Abstract
In eukaryotes, the three families of ATP-dependent DNA ligases are associated with specific functions in DNA metabolism. DNA ligase I (LigI) catalyzes Okazaki-fragment ligation at the replication fork and nucleotide excision repair (NER). DNA ligase IV (LigIV) mediates repair of DNA double strand breaks (DSB) via the canonical non-homologous end-joining (NHEJ) pathway. The evolutionary younger DNA ligase III (LigIII) is restricted to higher eukaryotes and has been associated with base excision (BER) and single strand break repair (SSBR). Here, using conditional knockout strategies for LIG3 and concomitant inactivation of the LIG1 and LIG4 genes, we show that in DT40 cells LigIII efficiently supports semi-conservative DNA replication. Our observations demonstrate a high functional versatility for the evolutionary new LigIII in DNA replication and mitochondrial metabolism, and suggest the presence of an alternative pathway for Okazaki fragment ligation.
INTRODUCTION
Vertebrates contain three different DNA ligase species (1,2). LigI is thought to play a central role in DNA replication. It is targeted to DNA replication factories through interaction with proliferating cell nuclear antigen (PCNA) (3–5) and a human cell line with mutated LIG1 alleles (6,7), 46BR, from an immunodeficient patient, shows defects in Okazaki fragment joining but nearly normal overall growth characteristics. A LIG1 knockout mouse harboring homozygous deletions of the 3′-end of the LIG1 gene is embryonic lethal (8). However, mouse embryonic fibroblast (MEF) cell lines generated from such embryos show defects in Okazaki fragment joining but normal proliferation patterns (8,9). Therefore, the essential role of LigI in DNA replication is still under debate.
The second family of ligases found in all eukaryotes is that of LigIV. The main function of LigIV is the ligation step during the repair of DNA double strand breaks (DSBs) by the canonical non-homologous end-joining (NHEJ) pathway. Through breast cancer gene 1 carboxy terminal (BRCT) motifs, LigIV interacts with X-ray cross complementing 4 (Xrcc4) and becomes integrated in a NHEJ pathway (to be termed here D-NHEJ), which in vertebrates also comprises XRCC4-like factor (XLF), and the DNA-dependent protein kinase (DNA-PK) complex, consisting of the Ku heterodimer and the catalytic subunit, DNA-PKcs (10,11). Although in the mouse LIG4 knockout is embryonic lethal (12–14), this lethality can be rescued by concomitant loss of p53 function and LIG4−/−/p53−/− MEFs are highly radiosensitive and show defects in D-NHEJ (14–16).
The LigIII family is newer evolutionary and restricted to vertebrates (2). Nuclear and mitochondrial versions of LigIIIα are ubiquitously synthesized from LIG3 mRNA by an alternative translation initiation mechanism (17,18). In addition, germ-cell-specific alternative splicing of the LigIIIα 3′-coding exon generates LigIIIβ (18,19). Nuclear LigIIIα interacts with Xrcc1 and functions in the short-patch subpathway of BER (20), the repair of single strand breaks (21,22) and a NER subpathway (23). There is also evidence that LigIIIα is a component of an alternative pathway of NHEJ functioning as a backup (B-NHEJ) to D-NHEJ (24,25). Deletion of LIG3 has consequences significantly more severe than deletion of either LIG1 or LIG4 and attempts to generate LIG3 knockout cells or animals had failed until recently (26). Recent work shows that loss of mitochondria ligase function underlies this lethality and that viability of a LIG3 knockout is rescued by other eukaryotic ligases targeted to this organelle, or by expressing prokaryotic homologs (27,28). Although these observations indicate that LIG3 is dispensable, they leave open the question as to whether LigI or LigIV substitute for important LigIII functions, and vice-versa, as a result of unknown functional redundancy among eukaryotic DNA ligases. Here, we employ the chicken B cell line, DT40, and powerful conditional targeting approaches to investigate the role of the different DNA ligases in DNA replication and to study the inherent functional flexibility built by evolution into the vertebrate ligase system.
MATERIAL AND METHODS
Cell culture and electroporation
DT40 cells were grown at 41°C in a mixture of D-MEM/F12 growth medium supplemented with 10% fetal bovine serum, 1% chicken serum, 50 µM β-mercaptoethanol in a humidified incubator supplemented with 5% CO2. All cells were routinely maintained in the logarithmic phase of growth. Stable transfectants were selected in 15 µg/ml of blasticidin S, 1 µg/ml mycophenolic acid or 1 µg/ml of puromycin, as appropriate. Targeted clones were screened by polymerase chain reaction (PCR) according to Arakawa et al. (29).
Two methods of electroporation were used to introduce DNA into DT40 cells. In the first protocol, electroporation was carried out with cells suspended in complete growth medium, and 107 cells were electroporated using the GenePulser-Xcell (BIORAD) at 25 µF and 700 V. In the second protocol, 106 cells were electroporated with program 21 in Buffer B using commercially available equipment and protocols (Amaxa).
Parental cell line
Mutants described here were derived from the DT40-Cre1 cell line that conditionally expresses Cre recombinase to allow genome editing, and v-myb to enhance gene conversion (29,30). Cre recombinase is expressed from a human β-actin promoter as a chimera, MerCreMer, between a mutated estrogen receptor (Mer), only responding to tamoxifen or 4-hydroxytamoxifen (4HT) (31), and Cre recombinase (29,30). In the absence of tamoxifen, MerCreMer is efficiently sequestered by heat shock proteins in the cytoplasm. This interaction is rapidly disrupted upon administration of 4HT and causes the translocation of the protein to the nucleus, where Cre exerts recombination activity at loxP sites. Cells are also AID−/− and carry a deletion in the pseudo V locus of the non-rearranged Ig light chain locus. Furthermore, the non-rearranged V-intervening-sequence is replaced by a rearranged VL light chain gene together with DsRed as a hypermutation reporter. Thus, the full description of the cell line is DT40-AID−/−IgLdual; and is referred here as DT40. This genetic background extends the utility of the mutants generated here to studies of gene conversion and hypermutation through analysis of inactivating mutations in the DsRed gene and will be reported elsewhere.
Conditional gene knockout using an especially designed mutant 3loxP system
Three loxP targeting vectors were generated as outlined in Figure 1 and Supplementary Figure S1C–E by cloning 5′, middle and 3′ arms of the genes of interest into a plasmid vector, p3loxBsr. After genomic integration of these vectors, clones in which all 3 loxP sites were retained in the targeted locus were identified by PCR (see Supplementary Table 1 for a list of all oligos used for PCR genotyping as part of this work). To generate conditional knockout alleles, the 3loxP targeted clones were cultured with 20 nM 4-HT for 2 h and were subcloned by limited dilution in duplicate. Blasticidin-sensitive subclones were further screened by PCR to ensure retention of the two loxP sites.
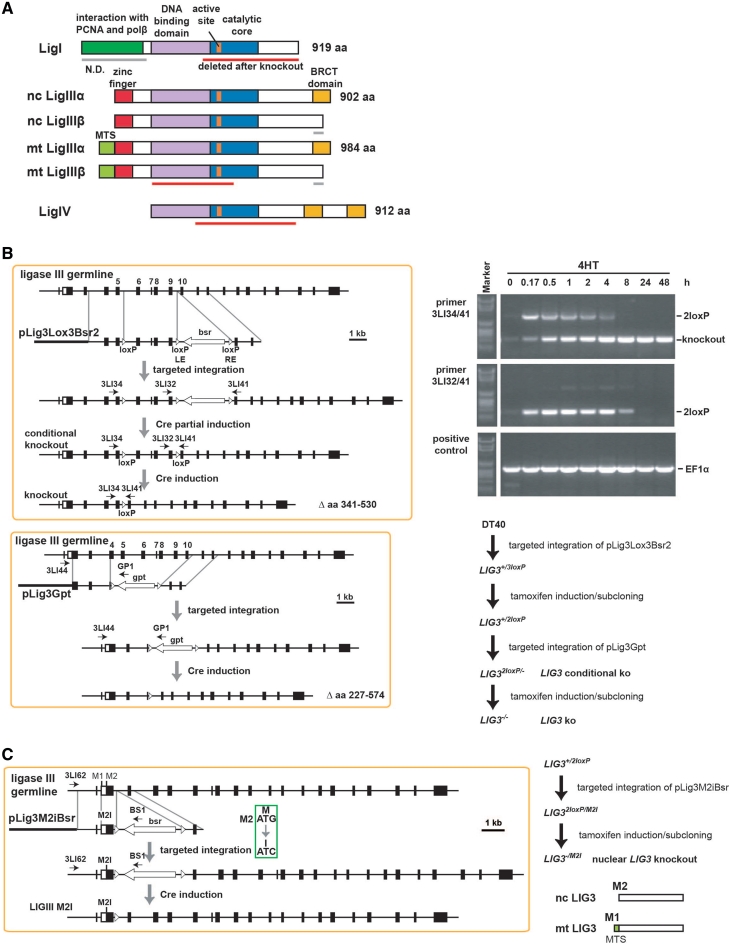
Figure 1.
Eukaryotic DNA ligases and targeting strategies. (A) Domain structure of chicken DNA ligases. Nc, nuclear; mt, mitochondrial. Red bars indicate regions deleted in the mutants generated. The β form of LigIII is inferred, as the corresponding exon could not be identified. (B) Vectors and approach taken to generate conditional and constitutive knockouts of DT40 LIG3 alleles. Gels show PCR products confirming genome editing steps. The steps followed to generate the indicated mutants are outlined in the lower right. (C) Approach to generate an allele expressing the mitochondrial version of LigIII.
Targeted integration of overexpression vectors
To ensure consistent expression of transgenes, we constructed an expression vector that can be targeted into a defined intergenic locus on chromosome 8, pChr8RsvLoxIresBsr. The vector integrates without destroying or grossly disturbing nearby genes. Transgene expression is driven by the Rous sarcoma virus (RSV) promoter and is followed by an internal ribosome binding site (IRES), the bsr selection marker and the SV40 polyA signal. The yeast LIG1 homolog, CDC9, the human LIG3α and the human M2I LIG3α genes were cloned into this vector and knocked into DT40 chromosome 8.
Targeting of LIG1, LIG3 and LIG4
The first LIG3 allele was targeted using the vector pLig3lox3Bsr2 shown in Figure 1B. One pair of loxP sites flanks the bsr gene and is placed between LIG3 exons 9 and 10, whereas a third loxP site is placed between exons 5 and 6. After stable integration of pLig3lox3Bsr2, clones resistant to blasticidin S were first selected and among those, clones with targeted integration identified by PCR. After removal of the selection marker using 4HT, the mutant LIG3+/2loxP was generated. The targeted LIG3 allele in this mutant has two loxP sites that allow the conditional removal of exons 6–9 upon treatment with 4HT; they encode the LigIII DNA binding domain and the 5′ half of the catalytic core including the active site (red bar in Figure 1A). The second allele of the LIG3 gene was knocked out in LIG3+/loxP using a vector deleting a segment starting from the middle of exon 4 and ending before exon 11 and inserting at the same time an in-frame stop codon into the exon 3, as well as the gpt (xanthine–guanine phosphoribosyl transferase) drug resistance cassette. Following selection in 1 µg/ml mycophenolic acid, targeted integration of pLig3Gpt was screened by PCR using primers 3LI44 and GP1. The preservation of the conditional knockout allele was checked in parallel using the primers 3LI34 and 3LI41. Limited exposure to 4HT allowed excision of the gpt drug resistance cassette, while the possible loss of the conditional allele was counter-selected by its lethal character.
DT40 mutants expressing the mitochondrial LigIII, were generated using the targeting vector pLig3M2IBsr (Supplementary Figure S1C). The vector introduces an inactivating mutation in the M2, while preserving M1, translation initiation site and is used to disrupt the second allele in LIG3+/2loxP. Following excision of the bsr drug resistance cassette from the second allele, a mutant is generated with one conditional allele and a second allele expressing constitutively only the mitochondrial form of LigIII (LIG32loxP/M2I).
Conditional targeting of the LIG1 gene was carried out in DT40 or LIG32loxP/− genetic background (Supplementary Figure S1D). In the bursal cDNA database, we identified an incomplete chicken LIG1 cDNA sequence encoding the 658 C-terminal amino acids. The amino acid sequence of chicken LigI derived from this cDNA, shared 66% identity and 81% similarity with the human protein. Therefore, we deduced the numbering of amino acids and exons of the entire chicken LIG1 locus from that of human and used it to construct appropriate targeting vectors. The first allele of LIG1 was targeted by the vector pLig1Lox3Bsr. The vector contains, in addition to the three loxP sites, a genomic fragment of LIG1 spanning exons 12–28. The Bsr drug resistance cassette spanned by two loxP sites is placed in the 3′-untranslated region of exon 28. Targeting of allele 1 with this vector and excision of the drug resistance cassette as described for LIG3, results in a conditional allele for LIG1. The second allele of LIG1 was targeted with the pLig1Puro targeting vector and was followed by recycling of the selection marker (Supplementary Figure S1D).
We employed a conventional knockout strategy to inactivate LigIV (Supplementary Figure S1E). The amino acid sequence of chicken LigIV has 75% identity and 85% similarity with the human enzyme. The chicken LIG4 gene does not contain introns and is composed of a single exon. The vector pLig4Bsr was therefore, designed to delete the entire coding sequence of one LIG4 allele. The vector pLig4Puro4 was designed to delete amino acids 185–614 of the second allele while inserting an in-frame stop codon after codon 184. Integration of this vector deletes the entire catalytic core including the LigIV active site. Following selection in 15 µg/ml blasticidin S and 1 µg/ml of puromycin, cells with both alleles disrupted were selected. Cre induction by tamoxifen allowed the removal of drug selection cassettes and the generation of LIG4−/− cells.
Reverse transcription reaction and real-time PCR
The RNA was prepared according to the protocol of the High Pure RNA Isolation Kit (Roche) with the exception that three million cells were used for total RNA isolation. RNA concentration was determined with a spectrophotometer (NanoDrop; Thermo Scientific).The cDNA was prepared from 1 µg total RNA by reverse transcription using the ‘Transcriptor First Strand cDNA Synthesis Kit’ (Roche) according to the manufacturer's instructions. This cDNA was used as input in real-time PCR reactions according to the protocol suggested in the LightCycler® FastStart DNA MasterPLUS SYBR Green I kit (Roche). Briefly, 1 µl of cDNA, 0.5 µl of sense and antisense primer solution, 2 µl of LightCycler® FastStart DNA MasterPLUS SYBR Green I Master Mix and 6 µl H2O were mixed together in a 10 µl reaction mixture in a LightCycler® Capillary. Capillaries were placed in the sample carousel of the LightCycler® (Roche). The settings for the thermal cycles were as follows: initial denaturation step 10 min at 95°C; 45 cycles of 10 s at 95°C, 5 s at 62°C and 10 s at 72°C. The primer sequences used were: Lig4-F: 5′-CCC CAT TAA CAG GCA GGA TA-3′; Lig4-R: 5′-CCA CGT TTG TCA GGC TTG TA-3′; TBP1-F: 5′-CAG CAC CAA CAG TCT GTC CA-3′; TBP1-R: 5′-GGG GCT GTG GTA AGA GTC TG-3′; Lig3-F: 5′-GAT GAC CCC AGT TCA GCC TA-3′; Lig3-R: 5′-GTG GGC TAC TTT GTG GGG AA-3′; Lig1-F7: 5′-CAT CTG CAA GAT AGG CAC TG-3′; Lig1-R7: 5′-CCC AAA TCG TCA CCA AAC AG-3′.
Analysis of cell cycle distribution by flow cytometry
Measurements of cell cycle distribution were carried out with an Epics XL-MCL (Beckman–Coulter) flow cytometer. Cells were collected by centrifugation and fixed in 70% cold ethanol for 24 h. Ethanol-fixed cells were spun down at 1500 rpm for 5 min and were resuspended in phosphate buffered saline (PBS) containing 40 µg/ml propidium iodide (PI) and 62 µg/ml RNase A. Samples were incubated at 37°C for 20 min and 20 000 cells were measured. The fraction of cells in the different phases of the cell cycle was calculated using the Wincycle® software.
Analysis of DNA replication by BrdU incorporation
Exponentially growing cells were incubated with 10 µM BrdU for 1 h and fixed in 70% ethanol. After washing with 0.9% NaCl, cell pellet was incubated for 10 min at 41°C in 0.5% pepsin. HCl solution (5.5 ml of 1 M HCl per 100 ml water). Subsequently, cells were then incubated in 2 M HCl for 20 min at room temperature and washed in 0.05% PBS–Tween 20 before incubation for 30 min at 4°C with an anti-BrdU antibody (1 : 50 dilution, Becton Dickinson) in the same solution. After washing with PBS–Tween 20 (0.5%)–bovine serum albumin (BSA) (1%), an fluorescein isothiocyanate (FITC) conjugated secondary antibody (1 : 100 dilution, Sigma) was applied for 30 min at 4°C in the same solution. Finally, cells were stained with PI and analyzed by flow cytometry. In the course of experiments using different DT40 mutants, we noticed mutation-dependent fluctuations among cells with S-phase DNA content in the fraction of those incorporating BrdU and therefore actively synthesizing DNA. For a quantitative evaluation of this effect, we defined the ‘Fraction of active S’ phase cells. The calculation of this parameter is graphically illustrated in Figure 4C (left) and consists of determining the fraction of BrdU positive cells in the region defined between the end of the G1 peak and the beginning of the G2 peak.
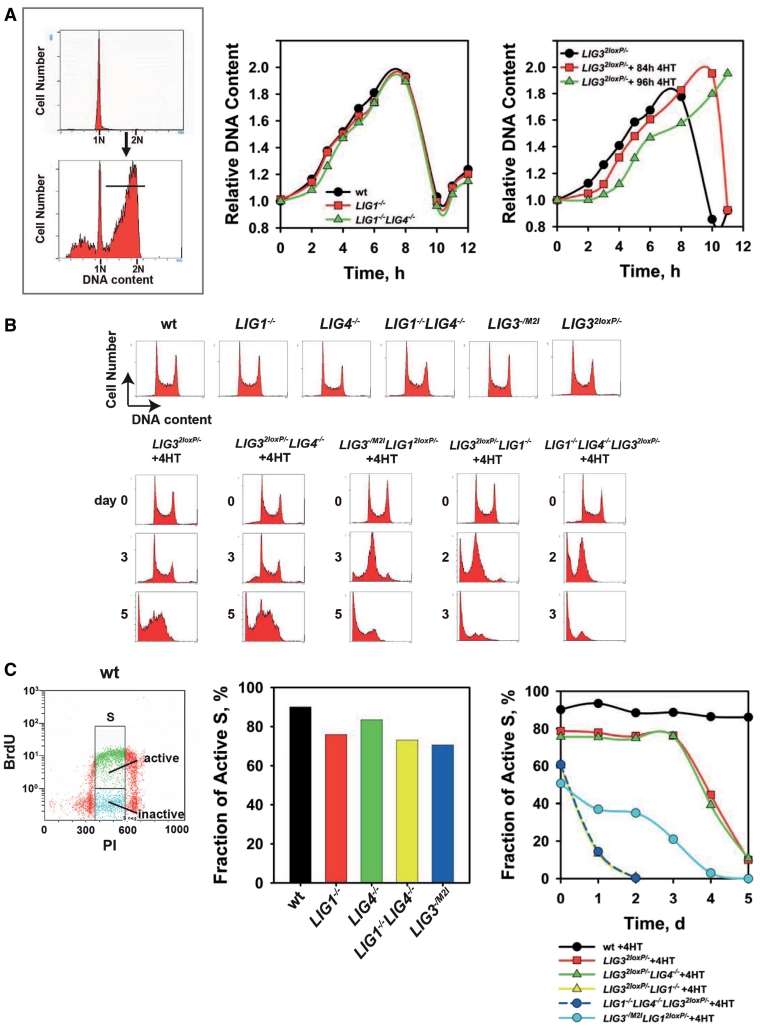
Figure 4.
Analysis of LigI and LigIII functions in DNA replication (A) The upper panel on the left shows a representative flow-cytometry histogram of G1-enriched cells obtained by centrifugal elutriation; the lower panel shows the same population 6 h after incubation at 41°C to allow progression through the cell cycle. The horizontal bar shows the subpopulation used to estimate progression through the cycle of the population median. Middle panel: progression through S-phase calculated by following the relative increase in DNA content for the median of the population described in A. Right panel: progression through S-phase of LIG32loxP/− cells treated with 4HT for 3.5 and 4 days. (B) Representative flow-cytometry histograms of the indicated mutants with or without 4HT incubation. (C) Left panel: representative dot plot of BrdU-labeled wt cells. The gates applied to calculate the active S-phase fraction are shown. Middle panel: fraction of actively replicating cells. Right panel: fraction of actively replicating cells at various times after treatment with 4HT.
Cell fractionation in the different phases of the cell cycle by centrifugal elutriation
For elutriation, cells were grown for 24 h from a concentration appropriately selected to reach ∼1.5 × 106 cells/ml. Cells (2–3 × 108) were collected and elutriated at 4°C using a Beckman JE-6 elutriation rotor and a Beckman J2-21M high-speed centrifuge at 25 ml/min (Beckman, Krefeld, Germany). Cells were loaded at 4500 rpm, and 250 ml fractions collected between 3400 and 2900 rpm at 100-rpm steps. Fractions of highly enriched cells in G1 were used for experiments. Cell-cycle analysis was carried out by flow cytometry as described above. For cell cycle analysis, cells were fixed in 70% ethanol and stained in PBS containing 62 mg/ml RNase A and 40 mg/ml propidium iodide at 37°C for 30 min.
Colony formation assay
For evaluation of colony forming ability DT40 cells were seeded in a medium containing 1.5% methylcellulose (MC) (Sigma, M0387). Cells were plated in 5 ml MC-growth medium in dilutions aiming at 100–200 colonies per dish, incubated for 10–14 days at 41°C and counted. To determine the surviving fraction after treatment with the poly-ADP-ribose-polymerase (PARP) inhibitor PJ-34, cells were plated in MC-growth medium containing PJ-34 at the indicated concentrations and were allowed to form colonies as described above. Typically, 200–1000 cells were plated aiming at 100–200 colonies per dish, depending on the mutant tested.
SDS–PAGE and western blotting
Protein gel electrophoresis under denaturating conditions was carried out using 10% polyacrylamide gels. For western blot analysis, proteins were transferred onto 0.2 µm Nitrocellulose or polyvinylidene fluoride (PVDF) membranes using an iBlot dry-transfer system (Invitrogen). Equal loading and transfer efficiency were monitored by Ponceau-S staining combined with immunodetection of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). After transfer, membranes were blocked with 5% non-fat dry milk in tris-buffered saline with Tween20 (TBS-T) (0.1% Tween 20, 150 mM NaCl, 25 mM Tris–HCl, pH 7.6) for 1–2 h at room temperature. Subsequently, membranes were incubated overnight at 4°C with primary antibody appropriately diluted in blocking buffer. After three washes for 10 min with TBS-T, membranes were incubated for 1 h with secondary antibody appropriately diluted in blocking buffer. Membranes were developed using the ECL+ chemiluminescence detection kit (GE Healthcare), as recommended by the manufacturer. Signals were visualized in a VersaDocTM imaging system (Bio-Rad) and analyzed using the ‘Quantity One’ software (Bio-Rad). The following primary antibodies were used: anti-Lig3 (1F3) mouse mAb (GeneTex); anti-GAPDH mouse mAb (Millipore). The secondary antibody was anti-mouse IgG conjugated with horseradish peroxidase (Cell Signaling).
Assay for Okazaki fragment ligation
To assay Okazaki fragment ligation, we modified an earlier described method (9). Briefly, human or mouse fibroblasts exponentially growing in 30 mm dishes were rinsed twice with warm serum-free medium and incubated for 10 min at 37°C in serum-free medium containing 2 µCi/ml [methyl-3H]-thymidine (25 Ci/mole). After this incubation, the growth medium was replaced with serum-free medium containing 2 mM thymidine and cells were collected for analysis after a further incubation at 37°C for different periods of time as indicated. Human or mouse cells were scraped in 1 ml ice cold PBS, pelleted by brief centrifugation and resuspended in 20 µl buffer A (10 mM Tris–HCl, pH 8.0, 50 mM NaCl, 0.1 M EDTA). Subsequently, 60 µl of molten 1.5% low-melting point agarose was added to the suspension, mixed and pipetted into cylindrical molds, which were immediately immersed in ice and kept for 5 min. DT40 cells were handled in a similar way, but because they grow in suspension, scraping was not required and incubation was at 41°C. The generated agarose plugs were incubated for 18 h at 50°C in 1 ml lysis buffer [buffer A supplemented with 2% (w/v) N-laurylsarcosine and 0.2 mg/ml proteinase K]. Subsequently, plugs were transferred to 5 ml buffer A for 1 h. Low molecular weight DNA that included 3H-labeled nascent DNA was electrophoresed under alkaline conditions in 1% agarose gels at 2 V/cm for 6 h at room temperature. After electrophoresis, the gel was soaked in neutralization buffer (1 M Tris–HCl, pH 7.6, 1.5 M NaCl) for 1 h and cut into 1 cm slices. Molecular weight markers run in parallel in the same gel allowed assignment of molecular weight range (0.2 to ≥23 kb) to each agarose slice. Slices containing DNA corresponding to different sizes were soaked in 0.1 M HCl for 1 h, melted by heating in a microwave oven and [3H] present was counted in a scintillation counter. The results are plotted as the percentage of cpm measured in agarose slices with DNA <2 kb compared with the total cpm in the corresponding lane.
Immunofluorescence microscopy
Approximately 106 cells were spun for 1 min at 1500 rpm on Poly-l-Lysine pre-treated coverslips using cytospin (Thermo Scientific) and fixed for 20 min in 2% paraformaldehyde. Fixed cells were washed with PBS, permeabilized for 5 min in solution P (100 mM Tris, 50 mM EDTA, 0.5% Triton X-100) and blocked overnight at 4°C in PBG solution (PBS, 0.5 % BSA, 0.2 % Gelatin).
To visualize γ-H2AX, coverslips were incubated for 90 min at room temperature with an anti-γ-H2AX mouse monoclonal antibody (JBW301, Upstate) diluted 1 : 200 in PBG solution. Coverslips were washed once in PBS and an anti-mouse IgG antibody, conjugated with AlexaFluor488 (Invitrogen) was added for 60 min at room temperature at 1 : 400 dilution in PBG solution. Cell nuclei were counter-stained for 30 min with DAPI (4′,6-Diamidin-2-phenylindol, 2 µg/ml, 0.1 M Tris, 0.1 M NaCl, 5 mM MgCl2, 0.05 % Triton X-100) and were washed once with PBS. Subsequently, coverslips were mounted on microscope slides using Prolong-Gold Antifade (Invitrogen). Samples were scanned with a ×40 objective in an automated analysis station equipped with a fluorescence microscope (Axio Imager Z2, Zeiss) and controlled by the Metafer software (MetaSystems). On average, 2500 cells were analyzed per sample using Metafer software. The DAPI intensity was used to distinguish between cells in G1 and G2 phases of the cell cycle. On average, the results of 850 G1 and 500 G2 cells per experiment were included to calculate the data shown here.
RESULTS
Targeting strategies and mutants
For an initial characterization of the DT40 LIG3, we carried out a phylogenetic tree analysis using available sequences and the neighbor joining method (Supplementary Figure S1A). As expected, LIG1 and LIG4 are found in all eukaryotes, whereas LIG3 is only found in vertebrates, including the chicken. Surprisingly, we uncovered a previously undetected LIG3 homolog in Drosophila. The six amino acids of the active site are highly conserved across species (Supplementary Figure S1B) and are essential for enzyme activity. Therefore, we constructed targeting vectors deleting a substantial portion of the enzyme core region, always including the enzyme active site (shown by red bars in Figure 1A).
Since blast algorithms failed to unambiguously define in the chicken LIG3 locus, the LigIIIβ-specific exon, we confined our studies to LigIIIα. We employed the Cre/LoxP system to develop the conditional gene targeting strategy summarized in Figure 1B. Thereby, we conservatively knocked-in three loxP-sites into one allele of the endogenous LIG3 gene and then excised only the blasticidin-S resistance cassette by limited dilution after a short treatment with 4-OH-tamoxifen (4HT). The generated allele contains two loxP sites, which flank LIG3 exons 6–9, encoding the DNA binding domain and the 5′-half of the catalytic core including the enzyme active site (Figure 1A). In the LIG3+/2loxP cells, the second allele is conventionally targeted as shown in Figure 1B (left lower panel) to generate the mutant LIG32loxP/− in which the conditional LIG3 allele can be excised by treatment with 4HT (Figure 1B, right upper and lower panels).
To separate the nuclear and mitochondrial functions of LigIII, we inactivated the M2 translation initiation site in the second allele of the LIG3+/2loxP mutant to generate LIG32loxP/M2I (Figure 1C). In this mutant, inactivation of the conditional LIG3 allele generates cells exclusively expressing mitochondrial (mt)LIG3 (LIG3−/M2I).
In order to generate complete LIG1 knockout in DT40, we decided to delete entirely the catalytic core of LIG1. This strategy minimizes problems arising from dominant negative effects potentially arising when truncated proteins are expressed from the targeted locus. Because we considered the possibility that LIG1 ‘null’ DT40 may not be viable, or may show only limited growth capacity, we employed conditional targeting strategies for LIG1 as well (Supplementary Figure S1C).
LIG4 was targeted using a conventional knockout strategy (Supplementary Figure S1D). Mutants defective in multiple ligases were generated for a comprehensive study of functional overlap and possible functional substitution between eukaryotic DNA ligases (Supplementary Figure S1E–G) (see Table 1 for a summary of mutants).
Table 1.
List of DT40 mutants generated and tested. For each mutant the Table shows its key features including viability and DNA replication proficiency
| Cell line | Feature | Viability | DNA replication |
|---|---|---|---|
| LIG12loxP/− | LIG1 conditional knockout | + | + |
| LIG1−/− | LIG1 knockout | + | + |
| LIG32loxP/− | LIG3 conditional knockout | + | + |
| LIG3−/−a | LIG3 knockout | − | − |
| LIG3−/M2I | nuclear LIG3 knockout | + | + |
| LIG4−/− | LIG4 knockout | + | + |
| LIG1−/− LIG4−/− | LIG1 knockout; LIG4 knockout | + | + |
| LIG32loxP/− LIG1−/− | LIG3 conditional knockout; LIG1 knockout | + | + |
| LIG3−/− LIG1−/−a | LIG3 knockout; LIG1 knockout | − | − |
| LIG32loxP /− LIG4−/− | LIG3 conditional knockout; LIG4 knockout | + | + |
| LIG3−/− LIG4−/−a | LIG3 knockout; LIG4 knockout | − | − |
| LIG3−/M2I LIG12loxP/− | nuclear LIG3 knockout; LIG1 conditional knockout | + | + |
| LIG3−/M2I LIG1−/−a | nuclear LIG3 knockout; LIG1 knockout | − | − |
| LIG3−/M2I LIG4−/− | nuclear LIG3 knockout; LIG4 knockout | + | + |
| LIG1−/− LIG4−/− LIG32loxP/− | LIG1 knockout; LIG4 knockout; LIG3 conditional knockout | + | + |
| LIG1−/− LIG4−/− LIG3−/−a | LIG1 knockout; LIG4 knockout; LIG3 knockout | − | − |
| LIG1−/− LIG4−/− LIG32loxP/M2I | LIG1 knockout; LIG4 knockout; nuclear LIG3 conditional knockout | + | + |
| LIG1−/− LIG4−/− LIG3−/M2Ia | LIG1 knockout; LIG4 knockout; nuclear LIG3 knockout | − | − |
| LIG32loxP/− Cdc9 | LIG3 conditional knockout overexpressing CDC9 (yeast LIG1) | + | + |
| LIG3−/− Cdc9 | LIG3 knockout overexpressing CDC9 (yeast LIG1) | + | + |
| LIG32loxP/− hLIG3α | LIG3 conditional knockout overexpressing human nuclear LIG3α | + | ND |
| LIG3−/− hLIG3αa | LIG3 knockout overexpressing human nuclear LIG3α | − | ND |
| LIG32loxP/− hM2I LIG3α | LIG3 conditional knockout overexpressing human mitochondrial LIG3α | + | ND |
| LIG3−/− hM2I LIG3α | LIG3 knockout overexpressing human mitochondrial LIG3α | + | ND |
aConditional allele deleted. Phenotype analyzed by 4HT treatment of parental cell line; ND: not determined.
LIG3, but not LIG1 or LIG4, is essential for DT40 survival
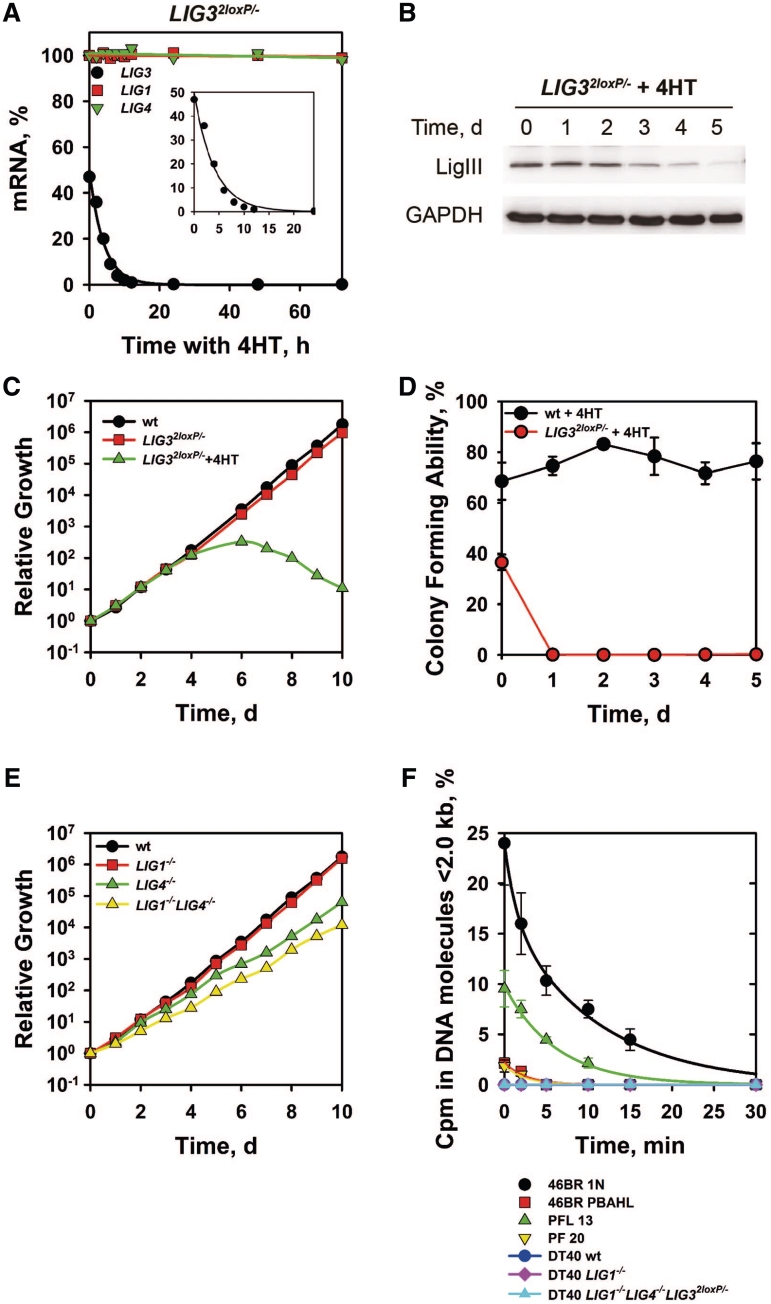
The mutant LIG32loxP/− converts to LIG3−/− by incubation with 4HT, which causes a rapid reduction of LIG3 mRNA (Figure 2A), from levels corresponding to 50% of wild-type (wt) to undetectable within 12 h. Deletion of LIG3 has no detectable compensatory effect on LIG1 or LIG4 mRNA levels (Figure 2A). LIG3 knockout is also associated with a marked reduction in LigIII protein after 3d (Figure 2B). Deletion of LIG3 has severe consequences for cell growth and viability with all cells dying after 5 days (Figure 2C) and the clonogenicity dropping to zero within 24 h (Figure 2D). We conclude that LIG3 is essential for survival in DT40.
Figure 2.
LIG3, but not LIG1 or LIG4, is essential for DT40 survival. (A) Determination of LIG3 mRNA level in LIG32loxP/− cells treated with 4HT. The mRNA levels of LIG1, LIG4 and LIG3 were measured by real-time PCR at different times after incubation with 4HT. (B) Western blot analysis of LigIII protein level in LIG32loxP/− cells treated with 4HT. A mouse monoclonal antibody, raised against human LigIII (clone 1F3), which efficiently recognizes the chicken LigIII was used. Protein loading was monitored with an anti-GAPDH antibody. (C and E) Growth kinetics of the indicated cell lines maintained in the exponential phase of growth by routine dilution in fresh growth medium. (D) Colony forming ability of wt and LIG32loxP/−DT40 cells as a function of time after incubation with 4HT. (F) Accumulation of Okazaki fragments in LigI-deficient human and mouse cells, as well as the indicated DT40 mutants. The graph shows the percentage of total radioactivity present in single-stranded DNA fragments <2.0 kb for each cell line.
In contrast to the observations following LIG3 knockout, LIG1 knockout generated by incubating with 4HT LIG12loxP/− cells, has no effect on cell growth (Figure 2E), despite rapid LIG1 mRNA depletion (Supplementary Figure S2A).
To examine whether LIG1 deletion is associated with more subtle defects in DNA replication, we analyzed Okazaki fragment ligation. Human 46BR-1N cells defective in LIG1 show the expected defect in Okazaki fragment ligation that is corrected in the LIG1 complemented counterpart, 46BR-PBAHL (Figure 2F). A similar, albeit 50% reduced, response is also detected in the LIG1−/− MEFs (PFL13), as compared with the wt (PF20). Surprisingly, an Okazaki fragment ligation defect is not detectable in LIG1−/− DT40. We conclude that in the chicken, the DNA replication functions of LigI can be assumed by LigIII or LigIV and be carried out with efficiency similar to LigI.
To investigate a possible function of LigIV in DNA replication, we compared the growth kinetics of LIG4−/− cells to those of the double mutant LIG1−/−LIG4−/−. Growth of LIG4−/− cells is slightly slower as compared with the wt, but growth of LIG1−/−LIG4−/− cells is not further affected (Figure 2E) and Okazaki fragment ligation remains intact in LIG32loxP/−LIG1−/−LIG4−/− cells (Figure 2F).
To examine whether in DT40 the deletion of LigI is associated with increased replication stress, we examined toxicity to the PARP inhibitor PJ-34. Cells of different genetic background were plated in MC-growth media containing the indicated drug concentrations and colony formation was evaluated. The results in Supplementary Figure S2B show similar PJ-34 toxicity, expressed as surviving fraction, for wt, LIG1−/− and LIG1−/−LIG4−/− cells.
As further test of replication stress in cells lacking LIG1, we analyzed γ-H2AX foci formation in non-irradiated DT40 cells of different genetic background. The results summarized in Supplementary Figure S2C show no increase in the number of foci per cell between wt, LIG1−/− and LIG1−/−LIG4−/− cells. Even separation between G1 and G2 cell data, and as direct consequence also of S-phase cell data, fails to uncover such trends (Supplementary Figure S2C). We conclude that in DT40, LigIII alone can efficiently substitute for all major DNA replication functions of LigI and can promote survival without detectable adverse consequences as a sole DNA ligase.
Synthetic lethality of LIG1 and nuclear LIG3 double knockout
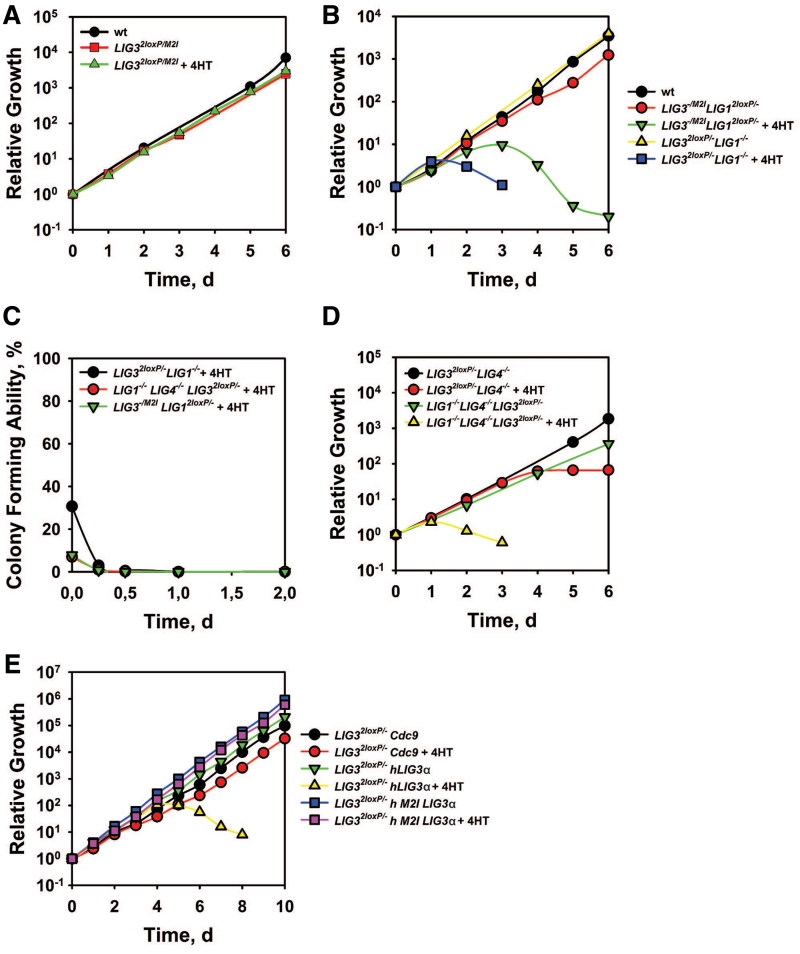
To examine whether nuclear LIG3 knockout is viable, we incubated LIG32loxP/M2I cells with 4HT. Notably, not only LIG32loxP/M2I, but also LIG3−/M2I cells are viable and grow as the wt (Figure 3A), despite reduced LIG3 mRNA levels (Supplementary Figure S2D). In LIG3−/M2I cells, LigI is the only remaining nuclear ligase with putative DNA replication functions and should therefore be essential for cell survival. We confirmed this postulate by incubating LIG3−/M2ILIG12loxP/− cells with 4HT and demonstrating cell death after 3 days (Figure 3B), associated with depletion in LIG1 mRNA levels (Supplementary Figure S2E). A role of LigI in DNA replication is also demonstrated by the practically immediate death of LIG32loxP/−LIG1−/−cells after incubation with 4HT (Figure 3B). Viability or lethality in the above mutants is also evident in parallel clonogenic assays (Figure 3C and Supplementary Figure S2F). We conclude that in the absence of nuclear LigIII, LigI is essential for DT40 survival and that there is functional overlap and substitution potential in DNA replication between LigI and LigIII.
Figure 3.
The role of mitochondria in DT40 cell viability. (A, B, D and E) Growth kinetics of indicated mutants in the presence or absence of 4HT. (C) Colony forming ability of indicated mutants incubated in the presence or absence of 4HT for the indicated periods of time.
Knockout of LIG4 does not further deteriorate the phenotype of LIG32loxP/− or of LIG32loxP/−LIG1−/− cells even after treatment with 4HT (Figure 3D), although the clonogenicity of the triple mutant is reduced (Figure 3C).
The mitochondria functions of LIG3 are essential for cell viability
The above results indicate that the lethality of LIG3−/− mutants is associated with defects in the mitochondrial function of the enzyme. To further examine this postulate, we generated LIG32loxP/− cells expressing the human nuclear and mitochondrial forms of LIG3α designed as previously reported (17) (Figure 1C), as well as the yeast LIG1 homolog, CDC9. LIG32loxP/− cells expressing nuclear hLigIIIα cease growing 4 days after treatment with 4HT despite strong expression of the protein (Supplementary Figure S3), whereas cells expressing the mitochondrial hLigIIIα are viable and grow in the presence of 4HT with kinetics similar to the untreated LIG32loxP/− cells (Figure 3E and Supplementary Figure S3). Notably, LIG32loxP/− cells expressing CDC9, which encodes for both nuclear and mitochondrial forms, are also viable in the presence of 4HT and show only a modest growth defect (Figure 3E). We conclude that the functions of the mitochondrial form of LigIII are essential for viability in DT40 cells, which is also in line with similar results in the mouse (27,28).
Analysis of LigI and LigIII function in DNA replication
DNA replication can be quantified by following progression into S-phase of G1 cells obtained by elutriation. This method minimizes the influence of dying cells, which generate ‘apparent’ reductions in growth kinetics. The flow-cytometry histograms (Figure 4A, left) allow estimates of cell cycle progression through comparison of the PI signal of the progressing-fraction-median to that of the initial G1 peak. G1 cells from LIG1−/−, LIG4−/− and LIG1−/−LIG4−/− cells complete the cell cycle with kinetics indistinguishable from the wt (Figure 4A middle). Similar results are obtained with untreated LIG32loxP/− cells (Figure 4A, right). On the other hand, G1 cells from LIG32loxP/− cell cultures incubated with 4HT for 84 h or 96 h show a delay in their progression through S-phase. The normal proliferation characteristics of LIG1−/−, LIG4−/−, LIG1−/−LIG4−/−, LIG32loxP/− and LIG3−/M2I mutants are associated with normal cell cycle profiles (Figure 4B), which is in line with the normal Okazaki-fragment ligation-characteristics presented above. After 4HT treatment, LIG3−/M2ILIG12loxP/− and LIG32loxP/−LIG1−/− cells show at first a severely reduced G2-fraction, probably reflecting problems in completing S-phase. At later times, cultures loose entirely the S-phase population and die by apoptosis as indicated by the sub-G1 population in the histograms. Knockout of LIG4 does not deteriorate this phenotype. We conclude that LIG1/LIG3-deficient cells die because of a defect in DNA replication that leads to apoptosis.
For a more detailed analysis, we measured DNA replication using two-parameter DNA/BrdU flow-cytometry. To compare the results obtained with the different mutants, we defined the parameter ‘Fraction of active S’-phase cells, which we calculated by dividing the number of BrdU-positive cells by the total number of cells, with both populations taken within the PI limits shown in Figure 4C (left).
The fraction of active S-cells is slightly decreased in LIG1−/− and LIG3−/M2I compared with wt cells (Figure 4C middle). LIG4−/− cells show a fraction of active S-cells similar to wt, while LIG1−/−LIG4−/− cells show a decrease compared with LIG4−/− that is similar in magnitude to that of LIG1−/− from the wt. The heterozygous state of LIG32loxP/− cells already causes a reproducible 10% drop in the fraction of active S-phase cells (Figure 4C, right); this effect is dramatically enhanced 3 days after deletion of LIG3 by 4HT incubation, and the fraction of active S-cells approaches zero at 5 days. The fraction of active S-cells in LIG3−/M2ILIG12loxP/− begins to decrease after 2 days and approaches zero at 4 days after treatment with 4HT. In LIG32loxP/−LIG1−/− cells, the fraction of active S-cells is 20% lower than in LIG32loxP/− cells and incubation with 4HT causes an immediate drop that reaches zero at 2 days. Knockout of LIG4 does not further deteriorate the phenotype of LIG32loxP/−LIG1−/− cells. These results suggest that LigIII has a function in DNA replication even in the presence of LigI. However, the strong apoptotic phenotype observed in DT40 cells depleted of LigIII prevents us from drawing firm conclusions. It remains possible that apoptosis induction even at mildly reduced levels of LigIII contributes to the generation of DNA replication inactive S-phase cells.
DISCUSSION
Our results demonstrate an impressive potential for LigIII to function as the primary ligase in DNA replication in the absence of LigI. Thus, a functional redundancy between LigI and LigIII in DNA replication is indicated. It is intriguing that such relationships developed only after the evolutionary appearance of LIG3, as LIG1 and LIG4 family members have well separated functions in lower eukaryotes. This functional flexibility is supported by the impressive substrate flexibility of LigIII (32).
The structural similarity between LigI and LigIII (33) offers a rationale for the functional overlap between the two enzymes. However, whereas LigIII fully supports Okazaki fragment ligation in DT40 cells (Figure 2F), this function is less efficiently supported in the human 46BR cell line (34); a significantly smaller but still discernible defect is observed in the mouse PFL13 cell lines (8) (Figure 2F). It is therefore, possible that species-specific differences in Okazaki fragment ligation underlie this effect. However, in human LigI-deficient cells, LigI protein encoding most of the catalytic core and other domains important for DNA replication is still expressed. Also, in the mouse knockout model some expression of LigI with catalytic core cannot be excluded. Therefore, it cannot be completely ruled out that dominant negative effects cause the observed defect in Okazaki fragment ligation, particularly in human cells, and that differences in the expression of such competing forms of LigI underlie the differences in the magnitude of the defect observed between mouse and human cells.
It is well established that LigI becomes incorporated in DNA replication centers through interaction with PCNA (3–5). It remains open how LigIII is incorporated in replication centers, but the interaction of the LigIII partner, XRCC1, with PCNA offers testable possibilities (35). It also remains to be established whether repair functions underlie Okazaki fragment ligation in LigI-deficient cells.
Similar to mouse systems (27,28), the lethality of LIG3 knockout can be averted in DT40 by expression of a mitochondria-specific enzyme (Figure 3A). Thus, whereas the nuclear-specific functions of LigIII are supported by other ligases, the mitochondria-specific functions are not. In other systems (28), cell survival for several generations is possible after LIG3 knockout. This raises the question as to why DT40 cells die after a reduction in LigIII levels. Mitochondria store apoptotic cell-suicide proteins (36), and it is possible that reduction in LigIII levels triggers their release causing thus the prompt apoptosis seen in this system.
Notably, the mitochondria-specific function of DT40 LigIII can be supported by the human mtLigIIIα. Even the LIG1 yeast homolog CDC9 rescues lethality suggesting that members of other ligase families also support mitochondria function if they carry an MTS. This has also been shown in mouse cells (27). Since in lower eukaryotes replication and maintenance of mitochondria DNA is part of LIG1 functions, it will be interesting to consider the evolutionary advantages that are associated with the transfer to LIG3 of this function in vertebrates.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
Bundes Ministerium für Bildung und Forschung (BMBF) (grants 02NUK001B and 02NUK005C). Funding for open access charge: BMBF.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs A. Lehmann and D. Melton for cells and Z. Storchova, G. Karras, G. Qin and T. Abe for suggestions on the manuscript.
REFERENCES
- 1.Lindahl T, Barnes DE. Mammalian DNA ligases. Annu. Rev. Biochem. 1992;61:251–281. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- 2.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardoso MC, Joseph C, Rahn H-P, Reusch R, Nadal-Ginard B, Leonhardt H. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J. Cell Biol. 1997;139:579–587. doi: 10.1083/jcb.139.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montecucco A, Savini E, Weighardt F, Rossi R, Ciarrocchi G, Villa A, Biamonti G. The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. EMBO J. 1995;14:5379–5386. doi: 10.1002/j.1460-2075.1995.tb00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes DE, Tomkinson AE, Lehmann AR, Webster ADB, Lindahl T. Mutations in the DNA ligase 1 gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69:495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 7.Webster ADB, Barnes DE, Arlett CF, Lehmann AR, Lindahl T. Growth retardation and immunodeficiency in a patient with mutations in the DNA ligase 1 gene. Lancet. 1992;339:1508–1509. doi: 10.1016/0140-6736(92)91266-b. [DOI] [PubMed] [Google Scholar]
- 8.Bentley DJ, Selfridge J, Millar JK, Samuel K, Hole N, Ansell JD, Melton DW. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammlian cell viability. Nat. Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- 9.Bentley DJ, Harrison C, Ketchen A-M, Redhead NJ, Samuel K, Waterfall M, Ansell JD, Melton DW. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J. Cell Sci. 2002;115:1551–1561. doi: 10.1242/jcs.115.7.1551. [DOI] [PubMed] [Google Scholar]
- 10.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 11.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 12.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng H-L, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 13.Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 14.Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Fergson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 15.Sekiguchi JM, Ferguson DO, Chen HT, Yang EM, Earle J, Frank K, Whitlow S, Gu Y, Xu Y, Nussenzweig A, et al. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc. Natl Acad. Sci. USA. 2001;98:3243–3028. doi: 10.1073/pnas.051632098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, Barnes DE, Lindahl T, McKinnon PJ. Defective neurogenesis resulting from DNA ligase IV deficiency requires Atm. Genes Dev. 2000;14:2576–2580. doi: 10.1101/gad.837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Jannotti RM, Klein SM, Bogenhagen DF. Two forms of mitochondrial DNA ligase III are produced in Xenopus laevis oocytes. J. Biol. Chem. 2001;276:48978–48987. doi: 10.1074/jbc.M107177200. [DOI] [PubMed] [Google Scholar]
- 19.Mackey ZB, Ramos W, Levin DS, Walter CA, McCarrey JR, Tomkinson AE. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell Biol. 1997;17:989–998. doi: 10.1128/mcb.17.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 21.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okano S, Lan L, Tomkinson AE, Yasui A. Translocation of XRCC1 and DNA ligase IIIα from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res. 2005;33:422–429. doi: 10.1093/nar/gki190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LHF, Fousteri MI. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III[alpha] in a cell-cycle-specific manner. Mol. Cell. 2007;27:311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 25.Simsek D, Brunet E, Wong SY-W, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puebla-Osorio N, Lacey DB, Alt FW, Zhu C. Early embryonic lethality due to targeted inactivation of dna ligase III. Mol. Cell Biol. 2006;26:3935–3941. doi: 10.1128/MCB.26.10.3935-3941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis A-K, Van Houten B, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, McKinnon PJ. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arakawa H, Hauschild J, Buerstedde J-M. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 30.Arakawa H, Lodygin D, Buerstedde J-M. Mutant lox P vectors for selectable marker recycle and conditional knockouts. BMC Biotechnol. 2001;1:7. doi: 10.1186/1472-6750-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Riesterer C, Ayrall AM, Sablitzky F, Littlewood TD, Reth M. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res. 1996;24:543–548. doi: 10.1093/nar/24.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomkinson AE, Mackey ZB. Structure and function of mammalian DNA ligases. Mutat. Res. 1998;407:1–9. doi: 10.1016/s0921-8777(97)00050-5. [DOI] [PubMed] [Google Scholar]
- 33.Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 34.Mackenney VJ, Barnes DE, Lindahl T. Specific function of DNA ligase I in simian virus 40 DNA replication by human cell-free extracts is mediated by the amino-terminal non-catalytic domain. J. Biol. Chem. 1997;272:11550–11556. doi: 10.1074/jbc.272.17.11550. [DOI] [PubMed] [Google Scholar]
- 35.Freie BW, Ciccone SLM, Li X, Plett A, Orschell CM, Srour EF, Hanenberg H, Schindler D, Lee S-H, Clapp DW. A role for the Fanconi anemia C protein in maintaining the DNA damage-induced G2 checkpoint. J. Biol. Chem. 2004;279:50986–50993. doi: 10.1074/jbc.M407160200. [DOI] [PubMed] [Google Scholar]
- 36.Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu. Rev. Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.