Abstract
Toward the expansion of the genetic alphabet of DNA, we present highly efficient unnatural base pair systems as an artificial third base pair for PCR. Hydrophobic unnatural base pair systems between 7-(2-thienyl)imidazo[4,5-b]pyridine (Ds) and 2-nitro-4-propynylpyrrole (Px) were fine-tuned for efficient PCR, by assessing the amplification efficiency and fidelity using different polymerases and template sequence contexts and modified Px bases. Then, we found that some modifications of the Px base reduced the misincorporation rate of the unnatural base substrates opposite the natural bases in templates without reducing the Ds–Px pairing selectivity. Under optimized conditions using Deep Vent DNA polymerase, the misincorporation rate was extremely low (0.005%/bp/replication), which is close to that of the natural base mispairings by the polymerase. DNA fragments with different sequence contexts were amplified ∼1010-fold by 40 cycles of PCR, and the selectivity of the Ds–Px pairing was >99.9%/replication, except for 99.77%/replication for unfavorable purine-Ds-purine motifs. Furthermore, >97% of the Ds–Px pair in DNA survived in the 1028-fold amplified products after 100-cycle PCR (10 cycles repeated 10 times). This highly specific Ds–Px pair system provides a framework for new biotechnology.
INTRODUCTION
Expansion of the genetic alphabet of DNA with artificial extra base pairs (unnatural base pairs) would have great potential for advancing the present genetic recombination systems to next generation biotechnologies. The genetic expansion by unnatural base pair systems could facilitate the site-specific incorporation of extra components into nucleic acids and proteins at desired positions, through the genetic information flow process comprising replication, transcription and translation (1,2). Researchers have chemically synthesized many unnatural base pairs and tested their abilities, and some of them function as a third base pair in in vitro biological processes (3–9). In addition, the attachment of functional groups to unnatural bases enables the site-specific functionalization of DNA and RNA molecules by replication and transcription (10–16). The capabilities of several unnatural base pairs have been demonstrated, and they have been utilized in diagnostics for target DNA detection and in basic research for site-specific labeling of nucleic acid molecules (9,17–21). For example, an unnatural base pair between isoguanine and isocytosine (22) is available as Plexor, in a multiplex real-time quantitative PCR detection platform (23–25).
Toward the next generation of biotechnology, the development of unnatural base pairs for specific PCR amplification is a seminal milestone, to allow the large-scale amplification of synthetic DNA fragments containing unnatural base pairs as a supply of DNA templates for use in further biological processes. Since the amplified DNA fragments are reused as templates in the following PCR amplification, high fidelity of the cognate base pairing is essential. If the selectivity of an unnatural base pairing, which is indicated by the incorporation selectivity (%) of the unnatural base substrates opposite their pairing partners in templates, is 99.9%/replication, then the amplified DNA would retain 96% of the unnatural base pair after 40 cycles of PCR (0.99940 = 0.961) in a theoretical exponential amplification. However, 99.0% selectivity of the unnatural base pairing would reduce the retention to 67% in the amplified products after 40 cycles of PCR (0.99040 = 0.669). Furthermore, for the high fidelity of unnatural base pairing, extremely low misincorporation rates of unnatural base substrates opposite the natural bases in templates are also required. Even if the misincorporation rate is 0.10%/natural bp/replication, up to 33% of the natural base pairs in a 10-base sequence flanked by PCR primers would be mutated to unnatural base pairs after 40 cycles (0.33 = 1 − [(1 − 0.10/100)10]40) in a theoretical exponential amplification. Thus, for the further development of unnatural base pair systems in PCR, the base pairing fidelity of both the high incorporation selectivity in cognate base pairings and the low misincorporation rate of unnatural bases opposite the natural bases, as well as the high amplification efficiency, are required to facilitate the advancement of the new biotechnology.
So far, various unnatural base pairs that function in PCR with 96−99.8% incorporation selectivity per replication or per PCR cycle have been reported (26–35), although the determination of the fidelity of unnatural base pairing depends on each researcher's method. Among them, we developed a hydrophobic unnatural base pair between 7-(2-thienyl)-imidazo[4,5-b]pyridine (Ds) and 2-nitro-4-propynylpyrrole (Px) for efficient PCR amplification (32). The Ds–Px pair was designed by strict shape-complementarity of the pairing surface, but both shapes are different from those of the natural bases. In addition, the Ds–Px pair lacks notable hydrogen bonds between the pairing bases, and thus the hydrophobic bases effectively exclude the non-cognate pairing with the natural bases. Furthermore, the Px base has the nitro group to electrostatically repel the mispairing with A (31). The oxygen of the nitro group of Px and the nitrogen of Ds, corresponding to the N3 position of the natural purine bases, are also important as proton acceptor residues, to interact with polymerases for efficient replication. By modifying a conventional sequencing method for the unnatural base pair, we determined the incorporation selectivity (>99%/replication) of the Ds–Px pairing in PCR with Deep Vent DNA polymerase, possessing a 3′ → 5′ exonuclease activity (exo+). In addition, we identified the efficiencies of the template sequence contexts of the natural bases around the unnatural base in PCR amplification. For example, in vitro selection experiments using a DNA library containing the unnatural base pair (32) and primer extension experiments using templates all combinations of 16 sequences of 3′-NDsN-5′ (N = A, G, C or T) revealed that the less efficient sequences contain 5′-YPxY-3′/3′-RDsR-5′ patterns (Morohashi, Kimoto, and Hirao, unpublished data), such as 5′-GTTPxCAT-3′/3′-CAADsGTA-5′ (32), in PCR amplification. However, the exact differences among these sequence variants, in terms of the efficiency and fidelity of the Ds–Px pairing in PCR, still require clarification, and its PCR conditions can be further optimized. Through the process of unnatural base pair development, we have expected to discover other potentially important parameters of the Ds–Px pair in PCR amplification.
Here, we present highly efficient, high-fidelity PCR systems involving the Ds–Px pair with optimized conditions for a variety of template sequence contexts. For this, we developed accurate determination methods for the fidelity of both the incorporation selectivity of the Ds–Px pairing and the misincorporation rates of the natural base replacement with the unnatural bases in DNA templates during PCR, as well as the amplification efficiency, by repeating 10 cycles of PCR to stay within exponential amplification. The Ds–Px pair survived through 100 cycles of PCR, and after 100 cycles of PCR (10 cycles of PCR repeated 10 times) under the optimized conditions, the DNA fragment containing the Ds–Px pair was amplified 1028-fold, and >97% of the Ds–Px pair was retained in the amplified products. Accordingly, the unnatural base pair selectivity was as high as 99.97%/replication. Furthermore, we found that some functional groups attached to the Px base reduced the misincorporation rates of the unnatural base substrate opposite the natural bases in templates, without reducing the amplification efficiency and the incorporation selectivity of the unnatural base pairing between Ds and modified Px. This Ds–Px pair system could be practically used for future genetic alphabet expansion technologies.
MATERIALS AND METHODS
General
DNA templates containing Ds were synthesized with an Applied Biosystems 392 DNA synthesizer, using CE phosphoramidite reagents for the natural and Ds bases (Glen Research). DNA fragments comprising only the natural bases were synthesized or purchased from Invitrogen. The primer labeled with FAM at the 5′-end was purchased from Invitrogen. DNA fragments were purified by gel electrophoresis. The syntheses of dDsTP, NH2-hx-dPxTP and Cy5-hx-dPxTP was described previously (30,32,36), and the syntheses of the other modified-dPxTPs are described in the Supplementary Data section. The following thermostable DNA polymerases were purchased: Deep Vent (exo+), Deep Vent (exo−), Vent (exo+), N9°, Phusion HF and Taq DNA pols from New England Biolabs; AccuPrime Pfx™ and Pfx50 DNA pols from Invitrogen, Pfu DNA pol from Promega; Pwo SY DNA pol from Roche; TITANIUM Taq DNA pol from Clontech.
PCR involving the Ds–Px pairing
PCR amplification (25 µl) of the 55-mer DNA fragments (5′-TTTCACACAGGAAACAGCTATGACGG-N7-CCCTATAGTGAGTCGTATTATC-3′; N7 = 5′-ATCDsTAT-3′ for Ds-temp 1, 5′-CCCDsTTG-3′ for Ds-temp 2, 5′-TACDsGTG-3′ for Ds-temp 3, 5′-ATGDsAAC-3′ for Ds-temp 4 and 5′-ATCCATT-3′ for Cont-temp) was performed by using 1 µM each 5′-primer (40-mer; 5′-CGTTGTAAAACGACGGCCAGGATAATACGACTCACTATAG-3′) and 3′-primer (24-mer; 5′-TTTCACACAGGAAACAGCTATGAC-3′) and each DNA pol at the manufacturer's recommended concentration (Deep Vent (exo+), 0.5 U; Deep Vent (exo−), 0.5 U; AccuPrime Pfx, 1.25 U; Pfx50, 2.5 U; Phusion HF, 0.5 U; Pfu, 0.625 U; Pwo SY, 1.25 U; Vent (exo+), 0.5 U; N9°, 0.5 U; TITANIUM Taq, 1×; Taq, 0.5 U), in the 1× reaction buffer accompanying each DNA pol, supplemented with 50 µM each dDsTP and modified-dPxTP, and 300 µM natural dNTPs, unless otherwise indicated. As for PCR with the AccuPrime Pfx DNA pol with final concentrations of 400 µM natural dNTPs, 100 µM natural dNTPs and 0.5 mM MgSO4 were further added to the 1× AccuPrime Pfx reaction mix, originally containing 300 µM natural dNTPs and 1 mM MgSO4. As for PCR with Deep Vent DNA pol (exo+) with final concentrations of 500 µM natural dNTPs, 2 mM MgSO4 was further added to the 1× ThermoPol reaction buffer originally containing 2 mM MgSO4. PCR conditions were 30 s at 94°C, 30 s at 45°C and 4 min at 65°C/cycle.
To screen DNA pols, we performed a 15-cycle PCR, by using 0.5 nM 55-mer DNA template and FAM-labeled 3′-primer. The amplified PCR products, together with a FAM-labeled 3′-primer (1 µM) as a marker, were analyzed by denaturing 15% polyacrylamide gel electrophoresis (PAGE). The full-length 75-mer DNA fragments on the gel were quantified by a bio-imaging analyzer LAS-4000 (FUJIFILM) in the SYBR-mode, and by comparing the band intensities with that of the marker, the amplification folds were determined. For the sequencing analysis of the amplified fragments to determine the retention rates of the Ds–Px pair, as described below, PCR was performed by using non-labeled primers, and the full-length PCR products were purified by denaturing 8% PAGE.
In the 40- and 100-cycle PCR amplifications, we repeated a 10-cycle PCR 4 or 10 times, to maintain exponential amplification conditions. In the first 10-cycle PCR, we used 2 nM DNA template, and in the following 10-cycle PCR amplifications, the amplified products were diluted 80- or 150-fold (5 µl) into a fresh PCR reaction mix (final dilutions: ×750 for PCR with Ds-temp 1, Ds-temp 3, and Cont-temp and ×400 for PCR with Ds-temp 4 for all substrates, except for ×400 for PCR with Ds-temp 1 for BPh-dPxTP or HBP-hx-dPxTP). To determine the fold amplification, the amplified PCR products after each 10-cycle PCR were analyzed by denaturing 15% PAGE, and the full-length 75-mer DNA fragments on the gel were quantified by the LAS-4000 analyzer after SYBR Green II staining. To determine the amplification fidelity, the amplified products after 10, 20, 40, 70 and 100 cycles of PCR were fractionated on an 8% denaturing gel, followed by an analysis of the retention rates of the unnatural base by sequencing (for Ds-temps 1, 3 and 4) and the misincorporation rate of the unnatural bases opposite the natural bases (for Cont-temp), as described below.
DNA sequencing
The cycle sequencing reaction (20 µl) was performed with the Cycle Sequencing mix (8 µl) from a BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems), containing 0.3 pmol of DNA fragments and 4 pmol of Sequencing primer (20-mer; 5′-CGTTGTAAAACGACGGCCAG-3′), in the presence of 4-propynyl-1-(β-d-ribofuranosyl)pyrrole-2-carbaldehyde 5′-triphosphate (dPa′TP) (40 pmol for Ds-temp 1 and 1 nmol for Ds-temp 2 and Ds-temp 3) or 1 nmol ddPa′TP. The sequencing cycling parameters were 25 cycles of 10 s at 96°C, 5 s at 50°C and 4 min at 60°C. The residual dye terminator was removed by passage through a CENTRI-SEP column (Princeton Separations), and the solutions were dried. The residues were resuspended in 4 µl formamide solution and analyzed by an ABI 377 DNA Sequencer, using a 6% polyacrylamide-6 M urea gel. The sequence data were analyzed with the Applied Biosystems PRISM sequencing analysis v3.2 software. The retention rates of the Ds–Px pair in the amplified PCR products were determined by comparing the sequencing patterns with the authentic sequencing patterns (see Supplementary Figures S1–S15).
Real-time qPCR
The PCR reaction (25 µl) was performed in 1× ThermoPol reaction buffer with 300 µM each natural dNTP, 50 µM dDsTP and NH2-hx-dPxTP, 1 µM each 5′-primer and 3′-primer, 30 000-fold diluted SYBR Green I (Lonza), 1000-fold diluted ROX (Invitrogen) as a reference dye, 0.5 unit of Deep Vent (exo+) DNA pol, and DNA template (Ds-temp 1, Ds-temp 4 or Cont-temp) at different concentrations (final concentrations: 20 pM, 2 pM, 200 fM and 20 fM) on a Mx3005P Real-time system (Agilent Technologies). Data collection was performed with the detection filters for SYBR and ROX, and the data were analyzed with the Mx3005P v4.10 software. The cycle thresholds were automatically set by the software, which gave the cycle threshold values (Ct).
Analysis of misincorporation rates of unnatural bases into natural bases
We first performed a 40- or 100-cycle PCR in the presence of modified-dPxTP by using Cont-temp as described earlier (first PCR), and the full-length amplified products after 10, 20, 40, 70 and 100 cycles of PCR were purified by denaturing 8% PAGE. The purified products (0.25 pmol) were subjected to seven extra cycles of PCR with the same concentration of the same DNA polymerase, but with Cy5-hx-dPxTP, instead of the modified-dPxTP (second PCR), to detect the modified Px base positions in the amplified DNA fragments. The conditions used in the second PCR were 1 µM each 5′-primer and FAM-labeled 3′-primer, 50 µM dDsTP, 5 µM Cy5-hx-dPxTP and 300 µM natural dNTPs and 2 mM MgSO4 for the Deep Vent DNA pol (exo+) or 400 µM natural dNTPs and 1.5 mM MgSO4 for the AccuPrime Pfx DNA pol. We also performed a 7-cycle PCR directly by using 10 nM (0.25 pmol) Ds-temp 1 and Cont-temp, to normalize the correct incorporation of Cy5-hx-dPxTP opposite Ds in the template during the second PCR and to subtract the background misincorporation of Cy5-hx-dPxTP during the second PCR, respectively. Under these conditions, we assumed that almost all of the Ds–Px pairs were retained after PCR amplification, since we confirmed the complete retention of the Ds–Px pair by a sequencing analysis of the amplified products of the 55-mer DNA template in the second PCR (data not shown). The second PCR products, together with a 75-mer single-stranded DNA (1 µM) as a marker, were analyzed by 15% denaturing PAGE. The fluorescent intensity of the full-length 75-mer DNA fragments on the gel was quantified with a bio-imager, FLA-7000 (GE-Healthcare) in the Cy5-mode and FAM-mode.
Misincorporations of the unnatural bases into the first PCR-amplified DNAs were calculated as follows. First, the amounts of the full-length amplified products for each sample (Nt[sample]), as double-stranded DNA, were determined from the FAM intensity. Next, the Cy5 intensity for each sample (ICy5[sample]) was normalized to the amounts of the DNA (ICy5[sample]/Nt[sample]). The background misincorporation ratio was then subtracted from the relative Cy5 intensity (ICy5[sample]/Nt[sample] − ICy5[Cont-temp]/Nt[Cont-temp]). Finally, each sample's misincorporation rate (M%) of the unnatural bases into the total amplified DNAs was calculated by the following formula: M(%) = (ICy5[sample]/Nt[sample] − ICy5[Cont-temp]/Nt[Cont-temp])/(ICy5[Ds-temp 1]/Nt[Ds-temp 1] − ICy5[Cont-temp]/Nt[Cont-temp]) × 100. We performed three independent experiments, and the obtained values were averaged. To calculate the misincorporation rates per bp per replication (F%), the misincorporation rates (M%) were plotted over the replication times [=log2 (Amplification fold)], and were fitted by linear regression with a high correlation coefficient (R > 0.99). The slope (S%) of the regression line corresponds to the averaged misincorporation rate per replication (%), and the misincorporation rate per bp per replication (F%) was calculated by the following formula: F = [1 − (1 − S/100)1/11] × 100.
RESULTS
PCR conditions for the Ds–Px pairing
We first examined the PCR conditions by using several DNA polymerases (DNA pols) and unnatural and natural base substrate combinations of dDsTP, NH2-hx-dPxTP (Figure 1) and natural base dNTPs at different concentrations. In addition, to determine the natural base sequence dependency around the unnatural base pair, we used four single-stranded Ds-containing DNA templates (55-mer, Ds-temps 1, 2, 3 and 4), as well as a control template (Cont-temp) comprising only the natural bases (Figure 2A). Our previous studies indicated that Ds-temp 1 containing 3′-TATDsCTA-5′, Ds-temp 2 containing 3′-GTTDsCCC-5′, and Ds-temp 3 containing 3′-GTGDsCAT-5′ are favorable templates, but Ds-temp 4 containing 3′-CAADsGTA-5′ (purine-Ds-purine) is an unfavorable one for efficient PCR amplification involving the Ds–Px pair by Deep Vent DNA polymerase (exo+) (32). In this study, we incorporated only one unnatural base into the DNA templates, to focus on the accurate characterization of the Ds–Px pairing. To reduce the chemical DNA synthesis error, we utilized 55-mer single-stranded DNA templates, and the total base lengths were extended by PCR using 40-mer and 24-mer PCR primers to facilitate DNA sequencing directly after PCR amplification, which converted the templates into double-stranded 75-mer DNA products. Throughout all of the experiments in the study, we employed a PCR cycle program involving 94°C for 30 s, 45°C for 30 s and 65°C for 4 min. After 15 cycles of PCR, the fold amplification of the PCR products (Supplementary Table S1) was determined by polyacrylamide gel electrophoresis (representative data are shown in Figure 2B–D). Besides the full-length PCR products, truncated products paused after the unnatural base incorporations were also observed on the gel. The unconsumed primers were partially degraded by the exonuclease activity of the polymerases during PCR.
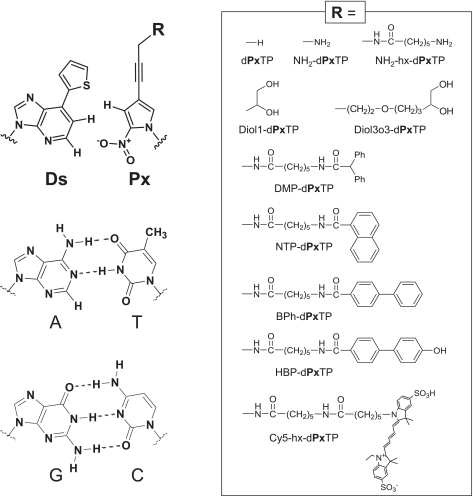
Figure 1.
Chemical structures of the unnatural Ds–Px pair and the natural A–T and G–C pairs. Functional groups (R), attached to the Px base via the propynyl linker, used in this study are summarized in the enclosed box.
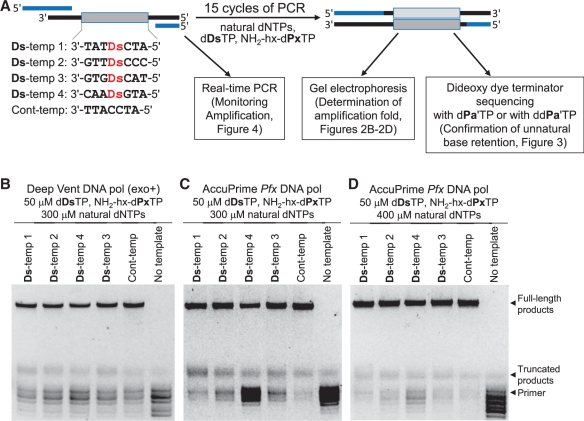
Figure 2.
PCR amplification involving the Ds–Px pairing with various DNA templates containing Ds for the determination of PCR conditions. (A) Scheme for PCR amplification experiments using 55-mer DNA fragments containing one Ds base, in the presence of dDsTP, NH2-hx-dPxTP and natural dNTPs. (B–D) Analyses by denaturing gel electrophoresis of PCR-amplified products after 15 cycles of PCR under different conditions by Deep Vent DNA pol (exo+) (B) and AccuPrime Pfx DNA pol (C,D), for the determination of the fold amplification at the end point.
The retention rates of the Ds–Px pair in the amplified DNA were determined by two sequencing methods of the Ds-containing DNA strands using conventional dideoxy dye-terminator sequencing, but either with a deoxyribonucleoside triphosphate of 4-propynylpyrrole-2-carbaldehyde (dPa′TP), another pairing partner of Ds, or with its dideoxyribonucleoside triphosphate (ddPa′TP) (30–32). Since no dye-terminator corresponding to the unnatural base is present in the sequencing, there is a gap at the unnatural base position on the sequencing pattern in the presence of dPa′TP. In another sequencing method in the presence of ddPa′TP, the sequencing reaction is stopped at the unnatural base position by incorporating ddPa′TP opposite Ds in DNA templates, and thus the following base peaks disappear. However, if some mutation from an unnatural to natural base occurs at the original unnatural base position during PCR, then the height of each following base peak increases, depending on the mutation rate in the DNA samples. The sequencing with dPa′TP is useful for confirming the unnatural base position, while that with ddPa′TP is appropriate for determining the unnatural base retention rate at the original position in the templates, by comparing the peak heights on the sequencing patterns with those obtained using standard authentic samples. We increased the determination accuracy of the retention rates by improving the peak height quantification (see Supplementary Figures S1 and S2).
Using the unnatural base substrate combination of dDsTP and NH2-hx-dPxTP, we screened 11 DNA polymerases, the Deep Vent (exo+), Deep Vent (exo−), AccuPrime Pfx, Pfx50, Phusion HF, Pfu, Pwo SY, Vent (exo+), N9° DNA pols from family B, and the TITANIUM Taq and Taq DNA polymerases from family A, by 15 cycles of PCR with 50 µM dDsTP and NH2-hx-dPxTP and 300 µM natural dNTPs. Representative gel and sequencing patterns of the PCR products are shown in Figures 2 and 3, respectively, and all of the data for the amplification folds and the retention rates of the Ds–Px pair in the amplified DNA products are summarized in Supplementary Table S1 and Supplementary Figures S3–S15. Although the sequencing patterns in the presence of dPa′TP for Ds-temp 4 did not give a clear gap corresponding to the unnatural base position (30), the retention rates of the Ds–Px pair in the PCR products were accurately determined from the sequencing patterns in the presence of ddPa′TP (Figure 3). In the PCR amplification of Ds-temps 1, 2 and 3 using the family B polymerases, most of the Ds–Px pair (>97%) was retained in the PCR products, although the fold amplification depended on the polymerase. In this analysis, >97% of the retention rate is the maximum detection limit. In the PCR amplification of the unfavorable Ds-temp 4, the retention rates of the Ds–Px pair and the amplification folds varied with the polymerase, but most of them were lower than those of Ds-temps 1, 2 and 3. Among them, the Deep Vent (exo+) (Figure 3A), AccuPrime Pfx (Figure 3B) and Pfx50 DNA pols (Figure 3C) exhibited high amplification folds and retention rates with all of the templates. In addition, increasing the concentration of the natural base substrates [dNTPs, 500 µM for Deep Vent (exo+) and 400 µM for AccuPrime Pfx] improved the amplification folds, especially those of Ds-temp 4, without a significant reduction in their retention rates. For example, in a 15-cycle PCR amplification by AccuPrime Pfx DNA pol with 50 µM dDsTP and NH2-hx-dPxTP and 400 µM natural dNTPs, the amplification folds were 1774 for Ds-temp 1 and 1503 for the unfavorable Ds-temp 4, and were only slightly lower than that (1955-fold) for Cont-temp. Under the same conditions, the retention rates of the Ds–Px pair in the amplified products were >97% for Ds-temp 1 and 93 ± 2% for Ds-temp 4. The dependency of the amplification folds on the natural base substrate concentrations suggests that the natural base incorporation after the unnatural base pairing is one of the rate limiting steps, and thus the amplification efficiency could be improved by increasing the natural base substrate concentrations. Figure 2C and D also support this suggestion, and the amounts of the truncated products were significantly reduced by increasing the concentrations of the natural base substrates.
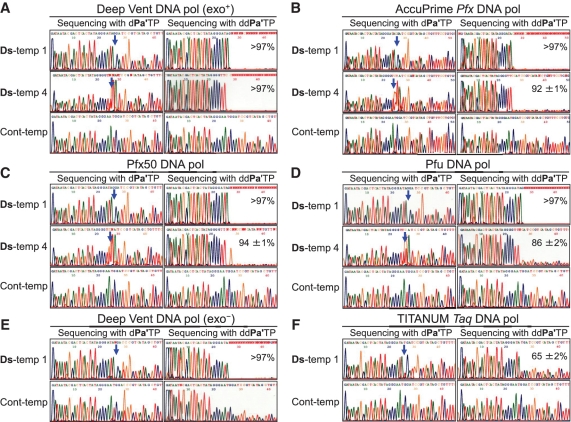
Figure 3.
Sequencing analysis of the PCR products after 15 cycles of PCR by each DNA polymerase: (A) Deep Vent DNA pol (exo+); (B) AccuPrime Pfx DNA pol; (C) Pfx50 DNA pol; (D) Pfu DNA pol; (E) Deep Vent DNA pol (exo−); (F) TITANIUM Taq DNA pol. Sequencing reactions were performed in the presence of dPa′TP (left panels) or ddPa′TP (right panels) in each figure. The blue arrow indicates the original unnatural base position. The percentages indicated in the right panels for Ds-temp 1 and Ds-temp 4 are the retention rates of the Ds–Px pair in the amplified products after 15 cycles of PCR. The details of the calculation method for the retention rates are provided in the Supplementary Data section.
Next, we examined the effect of the 3′ → 5′ exonuclease activity of the DNA polymerases on the unnatural base pair PCR amplification, using Deep Vent DNA pols with (exo+) and without the exonuclease activity (exo−). Deep Vent DNA pol (exo−) performed with relatively high efficiency (424-fold after 15 cycles of PCR) and selectivity (>97% retention rate of the unnatural base pair). However, the sequencing patterns in the presence of ddPa′TP of the PCR products using Ds-temp 1 and Cont-temp (Figure 3E) indicated a measurable problem with unnatural base misincorporation opposite the natural bases. As shown in the ddPa′TP sequencing using Ds-temp 1 and Cont-temp (Figure 3E), the peak heights corresponding to the natural bases gradually decreased, relative to those corresponding to the exonuclease-proficient pol (exo+) (Figure 3A). During the sequencing reaction, ddPa′TP may have been incorporated opposite multiple Ds positions, which resulted from the unnatural base misincorporations in place of the natural bases during PCR amplification, and the sequencing reaction was paused, depending on the unnatural base misincorporation rate by the ddPa′TP incorporation opposite Ds. Therefore, the 3′ → 5′ exonuclease activity of polymerases is important to remove the unnatural base substrates misincorporated opposite the natural bases during PCR.
We also examined PCR amplification using family A (Taq and TITANIUM Taq) DNA pols. The Taq and TITANIUM Taq DNA pols, which have intrinsically no 3′ → 5′ exonuclease activity, were ineffective, even in PCRs using Ds-temps 1 and 2 (Figure 3F and Supplementary Table S1). The selectivity of the Ds–Px pairing was much lower than that using the family B DNA pols. However, based on the sequencing pattern in the presence of ddPa′TP (Figure 3F), no gradual attenuation of each peak was observed, as compared to the pattern from the template obtained using Deep Vent DNA pol (exo−) (Figure 3E). Thus, the misincorporation rates of the unnatural base substrates opposite the natural bases are not very high when using Taq and TITANIUM Taq DNA pols. Indeed, our recent study showed that TITANIUM Taq DNA pol exhibited high selectivity and efficiency in a one-way incorporation of modified dPxTP opposite Ds in templates, and is useful for the unnatural base incorporation in a primer region in PCR (36). These results suggest that the TITANIUM Taq and Taq DNA pols efficiently incorporate dPxTP into DNA opposite Ds, but are not useful to incorporate dDsTP into DNA opposite Px.
Sequence dependence of the PCR amplification efficiency
We determined the exact amplification efficiency using the less effective template, Ds-temp 4, in comparison to those using Ds-temp 1 and Cont-temp, by real-time PCR. Four 10-fold dilutions (20 fM–20 pM) of each template were amplified by Deep Vent DNA pol (exo+) with 50 µM dDsTP and NH2-hx-dPxTP and 300 µM natural dNTPs, and the amplified products were monitored by the conventional SYBR Green dye detection system with a real-time PCR instrument (Figure 4A–C). Figure 4D shows the standard curves of the PCR amplification using each template, from which the amplification efficiencies (=[10(−1/slope)] − 1) of the PCRs using Ds-temp 1, Ds-temp 4 and Cont-temp [0.91, 0.80 and 1.04 (∼1.0), respectively] were determined. Although there are some differences among the template sequence contexts with or without the unnatural base, the values (0.80–0.91) of the amplification efficiency using the Ds-containing templates would be acceptable for the practical use of PCR involving the Ds–Px pair. Indeed, after 40 cycles of PCR starting from single-stranded DNA templates, the DNA fragments (as double strands) would theoretically be amplified 9.0 × 109-fold (=1.8039) for 0.80 and 9.1 × 1010-fold for 0.91, which coincide well with the experimental data, as discussed later (Table 1).
Figure 4.
Real-time quantitative PCR amplification plots of each DNA template [(A) Ds-temp 1, (B) Ds-temp 4, (C) Cont-temp] and their linear standard curve analysis (D). PCR was performed with Deep Vent DNA pol (exo+) in the presence of 50 µM dDsTP and NH2-hx-dPxTP, 300 µM natural dNTPs, and different concentrations of DNA templates (20 fM, 200 fM, 2 pM, 20 pM).
Table 1.
Amplification efficiencies and fidelities of the Ds–Px pairing in 40 cycles of PCR in different template sequence contexts, using dDsTP and NH2-hx-dPxTP
| DNA polymerase | DNA template | Natural substratesa (µM) | Unnatural substratesb (µM) | Amplification foldc | Amplification efficiency | Selectivityd (%/replication) | Misincorporation ratee (%/bp/replication) |
|---|---|---|---|---|---|---|---|
| Deep Vent (exo+) | Ds-temp 1 | 300 | 30 | 9.6 × 1010 | 0.91 | 99.92 | − |
| 40 | 9.0 × 1010 | 0.91 | 99.92 | − | |||
| 50 | 6.7 × 1010 | 0.89 | 99.92 | − | |||
| Ds-temp 3 | 30 | 6.6 × 1010 | 0.89 | 99.92 | − | ||
| 50 | 4.8 × 1010 | 0.88 | 99.91 | − | |||
| Ds-temp 4 | 30 | 1.4 × 1010 | 0.82 | 99.61 | − | ||
| 50 | 8.4 × 109 | 0.80 | 99.75 | − | |||
| Cont-temp | 30 | 5.7 × 1010 | 0.89 | − | 0.007 | ||
| 40 | 9.4 × 1010 | 0.91 | − | 0.010 | |||
| 50 | 7.2 × 1010 | 0.90 | − | 0.012 | |||
| AccuPrime Pfx | Ds-temp 1 | 400 | 30 | 7.7 × 1010 | 0.90 | 99.74 | − |
| 50 | 1.0 × 1011 | 0.91 | 99.88 | − | |||
| Ds-temp 3 | 30 | 1.0 × 1011 | 0.91 | 99.72 | − | ||
| 50 | 7.7 × 1010 | 0.90 | 99.89 | − | |||
| Ds-temp 4 | 30 | 1.7 × 1010 | 0.83 | 98.94 | − | ||
| 50 | 1.2 × 1010 | 0.81 | 99.22 | − | |||
| Cont-temp | 30 | 1.0 × 1011 | 0.91 | − | 0.011 | ||
| 40 | 1.4 × 1011 | 0.93 | − | 0.016 | |||
| 50 | 8.0 × 1010 | 0.90 | − | 0.023 | |||
| 600 | 50 | 1.0 × 1011 | 0.91 | − | 0.017 |
aConcentration of each natural base substrate (dNTPs), bConcentration of dDsTP and NH2-hx-dPxTP, cAmplification folds after 40 cycles of PCR, dSelectivity of the unnatural base pairing, eMisincorporation rate of the unnatural base substrates opposite the natural bases in templates.
We also found that the PCR amplification with Ds-temp 4 took a longer time to reach the exponential phase than those with Ds-temp 1 and Cont-temp (Figure 4). The y-intercept values in the standard curves were significantly different among the templates (41.73 using Ds-temp 1, 46.78 using Ds-temp 4 and 37.46 using Cont-temp) (Figure 4D). The Cont-temp value (37.46) is reasonable, relative to that in conventional real-time PCR detection, but the values (41.73 and 46.78) obtained using Ds-containing templates, especially Ds-temp 4, are relatively high. In general, the high y-intercept values result from the degradation of templates during PCR, but we confirmed the stability of the Ds-containing DNA templates under the PCR conditions. A plausible possibility is that the complex of the polymerase and the Ds-containing template, especially Ds-temp 4, may dissociate at low template concentrations. Therefore, increasing the initial concentration of the DNA template, as well as the substrate concentrations as mentioned earlier, might increase the fold amplification when using an unfavorable sequence context, such as Ds-temp 4.
100 cycles of PCR involving the Ds–Px pairing
Using the optimized PCR conditions, we performed 100 cycles of PCR amplification of Ds-temp 1 with dDsTP and NH2-hx-dPxTP by the Deep Vent (exo+) and AccuPrime Pfx DNA pols, to examine the abilities and limitations of the Ds–Px pair in PCR, as well as to increase the maximum detection limits (>97%) of the unnatural base pair retention rates in the amplified products. To maintain the exponential amplification through 100 cycles of PCR, we repeated the process of a 10-cycle PCR and the dilution of the PCR solution 10 times. Ds-temp 1 (50 fmol) was amplified by 10 cycles of PCR with 30 µM dDsTP and NH2-hx-dPxTP and 300 µM natural dNTPs with Deep Vent DNA pol (exo+) or with 50 µM dDsTP and NH2-hx-dPxTP and 400 µM natural dNTPs with AccuPrime Pfx DNA pol, and the PCR solution was diluted 750-fold for the following amplification (Figure 5A). From the gel electrophoresis of the products after 100 cycles of PCR (Figure 5C), Ds-temp 1 was amplified 5 × 1027-fold by Deep Vent DNA pol (exo+) and 2 × 1028-fold by AccuPrime Pfx DNA pol. The position of the Ds–Px pair in the amplified products was confirmed by sequencing in the presence of dPa′TP (left panels in Figure 5D and E). The retention rates of the Ds–Px pair in the amplified products were determined by sequencing in the presence of ddPa′TP (right panels in Figure 5D and E), and for the Ds–Px pair, >97% for Deep Vent DNA pol (exo+) and 88.4% for AccuPrime Pfx DNA pol were retained in the amplified DNA after 100 cycles of PCR. The selectivities of the Ds–Px pairing were determined from the amplification folds and the retention rates, and were as high as 99.97 and 99.87%/replication for Deep Vent DNA pol (exo+) and AccuPrime Pfx DNA pol, respectively.
Figure 5.
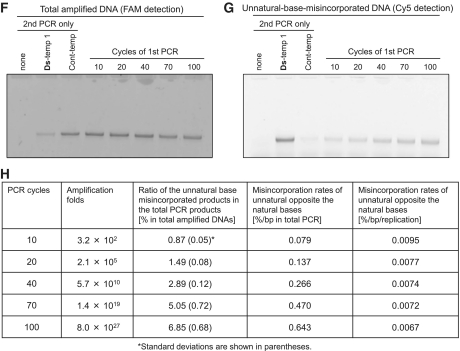
Amplification efficiency and fidelity assessments of the Ds–Px pairing by 40-cycle and 100-cycle PCR amplifications (repeated 10-cycle PCR). (A) Scheme of the 40- or 100-cycle PCR amplifications of the Ds-containing DNA templates using dDsTP and modified-dPxTP, for the determination of the fold amplification, efficiency and selectivity of the Ds–Px pairing in PCR. (B) Scheme of the 40- or 100-cycle PCR amplification of the DNA template comprising only the natural bases using dDsTP and modified-dPxTP, for the determination of the misincorporation rates of the unnatural base substrates opposite the natural bases. The results are summarized in Tables 1 and 2. (C) Gel electrophoresis of the amplification products after each 10-cycle PCR (total 100 cycles of PCR) of Ds-temp 1 by the Deep Vent DNA pol (exo+) using 1 µM each primer, 30 µM dDsTP and NH2-hx-dPxTP and 300 µM natural dNTPs. A portion of each 10-PCR solution before (lanes T) and after each 10-cycle PCR (lanes +) was analyzed by 15% polyacrylamide denaturing gel electrophoresis, followed by SYBR Green II staining. M: marker (75-mer single-stranded DNA). (D and E) Sequencing analysis of the PCR products after 10, 40, 70 and 100 cycles of PCR by Deep Vent DNA pol (exo+) (D) and AccuPrime Pfx DNA pol (E). Sequencing reactions were performed in the presence of dPa′TP (left panels) or ddPa′TP (right panels). The blue arrow indicates the original unnatural base position. The percentage indicated in the right panels is the retention rate of the Ds–Px pair in the amplified products. The method for the calculation of the retention rates is provided in the Supplementary Data section. (F–H) Determination of misincorporation rates of the unnatural base substrates opposite the natural bases in Cont-temp after 10, 20, 40, 70 and 100 cycles of PCR by the Deep Vent DNA pol (exo+) using 30 µM dDsTP and NH2-hx-dPxTP and 300 µM natural dNTPs (first PCR). The second PCR products with the FAM-labeled 3′-primer and Cy5-hx-dPxTP were analyzed by 15% denaturing PAGE, and the DNA fragments were detected with FAM fluorescence (F) for the quantification of the total PCR products and with Cy5 fluorescence (G) for the quantification of the unnatural base misincorporated products. (H) Misincorporation rates calculated from the data are summarized in the panel.
Another important consideration is the misincorporation rates of the unnatural base substrates opposite the natural bases in templates during PCR amplification. To determine the misincorporation rates, we developed another method (Figure 5B), in which the misincorporated PCR products were specifically labeled by a second PCR with fluorescent dye-linked dPxTP. The natural base template, Cont-temp, was amplified by 100 cycles of PCR in the presence of the examined unnatural base substrates, and the products were again amplified by 7 cycles of the second PCR with another combination of fluorescent dye-attached unnatural base substrates, Cy5-hx-dPxTP (36) (Figure 1) and dDsTP, using a fluorescein (FAM)-labeled primer. The amplified products were then analyzed by Cy5 and FAM double detection on the gel. The fluorescence of Cy5 represents the replacement of the misincorporated unnatural bases (see Figure 5G), and the fluorescence of FAM corresponds to the total amplified DNA fragments (see Figure 5F). During the 7-cycle PCR with dDsTP and Cy5-hx-dPxTP, these unnatural base substrates are also slightly misincorporated opposite the natural bases, and thus the initial DNA template was also amplified by the second PCR with dDsTP and Cy5-hx-dPxTP (see the Cont-temp lane in Figure 5G), to subtract the misincorporation rate of Cy5-hx-Px from those after the first and second PCR amplifications.
Figure 5F and G show the gel electrophoreses for determining the misincorporation rates of unnatural base substrates opposite the natural bases after 10-, 20-, 40-, 70- and 100-cycles of the first PCR using 30 µM dDsTP and NH2-hx-dPxTP and 300 µM natural dNTPs by Deep Vent DNA pol (exo+), and the data including the misincorporation rates (% in total amplified products, %/bp in total amplified products, %/bp/replication) are summarized in Figure 5H. After 100 cycles of PCR, Cont-temp was amplified 8 × 1027-fold. Although 6.85% of the amplified DNA fragments contained the unnatural base pairs, resulting from the unnatural base misincorporation, after 100 cycles of PCR, the misincorporation rate was only 0.0067%/bp/replication. This misincorporation rate corresponds to an error rate of 6.7 × 10−5 error/bp, which is very close to the intrinsic error rate among the natural bases by Deep Vent DNA pol (exo+) (∼2 × 10−5 error/bp, from the New England BioLabs information).
Fidelity of the Ds–Px pairing in 40 cycles of PCR
Next, we determined the fidelity of both the selectivity of the Ds–Px pairing and the misincorporation rate of the unnatural base substrates opposite the natural bases. We performed 40 cycles of PCR (10 cycles repeated four times) using various templates, Ds-temps 1, 3 and 4 and Cont-temp, with different concentrations of dDsTP, NH2-hx-dPxTP, the natural dNTPs, and the Deep Vent (exo+) and AccuPrime Pfx DNA pols. The amplification folds and efficiencies, the selectivities of the Ds–Px pairing, and the misincorporation rates under several PCR conditions are summarized in Table 1. When using 50 µM dDsTP and NH2-hx-dPxTP and 300 µM natural dNTPs, the PCR amplification folds of each template by Deep Vent DNA pol (exo+) were relatively high (6.7 × 1010-fold for Ds-temp 1, 4.8 × 1010-fold for Ds-temp 3 and 8.4 × 109-fold for Ds-temp 4), and from the amplification folds, the amplification efficiencies [amplification fold = (1 + amplification efficiency)n, where n = PCR cycles − 1 for single-stranded templates] were estimated as 0.89 for Ds-temp 1, 0.88 for Ds-temp 3 and 0.80 for Ds-temp 4. These efficiencies closely agreed with those (0.91 for Ds-temp 1 and 0.80 for Ds-temp 4) obtained by the real-time PCR experiments as mentioned earlier, confirming the accuracy of our determination methods. Furthermore, reducing the concentration of the unnatural base substrates, from 50 to 30 µM each, improved both the amplification folds and misincorporation rates without any significant loss of the Ds–Px pairing selectivity. The improved amplification efficiency might be a consequence of the reduced misincorporation rates opposite the natural bases. In PCR with 30 µM dDsTP, NH2-hx-dPxTP and 300 µM natural dNTPs for all template sequence contexts, the amplification efficiency, the selectivity and the misincorporation rate were 0.82–0.91 and 99.61–99.92%/replication, and 0.007%/bp/replication, respectively. The PCR amplification by the AccuPrime Pfx DNA pol was less selective, as compared to that by the Deep Vent DNA pol. However, it exhibited the highest amplification efficiency among the examined polymerases, and thus decreasing the number of PCR cycles could compensate for the selectivity loss when using the AccuPrime Pfx DNA pol.
Side-chain effects of the Px base on the Ds–Px pair fidelity
During the extensive experiments of PCR amplification using modified Px base substrates for generating new functional DNA molecules, we found that modifications of the Px base positively affected the fidelity of the Ds–Px pairing in PCR. One of the advantages of the Ds–Px pair is the capability of the modified Px base, as we used Cy5-hx-dPxTP to site-specifically incorporate it into DNA opposite Ds. A variety of functional groups can be attached to the Px base via the propynyl linker, and these modified dPxTPs are also site-specifically incorporated opposite Ds in replication. We synthesized several modified Px substrates by attaching amino (NH2-C1- and NH2-hx-dPxTPs), diol (Diol1- and Diol3o3-dPxTPs), and aromatic groups (DMP-, NTP-, BPh- and HBP-dPxTPs) with different linkers, as well as the non-modified dPxTP (Figure 1, see Supplementary Data section for the syntheses). The amplification efficiency and fidelity of the Ds–Px pairing using these modified dPxTP were assessed by 40 cycles of PCR with 50 µM dDsTP and modified dPxTP, 300 µM natural dNTPs, DNA templates (Ds-temps 1 and 4 and Cont-temp) and the Deep Vent DNA pol (exo+).
Table 2 shows the amplification fold and efficiency, the selectivity of the Ds–Px pairing, and the misincorporation rate for PCR amplification using each unnatural base pair combination of modified dPxTP and dDsTP. Most of the modified dPxTPs, except for the biphenyl modifications (BPh- and HBP-dPxTPs), exhibited relatively high efficiency (0.79–0.91, around 1010-fold amplification after 40 cycles of PCR) and high selectivity (>99.9%/replication) when using Ds-temp 1. In the PCR amplification of Ds-temp 4, the efficiency and selectivity with all of the modified dPxTPs were decreased but were still as high as 0.75–0.80 and 99.22–99.87%/replication, respectively.
Table 2.
Amplification efficiencies and fidelities of the Ds–Px pairing in 40 cycles of PCR in different template sequence contexts using several combinations of dDsTP and modified dPxTP by Deep Vent DNA pol (exo+)
| Modified dPxTP | DNA template | Amplification folda | Amplification efficiency | Selectivityb (%/replication) | Misincorporation ratec (%/bp/replication) |
|---|---|---|---|---|---|
| dPxTP | Ds-temp 1 | 4.3 × 1010 | 0.87 | 99.91 | − |
| Ds-temp 4 | 4.3 × 109 | 0.77 | 99.88 | − | |
| Cont-temp | 4.2 × 1010 | 0.87 | − | 0.019 | |
| NH2-C1-dPxTP | Ds-temp 1 | 3.4 × 1010 | 0.86 | 99.91 | − |
| Ds-temp 4 | 2.9 × 109 | 0.75 | 99.22 | − | |
| Cont-temp | 5.0 × 1010 | 0.88 | − | 0.006 | |
| NH2-hx-dPxTP | Ds-temp 1 | 6.7 × 1010 | 0.89 | 99.92 | − |
| Ds-temp 4 | 8.4 × 109 | 0.80 | 99.75 | − | |
| Cont-temp | 7.2 × 1010 | 0.90 | − | 0.012 | |
| Diol1-dPxTP | Ds-temp 1 | 6.2 × 1010 | 0.89 | 99.92 | − |
| Ds-temp 4 | 7.6 × 109 | 0.79 | 99.77 | − | |
| Cont-temp | 6.1 × 1010 | 0.89 | − | 0.005 | |
| Diol3o3-dPxTP | Ds-temp 1 | 9.0 × 1010 | 0.91 | 99.92 | − |
| Ds-temp 4 | 5.3 × 109 | 0.78 | 99.64 | − | |
| Cont-temp | 1.2 × 1011 | 0.92 | − | 0.003 | |
| DMP-hx-dPxTP | Ds-temp 1 | 1.7 × 1010 | 0.83 | 99.91 | − |
| Ds-temp 4 | 3.3 × 109 | 0.75 | 99.77 | − | |
| Cont-temp | 5.8 × 1010 | 0.89 | − | 0.011 | |
| NTP-hx-dPxTP | Ds-temp 1 | 6.9 × 109 | 0.79 | 99.91 | − |
| Ds-temp 4 | 3.3 × 109 | 0.75 | 99.87 | − | |
| Cont-temp | 4.4 × 1010 | 0.87 | − | 0.016 | |
| BPh-hx-dPxTP | Ds-temp 1 | 3.1 × 108 | 0.65 | 98.66 | − |
| Ds-temp 4 | 8.2 × 108 | 0.69 | 99.76 | − | |
| Cont-temp | 2.2 × 1010 | 0.84 | − | 0.011 | |
| HBP-hx-dPxTP | Ds-temp 1 | 5.5 × 108 | 0.68 | 98.97 | − |
| Ds-temp 4 | 1.2 × 109 | 0.71 | 99.83 | − | |
| Cont-temp | 5.2 × 1010 | 0.88 | − | 0.008 |
aAmplification folds after 40 cycles of PCR using 50 µM dDsTP, modified-dPxTP and 300 µM natural base dNTPs, bSelectivity of the unnatural base pairing, cMisincorporation rate of the unnatural base substrates opposite the natural bases in templates.
The biphenyl modifications (BPh- and HBP-dPxTPs) showed a unique sequence context dependency (Table 2). Although the amplification efficiency and selectivity were lower than those of the other modified Px substrates, these modifications worked better with Ds-temp 4 than Ds-temp 1 in PCR, even by the Deep Vent DNA pol (exo+). For example, in PCR using HBP-dPxTPs and dDsTP, the amplification efficiencies were 0.68 for Ds-temp 1 and 0.71 for Ds-temp 4, and the selectivities of the Ds–Px pairing were 98.97% for Ds-temp 1 and 99.83% for Ds-temp 4, with a relatively low misincorporation rate opposite the natural bases (0.008%/bp/replication).
We also found that Px modifications improved the fidelity of the Ds–Px pairing in PCR. As compared with the misincorporation rate using dPxTP (0.019%/bp/replication), all of the Px modifications reduced the misincorporation rates (0.003–0.016%/bp/replication). Among the modified dPxTPs, the selectivity of the Ds–Px pairing was less correlated with the misincorporation rate, and the diol-modified dPxTPs showed the lowest misincorporation rate (0.003–0.005%/bp/replication) even when using high concentrations (50 µM) of the unnatural base substrates, without any significant reductions in the amplification fold and the selectivity of the Ds–Px pairing. From the overall evaluation among these modified dPxTPs, the combination of Diol1-dPxTP and dDsTP was the best for PCR amplification, and exhibited the highest amplification efficiency (0.79–0.89) and selectivity (99.77–99.92%/replication) among all of the templates, and the lowest misincorporation rate (0.005%/bp/replication → 5 × 10−5 error/bp of error rate) opposite the natural base templates. These results suggest that the modification of unnatural bases protruding into the major groove side also affects the fidelity of the unnatural base pairing in replication.
DISCUSSION
We fine-turned the PCR conditions involving the Ds–Px pair as a third base pair and developed methods for assessing the amplification efficiency and fidelity of the unnatural base pairing. In the methods, we increased the accuracy of the determination of the PCR amplification efficiency and the fidelity of the Ds–Px pair by maintaining exponential amplification conditions, by repeating 10-cycle PCRs. The best combination for the Ds–Px pair PCR system is dDsTP and Diol1-dPxTP. Under the optimized conditions using 50 µM dDsTP and Diol1-dPxTP and 300 µM natural dNTPs by Deep Vent (exo+) pol, DNA fragments with different sequence contexts, except for purine-Ds-purine motifs, were amplified ∼1010-fold (amplification efficiency = 0.89) by 40 cycles of PCR and the selectivity of the cognate Ds–Px pairing was >99.9%/replication. Even in PCR with unfavorable sequence contexts containing purine-Ds-purine, the unnatural base pair selectivity was 99.77%/replication, with high amplification efficiency (0.79) similar to those of other sequence contexts. The diol modification of the Px base reduced the misincorporation rate of the unnatural base substrates opposite the natural bases in template, which was 0.005%/bp/replication. The low misincorporation rate of the Ds and Diol1-Px substrates (0.005%/bp/replication = 5 × 10−5 error/bp) is very close to the intrinsic mispairing error rate among the natural bases (∼2 × 10−5 error/bp) of Deep Vent DNA pol (exo+). Therefore, the ability of the unnatural base pairing is approaching those of the natural A–T and G–C pairings.
The Ds–Px pair in combination with dDsTP and NH2-hx-dPxTP also exhibited high efficiency and fidelity in PCR amplification using the Deep Vent (exo+) and AccuPrime Pfx DNA pols. The selectivities in PCR with Ds-temp 1 using 30 µM dDsTP and NH2-hx-dPxTP and 300 µM natural dNTPs were 99.92%/replication for Deep Vent (exo+) pol and 99.74% for AccuPrime Pfx DNA pol. Under the same conditions, the misincorporation rates were 0.007%/bp/replication for Deep Vent (exo+) pol and 0.011%/bp/replication for AccuPrime Pfx DNA pol. We demonstrated the great potential of the Ds–Px pair by performing 100 cycles of PCR using dDsTP and NH2-hx-dPxTP. Even in PCR using the unfavorable template sequence context by Deep Vent (exo+) pol, the amplification efficiency (0.82 for Ds-temp 4) and the selectivity (99.6%/replication for Ds-temp 4) adequately reached practical levels for PCR. Although the PCR amplification by the AccuPrime Pfx DNA pol was less selective than that by Deep Vent (exo+), the higher amplification efficiency is compensated by the shorter PCR cycles. In addition, the AccuPrime Pfx DNA pol might be useful for PCR amplification of DNA templates containing multiple unnatural bases. Furthermore, a variety of modified Px substrates can also be used for the Ds–Px pair PCR system, enabling the site-specific functionalization of DNA molecules.
The present analyses of PCR involving the Ds–Px pair suggest several points for the further development of unnatural base pair systems. Some of the points might also offer suggestions for the mechanisms of the natural base pairing in replication and the development of natural base pair PCR systems with high efficiency and fidelity. (i) The main role of the 3′ → 5′ exonuclease activity of Deep Vent DNA pol is to prevent the misincorporation of the unnatural base substrates opposite the natural bases. Thus, the polymerase proofreading mechanisms function for the recognition of non-cognate pairs between the natural and unnatural bases. In addition, mixed polymerases with and without exonuclease activity might improve the unnatural base pair efficiency and selectivity. Indeed, AccuPrime Pfx DNA pol contains two types of polymerases. (ii) One of the rate limiting steps is the natural base incorporation opposite its complementary base after the unnatural base pair, as well as the unnatural base pairing. Thus, increasing the concentration of the natural base substrates could improve the amplification efficiency without any loss of fidelity in PCR. (iii) The real-time PCR analyses suggested that the amplification fold is also affected by the initial template concentration, and a higher template concentration could increase the fold amplification with fewer PCR cycles. The less effective purine-Ds-purine sequences might reduce the stability of the polymerase and substrate complex, and thus increasing the concentration of each component is important for efficient PCR amplification for all template sequence contexts and multiple incorporations of the unnatural base pairs. (iv) Extra side chains attached to unnatural bases affect the fidelity of the unnatural base pairing. Among the side chain functional groups, some of them increase the unnatural base pair fidelity and others change the polymerase's preference for the sequence contexts.
Our studies of the Ds–Px pair in PCR have confirmed the utility of a third base pair as a replicative component with high efficiency and fidelity, besides the natural A–T and G–C base pairs, and further development of the unnatural base pair systems and applications to new biotechnologies are expected in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Table 1 and Supplementary Figures 1–15.
FUNDING
Funding for open access charge: Targeted Proteins Research Program and RIKEN Structural Genomics/Proteomics Initiative; National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan; and Grants-in-Aid for Scientific Research (KAKENHI 19201046 to I.H. and 20710176 to M.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Service RF. Molecular biology. Creation's seventh day. Science. 2000;289:232–235. doi: 10.1126/science.289.5477.232. [DOI] [PubMed] [Google Scholar]
- 2.Benner SA, Sismour AM. Synthetic biology. Nat. Rev. Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunziker J, Mathis G. DNA with artificial base pairs. Chimia. 2005;59:780–784. [Google Scholar]
- 4.Bergstrom DE. Unnatural nucleosides with unusual base pairing properties. Curr. Protoc. Nucleic Acid Chem. 2009;Chapter 1 doi: 10.1002/0471142700.nc0104s37. Unit 1.4. [DOI] [PubMed] [Google Scholar]
- 5.Krueger AT, Kool ET. Redesigning the architecture of the base pair: toward biochemical and biological function of new genetic sets. Chem. Biol. 2009;16:242–248. doi: 10.1016/j.chembiol.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kool ET. Synthetically modified DNAs as substrates for polymerases. Curr. Opin. Chem. Biol. 2000;4:602–608. doi: 10.1016/s1367-5931(00)00141-1. [DOI] [PubMed] [Google Scholar]
- 7.Henry AA, Romesberg FE. Beyond A, C, G and T: augmenting nature's alphabet. Curr. Opin. Chem. Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Hirao I. Unnatural base pair systems for DNA/RNA-based biotechnology. Curr. Opin. Chem. Biol. 2006;10:622–627. doi: 10.1016/j.cbpa.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 9.Kimoto M, Cox RS, III, Hirao I. Unnatural base pair systems for sensing and diagnostic applications. Expert Rev. Mol. Diagn. 2011;11:321–331. doi: 10.1586/erm.11.5. [DOI] [PubMed] [Google Scholar]
- 10.Tor Y, Dervan PB. Site-specific enzymatic incorporation of an unnatural base, N6-(6-aminohexyl)isoguanosine, into RNA. J. Am. Chem. Soc. 1993;115:4461–4467. [Google Scholar]
- 11.Hirao I. Placing extra components into RNA by specific transcription using unnatural base pair systems. Biotechniques. 2006;40:711–715. doi: 10.2144/000112187. [DOI] [PubMed] [Google Scholar]
- 12.Kimoto M, Endo M, Mitsui T, Okuni T, Hirao I, Yokoyama S. Site-specific incorporation of a photo-crosslinking component into RNA by T7 transcription mediated by unnatural base pairs. Chem. Biol. 2004;11:47–55. doi: 10.1016/j.chembiol.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 13.Kawai R, Kimoto M, Ikeda S, Mitsui T, Endo M, Yokoyama S, Hirao I. Site-specific fluorescent labeling of RNA molecules by specific transcription using unnatural base pairs. J. Am. Chem. Soc. 2005;127:17286–17295. doi: 10.1021/ja0542946. [DOI] [PubMed] [Google Scholar]
- 14.Moriyama K, Kimoto M, Mitsui T, Yokoyama S, Hirao I. Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. Nucleic Acids Res. 2005;33:e129. doi: 10.1093/nar/gni128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimoto M, Mitsui T, Harada Y, Sato A, Yokoyama S, Hirao I. Fluorescent probing for RNA molecules by an unnatural base-pair system. Nucleic Acids Res. 2007;35:5360–5369. doi: 10.1093/nar/gkm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hikida Y, Kimoto M, Yokoyama S, Hirao I. Site-specific fluorescent probing of RNA molecules by unnatural base-pair transcription for local structural conformation analysis. Nat. Protoc. 2010;5:1312–1323. doi: 10.1038/nprot.2010.77. [DOI] [PubMed] [Google Scholar]
- 17.Moser MJ, Marshall DJ, Grenier JK, Kieffer CD, Killeen AA, Ptacin JL, Richmond CS, Roesch EB, Scherrer CW, Sherrill CB, et al. Exploiting the enzymatic recognition of an unnatural base pair to develop a universal genetic analysis system. Clin. Chem. 2003;49:407–414. doi: 10.1373/49.3.407. [DOI] [PubMed] [Google Scholar]
- 18.Sherrill CB, Marshall DJ, Moser MJ, Larsen CA, Daudé-Snow L, Jurczyk S, Shapiro G, Prudent JR. Nucleic acid analysis using an expanded genetic alphabet to quench fluorescence. J. Am. Chem. Soc. 2004;126:4550–4556. doi: 10.1021/ja0315558. [DOI] [PubMed] [Google Scholar]
- 19.Prudent JR. Using expanded genetic alphabets to simplify high-throughput genetic testing. Expert Rev. Mol. Diagn. 2006;6:245–252. doi: 10.1586/14737159.6.2.245. [DOI] [PubMed] [Google Scholar]
- 20.Moser MJ, Ruckstuhl M, Larsen CA, Swearingen AJ, Kozlowski M, Bassit L, Sharma PL, Schinazi RF, Prudent JR. Quantifying mixed populations of drug-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 2005;49:3334–3340. doi: 10.1128/AAC.49.8.3334-3340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimoto M, Mitsui T, Yamashige R, Sato A, Yokoyama S, Hirao I. A new unnatural base pair system between fluorophore and quencher base analogues for nucleic acid-based imaging technology. J. Am. Chem. Soc. 2010;132:15418–15426. doi: 10.1021/ja1072383. [DOI] [PubMed] [Google Scholar]
- 22.Switzer C, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA. J. Am. Chem. Soc. 1989;111:8322–8323. [Google Scholar]
- 23.Moser MJ, Christensen DR, Norwood D, Prudent JR. Multiplexed detection of anthrax-related toxin genes. J. Mol. Diagn. 2006;8:89–96. doi: 10.2353/jmoldx.2006.050049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall DJ, Reisdorf E, Harms G, Beaty E, Moser MJ, Lee WM, Gern JE, Nolte FS, Shult P, Prudent JR. Evaluation of a multiplexed PCR assay for detection of respiratory viral pathogens in a public health laboratory setting. J. Clin. Microbiol. 2007;45:3875–3882. doi: 10.1128/JCM.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nolte FS, Marshall DJ, Rasberry C, Schievelbein S, Banks GG, Storch GA, Arens MQ, Buller RS, Prudent JR. MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J. Clin. Microbiol. 2007;45:2779–2786. doi: 10.1128/JCM.00669-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson SC, Sherrill CB, Marshall DJ, Moser MJ, Prudent JR. A third base pair for the polymerase chain reaction: inserting isoC and isoG. Nucleic Acids Res. 2004;32:1937–1941. doi: 10.1093/nar/gkh522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, Chen F, Chamberlin SG, Benner SA. Expanded genetic alphabets in the polymerase chain reaction. Angew. Chem. Int. Ed. Engl. 2010;49:177–180. doi: 10.1002/anie.200905173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA. Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res. 2007;35:4238–4249. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sismour AM, Lutz S, Park JH, Lutz MJ, Boyer PL, Hughes SH, Benner SA. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from Human Immunodeficiency Virus-1. Nucleic Acids Res. 2004;32:728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat. Methods. 2006;3:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 31.Hirao I, Mitsui T, Kimoto M, Yokoyama S. An efficient unnatural base pair for PCR amplification. J. Am. Chem. Soc. 2007;129:15549–15555. doi: 10.1021/ja073830m. [DOI] [PubMed] [Google Scholar]
- 32.Kimoto M, Kawai R, Mitsui T, Yokoyama S, Hirao I. An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res. 2009;37:e14. doi: 10.1093/nar/gkn956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malyshev DA, Seo YJ, Ordoukhanian P, Romesberg FE. PCR with an expanded genetic alphabet. J. Am. Chem. Soc. 2009;131:14620–14621. doi: 10.1021/ja906186f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malyshev DA, Pfaff DA, Ippoliti SI, Hwang GT, Dwyer TJ, Romesberg FE. Solution structure, mechanism of replication, and optimization of an unnatural base pair. Chemistry. 2010;16:12650–12659. doi: 10.1002/chem.201000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, Chen F, Alvarado JB, Benner SA. Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J. Am. Chem. Soc. 2011;133:15105–15112. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashige R, Kimoto M, Mitsui T, Yokoyama S, Hirao I. Monitoring the site-specific incorporation of dual fluorophore-quencher base analogues for target DNA detection by an unnatural base pair system. Org. Biomol. Chem. 2011;9:7504–7509. doi: 10.1039/c1ob06118f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.