Abstract
The current understanding of electron tunneling through proteins has come from work on systems where donors and acceptors are held at fixed distances and orientations. The factors that control electron flow between proteins are less well understood, owing to uncertainties in the relative orientations and structures of the reactants during the very short time that tunneling occurs. As we report here, the way around such structural ambiguity is to examine oxidation–reduction reactions in protein crystals. Accordingly, we have measured and analyzed the kinetics of electron transfer between native and Zn-substituted tuna cytochrome c (cyt c) molecules in crystals of known structure. Electron transfer rates [(320 s−1 for *Zn-cyt c → Fe(III)-cyt c; 2000 s−1 for Fe(II)-cyt c → Zn-cyt c+)] over a Zn–Fe distance of 24.1 Å closely match those for intraprotein electron tunneling over similar donor–acceptor separations. Our results indicate that van der Waals interactions and water-mediated hydrogen bonds are effective coupling elements for tunneling across a protein–protein interface.
Extensive experimental and theoretical investigations have elucidated the role of polypeptide structure in facilitating electron tunneling through proteins (1–13). Most of the definitive work has centered on molecules with fixed donor–acceptor distances and orientations, such as proteins covalently modified with redox-active units (1–5) or proteins that contain both donors and acceptors (6, 7). This work has established that the dependence of rate on distance is exponential (1, 2, 12, 13), as expected for a tunneling reaction (8), with decay constants in the 1.0- to 1.2-Å−1 range (1, 2, 13). It is likely, therefore, that the redox centers in these proteins are coupled electronically through the chemical-bond framework of the intervening medium (1–6, 8, 11).
Electron transfer (ET) between proteins is understood less well, as it involves at least three steps: (i) association of the donor and acceptor; (ii) electron tunneling within the donor–acceptor complex; and (iii) dissociation of the oxidized and reduced products (14–16). Because the dynamics of the first and the third steps obscure the electron tunneling reaction, many studies have focused on the ET properties of stable protein–protein complexes in solution (5, 15, 17). It has been difficult to interpret the results, however, as neither the donor–acceptor docking geometries nor the conformations of these complexes are known. Studies of kinetics and structure under the same conditions are needed to probe the interactions that promote electron tunneling between proteins.
A protein crystal containing photoactivatable donors and acceptors at specific lattice sites is an ideal medium for investigating the dependence of tunneling rates on structure.§,¶ In the crystal lattice of tuna cytochrome c (cyt c) (23), chains of cyt c molecules form helices with a 24.1-Å separation between neighboring metal centers (Fig. 1). All other metal–metal distances in the lattice are greater than 30 Å, with estimated electron tunneling times that are at least three orders of magnitude slower (2). Thus, the heme groups can be treated as ordered in a one-dimensional chain, separated by identical protein and solvent media. By doping Zn-cyt c into this lattice (Table 1), interprotein ET reactions can be triggered by laser excitation (Scheme S1). The triplet state of Zn-cyt c (*Zn-cyt c) is generated in high yield with 550- or 580-nm excitation (R1). This highly reducing excited state (E0 ≈ −0.8 V) reacts with Fe(III)-cyt c (E0 ≈ 0.25 V) to generate Fe(II)-cyt c and the Zn-cyt c cation radical, Zn-cyt c+ (E0 ≈ 0.9 V) (R2) (24). In a dark reaction, Zn-cyt c+ and Fe(II)-cyt c recombine to yield the ground-state species (R3). Of special interest is our finding that the rates of tunneling reactions across a protein–protein interface (R2, R3) closely match those for intraprotein ET over similar donor–acceptor separations.
Figure 1.
Stereoviews of cyt c crystal packing (space group P43). The asymmetric unit contains two molecules, with one related to a 43 axis at the unit cell origin and the other related to a second 43 axis at the unit cell center. The two screw axes run antiparallel to each other and are related by a pseudotwofold axis directed along the ab diagonal. This packing produces a 24.1-Å separation between the metal centers of adjacent molecules (Moli and Moli+1) within each screw axis. (a) View down the P43 axis. Different colors indicate pairs of cyt c molecules in the same asymmetric unit. White lines connect the iron centers that are separated by 24.1 Å. (b) View of heme orientation along the P43 axis. Broken lines connect the hemes that are involved in ET reactions.
Table 1.
X-ray data collection, refinement, and metal occupancy statistics for tuna Fe:Zn-cyt c structures
| Stoichiometry* | 2Fe:1Zn | 1Fe:2Zn |
| Residues | 103 + 1 heme | 103 + 1 heme |
| Water molecules | 488 | 488 |
| Symmetry group | P43 | P43 |
| Unit cell dimensions, Å | 74.18 × 74.18 × 35.54 | 74.36 × 74.36 × 35.70 |
| Resolution, Å | 30.0–1.5 | 30.0–2.0 |
| X-ray wavelength, Å | 1.54 | 1.28 |
| Completeness, % | 89.0 | 81.7 |
| 〈I/σI〉† | 19.3 | 17.1 |
| Rsym, %‡ | 6.6 | 6.2 |
| R, %§ | 22.9 | 22.0 |
| Free R, %¶ | 25.1 | 24.4 |
| rmsd bond, Å‖ | 0.008 | 0.008 |
| rmsd angle, °‖ | 1.4 | 1.4 |
| Fe:Zn occupancy** | 0.68:0.32 | 0.39:0.61 |
| Average temperature factor, Å2 | ||
| Main-chain atoms | 12.58 | 18.08 |
| Side-chain atoms | 12.74 | 18.12 |
| Water molecules | 23.46 | 29.98 |
| Metal–ligand bond distances, Å | ||
| His-18 | (2.00)‡‡ 2.00 | 2.00 |
| Met-80 | (2.27)‡‡ 2.44 | 2.50†† |
Approximate stoichiometry of the crystallization solution.
Intensity signal-to-noise ratio.
Rsym = ∑∑j|Ij − 〈I〉|/∑∑j|Ij|.
R = ∑∥Fobs| − |Fcalc∥/∑|Fobs| for all reflections (no σ cutoff).
Free R calculated against 8% of the reflections removed at random.
Root-mean-square deviations (rmsd) from ideal bond and angle restraints.
Relative occupancies of Fe and Zn as determined by multiwavelength anomalous diffraction experiments. For 2Fe:1Zn, different crystals were used for structure and metal-occupancy determinations. Values are averages of the two molecules in the asymmetric unit.
Fe(III)-only cyt c (Protein Data Bank ID code 3CYT).
The increase in the Met-80 bond length with increasing Zn occupancy indicates that for Zn-only cyt c, dMet-80 ≥ 2.50 Å.
Scheme 1.
Zn-cyt c redox photochemistry.
Materials and Methods
Crystallization Conditions.
Tuna heart cyt c (Sigma) was used as received. Zn- and Co-cyt c were prepared according to established procedures (24, 25). Zn:Fe-cyt c cocrystals were grown at room temperature in sitting or hanging drops. The reservoir solution contained 500 μl of 70–85% saturated (NH4)2SO4, 0.75 M NaCl, and 0.1 M NaPi (pH 6.0), and the drops were 2 μl of 4–10 mM protein and 2 μl of reservoir solution. Crystal growth is governed by the Fe(III) protein, which nucleates rapidly (few hours) to produce large rod-shaped crystals (500 × 50 × 50 μm), rather than the Zn protein, which crystallizes slowly (>2 weeks) in higher (NH4)2SO4 concentrations (≈85% saturated) to give bundles of thin needles (500 × 5 × 5 μm).
Structure Determination.
The structures of Fe:Zn-cyt c cocrystals were determined by refinement of a model from isomorphous crystals of tuna heart cyt c (Protein Data Bank ID code 3CYT) (23) against diffraction data processed with denzo (26). Rigid-body, simulated-annealing, positional, and thermal refinement with cns (27), amidst rounds of manual rebuilding, and water placement with xfit (28) produced the final models (Table 1). Superimposed, noninteracting Fe and Zn porphyrins were refined simultaneously for each cyt c molecule. Stereochemical restraints were removed from the heme axial ligand bonds in the later stages of refinement. Solvent-accessible surface area was calculated by using ms (29). Interatomic separations between 3.2 and 3.9 Å defined van der Waals contacts. Water molecules within 3.9 Å of both proteins were assigned as interfacial. Fe:Zn occupancies were defined by using multiwavelength anomalous diffraction data collected at the Stanford Synchrotron Radiation Laboratory BL-92 on crystals grown out of 2Fe:1Zn and 1Fe:2Zn solution stoichiometries. For each crystal, metal occupancy refinement with madphsref (30) was carried out against data sets collected at four energies [7124, 7135, 9666, and 9671 eV (1 eV = 1.602 × 10−19 J)] chosen to accentuate both the absorptive and dispersive components of Fe and Zn anomalous scattering.
Kinetics Measurements.
Crystals for transient absorption experiments were mounted and sealed inside 1 × 1 mm quartz capillaries in an anaerobic tent to prevent oxidative photodegradation of Zn-cyt c. If crystals were grown outside the tent, they were soaked in deaerated solutions for at least 2 days before kinetics measurements. Once deaerated, the crystals were resistant to photodegradation indefinitely. Transient absorption spectroscopy was carried out with a 75-W Xe-arc lamp probe light source, a microspectrophotometer to focus (≈100 μm diameter at the sample) and to collect the probe light, and a Nd:YAG pumped optical parametric oscillator (OPO) as the pump light source. Output from the OPO (550 or 580 nm) was used for excitation of Zn-cyt c. Large crystals [≈50 μm (width) × 50 μm (depth)] consistently exhibited reproducible kinetics; when only smaller crystals were available, they were clustered together to prevent excess stray probe light. Generally, no visible damage to crystals by laser excitation was observed even at high pulse energies (≥4 mJ). Owing to the intense absorption of *Zn-cyt c or Zn-cyt c+, extensive signal averaging was not necessary (≤25 shots at 470 or 675 nm). Kinetics were fit by a nonlinear least-squares routine.
Results and Discussion
We initially looked for ET by measuring the decay kinetics of *Zn-cyt c using transient absorption spectroscopy. *Zn-cyt c has intense absorption in the 450- to 500-nm range, whereas the ground-state molecule does not. In pure Zn-cyt c crystals, the excited-state decay could be fit satisfactorily to a monoexponential function with a rate constant of ≈80 s−1 = kint, intrinsic decay rate constant), similar to that measured in solution (Fig. 2a) (24). In Fe(III):Zn-cyt c cocrystals, the decay is considerably faster and better described by a biexponential function (Fig. 2 a and b). We assign the fast phase (kfast = 400 ± 100 s−1) to ET from *Zn-cyt c to Fe(III)-cyt c (R2), where the electron tunneling rate (kET = kfast − kint) is 320 s−1.‖ The slower phase (kslow = 70 ± 20 s−1) closely matches the intrinsic decay of *Zn-cyt c, which is consistent with a distribution of cyt c molecules in the crystals; a fraction of Zn-cyt c molecules is adjacent to only two other Zn molecules and hence decay without undergoing an ET process. Accordingly, the amplitude of the slow phase grows relative to the fast phase as the Zn fraction in the cocrystals increases (Fig. 2b).
Figure 2.
*Zn-cyt c decay kinetics monitored at 470 nm. Δ absorbances are normalized. (a) Pure Zn-cyt c (green) and 2Fe(III):1Zn-cyt c (red). (b) 2Fe(III):1Zn-cyt c (red) and 1Fe(III):1Zn-cyt c (purple). (c) Pure Zn-cyt c (green); Fe(II): Zn-cyt c (black); and Co(III): Zn-cyt c (blue). The second and third traces are offset for clarity.
As controls, we examined Fe(II):Zn-cyt c and Co(III):Zn-cyt c cocrystals—ET in the former case is disfavored thermodynamically, whereas in the latter case there is a large barrier, owing to a high Co(III/II) reorganization energy (>2.4 eV) (25). *Zn-cyt c decay in both cases was slow and monoexponential, with rate constants [68 s−1 for Fe(II) and 78 s−1 for Co(III)] that were essentially the same as those observed in pure Zn-cyt c crystals (Fig. 2c). Zemel and Hoffman (31) reported fast *Zn-porphyrin (*Zn-P) decay at high pulse energies attributable to triplet–triplet energy transfer in Zn hemoglobin (24.1-Å metal–metal separation). Although a similarly fast decay channel was apparent in pure Zn-cyt c crystals (>4 mJ per pulse), it was less than 20% of the total amplitude at the pulse energies (<800 μJ) used in our experiments. Moreover, no such power dependence of the excited-state decay was observed in Fe(III):Zn-cyt c cocrystals, indicating that the contribution of triplet–triplet energy transfer to the fast decay kinetics was negligible.
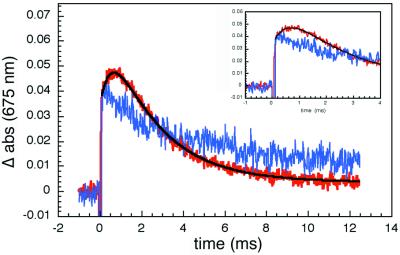
The search for ET products proved to be challenging. First, absorbance measurements in the Soret region are precluded in crystals, owing to high extinction coefficients (abs424 ≈ 65 for a 50-μm thick crystal), and second, Fe(II)-cyt c formation in the Q-band region is difficult to monitor, as the isosbestic point for Zn-cyt c and *Zn-cyt c (540 nm) coincides with that for Fe(II)- and Fe(III)-cyt c. Our efforts to detect Zn-cyt c+ were successful, because in the deep red region of the spectrum the molar absorbance of this cation radical greatly exceeds that of *Zn-cyt c (24). The transient kinetics probed at 675 nm reveal a prompt absorbance increase caused by *Zn-cyt c formation, followed by a slower rise corresponding to production of Zn-cyt c+ (Fig. 3). The time constant for the subsequent decay of the 675-nm absorbance matches that measured at 470 nm (Fig. 2), indicating that charge recombination (R3) is faster than charge separation (R2). A biexponential fit to the 675-nm data yields the following rate constants: 400 ± 100 s−1 (kET = 320 s−1) for R2 and 2,000 ± 500 s−1 for R3.
Figure 3.
Transient absorption (675 nm) kinetics. The prompt absorbance increase in pure Zn-cyt c crystals (blue) is caused by the generation of *Zn-cyt c. In 2Fe:1Zn-cyt c cocrystals (red), there is an additional rise attributable to the Zn-porphyrin cation radical, Zn-cyt c+. The negative signal following the laser flash results from Zn-cyt c fluorescence. (Inset) A view of early kinetics from 2Fe:1Zn-cyt c cocrystals showing the rounded feature.
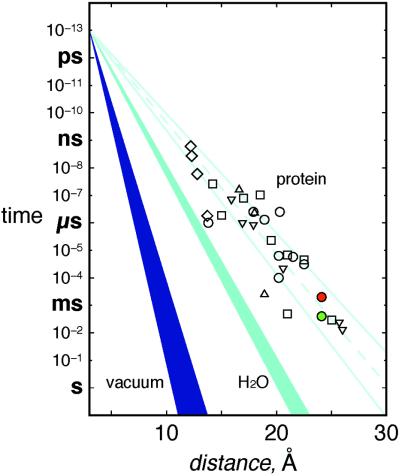
Rapid relay of electrons by redox enzymes necessarily involves short-lived, weakly bound protein–protein complexes. The recognition sites between proteins in such complexes tend to be smaller (<1,200 Å2) and include more water molecules than the interfaces between subunits in oligomeric proteins (32). In fact, the protein–protein interface between cyt c and CcP (770 Å2) (19) is very small compared with other interfaces; there are 17 van der Waals contacts and 13 water molecules (two of which form bridging hydrogen bonds across the interface) but only one direct hydrogen bond bridging the two proteins. The interprotein interactions in crystals of tuna cyt c are similar (Fig. 4): 760 Å2 of surface area is buried in an interface with 31 van der Waals contacts, 16 water molecules (3 bridging), and one direct hydrogen bond.** In addition, a heme vinyl group makes direct contacts across the interface in both the cyt c–cyt c and the CcP–cyt c complexes. Electron tunneling across hydrogen-bonded interfaces is well established (33–35), and the coupling across one or two water molecules (<5 Å) should not be much weaker than that over a comparable distance of peptide (36). Our finding that the ET rates for R2 and R3 fall well within the range that has been established for Ru proteins with similar donor–acceptor separations (Fig. 5; ref. 36) indicates that small interaction zones, such as that between Zn-cyt c and Fe-cyt c, are quite effective in mediating interprotein redox reactions.
Figure 4.
Stereoview of heme groups (brown) and the intervening protein and solvent medium. Residues below (a) and above (b) the heme plane on the left-hand side are shown separately. The side- and main-chain atoms of 14 residues on each molecule participate in the interface, burying 400 Å2 of solvent-accessible surface area on Moli (green) and 360 Å2 on Moli+1 (gray). Close contacts (black traces) in the interface include those between Ile-81i and Ile-75i+1, and the heme vinyli and Lys-55i+1. The side chain of Lys-55i+1 and the peptide carbonyl of Ile-81i form the only direct protein–protein hydrogen bond (yellow traces). Water-bridged hydrogen bonds link the main chain of Phe-82i to that of Lys-73i+1, the side chain of Asp-16i to that of Lys-55i+1, and the main chain of Ile-81i to both the main and side chains of Lys-55i+1. A series of two or more water molecules (blue spheres) mediate additional hydrogen bonds between interfacial residues.
Figure 5.
Tunneling timetable for ET in Ru-modified proteins (open symbols), water (light blue, β = 1.61–1.75 Å−1), and vacuum (dark blue, β = 3.0–4.0 Å−1) (adapted from ref. 36). Most coupling-limited electron tunneling times in proteins [cyt c (○); azurin (▿); cyt b562 (□); myoglobin (▵); and high-potential iron–sulfur protein (⋄)] fall in the 1.0- to 1.2-Å−1 wedge (pale blue solid lines; pale blue dashed line is the average β of 1.1 Å−1). Colored circles (*Zn-cyt c → Fe(III)-cyt c, green and Fe(II)-cyt c → Zn-cyt c+, red) are interprotein time constants.
Integrating photosensitizers into protein crystals provides a powerful tool for studying biochemical reaction dynamics. Indeed, the applications of this methodology could extend well beyond the bounds of interprotein ET kinetics. Zn-cyt c should be an excellent optical trigger for time-resolved x-ray crystallography, a technique that requires rapid and efficient initiation of a reaction throughout the x-ray beam cross section in the sample (37, 38). The long-lived, strongly reducing triplet excited state (φ*Zn-cyt c = 0.9) of Zn-cyt c leads to high quantum yields for ET (φET ≈ 0.7 in our system). Introduction of Zn-porphyrins thus creates opportunities to probe redox-induced structural changes and catalytic intermediates in protein crystals.
Acknowledgments
We thank D. C. Rees and the Stanford Synchrotron Research Laboratory for access to data collection facilities, and A. M. Bilwes for technical assistance and helpful discussions. B.R.C. acknowledges the Helen Hay Whitney Foundation for a postdoctoral fellowship. This work was supported by the National Science Foundation and the Arnold and Mabel Beckman Foundation.
Abbreviations
- ET
electron transfer
- cyt c
cytochrome c
- *Zn-cyt c
triplet excited state of Zn-substituted cyt c
- CcP
cyt c peroxidase
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes 1I54 (2Fe:1Zn-cyt c) and 1I55 (1Fe:2Zn-cyt c)].
Feher and colleagues (18) have measured the kinetics of photoinduced ET between the photosynthetic reaction center and cyt c2 (Rhodobacter sphaeroides) complexed in crystals. However, the low resolution (3.5 Å) of this structure and poor cyt c2 electron density did not allow a precise definition of the polypeptide and solvent medium between the electron donor (cyt c2 heme) and the acceptor (bacteriochlorophyll dimer).
Crystal structures of four redox protein complexes have been determined: cyt c-cyt c peroxidase (CcP; ref. 19); methylamine dehydrogenase-amicyanin (20); methylamine dehydrogenase-amicyanin-cyt c551I (21); and FMN-cytochrome P450BM-3 (22). The kinetics of electron tunneling reactions in these crystals have not been measured.
The rate constant for the fast phase increases (from 350 s−1 for 1Fe:2Zn up to 510 s−1 with 2Fe:1Zn cocrystals) as the relative Fe-cyt c concentration is raised. This observation is consistent with an increase in the fraction of Zn-cyt c molecules adjacent to two Fe-cyt c molecules (instead of one), whose excited-state decay rate (kfast, 2) should be faster by kET (kfast, 2 ≈ 2 kET + kint). Although the observed decay kinetics should be described by a triexponential function, it has not been possible to extract statistically significant values of kfast and kfast, 2 from fits to the single-crystal ET kinetics. Instead, increasing contributions from kfast, 2 lead to larger values for the kfast component in the biexponential fits.
Direct comparison of cyt c–cyt c and cyt c–CcP interfaces is limited by differences in the respective resolution of the structures (1.5 Å vs. 2.3 Å) and disorder in the cyt c–CcP complex that leads to high thermal factors for cyt c (〈B〉 = 51.7 Å2), interfacial water molecules (〈B〉 = 50.6 Å2), and interfacial CcP residues (〈B〉 = 35.3 Å2). The interfacial water molecules (〈B〉 = 19.2 Å2) and residues (〈B〉 = 10.6 Å2) in the cyt c–cyt c complex are well ordered.
References
- 1.Langen R, Chang I-J, Germanas J P, Richards J H, Winkler J R, Gray H B. Science. 1995;268:1733–1735. doi: 10.1126/science.7792598. [DOI] [PubMed] [Google Scholar]
- 2.Gray H B, Winkler J R. Annu Rev Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 3.Winkler J R, DiBilio A J, Farrow N A, Richards J H, Gray H B. Pure Appl Chem. 1999;71:1753–1764. [Google Scholar]
- 4.Winkler J R. Curr Opin Chem Biol. 2000;4:192–198. doi: 10.1016/s1367-5931(99)00074-5. [DOI] [PubMed] [Google Scholar]
- 5.Mei H, Wang K, Peffer N, Weatherly G, Cohen D S, Miller M, Pielak G J, Durham B, Millett F. Biochemistry. 1999;38:6846–6854. doi: 10.1021/bi983002t. [DOI] [PubMed] [Google Scholar]
- 6.Farver O, Pecht I. J Biol Inorg Chem. 1997;2:387–392. doi: 10.1007/s00775-013-1080-7. [DOI] [PubMed] [Google Scholar]
- 7.Dick L A, Malfant I, Kuila D, Nebolsky S, Nocek J M, Hoffman B M, Ratner M A. J Am Chem Soc. 1998;120:11401–11407. [Google Scholar]
- 8.Beratan D N, Betts J N, Onuchic J N. Science. 1991;252:1285–1288. doi: 10.1126/science.1656523. [DOI] [PubMed] [Google Scholar]
- 9.Onuchic J N, Beratan D N, Winkler J R, Gray H B. Annu Rev Biophys Biomol Struct. 1992;21:349–377. doi: 10.1146/annurev.bb.21.060192.002025. [DOI] [PubMed] [Google Scholar]
- 10.Beratan D N, Skourtis S S. Curr Opin Chem Biol. 1998;2:235–243. doi: 10.1016/s1367-5931(98)80065-3. [DOI] [PubMed] [Google Scholar]
- 11.Daizadeh I, Gehlen J N, Stuchebrukhov A A. J Chem Phys. 1997;106:5658–5666. [Google Scholar]
- 12.Moser C C, Keske J M, Warncke K, Farid R S, Dutton P L. Nature (London) 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 13.Williams R J P. J Biol Inorg Chem. 1997;2:373–377. [Google Scholar]
- 14.McLendon G, Hake R. Chem Rev. 1992;92:481–490. [Google Scholar]
- 15.Nocek J M, Zhou J S, De Forest S, Priyadarshy S, Beratan D N, Onuchic J N, Hoffman B M. Chem Rev. 1996;96:2459–2489. doi: 10.1021/cr9500444. [DOI] [PubMed] [Google Scholar]
- 16.Davidson V L. Acc Chem Res. 2000;33:87–93. doi: 10.1021/ar9900616. [DOI] [PubMed] [Google Scholar]
- 17.Pletneva E V, Fulton D B, Kohzuma T, Kostic N M. J Am Chem Soc. 2000;122:1034–1046. [Google Scholar]
- 18.Adir N, Axelrod H L, Beroza P, Isaacson R A, Rongey S H, Okamura M Y, Feher G. Biochemistry. 1996;35:2535–2547. doi: 10.1021/bi9522054. [DOI] [PubMed] [Google Scholar]
- 19.Pelletier H, Kraut J. Science. 1992;258:1748–1755. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Durley R, Poliks B J, Hamada K, Chen Z, Mathews F S, Davidson V L, Satow Y, Huizinga E, Vellieux F M, et al. Biochemistry. 1992;31:4959–4964. doi: 10.1021/bi00136a006. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Durley R C, Mathews F S, Davidson V L. Science. 1994;264:86–90. doi: 10.1126/science.8140419. [DOI] [PubMed] [Google Scholar]
- 22.Sevrioukova I F, Hazzard J T, Tollin G, Poulos T L. J Biol Chem. 1999;274:36097–36106. doi: 10.1074/jbc.274.51.36097. [DOI] [PubMed] [Google Scholar]
- 23.Takano T, Dickerson R E. Proc Natl Acad Sci USA. 1980;77:6371–6375. doi: 10.1073/pnas.77.11.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elias H, Chou M H, Winkler J R. J Am Chem Soc. 1988;110:429–434. [Google Scholar]
- 25.Sun J, Su C, Wishart J F. Inorg Chem. 1996;35:5893–5901. [Google Scholar]
- 26.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.McRee D. J Mol Graphics. 1992;10:44–46. [Google Scholar]
- 29.Connolly M L. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 30.Crane B R, Getzoff E D. Acta Crystallogr D. 1997;53:23–40. doi: 10.1107/S0907444996007263. [DOI] [PubMed] [Google Scholar]
- 31.Zemel H, Hoffman B M. J Am Chem Soc. 1981;103:1192–1201. [Google Scholar]
- 32.Lo Conte L, Chothia C, Janin J. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 33.de Rege P J F, Williams S A, Therien M J. Science. 1995;269:1409–1413. doi: 10.1126/science.7660123. [DOI] [PubMed] [Google Scholar]
- 34.Kirby J P, Roberts J A, Nocera D G. J Am Chem Soc. 1997;119:9230–9236. [Google Scholar]
- 35.Yang J, Seneviratne D, Arbatin G, Andersson A M, Curtis J C. J Am Chem Soc. 1997;119:5329–5336. [Google Scholar]
- 36.Ponce A, Gray H B, Winkler J R. J Am Chem Soc. 2000;122:8187–8191. [Google Scholar]
- 37.Ren Z, Bourgeois D, Helliwell J R, Moffat K, Srajer V, Stoddard B L. J Synchrotron Radiol. 1999;6:891–917. [Google Scholar]
- 38.Schlichting I. Acc Chem Res. 2000;33:532–538. doi: 10.1021/ar9900459. [DOI] [PubMed] [Google Scholar]