Abstract
The biological role of human DNA polymerase θ (POLQ) is not yet clearly defined, but it has been proposed to participate in several cellular processes based on its translesion synthesis capabilities. POLQ is a low-fidelity polymerase capable of efficient bypass of blocking lesions such as abasic sites and thymine glycols as well as extension of mismatched primer termini. Here, we show that POLQ possesses a DNA polymerase activity that appears to be template independent and allows efficient extension of single-stranded DNA as well as duplex DNA with either protruding or multiply mismatched 3′-OH termini. We hypothesize that this DNA synthesis activity is related to the proposed role for POLQ in the repair or tolerance of double-strand breaks.
INTRODUCTION
Human DNA polymerase θ (POLQ) has homologs throughout the higher eukaryotes but the in vivo role of POLQ is not clearly understood at this point (see ‘Discussion’ section below). The enzyme has a domain arrangement similar to the Drosophila melanogaster mus308 gene product and consists of an N-terminal helicase-like domain, a central spacer domain and a C-terminal polymerase domain (1). The N-terminus of the protein is assigned as a helicase domain based on sequence similarities to other known helicases although no helicase activity has yet been demonstrated for POLQ. The central domain has no known function and sequence predictions show no similarities to other proteins. The C-terminal portion of the enzyme is classified as an A-Family DNA polymerase based on its primary amino acid sequence (1).
Unlike most other A-Family DNA polymerases, POLQ is a low-fidelity enzyme with an error rate on par with that of the Y-Family polymerases κ and η (2,3) and is efficient at extending mismatched primer termini (4). POLQ is also very efficient at bypassing lesions such as abasic sites and thymine glycols, which are otherwise strong blocks to most other A-Family as well as B-Family replicative DNA polymerases (5,6). In addition, a domain within the polymerase has been shown to confer deoxyribophosphodiesterase (dRPase) activity to POLQ (7) and the enzyme has also been shown to exhibit ATPase activity in the presence of single-stranded DNA (1). The unique lesion bypass capacity of the enzyme has been proposed to arise from three insertion loops that are not present in other A-family DNA polymerases. Experiments with mutant forms of the polymerase in which each of the three loops had been deleted suggested that one of these loops acts as a processivity factor and the other two increase the efficiency of polymerization on undamaged templates and are required for translesion synthesis past an abasic site or a thymine glycol lesion (5). POLQ is highly efficient at incorporating nucleotides opposite abasic sites and then extending past the lesion. Under steady-state conditions, the efficiency of extension past an abasic site by the full-length enzyme was shown to be comparable with extension past a standard Watson–Crick base pair (6). The translesion synthesis capability of POLQ has been suggested to play a critical role in somatic hypermutation in immunoglobulin genes (3,8–12). However, there is conflicting evidence suggesting that such a function may be tissue specific or be only a minor part of POLQ's role in the cell (13). The dRP-lyase activity of POLQ and the observation that knockout of POLQ along with POLβ results in a deficiency in base excision repair in DT40 cells (14) suggest that POLQ may be involved in base excision repair in a manner similar to Pol λ, a polymerase that also possesses a dRPase domain (15) and is able to extend past abasic sites (16). Accumulating evidence suggests a role for POLQ in the repair or tolerance of double-strand breaks. POLQ deficient mouse bone marrow cells (17) and human tumor cells (18) show increased sensitivity to ionizing radiation and to low doses of bleomycin (17), both of which are known to produce double-strand breaks. Knockdown of POLQ in CH12 mouse B lymphoma cells increases their sensitivity to the double-strand break inducer etoposide (19) and chaos-1 mice [these mice have a serine to proline mutation at position 1932 in POLQ (20)] show high levels of micronuclei, indicating increased levels of chromosome breaks (21), both of which suggest a role for POLQ in the repair of double-strand breaks in mammalian cells. POLQ knockout mice also show a decreased viability when combined with a knockout in the ataxia telangiectasia mutated (ATM) gene (21). ATM is a crucial part of the double-strand break repair pathway in mammalian cells (22) providing further evidence that POLQ is involved in double-strand break repair. POLQ orthologs have also been suggested to be involved in interstrand cross-link repair, presumably through double-strand break intermediates (23). However, even in the case of double-strand breaks, it has been the translesion synthesis capacity of POLQ that is invoked as playing a role in the repair pathways of these devastating DNA lesions.
We show here that POLQ has the capacity to extend single-stranded oligonucleotides. The extension appears to be, in some cases, template independent, and there is a possibility that POLQ can utilize base pairing of only one to two nucleotides as well as multiply mismatched 3′ primer termini in a template-dependent reaction. Interestingly, in certain sequence contexts, POLQ extends a single-stranded oligonucleotide more efficiently than duplex DNA or duplex DNA with an abasic site in the template. Control experiments show that this same single-stranded oligonucleotide cannot serve as a substrate for other A- or B-family DNA polymerases. We hypothesize that this promiscuous DNA synthesis by POLQ is beneficial during double-strand break repair.
METHODS
Materials
Truncated versions of human POLQ enzymes (consisting of residues 1792–2590) were expressed and purified as previously described (5). A coomassie blue stained gel of the purified polymerases is available as Supplementary Figure S1. The DNA polymerase from bacteriophage RB69 was expressed and purified as previously described (24). Bacteriophage T7 DNA polymerase (both wild type and exonuclease deficient) was from GE Lifesciences while Vent DNA polymerase and the Klenow fragment of DNA polymerase I were from New England Biolabs. All DNA substrates were from Midland Certified Reagents or MWG Biofins and were purified by electrophoresis through 16% polyacrylamide gels followed by desalting on Sep Pak C18 cartridges (Waters Inc.). Duplex oligonucleotides were annealed by heating to 80°C and slow cooling in a buffer of 50 mM NaCl and 25 mM Tris–HCl (pH 7.5). Duplex oligonucleotides were subsequently separated from unannealed single stranded oligonucleotides by electrophoresis through a 16% nondenaturing polyacrylamide gel followed by elution of the duplexes in 50 mM NaCl and 20 mM Tris–HCl (pH 7.5). The sequences of all oligonucleotides are given in the figures. All primers were labeled at the 5′ end with tetrachlorofluorescein for subsequent visualization and quantification on a Typhoon 9400 Variable Mode Imager (GE Lifesciences). Ultrapure deoxyribonucleoside triphosphates were from GE Lifesciences.
Primer extension assays
100 nM polymerase was mixed with 250 nM DNA substrate in the reaction buffer [20 mM Tris–HCl (pH 8.8), 4% glycerol, 80 µg ml−1 bovine serum albumin and 0.1 mM EDTA]. Individual deoxyribonucleoside triphosphates (150 µM), or a mixture of all four at 150 µM each, were mixed with 10 mM MgCl2 in the same reaction buffer. For reactions using ribonucleotides, each base was present at a concentration of 1 mM. Reactions were initiated by mixing 50 µl of the polymerase:DNA solution with 50 µl of the dNTP:MgCl2 solution. At 0 s, 10 s, 30 s, 1 min, 5 min, 10 min and 30 min (unless otherwise noted), the reactions were quenched by mixing 5 µl aliquots in 15 µl of a stop solution consisting of 95% formamide, 20 mM EDTA and 0.1% xylene cyanol. Reaction products were electrophoresed on 16% denaturing polyacrylamide gels and then scanned on a Typhoon 9400 Variable Mode Imager at the Alexa 532 setting to excite the tetrachlorofluorescein tags at the 5′ end of the primer oligonucleotides.
Steady state kinetics
Reactions were performed as described for the primer extension assays except that 10 nM polymerase was used and individual deoxyribonucleoside triphosphates were used at varying concentrations ranging from 0.01 to 3 mM. The band intensities of extended primers and unextended primers were quantified using the ImageQuant 5.2 software provided with the Typhoon 9400. Time points were chosen in which primer extension was <20% and the ratio of extended primers to unextended primers was plotted against time to give a linear plot of product formation. The initial velocities of the reactions were calculated as the slopes of the lines for each concentration of dNTP and these were then plotted against dNTP concentrations and fit to a square hyperbola (V = (Vmax[dNTP])/(Km + [dNTP])).
RESULTS
Extension of single-stranded oligonucleotides
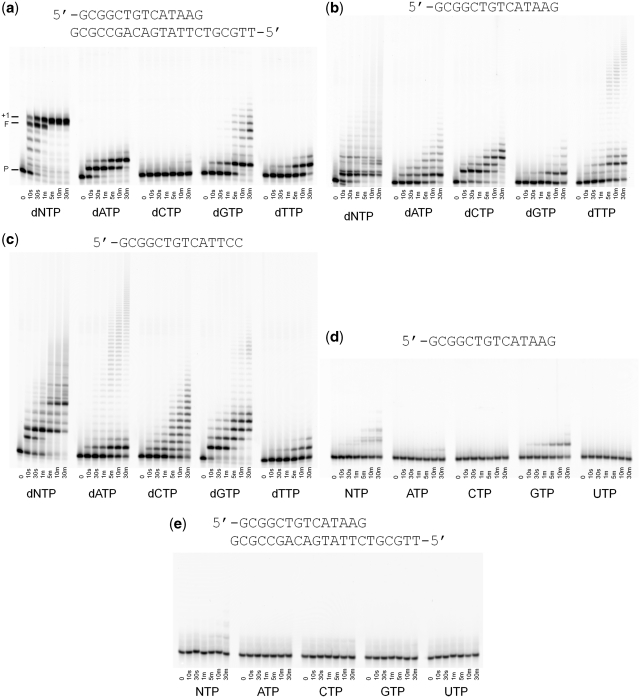
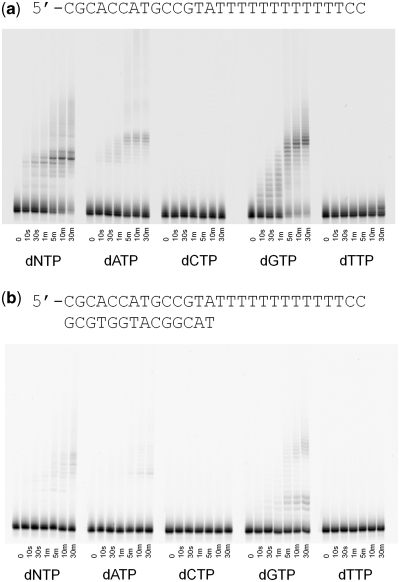
Previous work has shown that the full-length POLQ and the isolated polymerase domain have comparable properties when replicating undamaged and damaged DNA templates. The truncated version of POLQ used in this study (residues 1792–2590) retains the intrinsic translesion properties of the full-length enzyme (5) as well as the template-dependent DNA polymerase activity (Figure 1a). With primer/template DNA (Figure 1a), POLQ extends the primer to the end of the template and then adds a single nontemplated nucleotide to the end of the duplex as has been reported previously (6). By serendipity, we discovered that POLQ also was able to extend an end-labeled oligonucleotide with deoxyribonucleotides (Figure 1b) and essentially not with ribonucleotides (Figure 1d and e). Upon addition of all four dNTPs, a strong pause site was observed after addition of a single nucleotide followed by less intense bands at what appear to be the second and third positions (Figure 1b). We also observed a smearing that became progressively longer at longer time points. We attribute this smearing to be the result of random incorporation of nucleotides at the end of the primer strand. This hypothesis is supported by the fact that all four individual nucleotides are incorporated onto the 3′ end of the single stranded primer; even dCTP is readily incorporated even though no appreciable amount of dCTP was incorporated when a DNA template was present (compare Figure 1b with Figure 1a). When only dTTP was added to the reaction mix, most of the primers were extended by two or three nucleotides but a small population of primers continued to be extended out to about 30 nt after 30 min incubation. The extension profile of POLQ with single-stranded DNA appears to be dependent on the sequence at the 3′ end of the single stranded primer. When a primer strand ending with pyrimidines was used (Figure 1c), we observed a different pattern of pause sites when all four dNTPs were added. With this primer, there was very little extension with dTTP, unlike when the primer ended with purines (Figure 1b), and we instead saw a subset of the primers being extended out to about 30 nt after 30 min incubation with dATP.
Figure 1.
Primer extension assays. (a) Extension of properly paired duplex DNA. P denotes the original primer, F denotes full extension to the end of the template and +1 denotes the non-templated addition of a single nucleotide. Extension of single-stranded DNA with (b) purines at the 3′-end of the oligonucleotide, (c) pyrimidines at the 3′-end, (d) purines at the 3′-end of the oligonucleotide with 1 mM ribonucleotides. (e) Extension of duplex DNA with 1 mM ribonucleotides.
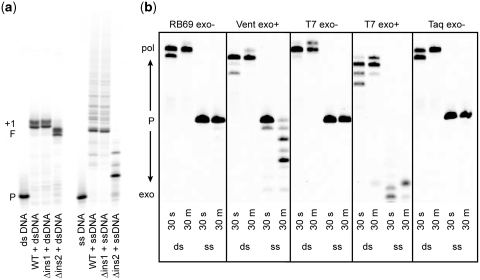
The apparent ability of POLQ to extend single-stranded DNA was quite unexpected considering that POLQ has been classified as a template-dependent, A-family DNA polymerase. Thus, we designed control experiments to validate our findings. The first was to confirm that the observed activity was a result of POLQ itself and not contaminating proteins. Two POLQ mutants in which protein loops have been deleted (5), and that have been purified identically to the wild type enzyme, were tested for their ability to extend single-stranded DNA (Figure 2a). The POLQ mutant in which loop 1 (residues 2149–2170) had been deleted was able to extend duplex DNA as well as single-stranded DNA. The mutant in which loop 2 (22,64–231,5) had been deleted was able to extend duplex DNA but was unable to efficiently extend single-stranded DNA beyond more than two nucleotides. This is in keeping with our previous work showing that loop 2 is required for translesion synthesis by POLQ (5) and demonstrates that the single-stranded DNA extension is intrinsic to POLQ. We next tested several other replicative DNA polymerases to see if the single-stranded substrate could be utilized by other polymerases that are known to be unable to synthesize DNA without a template strand. All of the polymerases tested were able to extend duplex DNA but showed no extension of the single-stranded primer and those with exonuclease activity rapidly degraded the single-stranded substrate (Figure 2b). This result suggests that POLQ is extending single-stranded DNA and that the substrate is not forming stable hairpins or self-complimentary structures that would provide a substrate for template-dependent DNA synthesis. If self-complementary structures are being formed, our oligonucleotide sequences dictate that they would only consist of 1 or 2 bp and extension of such primer ends would still in itself be an interesting property of POLQ. The third test was to examine the efficiency of single-stranded DNA as a substrate by POLQ. We measured the steady-state kinetic parameters of incorporation with three substrates; incorporation of dA opposite a templating T (sequence as in Figure 1a), incorporation of dA opposite a templating abasic site (sequence as in Figure 1a but in which the templating T has been replaced by the abasic site analog tetrahydrofuran) and incorporation of dG onto single-stranded DNA (sequence as in Figure 1c). The results of these experiments show a clear preference for extension of single-stranded DNA (Table 1). In the absence of a template strand, the maximum rate of product formation is ∼3-fold and 5-fold faster than for incorporation opposite a templating T and a templating abasic site, respectively. The apparent Km for the incoming nucleotide is ∼7-fold lower for non-templated primer extension compared with extension opposite a templating T and ∼20-fold lower than for incorporation opposite a templating abasic site. Thus, under our experimental conditions and in certain sequence contexts, the efficiency of nucleotide incorporation (Vmax/Km) in the absence of a specific templating strand appears to be about 20-fold greater than for incorporation opposite undamaged DNA and ∼100-fold greater than for incorporation opposite an abasic site.
Figure 2.
Control experiments. (a) Deletions of loop 1 (residues 2149–2170; Δins1) and loop 2 (residues 2264–2315; Δins2) were tested with double-stranded (ds) or single-stranded (ss) DNA and compared with the wild type polymerase (WT). The DNA templates were those used in Figure 1a and c, respectively, and all four nucleotides were provided. P indicates the original primer, F indicates extension to the end of the template with the duplex DNA and +1 indicates the single non-templated extension past the end of the template with duplex DNA. Reactions were quenched after 30 min. (b) Primer extension by other polymerases. Duplex DNA (ds) is the sequence in Figure 1a and single-stranded DNA (ss) is the sequence in Figure 1c. All four nucleotides were provided and time points were taken at 30 s and 30 min.
Table 1.
Steady kinetics parameters for primer extension by POLQ
| Incoming nucleotide: template | Vmax (nM s−1) | Km (μM) | Vmax/Km (nM s−1 μM−1) |
|---|---|---|---|
| dATP : T | 1.095 ± 0.09 | 110 ± 30 | 0.00995 |
| dATP : abasic site | 0.622 ± 0.07 | 394 ± 146 | 0.00157 |
| dGTP : no template | 3.08 ± 0.18 | 16 ± 5 | 0.1925 |
Extension of multiply mismatched termini
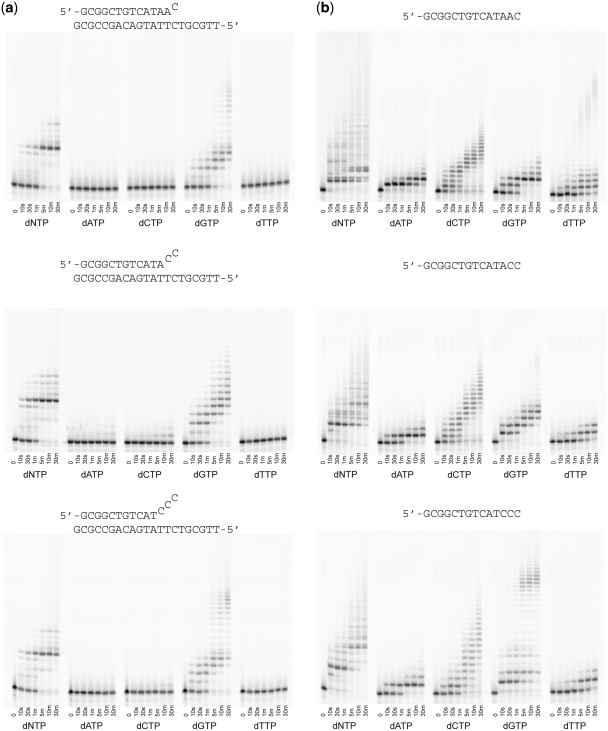
We next hypothesized that the ability of POLQ to synthesize DNA in what might be a nontemplated manner may allow the enzyme to extend multiply mismatched primer termini. POLQ has been previously shown to extend singly mismatched primer termini (4), but our working hypothesis was that as mismatches became longer in length, POLQ may extend them in a nontemplated manner. Figure 3a shows the results of incubating POLQ with oligonucleotide substrates containing one, two or three mismatched nucleotides at the primer terminus. These substrates had previously been used with the B-family replicative DNA polymerase from bacteriophage RB69 (25) and could not be extended in the absence of exonucleolytic proofreading activity that would remove the mismatches before template-dependent extension. However, POLQ shows no discrimination between the three lengths of mismatched primer termini as all three appear to be extended with equal efficiency. When all four dNTPs are present, POLQ rapidly extends the primer by what appears to be about 6 or 7 nt, which is the length one would expect if the polymerase were reading the template DNA, but at longer incubation times POLQ adds four more bases to the primer end. The same pattern of incorporation is seen for all three mismatch lengths used. Surprisingly, when dNTPs are provided individually, only dGTP is readily incorporated. Even dATP, which is the next correct nucleotide based on the sequence context, is not added to the primer end by POLQ in any appreciable amount. Thus, we observe a significant difference between POLQ incorporation when the primer terminus is correctly or incorrectly matched. With the matched primer (Figure 1a), POLQ was able to incorporate the correct base dATP as well as form mismatches with dGTP and dTTP. When the primer terminus consisted of mismatched bases, only dGTP was readily incorporated. We note that the band pattern between adding all four dNTPs and just dGTP are not the same with the mismatched primers. This suggests that after incorporation of one or more guanines, POLQ is able to incorporate other nucleotides but this has not yet been directly tested.
Figure 3.
Primer extension assays with mismatched primer termini. (a) Extension past single, double and triple mismatches. (b) Extension of the single-stranded primers as used in the mismatched primer experiments. The sequences are shown above each respective gel.
When extending a primer in a template-dependent manner, POLQ reaches the end of the template and then adds a single non-templated nucleotide as has been observed for many other polymerases (26) and no further extension is observed (Figure 1a). However, when a mismatch is extended, products longer than a single insertion past the end of the template are observed (Figure 3a). Extensions past these mismatches likely do not involve slippage events as such events have not previously been observed for translesion synthesis or mismatch extension by POLQ (4,6). While the predominant extension product of these mismatches appears to correspond with the length of single-stranded template, POLQ does not appear to be reading the sequence of nucleotides present in the template strand. With a single mismatch, incorporation of A opposite the next templating T is only barely detectable suggesting that POLQ is unable to recognize the sequence of the template strand when extending past a mismatched primer terminus. When full-length POLQ was previously tested against three different mismatches, extension appears to proceed to the end of the template but the band patterns are distinctly different depending on the nature of the mismatch [see Figure 1b in ref. (4)]. These results support our hypothesis that POLQ may not be directly reading the template strand when extending past a mismatch. Surprisingly, when only dGTP is supplied, POLQ readily incorporates up to 20 nt within the time frame of the experiment, which is well beyond the end of the template strand. Thus, POLQ appears to recognize the template strand when the possibility of making base pairs is present such as is the case when all four dNTPs are available but can switch to what appears to be nontemplated DNA synthesis when only one nucleotide, in this case dGTP, is available. Such an incorporation of runs of G was also observed with normal template-dependent synthesis in which dGTP appears to be incorporated without regard to the sequence of the template (Figure 1a).
We also tested each of the mismatch primers as single-stranded substrates in the absence of their complementary template strand (Figure 3b). When no template is provided, the same primer sequences that were used to form mismatched primer termini can now be extended by all four nucleotides. This suggests that the template strand in the previous experiment (Figure 3a) still has an effect on POLQ DNA synthesis even if it is not being directly read. These results also suggest that very subtle changes in the starting sequence can yield significant differences as to which nucleotides are added to single-stranded oligonucleotides. As shown in Figure 3b, when the sequence of the primer terminated in 5′-TAAC, the pattern of bands observed was similar to those seen when the primer terminated in 5′-TAAG (Figure 1b), including extension with dTTP out to a few dozen bases. When the number of terminal dC residues increased to two or three, the band pattern observed upon addition of all four dNTPs changed and long-range incorporation of dTTP was replaced by long-range extension with dGTP.
Extension of 3′ single-stranded overhangs
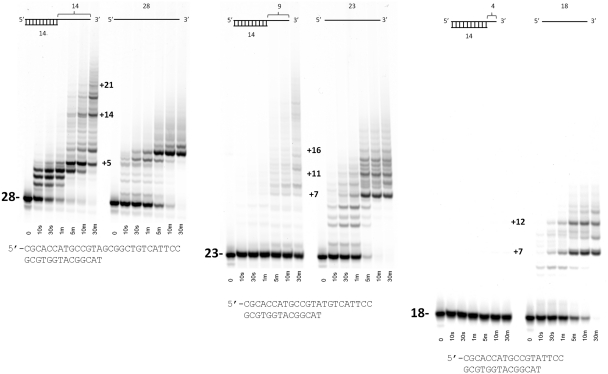
We next designed three sets of primer/template oligonucleotides with different lengths of 3′ single-stranded overhangs to mimic double-strand breaks and to determine if there was a minimum length of single-stranded DNA that could be extended by POLQ. All three sets of oligonucleotides had a 14-mer template annealed to a 28-mer primer (14 nucleotide 3′ overhang), a 23-mer primer (nine nucleotide 3′ overhang) or an 18-mer primer (four nucleotide 3′ overhang). All three overhung primers were extended with dNTPs and compared with the same reactions in which only the single-stranded primers were the substrate (Figure 4). These results show a clear preference for the length of the 3′-single-stranded overhang. When the overhang was 14 nt, the overhang was readily extended by POLQ. However, for the 9 nt overhang, only a small fraction was extended when compared with the same primer without double-stranded DNA. The shortest 3′ overhang of only 4 nt showed no detectable amount of non-templated extension while the primer alone was readily extended. Thus, the minimum length of single-stranded 3′-overhang DNA that is extendable by POLQ appears to be between 9 and 14 nt. We also note that the pattern of bands formed between the 14-mer 3′ overhang and the identical primer without a template are not the same. This suggests that even at 14 nt from the 3′-end of the DNA POLQ is still able to detect the presence of a duplex DNA. Interestingly, the overall length of non-templated DNA polymerization appears to be greater when the template strand is present as opposed to when it is just the 28-mer primer. Thus, there may even be a slightly stimulatory effect of the template strand on POLQ's non-templated extension of 3′-single stranded overhangs.
Figure 4.
Primer extension assays with duplex DNA with varying length of 3′ single-stranded overhangs. The DNA sequences are shown beneath each set of lanes. For each overhang length, the first set of lanes is the duplex DNA substrate and the second set is the single-stranded extension of the primer used without the 14-mer template. In each case, all four dNTPs were supplied. The approximate numbers of nucleotide incorporations for several of the more prominent bands are shown.
To ask if the sequence context of the 14-nt 3′-overhang could influence the polymerase activity, we modified the substrate and replaced all nucleotides except the last two Cs with Ts. This created an overhang with a homonucleotide run of 12 Ts ending with two Cs. This 3′-overhang was extended, albeit with lower efficiency when present in duplex DNA. The percentage of primers extended after 30 min were 77, 15 and 81% for incorporation of dNTPs, dATP and dGTP, respectively, with the single-stranded oligonucleotide (Figure 5a) compared with 14, 5 and 28% with the duplex oligonucleotides (Figure 5b). The sequence of this oligonucleotide is such that there is no opportunity to form two or more consecutive base pairs if the oligonucleotide were to self-anneal or form hairpins. In both cases, dGTP was preferentially incorporated over dATP.
Figure 5.
Primer extension assay with duplex DNA with a 3′ single-stranded overhang containing a homopolymeric run of thymines. (a) Extension of the primer alone. (b) Extension of the primer annealed to the template.
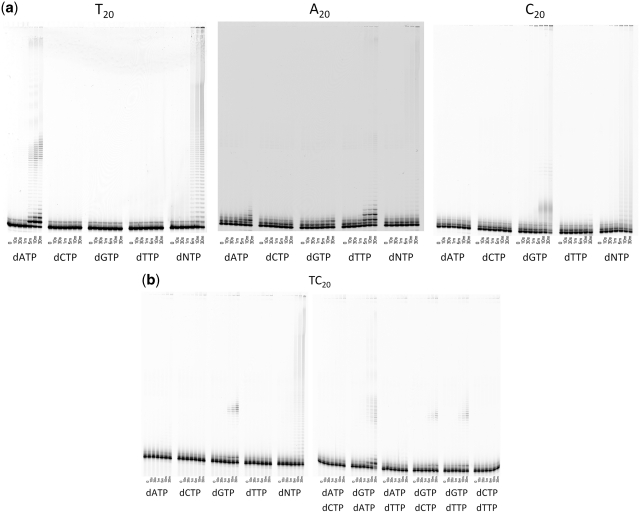
Extension with homopolymeric oligonucleotides
In an attempt to eliminate all possibility of self-annealing primers, we tested the ability of POLQ to extend homopolymeric runs of T, A or C (Figure 6a) and polyTC (Figure 6b). The reaction efficiency was greatly reduced with these substrates as observable levels of product did not accumulate until several minutes. Interestingly, the predominantly incorporated nucleotide was the complimentary nucleotide although detectable levels of dATP were incorporated onto the polyA substrate. With the polyTC substrate (Figure 6b), primer extension with a single nucleotide was only observed with dGTP or dGTP paired with any of the other three dNTPs. No extension was observed when dATP was provided as the only complimentary nucleotide. Interestingly, for all four homopolymeric substrates tried, the addition of all four nucleotides at the same time resulted in extremely long extensions of at least a few hundred nucleotides. These extensions also appear as ladders, not the smears observed with single-stranded primers composed of all 4 nt, and are longer than when the complimentary nucleotide is provided on its own.
Figure 6.
Primer extension assay with homopolymeric substrates. (a) Extension of polyT, polyA and polyC oligonucleotides. (b) Extension of polyTC; where two dNTPs are shown, these were added in equimolar amounts together in the reaction mix.
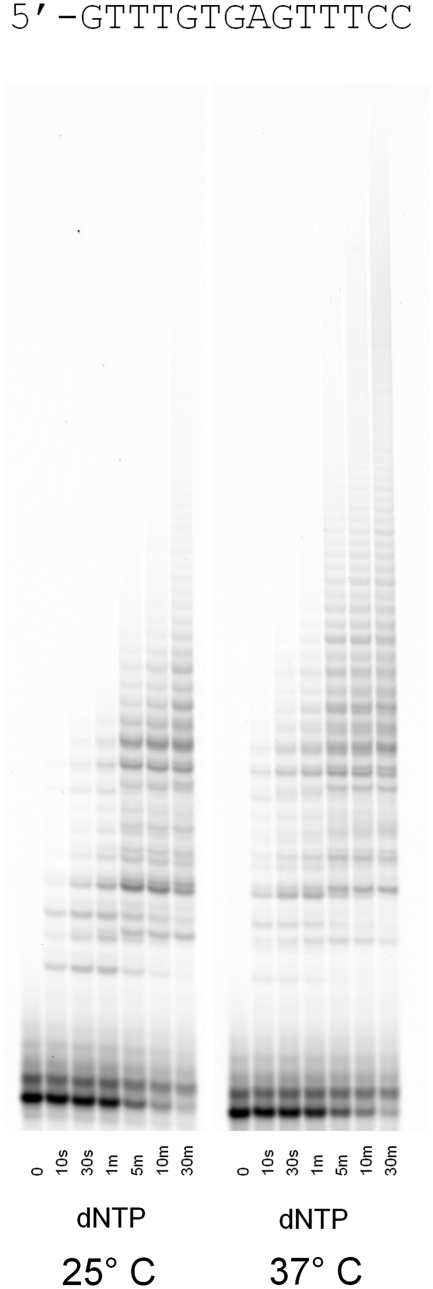
The experiments to this point suggested that POLQ may catalyze both template-independent and template-dependent DNA synthesis. Based on the pause sites in Figures 1, 4 and 5, there was a hypothetical chance that 2 nt of base pairing was sufficient to allow template-independent DNA synthesis by POLQ. To address whether there could be exclusive template-independent DNA synthesis in the presence of all four dNTPs, we designed an oligonucleotide that should not be able to form hairpins or allow more than 1 nucleotide to form a base pair. We found rapid and efficient extension at 25°C with incorporation of up to 6 nt within 10 s (Figure 7). Our hypothesis was that increasing the temperature to 37°C should destabilize any weak base pairing and thus inhibit template-dependent DNA synthesis. Instead, we found that POLQ was stimulated at 37°C, incorporating up to 12 nt within 10 s. This experiment, together with previous observations, strongly suggests that POLQ has the capacity to perform both template-independent and template-dependent DNA synthesis depending on the substrate presented to the enzyme.
Figure 7.
Primer extension assay with a single-stranded oligonucleotide designed to be unable to form any self-complimentary base pairs longer than a single nucleotide. The experiment was conducted at both 25 and 37°C and all four dNTPs were provided.
DISCUSSION
Mammalian cells have at least 15 different DNA polymerases that participate in DNA replication and repair. Most of the DNA polymerases are template-dependent DNA polymerases although many possess a transferase activity that allows the addition of one nontemplated nucleotide past the end of the DNA template. Other enzymes such as poly(A)polymerases and CCA-adding enzymes have the ability to extend RNA with multiple ribonucleotides in a template-independent reaction. However, these enzymes utilize specific amino acids in the active site to stabilize the incoming nucleotide by mimicking base pairing and thus selectively restrict which nucleotide can be added at the 3′-end of the substrate (27). Among the DNA polymerases, exceptions to template-dependent DNA synthesis are found in two X-family DNA polymerases, terminal deoxynucleotidyl transferase (TdT), which is completely template independent, and DNA polymerase μ (Pol μ), which shows a mixture of template dependence and independence depending on the reaction conditions. TdT creates diversity during immunoglobulin V(D)J recombination by adding random sequences between the DNA elements (28). Pol μ also adds one (or a few) random nucleotide(s) between the two protruding 3′-ends at the junction during nonhomologous end-joining (NHEJ) (29).
It has been proposed that the X-Family polymerases perform distinct cellular roles based on a gradient of template dependence (30). The single-stranded DNA extension property of POLQ complements TdT and Pol μ, positioning POLQ among the X-Family polymerases in the proposed gradient of template dependence in NHEJ. The terminal transferase activity of TdT and Pol µ depends on the presence of a protein loop that is proposed to interact with the DNA substrate and allow the enzymes to catalyze DNA extension in the absence of a template (31). Similarly, the translesion synthesis capability of POLQ has been shown to rely on protein loops (5) and we now show that non-templated DNA synthesis is also dependent on these same protein loops (Figure 2).
The ability of POLQ to extend DNA appears to have distinct properties from those observed for Pol µ and TdT. First, TdT is able to extend single-stranded DNA but is unable to perform template directed DNA synthesis and the terminal transferase activity of TdT can be stimulated by using cobalt as the catalytic metal ion in place of magnesium (32).On the other hand, POLQ can catalyze both nontemplated as well as template-directed DNA synthesis and its catalytic activity is strongly inhibited by replacing magnesium ions with cobalt ions (data not shown). Second, both TdT and Pol µ are able to efficiently incorporate both deoxyribonucleotides as well as ribonucleotides (33,34), while ribonucleotide incorporation onto the end of the single-stranded primer by POLQ was very inefficient and barely detectable with duplex DNA even at high concentrations of ribonucleotide and at extended reaction times (Figure 1d and e). Third, as shown in Figure 4, 3′ overhangs from duplex DNA become less usable as a substrate for POLQ as they become shorter. This is the opposite of what has been observed for Pol µ, where 3′ overhangs longer than only one or two nucleotides become progressively worse substrates for Pol µ (29). Interestingly, our results with the extension of mismatched primers in which POLQ appears to detect the template strand without reading its sequence are reminiscent of those made with Pol µ. In those experiments, it was demonstrated that abasic sites were bypassed in an apparently template-dependent manner but without regard for the actual sequence of the template beyond the abasic site lesion (35). Thus, it is possible that Pol μ and POLQ may have non-overlapping functions during NHEJ. However, in mice lacking Pol λ, Pol μ or both Pol λ and Pol μ, POLQ might step in explaining why those three mice only have a limited radiosensitivity (36).
POLQ is expressed in bone marrow where B cells undergo V(D)J recombination, and it was postulated that POLQ may play a role in this process (37). V(D)J recombination in immunoglobulin genes is carried out by a process that uses double-strand breaks as intermediates that are subsequently repaired by NHEJ pathways. Random nucleotide insertions provide variability in immunoglobulin genes and such insertions have been shown to be supplied by non-templated incorporation of nucleotides by TdT (38). In TdT knockout mice, a reduction in random nucleotide incorporation during V(D)J recombination of about 90% is observed, suggesting that TdT is the primary polymerase involved in the insertion of random nucleotide sequences during antigen-receptor diversification in mice (39). Although it is possible that POLQ plays some role in the creation of the remaining 10% of randomly incorporated nucleotides, our results suggest that it does not. Protein-mediated cleavage of hairpins during double-strand break repair at V(D)J recombination sites yields blunt ends or 3′ overhangs of only four nucleotides (40). Our results in Figure 4 show that 3′ overhangs of only four bases are very poor substrates for non-templated extension by POLQ and suggests that they are not likely to be an in vivo substrate for this enzyme directly, although further processing of these ends by exonucleolytic activity could conceivably provide the longer 3′ overhangs that we have shown to be efficient substrates for POLQ.
An alternative to playing a role in canonical NHEJ pathways is that POLQ may be involved in Ku independent microhomology-mediated end joining (MMEJ) reactions (41). Recent work on the POLQ ortholog Mus308 in Drosophila supports our hypothesis that single-stranded DNA extension by POLQ may play a role in double-strand break repair and may do so via MMEJ pathways (42). In that work, double-strand breaks were mimicked by the excision of transposable elements leaving behind 17-base 3′ single-stranded ends. These DNA ends are strikingly similar in length to the 14-base substrates that we have found to be highly extendable by POLQ. The authors of that study showed that random incorporation of nucleotides between the 3′ overhangs of the resulting double-strand breaks were significantly reduced in the absence of Mus308. Although Mus308 lacks the three protein insertion loops that are unique to POLQ and appear to be critical to POLQ's unique enzymatic activities, the organization of the enzyme in terms of an N-terminal helicase domain and C-terminal A-Family DNA polymerase suggests that the two enzymes may play a role in similar pathways in metazoan cells. In the model presented by McVey and colleagues (42), the Mus308 polymerase is only considered as a template-dependent polymerase that creates regions of microhomology at which the 3′ overhangs can anneal for subsequent gap-filling polymerization and ligation reactions. Our data suggests that POLQ may be able to directly extend 3′ overhangs (as long as they are of sufficient length) by the incorporation of apparently random sequences, which may be a different method of inserting random nucleotide sequences between flies and higher organisms in which POLQ homologs are found. The current model for MMEJ calls for 3′ single-stranded flaps to occur after annealing of the microhomologous regions. These flaps are removed by an exonuclease activity, but we find it very interesting that in our experiments mismatched primer ends of at least three nucleotides are readily extended by POLQ. It may be possible then that POLQ may play an additional role in the repair of double-strand breaks by utilizing as a substrate DNA in which these 3′ flaps are not removed. The significance of this finding in terms of double-strand break repair warrants further investigation.
While we propose that POLQ is able to extend single-stranded DNA in a non-templated manner, such activity is difficult to differentiate from error-prone template-dependent synthesis. This is especially noteworthy in light of our results with homopolymeric substrates in which most incorporation is with nucleotides complimentary to the substrate. Very similar results have been observed for DNA polymerase I from Sulfolobus solfataricus (a B-family DNA polymerase), in which it was proposed that this polymerase is able to add nontemplated bases to the end of a single-stranded template as well as stabilize poorly annealed ends to stimulate template-dependent synthesis (43). Interestingly, the terminal transferase activity proposed for that polymerase was only observed on oligonucleotides that were a minimum of 10–15 nucleotides in length, which is the minimum required length for our activity with POLQ. We note that some of our oligonucleotides may promote self-annealing more than others, for example the single-stranded template in Figure 1c could loop back onto itself to pair the two 3′ terminal C residues with two G residues. This would allow for the templated addition of dGTP, which happens to be the most efficiently incorporated nucleotide. However, we observe incorporation of several dozen A residues, which is difficult to explain by a simple realignment of a self-annealed oligonucleotide. For other oligonucleotides, such as those used in Figures 4, 5 and 7, no pairings of two or more continuous complimentary nucleotides are possible for any self-priming 3′-termini to allow for efficient template-directed synthesis. We also observe absolutely no extension of more than a single nucleotide past the end of the template when using properly base-paired duplex DNA. However, in every instance with single-stranded DNA, we observe extensions to lengths beyond those that would be expected if the polymerase were utilizing a looping back mechanism to allow for template-directed DNA synthesis. Our observation that ribonucleotides can be incorporated when single-stranded DNA is the substrate (Figure 1d) but not in duplex DNA (Figure 1e) provides further evidence that POLQ may be extending single-stranded DNA in a non-templated manner and not forming hairpins or otherwise self-annealing templates. If the single-stranded oligonucleotide were forming hairpins such that POLQ was utilizing a template strand, we would expect to see no incorporation of ribonucleotides as seen with duplex DNA. Finally, we have previously observed that the loop 2 deletion mutant is almost completely unable to add a single non-templated nucleotide at the end of duplex DNA [see Figure 3 in ref. (5)] and such incorporation is attributed to terminal transferase-like activity (26). The loop 2 deletion mutants subsequent difficulty in extending single-stranded DNA (Figure 2a) provides further evidence that such extension by wild type POLQ may be based on nontemplated terminal transferase activity. However, there still exists the formal possibility that POLQ is able to anneal single base pairs at the 3′-end of the primer, insert a single nucleotide, dissociate or melt the newly formed duplex DNA and reanneal a single base pair somewhere else in the sequence. Such an activity would yield a random sequence that would be indistinguishable from random non-templated nucleotide incorporation. The mechanism by which this would occur would necessarily be distinct from that in which primer/template DNA is the substrate and studies are underway to determine if such alternative processes exist with POLQ.
Based on our current findings, we propose that POLQ may be involved in double-strand break repair by extending the single-stranded 3′-ends of broken DNA molecules. One of the hallmarks of cancer is the occurrence of high levels of chromosomal rearrangements as a result of inaccurate repair of double-strand breaks. Therefore, it is noteworthy that POLQ is highly overexpressed in breast and colon cancers (37,44–46). POLQ overexpression in these cancers is also a negative prognostic marker, the higher the expression level of POLQ the more likely the patient is to die from the disease. Common thinking has been that high levels of POLQ increase the mutation rate in cancer cells due to its ability to bypass lesions such as abasic sites and thymine glycols as well as its inherent lack of fidelity (47). But perhaps the opposite is the case. In light of our current results, it is tempting to speculate that perhaps high levels of POLQ in cancer cells increase the survival of cancer cells by increasing their ability to cope with high levels of double-strand breaks. The activity of POLQ in these cells may be such that cancer cells undergo further rearrangements to their genomes as a result of enhanced double-strand break repair but these rearrangements may preclude cell death due to what would otherwise be broken, and thus unreplicable, genomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure S1.
FUNDING
Kempestiftelserna (M.H., A.E.S.-E., and E.J.); the Swedish Research Council (E.J.); the Swedish Cancer Society (E.J.); Smärtafonden (E.J.). Funding for open access charge: The Swedish Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The initial experiments that provided the impetus for the current work were performed under grant R01CA52040 from the National Cancer Institute to Drs. Susan S. Wallace and Sylvie Doublié at the University of Vermont (USA)
REFERENCES
- 1.Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebenek K, Joyce CM, Fitzgerald MP, Kunkel TA. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 1990;265:13878–13887. [PubMed] [Google Scholar]
- 3.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair. 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogg M, Seki M, Wood RD, Doublié S, Wallace SS. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J. Mol. Biol. 2011;405:642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5′-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, O-Wang J. DNA polymerase theta contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc. Natl Acad. Sci. USA. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, O-Wang J. DNA polymerases eta and theta function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J. Biol. Chem. 2007;282:17387–17394. doi: 10.1074/jbc.M611849200. [DOI] [PubMed] [Google Scholar]
- 10.Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, Kurosaki T, O-Wang J. Absence of DNA polymerase theta results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair. 2006;5:1384–1391. doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger Iii AJ, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6:1143–1148. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair. 2008;7:1603–1608. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, et al. Vertebrate POLQ and POL beta cooperate in base excision repair of oxidative DNA damage. Mol. Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J. Biol. Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 16.Blanca G, Villani G, Shevelev I, Ramadan K, Spadari S, Hubscher U, Maga G. Human DNA polymerases lambda and beta show different efficiencies of translesion DNA synthesis past abasic sites and alternative mechanisms for frameshift generation. Biochemistry. 2004;43:11605–11615. doi: 10.1021/bi049050x. [DOI] [PubMed] [Google Scholar]
- 17.Goff JP, Shields DS, Seki M, Choi S, Epperly MW, Dixon T, Wang H, Bakkenist CJ, Dertinger SD, Torous DK, et al. Lack of DNA polymerase theta (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat. Res. 2009;172:165–174. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins GS, Prevo R, Lee YF, Helleday T, Muschel RJ, Taylor S, Yoshimura M, Hickson ID, Bernhard EJ, McKenna WG. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010;70:2984–2993. doi: 10.1158/0008-5472.CAN-09-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ukai A, Maruyama T, Mochizuki S, Ouchida R, Masuda K, Kawamura K, Tagawa M, Kinoshita K, Sakamoto A, Tokuhisa T, et al. Role of DNA polymerase theta in tolerance of endogenous and exogenous DNA damage in mouse B cells. Genes Cells. 2006;11:111–121. doi: 10.1111/j.1365-2443.2006.00922.x. [DOI] [PubMed] [Google Scholar]
- 20.Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol. Cell Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 23.Muzzini DM, Plevani P, Boulton SJ, Cassata G, Marini F. Caenorhabditis elegans POLQ-1 and HEL-308 function in two distinct DNA interstrand cross-link repair pathways. DNA Repair. 2008;7:941–950. doi: 10.1016/j.dnarep.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 24.Hogg M, Wallace SS, Doublié S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogg M, Aller P, Konigsberg W, Wallace SS, Doublié S. Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J. Biol. Chem. 2007;282:1432–1444. doi: 10.1074/jbc.M605675200. [DOI] [PubMed] [Google Scholar]
- 26.Clark JM. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Just A, Butter F, Trenkmann M, Heitkam T, Morl M, Betat H. A comparative analysis of two conserved motifs in bacterial poly(A) polymerase and CCA-adding enzyme. Nucleic Acids Res. 2008;36:5212–5220. doi: 10.1093/nar/gkn494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilfillan S, Benoist C, Mathis D. Mice lacking terminal deoxynucleotidyl transferase: adult mice with a fetal antigen receptor repertoire. Immunol. Rev. 1995;148:201–219. doi: 10.1111/j.1600-065x.1995.tb00099.x. [DOI] [PubMed] [Google Scholar]
- 29.Davis BJ, Havener JM, Ramsden DA. End-bridging is required for pol mu to efficiently promote repair of noncomplementary ends by nonhomologous end joining. Nucleic Acids Res. 2008;36:3085–3094. doi: 10.1093/nar/gkn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Juarez R, Ruiz JF, Nick McElhinny SA, Ramsden D, Blanco L. A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 2006;34:4572–4582. doi: 10.1093/nar/gkl457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato KI, Goncalves JM, Houts GE, Bollum FJ. Deoxynucleotide-polymerizing enzymes of calf thymus gland. II. Properties of the terminal deoxynucleotidyltransferase. J. Biol. Chem. 1967;242:2780–2789. [PubMed] [Google Scholar]
- 33.Boule JB, Rougeon F, Papanicolaou C. Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J. Biol. Chem. 2001;276:31388–31393. doi: 10.1074/jbc.M105272200. [DOI] [PubMed] [Google Scholar]
- 34.Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Covo S, Blanco L, Livneh Z. Lesion bypass by human DNA polymerase mu reveals a template-dependent, sequence-independent nucleotidyl transferase activity. J. Biol. Chem. 2004;279:859–865. doi: 10.1074/jbc.M310447200. [DOI] [PubMed] [Google Scholar]
- 36.Ramsden DA. Polymerases in Nonhomologous End Joining: building a Bridge over Broken Chromosomes. Antioxid. Redox Signal. 2011;14:2509–2519. doi: 10.1089/ars.2010.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, et al. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 38.Gilfillan S, Benoist C, Mathis D. Mice lacking terminal deoxynucleotidyl transferase: adult mice with a fetal antigen receptor repertoire. Immunol. Rev. 1995;148:201–219. doi: 10.1111/j.1600-065x.1995.tb00099.x. [DOI] [PubMed] [Google Scholar]
- 39.Cabaniols JP, Fazilleau N, Casrouge A, Kourilsky P, Kanellopoulos JM. Most alpha/beta T cell receptor diversity is due to terminal deoxynucleotidyl transferase. J. Exp. Med. 2001;194:1385–1390. doi: 10.1084/jem.194.9.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan SH, Yu AM, McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuo Z, Lin HK, Trakselis MA. Strand Annealing and Terminal Transferase Activities of a B-family DNA Polymerase. Biochemistry. 2011;50:5379–5390. doi: 10.1021/bi200421g. [DOI] [PubMed] [Google Scholar]
- 44.Lemée F, Bergoglio V, Fernandez-Vidal A, Machado-Silva A, Pillaire M-J, Bieth A, Gentil C, Baker L, Martin A-L, Leduc C, et al. DNA polymerase Θ up-regulation is associated with poor survival in breast cancer, perturbs DNA replication and promotes genetic instability Proc. Natl Acad. Sci. USA. 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pillaire MJ, Selves J, Gordien K, Gourraud PA, Gentil C, Danjoux M, Do C, Negre V, Bieth A, Guimbaud R, et al. A 'DNA replication' signature of progression and negative outcome in colorectal cancer. Oncogene. 2010;29:876–887. doi: 10.1038/onc.2009.378. [DOI] [PubMed] [Google Scholar]
- 46.Higgins GS, Harris AL, Prevo R, Helleday T, McKenna WG, Buffa FM. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget. 2010;1:175–184. doi: 10.18632/oncotarget.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat. Rev. Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.