destabilization induced in a DNA duplex influenced by parameters such as base-pairing disruption, bending, and flexibility is correlated with XPC-RAD23B binding affinity and overall NER efficiency (23,45). We investigated whether the binding affinity of XPC-RAD23B correlated with the efficiency of GG-NER for dA-ALII adducts in the matched and mismatched duplexes used for the NER experiments. 44-mer oligonucleotides containing a dA-ALII residue in the same sequence context that was used in the NER experiments (Table 2) were annealed to 5′-Cy5-labeled complementary strands and incubated with increasing concentrations of XPC-RAD23B in the presence of a 3-fold excess of a competitor (25,46), and bound and unbound fractions were separated on a 5% native PAGE gel (Figure 5A and Supplementary Figure S4).
Figure 5.
Determination of binding affinities of XPC-RAD23B binding to dG-ALII and dG-AAF containing oligonucleotides. (A) Lesion-containing 44-mer oligonucleotides (4 nM) were annealed to a complement containing a 5′ Cy5 fluorescent label and incubated with different concentrations (0–125 nM) of XPC-RAD23B for 30 min. The reactions were analyzed on a 5% native polyacrylamide gel and visualized on a Typhoon imaging system. Two representative gels using C209-ALII and C209-ALII with mismatch are shown. (B) Determination of Kd values: the percentage of bound (DNA+XPC-RAD23B) and non-bound fractions (DNA) were determined using Image Quant TL. Kd were calculated from at least three independent experiments and the data analyzed using sigma plot. Standard deviations are indicated by error bars.
Comparison of the binding affinities of C209 and C209-ALII revealed that the presence of dA-ALII had no effect on binding affinity of XPC-RAD23B (Figure 5 and Table 3). However, when C209 and C209-ALII were placed opposite a three base pair mismatch, the Kd values of both substrates are decreased to similar levels by about 2-fold, similar to the binding affinity of dG-AAF (Table 3). This result showed that the presence of a mismatch significantly affected the binding affinity of XPC-RAD23B, while the presence of dA-ALII did not. Similar results were obtained in binding experiments with the SI6 sequence, with the exception that the SI6-dA-ALII oligonucleotide had a binding affinity to XPC-RAD23B intermediate of that of C209 and the mismatch-containing sequences. Overall the binding affinity of XPC-RAD23B was proportional to the efficiency of NER, suggesting that dA-ALII adducts in DNA are refractory to GG-NER because they fail to bind the damage sensor XPC-RAD23B with higher affinity than non-damaged DNA.
Table 3.
Dissociation constants (Kd) of XPC-RAD23B bound to dA-ALII and dG-AAF-containing oligonucleotides
| Sequence | Kd (nM) |
|---|---|
| NarI-AAF | 29 ± 3 |
| C209 | 60 ± 2 |
| C209-ALII | 59 ± 3 |
| C209 with mismatch | 31 ± 5 |
| C209-ALII with mismatch | 26 ± 10 |
| SI6 | 60 ± 3 |
| SI6-ALII | 44 ± 4 |
| SI6 with mismatch | 27 ± 4 |
| SI6-ALII with mismatch | 17 ± 8 |
DISCUSSION
Nucleotide excision repair (NER) excises a variety of bulky lesions from DNA. Such DNA adducts are recognized with varying efficiency by the XPC-RAD23B protein in the GG-NER pathway. DNA lesions that are repaired with high efficiency, such as dG-AAF adducts, 6–4 photoproducts or benzo[a]pyrene (BPDE) adducts in most contexts, distort locally and/or destabilize DNA. In contrast, adducts that destabilize duplexes to a lesser degree, such cyclopyrimidine dimers (CPDs) or BPDE in some sequence contexts, are repaired with low efficiency (47–51). A second NER pathway, TC-NER, is initiated when a RNA polymerase stalls at a lesion during transcription and is dependent on the size or structure of the lesion, but not on the destabilization it induces in the duplex. Therefore, lesions like CPDs are repaired efficiently by TC-NER in the transcribed strand of active genes, while being relatively persistent in other parts of the genome (18,19).
In the course of our studies of aristolochic acid toxicity and mutagenicity, we discovered that A : T to T : A transversions in the TP53 gene of AA-induced tumors mapped almost exclusively to the non-transcribed strand, suggesting that AL-DNA adducts are repaired at much higher rates by TC-NER than by GG-NER (14). In this study, we determined the cytotoxicity of AA and AL-DNA adduct levels in cells with deficiencies in GG-NER, TC-NER or both, and investigated why AL-DNA adducts might escape GG-NER.
Cellular studies support the proposal that dA-ALII adducts are resistant to GG-NER
We exposed cells with defects in the genes involved in GG-NER (XPC), TC-NER (CSA, CSB) or both (XPA, XPD, XPG) to determine if adduct levels support the notion that dA-AL adducts are repaired by TC-NER, while being resistant to GG-NER. Although we observed large variation in adduct levels and in AA toxicity among various cell lines, likely due to the known significant differences in residual NER levels (52), our studies allow us to conclude that AL-DNA adducts are primarily repaired by TC-NER and are resistant to GG-NER. The XPA-deficient cells XP2OS showed the most striking cytotoxic and genotoxic response, consistent with the severe XP phenotype associated with this cell line, caused by an almost complete defect in GG-NER and TC-NER. In contrast, dA-AL adduct levels and AA-induced cytotoxicity did not increase in the XPC-deficient cell line XP4PA. This cell line has a similar low level of GG-NER as XP2OS, but is fully proficient in TC-NER, since the XPC protein operates exclusively in GG-NER. However, cells deficient in the TC-NER gene CSA displayed increased levels of dA-ALII adducts, supporting in turn the proposition that these adducts are excised by TC-NER. The patterns observed in other cell lines, including those with mutations in XPD and XPG, were intermediate, consistent with the known milder NER phenotypes in these cells (53,54).
In CSB-deficient cell lines, effects on AL-DNA adduct accumulation were more pronounced only at the highest AAII dose. Intriguingly, despite similarity at low doses adduct levels, CSB complementation leads to increased cytotoxicity to AAII and cisplatin as compared to the control cells. The loss of sensitivity to AAII of CSB-deficient cells complemented with the mutant CSB protein E646Q (36,55) supports that observation. Overexpression of repair-proficient CSB may lead to a rapid accumulation of DNA repair intermediates that may not be immediately processed by downstream NER elements.
Mechanistic basis for inefficient NER of dA-ALII adducts
We found that dA-ALII adducts were not repaired by GG-NER in vitro under standard conditions in several sequence contexts and that the lack of repair correlated with a failure of the damage sensor XPC-RAD23B to bind these lesions specifically. Similar to previous observations for CPDs and other non-distorting lesions (25,56,57), positioning dA-AL adducts in the context of a mismatch restored NER activity, suggesting that lack of thermodynamic destabilization is responsible for the failure of XPC-RAD23B to bind and trigger NER at dA-AL lesions. These observations lend further support for the current model of substrate recognition in NER positing that local thermodynamic destabilization of duplexes by lesions is recognized by XPC-RAD23B, initiating NER, followed by verification of the presence of a bulky lesion by the XPD helicase subunit of TFIIH, with the help of other factors (58,59). The availability of an NMR structure of a dA-ALII residue in DNA (Lukin, M. et al., accompanying paper) provides some insight into why this lesion may fail to bind XPC-RAD23B. The ALII moiety is intercalated in the duplex and takes up the position of the T residue paired to A. The T residue is displaced from the helical stack, assuming a defined position in the major groove. The planar ALII polycyclic ring system engages in substantial stacking interactions with the two neighboring bases. Therefore, although dA-ALII adducts cause some destabilization of the duplex, it appears that the strong stacking interactions and stable structure of the T residue opposite the lesion disfavor binding of XPC-RAD23B by blocking insertion of the principal recognition motif of XPC, a β-hairpin, into the duplex (27,60). It will be interesting to determine how dG-ALII adducts influence structural features of DNA duplexes and how they interact with NER proteins to provide a molecular basis for shorter persistence of these adducts in cells. It would also be desirable to demonstrate more directly that dA-ALII adducts are repaired by TC-NER, but TC-NER is notoriously difficult to study in vitro (61), and such studies are beyond the scope of the present manuscript.
In conclusion, our studies provide a mechanistic basis for the lack of repair of dA-ALII adducts by GG-NER and explain why such adducts persist in tissues of humans decades after exposure, and why dA mutations in patients with AA-induced upper urothelial cancer are almost exclusively located on the non-transcribed strand of the TP53 tumor suppressor gene.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
National Institutes of Health (NIH) (grants ES004068 to A.P.G. and GM080454 to O.D.S.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Adebanke Fagbemi for a gift of purified XPC-RAD23B, Irina Zaitseva for technical assistance and Dmitry Zharkov (Institute of Chemical Biology and Fundamental Medicine, Novosibirsk, Russia) for valuable discussions and critique of the manuscript.
REFERENCES
- 1.Debelle FD, Vanherweghem JL, Nortier JL. Aristolochic acid nephropathy: a worldwide problem. Kidney Int. 2008;74:158–169. doi: 10.1038/ki.2008.129. [DOI] [PubMed] [Google Scholar]
- 2.National Toxicology Program. Aristolochic Acids. Rep. Carcinogens. 2011;12:45–49. [PubMed] [Google Scholar]
- 3.Kumar V, Poonam PAK, Parmar VS. Naturally occurring aristolactams, aristolochic acids and dioxoaporphines and their biological activities. Nat. Product Rep. 2003;20:565–583. doi: 10.1039/b303648k. [DOI] [PubMed] [Google Scholar]
- 4.Vanherweghem JL, Debelle F, Muniz Martinez MC, Nortier J. In: Clinical Nephrotoxins. 2nd edn. De Broe ME, Porter GA, Bennet WM, Verpooten GA, editors. Netherlands: Dordrecht Kluwer; 2003. pp. 588–601. [Google Scholar]
- 5.Schmeiser HH, Bieler CA, Wiessler M, van Ypersele de Strihou C, Cosyns JP. Detection of DNA adducts formed by aristolochic acid in renal tissue from patients with Chinese herbs nephropathy. Cancer Res. 1996;56:2025–2028. [PubMed] [Google Scholar]
- 6.Nortier JL, Martinez MC, Schmeiser HH, Arlt VM, Bieler CA, Petein M, Depierreux MF, De Pauw L, Abramowicz D, Vereerstraeten P, et al. Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi) N. Engl. J. Med. 2000;342:1686–1692. doi: 10.1056/NEJM200006083422301. [DOI] [PubMed] [Google Scholar]
- 7.Grollman AP, Scarborough J, Jelaković B. In: Advances in Molecular Toxicology. Fishbein JC, editor. Vol. 3. Elsevier, Amsterdam: The Netherlands; 2009. pp. 211–222. [Google Scholar]
- 8.Stiborova M, Frei E, Sopko B, Sopkova K, Markova V, Lankova M, Kumstyrova T, Wiessler M, Schmeiser HH. Human cytosolic enzymes involved in the metabolic activation of carcinogenic aristolochic acid: evidence for reductive activation by human NAD(P)H:quinone oxidoreductase. Carcinogenesis. 2003;24:1695–1703. doi: 10.1093/carcin/bgg119. [DOI] [PubMed] [Google Scholar]
- 9.Bieler CA, Stiborova M, Wiessler M, Cosyns JP, van Ypersele de Strihou C, Schmeiser HH. 32P-post-labelling analysis of DNA adducts formed by aristolochic acid in tissues from patients with Chinese herbs nephropathy. Carcinogenesis. 1997;18:1063–1067. doi: 10.1093/carcin/18.5.1063. [DOI] [PubMed] [Google Scholar]
- 10.Schmeiser HH, Scherf HR, Wiessler M. Activating mutations at codon 61 of the c-Ha-ras gene in thin-tissue sections of tumors induced by aristolochic acid in rats and mice. Cancer Lett. 1991;59:139–143. doi: 10.1016/0304-3835(91)90178-k. [DOI] [PubMed] [Google Scholar]
- 11.Arlt VM, Stiborova M, Schmeiser HH. Aristolochic acid as a probable human cancer hazard in herbal remedies: a review. Mutagenesis. 2002;17:265–277. doi: 10.1093/mutage/17.4.265. [DOI] [PubMed] [Google Scholar]
- 12.Attaluri S, Bonala RR, Yang IY, Lukin MA, Wen Y, Grollman AP, Moriya M, Iden CR, Johnson F. DNA adducts of aristolochic acid II: total synthesis and site-specific mutagenesis studies in mammalian cells. Nucleic Acids Res. 2010;38:339–352. doi: 10.1093/nar/gkp815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, et al. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc. Natl Acad. Sci. USA. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moriya M, Slade N, Brdar B, Medverec Z, Tomic K, Jelakovic B, Wu L, Truong S, Fernandes A, Grollman AP. TP53 mutational signature for aristolochic acid: an environmental carcinogen. Int. J. Cancer. 2011;129:1532–1536. doi: 10.1002/ijc.26077. [DOI] [PubMed] [Google Scholar]
- 15.Fernando RC, Schmeiser HH, Scherf HR, Wiessler M. Formation and persistence of specific purine DNA adducts by 32P-postlabelling in target and non-target organs of rats treated with aristolochic acid I. IARC Sci. Publ. 1993:167–171. [PubMed] [Google Scholar]
- 16.Grollman AP, Jelakovic B. Role of environmental toxins in endemic (Balkan) nephropathy. October 2006, Zagreb, Croatia. J. Am. Soc. Nephrol. 2007;18:2817–2823. doi: 10.1681/ASN.2007050537. [DOI] [PubMed] [Google Scholar]
- 17.Jelakovic B, Karanovic S, Vukovic L, Miller F, Edwards K, Nikolic J, Tomic K, Slade N, Brdar B, Turesky R, et al. Aristolactam-DNA adducts in the renal cortex: biomarkers of environmental exposure to aristolochic acid. Kidney Int. 2011 doi: 10.1038/ki.2011.371. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillet LC, Schärer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 19.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 20.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 21.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 22.Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 24.Dip R, Camenisch U, Naegeli H. Mechanisms of DNA damage recognition and strand discrimination in human nucleotide excision repair. DNA Repair. 2004;3:1409–1423. doi: 10.1016/j.dnarep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem. Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 28.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 29.Damsma GE, Alt A, Brueckner F, Carell T, Cramer P. Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nat. Struct. Mol. Biol. 2007;14:1127–1133. doi: 10.1038/nsmb1314. [DOI] [PubMed] [Google Scholar]
- 30.Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl Acad. Sci. USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tornaletti S. Transcription arrest at DNA damage sites. Mutat. Res. 2005;577:131–145. doi: 10.1016/j.mrfmmm.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 32.Djukanovic L, Radovanovic Z. In: Clinical Nephrotoxins. 2nd edn. De Broe ME, Porter GA, Bennet WM, Verpooten GA, editors. Netherlands: Dordrecht Kluwer; 2003. pp. 588–601. [Google Scholar]
- 33.Kucab JE, Phillips DH, Arlt VM. Linking environmental carcinogen exposure to TP53 mutations in human tumours using the human TP53 knock-in (Hupki) mouse model. FEBS J. 2010;277:2567–2583. doi: 10.1111/j.1742-464X.2010.07676.x. [DOI] [PubMed] [Google Scholar]
- 34.O'Neil MJ, Heckelman PE, Koch CB, Roman KJ. An Encyclopedia of Chemicals, Drugs and Biologicals. 14th edn. Merck: NJ, Whitehouse Station; 2006. [Google Scholar]
- 35.Gillet LC, Alzeer J, Schärer OD. Site-specific incorporation of N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) into oligonucleotides using modified `ultra-mild' DNA synthesis. Nucleic Acids Res. 2005;33:1961–1969. doi: 10.1093/nar/gki335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balajee AS, Proietti De Santis L, Brosh RM, Jr, Selzer R, Bohr VA. Role of the ATPase domain of the Cockayne syndrome group B protein in UV induced apoptosis. Oncogene. 2000;19:477–489. doi: 10.1038/sj.onc.1203372. [DOI] [PubMed] [Google Scholar]
- 37.Shibutani S, Kim SY, Suzuki N. 32P-postlabeling DNA damage assays: PAGE, TLC, and HPLC. Methods Mol. Biol. 2006;314:307–321. doi: 10.1385/1-59259-973-7:307. [DOI] [PubMed] [Google Scholar]
- 38.Masutani C, Araki M, Sugasawa K, van der Spek PJ, Yamada A, Uchida A, Maekawa T, Bootsma D, Hoeijmakers JH, Hanaoka F. Identification and characterization of XPC-binding domain of hHR23B. Mol. Cell Biol. 1997;17:6915–6923. doi: 10.1128/mcb.17.12.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shivji MK, Moggs JG, Kuraoka I, Wood RD. Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol. Biol. 1999;113:373–392. doi: 10.1385/1-59259-675-4:373. [DOI] [PubMed] [Google Scholar]
- 40.Furuta T, Ueda T, Aune G, Sarasin A, Kraemer KH, Pommier Y. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res. 2002;62:4899–4902. [PubMed] [Google Scholar]
- 41.Jaeken J, Klocker H, Schwaiger H, Bellmann R, Hirsch-Kauffmann M, Schweiger M. Clinical and biochemical studies in three patients with severe early infantile Cockayne syndrome. Hum. Genet. 1989;83:339–346. doi: 10.1007/BF00291378. [DOI] [PubMed] [Google Scholar]
- 42.Protic-Sabljic M, Seetharam S, Seidman MM, Kraemer KH. An SV40-transformed xeroderma pigmentosum group D cell line: establishment, ultraviolet sensitivity, transfection efficiency and plasmid mutation induction. Mutat. Res. 1986;166:287–294. doi: 10.1016/0167-8817(86)90028-3. [DOI] [PubMed] [Google Scholar]
- 43.Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 44.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Schärer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Isaacs RJ, Spielmann HP. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair. 2004;3:455–464. doi: 10.1016/j.dnarep.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Trego KS, Turchi JJ. Pre-steady-state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination. Biochemistry. 2006;45:1961–1969. doi: 10.1021/bi05196t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai Y, Kropachev K, Xu R, Tang Y, Kolbanovskii M, Kolbanovskii A, Amin S, Patel DJ, Broyde S, Geacintov NE. Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion. J. Mol. Biol. 2010;399:397–409. doi: 10.1016/j.jmb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, et al. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J. Mol. Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. EMBO J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szymkowski DE, Lawrence CW, Wood RD. Repair by human cell extracts of single (6–4) and cyclobutane thymine-thymine photoproducts in DNA. Proc. Natl Acad. Sci. USA. 1993;90:9823–9827. doi: 10.1073/pnas.90.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai Y, Patel DJ, Broyde S, Geacintov NE. Base sequence context effects on nucleotide excision repair. J. Nucleic Acids. 2010;2010:1–9. doi: 10.4061/2010/174252. doi:10.4061/2010/174252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cleaver JE, Thompson LH, Richardson AS, States JC. A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum. Mutat. 1999;14:9–22. doi: 10.1002/(SICI)1098-1004(1999)14:1<9::AID-HUMU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 53.Takayama K, Salazar EP, Lehmann A, Stefanini M, Thompson LH, Weber CA. Defects in the DNA repair and transcription gene ERCC2 in the cancer-prone disorder xeroderma pigmentosum group D. Cancer Res. 1995;55:5656–5663. [PubMed] [Google Scholar]
- 54.Nouspikel T, Lalle P, Leadon SA, Cooper PK, Clarkson SG. A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc. Natl Acad. Sci. USA. 1997;94:3116–3121. doi: 10.1073/pnas.94.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Brosh RM, Jr, Balajee AS, Selzer RR, Sunesen M, Proietti De Santis L, Bohr VA. The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol. Biol. Cell. 1999;10:3583–3594. doi: 10.1091/mbc.10.11.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl Acad. Sci. USA. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: directional binding of the XPC complex and DNA strand scanning. Mol. Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 58.Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair. 2011;10:673–683. doi: 10.1016/j.dnarep.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, et al. Probing for DNA damage with beta-hairpins: Similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair. 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camenisch U, Trautlein D, Clement FC, Fei J, Leitenstorfer A, Ferrando-May E, Naegeli H. Two-stage dynamic DNA quality check by xeroderma pigmentosum group C protein. EMBO J. 2009;28:2387–2399. doi: 10.1038/emboj.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laine JP, Egly JM. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 2006;25:387–397. doi: 10.1038/sj.emboj.7600933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.