Abstract
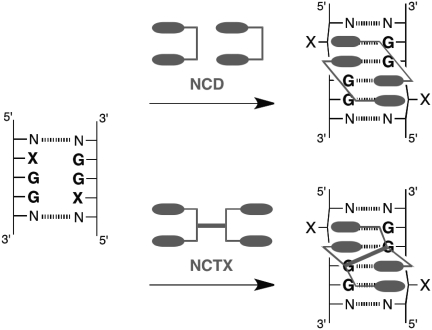
A naphthyridine carbamate dimer (NCD) is a synthetic ligand for DNA containing a CGG/CGG sequence. Although NCD can bind selectively and tightly to a CGG/CGG sequence, the highly cooperative 2:1 binding mode has hampered precise analysis of the binding. We describe herein the synthesis of a series of naphthyridine tetramers consisting of two NCD molecules connected with various linkers to seek a ligand that binds to a CGG/CGG sequence exclusively with a 1:1 stoichiometry. Among the tested ligands, NCTB and Z-NCTS, which have linker moieties with restricted conformational flexibility [biphenyl and (Z)-stilbene linker, respectively], gave the exclusive formation of a 1:1 ligand–CGG/CGG complex. The (Z)-stilbene linker in Z-NCTS was designed to have pre-organized conformation appropriate for the binding and, in fact, resulted in the highest binding affinity. Thermodynamic parameters obtained by isothermal titration calorimetry indicated that the stronger binding of Z-NCTS was attributed to its lower entropic cost. The present study provides not only a novel 1:1 binding ligand, but also valuable feedback for subsequent molecular design of DNA and RNA binding ligands.
INTRODUCTION
Small molecular probes that bind specific DNA sequences and structures are promising molecular tools for a wide range of DNA-based biotechnology. Pyrrole–imidazole polyamide is a sequence-specific DNA binder rationally developed from natural minor groove binders, and has been studied for the regulation of gene expression and applied to DNA nanotechnology (1–3). Besides the sequence-specific ligands for regular double-stranded DNA, much effort has been devoted to develop artificial ligands targeting a variety of local DNA and RNA structures (4–27), such as multistranded helices (4–6), bulges (12,26), mismatches (9–13), loops (22), and repetitive motifs (15–17,25). Since these local structures are related closely to natural and artificial functions of nucleic acids, ligands binding to the structures are useful molecular tools for detection and control of the functions. However, despite the effort, rational design of the ligands remains complicated.
We have demonstrated that small synthetic ligands can recognize a specific DNA mismatch through complementary hydrogen bonding in conjugation with stacking and electrostatic interactions (11). Based on this rational design, we have developed a series of synthetic ligands that bind selectively to characteristic local DNA structures, such as single nucleotide bulges, mismatched base pairs and trinucleotide repeats (11–22). Synthetic ligands for abasic sites and repetitive motifs have also been developed by other groups (23–26). N-Acyl-2-amino-1,8-naphthyridine, which has a hydrogen-bonding surface fully matched with that of guanine base, is a key component of the synthetic ligands (28–30). Dimeric naphthyridine derivatives bind selectively to a GG mismatch, where each of the two naphthyridine moieties recognizes two mispaired guanine bases (13,18). While the mismatch binding ligand (MBL) has been studied as a diagnostic tool for SNPs (13,14), trinucleotide repeats (15–17), telomeric repeats (20,21) and as a molecular glue for controlling DNA hybridization (31–35), MBLs with enhanced selectivity and affinity are still needed for practical applications.
A naphthyridine carbamate dimer (NCD) is one of the highest affinity ligands for a GG mismatch (Figures 1 and 2) (18). In particular, the GG-mismatch 5′-adjacent to guanine, in a 5′-XGG-3′/5′-XGG-3′ sequence (XGG/XGG, where X = A, T, C and G), is the most preferential binding site for NCD (17,31). NMR analysis of the NCD–CGG/CGG complex revealed that two NCD molecules are involved in order to recognize four guanine bases in the CGG/CGG sequence, and the nucleotide C was flipped out from the π stack (Figure 1, top, X = C) (19). Although the exclusive formation of a 2:1 ligand–DNA complex is characteristic for MBLs (11), improved MBLs with a 1:1 binding mode to the CGG/CGG sequence are desirable for the following reasons. First, the complex binding behavior of standard MBLs hampers the quantitative analysis necessary for improved molecular design. The binding of two NCD molecules to a XGG/XGG sequence is a highly positively cooperative process, but the actual binding process remains unclarified. Second, reducing stoichiometry of ligands by covalent linking of two ligand molecules has been used as an effective approach to enhance the binding selectively and affinity (36,37). Third, a specific MBL–DNA pair provides a potential platform for noncovalent functionalization of DNA (38–46). When an NCD bearing an additional functional unit is used for this purpose, two functional units are inevitably introduced into the DNA because of the 2:1 binding mode (45,46). The presence of two functional units in close proximity may cause unintended interactions leading to impaired functions and destabilization of the complex. MBLs that bind to the target sequence exclusively with a 1:1 stoichiometry are needed to address these issues.
Figure 1.
Schematic illustration of the binding of dimeric naphthyridine (top, NCD) and tetrameric naphthyridine derivatives (bottom, NCTXs) to a XGG/XGG sequence.
Figure 2.
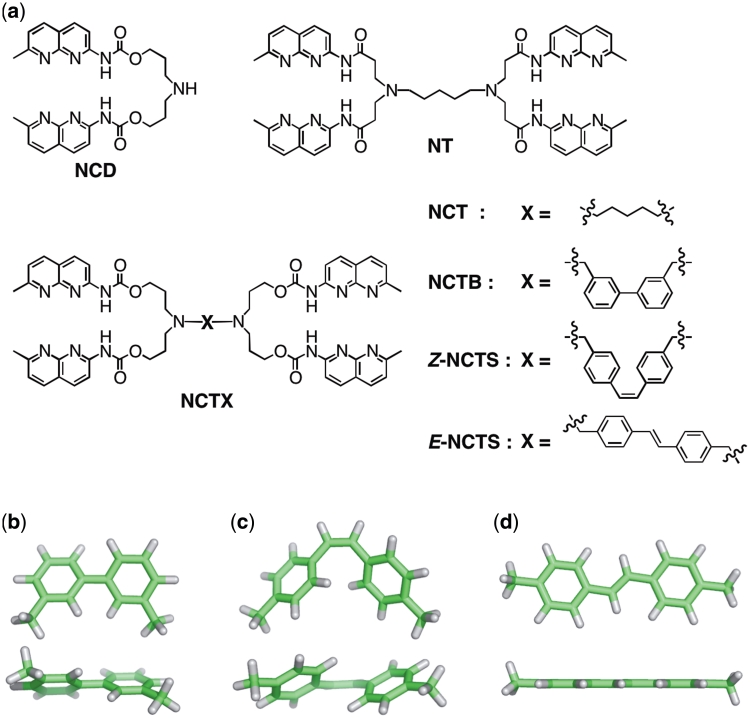
(a) Mismatch binding ligand used in this study. (b–d) Energy minimized structures [B3LYP/6-31 G(d)] of the linker moieties. (b) 3,3′-dimethylbiphenyl in NCTB; (c) (Z)−4,4′-dimethylstilbene in Z-NCTS; (d) (E)−4,4′-dimethylstilbene in E-NCTS.
In principle, an MBL showing a 1:1 binding stoichiometry can be designed by covalent linking of two MBLs having a 2:1 binding mode (Figure 1, bottom). Herein, we report the design and evaluation of novel ligands possessing two NCD molecules linked by four different units (NCTXs, Figure 2). Z-NCTS possessing a (Z)-stilbene unit with a rigid and pre-organized conformation tightly bound to the CGG/CGG sequence exclusively with 1:1 binding stoichiometry and a dissociation constant (Kd) of 98 nM. Quantitative thermodynamic analysis revealed that the strong binding of Z-NCTS was attributed to the decreased total entropic cost. These findings provide valuable insights for the development of high-affinity ligands for various DNA and RNA structures.
MATERIALS AND METHODS
Synthesis of NCTB
To a solution of NCD (170 mg, 0.34 mmol) in anhydrous N,N-dimethylformamide (DMF, 4.1 ml) were successively added KI (54 mg, 0.33 mmol), 3,3′-bis(bromomethyl)biphenyl (48) (55 mg, 0.16 mmol) and K2CO3 (54 mg, 0.39 mmol), and stirred at ambient temperature for 3 h. The reaction mixture was diluted with saturated NaHCO3 and extracted with AcOEt. The organic layer was successively washed with water and brine, dried over MgSO4, filtered, and evaporated in vacuo. The crude product was purified by column chromatography on silica gel (CHCl3/MeOH = 25:1) to give NCTB (64 mg, 34%) as a white powder: 1H NMR (400 MHz, CDCl3) δ 8.22 (d, 4 H, J = 8.8 Hz), 8.04 (d, 4 H, J = 8.8 Hz), 7.89 (d, 4 H, J = 8.32 Hz), 7.78 (br, 4 H), 7.49 (s, 2 H), 7.40 (d, 2 H, J = 7.56 Hz), 7.30 (t, 2 H, J = 7.56 Hz), 7.26 (s, 2 H), 7.17 (d, 4 H, J = 8.08 Hz), 4.27 (t, 8 H, J = 6.5 Hz), 3.56 (s, 4 H), 2.63 (s, 12 H), 2.54 (t, 8 H, J = 6.6 Hz), 1.86 (m, 8 H, J = 6.46 Hz); 13C NMR (150 MHz, CDCl3) δ 162.9, 154.7, 153.5, 153.2, 141.0, 139.8, 138.8, 136.3, 128.7, 127.6, 127.4, 125.9, 121.1, 117.9, 112.8, 64.0, 58.9, 50.0, 26.7, 25.5; HRMS (ESI, positive-ion mode, MeOH) m/e calcd for C66H68N14NaO8 [M+Na]+ 1207.5242, found 1207.5241.
Synthesis of Z-NCTS
To a solution of NCD (171 mg, 0.34 mmol) and (Z)−4,4’-diformylstilbene (47) (35 mg, 0.15 mmol) in CHCl3 (1.5 ml) and MeOH (9.2 ml) was added acetic acid (27 µl, 0.47 mmol) to adjust the pH at 6, and the reaction mixture was stirred at ambient temperature for 10 min. To this solution was added NaBH3CN (21 mg, 0.33 mmol) dissolved in MeOH. After being stirred at ambient temperature for 46 h, the solvent was evaporated in vacuo and the residue was diluted with saturated NaHCO3 and extracted with CHCl3. The organic layer was washed with brine, dried over anhydrous MgSO4, filtered and evaporated in vacuo. The crude product was purified by column chromatography on silica gel (CHCl3/MeOH = 10:1) to give Z-NCTS (34 mg, 18%) as a pale yellow powder: 1H NMR (400 MHz, CDCl3) δ 8.26 (d, 4 H, J = 8.56 Hz), 8.09 (d, 4 H, J = 8.76 Hz), 7.94 (d, 4 H, J = 8.08 Hz), 7.72 (br, 4 H), 7.21 (d, 4 H, J = 8.32 Hz), 7.13 (d, 4 H, J = 12.7 Hz), 7.11 (d, 4 H, J = 12.7 Hz), 6.27 (s, 2 H), 4.25 (t, 8 H, J = 6.5 Hz), 3.49 (s, 4 H), 2.72 (s, 12 H), 2.52 (t, 8 H, J = 6.6 Hz), 1.85 (m, 8 H, J = 6.7 Hz); 13C NMR (150 MHz, CDCl3) δ 163.0, 154.8, 153.5, 153.2, 138.9, 138.2, 136.3, 136.1, 129.7, 128.8, 128.6, 121.1, 118.0, 112.8, 64.1, 58.6, 50.1, 26.7, 25.5; HRMS (ESI, positive-ion mode, MeOH) m/e calcd for C68H70N14NaO8 [M + Na]+ 1233.5399, found 1233.5399.
Synthesis of E-NCTS
To a solution of NCD (150 mg, 0.30 mmol) in anhydrous DMF (4 ml) were added (E)−4,4′-dibromomethylstilbene (49) (24 mg, 0.065 mmol) and stirred at ambient temperature for 2.5 h. The reaction mixture was diluted with saturated NaHCO3 and extracted with AcOEt. The organic phase was successively washed with water and brine, dried over MgSO4, filtered, and evaporated in vacuo. The crude product was purified by column chromatography on silica gel (CHCl3/methanol = 30:1) to give E-NCTS (14 mg, 18%) as a white powder: 1 H-NMR (400 MHz, CDCl3) δ = 8.24 (d, 4 H, J = 8.68 Hz), 8.04 (d, 4 H, J = 8.72 Hz), 7.87 (d, 4 H, J = 8.24 Hz), 7.70 (br, 4 H), 7.17 (8 H), 7.16 (d, 4 H, J = 8.28 Hz), 6.63 (s, 2 H), 4.27 (t, 8 H, J = 6.4 Hz), 3.51 (s, 4 H), 2.70 (s, 12 H), 2.55 (t, 8 H, J = 6.6 Hz), 1.87 (m, 8 H, J = 6.4 Hz); 13C-NMR (100 MHz, CDCl3) δ = 163.0, 154.7, 153.4, 153.1, 139.0, 138.5, 136.3, 136.1, 128.9, 127.5, 126.3, 121.1, 117.9, 112.6, 63.9, 58.5, 49.9, 26.6, 25.6; HRMS (ESI) m/e calcd for C68H70N14NaO8 [(M + Na)+] 1233.5399, found 1233.5386.
Melting temperature (Tm) measurements
The sample solutions were prepared by mixing DNA duplex (4.5 µM) and ligand (9.1 µM) in 10 mM sodium cacodylate buffer (pH 7.0) and 0.1 M NaCl containing 0.1% (v/v) Tween-20. Thermal denaturation profiles were recorded on a SHIMADZU UV-2550 UV-Vis spectrophotometer equipped with SHIMADZU TMSPC−8 temperature controller. The absorbance of the samples was monitored at 260 nm from 2°C to 80°C with heating rate of 1°C/min. Tm values were calculated by using the median method.
CSI-TOF-MS measurements of ligand–DNA complex
Samples were prepared by mixing DNA duplex (20 µM) and ligand (40 µM) in 50% MeOH in water containing 0.1 M ammonium acetate. Mass spectra were obtained with JEOL AccuTOF JMS-T100N mass spectrometer in the negative mode (orifice 1 voltage = −60 V). Spray temperature was fixed at −10°C with a sample flow rate of 20 µl/min. Nitrogen gas was used as a desolvation gas as well as a nebulizer.
Circular dichroism measurements
Circular dichroism (CD) measurements were carried out on a J-725 CD spectropolarimeter (JASCO) using a 1.0 cm path length cell. CD spectra change of DNAs were measured while titrating with ligand at ambient temperature in 10 mM sodium cacodylate (pH 7.0) containing 0.1 M NaCl and 0.1% (v/v) Tween-20.
Isothermal titration calorimetry measurements
Isothermal titration calorimetry (ITC) measurements were carried out with a VP-ITC calorimeter (MicroCal, Northampton, MA, USA). Since the solubility of NCTXs toward aqueous buffered solutions is not high enough for standard titration experiments, a reverse titration method was employed. A solution of ODN3, 5′-d(CTAA CGG AATGTG TTTT CACATT CGG TTAG)-3′, (58.2 µM, 290 µl) in 10 mM sodium cacodylate (pH 7.0) containing 0.1 M NaCl and 0.1% (v/v) Tween 20 was titrated into a solution of ligand (10 or 20 µM, 1.4 ml) in the same buffer. Titrations typically consisted of 29 × 10 µl injections of 24 s duration each, with 3 min spacing between injections. All experiments were performed at 25°C. Prior to fitting analysis, data point from the first injection was removed, and the reaction heat data were corrected by subtracting the heat of dilution obtained from injection of DNA solution into a buffer solution. Binding isotherms were fitted to the Origin models by least-squares analysis (Origin 5.0, MicroCal).
Calculation of structures
Molecular modeling studies were carried out using the MacroModel 9.1 molecular modeling package. Initial structures were constructed manually from the NMR-structure we determined previously (15). The resulting ligand–DNA complex was subjected to energy minimization using the AMBER* force field with the extended cutoff for non-bonded interactions (cutoff for van der Waals, electrostatic, and hydrogen bonding were 8, 20 and 4 Å, respectively) (50). Energy minimization was performed by the Polak–Ribier Conjugate Gradient (PRCG) method with a convergence threshold of 0.05. The solvent effects are simulated using the analytical Generalized-Born/Surface-Area (GB/SA) model with the constant dielectric treatment.
RESULTS
Synthesis of tetrameric naphthyridine derivatives
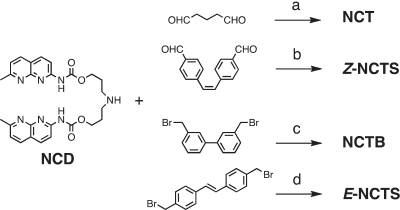
We synthesized a series of tetrameric naphthyridine derivatives (NCTXs, Figure 2), which consist of two NCD molecules connected by different linkers, such as a five-methylene linker (NCT), a biphenyl linker (NCTB), and (Z)- and (E)-stilbene (Z-NCTS and E-NCTS, respectively) linkers. These linkers provide different orientation and conformational flexibility for the four naphthyridines, which should be important for the ligand binding (Figure 2b–d). The (Z)-stilbene linker in Z-NCTS was designed to place the naphthyridines in preorganized positions appropriate for the binding. Synthesis of NCT (22) and Z-NCTS was achieved by reductive amination of glutaraldehyde and (Z)-4,4′-diformylstilbene (47), respectively, with the secondary amino group of NCD (Scheme 1). NCTB and E-NCTS were synthesized from 3,3′-bis(bromomethyl)biphenyl (48) and (E)−4,4′-bis(bromomethyl)stilbene (49) using a nucleophilic substitution reaction with two NCD molecules. NT, where naphthyridines are connected with amide linkages instead of carbamate linkages in NCD, was synthesized as reported previously (21). All ligands contain two sp3 nitrogen atoms that are protonated at physiological condition. The positively charged amino group increases not only the binding affinity to negatively charged DNA and the solubility in water. Because NCTXs have low solubility in water, they were used as corresponding hydrochlorides in an aqueous buffered solution containing the surfactant Tween 20.
Scheme 1.
Synthesis of NCTXs. (a) NaBH3CN, MeOH, CHCl3, AcOH, 23%; (b) NaBH3CN, MeOH, CHCl3, AcOH, 18%; (c) KI, K2CO3, DMF, 34%; (d) DMF, 18%.
Binding of NCTXs to a CGG/CGG sequence
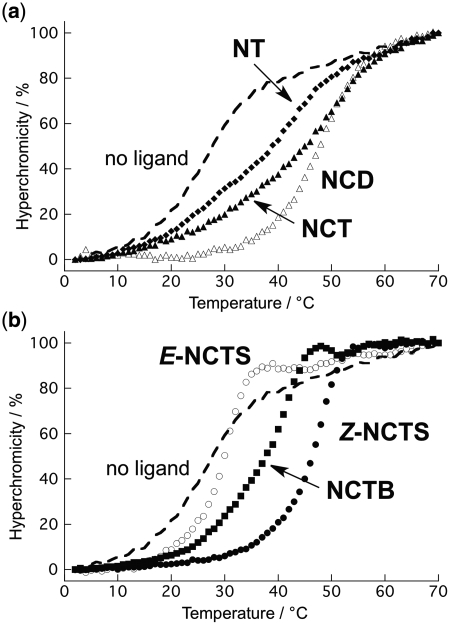
Binding of the ligands to the GG-mismatch DNA was evaluated using melting temperatures (Tm) for the target DNA duplexes, which provides qualitative information regarding binding affinity and selectivity. Tms of the 11-mer duplex 5′-(CTAA CGG AATG)-3′/5′-(CATT CGG TTAG)-3′ (ODN1/2) containing a CGG/CGG sequence were measured in the presence of NCTXs (Figure 3). All NCTXs except for E-NCTS selectively stabilized the GG-mismatch DNA duplex (Table 1). Among the tested ligands, Z-NCTS provided the highest Tm (47.6°C, ΔTm = 21.4°C), which was comparable with the Tm in the presence of a twofold concentration of NCD (ΔTm = 18.6°C) (18). While NCTXs showed moderate stabilization of the DNAs containing GA and GT mismatches, Z-NCTS showed the highest selectivity for the GG mismatch; i.e. the smaller ΔTm for the GA and GT mismatches and the highest ΔTm for the GG mismatch. The selectivity was enhanced at higher concentrations of NaCl (200 and 400 mM, Supplementary Figure S1), suggesting nonspecific electrostatic interactions were suppressed under the conditions. Notably, thermal melting profiles for NCTB and Z-NCTS exhibited a sharp melting transition from double-stranded DNA (dsDNA) to single-stranded DNA (ssDNA) (Figure 3b). This observation is in stark contrast to the protracted transition observed for NT and NCT (Figure 3a), suggesting highly cooperative dissociation of the four naphthyridine–guanine pairs in the melting of Z-NCTS and NCTB complexes.
Figure 3.
Thermal melting profiles of DNA duplex ODN1/2 containing GG mismatch (4.5 µM) in the presence of NCTXs (9.1 µM). The absorbance at 260 nm was measured in 10 mM Na·cacodylate buffer (pH 7.0) containing 0.1 M NaCl and 0.1% (v/v) Tween 20. (a) Plots in the presence of NT (filled diamond), NCT (filled triangle) and NCD (open triangle). (b) Z-NCTS (filled circle), E-NCTS (open circle) and NCTB (filled square). The melting curve in the absence of ligand is shown as a dotted line.
Table 1.
Melting temperature (Tm/°C) of DNA duplexes in the presence of a series of tetrameric naphthyridine ligands
| X-Y | Tm0a | NCT |
NCTB |
Z-NCTS |
E-NCTS |
NT |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tmb | ΔTmc | Tmb | ΔTmc | Tmb | ΔTmc | Tmb | ΔTmc | Tmb | ΔTmc | ||
| G-G | 26.2 | 44.5 | 18.3 | 39.5 | 13.3 | 47.6 | 21.4 | 29.8 | 3.6 | 39.7 | 13.5 |
| (0.3) | (1.2) | (0.6) | (0.3) | (0.3) | (0.3) | ||||||
| A–A | ndd | 24.7 | ndd | ndd | ndd | 23.1 | ndd | ndd | ndd | – | – |
| (0.2) | (1.1) | ||||||||||
| A–C | 17.9 | 18.3 | 0.4 | 23.4 | ndd | ndd | ndd | ndd | ndd | – | – |
| (1.1) | (0.6) | (0.6) | |||||||||
| C–C | 20.4 | 22.2 | 1.8 | 22.1 | 1.7 | 22.3 | 1.9 | 25.1 | 4.7 | – | – |
| (1.3) | (0.9) | (1.7) | (1.3) | (0.9) | |||||||
| G–A | 28.3 | 36.0 | 7.7 | 36.1 | 7.8 | 34.3 | 6.0 | 30.4 | 2.1 | 27.7 | −0.6 |
| (0.3) | (1.0) | (1.0) | (1.0) | (0.7) | (0.2) | ||||||
| G–T | 29.3 | 36.9 | 7.6 | 37.2 | 7.9 | 34.9 | 5.6 | 30.3 | 1.0 | 26.1 | −3.2 |
| (0.2) | (0.7) | (1.0) | (0.3) | (1.0) | (1.2) | ||||||
| T–C | 20.5 | 23.3 | 2.8 | 24.6 | 4.1 | 23.5 | 3.0 | ndd | ndd | – | – |
| (1.1) | (0.6) | (0.7) | (0.7) | ||||||||
| T–T | 26.4 | 24.8 | −1.5 | 27.3 | 0.9 | 28.6 | 2.2 | 28.0 | 1.6 | – | – |
| (0.8) | (0.8) | (1.1) | (0.7) | (0.9) | |||||||
Thermal melting curves were measured for ODNs, d(CTAA CXG AATG)/d(CATT CYG TTAG), (4.5 µM) in 10 mM Na·cacodylate buffer (pH 7.0) containing 0.1 M NaCl and 0.1% (v/v) Tween 20. Tm values (°C) were calculated by median method and mean of at least three experiments. Values in parentheses represent standard deviations.
aTm values of DNA duplexes in the absence of ligand.
bTm values in the presence of ligand (9.1 µM).
cΔTm is calculated as the difference between Tm0 and Tm.
dTm value could not be determined due to the unclear transition.
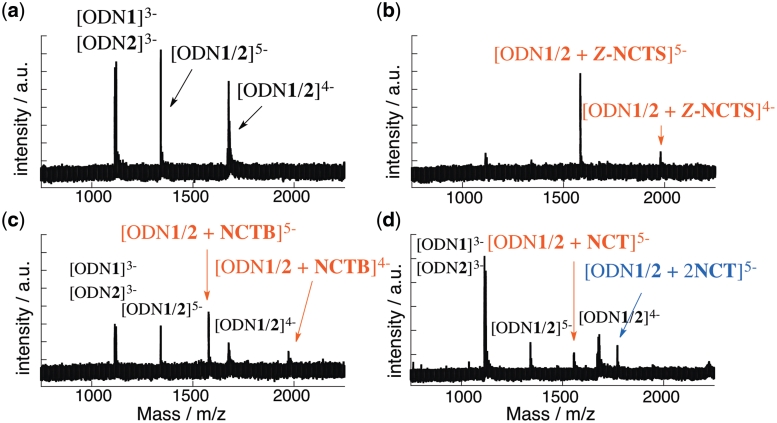
The increased Tm values in the presence of the NCTXs indicate the formation of a stable complex with the CGG/CGG sequence. Cold-spray ionization time-of-flight mass spectrometry (CSI-TOF MS) measurements revealed the preferred binding stoichiometry of the stable complex formation (Figure 4). In the absence of ligands, ions corresponding to 3–ions of the ssDNAs ([ODN1]3−, m/z: found 1120.5, calcd 1120.5; [ODN2]3−, m/z: found 1114.5, calcd 1114.5), 4-ions ([ODN1/2]4−, m/z: found 1677.0, calcd 1676.8), and 5-ions ([ODN1/2]5−, m/z: found 1341.4, calcd 1341.4) of the dsDNA were observed. In the presence of two molar equivalents of Z-NCTS, new ion peaks at m/z 1680.0 and 1583.8 corresponding to the 4- and 5-ions of the Z-NCTS–ODN1/2 complex with a 1:1 stoichiometry (calcd 1979.3 and 1583.4, respectively) were observed (Figure 4b). No other ion peaks corresponding to complexes with a different stoichiometry and almost complete disappearance of ions corresponding to free dsDNA under the binding conditions indicated exclusive formation of a 1:1 complex. NCTB also produced a 1:1 complex (NCTB–ODN1/2), although a substantial amount of the DNA remained unbound under the same conditions (Figure 4c). In contrast, multiple ions with different stoichiometries were observed in the presence of NCT (Figure 4d). This is most likely because the flexible methylene linker allowed NCT to bind to the DNA in different binding modes, whereas the rigid linkers in Z-NCTS and NCTB provided an unambiguous 1:1 binding mode. Having discovered the highest affinity of this group of molecules to the target CGG/CGG with exclusive 1:1 complex formation, we focused our attention on the binding of Z-NCTS.
Figure 4.
CSI-TOF MS of DNA duplex ODN1/2 (20 µM) in the presence of Z-NCTS, NCTB and NCT. Sample solutions in 50% aqueous methanol and 0.1 M ammonium acetate were cooled at −10°C during the injection with a flow rate of 20 µl/min. Key: (a) In the absence of ligand; (b) 40 µM Z-NCTS; (c) 40 µM NCTB; (d) 40 µM NCT.
Thermodynamic analysis of Z-NCTS binding
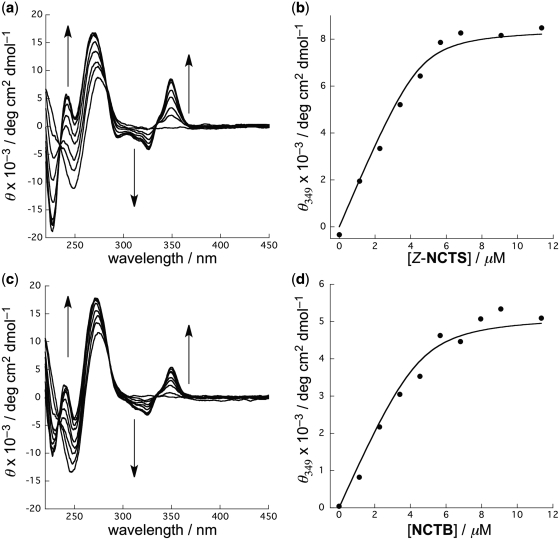
CD titration studies were conducted to determine the binding of Z-NCTS to the CGG/CGG sequence. The CD spectra of the CGG/CGG-containing DNA ODN1/2 were measured while titrating with Z-NCTS (Figure 5a). Upon addition of Z-NCTS, induced CD bands appeared at wavelengths >300 nm. The intensity of the bands increased with increasing concentration of Z-NCTS with isodichroic points at 235, 285 and 335 nm, indicating that a unique binding equilibrium exists between the free and bound states of ODN1/2 and Z-NCTS. The association constant (Ka) of Z-NCTS for the CGG/CGG sequence was calculated to be 4.4 × 106 M−1 using a least-squares curve fitting analysis of the observed data to a 1:1 binding isotherm (Figure 5b). CD titration experiments were also conducted with NCTB, and provided a similar CD spectral change (Figure 5c). The induced CD bands observed for Z-NCTS and NCTB corresponding to the absorption of the 1,8-naphthyridine moieties were almost superimposable, suggesting that the spatial alignment of the four naphthyridine–guanine pairs are similar for the two complexes. As expected from the Tm and CSI-TOF MS data, the Ka value for NCTB (2.8 × 106 M−1) was smaller than for Z-NCTS.
Figure 5.
CD titration experiments for the binding of Z-NCTS and NCTB to the CGG/CGG sequence. (a and c) CD spectra of ODN1/2 (4.5 µM) were measured while titrating with (a) Z-NCTS or (c) NCTB at 0, 1.14, 2.27, 3.41, 4.54, 5.68, 6.81, 9.08 and 11.4 µM. (b and d) The ellipicities at 349 nm were plotted against the concentration of (b) Z-NCTS or (d) NCTB. The solid line represents the best fits of the data to a 1:1 binding isotherm.
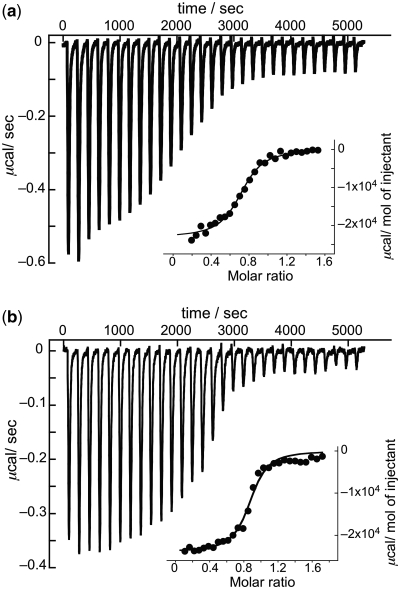
To further characterize the binding, isothermal titration calorimetry (ITC) measurements were conducted for NCTXs. The ITC profiles obtained by injection of hairpin DNA containing a CGG/CGG sequence (ODN3, 5′-d(CTAA CGG AATGTG TTTT CACATT CGG TTAG)-3′) into a solution of NCTXs are shown in Figure 6. The 1:1 stoichiometry of the binding of Z-NCTS and NCTB to the CGG/CGG sequence enabled us to determine the thermodynamic parameters of their complexes (Figure 6b and a). Fitting the data for Z-NCTS and NCTB to a single set of identical binding sites provided Ka values of 1.0 × 107 M−1 and 3.4 × 106 M−1, respectively (Table 2). The Ka values obtained by ITC were roughly consistent with those obtained using CD titration experiments. The larger Ka value for Z-NCTS compared with that for NCTB is attributed to the lower total entropic cost upon the complex formation. In contrast, the calorimetric isotherms obtained for NCT and NCD were not well fitted to the binding models available for analysis (Supplementary Figure S2). This is most likely because of the presence of multiple binding stoichiometries and the positively cooperative binding.
Figure 6.
Calorimetric isotherms for NCTX-binding. (a) Titration of 58.2 µM ODN3 with 10 µM NCTB at 25°C. Every peak represents the heat released by an each injection. Inset: the corrected molar heat per injection with the best fit curve for a single set of identical binding sites model. (b) Titration with 10 µM Z-NCTS.
Table 2.
Thermodynamic parameters for NCTB and Z-NCTS-binding to a CGG/CGG sequence
| Kaa | nb | Kab | Kb | ΔHb | TΔSb | ΔGb | |
|---|---|---|---|---|---|---|---|
| 106M−1 | 106M−1 | nM | /kcal/mol | /kcal/mol | /kcal/mol | ||
| NCTB | 2.8 | 1.25 | 3.4 | 290 | −21.3 | −12.5 | −8.9 |
| (0.14) | (2.19) | (3.23) | (3.28) | (0.45) | |||
| Z-NCTS | 4.4 | 1.22 | 10.2 | 98 | −19.6 | −10.1 | −9.5 |
| (0.13) | (3.02) | (0.91) | (0.97) | (0.2) |
aKa values determined by CD titration experiments.
bThermodynamic parameters at 298 K obtained from ITC data by fitting with a single set of identical sites. Values in parentheses represent standard deviations of three replicates.
DISCUSSION
We have synthesized a series of ligands that have four naphthyridines as recognition elements connected by different linkers. The structure of the linker is an important determinant of the ligand-binding characteristics. The ligands we discussed here can be classified into two groups: ligands with a flexible linker (NT and NCT) and those with a rigid linker (NCTB, E-NCTS and Z-NCTS).
Ligand with a flexible linker
The first choice of a linker for a ligand design is usually a flexible linker, because the conformational flexibility may allow an induced fit upon binding to the target. Although we have reported that NT and NCT containing flexible methylene linkers could uniquely interact with human telomeric repeats d(TTAGGG)n (21) and DNA hairpin loops (22), respectively, their interactions with DNA GG mismatches remained to be studied. Thermal melting experiments showed that both NT and NCT increased the Tm of DNA containing a CGG/CGG sequence by 13.5°C and 18.7°C, respectively (Figure 3a). A difference in the linker structure connecting two naphthyridines was found responsible for the higher ΔTm for NCT complexes compared with that for NT complexes. The carbamate linkage in NCT has expanded π surfaces favorable for the stacking interaction, and the longer linker may offer a better opportunity for adopting the spatial arrangement of naphthyridines in the complex (18,51). Indeed, the ΔTm observed for NCT (18.7°C) is comparable with the highest ΔTm value (21.4°C) obtained for Z-NCTS. A major drawback of the flexible linker in NCT is that multiple binding modes and kinetics are involved in the binding process because of the large conformational flexibility. CSI-TOF MS measurements clearly showed that the NCT–CGG/CGG complex consisted of a mixture of at least two different binding stoichiometries (Figure 4d). Another possible drawback is the large entropic cost because of the loss of degrees of freedom upon binding. The entropic contribution to the binding affinity, however, could not be estimated in the NCT binding, because its multiple binding modes preclude quantitative analysis of the NCT binding (Figure S2).
Ligand with a rigid linker
NCTB and NCTS have linker moieties with restricted conformational flexibility (Figure 2b–d). Among the tested ligands, the highest and lowest ΔTm values were obtained for Z-NCTS (21.4°C) and E-NCTS (3.6°C), respectively, whereas the ΔTm value for NCTB was modestly (13.3°C) between those for the two NCTS ligands (Figure 3b). The ligands with restricted conformational freedom provided two extremes of binding behavior. Z-NCTS was designed to have a preorganized conformation for binding to the GG mismatch, where the bent (Z)-stilbene linker can place the four naphthyridines in a favorable position and orientation for the binding. On the other hand, location and orientation of naphthyridines led by the extended conformation of the (E)-stilbene linker in E-NCTS completely abolished the binding. A marked preference for the Z configuration in the binding has been observed for naphthyridines connected with an azobenzene linker, where (Z)-azobenzene stabilized the mismatch duplex, but (E)-azobenzene did not (33–35). Quantitative analysis of the binding of the (Z)-azobenzene derivative is hindered by the difficulty in isolating pure (Z)-azobenzene derivatives because of their thermal and light sensitivity. Z-NCTS possessing a stable stilbene linker allowed detailed analysis of the preorganized linker conformation effects on the binding.
The thermal melting profile of Z-NCTS showed a sharp melting transition compared with those observed for NT and NCT, suggesting a cooperative dissociation of the four naphthyridine–guanine pairs and the hybridized DNAs (Figure 3b). CSI-TOF MS measurements revealed that the binding of Z-NCTS and NCTB resulted in the exclusive formation of a 1:1 ligand–dsDNA complex (Figure 4). These data are in contrast to the smooth melting transition and multiple binding modes observed for the ligands with flexible linkers, NCT and NT. These findings support the hypothesis that Z-NCTS adopts a preorganized conformation favorable for the 1:1 binding.
The unique 1:1 binding of Z-NCTS and NCTB to the CGG/CGG sequence enabled us to analyze thermodynamic parameters of the ligand binding (Figure 6). The quantitative data from ITC measurements indicated that the stronger binding of Z-NCTS (Ka = 1.0 × 107 M−1) compared with NCTB (3.4 × 106 M−1) was mainly attributed to its lower entropic cost. Since global structures of Z-NCTS- and NCTB–CGG/CGG complexes are expected to be very similar from CD spectra (Figure 5) and modeling studies as described below (Figure 7), we have focused on local entropic changes about the ligand structures. Biphenyl has a rotational degree of freedom conferred by the internal phenyl–phenyl linkage, which will be lost in the bound state. On the other hand, the rotation about the central double bond is restrained in the stilbene linker, and the (Z)−4,4′-stilbene linkage in Z-NCTS maintains the four recognition elements in restricted orientations and locations. The preorganized conformation of Z-NCTS may contribute to its strong binding through the reduced entropic loss.
Figure 7.
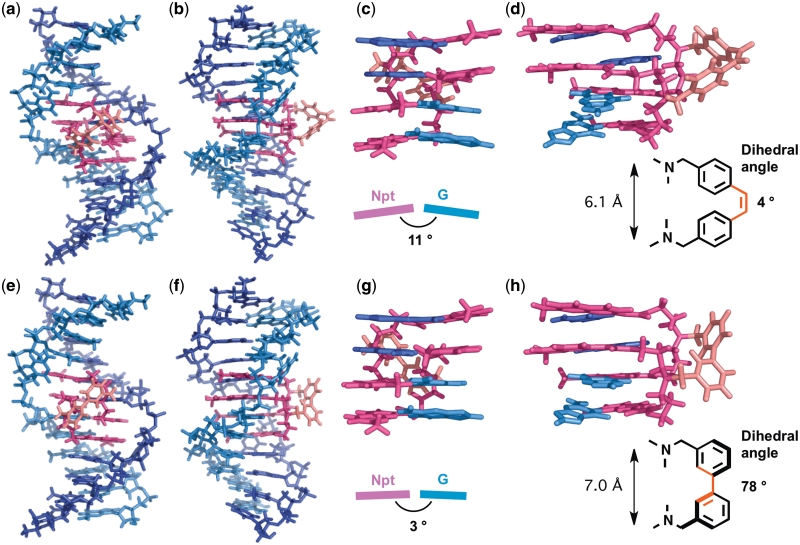
Molecular modeling of the complex between the CGG/CGG sequence and tetrameric naphthyridine ligands, Z-NCTS (a–d) and NCTB (e–h). The model structures were optimized by use of the AMBER* force field in water with MacroModel Version 9.1. Overall structures viewed from the major groove (a and e) and side (b and f). The NCD and linker parts in the ligands are represented in magenta and red, respectively. Expanded views of the four naphthyridine-guanine (red–blue) pairs from the minor groove (c and g) and side (d and h). Buckle angles of naphthyridine-guanine base pairs (c and g), dihedral angles between two phenyl rings (d and h), and distance between two nitrogen atoms in the linkers (d and h) are shown.
Although the binding affinity of NCTB was lower than that of Z-NCTS, the NCTB binding provided a more favorable exothermic enthalpy change, indicating that the NCTB–CGG/CGG complex is structurally more favorable than the Z-NCTS–CGG/CGG complex. To obtain further insight into the structures of the complexes, we performed computer-modeling studies of the NCTB– and Z-NCTS–CGG/CGG complexes using a molecular mechanics calculation (Figure 7). The overall structures were quite similar to each other as expected from CD studies. In both structures, four guanines in the CGG/CGG sequence were hydrogen bonded to four naphthyridine moieties in a single ligand molecule. Focusing on the linker structures, the (Z)−4,4′-stilbene linker of Z-NCTS has a smaller dihedral angle (4°) between its two phenyl rings than the 3,3′-biphenyl linker of NCTB (78°) (Figure 7d, h). This difference resulted in the different distances between two NCD moieties and a kink in the naphthyridine–guanine pseudo-base pair in the Z-NCTS–CGG/CGG complex, represented by larger buckle angles (Figure 7c, κ = 11°) than for NCTB (Figure 7g, κ = 3°). Molecular dynamics simulations also support the larger structural distortion in Z-NCTS–CGG/CGG complex (Supplementary Figure S3). The perturbations of the binding site agree with the lower enthalpy gain observed for Z-NCTS binding compared with NCTB. These data suggested that the molecular structure of Z-NCTS still has room for further improvement in its binding affinity. A novel ligand mimicking a preorganized alignment of four naphthyridine units in the NCTB–CGG/CGG complex would have both entropically and enthalpically favorable binding thermodynamics.
CONCLUSION
We synthesized a series of naphthyridine tetramers that consist of two NCD molecules connected by various linkers to search for a ligand that binds to a CGG/CGG sequence exclusively with a 1:1 stoichiometry. The linker structure had significant effects on the binding properties of the ligands, and we found that Z-NCTS possessing a rigid (Z)-stilbene linker bound to the CGG/CGG sequence with high affinity and exclusive 1:1 binding stoichiometry. This binding mode allowed us to perform a precise thermodynamic analysis of the Z-NCTS binding, suggesting that the pre-organized conformation of Z-NCTS enhanced its binding by lowering the entropic cost. Compared with NCD having a 2:1 binding stoichiometry, Z-NCTS provides the following benefits resulting from the 1:1 binding: (i) quantitative analysis based on the simple 1:1 binding mode; (ii) high binding affinity; and (iii) facile introduction of a single functional unit per CGG/CGG site on the DNA scaffold. The present study provides not only a novel 1:1 binding ligand useful for DNA-based technologies, but also valuable feedback for subsequent design of DNA and RNA binding ligands.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–S3.
FUNDING
The Japan Society for the Promotion of Science [JSPS; Grant in Aid for Scientific Research on Innovative Areas (21200042) and Grant in Aid for Scientific Research (S) (18105006)]; Sumitomo Foundation and National Institute of Biomedical Innovation (10–22). Funding for open access charge: JSPS.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Dervan PB. Molecular recognition of DNA by small molecules. Biorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 2.Sugiyama H, Bando T. Synthesis and biological properties of sequence-specific DNA-alkylating pyrrole-imidazole polyamides. Acc. Chem. Res. 2006;39:935–944. doi: 10.1021/ar030287f. [DOI] [PubMed] [Google Scholar]
- 3.Tse WC, Boger DL. Sequence-selective DNA recognition: natural products and nature's lessons. Chem. Biol. 2004;11:1607–1617. doi: 10.1016/j.chembiol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 5.Rezler EM, Bearss DJ, Hurley LH. Telomere inhibition and telomere disruption as processes for drug targeting. Annu. Rev. Pharmacool. Toxicol. 2003;43:359–379. doi: 10.1146/annurev.pharmtox.43.100901.135733. [DOI] [PubMed] [Google Scholar]
- 6.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannon MJ. Supramolecular DNA recognition. Chem. Soc. Rev. 2007;36:280–295. doi: 10.1039/b606046n. [DOI] [PubMed] [Google Scholar]
- 8.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 9.Jackson BA, Barton JK. Recognition of DNA base mismatches by a rhodium intercalator. J. Am. Chem. Soc. 1997;119:12986–12987. [Google Scholar]
- 10.Pierre VC, Kaiser JT, Barton JK. Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc. Natl Acad. Sci. USA. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakatani K. Recognition of Mismatched Base Pairs in DNA. Bull. Chem. Soc. Jpn. 2009;82:1055–1069. [Google Scholar]
- 12.Nakatani K, Sando S, Saito I. Recognition of a single guanine bulge by 2-acylamino-1,8-naphthyridine. J. Am. Chem. Soc. 2000;122:2172–2177. [Google Scholar]
- 13.Nakatani K, Sando S, Saito I. Scanning of guanine-guanine mismatches in DNA by synthetic ligands using surface plasmon resonance. Nat. Biotechnol. 2001;19:51–55. doi: 10.1038/83505. [DOI] [PubMed] [Google Scholar]
- 14.Takei F, Igarashi M, Hagihara M, Oka Y, Soya Y, Nakatani K. Secondary-Structure-Inducible Ligand Fluorescence Coupled with PCR. Angew. Chem. Int. Ed. Engl. 2009;48:7822–7824. doi: 10.1002/anie.200902449. [DOI] [PubMed] [Google Scholar]
- 15.Nakatani K, Hagihara S, Goto Y, Kobori A, Hagihara M, Hayashi G, Kyo M, Nomura M, Mishima M, Kojima C. Small-molecule ligand induces nucleotide flipping in (CAG)(n) trinucleotide repeats. Nat. Chem. Biol. 2005;1:39–43. doi: 10.1038/nchembio708. [DOI] [PubMed] [Google Scholar]
- 16.He H, Hagihara M, Nakatani K. A Small Molecule Affecting the Replication of Trinucleotide Repeat d(GAA)(n) Chem. Eur. J. 2009;15:10641–10648. doi: 10.1002/chem.200901088. [DOI] [PubMed] [Google Scholar]
- 17.Peng T, Nakatani K. Binding of naphthyridine carbamate dimer to the (CGG)(n) repeat results in the disruption of the G-C base pairing. Angew. Chem. Int. Ed. Engl. 2005;44:7280–7283. doi: 10.1002/anie.200502282. [DOI] [PubMed] [Google Scholar]
- 18.Peng T, Murase T, Goto Y, Kobori A, Nakatani K. A new ligand binding to G-G mismatch having improved thermal and alkaline stability. Bioorg. Med. Chem. Lett. 2005;15:259–262. doi: 10.1016/j.bmcl.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Nomura M, Hagihara S, Goto Y, Nakatani K, Kojima C. NMR structural analysis of the GG mismatch DNA complexed with naphthyridine-dimer. Nucleic Acids Symp. Ser. 2005;49:213–214. doi: 10.1093/nass/49.1.213. [DOI] [PubMed] [Google Scholar]
- 20.Nakatani K, Hagihara S, Sando S, Sakamoto S, Yamaguchi K, Maesawa C, Saito I. Induction of a remarkable conformational change in a human telomeric sequence by the binding of naphthyridine dimer: Inhibition of the elongation of a telomeric repeat by telomerase. J. Am. Chem. Soc. 2003;125:662–666. doi: 10.1021/ja027055g. [DOI] [PubMed] [Google Scholar]
- 21.Goto Y, Hagihara S, Hagihara M, Nakatani K. Small-molecule binding to the nonquadruplex form of the human telomeric sequence. ChemBioChem. 2007;8:723–726. doi: 10.1002/cbic.200600564. [DOI] [PubMed] [Google Scholar]
- 22.Hong C, Hagihara M, Nakatani K. Ligand-assisted complex formation of two DNA hairpin loops. Angew. Chem. Int. Ed. 2011;50:4390–4393. doi: 10.1002/anie.201100075. [DOI] [PubMed] [Google Scholar]
- 23.Yoshimoto K, Nishizawa S, Minagawa M, Teramae N. Use of abasic site-containing DNA strands for nucleobase recognition in water. J. Am. Chem. Soc. 2003;125:8982–8983. doi: 10.1021/ja029786m. [DOI] [PubMed] [Google Scholar]
- 24.Zhao CX, Dai Q, Seino T, Cui YY, Nishizawa S, Teramae N. Strong and selective binding of amiloride to thymine base opposite AP sites in DNA duplexes: simultaneous binding to DNA phosphate backbone. Chem. Commun. 2006:1185–1187. doi: 10.1039/b516575j. [DOI] [PubMed] [Google Scholar]
- 25.Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc. Natl Acad. Sci. USA. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ong HC, Arambula JF, Rao Ramisetty S, Baranger AM, Zimmerman SC. Molecular recognition of a thymine bulge by a high affinity, deazaguanine-based hydrogen-bonding ligand. Chem. Commun. 2009;2009:668–670. doi: 10.1039/b817733n. [DOI] [PubMed] [Google Scholar]
- 27.Oleksi A, Blanco AG, Boer R, Uson I, Aymami J, Rodger A, Hannon MJ, Coll M. Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate. Angew. Chem. Int. Ed. Engl. 2006;45:1227–1231. doi: 10.1002/anie.200503822. [DOI] [PubMed] [Google Scholar]
- 28.Murray TJ, Zimmerman SC. 7-Amido-1,8-naphthyridines as hydrogen-bonding units for the complexation of guanine derivatives - the role of 2-alkoxyl groups in decreasing binding-affinity. Tetrahedron Lett. 1995;36:7627–7630. [Google Scholar]
- 29.Feibush B, Saha M, Onan K, Karger B, Giese R. HPLC separation of DNA adducts based on hydrogen-bonding. J. Am. Chem. Soc. 1987;109:7531–7533. [Google Scholar]
- 30.Hamilton AD, Pant N. Nucleotide base recognition - ditopic binding of guanine to a macrocyclic receptor containing naphthyridine and naphthalene units. J. Chem. Soc. Chem. Comm. 1988;1988:765–766. [Google Scholar]
- 31.Peng T, Dohno C, Nakatani K. Mismatch-binding ligands function as a molecular glue for DNA. Angew. Chem. Int. Ed. Engl. 2006;45:5623–5626. doi: 10.1002/anie.200601190. [DOI] [PubMed] [Google Scholar]
- 32.Peng T, Dohno C, Nakatani K. Bidirectional control of gold nanoparticle assembly by turning on and off DNA hybridization with thermally degradable molecular glue. ChemBioChem. 2007;8:483–485. doi: 10.1002/cbic.200700005. [DOI] [PubMed] [Google Scholar]
- 33.Dohno C, Uno SN, Nakatani K. Photoswitchable molecular glue for DNA. J. Am. Chem. Soc. 2007;129:11898–11899. doi: 10.1021/ja074325s. [DOI] [PubMed] [Google Scholar]
- 34.Uno S, Dohno C, Bittermann H, Malinovskii VL, Häner R, Nakatani K. A light-driven supramolecular optical switch. Angew. Chem. Int. Ed. Engl. 2009;48:7362–7365. doi: 10.1002/anie.200903251. [DOI] [PubMed] [Google Scholar]
- 35.Dohno C, Yamamoto T, Nakatani K. Photoswitchable unsymmetrical ligand for DNA hetero-mismatches. Eur. J. Org. Chem. 2009;2009:4051–4058. [Google Scholar]
- 36.Mrksich M, Dervan PB. Design of a covalent peptide heterodimer for sequence-specific recognition in the minor-groove of double-helical DNA. J. Am. Chem. Soc. 1994;116:3663–3664. [Google Scholar]
- 37.Nakatani K, Sando S, Kumasawa H, Kikuchi J, Saito I. Recognition of guanine-guanine mismatches by the dimeric form of 2-amino-1,8-naphthyridine. J. Am. Chem. Soc. 2001;123:12650–12657. doi: 10.1021/ja0109186. [DOI] [PubMed] [Google Scholar]
- 38.Best TP, Edelson BS, Nickols NG, Dervan PB. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc. Natl Acad. Sci. USA. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rucker VC, Foister S, Melander C, Dervan PB. Sequence specific fluorescence detection of double strand DNA. J. Am. Chem. Soc. 2003;125:1195–1202. doi: 10.1021/ja021011q. [DOI] [PubMed] [Google Scholar]
- 40.Ihara T, Uemura A, Futamura A, Shimizu M, Baba N, Nishizawa S, Teramae N, Jyo A. Cooperative DNA probing using a beta-cyclodextrin-DNA conjugate and a nucleobase-specific fluorescent ligand. J. Am. Chem. Soc. 2009;131:1386–1387. doi: 10.1021/ja809023n. [DOI] [PubMed] [Google Scholar]
- 41.Cohen JD, Sadowski JP, Dervan PB. Addressing single molecules on DNA nanostructures. Angew. Chem. Int. Ed. Engl. 2007;46:7956–7959. doi: 10.1002/anie.200702767. [DOI] [PubMed] [Google Scholar]
- 42.Cohen JD, Sadowski JP, Dervan PB. Programming multiple protein patterns on a single DNA nanostructure. J. Am. Chem. Soc. 2008;130:402–403. doi: 10.1021/ja0772400. [DOI] [PubMed] [Google Scholar]
- 43.Dohno C, Atsumi H, Nakatani K. Ligand inducible assembly of a DNA tetrahedron. Chem. Commun. 2011;47:3499–3501. doi: 10.1039/c0cc05473a. [DOI] [PubMed] [Google Scholar]
- 44.Shelke SA, Sigurdsson ST. Noncovalent and site-directed spin labeling of nucleic acids. Angew. Chem. Int. Ed. Engl. 2010;49:7984–7986. doi: 10.1002/anie.201002637. [DOI] [PubMed] [Google Scholar]
- 45.Maekawa K, Nakazawa S, Atsumi H, Shiomi D, Sato K, Kitagawa M, Takui T, Nakatani K. Programmed assembly of organic radicals on DNA. Chem. Commun. 2010;46:1247–1249. doi: 10.1039/b913061f. [DOI] [PubMed] [Google Scholar]
- 46.Atsumi H, Maekawa K, Nakazawa S, Shiomi D, Sato K, Kitagawa M, Takui T, Nakatani K. Noncovalent assembly of TEMPO radicals pair-wise embedded on a DNA duplex. Chem. Lett. 2010;39:556–557. [Google Scholar]
- 47.Bosanac T, Wilcox CS. Precipiton reagents: precipiton phosphines for solution-phase reductions. Org. Lett. 2004;6:2321–2324. doi: 10.1021/ol049369+. [DOI] [PubMed] [Google Scholar]
- 48.Utley JHP, Gao YP, Gruber J, Lines R. Electrochemical route to xylylene polymers and copolymers via cathodically generated quinodimethanes - preparative and structural aspects. J. Mater. Chem. 1995;5:1297–1308. [Google Scholar]
- 49.Soomro SA, Benmouna R, Berger R, Meier H. Preparation and photochemistry of dendrimers with isolated stilbene chromophores. Eur. J. Org. Chem. 2005;2005:3586–3593. [Google Scholar]
- 50.Weiner SJ, Kollman PA, Nguyen DT, Case DA. An all-atom force field for simulation of protein and nucleic acids. J. Comput. Chem. 1986;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 51.Dohno C, Uno SN, Sakai S, Oku M, Nakatani K. The effect of linker length on binding affinity of a photoswitchable molecular glue for DNA. Biorg. Med. Chem. 2009;17:2536–2543. doi: 10.1016/j.bmc.2009.01.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.