Abstract
Yeast histone mRNAs are polyadenylated, yet factors such as Rrp6p and Trf4p, required for the 3′-end processing of non-polyadenylated RNAs, contribute to the cell cycle regulation of these transcripts. Here, we investigated the role of other known 3′-end processing/transcription termination factors of non-polyadenylated RNA in the biogenesis of histone mRNAs, specifically the Nab3p/Nrd1p/Sen1p complex. We also re-evaluated the polyadenylation status of these mRNAs during the cell cycle. Our analysis reveals that yeast histone mRNAs have shorter than average PolyA tails and the length of the PolyA tail varies during the cell cycle; S-phase histone mRNAs possess very short PolyA tails while in G1, the tail length is relatively longer. Inactivation of either Sen1p or Rrp6p leads to a decrease in the PolyA tail length of histone mRNAs. Our data also show that Sen1p contributes to 3′-end processing of histone primary transcripts. Thus, histone mRNAs are distinct from the general pool of yeast mRNAs and 3′-end processing and polyadenylation contribute to the cell cycle regulation of these transcripts.
INTRODUCTION
The production of histones, the core components of the nucleosome, is a highly regulated process in all eukaryotic cells. Progression through the cell cycle requires the precise temporal expression of histone mRNAs and proteins, with maximum expression occurring in the S-phase of the cell cycle, coincident with DNA synthesis. This precise expression requires stringent regulation at the levels of gene transcription, transcript processing, nuclear export and both histone mRNA and protein degradation (1,2).
Despite evolutionary conservation of the temporal expression of histone mRNAs during the cell cycle, an intriguing structural difference exists between histone mRNAs in fungi, protozoa, plants and metazoans. While replication-dependent histone mRNAs in lower eukaryotes and plants possess 3′ PolyA tails, those in higher eukaryotes lack PolyA tails and instead contain a highly conserved stem-loop structure at the 3′-end of the mRNA. Regulation of 3′-end processing of the mammalian histone mRNAs contributes significantly to the cell cycle expression of these mRNAs (3). Processing of the 3′-ends of these transcripts requires a number of unique factors including a stem-loop binding protein (SLBP), the U7 snRNP and a zinc-finger-like protein ZFP (1), all of which are absent in lower eukaryotes, with the exception of the identification in silico of a protein sharing sequence homology with SLBP in a number of protozoal species (4).
Histone mRNAs in the yeast Saccharomyces cerevisiae have been described as being polyadenylated (5). We have previously shown that components of the 3′-end processing and polyadenylation machinery, specifically CFIA, are required for the biogenesis of yeast histone mRNAs (6). Additionally, Rrp6p, a component of the nuclear exosome, a complex of 3′–5′ exoribonucleases, contributes to the cell cycle regulation of these mRNAs (6). Deletion of RRP6 leads to continued accumulation of the HTB1 histone mRNA during the S-phase of the cell cycle and results in a delay in the S- to G2-phase transition. A role for the TRAMP (Trf4/5p, Air1/2p, Mtr4p) complex in histone mRNA regulation has also been identified (7). The nuclear exosome and the TRAMP complex primarily function in the post-transcriptional processing of non-coding, non-polyadenylated RNAs such as 5.8S rRNA, small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). They also form part of a surveillance system that degrades aberrantly synthesized or processed mRNAs, tRNAs and rRNAs (8). Both routine processing of the 3′-ends of sn/snoRNAs and the degradation of aberrant transcripts start by the addition of a short PolyA tail mediated by the PolyA polymerase activity (Trf4p or Trf5p) present in the TRAMP complex (9,10) or by the canonical PolyA polymerase (11,12). The polyadenylated sn/snoRNAs are then trimmed to generate the correct 3′-end, while aberrant transcripts are degraded by the exosome. The exosome, and more specifically Rrp6p, also play a role in the biogenesis of some mRNAs such as Cth2 and Nab2 (13,14).
How the exosome discriminates between transcripts to be processed or degraded is currently not known although this most likely involves competition between the 3′-end processing machinery and the exosome for substrates, regulated by auxiliary cofactors. One such group of cofactors is the Nab3p/Nrd1p/Sen1p complex. This complex primarily plays an essential role in the 3′-end processing and transcription termination of sn/snoRNAs, mediated in part by interactions of Nrd1p with the C-terminal domain (CTD) of Pol II and with the exosome (15,16). Nrd1p has also been shown to mediate transcription termination of some protein encoding genes (17) and Sen1p can influence the RNA Polymerase II occupancy on protein encoding genes with an ORF size of <600 bp (18). Thus, the picture emerging is one of cross-communication between the 3′-end processing/transcription termination machinery of both polyadenylated and non-polyadenylated mRNAs.
The involvement of factors such as Rrp6p and Trf4p in the regulation of yeast histone mRNA levels prompted us to explore the role of the Nab3p/Nrd1p/Sen1p complex in the biogenesis of yeast histone RNAs and to re-evaluate the polyadenylation status of these mRNAs. Our analysis reveals that in asynchronous cells, histone mRNAs have shorter than average PolyA tails. Inactivation of either Sen1p or Rrp6p leads to a decrease in the average PolyA tail length of histone mRNAs and Sen1p is required for efficient 3′-end processing of the transcripts. Additionally, we show that the histone mRNA PolyA tail length varies during the cell cycle; in S-phase transcripts possess very short PolyA tails, while in G1 the tail length is relatively longer. HTB1 transcripts appear to be retained in the nucleus in G1. The reduction in the steady state levels of histone mRNAs after S-phase involves to a large extent active deadenylation and degradation of the mRNAs in a 3′–5′ direction although concomitant 5′ to 3′ nuclease activity cannot be ruled out. The results show for the first time that histone mRNAs are distinct from the general pool of mRNAs in yeast cells and are regulated during the cell cycle by 3′-end processing and polyadenylation. The results further strengthen the connectivity between transcription termination and 3′-end processing of PolyA and non-PolyA RNAs.
MATERIALS AND METHODS
Strains
The yeast strains used in this study are listed in Table 1. Strains were cultured in YEPD medium [1% (w/v) yeast extract, 2% (w/v) bacto peptone, 2% (w/v) dextrose] to an optical density (OD660nm) of 0.6–1.8. The sen1, nab3-11 and nrd1-5 strains were kindly provided by David Brow, University of Wisconsin, USA. The rrp6KO strain was previously provided by Domenico Libri, CNRS, France.
Table 1.
Strains
| Strain | Genotype |
|---|---|
| 46a | MATa, cup1, Δura3, his3, trp1, lys2, ade2, leu2 |
| 46α | MATα, cup1, Δura3, his3, trp1, lys2, ade2, leu2 |
| 46a sen1-E159K | MATa, cup1, Δura3, his3, trp1, lys2, ade2, leu2, sen1-E159K |
| 46a nrd1-5 | MATa, cup1, Δura3, his3, trp1, lys2, ade2, leu2, nrd1-V368G |
| 46α nab3-11 | MATα, cup1, Δura3, his3, trp1, lys2, ade2, leu2, NAB3:: KANMX [pRS313-NAB3-11(F317L, P374T)] |
| W303.rrp6KO | MATα, his3, leu2, ura3, ade2, trp1, rrp6::KAN |
RNA extraction
Total cellular RNA from yeast strains was extracted and DNase I-treated as previously described (19). Cytoplasmic and nuclear fractions were prepared using a protocol adapted from the Hahn laboratory (http://labs.fhcrc.org/hahn/Methods/biochem_meth/polii_nuc_ext.html). The cell lysis buffer was supplemented with the ribonuclease inhibitor Vanadyl ribonucleoside vanadyl complex, at a final concentration of 1 mM, (Sigma Chemical Co., Poole, UK). Following lysis, cellular debris was removed by two rounds of low speed centrifugation at 4000 rpm in a Sorvall SS-34 rotor. The nuclei were pelleted from the supernatant fraction by centrifugation at 10 000 rpm for 20 min. The integrity of the nuclear/cytoplasmic fractionation was ascertained by semi-quantitative RT–PCR analysis of a predominantly nuclear transcript, snR37. RNA was extracted from the cytoplasmic supernatant and nuclear pellet fractions as previously described (19). The RNA was treated with DNase 1 (10 U/ml), in the presence of Placental Ribonuclease Inhibitor (40 U/mL), to remove any contaminating DNA, and the RNA was recovered through binding and elution from silica columns (Epoch Biolabs, TX, USA) as per manufacturer's instructions.
Reverse–transcription of RNA and PCR amplification
DNase-treated RNA samples (2 μg) were reverse transcribed in a 20 µl reaction using ImProm-II™ system (Promega Co., WI, USA) as per manufacturer's instructions. In addition to the gene-specific reverse primers, a random nonamer was also included in the cDNA synthesis reaction. PCR reactions were carried out as previously described (6), generally using one-tenth of the cDNA reaction unless otherwise indicated in the figure legends and forward and reverse gene-specific primers at a final concentration of 125 nM. The primers used for cDNA synthesis and PCR amplification are listed in Table 2. For semi-quantitative reverse–transcription-PCR (RT–PCR), reactions were carried out as described above except that the concentrations of RNA and/or cDNA were varied and only 20 rounds of amplification were carried out to ensure that the reactions were in the log phase. The relative efficiency of RT–PCR was verified by spiking the cDNA synthesis reaction with 400 pg MS2 viral genomic RNA (Roche Diagnostics Ltd, UK) and appropriate primers (Table 2). RT–PCR products were separated on a 2% Tris–Boric acid–EDTA (TBE) agarose gel and transferred to a nylon membrane. The membranes were processed as previously described (19) and hybridized with 5 ng of digoxygenin-UTP HTB1 probe prepared by PCR-amplification using reverse primer 7 and the forward primer For1 (Table 2). Following hybridization, the membranes were processed as previously described (19).
Table 2.
Primers
| Oligonucleotides | Sequence 5′–3′ |
|---|---|
| HTB1 For1343–351 | GTCTCTGAAGGTACTAGAGCTG |
| HTB1 R1473–489 | GGCCGCGGATTAAACTATTATACAA |
| HTB1 R2551–573 | TAAGCGCATTCCCTCTATGAGAC |
| HTB1 R3620–646 | TTTCTGAGTCATTAATAAGCAACACTA |
| HTB1 R4705–733 | CGAATTAAAATTTGAGGAAAAATCTAGTA |
| HTB1 R5788–810 | GAAGTTAATCACAACAGAGGGTT |
| HTB1 R6870–896 | TTTTATACGTGCGTATTCTATTGTTCA |
| HTB1 R7979–997 | TGAAGTGCAGCTGACGATC |
| HTB1 Rev B | b-GCTTGAGTAGAGGAAGAGTACTTGG TAACAG |
| HTB1 For1–22 | GGCGCATGTCTGCTAAAGCCGAAAAG |
| Oligo C primer | CGAGGACTCGAGCTCAAGCCCCCCCCCC CCCCC |
| HTB1 For16–32 | GAAAAGAAACCAGCCTCC |
| HTB1 Rev368–388 | GAGTAGAGGAAGAGTACTTGG |
| ACT1 For | CCGCCTGAATTAACAATGGATTCTGGTA |
| ACT1 Rev | CTCTCAATTCGTTGTAGAAGGT |
| M13 Forward | TGTAAAACGACGGCCAGT |
| M13 Reverse | CAGGAAACAGCTATGACC |
| snR13For16–40 | CGTCTCTATATCTTTTGTTAATTGG |
| snR13Rev180–103 | GGTAGCTTGAGTTTTTCCACACCG |
| snR13 Rev2172–194 | CGCATTATATATGCAGCGCTACG |
| snR13 Rev3266–291 | CTTCGTCACTTTCGTTTTTTTACAAG |
| 18S For | TTGCTGGTTATCCACTTCTTAGA |
| 18S Rev | TTCACAAGATTACCAAGACCTCTC |
| MS2 For | GGAGTGTTTACAGTTCCGAAGAAT |
| MS2 Rev | ATCGGATGCAGACGATAAGTCT |
| snR37-For | CATTATACCATAGAAACCTAAAGATTAGG |
| snR37-Rev | ATCATGAAGAAGAGTCGCTTAGG |
Hybrid selection of HTB1 mRNA
DNase-treated total RNA (2 mg) was denatured at 65°C in the presence of HTB1 biotinylated primer (RevB; 600 nM; Table 2) and renatured slowly to 22°C following addition of SSC to a final concentration of 5× SSC (750 mM NaCl, 0.075 mM sodium citrate). The biotin primer-HTB1 mRNA hybrid was captured with 100 µl of prewashed streptavidin-paramagnetic beads (1 mg/ml; Kisker Biotech Gmbh, Germany) at room temperature (10 min with gentle inversion every 1–2 min) using a magnetic stand. Following four washes (300 µl, 0.5× SSC), the bound HTB1 mRNA was eluted from the beads using 250 µl of RNase-free water at 65°C. The eluted mRNA was lyophilized and resuspended in 25 µl of RNase-free water.
G-tailing and cDNA cloning of HTB1 mRNA
The purified HTB1 mRNA was G-tailed using Yeast Poly(A) Polymerase (USB Corporation, OH, USA) at 37°C for 30 min in a reaction volume of 25 µl containing 0.5 mM rGTP, 750 U of Yeast Poly(A) Polymerase and 10 µl of hybrid selected mRNA using the buffer supplied by the manufacturer. The reaction was stopped by the addition of 700 µl of 4M guanidinium thiocyanate in 20 mM Potassium acetate (pH 5.5) and the RNA was purified on a silica spin column as described above. RNA was eluted in 30–60 µl RNase-free water at 65°C, lyophilized and resuspended in 10 µl RNase-free water. cDNAs were prepared from the total pool of eluted RNAs as described above using the Oligo C reverse primer (Table 2) in a 20 µl reaction and one-tenth of the cDNA was PCR amplified with the HTB1 For1–22 and Oligo C primers (Table 2; 125 µM final concentration). The PCR products were cloned using a TOPO TA cloning kit (Life Technologies Invitrogen Corp., CA, USA) as per manufacturer’s instructions. Colonies were screened by direct colony PCR using M13 forward and reverse primers (Table 2). Plasmid DNA was extracted from colonies showing PCR products in the expected size range of >500 nt. Direct sequencing of the plasmids using M13 forward and reverse primers (Table 2) was outsourced to GATC Biotech, Germany. Statistical analysis of the HTB1 PolyA tail length was carried out using the software Prism 5.0.
Cell cycle synchronisation of yeast cells
Yeast cells were grown in YEPD to an OD660nm of 0.4–0.6. The α-mating factor was added to give a final concentration of 2 μg/ml and the culture was incubated for up to 3 h until ∼90% of the cells were arrested in G1 phase (shmooed). The α-mating factor was removed by centrifugation and the cell pellets were washed twice in excess sterile distilled water (4°C) and maintained on an ice–water bath. The cell pellets were resuspended in the original volume of prewarmed YEPD and incubated at 25°C. Samples (200 ml) were collected at 15-min intervals, centrifuged at 4000 rpm for 5 min and frozen a −70°C.
Flow cytometry analysis of cell cycle progression
Following α-factor removal, ∼1.0 × 107 cells were collected as above and fixed in 95% (v/v) ethanol and stored at 4°C for up to 3 days. The samples were processed as previously described (6). Fluorescence activated cell sorting (FACS) was carried out on a Beckman Coulter Epics XL Flow Cytometer. The data was analysed using WinMDI software (http://facs.scripps.edu). FACS analysis of asynchronous cells was carried out in the same manner. Cell populations were gated to quantify the number of 1n and 2n cells.
RESULTS
Sen1p is required for 3′-end processing of histone mRNAs
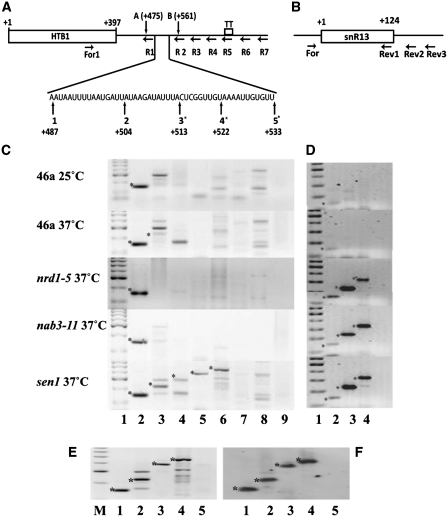
A number of independent studies have demonstrated that histone mRNA levels are influenced by components of both the nuclear exosome and the TRAMP complex (6,7). Since both complexes interact with the Nab3p/Nrd1p/Sen1p complex (15,20), we first tested if these factors might be exerting a similar affect on histone 3′-end processing and transcription termination. We have previously demonstrated that the majority of HTB1 transcripts (up to 80%) terminate within 280 bases of the most frequently used 3′-end cleavage site [Figure 1A, transcription termination region; TT; (19)]. Inefficient 3′-end processing or transcription termination leads to read-through transcription beyond the normal 3′-end cleavage sites (21). To capture unprocessed HTB1 read-through transcripts, RT–PCR was carried out using primers (R1–R7), complementary to sequences downstream of the known 3′-end cleavage sites, and a second primer (For 1) complementary to a region in the open reading frame (Figure 1A, Table 2). In wild-type (WT) cells, grown at the permissive temperature, correctly cleaved and polyadenylated transcripts were detected using primer R1 (Figure1C, lane 2; panel 46a, 25°C, band denoted by a asterisk). No HTB1-specific read-through products were detected using primers R2–R7 that are located downstream of the 3′-end cleavage site (Figure 1C, lanes 3–8) with the exception of a few non-specific PCR products which on sequencing were identified as originating from non-specific priming from rRNAs (data not shown).
Figure 1.
(A) Identification of read-through primary transcripts of HTB1. Schema of the HTB1 gene transcription unit. The location of the primers used for cDNA synthesis (R1–R7) is shown, as is the forward primer (For1) used for PCR amplification of the cDNAs. The open reading frame 1–397 is designated by a black rectangle. The region of transcription termination (TT) is indicated. The expanded nucleotide sequence shows the location of the major 3′-end cleavage sites for HTB1. Those marked with asterisks have been identified previously (19,23). Newly identified cleavage sites A and B are also shown (Table 3). (B) Schema of snR13 gene. Primers used for cDNA synthesis (Rev1-3) are shown, as is the location of the forward primer (For) for PCR amplification. (C) RT–PCR analysis of HTB1 mRNA from the strains, shown on the left, grown at either 25° or 37°C. Lane 1; molecular weight marker, lanes 2–8; cDNAs were prepared using reverse primers R1–R7, respectively and amplified with the common forward primer For1, lane 9; cDNA from R1 in the absence of reverse transcriptase. Asterick represents HTB1-specific PCR products of the expected size. (D) RT–PCR analysis of snR13 RNA. Lane 1; molecular weight marker, 2–4; cDNAs were prepared using reverse primers Rev1, Rev2 and Rev3, respectively and amplified with the common forward primer, For. (E) RT–PCR analysis of HTB1 mRNA from sen1 cells grown at 37°C for 2 h. Lane M; molecular weight marker, lanes 1–4; PCR template cDNAs prepared using reverse primers R1, R2, R4 and R5, respectively, lane 5; cDNAs prepared using R1 in the absence of reverse transcriptase. (F) Southern blot of gel shown in E hybridized with a HTB1-specific probe.
A similar pattern of RT–PCR products was also detected from RNA extracted from WT cells incubated at 37°C for 2 h (Figure 1C, panel 46a, 37°C) except that a small amount of read-through product was detected with primer R2 (Figure 1C, panel 46a, 37°C lane 3, denoted by a asterisk). This indicates that 3′-end processing per se was not adversely affected by the conditions required to inactivate temperature sensitive (ts) mutants. Similarly, RNA extracted from the ts-mutants nrd1-5 or nab3-11 (Table 1), following incubation at the non-permissive temperature, did not lead to increased read-through beyond the 3′-end cleavage sites (Figure 1C, panels nrd1-5 and nab3-11, 37°C, respectively). However, in the sen1 ts-mutant (Table 1) incubated at the non-permissive temperature (37°C), HTB1 read-through transcripts up to 280 nt downstream of the 3′-end cleavage sites were detected using reverse primers R1-R5 (Figure 1C; panel sen1 37°C, bands denoted by a asterisk, lanes 2–6). Transcription beyond the previously mapped transcription termination region (TT) was not observed with reverse primers R6-R7 (lanes 7 and 8) suggesting a 3′-end processing rather than a transcription termination defect as a result of inactive Sen1p. To confirm that these primary read-through transcripts were HTB1-specific, the relevant RT-PCR products from the sen1 ts mutant were Southern blotted and hybridized to a digoxygenin-labeled HTB1 probe spanning from the forward primer (For1) to the farthest reverse primer 7 (R7; Table 2 and Figure 1A). As shown in Figure 1E and F, only products of the expected sizes were detected by the HTB1-specific probe. DNA sequencing of the RT–PCR products confirmed the conclusions of Southern blot analysis (data not shown). Furthermore, Sen1p inactivation led to similar read-through transcription for a number of the other core histone RNAs including HTA1, HTB1, HTB2, HHF1 and HHT1 (data not shown), indicating the Sen1p is influencing the 3′-end processing of all core histone mRNAs.
To verify this finding and to ensure that all three temperature sensitive mutants had been sufficiently inactivated by incubation at the non-permissive temperature, the effects of each mutant on the 3′-end formation of the small nucleolar RNA snR13 was also examined. The 3′-ends of snoRNAs, such as snR13 are generated by specific transcription termination and have previously been shown to be sensitive to mutations of all three genes encoding the components of Nab3p/Nrd1p/Sen1p complex (22). Read-through transcripts of snR13 were detected at the non-permissive temperature in all three mutants (Figure 1D, panels nrd1-5, nab3-11 and sen1) using primers located downstream of the 3′-end of the snR13 gene (Figure 1B, primers snR13 Rev2 and Rev3, bands denoted by a asterisk), while only mature transcripts were evident in the WT cells incubated at both 25 and 37°C (Figure 1D, panels 46a 25 and 37°C). Taken together, the data clearly shows that Sen1p contributes to the 3′-end processing of histone mRNAs independent of its role in the Nab3p/Nrd1p/Sen1p complex.
Mutations in Sen1p affect cell cycle progressio
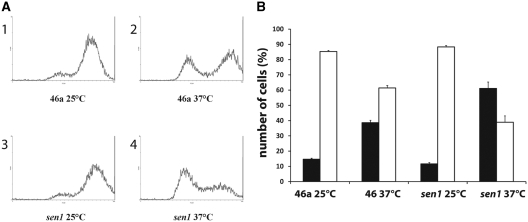
We have previously shown that deletion of RRP6 leads to continued accumulation of HTB1 mRNAs in S-phase of the cell cycle and results in a delay of entry of cells into G2 phase (6). Likewise mutations in TRF4, a component of the TRAMP complex, resulted in elevated levels of histone mRNAs and also displayed cell cycle progression defects (7). We therefore next asked if cell cycle progression is affected in sen1 mutants. Both sen1 and its parental wild-type strain were grown asynchronously at both the permissive and non-permissive temperatures and analysed by flow cytometry to identify the percentage of replicating cells. As shown in Figure 2A and B, ∼80% of WT cells grown at the permissive temperature are actively replicating and present in the 2 n population. Following incubation of the WT cells at the non-permissive temperature, the number of actively replicating cells decrease to ∼60%. At the permissive temperature, sen1 cells display DNA replication levels similar to WT cells. However, at the non-permissive temperature, the percentage of replicating cells drops significantly, concomitant with an increase in the number of 1n cells. These results indicate that, as with deletion of RRP6 and mutations in TRF4, mutations in SEN1 interfere with the efficient progression of the cell cycle.
Figure 2.
(A) Flow cytometry analysis of asynchronous cultures of sen1. Wild-type (46a; panels 1 and 2) or sen1 (panels 3 and 4) cells were grown at the permissive (25°C; panels 1 and 3) and non-permissive temperature (37°C; panels 2 and 4) and stained with propidium iodide as previously described (6). The cells were passed through a flow cytometer (Beckman-Coulter Epic). The pattern of cell sorting is indicated for each of the strains. (B) The flow-sorted cells were gated to distinguish 1n from 2n cells. Black columns; 1n, white columns; 2n. The results represent the average cell counts from four independent experiments and the standard error for each sample is shown.
Sen1p and Rrp6p influence the Poly A tail length of HTB1 transcripts
The finding that Sen1p contributes to histone mRNA 3′-end processing and cell cycle progression, prompted us to re-evaluate the polyadenylation status of yeast histone mRNAs, which was first established in 1980 (5). We have previously shown that histone mRNAs are poorly retained on oligo dT or oligo U matrices compared to other mRNAs such as ACT1 (S. Campbell and U. Bond, unpublished results), suggesting that the PolyA status of these transcripts may be different from the general population of mRNAs.
To examine the PolyA tail length, HTB1 mRNAs from WT, sen1 and Δrrp6 grown at the permissive and non-permissive temperatures were first enriched from the total RNA pool by hybrid selection using a biotinylated antisense HTB1 oligonucleotide, (Table 2; RevB). This selection process generated a RNA population enriched in HTB1 transcripts but still contained small amounts of other RNAs as evidenced by the presence of trace amounts of ACT1 RNA and rRNA in this pool (data not shown). The selected RNA was G-tailed using PolyA polymerase and cDNAs were synthesized using an oligo C anchor reverse primer and the cDNA was PCR amplified with the addition of a HTB1 specific primer inclusive of the start codon (Table 2; For1–22). DNA sequence analysis of the cloned cDNAs indicated that ∼70% were HTB1-specific, thus demonstrating the enrichment of HTB1 transcripts by this method.
Sequence analysis of the cloned HTB1 cDNAs allowed for the identification of the major cleavage sites and quantification of the PolyA tail length in the WT and mutant strains. An average of 8–10 clones were analysed per mutant, with the exception of sen1 for which 16 clones were analysed. In WT cells, four major cleavage sites were identified for HTB1 mRNAs (sites 2–5) with site 4 being the preferred site (Figure 1A and Table 3). Three of the four cleavage sites (indicated by a asterisk in Figure 1A) have previously been identified by S1 mapping and/or cDNA cloning of HTB1 mRNAs, although their relative usage had not been determined (19,23). For comparison with the ts-mutants, the pattern of cleavage in WT cells incubated for 2 h at 37°C was also examined. At this temperature, cleavage site 4 was almost exclusively used and one additional cleavage site was identified (Figure 1A, site 1; Table 3), indicating that incubation at higher temperatures does not significantly alter 3′-end cleavage site selection. In the sen1 mutant, site 4 remained the predominant cleavage site at the permissive temperature while cleavage sites 1–4 were used at the non-permissive temperature (Table 3). Despite the identification of read-through primary transcripts in sen1 at the non-permissive temperature (Figure 1B), only 6% of cDNAs had 3′-ends further downstream of the canonical cleavage sites (at site B, Figure 1A, Table 3) and no transcripts corresponding to the extended read-through products were detected in the steady state pool of HTB1 mRNAs suggesting that sen1 is exerting an effect on the efficiency of 3′-end processing but not on 3′-end cleavage site selection.
Table 3.
Cleavage site usage (%)
| Cleavage site | WT 30°C | WT 37°C | sen1 25°C | sen1 37°C | Δrrp6 30°C |
|---|---|---|---|---|---|
| 1 (+487) | 10 | 62.5 | 10 | ||
| 2 (+504) | 25 | 10 | 12.5 | 20 | |
| 3 (+513) | 12.5 | 10 | 37.5 | ||
| 4 (+522) | 37.5 | 70 | 100 | 37.5 | 60 |
| 5 (+533) | 25 | ||||
| A (+475) | 10 | ||||
| B (+561) | 6.25 |
Since we have previously shown that Rrp6p can influence histone mRNA biogenesis during the cell cycle (6), we also examined the 3′-end cleavage sites in the Δrrp6 background. Again the majority of the transcripts were cleaved at site 4 with a small percentage using sites 1 and 2 (Table 3). As with the sen1 mutant, a small percentage (10%) of transcripts had a 3′-end at a site other than the canonical cleavage sites (at site A; Figure 1A and Table 3).
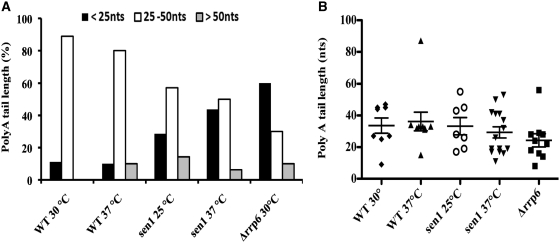
In addition to identifying the major 3′-ends of HTB1 mRNAs, sequence analysis of the G-tailed cDNA clones allowed us to examine the PolyA tail length on the transcripts. The sequence analysis identified two distinct populations of RNAs based on PolyA tail length in the asynchronous population. In WT cells, the majority of HTB1 RNAs contain PolyA tails of between 25 and 50 nt while a small proportion (10%) have tails <25 nt (Figure 3A). The mean HTB1 mRNA PolyA tail length was 33 nt. Thus, HTB1 transcripts appear to have much shorter PolyA tails than the general pool of mRNAs in WT cells, which have PolyA tails of ∼70–100 nt (24). In the case of the sen1 mutant, a different distribution of PolyA tail length was observed. At the permissive temperature, we observe an increase in the percentage of RNAs with polyA tails of <25 nt and a further increase in this pool at the non-permissive temperature (45% compared to 10% in WT cells; Figure 3A). The clear trend toward shorter PolyA tails in sen1 cells is even more apparent when the distribution of PolyA tails on cDNA clones is examined in more detail (Figure 3B). A bimodal distribution about the mean is observed in all samples with the exception of WT 37°C. The median HTB1 PolyA tail length in WT cells is 36 and 33 nt at 25 and 37°C, respectively while in sen1, the median tail length is 28 and 26 nt at 25 and 37°C, respectively. Furthermore, the median PolyA tail length in the 1st quartile of the population is 17 nt in sen1 compared to 32 nt in WT cells at the non-permissive temperature. Thus, there is a substantial increase in the pool of shorter tailed transcripts in sen1 cells (P = 0.08).
Figure 3.
PolyA tail length of HTB1 mRNAs. (A) Analysis of the polyA tail length of G-tailed cloned cDNAs from WT, sen1, and Δrrp6 cells. Cells were grown at the permissive (25–30°C) or non-permissive temperatures (37°C) prior to RNA extraction. The percentage of clones with polyA tail in the size range 25–50 nt; White columns, <25 nt; Black columns, >50 nt; Gray columns. (B) Scatter plot of Poly A tail length of HTB1 cDNA clones. The PolyA tail length for each cDNA clone is shown for each mutant. The mean tail length (nt) with standard error is shown. WT 30°C; filled diamond, WT 37°C; filled triangle, sen1 25°C; open circle, sen1 37°C inverted filled triangle, Δrrp6; filled square. Only cDNAs with canonical 3′-end cleavage sites were included in the analysis.
A pool of transcripts bearing tails <25 nt is also apparent in Δrrp6 cells. Here up to 60% of transcripts had PolyA tails <25 nt (Figure 3A). A median HTB1 PolyA tail length of 23 nt and a 1st quartile tail length of 16 nt is observed in this mutant. HTB1 transcripts with PolyA tails longer than 50 nt are relatively rare and were only encountered in a small percentage of cDNAs in WT cells incubated at 37°C and in sen1 and rrp6 mutant cells (Figure 3A and B).
PolyA tail length of HTB1 mRNAs varies during the cell cycle
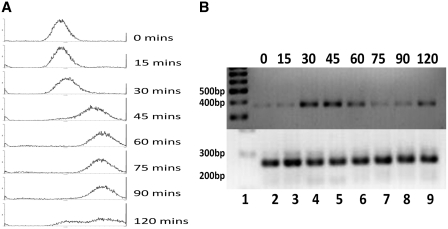
The identification of distinct pools of HTB1 transcripts with different PolyA tail lengths in asynchronous cells, specifically in the sen1 and Δrrp6 mutants, prompted us to examine the PolyA tail length on HTB1 transcripts at different stages of the cell cycle. Yeast cells were synchronized at the G1 checkpoint by incubation with α-factor and RNA was isolated from samples taken every 15 min following α-factor removal. Cell cycle progression was monitored by FACS (Figure 4A) and the levels of HTB1 mRNAs were determined at each stage of the cell cycle (Figure 4B). Increased levels of HTB1 were evident at 30, 45 and 60 min, coinciding with DNA synthesis (Figure 4A) and thereafter decrease, as cells enter into G2-phase (Figure 4B). We have previously shown that HTB1 mRNA levels increase 4- to 5-fold as cells switch from G1 to S-phase (6,19).
Figure 4.
Cell cycle analysis of HTB1 mRNA expression. (A) WT (46a) cells were synchronized with α-factor. Following synchronization, samples were collected every 15 min and analysed by flow cytometer. (B) RNA was extracted from each sample and HTB1 mRNA levels determined by semi-quantitative RT–PCR (top panel). As an internal control, the levels of ACT1 mRNA were also analysed (bottom panel). Lane 1; molecular weight marker, the 200, 300, 400 and 500 bp bands are indicated, lane 2–9, RT–PCR products from RNA extracted at the time intervals in min as shown above the panels using the primers HTB1 For16–32 and Rev368–388 and ACT1 For1 and Rev1 (Table 2).
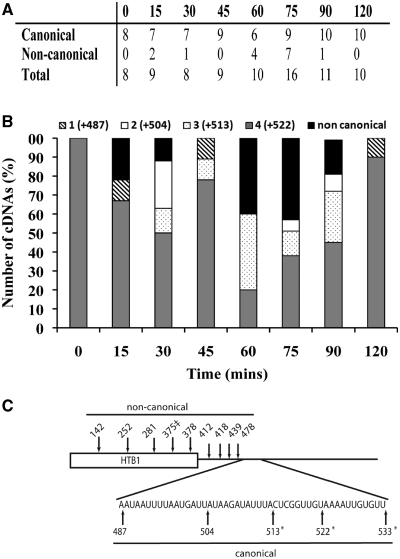
The cleavage site usage and the PolyA tail length of HTB1 mRNAs from the cell cycle staged samples were determined as described above. In total 81 individual cDNA clones were analysed, ∼8–10 per time point (Figure 5A). The HTB1 mRNA population in cells captured in G1 (0 min) appears extremely homogeneous, with cleavage site 4 being exclusively used (Figure 5B). As cells enter S-phase (30–60 min) there is an increase in the diversity of 3′-end cleavage sites, with a decrease in the usage of site 4 and an increase in the use of sites 1–3. The decrease in HTB1 mRNA levels detected at 75 min (Figure 4B) is accompanied by an increase in transcripts with upstream non-canonical 3′-ends (other than sites 1–5; Figure 5A and C). By 75 min, almost 50% of cDNAs represent RNAs with 3′-ends further upstream of the canonical cleavage sites (Figure 5C). A small amount of transcripts with non-canonical 3′-ends are also observed at 15 and 30 min (Figure 5B). By 90–120 min, when mitosis has occurred, cell cycle synchrony is decreased and therefore 3′-end usage at these latter stages are difficult to interpret, although cleavage site usage appears to revert back to an almost exclusive use of site 4, similar to what is observed at 0 min.
Figure 5.
The 3′-end cleavage site usage of HTB1 mRNAs during the cell cycle. (A) Number of G-tailed HTB1 cDNAs analysed at 15-min intervals during the cell cycle. The number of cDNAs displaying canonical and non-canonical 3′-ends is also shown. (B) Location of the 3′-ends of HTB1 cDNAs as a percentage of total at intervals (in min) following release from α-factor. Diagonal stripe; site 1, white; site 2, dots; site 3, gray; site 4, black; non-canonical cleavage sites. (C) Schematic of HTB1 mRNA showing the major canonical and non-canonical 3′-ends detected during the cell cycle. ‡denotes the 3′-end of a cDNA containing two additional non-encoded A residues.
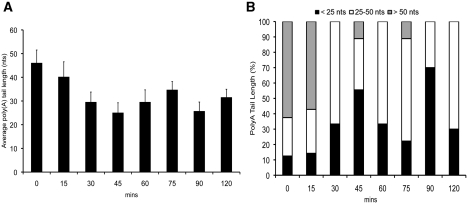
We also observed that the PolyA tail length varies significantly as cell progress through the cell cycle. Compared to the asynchronous cell population, the PolyA tails on HTB1 mRNAs in G1 cells (0 min) are relatively long with a mean length of 46 nt (Figure 6A). As shown in Figure 3 above, tails of >50 nt are rarely encountered in asynchronous cells, yet in G1 >60% of RNAs have tails >50 nt (Figure 6B). The mean tail length of HTB1 mRNAs decreases as cells progress through the cell cycle (Figure 6A) from 46 nt at 0 min to 22 nt at 45 min. The median tail length was 54 and 19 nt at these time points, respectively. The percentage of RNAs (cleaved at the canonical 3′-ends) with PolyA tails <25 nt increases from 10% to 55%, respectively between 0 and 45 min post cell cycle arrest and thereafter decreases with the exception of the 90-min sample (Figure 6B) where the cell population is becoming asynchronous thus making it difficult to assess the true relevance of lengths of PolyA tails in these latter samples.
Figure 6.
The PolyA tail length of HTB1 varies during the cell cycle. (A) Average PolyA tail length (nt) of cDNAs isolated from RNA extracted at intervals (min) following release from α-factor. Error bars represent the standard error of the sample population. (B) Percentage of HTB1 RNAs with PolyA tails <25 nt (black columns), 25–50 nt (white columns) and >50 nt (gray columns) at time intervals after α-factor release. RNAs with canonical 3′-ends only were included in the analysis.
As mentioned above, a substantial proportion of HTB1 RNAs at times 60 and 75 min have non-canonical 3′-ends, all of which lie upstream of the normal cleavage sites (Figure 5A and B) and therefore most likely represent degradation intermediates. The vast majority of these (>90%) do not contain polyA tails. One of the clones from the 75-min sample contained two additional non-encoded A residues at the 3′-end (Figure 5C, indicated by double dagger).
HTB1 transcripts in G1 cells are predominately nuclear
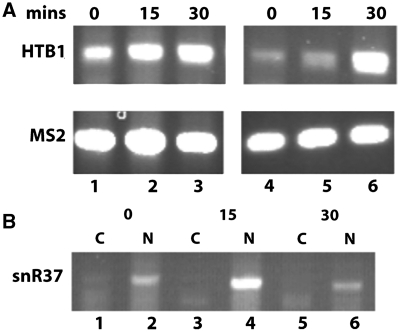
Our analysis of HTB1 levels during the cell cycle revealed a correlation between PolyA tail length and the steady state levels of HTB1 transcripts. At early stages of the cell cycle (0–15 min) when HTB1 steady state levels are low (Figure 4), transcripts bearing long PolyA tails are observed. As cells enter S-phase (30–45 min), HTB1 transcript levels increase and their PolyA tail length decreases. Since histones are preferentially synthesized during S-phase, concomitant with DNA replication, we questioned whether the longer polyA tailed HTB1 transcripts are retained in the nucleus during G1. To test this, WT cells were synchronized with α-factor and cytoplasmic and nuclear fractions were prepared from samples taken at 0, 15 and 30 min after release from α-factor arrest as described in the Methods section. To directly compare the levels of HTB1 transcripts in the nuclear and cytoplasmic fractions, semi-quantitative RT–PCR was carried out using equal amounts of total RNA from each fraction. As shown in Figure 7A, the level of HTB1 mRNA detected in the nuclear fraction at both 0 and 15 min is significantly higher than the level in the cytoplasmic fraction (compare lanes 1 and 2 with lanes 4 and 5, respectively). By 30 min, HTB1 levels in the cytoplasmic fraction have increased significantly (lane 6). The same pattern of HTB1 nuclear retention at 0 and 15 min was observed in multiple independent experiments. PCR analysis of cDNAs prepared in the presence or absence of reverse transcriptase and using primers for HTB1 or 18S rRNA indicated that the higher levels detected in the nucleus at 0 and 15 min do not result from contaminating nuclear DNA (data not shown). The quality of the nuclear/cytoplasmic fractionation was monitored by analyzing the levels of the nucleolar snRNA, snR37, which was completely retained in the nuclear fractions at the three time points examined (Figure 7B, lanes 2, 4 and 6). Real-time RT–PCR analysis quantified the fold-difference of HTB1 transcripts between the cytoplasmic and nuclear fractions at 4.5- and 6.5-fold for 0 and 15 min, respectively (data not shown).
Figure 7.
Semi-quantitative RT–PCR analysis of HTB1 mRNA in nuclear and cytoplasmic fractions at early stages of the cell cycle. (A) Reverse transcription was carried out with nuclear or cytoplasmic total RNA isolated from WT cells at 0, 15 and 30 min. following removal of α-factor (‘Materials and Methods’ section). The reactions were spiked with 400 pg of MS2 RNA to measure the relative efficiency of RT–PCR in the nuclear and cytoplasmic fractions. Lanes 1–3; nuclear RNA isolated 0, 15 and 30 min post α-factor removal respectively, lanes 4–6, corresponding cytoplasmic RNA fractions from 0, 15 and 30 min post α-factor removal. Top panel; RT–PCR using HTB1 primers (271 bp) and one tenth of the original cDNA, bottom panel; amplification of MS2 RNA (219 bp) from the same reaction using MS2 primers (Table 2) and cDNA diluted 1 : 80. Only 20 rounds of PCR amplification were carried out. (B) Distribution of snR37 transcript in the nuclear and cytoplasmic fractions. The cDNA samples used in A above were PCR amplified using primers specific to snR37. Lanes 1, 3 and 5; cytoplasmic fractions (C) lanes 2, 4 and 6; nuclear fractions (N). The time points at which the RNA was isolated post α-factor removal are indicated above the lanes.
DISCUSSION
As a continuation of our previous studies, here we investigated if the Nab3p/Nrd1p/Sen1p complex contributes to the biogenesis of yeast histone mRNAs. Our results demonstrate that Sen1p, plays a role in 3′-end formation of histone mRNAs independent of its association with Nrd1p and Nab3p. Additionally, inactivation of Sen1p leads to the accumulation of histone transcripts with very short PolyA tails.
In addition to its central role in 3′-end processing and transcription termination of non-polyadenylated sn/snoRNAs as part of the Nab3/Nrd1/Sen1p complex (25–27), recent studies have demonstrated an expanded role for Sen1p in other cellular processes. Sen1p is found associated with a number of protein encoding genes, particularly those transcribing small PolyA-dependent transcripts (18) and in association with Nrd1p/Nab3p, can provide a ‘fail safe’ transcription termination mechanism for PolyA-dependent transcripts in the event of incorrect or inefficient 3′-end processing (17). Such transcripts are then rapidly degraded by the nuclear exosome. In the case of histone mRNAs, Sen1p appears to be influencing the 3′-end processing step under conditions when general PolyA-dependent 3′-end processing/transcription termination is not impaired. Since histone mRNA metabolism is temporally regulated during the cell cycle, it is possible that Sen1p may exert its influence on histone mRNA biogenesis in a cell cycle stage-specific manner. Mischo et al. (28) recently demonstrated a role for Sen1p in the resolution of DNA/RNA hybrid structures, referred to as R-loops. Such structures form preferentially at regions of transcription termination possibly due to changes in the processivity of RNA Polymerase II. It has been suggested that the helicase activity of Sen1p may promote transcription termination through its role in resolving R-loops and thereby release the RNA transcript from the site of transcription. Genetic interactions between SEN1 and genes involved in homologous recombination and DNA repair suggests that Sen1p also plays a broader role in maintaining genome integrity (26,28). Our finding that Sen1p influences 3′-end processing of histone mRNAs and Poly A tail length, adds to the growing list of activities of this DNA/RNA helicase.
The polyA tail length of histone mRNAs was also affected by deletion of RRP6, therefore it is possible that both Rrp6p and Sen1p are required for efficient 3′-end formation of histone mRNAs and defects in either protein leads to inefficient recruitment or activity of PolyA polymerase leading to transcripts with shorter PolyA tails. A previously identified genetic association between PAP1, the gene encoding PolyA polymerase, and RRP6 lends support to this view (29).
Inactivation of Sen1p alters the progression of cell cycle
A further link between Rrp6p and Sen1p in histone mRNA biogenesis was uncovered in our finding that, as with deletion of RRP6, mutations in SEN1 also influences cell cycle progression. These effects on cell cycle progression may be a secondary effect due to the influence of Sen1p on many aspects of cell cycle regulation, particularly in light of its role in maintenance of genome integrity (28) and preventing DNA damage (26), however, its concomitant role in histone mRNA biogenesis suggests a more immediate link between cell cycle progression and histone mRNA biogenesis. Thus, there is growing body of evidence for a link between the tightly controlled histone mRNA metabolism and cell cycle progression, leading us to postulate that histone mRNA levels within the cell may act as a checkpoint for cell cycle progression.
PolyA tail length of HTB1 varies during the cell cycle
We also re-evaluated the polyadenylation status of the histone mRNA HTB1 both in asynchronous and synchronized populations of yeast cells. Previous analysis had indicated that histone mRNAs are polyadenylated although tail length was not determined (5,23). Here we find that HTB1 mRNAs contain PolyA tails which are much shorter than the average tail length of the general yeast mRNA pool, which contain tails of ∼70–100 nt. The shorter tail length of HTB1 mRNAs is consistent with data from a genome-wide study of PolyA tail length in S. cerevisiae, which demonstrated an enrichment of transcripts encoding cell cycle related functions in a pool of RNAs with short PolyA tails (30).
We also found that the Poly A tail length of HTB1 transcripts varies as cells progress through the cell cycle. Three distinct cell cycle-stage dependent pools of transcripts were identified. In G1, HTB1 transcripts bear longer PolyA tails (mean length 46 nt). As cells proceed into S-phase, the PolyA tail length decreases (mean length 22 nt) and finally as cells enter G2-phase, transcripts with no Poly A tails with non-canonical 3′-ends are found.
The longer tailed transcripts encountered at the early stages of the cell cycle correspond to the period when HTB1 steady state levels are relatively low. It is possible that longer tailed transcripts encountered in G1 phase are not export-competent and are degraded in the nucleus via the exosome mediated nuclear surveillance mechanism. Since inactivation of Sen1p leads to accumulation of shorter-tailed transcripts, it is possible that such nuclear degradation of longer-tailed transcripts is linked to its role in histone mRNA 3′-end processing. Our finding that HTB1 transcripts are predominately nuclear in G1 phase supports this argument. Further detailed genetic studies will be required to elucidate the exact molecular mechanism of this process.
Paradoxially and contrary to convention, the accumulation of transcripts with shorter PolyA tails is correlated with increased steady state levels of HTB1 in S-phase. It is possible that this shift in PolyA tail length results from an alteration in 3′-end processing/transcription termination of histone genes to generate nuclear export competent transcripts, thus ensuring the presence of histone transcripts in the cytoplasm in S-phase when translation of histones is at its maximum. In fact we did observe higher levels of HTB1 mRNA in the cytoplasmic fraction in S-phase compared to the levels observed in the G1 phase. Alternatively, since polyA tail shortening is associated with active translation of mRNAs (31), the observed shortening of PolyA tails in HTB1 mRNA in S-phase may be a consequence of their association with polysomes.
The identification of transcripts with non-canonical 3′-ends upstream of the canonical 3′-end cleavage sites after S-phase, points to active degradation of histone mRNAs in a 3′–5′ direction. Of the cDNA clones containing non-canonical 3′-ends from the 60–90 min timepoints, only one had non-encoded A residues, a hallmark of nuclear degradation via the exosome, therefore the bulk of this degradation is most likely occurring in the cytoplasm. Due to the cloning method employed in this study, whereby a single forward primer located at nucleotides 1–22 of the HTB1 ORF was used for amplification of G-tailed cDNAs, we were not able to examine the 5′ untranslated region (UTR) of the transcripts. Therefore, we cannot rule out a role of concomitant 5′-3′ exonucleolytic degradation as part of the turnover of HTB1 transcripts at this stage of the cell cycle. Since both 5′–3′ and 3′–5′ degradation of mRNAs occur simultateously during general mRNA degradation, transcripts with shortened 5′-ends may also be present after S-phase. Further studies will be required to unravel the contibution of both 5′ to 3′ and 3′ to 5′ degradation of histone mRNAs at this stage of the cell cycle. Recent studies have identified histone mRNAs as a category of transcripts particularly sensitive to mutations in CCR4, the major deadenylase required for the initialisation of cytoplasmic degradation of mRNAs (32). While playing a major role in cytoplasmic mRNA degradation, the CCR4-NOT complex has recently been shown to physically interact with both TRAMP and the exosome complex, indicating that this deadenylase complex may also play an important role in nuclear mRNA degradation (32). Separately, a role for CCR4 in cell cycle progression has also been identified (33).
Finally, it is interesting to note that deletions or mutations in Senataxin, the human homologue of Sen1p, are associated with the development of the neurological disorder ataxia-ocular apraxia 2 (AOA2) (34). Likewise, mutations in a protein termed NPAT (nuclear protein ataxia-telangiectasia), which is required for the recruitment of specific 3′-end processing factors to replication-dependent histone genes in human cells (35), are associated with the autosomal recessive disease ataxia-telangiectasia, AOA1, which leads to cerebellar ataxia and oculocutaneous telangiectasiain. Both AOA1 and AOA2 share several clinical features, but can be distinguished by the age of onset. The observation that both the yeast homologue of Senataxin, Sen1p and NPAT play important roles in regulating histone mRNA 3′-end processing, may suggest a role for cell cycle dependent histone mRNA metabolism in the development of ataxia-related diseases.
FUNDING
Funding for open access charge: Science Foundation Ireland (06/RFP/GEN/060 to U.B.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank David Brow and Domenico Libri for kindly supplying yeast mutant strains.
REFERENCES
- 1.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osley MA. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 3.Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Davila Lopez M, Samuelsson T. Early evolution of histone mRNA 3′ end processing. RNA. 2008;14:1–10. doi: 10.1261/rna.782308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahrner K, Yarger J, Hereford L. Yeast histone mRNA is polyadenylated. Nucleic Acids Res. 1980;8:5725–5737. doi: 10.1093/nar/8.23.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canavan R, Bond U. Deletion of the nuclear exosome component RRP6 leads to continued accumulation of the histone mRNA HTB1 in S-phase of the cell cycle in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:6268–6279. doi: 10.1093/nar/gkm691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reis CC, Campbell JL. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics. 2007;175:993–1010. doi: 10.1534/genetics.106.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebreton A, Seraphin B. Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim. Biophys. Acta. 2008;1779:558–565. doi: 10.1016/j.bbagrm.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 9.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 10.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell. Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 11.Lemay JF, D'Amours A, Lemieux C, Lackner DH, St-Sauveur VG, Bahler J, Bachand F. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol. Cell. 2010;37:34–45. doi: 10.1016/j.molcel.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Grzechnik P, Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol. Cell. 2008;32:247–258. doi: 10.1016/j.molcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciais D, Bohnsack MT, Tollervey D. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res. 2008;36:3075–3084. doi: 10.1093/nar/gkn160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth KM, Byam J, Fang F, Butler JS. Regulation of NAB2 mRNA 3′-end formation requires the core exosome and the Trf4p component of the TRAMP complex. RNA. 2009;15:1045–1058. doi: 10.1261/rna.709609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 16.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol. Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 19.Campbell SG, Li Del Olmo M, Beglan P, Bond U. A sequence element downstream of the yeast HTB1 gene contributes to mRNA 3′ processing and cell cycle regulation. Mol. Cell. Biol. 2002;22:8415–8425. doi: 10.1128/MCB.22.24.8415-8425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rondon AG, Mischo HE, Proudfoot NJ. Terminating transcription in yeast: whether to be a ‘nerd’ or a ‘rat’. Nat. Struct. Mol. Biol. 2008;15:775–776. doi: 10.1038/nsmb0808-775. [DOI] [PubMed] [Google Scholar]
- 21.Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 22.Ursic D, Chinchilla K, Finkel JS, Culbertson MR. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 2004;32:2441–2452. doi: 10.1093/nar/gkh561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl Acad. Sci. USA. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viphakone N, Voisinet-Hakil F, Minvielle-Sebastia L. Molecular dissection of mRNA poly(A) tail length control in yeast. Nucleic Acids Res. 2008;36:2418–2433. doi: 10.1093/nar/gkn080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkel JS, Chinchilla K, Ursic D, Culbertson MR. Sen1p performs two genetically separable functions in transcription and processing of U5 small nuclear RNA in Saccharomyces cerevisiae. Genetics. 2010;184:107–118. doi: 10.1534/genetics.109.110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prakash S, Johnson RE, Washington MT, Haracska L, Kondratick CM, Prakash L. Role of yeast and human DNA polymerase eta in error-free replication of damaged DNA. Cold Spring Harb. Symp. Quant. Biol. 2000;65:51–59. doi: 10.1101/sqb.2000.65.51. [DOI] [PubMed] [Google Scholar]
- 27.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat. Rev. Mol. Cell. Biol. 2011;12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burkard KT, Butler JS. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 2000;20:604–616. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13:982–997. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 32.Azzouz N, Panasenko OO, Colau G, Collart MA. The CCR4-NOT complex physically and functionally interacts with TRAMP and the nuclear exosome. PLoS One. 2009;4:e6760. doi: 10.1371/journal.pone.0006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westmoreland TJ, Marks JR, Olson JA, Jr, Thompson EM, Resnick MA, Bennett CB. Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae. Eukaryot. Cell. 2004;3:430–446. doi: 10.1128/EC.3.2.430-446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, Tranchant C, Aubourg P, Tazir M, Schols L, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- 35.Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep. 2009;10:894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]