Abstract
CRISPR loci are essential components of the adaptive immune system of archaea and bacteria. They consist of long arrays of repeats separated by DNA spacers encoding guide RNAs (crRNA), which target foreign genetic elements. Cbp1 (CRISPR DNA repeat binding protein) binds specifically to the multiple direct repeats of CRISPR loci of members of the acidothermophilic, crenarchaeal order Sulfolobales. cbp1 gene deletion from Sulfolobus islandicus REY15A produced a strong reduction in pre-crRNA yields from CRISPR loci but did not inhibit the foreign DNA targeting capacity of the CRISPR/Cas system. Conversely, overexpression of Cbp1 in S. islandicus generated an increase in pre-crRNA yields while the level of reverse strand transcripts from CRISPR loci remained unchanged. It is proposed that Cbp1 modulates production of longer pre-crRNA transcripts from CRISPR loci. A possible mechanism is that it minimizes interference from potential transcriptional signals carried on spacers deriving from A-T-rich genetic elements and, occasionally, on DNA repeats. Supporting evidence is provided by microarray and northern blotting analyses, and publicly available whole-transcriptome data for S. solfataricus P2.
INTRODUCTION
Archaeal CRISPR-based immune systems provide a defence against invading genetic elements, primarily viruses and conjugative plasmids, and they fall into three main types, the DNA-targeting CRISPR/Cas systems where CRISPR loci and cas genes are invariably linked on the genome, and the DNA-targeting CRISPR/Csm and RNA-targeting CRISPR/Cmr systems for which the csm and cmr gene cassettes are often uncoupled from CRISPR loci (1–4). Almost all archaea carry CRISPR-based defence systems and crenarchaea often exhibit a complex mixture of different types (5,6). The CRISPR locus is an essential functional component of all these systems and consists of a long leader region followed by up to about 100 spacer-repeat units. Spacer sequences originating from foreign genetic elements are about 30–40 bp long and the interspaced identical direct repeats are ∼25–37 bp in length; both tend to be conserved in length for a given CRISPR locus (2,7–9).
All CRISPR-based immune systems are basically modular with three primary functions: (i) adaptation that involves excision of DNA from invading DNA genetic elements and integration of the DNA as a new spacer in a CRISPR locus at or near the leader, (ii) generation and processing of CRISPR transcripts to yield mature crRNAs, and (iii) interference of the genetic element by targeting and cleavage via a crRNA–protein complex (10). A few Cas, Cmr and Csm proteins have been assigned roles associated with each of these functional steps on the basis of predictions from bioinformatical or crystal structure analyses and, less commonly, experiments (2,4).
The crenarchaeal genus Sulfolobus, in particular, has yielded novel insights into these CRISPR-based systems. Sulfolobus species generally carry complex and diverse systems including DNA-targeting CRISPR/Cas and CRISPR/Csm and RNA targeting CRISPR/Cmr, sometimes encoded in multiple copies in a given species (5,7,11). Moreover, many novel viruses have been characterized for Sulfolobus and the related genus Acidianus which have recently been classified into seven new viral families with several remaining unclassified (12,13), and plasmids with an archaea-specific conjugative apparatus have also been identified (14). These provide a major advantage for studying the interplay between genetic elements and host CRISPR-based systems. For example, numerous virus and conjugative plasmid sequence matches to CRISPR spacers were used to demonstrate that the uptake of DNA from invading genetic elements was essentially a random process (15). Moreover, it was recently shown that by employing vectors carrying viral genes or sequences matching CRISPR spacers under selection, one can induce different sized CRISPR deletions which all include the matching spacer (16), and these genetic systems were also used to study sequence stringency requirements for DNA targeting by crRNAs (16,17).
Our understanding of mechanisms of transcriptional regulation of CRISPR loci is still at an early stage. In enterobacteria transcription of CRISPR loci and associated cas genes is silenced by the H-NS regulator and activation of the system requires an anti-silencer (18–20). Moreover, a bacteriophage EPV1 was characterized in a metagenomic study encoding the H-NS protein which could inactivate the host defence system upon viral infection (21). For Sulfolobus, the complexity and diversity of the CRISPR systems, and the presence of putative transcriptional regulators associated with the different genetic modules, would require multiple regulatory mechanisms. For example, the putative provirus M164 was found integrated into the gene of the putative transcriptional regulator Csa3 associated with the gene cassette encoding proteins involved in adaptation (22). The simplest way to inactivate the diverse systems would be to inhibit production of crRNAs. However, it has been shown for Sulfolobus, and other hyperthermophilic archaea, that pre-crRNAs are generated constitutively in the absence of invading foreign DNA elements (7,11,23–25) and, currently, there is no evidence to indicate that the level of pre-crRNA transcripts increases when genetic elements enter cells. Presumably, this reflects that the CRISPR immune system can respond rapidly to the continual exposure to a wide variety of foreign genetic elements that frequent these extreme natural environments (12). The regulatory difference between pre-crRNA regulation in the enterobacteria and Sulfolobus could also reflect that the diverse and complex CRISPR-based systems of Sulfolobus and other archaea are actively involved in maintaining relatively low levels of viruses intracellularly (12,13).
Only one protein to date has been shown to bind directly to a CRISPR locus. That is the Sulfolobus solfataricus P2 protein Sso0454 (formerly SRSR repeat-binding protein) that exhibits a triple internal repeat sequence and binds specifically to DNA repeats of CRISPR loci of S. solfataricus and the Sulfolobus conjugative plasmid pNOB8 (26). It protects the repeat and repeat–spacer junctions against endonuclease attack and induces a distortion at the centre of the DNA repeat (26). Homologues of Cbp1 are found primarily amongst members of the acidothermophilic Sulfolobales but have also been detected in genomes of the hyperthermophilic Desulfurococcales (27). The cbp1 gene is not physically linked on chromosomes to either CRISPR loci or CRISPR-associated proteins, which suggests that it also has other cellular target sites.
Prior to the discovery of CRISPR transcription, Cbp1 was considered to be involved in chromosomal packaging of CRISPR loci (26) but the detection of a range of intermediate processed CRISPR transcripts (pre-crRNAs) in Archaeoglobus fulgidus and S. solfataricus (23,24) raised the possibility of a transcriptional role for the protein. CRISPR loci generally appear to be transcribed as single transcripts from a promoter within the leader region (7,11) followed by processing within repeat sequences by Cas6 to yield mature crRNAs, carrying part of the repeat and all or most of the spacer sequence (28–30). It was also shown for S. acidocaldarius that transcripts are produced from reverse CRISPR strands for each of the five CRISPR loci present (11) and evidence for the formation of reverse-strand transcripts from the six CRISPR loci of S. solfataricus P2 was also provided by a whole-transcriptome analysis (31). The functional significance, if any, of these reverse-strand transcripts remains unclear but potentially they can base pair with crRNAs and impede their interference reactions.
Sequence-specific DNA-binding proteins are often transcription factors but, since Cbp1 can potentially bind to all repeats of CRISPR loci within a cell, in total 208 repeats in S. islandicus REY15A and 423 in S. solfataricus P2 (11,16), it is not a typical transcriptional regulator. In order to gain some insight into its function(s), we exploited the recently developed genetic systems for S. islandicus REY15A and S. solfataricus P2 as well as a microarray for the latter organism. Cbp1 knockout and overexpression mutants were generated and transcriptional properties of the CRISPR loci were examined for the mutants (16,32).
MATERIALS AND METHODS
Strains, media and constructs
Experiments were performed with S. islandicus E233S carrying a large deletion within the pyrEF genes and a complete lacS gene deletion. S. solfataricus InF1 was also employed carrying an inactivated pyrF gene. Cells were cultured at 75–78°C in the complex medium TYS or in the selective media SCVy, GCVy or ACVy (5,32). To arrest transcription, actinomycin D was added to the culture at 20 µg/ml and samples were taken at different time points (33).
Plasmid pK454 was used to generate the cbp1 gene deletion in S. islandicus E233S. It was constructed by a triple ligation of pHZ1, the L arm (amplified using primers 5′-ttggatCCATTGACAAACCTAAAATAATCCCT-3′ and 5′-ttctgcagAAGCATTCTACGAACCCTAGAGTAACTT-3′), and the R arm (amplified using 5′-ttctgcagTGCAAAAGAACTTAACATTTCCACTAAT-3′ and 5′-ttgtcgacGCACATAGGACACCTAATACCATTCAT-3′) before transforming into S. islandicus E233S to produce first pop-in transformants and then the deletion mutant (16,32). Primers 5′-GAAATCCCAACAGTAACCCACC-3′ and 5′-GCATGTCATGCTTAGGAGAAACG-3′ were used to confirm the recombination events.
Plasmid pC454 was used to complement the Cbp1 deletion mutant and was constructed by inserting the SOE-PCR product into pHZ1 (32). Briefly, primers CompLf 5′-ttgcatgCCATTGACAAACCTAAAATAATCCCT-3′ and CompLr 5′-CCTAGATTATATTTCTTAAAAATTCTCAAT-3′ were employed to produce fragment L, and fragment R was generated using CompRf 5′-ATTGAGAATTTTTAAGAAATATAATCTAGG-3′ and CompRr 5′-ttgtcgacGCACATAGGACACCTAATACCATTCAT-3′. The two products were mixed and amplified by primer CompLf and CompRr. The fused fragment, with a single nucleotide A to G change in the cbp1 gene was purified, digested by SphI and SalI, repurified and ligated into pHZ1.
Expression, purification and detection of Cbp1
The cbp1 gene of S. islandicus was amplified using primers SisCbpF 5′-TTTGGAATTCCATATGGTGAGTGAAGAAGAAATAATTGAAAAAGTTAAGAAAATG-3′ and SisCbpHR 5′-TTTCCGCTCGAGAGCAGATGTGGGAGAAGATTCACGAA-3′. The PCR product was inserted into a pET28a vector (Novagen, Darmstadt, Germany) using XhoI and NdeI (Fermentas, St Leon-Rot, Germany) and the stop codon was replaced by codons for a C-terminal hexameric His-tag. The resulting clone was inoculated into LB broth carrying kanamycin and chloramphenicol and cultured at 37°C to A600 = 0.6. The protein was expressed overnight at room temperature in BL21 (DE3) pLYSs cells (Promega, Madison, USA) and expression was induced by adding isopropylthio-β-d-galactoside (IPTG) at 0.5 mM.
The cbp1 gene of S. islandicus was amplified using primers XCbpSisF 5′-tttcgcgacatATGAGTGAAGAAGAAATAATTGAAAAAGT-3′ and XCbpSisR 5′-tttaggcctCTAAGCAGATGTGGGAGAAGATTC-3′ for expression in S. islandicus. Primer XCbpSsoF 5′-tttcgcgaCATATGAGCGAGGAAGAAAACATTGAAAAAGT-3′ and XCbpSisR were employed to amplify the cbp1 gene of S. solfataricus for expression in S. solfataricus. The modified pEXA vector pSeSD was used for protein overexpression (16,34).
Cbp1 protein were detected by western blots using antibodies derived from Escherichia coli-expressed Cbp1 (Innovagen AB, Lund, Sweden) and an alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Invitrogen, Paisley, UK), using the 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (NBT) reagent (Sigma-Aldrich, Munich, Germany).
DNA band-shift assay
A 157-bp DNA fragment carrying a repeat–spacer-repeat sequences and short-flanking regions was amplified from S. islandicus by PCR using oligonucleotide pairs Sis2rptF 5′-TTCCTCCATCTTCATCTTCACCACC-3′ and Sis2rptR 5′-TCTTCTTTGTCATCTTCGCAGTCGC-3′ and [32P]-5′-end labelled using T4 polynucleotide kinase (Fermentas). Cbp1 (35 nM) was incubated with 7 nM [32P] 5′-end labelled CRISPR-2 r substrate in DNA binding buffer (10 mM Tris–Cl, pH 7.6, 150 mM KCl, 2 mM DTT, 10% glycerol) for 20 min at 50°C before loading on an 8% polyacrylamide gel. To test for binding specificity, the unlabelled 157 bp CRISPR-2r substrate was used as specific competitor and a 179-bp recA gene PCR product was used as unspecific competitor. After cooling to room temperature, 3 µl loading buffer (10 mM Tris–Cl, pH 7.6, 1 mM EDTA, 50% glycerol, 0.5% bromophenol blue) was added and samples were fractionated in a prerun 8% polyacrylamide gel containing 89 mM Tris–Cl, 25 mM taurine, 0.5 mM EDTA, pH 8.9 and autoradiographed. The competition assay of S. solfataricus Cbp1 to different CRISPR repeats followed the same experimental conditions. For amplification of the 148 bp CRISPRSs-2r (A + B) DNA, primers 5′-CTCCGCAACTTCATCAATAGTG-3′ and 5′-GAGTTGCGGGCACTTTATGACAG-3′ were used, and the 151 bp CRISPRSs-2r (C + D) DNA was amplified using primers 5′-CGGACACTGGTATAAACATGC-3′ and 5′-CATCTGGGGCATATTGTACTG-3′.
RNA preparation and northern blotting
Total cellular RNA was prepared as described earlier (20). For northern blotting, 15 µg RNA was mixed with the same volume of Gel Loading Buffer II or NorthernMax-Gly Sample Loading Buffer (Applied Biosystems/Ambion, Austin, USA) and fractionated in a 6% polyacrylamide gel containing 7 M urea and 90 mM Tris-borate, 2 mM EDTA, pH 8.3, or a 1.5% agarose gel in 10 mM PIPES, 30 mM Bis–Tris, 1 mM EDTA, pH 8.0 with 0.1–2.0 or 0.5–10 kb RNA ladders (Invitrogen) or a 50- to 1000-nt RNA ladder (New England Biolabs, Boston, USA).
Procedures for transferring and immobilizing RNA on nylon membranes, prehybridizing, end-labelling of complementary nucleotides, hybridization and film exposure followed earlier protocols (11). Probes were stripped from hybridized membranes with 0.5% SDS for 1 h and membranes were reused for hybridization when no residual radioactivity was detected. Oligonucleotide probes used were as follows: repeat of loci 1 and 2 of S. islandicus pre-crRNA—5′-CTTTCAATTCTATAGTAGATTAGC-3′; spacer 10, locus 2—5′-GCCCCCATTATACAATATCTACG-3′; S. solfataricus spacer 28, locus A—5′-TTGAAAGATTTGAACGTTAGCGAG-3′; spacer 24, locus B—5′-GGAGGGTGAGACAATGAAGGTTAC-3′; spacer 1, locus C—5′-GCAACACAAGAGGCTAGTAAGGTTG-3′; repeat of loci A + B—5′-CTTTCAATTCCTTTTAGGATTAATC-3′, and repeat of loci C + D—5′-CTTTCAATTCTATAAGAGATTATC-3′.
Localizing CRISPR locus deletions
PCR products were obtained across CRISPR loci 1 and 2 of S. islandicus REY15A using premixed Ex Taq (Takara, Otsu, Japan) following the manufacturer's protocol with 75–100 ng genomic DNA in 10 µl. To amplify S. islandicus CRISPR loci 1 and 2, respectively, we used C1F 5′-AGCTTGCTTACCTCAAGGTACTTTACGT-3′ and C1R 5′-TTAATAAACGACGATTTTCCTCTTGAT-3′, and C2F 5′-AGGATAGCGAAGTCGTAGAGTTTGGAT-3′ and C2R 5′-TAACGCACGGTATTGAAACTTCTCATC-3′. Purified PCR products were sequenced either directly (Eurofins MWG Operon, Ebersberg, Germany) or after cloning using CloneJET™ PCR Cloning Kit (Fermentas).
Analysis of microarray and transcriptome data for S. solfataricus P2
S. solfataricus P2 microarrays were designed by the Sulfolobus genome chips consortium and manufactured by Ocimum Biosolutions (Hyderabad, India). They carried 3042 gene probes and several sets of probes against crenarchaeal viral genomes and plasmids (35). Microarray hybridizations were performed as described (35) except that the CyScribe Post-Labeling Kit (GE Healthcare) was used for labelling. Data analyses were conducted by ImaGene (BioDiscovery, CA, USA) using default settings. A dye swap was performed for each time point and values were averaged. CLC Genomic Workbench (Aarhus, Denmark) was employed for analysing the raw CRISPR transcriptome sequencing data (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?study=SRP001461). Strand-specific sequences from three different cDNA libraries were analysed and the parameters were set such that the only perfect matches of the 36 nt reads to the genome were taken.
RESULTS
Sulfolobus islandicus Cbp1 binds specifically to CRISPR repeats
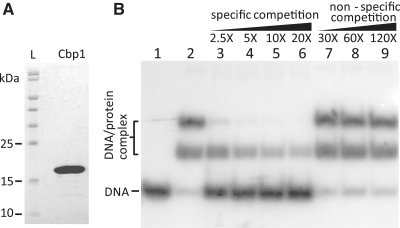
Cbp1 of S. islandicus REY15A shows 93% sequence identity to Sso0454 of S. solfataricus and was expressed in E. coli and purified (Figure 1A). DNA repeat binding activity was assayed using a [32P] 5′-end labelled 157 bp repeat–spacer–repeat construct with short flanking sequences (CRISPR-2r). Electrophoretic band-shift assays showed the formation of two retarded bands consistent with Cbp1 binding to one or both repeats of the substrate (Figure 1B, lane 2). Competition experiments were performed using either unlabelled CRISPR-2r DNA as specific competitor or a 179-bp DNA fragment amplified from a recA gene of S. islandicus as an unspecific competitor. Only the unlabelled CRISPR-2r competed strongly over the range 2.5- to 20-fold molar excess (lanes 3–6 compared with lane 2) showing a progressive transition from the upper to the lower bands thereby providing support for a specific Cbp1–DNA interaction (Figure 1B).
Figure 1.
Cbp1 purification and DNA binding. (A) Electrophoresis of purified Cbp1 protein in a 12.5% SDS–PAGE stained with Coomassie blue. (B) Competition assay of Cbp1 with CRISPR-2 r DNA. Cbp1 (35 nM) was incubated with 7 nM [32P] 5′-end labelled 157 bp CRISPR-2r substrate in 10 mM Tris–Cl, pH 7.6, 150 mM KCl, 2 mM DTT, 10% glycerol at 50°C for 20 min before loading on an 8% polyacrylamide gel (see ‘Materials and Methods’ section). Binding specificity was tested by competing with unlabelled CRISPR-2r as specific competitor (lanes 3–6), and a 179 bp DNA fragment amplified from the recA gene S. islandicus as unspecific competitor (lanes 7–9). Lane 1—CRISPR-2r DNA, lane 2—Cbp1-DNA complex alone. Molar excesses of competitor DNA are indicated for lanes 3–9.
Generation of a deletion mutant, a complemented strain and overexpression vectors for Cbp1
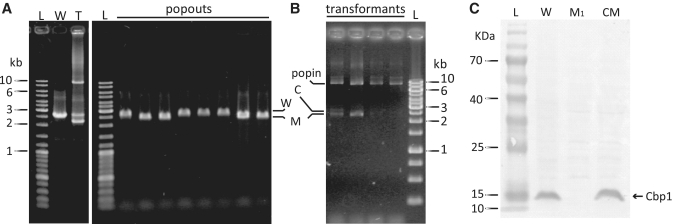
In order to test for Cbp1 function, we first generated a Cbp1-minus mutant of S. islandicus REY15A using a ‘pop in-pop out’ gene targeting method (32). Plasmid pK454 was constructed and then transformed to produce pop in transformants and then Cbp1 deletion mutants (see ‘Materials and Methods’ section). To minimize possible detrimental effects on expression of flanking genes, part of the cbp1 gene, encoding the N-terminal 38 amino acids, was retained after the knockout. Deletion mutants were identified by colony PCR for four of the eight colonies that formed on counter-selective plates (Figure 2A). Growth rates and morphologies of the Cbp1-minus cells were indistinguishable from those of wild-type cells indicating that Cbp1 is not essential for cell viability (data not shown).
Figure 2.
Generation of pop in-pop out Cbp1-minus mutant and a complementing Cbp1 mutant. (A) Analysis of pop-in and pop-out transformants of pK454 by PCR. (B) Analysis of the transformants of pC454 by PCR. C is the complemented band. (C). Western blot of the Cbp1 protein in S. islandicus. L—protein size ladders, W—wild-type, T—pop-in transformant, M—deletion mutant, CM—complemented mutant.
Next, a complemented strain for the deletion mutant was produced using the same strategy as for the deletion mutant (Figure 2B). It carried a single A–G mutation, confirmed by PCR amplification and sequencing that resulted in conversion of the lysine-86 codon from AAA to AAG (data not shown). Both codons occur frequently such that the translation efficiency should not be affected. A western blot analysis was employed to verify that Cbp1 was only expressed from the wild-type and Cbp1 complemented strains, and it was also demonstrated that the expression level from the complemented strain was similar to the wild-type (Figure 2C).
The cbp1 gene from S. islandicus was cloned into the pSeSD overexpression vector and transformed into S. islandicus E233S. A similar overexpression construct was also generated for the cbp1 gene of S. solfataricus (sso0454) and was transformed into S. solfataricus lnF1. The cbp1 genes were preceded by an araS promoter such that Cbp1 expression could be controlled in both species by using different carbon sources (16,34).
Active foreign DNA interference in the S. islandicus Cbp1-minus mutant
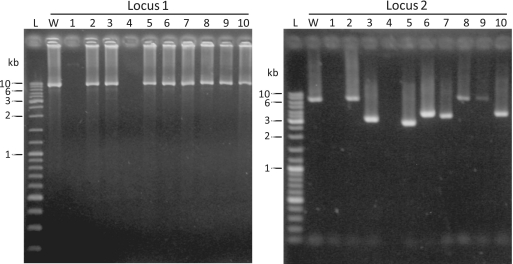
First, we tested whether foreign DNA interference by the CRISPR/Cas system was inhibited by the absence of Cbp1. A vector was employed carrying spacer 45 of CRISPR locus 2, a CC protospacer adjacent motif (PAM) and pyrE/F genes and it was transformed into the uracil auxotrophic Cbp1-minus S. islandicus E233S cells. A very low transformation efficiency was observed compared with non-targeted plasmids that is consistent with active DNA targeting (16). Ten transformants were cultured and PCR products were generated for CRISPR locus 2 carrying the matching spacer 45 and for non-targeted CRISPR locus 1 as a control. The results demonstrate that deletions occurred in locus 2 in up to eight transformants, including the apparent loss of CRISPR loci 1 and 2 in transformants 1 and 4 (Figure 3). PCR products from transformants 5 and 6 were sequenced and showed deletions from repeats 1–62 and 7–56, respectively, in locus 2 which both included the matching spacer 45. These results indicate that the DNA interference system is still active in the absence of Cbp1 (16).
Figure 3.
Testing for DNA-targeting activity of the CRISPR/Cas system of the Cbp1-minus mutant of S. islandicus. The transformed plasmid carried a target for the crRNA from spacer 45 of CRISPR locus 2 and PCR results are shown from the viable transformants for loci 1 and 2. All except transformant 2 show evidence of deletions in locus 2. Transformants 1 and 4 appear to have lost both CRISPR loci. DNA size markers are shown on the left.
Reduced pre-crRNA levels in the Cbp1-minus mutant
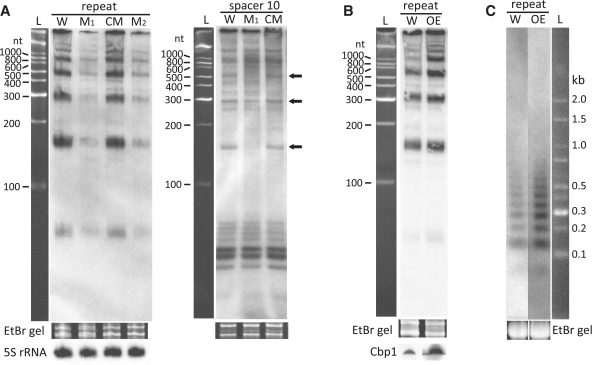
To test for a possible transcriptional role, we estimated the pre-crRNA and mature crRNA transcript levels for the Cbp1-minus mutant. Northern blots were obtained using a probe specific for the identical repeat sequences of CRISPR loci 1 and 2 and another for spacer 10 of CRISPR locus 2. In each experiment, processed intermediates generated a typical Sulfolobus pre-crRNA ladder corresponding to multiples of two to three spacer-repeat units (24) (Figure 4).
Figure 4.
Northern blot analyses of pre- and mature crRNAs of S. islandicus. (A) Pre-crRNAs present in RNA extracts from wild-type cells (W), Cbp1-minus mutants (M1 and M2) and the Cbp1 complemented mutant (CM) on probing against repeats of loci 1 and 2 and against spacer 10 of locus 2. Arrows indicate pre-crRNA bands of decreased intensity in the mutant sample. (B) Probing of repeats of the wild-type (W) and Cbp1 overexpression mutant (OE). The Western blot below shows the enhanced expression level of Cbp1. Cells were grown in sucrose medium (SCVy). (C) Northern analysis of pre-crRNAs separated on an agarose gel. Ethidium bromide (EtBr) staining of the gel prior to membrane transfer supports comparable loading levels and RNA integrity. L—DNA size ladders.
The results with the repeat probe revealed a strong reduction in yields of pre-crRNA products for two isolated Cbp1-minus mutant clones (M1 and M2) compared to wild-type and Cbp1 complemented strains, consistent with an overall reduction in the level of longer CRISPR transcripts (Figure 4A). The results with the spacer 10 probe also showed the presence of relatively weaker intermediate bands at ∼180, 300 and 540 nt but strong mature crRNA bands were still present consistent with the demonstration that foreign DNA targeting remains active (Figure 4A). The uppermost bands >600 nt may result partially from cross-hybridization effects with the spacer probe.
These results suggest that in the absence of Cbp1 there is a reduced overall level of longer pre-crRNA transcripts generated from loci 1 and 2, but the reduction could also reflect that the absence of Cbp1 leads to a destabilization of pre-crRNAs. Therefore, we tested for pre-crRNA stability employing actinomycin D that actively blocks transcription in Sulfolobus cells for up to 2 h (33). The relative yields of pre-crRNA and mature crRNAs in the Cbp1-minus mutant and wild-type cells were monitored by northern blotting using a repeat probe. Yields of the RNA products remained essentially constant over a 2-h period for both mutant and wild-type (Supplementary Figure S1) indicating that the absence of Cbp1 did not affect the stability of the pre-crRNAs. The experiment was also repeated by probing spacer 1 of CRISPR locus 2 for the wild-type and Cbp1-minus mutant. This showed that the yields of the mature crRNAs were also unchanged over the 2-h period (Supplementary Figure S1). These combined results support the conclusion that production of longer pre-crRNAs is reduced in the absence of Cbp1 but they also establish that the pre- and mature crRNAs have relatively long half-lives. For the mature crRNAs, this may reflect that they are stabilized by complexing with the Csa2 (Cas7) protein or Cmr proteins (2,25,34).
Overexpression of Cbp1 produces increased levels of pre-crRNAs
The preceding results suggest that the presence of repeat-bound Cbp1 enhances production of longer CRISPR transcripts from the leader because transcripts initiating and terminating within spacers of a CRISPR locus would tend to generate additional irregularly sized intermediate pre-crRNA products. Since it was estimated for S. solfataricus P2 that there are insufficient Cbp1 copies to saturate all the CRISPR repeats (26), we reasoned that overexpression of Cbp1 should increase levels of longer pre-crRNA transcripts. Therefore, we introduced the Cbp1 overexpression vector into S. islandicus E233S and examined the yields of larger pre-crRNAs relative to those of wild-type cells by northern blot analysis. The results demonstrated a significant increase in the yields of the larger (>600 nt) pre-crRNA products (Figure 4B) consistent with increased coverage of CRISPR repeats by Cbp1 leading to higher yields of larger pre-crRNAs. Since unprocessed CRISPR transcripts could be 6–7000 nt in length, the approximate size range of pre-crRNAs was estimated by separating RNAs from the overexpressed Cbp1 strain in agarose gels prior to northern blotting. Most pre-crRNAs fell within the size range 60–1000 nt (Figure 4C).
Microarray analysis of selected crRNAs in S. solfataricus
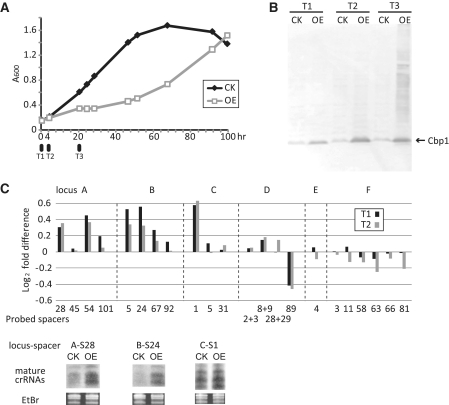
Since overexpression of Cbp1 produced higher yields of larger pre-crRNAs (Figure 4B), we exploited an available microarray carrying probes against all predicted ORFs and selected spacers of CRISPR loci A to F of the closely related species S. solfataricus P2 (11). No gene knockout system is available for this strain but we could overexpress Cbp1. Samples were collected at 0, 4 and 20 h after changing to an arabinose medium when transformants carrying empty arabinose-inducible expression vectors, or Cbp1 overexpression vectors, were selectively cultured (Figure 5A). Western blots performed on transformants from each culture showed enhanced Cbp1 expression at 0 h that increased strongly after 4 h and remained at a similar level after 4 h (Figure 5B). Enhanced expression at the first time-point reflects leakiness of the strong arabinose promoter (36).
Figure 5.
Probing for pre-crRNAs and crRNAs and on a S. solfataricus P2 microarray. (A) Overexpression of Cbp1 resulted in growth retardation. Cell cultures in exponential growth phase were changed from glucose medium (GCVy) to arabinose medium (ACVy) at 0 h (T1) for induction of Cbp1 expression. CK—transformants carrying the empty vector and OE—overexpression of Cbp1. (B). Western blot of the Cbp1 at time intervals of 0, 4 and 20 h. (C) Differential hybridization yields of pre-crRNAs and mature crRNAs in the Cbp1 overexpression mutant relative to wild-type cells. The microarray carried oligonucleotide probes specific for selected spacer sequences of the CRISPR loci A–F. Northern blot analysis results showing band intensities of mature crRNAs that are consistent with the microarray results.
Random cDNAs were generated from crRNAs of the six CRISPR loci and hybridized to the microarray. Relative hybridization yields for the overexpression mutant and wild-type were estimated in duplicate samples probing for sequences of 22 individual crRNAs. We focused on the time points 0 and 4 h because strong growth retardation occurred after 4 h. Seven crRNA transcripts showed significant changes, six with increased expression in the overexpression mutant and one with decreased expression while the remaining eight changes were considered insignificant (Figure 5C). The results from loci E and F which carry a defect CRISPR promoter and lack a leader region, respectively (11), exhibited low transcript levels presumed to derive from internal promoters (Figure 5C). Northern blot analyses of selected crRNAs confirmed the enhanced levels of mature crRNAs in the Cbp1-overexpressed strain for locus A/spacer 28, locus B/spacer 24 and locus C/spacer 1 (Figure 5C).
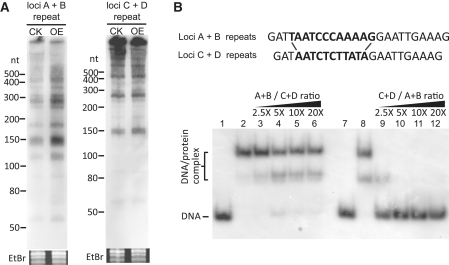
Furthermore, northern blotting using probes against the repeats, revealed that pre-crRNAs of loci A and B were more strongly induced than those of C and D (Figure 6A). We tested for the relative binding strength of Cbp1 to the two repeats, which differ in sequence at five positions and in length by 1 bp, in a competition experiment (Figure 6B). The results showed that the C + D repeat displaced Cbp1 from the A + B repeat consistent with the protein binding more strongly to the former repeat (Figure 6B). This in turn provides a rationale for the result seen in Figure 6A. Under normal cellular conditions, Cbp1 will preferentially saturate CRISPR loci C + D while Cbp1 overexpression will lead to the additional saturation of loci A + B, consistent with strongly enhanced crRNA yields observed for these loci.
Figure 6.
Effect of S. solfataricus Cbp1 on transcription of CRISPR repeats with different sequences. (A) Northern blot analysis of pre-crRNA transcribed from loci A and B, and from loci C and D using the repeat probes. CK—transformants carrying the empty vector and OE—overexpression of Cbp1. (B) Competition assays of Cbp1 with different CRISPR repeat sequences. Experimental conditions were as for Figure 1B. Lanes 1 and 2: [32P] 5′-end labelled CRISPR(C + D)-2r DNA alone and complexed with Cbp1, respectively. Lanes 7 and 8: [32P] 5′-end labelled CRISPR(A + B)-2r alone and complexed with Cbp1, respectively. Molar excesses of added unlabelled competitor DNA are indicated for lanes 3–6 and lanes 9–12.
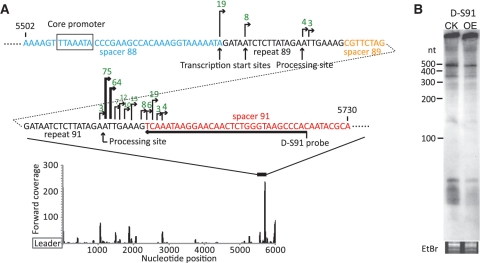
The major decrease in the level of transcripts observed for locus D/spacer 89 (Figure 5C) correlates with an exceptionally high level of transcripts observed in this region for the wild-type strain probably due to a strong promoter located in spacer 88 (Figure 7A). The northern blot result confirms that for the overexpression strain there is a decrease in the level of crRNA transcripts downstream from this site (Figure 7B). These results suggest that Cbp1 has a modulating effect on pre-crRNA transcription and does not simply enhance transcription but can also reduce transcription at abnormally highly transcribed internal CRISPR sites.
Figure 7.
The effect of Cbp1 on transcription from an internal promoter in CRISPR locus D of S. solfataricus P2. (A) Forward read coverage of CRISPR locus D and detailed results at sites with an exceptionally high level of 5′-ends. Only sites with three or more supporting sequence reads are shown. (B) Northern blot analysis of the locus D spacer 91 in the wild-type strain with empty vector (CK) and the Cbp1 overexpession strain (OE).
In addition to affecting transcription of pre-crRNAs, overexpression of Cbp1 also produced other minor transcriptional changes at time points T1 and T2 (Figure 5A) for a few genes most of which encode conserved hypothetical proteins (data not shown). The single exception was the sso1101 gene encoding a putative transcription regulator. Transcription was repressed ∼3-fold and 2-fold, respectively, at time points T1 and T2. Sso1101 is one of the few proteins that exhibits enhanced expression on biofilm formation for diverse Sulfolobus species (37).
Reverse-strand transcripts from CRISPR loci of S. solfataricus P2
In an earlier study, reverse-strand transcripts covering a range of sizes were detected from the five CRISPR loci of S. acidocaldarius by northern blot analyses (7,11). More recently, a high-coverage sequencing study of the transcriptome of S. solfataricus P2 was performed (31) and the raw data are publicly available (see ‘Materials and Methods’ section). 5′-ends of both leader and reverse-CRISPR transcripts, constituting either start or cleavage sites, were identified for the six CRISPR loci. Loci A–D carry strong leader promoters and most 5′-ends were located on the leader strand and correspond to cleavage sites within repeats. Loci E and F exhibit defective transcription from the leader and while locus E (seven spacers) showed very few 5′-ends, for the larger locus F (88 spacers) 5′-ends were distributed fairly evenly along both strands. Representative results for these two groups of loci are shown for loci C and F, respectively, in Figure 8A. Corresponding results for both leader and reverse strands of the other loci A, B and D are given in Supplementary Figure S2.
Figure 8.
Reverse-strand transcripts from loci C and F of S. solfataricus P2 (red peaks). Overexpression of Cbp1 did not affect opposite strand transcription of CRISPR loci. (A) Read coverage of CRISPR loci C and F, and the detailed results around the highly transcribed region of the reverse strand. Only sites with three or more supporting reads are shown. (B) Northern analysis of the high level reverse strand transcript (RT) in the wild-type strain with empty vector (CK) and Cbp1 overexpressed mutant (OE) for locus C (C-RT) and locus F (F-RT). Locations of the probed sequences are indicated in (A). Transcript lengths were estimated using RNA size markers.
5′-end locations and the number of reads with identical 5′-ends are shown for the two main peaks of reverse-strand transcripts, located distal from the leader region. They are associated with repeat–spacer 29 of locus C and with repeat–spacer 84 of locus F (Figure 8A). In order to gain some insight into the size of the reverse-strand transcripts generated and whether they are influenced by Cbp1 overexpression, northern blot analyses were performed by probing the transcripts in loci C and F adjacent to the illustrated start sites (Figure 8A). The results show discrete RNA bands of ∼100 and 170 nt for locus C and strong larger bands, while a range of band sizes >100 nt was observed for locus F (Figure 8B). Thus the increased expression of Cbp1 had no apparent influence on the size or yields of these reverse-strand transcripts.
DISCUSSION
We provide evidence for Cbp1 modulating transcription of CRISPR loci from the leader. We still know little about the stepwise processing of large CRISPR transcripts, and the mechanisms may vary for different CRISPR-based systems (11,23,24), Moreover, the yields of individual mature crRNAs are non-uniform (see, for example, Figure 7A). Nevertheless, the data presented are consistent with a model in which Cbp1 inhibits internal initiation and termination at putative archaeal transcriptional motifs located within spacers, and occasionally within repeats, of CRISPR loci. Such a function could be especially important for the acidothermophilic Sulfolobales and their genetic elements which carry A + T-rich (∼65%) genomes. The frequency of possible archaeal-specific promoter motifs (hexameric TATA-like sequences) and archaeal terminator motifs (T-rich pyrimidine sequences) is likely to be high (38,39). An estimate of the number of such motifs in the 4800 spacers contained in sequenced genomes of the Sulfolobales indicated that a large fraction carried potential promoter motifs and a smaller fraction of terminator motifs (3) and there is an additional promoter motif (ATTAAT) within the repeats of loci A and B of S. solfataricus P2. Even if most of these motifs are ineffective or weakly effective, collectively they could severely impede generation of long CRISPR transcripts of up to about 7000 nt. The demonstration by footprinting studies that Cbp1 partially protects the ends of the spacer regions (26) provides a potential mechanism for the transcriptional modulation. To test this model further, we plan to generate knockout mutants of the CRISPR pre-crRNA processing enzyme Cas6 and study the effect of Cbp1 on the formation of primary CRISPR transcripts.
There are no obvious precedents for this type of transcriptional modulation but there could be a mechanistic link to the eukaryotic THO complex consisting of four protein subunits. This complex has been implicated in inhibiting formation of aberrant DNA structures during transcription of genes containing repeat sequences which might otherwise impede polymerase progression or lead to increased recombination (40). This potential mechanistic similarity also suggests a possible secondary role for Cbp1. Repeat-bound Cbp1 could reduce the likelihood of recombination occurring between CRISPR repeats which might lead to deletions within CRISPR loci that do occur periodically (7,11) and can be induced by vectors carrying matching protospacers (16,41).
The cbp1 gene is not linked directly to the CRISPR-based gene cassettes in Sulfolobus chromosomes which suggests that it has other cellular functions. There is a precedent for this with the bacteria-specific RNase III endonuclease. It contributes to important cellular RNA processing functions but is also essential for processing of bacteria-specific type II CRISPR RNAs (42). A more general role for Cbp1 in inhibiting recombination between repeat sequences mentioned earlier is one possibility. Another is the potential link of Cbp1 to biofilm formation. Enhanced Cbp1 transcription produced a significant reduction in Sso1101 expression and this is one of very few proteins that are overexpressed during biofilm formation in diverse Sulfolobus species (37). If Cbp1 were to be overexpressed on viral infection biofilm formation might be inhibited in order to reduce mixing of uninfected with infected cells.
Cbp1 binds specifically to a range of similar repeat sequences associated with different CRISPR loci of Sulfolobus and it is likely that the more conserved repeat sequence at the distal end from the leader provides the main binding specificity (26, our unpublished data). Our results show that Cbp1 binds more strongly to the most common family I repeats of CRISPR loci C + D that dominate in the Sulfolobales and in other crenarchaea than to the less common family II repeats of loci A + B (11). Given that CRISPR/Cas and Cmr modules have been shown to exchange intercellularly for S. islandicus species (6), Cbp1 could influence which types of CRISPR loci are retained in the cell and also, explain the predominance of the family I repeats amongst the Sulfolobales (11).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1 and S2.
FUNDING
Danish Natural Science Research Council (grant no. 272-08-0391); Danish Research Council for Technology and Production (grant no. 274-07-0116); Danish National Research Foundation. Funding for open access charge: Danish Natural Science Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Shiraz A. Shah for helpful discussions on genome analyses and the referees for their critical insights.
REFERENCES
- 1.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011;14:1–7. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrett RA, Shah SA, Vestergaard G, Deng L, Gudbergsdottir S, Kenchappa CS, Erdmann S, She Q. CRISPR-based immune systems of the Sulfolobales – complexity and diversity. Bioch. Soc. Trans. 2011;39:51–57. doi: 10.1042/BST0390051. [DOI] [PubMed] [Google Scholar]
- 4.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo L, Brügger K, Liu C, Shah SA, Zheng H, Zhu Y, Wang S, Lillestøl RK, Chen L, Frank J, et al. Genome analyses of Icelandic strains of Sulfolobus islandicus: model organisms for genetic and virus-host interaction studies. J. Bacteriol. 2011;193:1672–1680. doi: 10.1128/JB.01487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah SA, Garrett RA. CRISPR/Cas and Cmr modules – mobility and evolution of an adaptive immune system. Res. Microbiol. 2011;162:27–38. doi: 10.1016/j.resmic.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Lillestøl RK, Redder P, Garrett RA, Brügger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grissa I, Vergnaud G, Pourcel C. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2008;36:145–148. doi: 10.1093/nar/gkn228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath P, Romero DA, Coûté-Monvoisin A-C, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrett RA, Shah SA, Vestergaard G. Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol. 2011;19:549–556. doi: 10.1016/j.tim.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Lillestøl RK, Shah SA, Brügger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 2009;72:259–272. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- 12.Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 2006;11:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence CM, Menon S, Eilers BJ, Bothner B, Khayat R, Douglas T, Young MJ. Structural and functional studies of archaeal viruses. J. Biol. Chem. 2009;284:12599–12603. doi: 10.1074/jbc.R800078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greve B, Jensen S, Brügger K, Zillig W, Garrett RA. Genomic comparison of archaeal conjugative plasmids from Sulfolobus. Archaea. 2004;1:231–239. doi: 10.1155/2004/151926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah SA, Hansen NR, Garrett RA. Distributions of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Trans. Biochem. Soc. 2009;37:23–28. doi: 10.1042/BST0370023. [DOI] [PubMed] [Google Scholar]
- 16.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manica A, Zebec Z, Teichmann D, Schleper C. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol. Microbiol. 2011;80:481–491. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- 18.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterisation of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 19.Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernández AL, Vázquez A, Olvera L, Gutiérrez-Ríos RM, Calva E, Hernández-Lucas I. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS and LRP in Salmonella enterica serovar Typhi. J. Bacteriol. 2011;193:2396–2407. doi: 10.1128/JB.01480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, Wurm R, Raine A, Mescher M, Van Heereveld L, et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 2010;77:1380–1393. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- 21.Skennerton CT, Angly FE, Breitbart M, Bragg L, He S, McMahon KD, Hugenholltz P, Tyson GW. Phage encoded H-NS: a potential achilles heel in the bacterial defence system. PLoS One. 2011;6:e20095. doi: 10.1371/journal.pone.0020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah SA, Vestergaard G, Garrett RA. CRISPR/Cas and CRISPR/Cmr immune systems of archaea. In: Marchfelder A, Hess W, editors. Regulatory RNAs in Prokaryotes. Vienna, New York: Springer; 2011. pp. 163–181. [Google Scholar]
- 23.Tang T-H, Bachellerie J-P, Rozhdestvensky T, Bortolin M-L, Huber H, Drungowski M, Elge T, Brosius J, Hüttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl Acad. Sci. USA. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang T-H, Polacek N, Zywicki M, Huber H, Brügger K, Garrett R, Bachellerie JP, Hüttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 25.Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:1–8. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng X, Brügger K, Shen B, Chen L, She Q, Garrett RA. Genus-specific protein binding to the large clusters of DNA repeats (Short Regularly Spaced Repeats) present in Sulfolobus genomes. J. Bacteriol. 2003;185:2410–2417. doi: 10.1128/JB.185.8.2410-2417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brügger K, Chen L, Stark M, Zibat A, Redder P, Ruepp A, Awayez M, She Q, Garrett RA, Klenk H-P. The genome of Hyperthermus butylicus: a sulphur-reducing, peptide fermenting, neutrophilic crenarchaeote growing up to 108°C. Archaea. 2007;2:127–135. doi: 10.1155/2007/745987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Develop. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurtzel O, Sapra R, Chen F, Zhu YW, Simmons BA, Sorek R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng L, Zhu H, Chen Z, Liang YX, She Q. Unmarked gene deletion and host-vector system for the hyperthermophilic archaeon Sulfolobus islandicus. Extremophiles. 2009;13:735–746. doi: 10.1007/s00792-009-0254-2. [DOI] [PubMed] [Google Scholar]
- 33.Bini E, Dikshit V, Dirksen K, Drozda M, Blum P. Stability of mRNA in the hyperthermophilic archaeon Sulfolobus solfataricus. RNA. 2002;8:1129–1136. doi: 10.1017/s1355838202021052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copié V, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J. Biol. Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortmann AC, Brumfield SK, Walther J, McInnerney K, Brouns SJ, van de Werken HJ, Bothner B, Douglas T, van der Oost J, Young MJ. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J. Virol. 2008;82:4874–4883. doi: 10.1128/JVI.02583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng N, Xia Q, Chen Z, Liang YX, She Q. An upstream activation element exerting differential transcriptional activation on an archaeal promoter. Mol. Microbiol. 2009;74:928–939. doi: 10.1111/j.1365-2958.2009.06908.x. [DOI] [PubMed] [Google Scholar]
- 37.Koerdt A, Orell A, Pham TK, Mukherjee J, Wlodkowski A, Karunakaran E, Biggs CA, Wright PC, Albers SV. Macromolecular fingerprinting of Sulfolobus species in biofilm: a transcriptomic and proteomic approach combined with spectroscopic analysis. J. Proteome Res. 2011;10:4105–4119. doi: 10.1021/pr2003006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torarinsson E, Klenk HP, Garrett RA. Divergent transcriptional and translational signals in Archaea. Environ. Microbiol. 2005;7:47–54. doi: 10.1111/j.1462-2920.2004.00674.x. [DOI] [PubMed] [Google Scholar]
- 39.Santangelo TJ, Cubonova L, Skinner KM, Reeve JN. Archaeal intrinsic transcription termination in vivo. J. Bacteriol. 2009;191:7102–7108. doi: 10.1128/JB.00982-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voynow V, Verstrepen KJ, Jansen A, Runner VM, Buratowski S, Fink GR. Genes with internal repeats require the THO complex for transcription. Proc. Natl Acad. Sci. USA. 2006;103:14423–14428. doi: 10.1073/pnas.0606546103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dyall-Smith M. Dangerous weapons: a cautionary tale of CRISPR defence. Mol. Microbiol. 2011;79:3–6. doi: 10.1111/j.1365-2958.2010.07451.x. [DOI] [PubMed] [Google Scholar]
- 42.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.