Abstract
Many cells and double-stranded DNA (dsDNA) viruses contain an AAA+ ATPase that assembles into oligomers, often hexamers, with a central channel. The dsDNA packaging motor of bacteriophage phi29 also contains an ATPase to translocate dsDNA through a dodecameric channel. The motor ATPase has been investigated substantially in the context of the entire procapsid. Here, we report the sequential action between the ATPase and additional motor components. It is suggested that the contact of ATPase to ATP resulted in its conformational change to a higher binding affinity toward dsDNA. It was found that ATP hydrolysis led to the departure of dsDNA from the ATPase/dsDNA complex, an action that is speculated to push dsDNA to pass the connector channel. Our results suggest that dsDNA packaging goes through a combined effort of both the gp16 ATPase for pushing and the channel as a one-way valve to control the dsDNA translocation direction. Many packaging models have previously been proposed, and the packaging mechanism has been contingent upon the number of nucleotides packaged per ATP relative to the 10.5 bp per helical turn for B-type dsDNA. Both 2 and 2.5 bp per ATP have been used to argue for four, five or six discrete steps of dsDNA translocation. Combination of the two distinct roles of gp16 and connector renews the perception of previous dsDNA packaging energy calculations and provides insight into the discrepancy between 2 and 2.5 bp per ATP.
INTRODUCTION
Most cells and dsDNA viruses contain at least one AAA+ (ATPases associated with diverse cellular activities) protein that possesses a common adenine nucleotide-binding fold. A typical characteristic of the AAA+ family is the coupling of chemical energy by the ATPase, derived from the adenosine triphosphate (ATP) hydrolysis to mechanical motion using force exerted on a substrate, such as dsDNA. This process usually requires a conformational change on the AAA+ protein that assembles into oligomers, often hexamers, forming a ring-shaped structure with a central channel (1–8).
Double-stranded DNA (dsDNA) viruses package their genomic dsDNA into a pre-formed protein shell, deemed procapsid, during maturation (9,10). This entropically unfavorable process is accomplished by a nanomotor which also uses ATP as an energy source (11–14). In general, the dsDNA packaging motor involves a protein channel and two packaging molecules, with the larger molecule serving as part of the ATPase complex and the smaller one being responsible for dsDNA binding and cleavage (11,15). Besides the well-characterized connector channel core, the motor of bacterial virus phi29 involves an ATPase protein gp16 (11,16–22) and a hexameric packaging RNA ring (15,23,24). In 1998, Guo et al. (15) first proposed that the mechanism of the intriguing viral dsDNA packaging motor resembles the action of other AAA+ proteins which form a hexameric ring to translocate dsDNA using ATP as an energy source [see discussion in Ref. (15)]. This motor is of particular interest to nanotechnology in that, it is both simple in structure and can be assembled in vitro using purified components. The elegant design of the 30-nm nanomotor, one of the strongest motors (25) assembled in vitro to date (26), has instigated the re-engineering of an imitative packaging motor for several applications. Previous reports indicate that phi29 nanomotor possesses packaging efficiencies up to 90% and the ability to switch off packaging through the addition of a non-hydrolyzable ATP derivative, γ-S-ATP (11,16,27). The latter attribute has enabled single-molecule measurements of motor velocities and force against an external load using an optical trap, contributing to the evidence of a stalling force up to 57 pN (25). About 20 years ago, Guo et al. (11) determined that one ATP was used to package 2 bp of dsDNA, and later, the same group demonstrated the sequential action of motor components (28). Recently, Moffitt and coworkers (29) confirmed the sequential action mechanism of motor components. They calculated that during this process each ATP that is hydrolyzed, led to about 2.5 bp of dsDNA translocation (29). Clarification of such discrepancy will help to illuminate the mechanism of motor action.
The packaging of 19.3-kb dsDNA into a confined procapsid is entropically unfavorable and requires a large amount of energy. The packaged dsDNA undergoes ∼30- to 100-fold decrease in volume as opposed to pre-packaging (30). Previous results suggest that ATPase activity of gp16 is dsDNA-dependent and may be stimulated by pRNA (20–22,31). It has also been shown that maximal ATPase activity was generated in the presence of all packaging components, including the procapsid and all its constituents (11,21,31).
The dsDNA packaging motor is well characterized in bacteriophage phi29, however gp16 (the ATPase protein) has long been an enigma. This protein tends to form aggregates in solution, which has negative consequences including the hindrance of the study and application of the protein, as well as contributing to contradictory data regarding ATPase activity, ATP/dsDNA/pRNA binding location and stoichiometry studies. Re-engineering techniques have both increased the solubility of this protein (17,18), as well as served to provide a fluorescent arm (32) that facilitates identification and application.
The mechanism of ATP hydrolysis is important and ubiquitous, but the mechanism of energy conversion from ATP hydrolysis to physical motion remains elusive. The mechanism in which gp16 uses ATP to drive the motor is still not well understood. However, it is well known that both pRNA and gp16 play roles in the packaging of 19.3 kb of gp3-dsDNA into a pre-formed procapsid during maturation. Surprisingly, we also found that gp16 alone is capable of fastening itself to dsDNA and releasing this dsDNA through ATP hydrolysis, independent of pRNA and the procapsid. Furthermore, the sequence in which the dsDNA binds to ATP was studied, revealed an important phenomenon in the packaging mechanism. Moreover, dsDNA passes through the portal protein during its translocation into the procapsid and it has been speculated that the channel plays a role in the process (33).
MATERIALS AND METHODS
Expression and purification of eGFP-gp16 in Escherichia coli
The engineering of Enhanced green fluorescent protein (eGFP)-gp16 was published by Lee et al. (32). eGFP-gp16 was expressed and purified as described previously (18,19,32) with minor modifications. Briefly, the protein was over-expressed in E. coli BL21(DE3) with induction of 0.4 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). The bacterial cells were harvested and resuspended in His-binding buffer (20 mM Tris–HCl, pH 7.9, 500 mM NaCl, 15% glycerol, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) and 0.1% Tween-20). The cells were then lysed by passing though French Press and the lysate was clarified by centrifugation. Then, 0.1% Polyethylenimine (PEI) was added to the clarified lysate to remove nucleotides and other proteins. Homogeneous eGFP-gp16 was purified by one-step Ni-resin chromatography.
Electrophoretic mobility shift assay
The samples were prepared in 20 μl buffer A (20 mM Tris–HCl, 50 mM NaCl, 1.5% glycerol, 0.1 mM Mg2+). Typically, 1.78 μM eGFP-gp16 was mixed with 7.5 ng/μl 40 bp Cy3-dsDNA at various conditions. The samples were incubated at ambient temperature for 20 min and then loaded onto a 1% agarose gel (0.5 Tris-Borate (TB): 44.5 mM Tris, 44.5 mM boric acid) for electrophoresis for 2 h under 80 V at 4°C. The eGFP-gp16 and Cy3-dsDNA in the gel was analyzed by fluorescent LightTools Whole Body Imager using 488 nm and 540 nm wavelengths for eGFP and cy3, respectively.
Forster resonance energy transfer
Forster resonance energy transfer (FRET) samples were analyzed using Horiba Jobin Yvon FluoroHub at excitation wavelength of 480 nm and the emission spectra was scanned from 500 to 650 nm with 5 nm slits at both excitation and emission. Samples were prepared in an appropriate cuvette volume (typically 50 µl) and allowed to incubate at room temperature for at least 5 min prior to excitation in order to allow reaction to fully catalyze.
Sucrose gradient sedimentation
Sucrose was diluted at 5 and 20% (w/v) using a dilution buffer (50 mM NaCl, 25 mM Tris pH 8.0, 2% glycerol, 0.01% Tween-20, 0.1 mM MgCl2) and a gradient was made using protocols established by BioComp Gradient Maker. Samples were subsequently gently added to the top of the gradient as not to disrupt the formed gradient, balanced and placed in Beckman Optima L Preparative Ultracentrifuge for 5.5 h, 35 000 rpm, 4°C. Samples were then fractionated directly from the bottom of the tube and analyzed by a Biotek Synergy 4 microplate reader at both GFP and cy3 wavelengths.
Kinetic assay
The conversion of 7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl)coumarin (MDCC) emission to an enzymatic kinetic equation has previously been reported (20).
Phage assembly activity inhibition and isolation of partially filled procapsids
The phage assembly assay has been previously described (34). To isolate the partially filled procapsid, γ-S-ATP was added to the packaging reaction buffer and the reaction was added to the top of a sucrose gradient and centrifuged for an extended period. The gradient was subsequently fractionated and variations of packaging components were again added to the individual fractions including ATP, adenosine diphosphate (ADP) or adenosine monophosphate (AMP). The fractions were then plated as performed in the phage assembly assay and tested for viral activity.
RESULTS
γ-S-ATP, a non-hydrolyzable ATP analog, promotes binding of gp16 to dsDNA
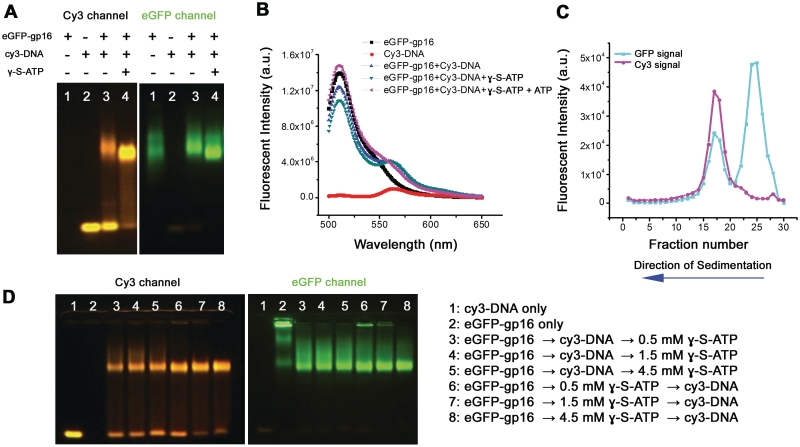
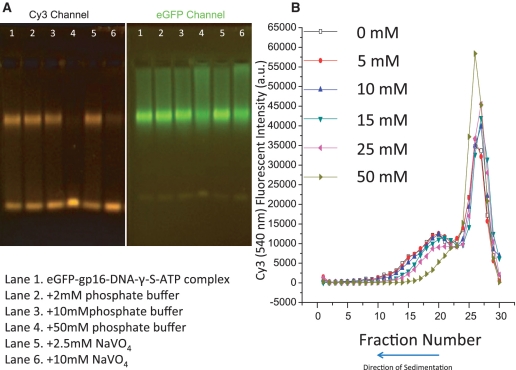
The conditions in which gp16 interacted with dsDNA were investigated. It was immediately discovered that the ATPase was capable of binding to dsDNA in the absence of pRNA and other motor components. An electrophoretic mobility shift assay (EMSA) was employed to study the interaction. Fusion of an eGFP tag at the N-terminus of gp16 did not affect its biological activity (32), but provided a fluorescent marker for detection. In Figure 1A, the binding between eGFP-gp16, dsDNA and γ-S-ATP was explored. Two different fluorescent filters, fluorescein isothiocyanate (FITC) and cy3, were used to visualize the protein and dsDNA, respectively. It was hypothesized that γ-S-ATP would lock gp16 onto dsDNA and our results proved this phenomenon. dsDNA was bound by eGFP-gp16 in the absence of the nucleotide (Figure 1A, lane 3). However, stronger binding of gp16 to dsDNA was observed when gp16 was incubated with γ-S-ATP (Figure 1A, lane 4). To further validate the finding, two different assays were utilized. The FRET analysis revealed an increase of energy transfer from eGFP-gp16 to Cy3-dsDNA when γ-S-ATP was included (Figure 1B, blue curve), to the sample in the absence of γ-S-ATP (green curve). FRET decreased significantly upon addition of excess ATP to the gp16/dsDNA/γ-s-ATP complex (teal curve). Furthermore, when γ-S-ATP was included in the mixture, sedimentation studies utilizing a 5–20% sucrose gradient revealed that gp16–dsDNA complex was highly prevalent as indicated by overlap in the eGFP and cy3 wavelength spectra (Figure 1C). These results suggested that the gp16/dsDNA complex is stabilized through addition of the non-hydrolyzable ATP substrate. Furthermore, the data suggested that gp16 possesses both a dsDNA binding domain and a motif to bind ATP.
Figure 1.
Demonstration of gp16 fastening to fluorescent dsDNA after incubation with non-hydrolyzable ATP derivative through (A) EMSA, (B) FRET and (C) sucrose gradient sedimentation; (D) EMSA to demonstrate efficiency of binding when ATP substrate is added before dsDNA.
ATP induced a conformational change in gp16 that led to increased binding affinity of gp16 to dsDNA
In order to continue with the previous findings, the mechanism of gp16 action in relation to ATP was studied. It has been extensively reported that gp16 is a dsDNA-dependent ATPase (11,20), that dsDNA stimulates the ATPase activity of gp16 (11,20,21) and that the phi29 dsDNA packaging motor uses one ATP to translocate 2 (11) or 2.5 (12,29) bp of dsDNA into the prohead. However, the interaction of gp16 with dsDNA or ATP and the sequential action of these three components was previously unidentified. In order to tackle this question, eGFP-gp16 was mixed with either dsDNA or γ-S-ATP and allowed to incubate. Subsequently, the remaining, missing component was added to the mixture and again allowed to incubate. All samples were subjected to an EMSA and imaged. The band representative of gp16 in complex with dsDNA appeared significantly sharper when γ-S-ATP was incubated first (Figure 1D, lanes 6,7 and 8) than when dsDNA was initially added (Figure 1D, lanes 3–5). FRET and gradient sedimentation also revealed the same phenomenon (data not shown). All these data support the speculation that gp16 binds first to γ-S-ATP to increase the binding affinity to dsDNA, and that γ-S-ATP is capable of fastening gp16 to dsDNA to form a more stable complex.
Gp16 departed from dsDNA following ATP hydrolysis
It has been reported that gp16 is a dsDNA-dependent ATPase of the phi29 dsDNA packaging motor (11,20–22,35), providing energy to the motor by hydrolyzing ATP into ADP and inorganic phosphate. For further applications, it is important to elucidate the mechanism in which ATP hydrolysis is related to the motion of motor components for the translocation of dsDNA.
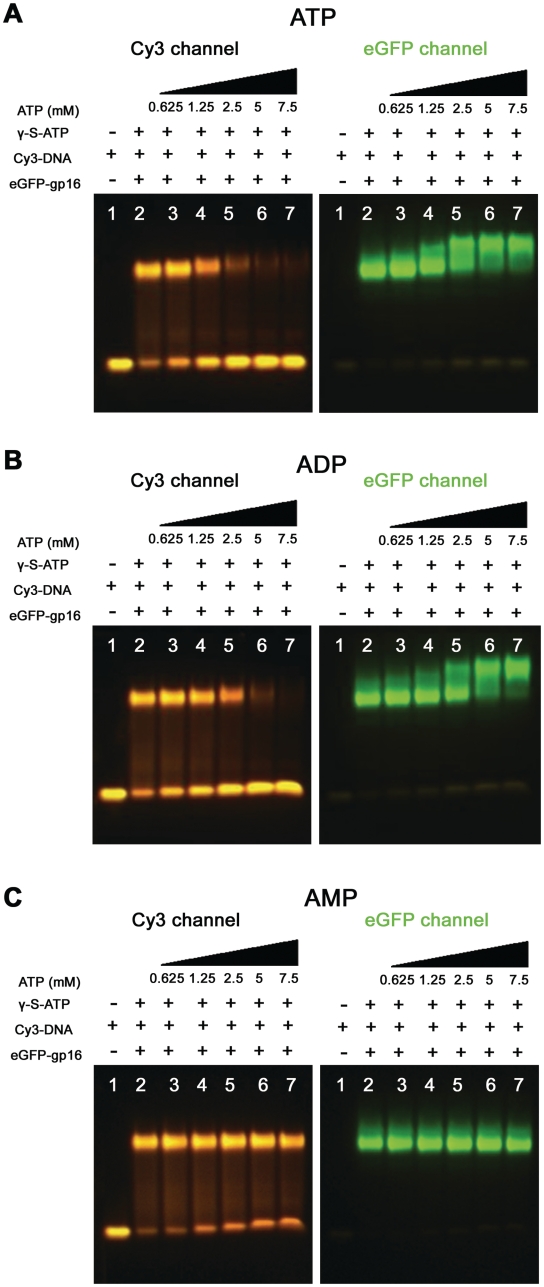
As aforementioned, γ-S-ATP stalled and fastened the gp16/dsDNA complex. It was subsequently found that hydrolysis of ATP led to the release of dsDNA from gp16 (Figures 2 and 3). When increasing amounts of ATP was added to the gp16/dsDNA/γ-S-ATP complex, the band representing the gp16/dsDNA complex disappeared (Figure 2A). ADP had a lesser effect on dsDNA release (Figure 2B), whereas AMP was unable to release dsDNA from gp16 (Figure 2C). This same phenomenon was observed in the previous FRET assay (Figure 1B, teal curve).
Figure 2.
EMSA showing release of dsDNA from gp16 after addition of increasing amounts of (A) ATP; (B) ADP had lower efficiency than ATP in the release of dsDNA; (C) AMP is unable to release gp16 from dsDNA.
Figure 3.
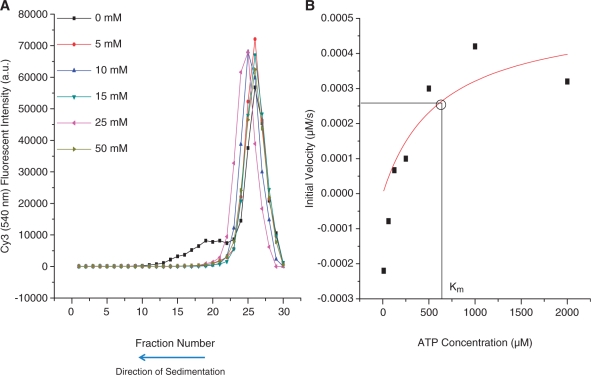
Effect of ATP on Gp16/dsDNA/γ-S-ATP complex. Complex was formed and subjected to increasing amounts of ATP and assayed by (A) sucrose gradient sedimentation and (B) fluorescent assay using a inorganic phosphate binding substrate to determine kinetics of ATP hydrolysis.
The release of dsDNA from gp16/dsDNA/γ-S-ATP complex by ATP and ADP was also demonstrated by sucrose gradient sedimentation. Binding of eGFP-gp16 to dsDNA was evidenced by a shift in the dsDNA profile in the presence of γ-S-ATP and in the absence of ATP (Figure 3A, black curve). With low concentrations of ATP however, gp16 and dsDNA existed as a free molecule in solution with slower sedimentation rates (Figure 3A, colored curves). To investigate the mechanism further, gp16/dsDNA/γ-S-ATP complex was purified using the sucrose gradient and subjected to an ATP hydrolysis kinetic assay. The hydrolysis of ATP to ADP and inorganic phosphate was confirmed by the demonstration that purified gp16/dsDNA/γ-S-ATP complex was able to hydrolyze ATP after addition of ATP to the purified complex (Figure 3B). Simply, a fluorescent molecule 7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl)coumarin-phosphate binding protein (MDCC-PBP) undergoes a conformational change after binding to inorganic phosphate which gives off fluorescence emission. The increase in fluorescence emission can be correlated to the hydrolysis of ATP to ADP with simple calculations. The results suggested that the hydrolysis of ATP led to the release of dsDNA from the gp16.
Evaluation of the findings on γ-S-ATP, ATP, ADP, AMP and gp16 interaction using the active ATP-driven dsDNA packaging motor
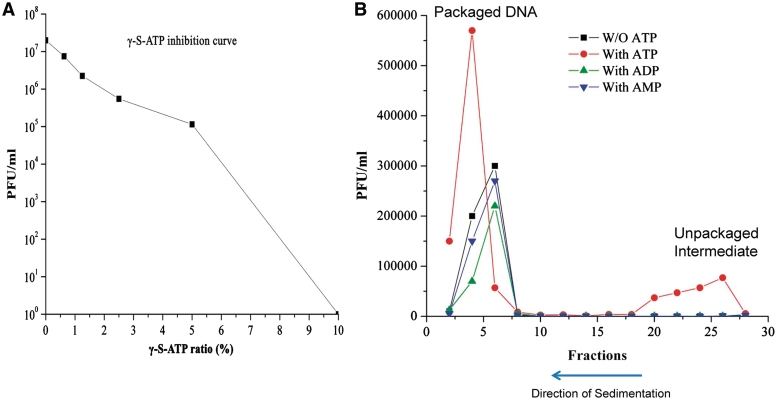
All previous experiments involving the interaction between gp16 and ATP or its derivatives were carried out in the procapsid-free system. Even though the previous experiments were derived from precursor proteins of the active motor, it is important to relate this interaction to an active motor involving the procapsid. A dsDNA packaging and viral assembly assay was performed in which purified motor components were added together and allowed to form an active virion in the presence and absence of γ-S-ATP, ATP, ADP, AMP and gp16 imitating their interaction in the procapsid-free system. When γ-S-ATP was added with an increased ratio to ATP, dsDNA packaging and viral assembly was gradually blocked (Figure 4A). The data agrees perfectly with that from the procapsid-free system showing that γ-S-ATP blocked the dissociation of dsDNA from gp16 (Figure 1A). The partially filled procapsids, incubated with optimal γ-S-ATP concentration, were then isolated in a sucrose gradient and subjected to addition of other motor components including ATP, ADP and AMP in the phage assembly assay. The partially filled procapsids were able to be converted into infectious virion when ATP was added (Figure 4B, red curve), agreeing with the data from the procapsid-free system in which ATP promoted the departure of dsDNA from the fastened gp16/dsDNA complex (Figure 2A). When ATP was replaced by ADP or AMP, active phages were not produced (Figure 4B), again in accordance with the data from the procapsid-free system showing that ADP and AMP did not allow easy departure of dsDNA from the gp16/dsDNA complex (Figure 2B and C). After adding further eGFP-gp16, pRNA, gp9-14 and ATP, the partially filled procapsids were able to recommence packaging and form an assembled, active bacteriophage.
Figure 4.
Isolation of the partially filled procapsid to determine effect of ATP, ADP and AMP on viral assembly. (A) Decrease of phage assembly activity by introduction of non-hydrolyzable ATP derivative. (B) Activity of isolated partially filled procapsids after sucrose gradient sedimentation.
Inorganic phosphate inhibited gp16 binding to dsDNA and elicited dsDNA discharge
Now that we have provided conclusive evidence that hydrolysis of ATP by gp16 to ADP and inorganic phosphate is the catalytic step leading to the translocation of dsDNA, the question remains as to which of the two resulting products, ADP and Pi, departs and which component remains bound to gp16? That is, does the gp16 conformational change result from a departure of ADP or Pi from gp16?
Inorganic phosphate is one of the products of the energy-producing reaction when ATP is hydrolyzed to ADP. A gel retardation assay revealed that in the presence of high concentrations of phosphate, dsDNA was released from the gp16/dsDNA/γ-S-ATP complex (Figure 5A, lane 4). It was also found that the presence of inorganic phosphate prevented the formation of gp16/dsDNA complex, either in the presence of γ-S-ATP or ATP. The gp16/dsDNA/γ-S-ATP complex was incubated with the phosphate analog sodium vanadate, which also proved to inhibit binding of gp16 to dsDNA (Figure 5A, lane 6). A sucrose sedimentation gradient assay revealed concurring results. In total, 50 mM phosphate was able to completely inhibit the formation of gp16/dsDNA complex or dissociate the complex into free gp16 and dsDNA, as evidenced by the shift of the dsDNA peak in the sucrose gradient (Figure 5B). It is interesting to find that ADP also stimulated the release of gp16 from the gp16/dsDNA complex, albeit with lower efficiency (Figure 2B). All these results questioned whether the inorganic phosphate by itself can compete with the ATPase center for ATP binding and whether the gp16/Pi or gp16/ADP complex remains as the final product after gp16 propels dsDNA forward.
Figure 5.
Effect of phosphate and phosphate derivative sodium vanadate on gp16/dsDNA/γ-S-ATP complex formation. Release of eGFP-gp16 from dsDNA complex by adding excess amounts of phosphate, assayed by EMSA (A) and sucrose gradient (B), as well as excess of phosphate derivative sodium vanadate (lanes 5 and 6 of A).
DISCUSSION
EMSA, sucrose gradient sedimentation, FRET and kinetic studies provided evidence to support the hypothesis that gp16 first interacts with ATP or the ATP analog γ-S-ATP and then with dsDNA. eGFP-gp16 was first incubated with either γ-S-ATP or short cy3-dsDNA and subsequently incubated with the third component, either dsDNA or γ-S-ATP (Figure 1D). The mixtures were then subjected to a low percentage agarose gel to evaluate the gp16/dsDNA complex. A significantly sharper cy3 band was detected corresponding to gp16/dsDNA complex in samples in which γ-S-ATP was added first. This data indicated that more gp16/dsDNA complex had formed under those conditions.
To further study the γ-S-ATP function in fastening gp16 to dsDNA, we carried out three different experiments. Gp16 was incubated with dsDNA and γ-S-ATP. The sample was then applied to a 5–20% sucrose gradient, fractionated and analyzed for eGFP (representing gp16) and cy3 (representing dsDNA) signal (Figure 1C). Profile-overlay analysis indicated that, in the presence of γ-S-ATP, gp16 tightly bound to dsDNA despite large centrifugal force and dilution factor. Similar samples from agarose gel electrophoresis revealed a much stronger gp16/dsDNA complex band when γ-S-ATP was present. Finally, FRET was applied to measure the energy transfer and distance between eGFP-gp16 (donor fluorophore) and cy3-dsDNA (acceptor fluorophore). Our data illustrated that higher energy transfer was present with the addition of γ-S-ATP, which provided indirect evidence that the protein formed a complex with dsDNA. In combination, these assays contributed to the principle that the motor complex can be stalled by addition of a non-hydrolyzable ATP substrate (16,20) but more importantly, they expanded our understanding of the phi29 packaging mechanism.
After gp16/dsDNA complex was formed through addition of γ-S-ATP, it was critical to understand what compounds were capable of dissociating gp16 from dsDNA. Again using an EMSA, gp16–dsDNA complex was allowed to form by incubating eGFP-gp16, cy3-dsDNA and γ-S-ATP together, but this time, adenosine monophosphate, adenonsine diphosphate and adenosine triphosphate were subsequently added before electrophoresis. Figure 2 clearly shows the concentration of tri-, di- and monophosphate at which the complex dissociates. ATP had the highest efficiency in promoting the kicking away of dsDNA from gp16, ADP had a lesser effect, but AMP had no effect. The same concept was used in Figure 3A in which gp16/dsDNA complex was pre-formed and subsequently incubated with varying concentrations of ATP. The samples were then added on top of a linear sucrose gradient and centrifuged for an extended period of time. In the absence of ATP, complex formed, but even in low concentrations of ATP, gp16 is released from its dsDNA substrate. Furthermore, a kinetic study was applied in which ATP was added to a gp16/dsDNA complex previously purified by a sucrose gradient. A fluorescent substrate was used to detect the release of inorganic phosphate in solution after ATP was hydrolyzed and the maximum velocity and Michaelis–Menten constant were calculated after varying concentrations of ATP were assayed. The curve is representative of all kinetic enzymes and again proves that ATP can be hydrolyzed and released even after gp16/dsDNA complex has formed.
To relate the above observed phenomena to a functional phage, an assay was performed in which the partially filled procapsids formed via incubation with γ-s-ATP was isolated in a sucrose gradient and subjected to a phage assembly assay with addition of either ATP, ADP and AMP. A similar assay has previously been performed (36). The results are consistent with the data from the procapsid-free system and provide indirect support for the conclusion that γ-S-ATP enhanced the binding of gp16 to dsDNA and that ATP hydrolysis promoted the departure of dsDNA from the gp16/dsDNA complex.
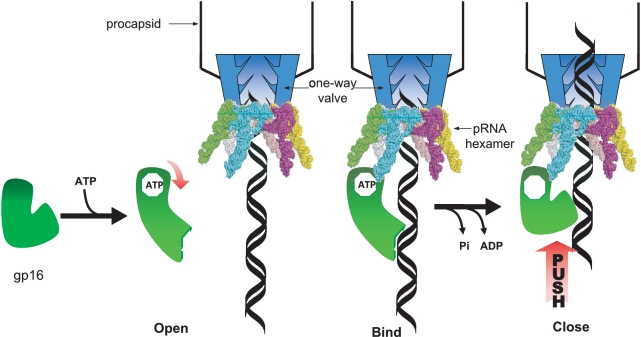
The dsDNA packaging mechanism is a universal biological phenomenon for dsDNA viruses including herpes viruses, pox viruses, adenoviruses and other dsDNA bacteriophages. The mechanism of packaging has provoked interest among virologists, bacteriologists, biochemists and especially researchers involved in nanotechnology; however, the actual mechanism remains elusive. In the past, many models have been proposed to interpret the mechanism of motor action including the (i) Gyrase-driven supercoiled and relaxation (37), (ii) force of osmotic pressure, (iii) Ratchet mechanism (38), (iv) Brownian motion (39), (v) 5- or 6-fold mismatch connector rotating thread (40), (vi) supercoiled dsDNA wrapping (41), (vii) sequential action of motor components (28,29), (viii) electro-dipole within central channel (42), (ix) connector contraction hypothesis (43,44) and (x) dsDNA torsional compression translocation mechanism (14,45). Based on our results, a new sketch has been developed to describe the mechanism of dsDNA packaging. We coined the ‘Push through a One-way Valve’ mechanism described as a combined effort between the gp16 ATPase which provides energy for pushing and the connector channel for one-way control. We believe that gp16 possesses at least four binding motifs for pRNA, dsDNA, adenosine and phosphate (the P loop). In our theory, at any given time, gp16 alone is able to bind to dsDNA but with low affinity. Upon binding to ATP or its derivatives (γ-S-ATP) however, gp16 undergoes a conformational change which promotes the binding to dsDNA. In this conformation, gp16 tightly binds to dsDNA in order to eliminate slipping in the packaging process. However, in order to generate energy, gp16 cleaves the gamma phosphate of ATP, producing a force from which gp16 switches to a relaxed conformation propelling the dsDNA unidirectionally into the procapsid using pRNA as a fulcrum. In this relaxed form, the binding site is unoccupied until a new ATP molecule is introduced to gp16 to restart the cycle (Figure 6).
Figure 6.
The ‘Push through a One-way Valve’ mechanism in phi29 dsDNA packaging. Schematic of dsDNA packaging mechanism termed ‘push through a one-way valve’. ATP binds to gp16, promoting gp16 binding to dsDNA. ATP hydrolysis induced a force or conformational change to push dsDNA translocation into the connector channel, which is a one-way valve that only allows dsDNA to enter but not exit the procapsid during dsDNA packaging.
In many packaging motors, the ATPase acts to rid itself of the phosphate but continues to clutch the ADP (12,46). The data shown in this report clearly shows that both ADP and phosphate can release gp16 from its substrate dsDNA. Our results suggest that ADP competes for the binding pocket better than inorganic phosphate, so it is assumed that gp16 has higher affinity for ADP than inorganic phosphate. This dictates that inorganic phosphate is expelled first, as observed in other phages, but also suggests that ADP is released from the pocket to allow the cycle to restart. The data is unable to clarify which expulsion step catalyzes the motor action, but that both steps are required to generate a new cycle.
Recently, our group discovered that the channel of phi29 dsDNA packaging motor exercises a one-way traffic mechanism of dsDNA translocation from the N-terminal external end to the C-terminal internal end, but blocked dsDNA to exit (47). Therefore, we concluded that phi29 dsDNA packaging went through a schematic marching mechanism via a unique mechanism by pushing through a one-way channel valve of the dsDNA packaging motor (Figure 6). The mechanism of providing force through a one-way valve agrees with the findings of Ray et al. (48,49) and Dixit et al. (50) in bacteriophage T4 that dsDNA was compressed if the portal entrance was blocked at the front end. The authors interpreted that the force for the compression is due to the torsional force from coiled dsDNA, but relates to our idea that dsDNA is rotated into the portal (15). Our suggested mechanism is also validated through the discovery in T4 in which it was determined that both ends of dsDNA remain in the portal of the procapsid during the packaging process (48,49). If the motor functioned by pulling dsDNA within the procapsid rather than pushing by gp16, one end of dsDNA would be required to be internalized for packaging to commence. Finally, this mechanism agrees with Bustamante et al. who clearly confirmed that dsDNA is processed by an unknown dsDNA-contacting component in one strand (62).
The stoichiometry of gp16 has not been fully addressed in this proposed mechanism, but in 1998, Guo et al.(15) proposed that the mechanism of dsDNA packaging is similar to the hexameric AAA+ ATPase family that has many functions but also acts to translocate dsDNA during dsDNA replication and repair. Many well-characterized dsDNA tracking motors (51–55) and other ATPases within the AAA+ family (15) possess an even-numbered protein structure. Furthermore, such phages as phi12 (56,57) and others have proved to possess a hexameric ATPase. Since the pRNA of phi29 has been determined to be hexameric (15,24,58), this raises speculation that gp16 might be similar to the AAA+ family and also exist as a hexamer.
Twenty years ago, Guo et. al.(11) determined that one ATP was used to package 2 bp of dsDNA. The stoichiometry of one ATP for two base pairs of dsDNA was also subsequently confirmed by the T3 system (59). This information has been utilized substantially by biochemists and biophysicists to interpret the mechanism of motor action(10,12,39,57,60,61). Recently, Bustamante and coworkers (29) reported the packaging of 2.5 bp per ATP using single molecule analysis through tweezer-based experiments. Many packaging models have previously been proposed, and the packaging mechanism has been contingent upon the number of nucleotides packaged per ATP. Currently, the motion mechanism is interpreted based on structural and biophysical properties of the dodecameric channel and the B-type dsDNA linking number of 10.5 bp per helical turn. It is logical that a specific number of ATP is required to translocate a definite number of dsDNA if gp16 and connector are an integrated, concrete motor structure. From our results, it was revealed that the dsDNA packaging task is carried out by two different steps by two separate components: gp16 for active pushing and the channel serving as a one-way valve to control the direction. Currently, the debate is whether 2 (11) or 2.5 (29) bp are packaged per ATP and whether the motor ATPase is a tetramer, pentamer or hexamer for four (61), five (50) or six (11) discrete steps of motor action. The calculation of ATP and dsDNA ratio related to the linking number of B-type dsDNA would have been useful to interpret the motor mechanism if only one motor protein, either gp16 or connector, plays a determinative role in dsDNA translocation speed. However, as reported here, the pushing force is from the ATPase gp16, but the dsDNA translocation speed is most likely also affected by the connector channel. Temporary pauses and slips have been reported to occur during translocation (12,25,62,63). The calculated translocation rate resulted from two uncoordinated force generating factors, gp16 and connector, will make it impossible to obtain a definitive and reproducible number of base pairs per ATP consumed. The translocation rate generated by gp16 is altered by the channel valve since the temporary pause or slip (25) of the dsDNA during translocation through the channel will negatively affect the speed. The newly demonstrated mechanism of dsDNA packaging demonstrated here can address the discrepancy between the 2 and 2.5 bp per consumed ATP debate. The difference depends on the experimental conditions that can be varied. Finding of the combination of the two distinct roles of gp16 and connector renews the perception of previous dsDNA packaging energy calculations and provides insight into the mechanism of motor action (Figure 6).
FUNDING
National Institutes of Health (grants GM059944, EB012135). Funding for open access charge: NIH (grants GM059944, EB012135).
Conflict of interest statement. PG is a co-founder of Kylin Therapeutics Inc, and Biomotor and Nucleic Acid Nanotechnology Dev, LTD.
ACKNOWLEDGEMENTS
We would like to thank Mathieu Cinier and Jia Geng for help in preparation of figures; Gian Marco De Donatis and Farzin Haque for their helpful insight.
REFERENCES
- 1.Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J. Struct. Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Frickey T, Lupas AN. Phylogenetic analysis of AAA proteins. J. Struct. Biol. 2004;146:2–10. doi: 10.1016/j.jsb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 3.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA plus ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 5.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Ann. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 6.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 7.Wang J. Nucleotide-dependent domain motions within rings of the RecA/AAA(+) superfamily. J. Struct. Biol. 2004;148:259–267. doi: 10.1016/j.jsb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo PX, Lee TJ. Viral nanomotors for packaging of dsDNA and dsRNA. Mol. Microbiol. 2007;64:886–903. doi: 10.1111/j.1365-2958.2007.05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 11.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage ϕ29. J. Mol. Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 12.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 13.Hwang Y, Catalano CE, Feiss M. Kinetic and mutational dissection of the two ATPase activities of terminase, the DNA packaging enzyme of bacteriophage lambda. Biochemistry. 1996;35:2796–2803. doi: 10.1021/bi952322z. [DOI] [PubMed] [Google Scholar]
- 14.Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys. J. 2007;93:L17–L19. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo P, Zhang C, Chen C, Trottier M, Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 16.Guo P, Peterson C, Anderson D. Initiation events in in vitro packaging of bacteriophage ϕ29 DNA-gp3. J. Mol. Biol. 1987;197:219–228. doi: 10.1016/0022-2836(87)90120-3. [DOI] [PubMed] [Google Scholar]
- 17.Huang LP, Guo P. Use of acetone to attain highly active and soluble DNA packaging protein gp16 of phi29 for ATPase assay. Virology. 2003;312:449–457. doi: 10.1016/s0042-6822(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 18.Huang LP, Guo P. Use of PEG to acquire highly soluble DNA-packaging enzyme gp16 of bacterial virus phi29 for stoichiometry quantification. J. Virol. Methods. 2003;109:235–244. doi: 10.1016/s0166-0934(03)00077-6. [DOI] [PubMed] [Google Scholar]
- 19.Lee TJ, Guo P. Interaction of gp16 with pRNA and DNA for genome packaging by the motor of bacterial virus phi29. J. Mol Biol. 2006;356:589–599. doi: 10.1016/j.jmb.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 20.Lee TJ, Zhang H, Liang D, Guo P. Strand and nucleotide-dependent ATPase activity of gp16 of bacterial virus phi29 DNA packaging motor. Virology. 2008;380:69–74. doi: 10.1016/j.virol.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibarra B, Valpuesta JM, Carrascosa JL. Purification and functional characterization of p16, the ATPase of the bacteriophage phi29 packaging machinary. Nucleic Acids Res. 2001;29:4264–4273. doi: 10.1093/nar/29.21.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimes S, Anderson D. RNA dependence of the bateriophage phi29 DNA packaging ATPase. J. Mol. Biol. 1990;215:559–566. doi: 10.1016/s0022-2836(05)80168-8. [DOI] [PubMed] [Google Scholar]
- 23.Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 24.Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 26.Guo P, Grimes S, Anderson D. A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage phi29. Proc. Natl Acad. Sci. USA. 1986;83:3505–3509. doi: 10.1073/pnas.83.10.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koti JS, Morais MC, Rajagopal R, Owen BA, McMurray CT, Anderson D. DNA packaging motor assembly intermediate of bacteriophage phi29. J. Mol. Biol. 2008;381:1114–1132. doi: 10.1016/j.jmb.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Guo P. Sequential action of six virus-encoded DNA-packaging RNAs during phage phi29 genomic DNA translocation. J. Virol. 1997;71:3864–3871. doi: 10.1128/jvi.71.5.3864-3871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Earnshaw WC, Casjens SR. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- 31.Shu D, Guo P. A Viral RNA that binds ATP and contains an motif similar to an ATP-binding aptamer from SELEX. J. Biol. Chem. 2003;278:7119–7125. doi: 10.1074/jbc.M209895200. [DOI] [PubMed] [Google Scholar]
- 32.Lee TJ, Zhang H, Chang CL, Savran C, Guo P. Engineering of the fluorescent-energy-conversion arm of phi29 DNA packaging motor for single-molecule studies. Small. 2009;5:2453–2459. doi: 10.1002/smll.200900467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng J, Fang H, Haque F, Zhang L, Guo P. Three reversible and controllable discrete steps of channel gating of a viral DNA packaging motor. Biomaterials. 2011;32:8234–8242. doi: 10.1016/j.biomaterials.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CS, Guo P. A highly sensitive system for the in vitro assembly of bacteriophage phi29 of Bacillus subtilis. Virology. 1994;202:1039–1042. doi: 10.1006/viro.1994.1434. [DOI] [PubMed] [Google Scholar]
- 35.Lee TJ, Schwartz C, Guo P. Construction of bacteriophage phi29 DNA packaging motor and its applications in nanotechnology and therapy. Ann. Biomed. Eng. 2009;37:2064–2081. doi: 10.1007/s10439-009-9723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shu D, Guo P. Only one pRNA hexamer but multiple copies of the DNA-packaging protein gp16 are needed for the motor to package bacterial virus phi29 genomic DNA. Virology. 2003;309:108–113. doi: 10.1016/s0042-6822(03)00011-4. [DOI] [PubMed] [Google Scholar]
- 37.Khan SA, Hayes SJ, Wright ET, Watson RH, Serwer P. Specific single-stranded breaks in mature bacteriophage T7 DNA. Virology. 1995;211:329–331. doi: 10.1006/viro.1995.1411. [DOI] [PubMed] [Google Scholar]
- 38.Serwer P. Models of bacteriophage DNA packaging motors. J. Struct. Biol. 2003;141:179–188. doi: 10.1016/s1047-8477(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 39.Astumian RD. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- 40.Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl Acad. Sci. USA. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grimes S, Anderson D. The bacteriophage phi29 packaging proteins supercoil the DNA ends. J. Mol. Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- 42.Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high- resolution structure of the bacteriophage phi29 connector particle. J. Mol. Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 43.Hou X, Yang F, Li L, Song Y, Jiang L, Zhu D. A biomimetic asymmetric responsive single nanochannel. J. Am. Chem. Soc. 2010;132:11736–11742. doi: 10.1021/ja1045082. [DOI] [PubMed] [Google Scholar]
- 44.Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J. Mol. Biol. 1995;245:635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- 45.Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage T4 terminase by DNA structure. J. Mol. Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lisal J, Kainov DE, Bamford DH, Thomas GJ, Jr, Tuma R. Enzymatic mechanism of RNA translocation in double-stranded RNA bacteriophages. J. Biol. Chem. 2004;279:1343–1350. doi: 10.1074/jbc.M309587200. [DOI] [PubMed] [Google Scholar]
- 47.Jing P, Haque F, Shu D, Montemagno C, Guo P. One-way traffic of a viral motor channel for double-stranded DNA translocation. Nano Lett. 2010;10:3620–3627. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010;398:224–232. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ray K, Ma J, Oram M, Lakowicz JR, Black LW. Single-molecule and FRET fluorescence correlation spectroscopy analyses of phage DNA packaging: colocalization of packaged phage T4 DNA ends within the capsid. J. Mol. Biol. 2010;395:1102–1113. doi: 10.1016/j.jmb.2009.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dixit A, Ray K, Lakowicz JR, Black LW. Dynamics of the T4 bacteriophage DNA packasome motor endonuclease VII resolvase release of arrested Y-DNA substrates. J. Biol. Chem. 2011;286:18878–18889. doi: 10.1074/jbc.M111.222828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hess H, Vogel V. Molecular shuttles based on motor proteins: Active transport in synthetic environments. J. Biotechnol. 2001;82:67–85. doi: 10.1016/s1389-0352(01)00029-0. [DOI] [PubMed] [Google Scholar]
- 52.Lowe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol. Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skordalakes E, Berger JM. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell. 2006;127:553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 54.Matias PM, Gorynia S, Donner P, Carrondo MA. Crystal structure of the human AAA+ protein RuvBL1. J. Biol. Chem. 2006;281:38918–38929. doi: 10.1074/jbc.M605625200. [DOI] [PubMed] [Google Scholar]
- 55.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 56.Lisal J, Tuma R. Cooperative mechanism of RNA packaging motor. J. Biol. Chem. 2005;280:23157–23164. doi: 10.1074/jbc.M502658200. [DOI] [PubMed] [Google Scholar]
- 57.Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 58.Xiao F, Zhang H, Guo P. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res. 2008;36:6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morita M, Tasaka M, Fujisawa H. DNA packaging ATPase of bacteriophage T3. Virology. 1993;193:748–752. doi: 10.1006/viro.1993.1183. [DOI] [PubMed] [Google Scholar]
- 60.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 61.Roos WH, Ivanovska IL, Evilevitch A, Wuite GJL. Viral capsids: mechanical characteristics, genome packaging and delivery mechanisms. Cell. Mol. Life Sci. 2007;64:1484–1497. doi: 10.1007/s00018-007-6451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu J, Moffitt J, Hetherington CL, Bustamante C, Oster G. Mechanochemistry of a viral DNA packaging motor. J. Mol. Biol. 2010;400:186–203. doi: 10.1016/j.jmb.2010.05.002. [DOI] [PubMed] [Google Scholar]