Abstract
The 5′-untranslated region (5′-UTR) of the genomic RNA of human immunodeficiency viruses type-1 (HIV-1) and type-2 (HIV-2) is composed of highly structured RNA motifs essential for viral replication that are expected to interfere with Gag and Gag-Pol translation. Here, we have analyzed and compared the properties by which the viral 5′-UTR drives translation from the genomic RNA of both human immunodeficiency viruses. Our results showed that translation from the HIV-2 gRNA was very poor compared to that of HIV-1. This was rather due to the intrinsic structural motifs in their respective 5′-UTR without involvement of any viral protein. Further investigation pointed to a different role of TAR RNA, which was much inhibitory for HIV-2 translation. Altogether, these data highlight important structural and functional differences between these two human pathogens.
INTRODUCTION
Most eukaryotic mRNAs contain a 5′-terminal m7GpppN cap structure (where N is any nucleotide) that is incorporated during transcription (1). Cap-dependent translation begins with the recognition of the 5′ cap structure by the eIF4F multimeric complex, which is composed of the cap binding protein eIF4E, the ATP-dependent RNA-helicase eIF4A and the scaffold protein eIF4G (2). Then, a 43S complex composed of the 40S ribosomal subunit and the tRNA initiator anchored to a set of initiation factors such as eIF2, eIF3, eIF1 and eIF1A is loaded onto the mRNA via multiple protein/protein and protein/RNA interactions. This results in the formation of the 48S pre-initiation complex, which scans the 5′-untranslated region (5′-UTR) until the initiation codon (generally an AUG) is encountered (2–4). Several features within the 5′-UTR such as, upstream AUGs, nucleotide composition and the presence of secondary structures have a strong impact in 48S pre-initiation complex recruitment and, consequently, translational efficiency (5–7).
More than 20 years ago, study of Picornavirus translation has defined an alternative mechanism of ribosome recruitment, which is mediated by cis-acting RNA elements called Internal Ribosome Entry Sites (IRES) (8,9). These IRES elements allow cap-independent internal binding of the 43S pre-initiation complex onto the mRNA molecule independently from the cap structure (10). To date, IRES have been found in many viral and cellular mRNAs and were shown to be functional during inhibition of cap-dependent translation (10,11).
Human immunodeficiency viruses types-1 and -2 (HIV-1 and HIV-2) belong to the Lentivirus genus of the Retroviridae family and contain a capped and polyadenylated positive-stranded RNA molecule as genome. This genomic RNA (gRNA) is used both as template for translation of Gag and Gag-Pol precursors and as viral genome for viral particle assembly (12). The 5′-UTR present within the genomic RNA of both HIVs is organized in highly structured RNA motifs that contain signals required for most of the viral processes including trans-activation of transcription (TAR), reverse transcription (PBS), splicing (SD), dimerization (DIS) and packaging (ψ) (13–16). Interestingly, while the presence of these signals is conserved between HIV-1 and HIV-2, the length and folding of the 5′-UTR of both viruses strongly differs (13–15). These differences are particularly obvious at the level of the structure of the trans-activation response element (TAR), which is organized in a single stem–loop in HIV-1, while it is much longer and folded in a forked structure in HIV-2 (13,16–23). Although the TAR RNA element of HIV-1 was shown to impair ribosome entry at the 5′-end in cell-free in vitro systems, it was recently shown that ribosomal scanning along the 5′-UTR can take place in cultured cells (24–27). However, protein synthesis from the HIV-1 can also occur via an IRES element that functions under specific conditions such as G2/M phase of the cell cycle or during cellular stress (28–30). Interestingly, IRES elements have also been mapped within the Gag coding region of HIV-1 and HIV-2 indicating that genomic RNA translation is a complex process tightly controlled by different RNA motifs (31,32). In HIV-2, the Gag coding region was shown to contain three independent IRESes able to recruit three independent pre-initiation complexes in the complete absence of the 5′-UTR (32–34). This ability relies in the capacity of RNA structures present within the Gag coding region to directly recruit the 40S ribosomal subunit and eIF3 (35). Despite the fact that an IRES was also identified within the HIV-1 Gag coding region, the molecular mechanism of ribosome recruitment by this RNA element has not been investigated yet (31).
Given the strong differences in the structure of the 5′-UTR of both human immunodeficiency viruses and the ability to drive ribosome recruitment via multiple mechanisms, we have analyzed the translational properties of both gRNAs in cultured cells. Results presented here show that the HIV-2 5′-UTR and specifically the TAR RNA structure slow translation resulting in low levels of Gag production. This sharply contrasts with protein synthesis from the HIV-1 gRNA, which occurs very efficiently despite the presence of a structured 5′-UTR.
MATERIALS AND METHODS
DNA constructs
pNL4-3-Renilla construct was obtained by inserting a HA-tagged Renilla luciferase-coding region in the SpeI site present in the Gag coding region (1507–1512) of the infectious HIV-1 molecular clone pNL4-3 (36). Similarly, pRod10-Renilla was obtained by inserting the HA-tagged Renilla luciferase-coding region in the HindIII site present in the Gag coding region (1457–1462) of the infectious molecular clone pRod10 (37). All pRenilla-derived constructs were obtained by inserting the selected region between the PvuII–BamHI sites of the pRenilla vector as was previously described (38–40). The 5′-UTR and the 5′-UTR-Gag sequences were obtained by PCR using the pNL4-3 and pROD10 proviral DNA as template (36,37). The human pRenilla-globin vector was previously described (40). cDNA sequences for the human GAPDH, NADH and cyclin D2 5′-UTR were obtained by RT–PCR using HeLa cells total RNA as template. The Line-1 5′-UTR was obtained from the pJM101f L1.3 vector (kindly provided by Dr Gael Cristofari, Université de Nice, France). HIV-1/HIV-2, HIV-2/HIV-1 and HIV-2 spliced constructs were obtained by a two-step PCR strategy. All constructed clones were verified by sequencing.
In vitro transcription
In vitro transcription was carried as previously described (38–40). Briefly, pRenilla-derived vectors containing a 100 nt long poly(A) tail were linearized by EcoRI digestion. Capped mRNAs were obtained using 1 µg of linear DNA template; 80 U of T7 RNA polymerase (Promega); 40 U of RNAsin (Promega); 10 mM of rCTP, rUTP, rATP; 0.36 mM rGTP; 30 mM DTT in transcription buffer [40 mM Tris–HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine and 10 mM NaCl] and 1.24 mM of m7GpppG cap analogue (New England Biolabs). Transcription reaction was carried out at 37°C for 2 h and mRNAs were DNase treated and precipitated with 1 volume of ammonium acetate 5 M (2.5 M final concentration). The integrity of mRNAs was checked on agarose gel electrophoresis and their concentration was quantified by spectrophotometry at 260 nm using Nanodrop (NanoDrop Technologies, Wilmington, DE, USA).
Cell culture, DNA and RNA transfection
T-lymphocytes (Molt-4 cells) were maintained in RPMI growth media (Gibco, BRL) supplemented with 10% FCS and 1% l-glutamine at 37°C in a 5% CO2 atmosphere. Cells were transfected with proviral-Renilla DNAs using the GeneJuice® Transfection Reagent (Novagen, EMD Chemicals Inc.) as indicated by the supplier. Cells extracts were prepared at 24 h post-transfection and used either for Renilla activity or RNA extraction and RT–qPCR. HeLa cells were maintained in DMEM growth media (Gibco, BRL) supplemented with 10% FCS at 37°C and 5% CO2. DNA transfection was carried out using JetPEI™ (PolyPlus Transfection) following supplier's indications. Cells or supernatants were recovered at 24 h post-transfection and used either for Renilla activity, RNA extraction and RT–qPCR, reverse transcriptase (RT) assay or western blot. For RNA transfection, HeLa, Cos7 and 3T3 cells were maintained in DMEM growth media (Gibco, BRL) supplemented with 10% FCS at 37°C and 5% CO2. PBMCs-derived human macrophages were obtained and maintained as previously described (41). mRNA transfection was carried out using Mirus mRNA transfection system (Mirus Biosciences) as previously described (39). mRNA-transfected cells were incubated for 3 h at 37°C, in a 5% CO2 atmosphere prior to Renilla activity analysis. When appropriate, cells were treated with 0.5 mM sodium arsenite for 30 min at 37°C and 5% CO2 and cells extracts were prepared for western blot analysis.
RT assay
Supernatants from HeLa cells transfected with proviral DNA were spin down at 10 000 rpm for 30 s to eliminate cellular debris and then ultracentrifugated in a Beckman TL-100 rotor at 75 000 rpm during 1 h at 4°C. Viral particles were resuspended in RT buffer [60 mM Tris–HCl (pH 8.0), 180 mM KCl, 6 mM MgCl2, 0.6 mM EGTA (pH 8.0), 0.12% Triton X-100, 6 mM DTT, 12 µg/ml poly rA and 6 µg/ml oligo dT] and RT activity was determined as described previously (42).
In vitro translation and complementation assays
All mRNAs were translated in 50% of Untreated Rabbit Reticulocyte System (Promega Co., Madison, WI, USA) in the presence of KCl (75 mM), MgCl2 (0.5 mM), 20 µM of each amino acid for 30 min at 30°C as previously described (40). When corresponded mRNAs were pre-incubated for 5 min at 37°C with hypotonic buffer [10 mM HEPES-KOH (pH 7.6); 10 mM KOAc; 0.5 mM MgOAc and protease inhibitor cocktail (Roche)] or HeLa cells extracts corresponding to 15 µg of total protein.
Renilla activity
Renilla activity was measured using the Renilla Luciferase Assay System (Promega Co., Madison, WI, USA) in a Veritas Luminometer (TurnerBiosystems) as previously described (39).
Gag-Renilla-specific activity analysis
HIV-1 and HIV-2 Gag–Renilla fusion proteins were expressed in the Flexi® Rabbit Reticulocyte System (Promega Co., Madison, WI, USA) as described above in the presence of 0.6 mCi of [35S]-methionine (Perkin Elmer) and Renilla activity was measured as indicated above. Equivalent Gag–Renilla activities were precipitated with 100% TCA (1 volume TCA for 4 volumes of protein) for 10 min on ice and washed three times with acetone. TCA-precipitated radioactivity was quantified using the FLA-5000 PhosphorImager (Fujifilm).
Western blot analysis
Cells extracts were subjected to 10% SDS–PAGE and transferred to PVDF membrane (Amersham Biosciences, UK Ltd). The membrane was blocked for at least 2 h with 5% dried milk in TBST 1× and incubated overnight at 4°C with a 1/1000 dilution of antibodies directed against SIVmac Gag (43), eIF2α-P (US Biological) or actin (Santa Cruz Biotechnologies). After three washes with TBST 1×, an antirabbit horseradish peroxidase (HRP)-coupled secondary antibody was added and incubated for 1 h at RT. The membrane was washed three times with TBST and HRP was revealed by using SuperSignal® West Pico Chemiluminiscent Substrate (Pierce, Rockford, Il, USA).
RNA extraction and RT–qPCR
RNA extraction and RT–qPCR were performed exactly as previously described (44). Briefly, HeLa cells transfected with Renilla based in vitro transcribed mRNAs were washed intensively with PBS and lysed with 200 µl of RLNa buffer [10 mM Tris–HCl (pH 8.0), 10 mM NaCl, 3 mM MgCl2, 1 mM DTT, 0.5% NP40 and 15 U/ml of RNaseOUT (Invitrogen Co.)]. Whole-cell extracts were recovered and RNA extraction was carried out by adding 1 ml of TRIzol® Reagent (Invitrogen Co.) as indicated by the manufacturer. Extracted RNAs (200 ng) were reverse-transcribed using the qScript™ Flex cDNA kit (Quanta Biosciences). For quantitative PCR, a 20 µl reaction mix was prepared with 5 µl of template cDNAs (previously diluted to 1/10), 10 µl of MESA green SYBR premix (Eurogentec), 0.2 µM of sense and antisense primers and subjected to amplification using a fluorescence thermocycler (Applied Biosystems 7000 Real-time PCR, Foster City, CA, USA). The housekeeping gene GAPDH was amplified in parallel to serve as a control reference. Relative copy numbers of Renilla luciferase cDNAs were compared to GAPDH using x-DCt (where x corresponds to the experimentally calculated amplification efficiency of each primer couple). Primers used to quantify Renilla and GAPDH mRNAs by qPCR were described previously (44).
RNA secondary structure predictions
Secondary structures were predicted online using the mfold software (45) available at the unfold server (http://mfold.rna.albany.edu/).
RESULTS
Low Gag synthesis from the HIV-2 provirus in human cells
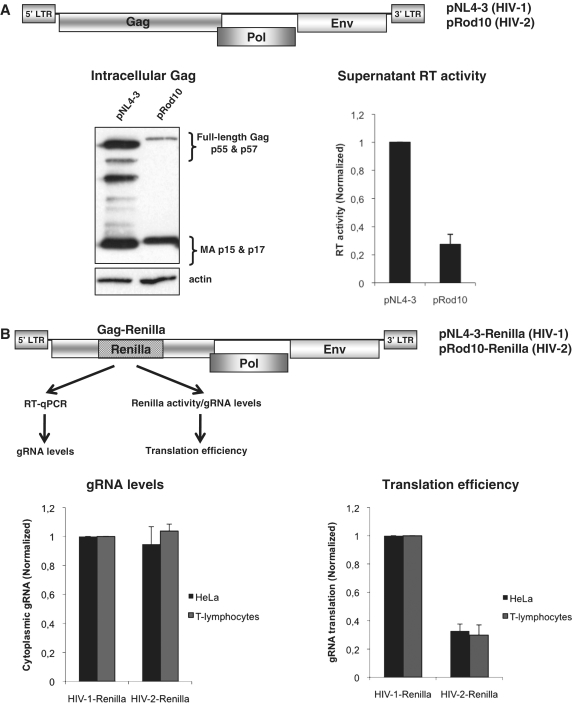
The Gag polyprotein is the major structural component of the retroviral particle and around 5000 molecules of this polyprotein are contained in an immature virion (46). However, up to 80% of the Gag polyprotein produced could undergo degradation in the cytoplasm (47), placing efficient Gag synthesis as a critical step to ensure viral production and the spread of infection. Although HIV-1 and HIV-2 are closely related, they strongly differ in their replication rates and viral production (48,49). These data prompted us to study the levels of Gag expression from the HIV-1 pNL4-3 (36) and HIV-2 pRod10 (37) molecular clones in cultured cells (Figure 1). As a first approach, we analyzed the intracellular levels of Gag polyprotein in extracts derived from HIV-1 and HIV-2 expressing HeLa cells by using an antibody raised against the matrix portion of the SIVmac Gag (43). As observed, this antibody recognized both Gag proteins and its processed forms (Figure 1A, see western blot). However, results obtained showed that cells expressing HIV-1 accumulate much more intracellular Gag than cells expressing HIV-2 suggesting that HIV-2 Gag could be produced at lower levels. Similar results were obtained with another antibody recognizing the capsid region of Gag (data not shown). Then, to rule out the possibility that lower intracellular levels of HIV-2 Gag reflect a higher rate of viral particles release, we measured the RT activity in the supernatant of transfected cells. This comparison could be possible as both recombinant viral enzymes were shown to possess similar specific activities (50). Results obtained showed that cells transfected with the HIV-1 molecular clone produced more than threefold RT activity than cells transfected with the HIV-2 clone suggesting that lower Gag production in HIV-2 expressing cells is not related to the release of higher amounts of HIV-2 Gag to the supernatant (Figure 1A, left graph). However, these results could reflect a different affinity of the SIV Gag antibody or even differences in the specific activities of the virion-associated RT enzymes rather than differences at the transcriptional or post-transcriptional level. Thus, to avoid any of the bias mentioned above and to perform a direct comparison between these two viruses, we have developed a quantitative system allowing us to directly analyze Gag production and gRNA levels by constructing HIV-1 and HIV-2-derived proviral molecular clones in which the Renilla luciferase reporter gene was inserted in frame within the Gag coding region (Figure 1B and ‘Materials and Methods’ section). These modified proviruses are transcribed, spliced, exported and translated as the wild-type proviruses but they are defective in budding (data not shown). Thus, by using these Renilla proviruses we were able to compare side by side both viruses under similar conditions and have a better estimation of translational efficiency from the gRNA by measuring the enzymatic activity of the Gag–Renilla fusion normalized to the amount of gRNA (determined by RT–qPCR against the Renilla ORF). Transfection of these modified proviral DNAs in human T-lymphocytes or HeLa cells revealed that translational efficiency (Gag–Renilla activity normalized by gRNA) from the HIV-1 gRNA was three- to fourfold higher than that of HIV-2 gRNA (Figure 1B, left graph). Quantification of the gRNA levels by RT–qPCR showed no significant differences between HIV-1 and HIV-2 indicating that they could be present at similar levels within the cells (Figure 1B, right graph). It should be noted that differences in translation efficiency were similar in T-lymphocytes and HeLa cells suggesting that lower HIV-2 gRNA translation with respect to HIV-1 occurs in human cells that are permissive for viral replication.
Figure 1.
Low HIV-2 Gag production in human cells. (A) Wild-type pNL4.3 (HIV-1) and pROD10 (HIV-2) proviral plasmids (upper scheme, regulatory and accessory proteins were omitted for simplicity) were transfected in HeLa cells and cells extracts from cells expressing HIV-1 and HIV-2 were prepared and probed against Gag by western blot as described in ‘Materials and Methods’ section. In parallel, viral production was determined by measuring the RT activity present in supernatant of transfected cells as described in ‘Materials and Methods’ section (left graph). (B) Schematic representation of the HIV-1 and HIV-2 proviral-Renilla DNA constructs used in this study (regulatory and accessory proteins were omitted for simplicity). They correspond to pNL4.3 (HIV-1) and pROD10 (HIV-2) proviral plasmids in which the Renilla reporter gene was inserted in frame within the Gag coding region as described in ‘Materials and Methods’ section. gRNA levels (left graph) and the translation efficiency from the gRNA (right graph) were determined in T-lymphocytes (black bars) and HeLa cells (gray bars) at 24 hpt as indicated. Results are normalized to values obtained for pNL4.3-Renilla (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three duplicate independent experiments.
Taken together, these results indicate that HIV-2 produces lower Gag levels than HIV-1 during a single round replication cycle probably due to a lower translational efficiency of its gRNA.
Translation of an HIV-2 gag mRNA is inefficient
Although results present above suggest that HIV-2 gRNA translation is less efficient than that of HIV-1 during a single round replication cycle, the molecular processes that precede gRNA translation (i.e. transcription, 3′-end processing and nuclear export) are complex and controlled by several viral and cellular proteins (51). These processes are not fully understood and it is unknown whether they occur at similar rates in both viruses making very difficult to compare gRNA translation in the context of a full-length provirus. Thus, an important challenge at this stage was to determine the most suitable experimental strategy to better evaluate the gRNA translational control in a cellular context. To focus exclusively on translation, we decided to use an mRNA transfection strategy that allowed us to study different in vitro generated transcripts directly in the cytoplasm of transfected cells. Compared to other mRNA transfection strategies, our system was very efficient with a wide range of mRNAs and allowed us to measure protein product as early as 3 h post-transfection using very low levels of transfected mRNAs (0.125 pmol, corresponding to 134 ng of the larger RNA used in this study). This is important as it allowed us to have a rapid estimation of protein synthesis rather than a cumulative effect of mRNA and protein product accumulation and/or degradation.
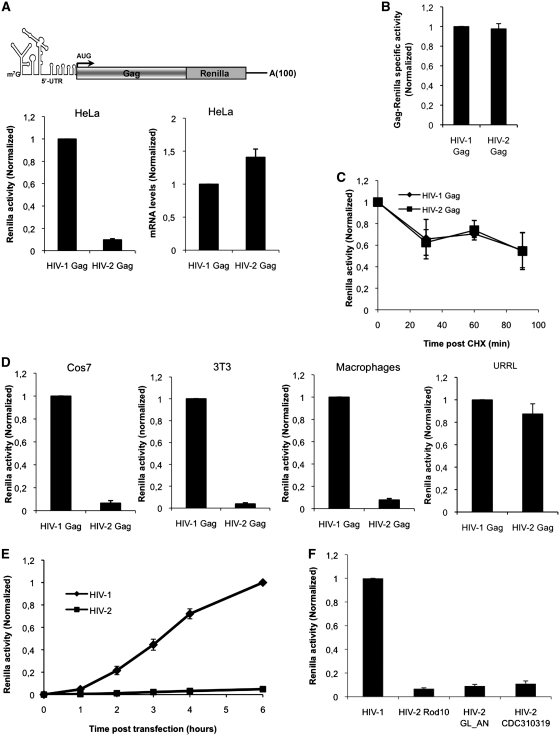
Thus, we first designed transcripts that contained all the viral elements involved in translation; these are the complete viral 5′-UTR (starting at the +1 site of transcription, HIV-2 Rod10 and HIV-1 NL4-3 strains) followed by the entire Gag coding region, which were appended in frame to the Renilla luciferase reporter gene which serves as a readout of Gag protein expression. These in vitro transcribed gag mRNAs contained an m7GpppG cap structure at their 5′-end and a poly (A) tail of 100 residues at the 3′ extremity as was previously described (40) (see cartoon in Figure 2A). Equimolar levels of HIV-2 and HIV-1 gag mRNAs were transfected in HeLa cells and Gag expression was measured by reading Renilla activity at 3 h post mRNA transfection. This revealed that Gag expression from the HIV-2 gag mRNA was much lower when compared to HIV-1 (Figure 2A, left graph). RT–qPCR quantification of transfected mRNAs revealed no major differences in their intracellular levels indicating that differences in Gag expression might not reflect differences in transfection efficiency or stability of both gag mRNAs (Figure 2A, right graph). This is not surprising as the HIV-1 and HIV-2 genomic RNAs were shown to be stable for several hours in the cytoplasm of expressing cells (52). To rule out the possibility that lower Gag–Renilla levels from cells transfected with the HIV-2 gag mRNA were due to a lower specific activity of the HIV-2 Gag–Renilla fusion protein, we generated HIV-1 and HIV-2 35S-labeled Gag–Renilla in vitro and volumes corresponding to equivalent activities for each Gag–Renilla product were precipitated with TCA and the associated radioactivity was quantified. As observed, no major differences were observed indicating that both Gag–Renilla fusions have similar specific activities (Figure 2B). As an additional control, we added two HA tags at the C-terminus of each Gag–Renilla fusion and we detected proteins by western blotting after mRNA transfection (data not shown). In agreement with results obtained using the Renilla activity as a measure, while HIV-1 Gag–Renilla product was easily detected by western blot, HIV-2 Gag–Renilla was barely detected and only a faint band was only observed at longer expositions (data not shown). Then, to rule out the possibility that differences in HIV-1 and HIV-2 Gag expression could reflect a faster degradation of the HIV-2 Gag–Renilla product, we analyzed the stability of both Gag–Renilla fusion proteins (Figure 2C). For this, HIV-1 and HIV-2 gag mRNAs were transfected as in Figure 1A and the translation inhibitor cycloheximide (CHX) was added at 3 h post mRNA transfection. We observed that both Gag–Renilla fusion proteins decayed at similar rates and thus presented the same stability indicating that differences in Gag expression occur at the level of translation. We were then interested to determine whether lower expression from the HIV-2 gag mRNA compared to HIV-1 could occur in other cell types. For this, we transfected both gag mRNAs into African green monkey fibroblasts (Cos7), murine fibroblasts (3T3) or peripheral blood mononuclear cells (PBMC)-derived human macrophages and Gag–Renilla expression was determined (Figure 2D). As it could be observed, no cellular tropism was evidenced and lower HIV-2 Gag–Renilla expression was reported in all cell types tested including primary human macrophages which are natural hosts for both human viruses. Interestingly, these differences were not observed in the untreated rabbit reticulocyte lysate (URRL) in vitro system (40), suggesting that host factors and/or the cellular environment rather than differences in the specific activity of the Gag–Renilla fusion proteins are critical in regulating such differences (see below).
Figure 2.
Translation of the HIV-2 gag mRNA is inefficient in cells. (A) About 0,125 pmoles of in vitro transcribed HIV-2 and HIV-1 gag mRNAs (see cartoon on the graph) were transfected into HeLa cells as described in ‘Materials and Methods’ section. Renilla activity (graph on the left) and transfected mRNA levels (graph on the right) were measured at 3 hpt. Results were normalized to values obtained for HIV-1 gag mRNA (HIV-1 Gag, arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments. (B) Specific activities of in vitro translated 35S-labeled HIV-1 and HIV-2 Gag–Renilla fusion proteins were analyzed as described in ‘Materials and Methods’ section. Results were first normalized by the methionine content and then to values obtained for HIV-1 Gag–Renilla (HIV-1 Gag, arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent experiments. (C) About 0.125 pmol of HIV-2 and HIV-1 gag mRNAs were transfected in HeLa cells and 3 hpt CHX was added at 100 µg/ml. Renilla activity was analyzed at 0, 30, 60 and 90 min post CHX treatment as indicated in the figure. Results are normalized to values obtained at time 0 (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments. (D) About 0.125 pmol of HIV-2 and HIV-1 gag mRNAs were transfected in Cos7 cells, 3T3 cells or human macrophages or translated in vitro in the URRL system and Renilla activity was measured at 3 hpt (transfected cells) or 30 min (in vitro translation). Results were normalized to values obtained for HIV-1 gag mRNA (HIV-1 Gag, arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments. (E) About 0.125 pmol of HIV-2 and HIV-1 gag mRNAs were transfected in HeLa cells and Renilla activity was measured at 0, 1, 2, 3, 4 and 6 hpt. Results are normalized to values obtained for HIV-1 gag mRNA (arbitrary set to 1 at 6 hpt point time) and expressed as mean ± SD corresponding to values obtained in three independent duplicated experiments. (F) About 0.125 pmol of gag mRNAs derived from different HIV-2 strains were transfected in HeLa cells and Gag–Renilla expression was compared to HIV-1. Renilla activity was measured at 3 hpt. Results are normalized to values obtained for HIV-1 (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments.
Then, we measured the kinetics of Gag expression from HIV-2 and HIV-1 gag mRNAs over time (Figure 2E). Once again, a much lower level of protein synthesis from the HIV-2 gag mRNA was observed at each time point suggesting that differences in Gag expression between HIV-2 and HIV-1 were not due to a delayed kinetics of HIV-2 translation.
Finally and in order to rule out the possibility that inefficient translation could reflect a specific feature of the HIV-2 strain used in these experiments (HIV-2 Rod10), we analyzed translation from HIV-2 gag mRNA constructs carrying sequences derived from other isolates. For this, we cloned the 5′-UTR-Gag region from the HIV-2 molecular clone GL-AN, which is able to replicate in a wide variety of lymphocytic cell lines of human and simian origins (53). We also used sequences from the primary isolate HIV-2 CDC310319 (54) that were amplified by using proviral DNA obtained from infected T-lymphocytes (our unpublished results). As observed with the Rod10 strain, low Gag expression was reported with these two HIV-2 isolates indicating that inefficient Gag synthesis (with respect to HIV-1 NL4-3) is a conserved feature among HIV-2 isolates (Figure 2F).
Taken together, these results indicate that differences in HIV-1 and HIV-2 Gag synthesis may reflect intrinsic differences between both gag mRNA sequence.
The 5′-UTR determines lower Gag expression from the HIV-2 gag mRNA
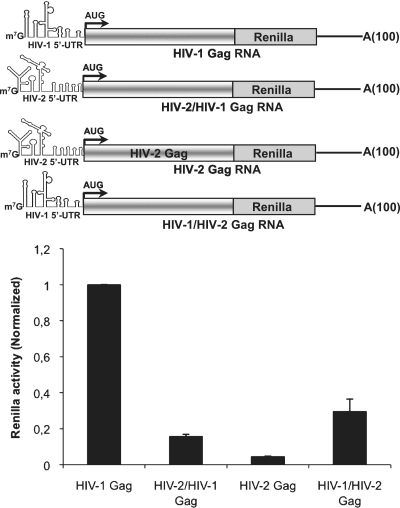
Then, we went on to characterize the RNA determinants that could account for low translation of the HIV-2 gag mRNA. Interestingly, we have previously shown that an HIV-2 gag mRNA lacking the entire 5′-UTR (leaderless RNA) showed enhanced Gag protein expression in vitro in a mechanism driven by the IRES located within the Gag coding region (32–34). These previously published observations suggest that ribosomal entry from the Gag coding region can be competed with ribosomal entry from the 5′-UTR. To address this hypothesis, we designed chimeric gag mRNAs in which the 5′-UTR of each type of virus was appended to the complete Gag coding region of the other (see cartoons in Figure 3). These 5′-UTR chimeric gag mRNAs were transfected in HeLa cells and Gag expression was determined by measurement of the Renilla activity at 3 h post mRNA transfection. Interestingly, when the HIV-2 5′-UTR was driving HIV-1 Gag synthesis, translation was dramatically reduced (by about 6-fold) compared to the wild-type HIV-1 gag mRNA (Figure 3). Conversely, when the HIV-1 5′-UTR was driving HIV-2 Gag synthesis, translation was strongly increased (by 6-fold), suggesting that specific sequences and/or RNA structures present within the HIV-2 5′-UTR may interfere with translation. It should be noted that replacement of the HIV-2 5′-UTR was not sufficient to reach a level of Gag synthesis that would be comparable to HIV-1 suggesting that the HIV-2 5′-UTR might compete with the IRESes present within the Gag coding region for ribosome recruitment (see below).
Figure 3.
The HIV-2 5′-UTR impairs Gag expression. About 0.125 pmol of chimeric gag mRNAs in which the 5′-UTR of one virus was appended to the Gag coding region of the other (see cartoon) were transfected in HeLa cells and Renilla activity was measured at 3 hpt. In the cartoon, HIV-2/HIV-1 Gag refers to the RNA that harbors the HIV-2 5′-UTR followed by the HIV-1 Gag coding region. HIV-1/HIV-2 RNA refers to the Renilla RNA that harbors the HIV-1 5′-UTR followed by the HIV-2 Gag coding region. Results were normalized to values obtained for wild-type HIV-1 gag mRNA (HIV-1 Gag, arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments.
Taken together, these results strongly indicate that the 5′-UTR of HIV-2 interferes with efficient ribosome recruitment onto the gag mRNA.
Inefficient translation driven by the HIV-2 5′-UTR
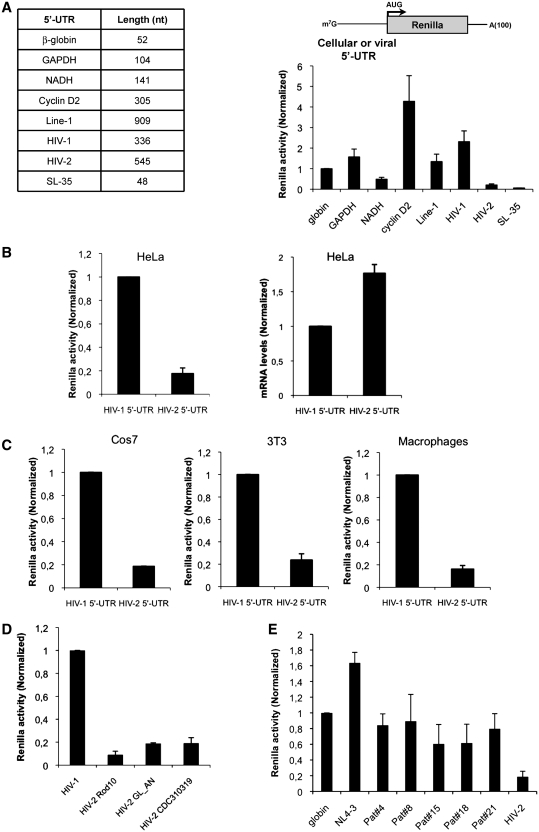
Results presented above suggest that inefficient translation of HIV-2 gag mRNA could be attributed to a negative effect of the 5′-UTR rather than any effect of the Gag coding region. To further confirm these observations, we have compared translation driven by the 5′-UTR of both human viruses in the absence of any further viral sequences (see cartoon, Figure 4A). To place our comparison in a more physiological context, we also included Renilla mRNAs bearing the 5′-UTR from different cellular transcripts. These cellular 5′-UTRs were selected on the basis of their length and complexity and included human β-globin, GAPDH, NADH, cyclin D2 and Line-1. As such, while the human β-globin 5′-UTR is short (52 nt long) and relatively unstructured, the length and complexity increased through GAPDH, NADH, cyclin D2 to Line-1 5′-UTR, which is longer (909 nt long), GC-rich and highly structured (see table on the left of Figure 4A). To our surprise, translation driven by the HIV-1 5′-UTR was very efficient and even higher than that of classical cellular 5′-UTR (Figure 4A). This contrasts with translation driven by the HIV-2 5′-UTR which was extremely low and only comparable to that of a Renilla transcript containing a 48 nt-long synthetic 5′-UTR forming an inhibitory stem–loop structure of −35 kcal/mol (SL-35 in Figure 4A). Similar to results obtained with gag mRNAs in Figure 2A, lower translation driven by the HIV-2 5′-UTR in HeLa cells (Figure 3B, left graph) may not reflect differences in transfected mRNA levels (Figure 4B, right graph) and were independent of the cell type (Figure 4C) and the HIV-2 strain used (Figure 4D) confirming that the HIV-2 5′-UTR is less efficient than that of HIV-1 to drive protein synthesis. However, it could be argued that high translation conferred by the HIV-1 5′-UTR was a peculiar feature of the NL4-3 molecular clone. Thus, 5′-UTRs from different HIV-1 clinical isolates obtained from a cohort of patients from the Croix Rousse Hospital in Lyon (France) were analyzed and compared to HIV-2 (Figure 4E). Interestingly, translation driven by the 5′-UTR of HIV-1 primary isolates was comparable to that of β-globin and NL4-3 and always higher than protein synthesis driven by the HIV-2 5′-UTR.
Figure 4.
The HIV-2 5′-UTR interferes with ribosome recruitment. (A) About 0.125 pmol of Renilla RNAs in which translation was driven by different 5′-UTR (see table on the left) were transfected in HeLa cells as described in ‘Materials and Methods’ section. Renilla activity was measured at 3 hpt. Results were normalized to values obtained for human β-globin 5′-UTR RNA (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments. (B) About 0.125 pmol of 5′-UTR-Renilla RNAs (see cartoon on the graph) were transfected in HeLa cells. Renilla activity and transfected RNA levels were determined at 3 hpt as described in ‘Materials and Methods’ section. Results were normalized to values obtained for HIV-1 5′-UTR RNA (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments. (C) About 0.125 pmol of 5′-UTR-Renilla RNAs (see cartoon on the graph) were transfected in Cos7 and 3T3 cells or human macrophages as described in ‘Materials and Methods’ section. Renilla activity was measured at 3 hpt. Results were normalized to values obtained for HIV-1 5′-UTR RNA (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments. (D) About 0.125 pmol of Renilla mRNAs containing the 5′-UTR derived from different HIV-2 strains were transfected in HeLa cells and Renilla expression was compared to HIV-1. Renilla activity was measured at 3 hpt. Results were normalized to values obtained for HIV-1 5′-UTR RNA (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments. (E) About 0.125 pmol of Renilla RNAs in which translation was driven by the 5′-UTR of different naturally occurring HIV-1 isolates (indicated as Pat#4, 8, 15, 18, 21) together with β-globin, HIV-1 NL4-3 and HIV-2 Rod10 (used as control references) were transfected in HeLa cells as described in ‘Materials and Methods’ section. Renilla activity was measured at 3 hpt. Results were normalized to values obtained for human β-globin 5′-UTR RNA (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments.
Taken together, these results confirm that the 5′-UTR present within the HIV-2 gRNA is very inefficient in support translation.
The TAR RNA structure interferes with HIV-2 translation
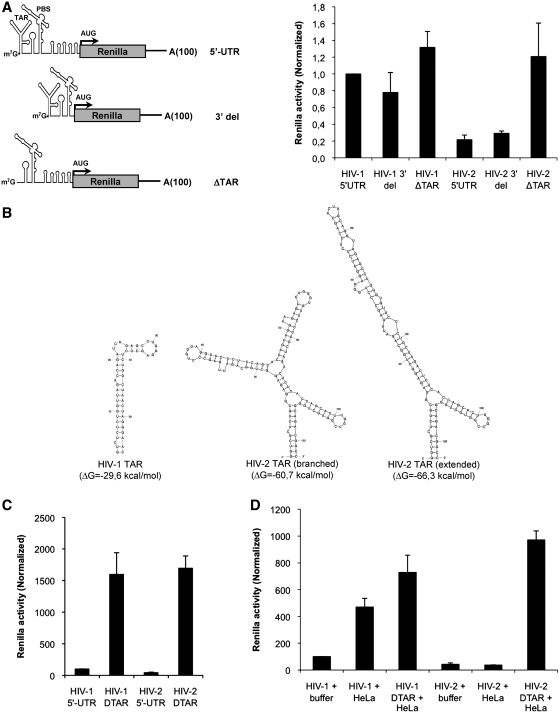
We then wanted to determine whether lower translation of HIV-2 could be attributed to any specific RNA determinant present within the 5′-UTR. For this, we have introduced 5′ and 3′ deletions within both viral 5′-UTRs and analyzed expression of the resulting Renilla transcripts (see cartoon in Figure 5). It should be mentioned that all these transcripts contains the same context surrounding the AUG start codon to avoid any bias due to differences in start site recognition. As observed in Figure 5, a mutant in which a 3′ deletion was introduced from the end of PBS to the AUG start codon (called 3′ del) did not significantly alter expression driven by the 5′-UTR of HIV-1 or HIV-2 indicating that the 3′ end of the HIV-2 5′-UTR is not responsible for inefficient translation. However, when the TAR RNA element was deleted from the HIV-2 5′-UTR, translation was massively increased rendering Renilla expression as efficient as that driven by the HIV-1 5′-UTR indicating that HIV-2 TAR interferes with translation (compare HIV-1 5′-UTR and HIV-2 ΔTAR in Figure 5A). Several studies have determined the conformation of the HIV-1 and HIV-2 TAR structures (13,16–23). As such, while the HIV-1 TAR structure forms a single stem loop, the HIV-2 TAR structure can adopt two different conformations that are longer and more complex than that of HIV-1 (Figure 5B). Nevertheless, the stability of such structures could be sufficient to interfere with translation (55–57). Indeed, HIV-1 TAR was shown to be a potent inhibitor of translation in cell-free systems and Xenopus oocytes (24,26,27). However, such an effect has been shown to be much smaller in intact cells (58–60). Interestingly, we observed that deletion of HIV-1 TAR structure in cells did not have a strong effect on translation, thus pointing out for a specific role of HIV-2 TAR in inhibiting translation (Figure 5A). The poor inhibitory effect of HIV-1 TAR that we observed in cells compared to previous studies could be mostly attributed to the context on which its effects were analyzed. Most of the previous studies were based on the addition of a TAR structure-containing region (usually nucleotides +1 to +81, +111 or +282 from the HIV-1 5′-UTR) in front of a reporter gene (CAT for instance). Then, translational efficiency of these TAR-containing mRNAs was compared with translation of CAT mRNAs that are devoid of any RNA secondary structure (24,26,27). In this present study, we have compared the effect of TAR deletion from the viral 5′-UTR. As such, TAR deleted constructs do still contains an important region of the viral 5′-UTR. These differences in experimental design may explain some of the functional differences that we observed.
Figure 5.
TAR interferes with ribosome recruitment onto the HIV-2 gRNA. (A) About 0.125 pmol of Renilla mRNAs containing the wild-type viral 5′-UTR or 5′ and 3′ deletions (see cartoon on the graph) were transfected in HeLa as described in ‘Materials and Methods’ section. Renilla activity was measured at 3 hpt. Results were normalized to values obtained for HIV-1 5′-UTR RNA (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments. (B) Secondary structure prediction and stability of HIV-1 and HIV-2 TAR RNA structure using the mfold software as indicated in ‘Materials and Methods’ section. (C) The untreated rabbit reticulocyte lysate was programmed with 0.125 pmol of wild-type or ΔTAR Renilla mRNAs and Renilla activity was measured after 30 min. Results were normalized to values obtained for HIV-1 5′-UTR RNA (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained at least in three independent experiments. (D) The untreated rabbit reticulocyte lysate was programmed with 0.125 pmol of wild-type or ΔTAR Renilla mRNAs in the presence of buffer or HeLa cells extracts as described in ‘Materials and Methods’ section. Renilla activity was measured after 30 min. Results were normalized to values obtained for HIV-1 5′-UTR RNA in the presence of buffer (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained at least in three independent experiments.
However, we were still surprised by this result and we wanted to analyze it further. Thus, we used the Renilla mRNAs bearing the wild type and the ΔTAR 5′-UTRs of HIV-1 and HIV-2 to program in vitro translation reactions in the untreated rabbit reticulocytes lysate (40). In agreement with previous observations, the TAR structures of both HIV-1 and HIV-2 were indeed potent inhibitors of translation in vitro (Figure 5C). The discrepancies between results observed in cells and in vitro could be attributed to the absence or limiting concentration of cellular factors such as Lupus autoantigen, Staufen1 or TRBP, which are able to stimulate HIV-1 translation by acting on TAR (61–63). Thus, to determine whether the lack of a strong inhibitory effect observed with HIV-1 TAR in cells could be attributed to the action of cellular factors, we complemented in vitro translation reactions with cell extracts derived from HeLa cells. Strikingly, addition of HeLa cell extracts to the rabbit reticulocyte lysate nicely recapitulates what was observed in cells. Whereas translation driven by the HIV-1 5′-UTR was stimulated by 5-fold by the addition of HeLa cells extract, HIV-2 5′-UTR driven translation was rather inhibited by 2-fold (Figure 5D). As such, while the inhibitory effect of HIV-1 TAR in vitro was almost completely lost in the presence of HeLa cells extracts, HIV-2 TAR still exerted a strong repression of translation.
Taken together, these results suggest that HIV-1 may have evolved the use of host factors to overcome the inhibitory effect of TAR on translation while HIV-2 TAR did not.
HIV-2 TAR stability impairs ribosome recruitment
Results presented above prompted us to investigate the mechanism by which HIV-2 TAR could affect translation. One possibility would be that inhibition was exerted through the induction eIF2α phosphorylation due to TAR-dependent activation of PKR (64,65). Another possibility is that the higher stability of the HIV-2 TAR structure interferes with entry of initiation complexes at the 5′-end.
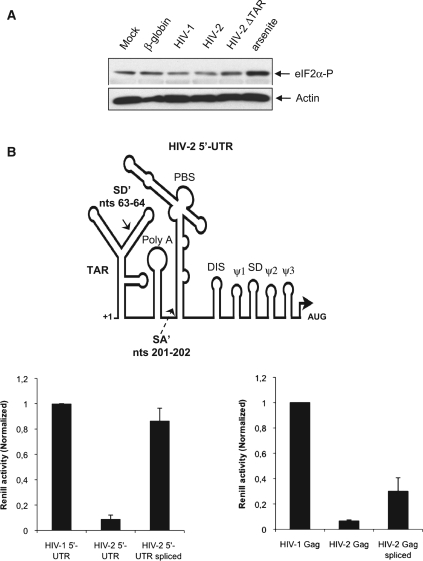
To test the first hypothesis, phosphorylated eIF2α was visualized by western blot in extracts derived from cells previously transfected with a control Renilla mRNA (β-globin-Renilla) or with the Renilla mRNAs carrying the HIV-1 5′-UTR, HIV-2 5′-UTR or HIV-2 ΔTAR (Figure 6A, lanes 2–5). As controls mock transfected cells untreated or treated with sodium arsenite were also included (Figure 6A, lanes 1 and 6, respectively). Although phosphorylated eIF2α was detected after transfection of all the mRNA tested, these levels were similar to the basal levels of phosphorylated eIF2α observed in mock transfected cells and lower to that observed in cells treated with sodium arsenite suggesting that induction of eIF2α phosphorylation was not dependent on HIV-2 TAR (Figure 6A, compare lanes 1–5 with line 6). Moreover, it should be mentioned that treating the cells with sodium arsenite virtually killed translation driven by the β-globin or the HIVs 5′-UTRs indicating that phosphorylation of eIF2α could not benefit translation of any of the mRNAs used in this study (data not shown).
Figure 6.
HIV-2 TAR stability impairs ribosome recruitment. (A) HeLa cells were transfected with equal molar quantities of Renilla mRNAs as indicated in the figure and cells extracts were prepared at 3 hpt for western blot analyses. In parallel, cells extracts from mock transfected cells or mock transfected cells treated with sodium arsenite during 30 min were also prepared and used as controls. (B) HeLa cells were transfected with 0.125 pmol of a control RNA or with HIV-1 5′-UTR, HIV-2 5′-UTR or HIV-2 ΔTAR-Renilla RNAs and whole cellular extracts were prepared at 3 hpt and probed against eIF2α, eIF2α phosphorylated and PABP by western blot as described in ‘Materials and Methods’ section. (C) Schematic representation not at scale of the HIV-2 5′-UTR indicating the position of the alternative splice donor (SD′) and splice acceptor (SA′) that results in the removal of the 5′-UTR intron described by Lodmell et al. (67). About 0.125 pmol of wild-type or 5′-UTR spliced HIV-2 Renilla mRNA (left graph) or 5′-UTR spliced HIV-2 Gag–Renilla mRNA (right graph) were transfected in HeLa cells together with the corresponding HIV-1 Renilla mRNA and Renilla activity was measured at 3 hpt. Results were normalized to values obtained for HIV-1 mRNAs (arbitrary set to 1) and expressed as mean ± SD corresponding to values obtained in three independent duplicate experiments.
As showed in Figure 5B, and references herein, HIV-2 and HIV-1 5′-UTRs strongly differ in the length and predicted folding of their TAR structures. Although both motifs are expected to be in close proximity to the cap structure, the much stronger stability of HIV-2 TAR could be responsible for reduced translational efficiency. However, the HIV-2 5′-UTR contains a 140 nt intron that is removed in multiple spliced transcripts (66). Interestingly, Lodmell and colleagues (67) recently showed that this intron could also be removed from the gRNA 5′-UTR resulting in an mRNA with enhanced translation. Removal of this intron results in the destabilization of the TAR RNA structure as at least half of its sequence is contained in the intron (see cartoon in Figure 6B). Given the results obtained so far by our group and these new data from Lodmell's lab, we created a Renilla mRNA carrying the spliced version of the 5′-UTR present within the HIV-2 gRNA as recently described (67). This HIV-2 5′-UTR spliced mRNA was transfected in HeLa cells and translational efficiency was compared to that of Renilla mRNAs carrying the wild-type HIV-2 and HIV-1 5′-UTRs (Figure 6B, left panel). Similar to results obtained with the HIV-2 ΔTAR Renilla RNA and in agreement with previous results (67), destabilization of HIV-2 TAR resulted in an enhanced translation that reached the high levels of the HIV-1 5′-UTR, confirming that the higher stability of the HIV-2 TAR structure strongly interferes with ribosome recruitment at the 5′-end.
Then, we wanted to determine the effects that such a splicing event could have in the context of an HIV-2 gag mRNA (Figure 6B, right panel). Interestingly, and in agreement with data obtained with the HIV-1/HIV-2 Gag chimera (Figure 3), removal of the 5′-UTR intron resulted in 5-fold stimulation of Gag expression. However, this enhancement was not sufficient to reach the levels of HIV-1 Gag production further indicating that ribosome recruitment at the 5′-end may interfere with the IRES located within the Gag coding region.
DISCUSSION
The molecular mechanism controlling HIV-1 translation has been a matter of debate for many years (68). While some people have found that cap-dependent ribosomal scanning is the major mechanism taking place (25,69,70), others have demonstrated the presence of a functional IRES element within the 5′-UTR (28–30,71,72) and the Gag coding region (31). In the case of HIV-2, IRES elements have only been described within the Gag coding region (32–35) and the role of the 5′-UTR remained largely unknown. Although a large number of these studies have focused on the molecular mechanism of ribosome recruitment (i.e. IRES-driven versus cap-dependent) none of them have investigated how overall translational efficiency of these two related viruses would be compared. This is probably due to the fact that a side-by-side comparison is technically difficult to perform in the context of viral infection as the viral life cycle and the nature of the proteins produced by each virus are very different. To circumvent some of these problems, we have used a very efficient mRNA transfection strategy of different synthetic constructs carrying the different RNA elements that have been shown to be involved in gRNA translation (i.e. the 5′-UTR and the Gag coding region).
Results presented herein show that expression from the HIV-1 gag mRNA was higher than that of the HIV-2 gag mRNA in different cell lines including T-lymphocytes and PBMC-derived human macrophages (Figures 1 and 2). Importantly, no evident differences in the levels of transfected mRNAs or the specific activity and stability of both Gag–Renilla fusion proteins were observed indicating that differences in expression between HIV-1 and HIV-2 gag mRNAs were exclusively at the level of translation. Switching the 5′-UTR of both gag mRNAs and the use of reporter constructs bearing the 5′-UTR in the absence of any further viral coding region showed that lower translational efficiency of the HIV-2 gag mRNA was merely due to the presence of its highly structured 5′-UTR (Figures 3 and 4). This was confirmed by deletion experiments pointing out a specific role of the HIV-2 TAR RNA structure (Figure 5). We observed that complementation of in vitro translation reactions with HeLa cells extracts was rather inhibitory for translation driven by the HIV-2 5′-UTR (Figure 5), indicating that host factors may contribute to the low translation observed from the HIV-2 gRNA in intact cells. Moreover, we observed that swapping the 5′-UTR (Figure 3) or destabilizing TAR (Figure 6) did not result in HIV-2 Gag levels resembling those observed with HIV-1 despite similar properties of both Gag–Renilla fusions. Interestingly, the activity of the HIV-2 gag IRES was increased by several folds when the entire 5′-UTR was removed from the mRNA (32–34). These results suggest that ribosomal entry at the 5′ end could potentially interferes with ribosome recruitment from the Gag IRES and both mechanisms may be tightly regulated during viral replication.
Although a strong inhibitory effect of TAR was seen for both viral mRNAs in vitro, such a strong effect of TAR was only seen with HIV-2 in intact cells in a mechanism that was dependent on its intrinsic stability rather than PKR activation (Figures 5 and 6). As such, protein synthesis driven by the HIV-1 5′-UTR was very efficient and even higher than translation driven by short and unstructured cellular 5′-UTR such as β-globin, GAPDH or NADH (Figure 4). Several host RNA-binding proteins including Staufen1, La autoantigen and TRBP were shown to stimulate HIV-1 translation by acting at the level of the TAR RNA structure, and thus may contribute to alleviate TAR interference (61–63). In fact, complementation of the rabbit reticulocyte lysate in vitro system with HeLa cells extracts resulted in an important loss of inhibition by TAR and the concomitant stimulation of HIV-1 translation (Figure 5). Interestingly, addition of HeLa cells extracts to in vitro translation reactions had no the same effect on HIV-2 translation. To date, it is unknown whether cellular factors participate in the regulation of HIV-2 translation through TAR. However, destabilization of the TAR structure by removal of an intron present within the 5′-UTR (66,67) and the use IRES elements within the Gag coding region (32–35) confers two alternative ways to overcome TAR constraints and allow Gag and Gag-Pol production. Thus, the direct binding of eIF3 and the 40S ribosomal subunit to the Gag IRES (35) may allow the full-length unspliced HIV-2 gRNA to be associated with polysomes. Nevertheless, the poor translation of the HIV-2 genomic RNA provides an explanation for limited Gag production observed during viral replication (Figure 1) and may be required for cis co-translational packaging of the gRNA (73,74). Interestingly, the lack of splicing event in the HIV-2 5′-UTR or mutations in TAR from the closely related virus SIVmac were shown to not impact viral replication despite its ability to increase translation of a reporter gene (67,75). Moreover, the HIV-2 5′-UTR spliced gRNA was shown to be excluded from the viral particles despite the presence of all packaging signals (67) (and our unpublished data), further supporting a physiological role for slowed translation and gRNA packaging during the HIV-2 replication cycle.
FUNDING
Conicyt Chile, ANRS fellowship to RSR, ANRS project grant, Sidaction to OM, FRM to TL. This work was supported by a ‘contrat d'interface’ with HCL. Funding for open charge: INSERM
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-2CDC310319 from S.W. and M.R. and SIVmac251 Gag Monoclonal Antibody (KK59) from K.K. and C.P. Authors are especially grateful of S.M. (University of Sussex, UK) for anti eIF4GI and PABP antibodies, G.C. (Université de Nice, France) for pJM101f L1.3 vector and F.U. (University of Tokushima, Japan) for HIV-2 pGL-AN molecular clone. Authors wish to specially thank Professor Jean-Luc Darlix for critical reading of the manuscript and helpful comments. R.S.R performed the experiments. R.S.R. and T.O. designed the experiments and wrote the article. T.L., P.R., E.P.R., D.D. and O.M. helped with experimental procedures. A.C. provided PBMCs-derived human macrophages and helped with discussion. M.-A.T. and P.A. provided naturally occurring HIV-1 sequences isolated from French patients.
REFERENCES
- 1.Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prevot D, Darlix JL, Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol. Cell. 2003;95:141–156. doi: 10.1016/s0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 5.Abaeva IS, Marintchev A, Pisareva VP, Hellen CU, Pestova TV. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 2011;30:115–129. doi: 10.1038/emboj.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell. Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 10.Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta. 2009;1789:542–557. doi: 10.1016/j.bbagrm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 12.Butsch M, Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 2002;76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 14.Damgaard CK, Dyhr-Mikkelsen H, Kjems J. Mapping the RNA binding sites for human immunodeficiency virus type-1 gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 1998;26:3667–3676. doi: 10.1093/nar/26.16.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCann EM, Lever AM. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J. Virol. 1997;71:4133–4137. doi: 10.1128/jvi.71.5.4133-4137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 17.Dirac AM, Huthoff H, Kjems J, Berkhout B. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res. 2002;30:2647–2655. doi: 10.1093/nar/gkf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pachulska-Wieczorek K, Purzycka KJ, Adamiak RW. New, extended hairpin form of the TAR-2 RNA domain points to the structural polymorphism at the 5′ end of the HIV-2 leader RNA. Nucleic Acids Res. 2006;34:2984–2997. doi: 10.1093/nar/gkl373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purzycka KJ, Pachulska-Wieczorek K, Adamiak RW. The in vitro loose dimer structure and rearrangements of the HIV-2 leader RNA. Nucleic Acids Res. 2011;39:7234–7248. doi: 10.1093/nar/gkr385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berkhout B, Schoneveld I. Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic Acids Res. 1993;21:1171–1178. doi: 10.1093/nar/21.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SenGupta DN, Berkhout B, Gatignol A, Zhou AM, Silverman RH. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA. 1990;87:7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berkhout B, Arts K, Abbink TE. Ribosomal scanning on the 5′-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res. 2011;39:5232–5244. doi: 10.1093/nar/gkr113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geballe AP, Gray MK. Variable inhibition of cell-free translation by HIV-1 transcript leader sequences. Nucleic Acids Res. 1992;20:4291–4297. doi: 10.1093/nar/20.16.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkin NT, Cohen EA, Darveau A, Rosen C, Haseltine W, Sonenberg N. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988;7:2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gendron K, Ferbeyre G, Heveker N, Brakier-Gingras L. The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res. 2011;39:902–912. doi: 10.1093/nar/gkq885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monette A, Ajamian L, Lopez-Lastra M, Mouland AJ. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J. Biol. Chem. 2009;284:31350–31362. doi: 10.1074/jbc.M109.048736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbreteau CH, Weill L, Decimo D, Prevot D, Darlix JL, Sargueil B, Ohlmann T. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- 33.Ricci EP, Herbreteau CH, Decimo D, Schaupp A, Datta SA, Rein A, Darlix JL, Ohlmann T. In vitro expression of the HIV-2 genomic RNA is controlled by three distinct internal ribosome entry segments that are regulated by the HIV protease and the Gag polyprotein. RNA. 2008;14:1443–1455. doi: 10.1261/rna.813608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weill L, James L, Ulryck N, Chamond N, Herbreteau CH, Ohlmann T, Sargueil B. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 2010;38:1367–1381. doi: 10.1093/nar/gkp1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Locker N, Chamond N, Sargueil B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. 2011;39:2367–2377. doi: 10.1093/nar/gkq1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clavel F, Guyader M, Guetard D, Salle M, Montagnier L, Alizon M. Molecular cloning and polymorphism of the human immune deficiency virus type 2. Nature. 1986;324:691–695. doi: 10.1038/324691a0. [DOI] [PubMed] [Google Scholar]
- 38.Ricci EP, Limousin T, Soto-Rifo R, Allison R, Poyry T, Decimo D, Jackson RJ, Ohlmann T. Activation of a microRNA response in trans reveals a new role for poly(A) in translational repression. Nucleic Acids Res. 2011;39:5215–5231. doi: 10.1093/nar/gkr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dieterich K, Soto Rifo R, Faure AK, Hennebicq S, Ben Amar B, Zahi M, Perrin J, Martinez D, Sele B, Jouk PS, et al. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat. Genet. 2007;39:661–665. doi: 10.1038/ng2027. [DOI] [PubMed] [Google Scholar]
- 40.Soto Rifo R, Ricci EP, Decimo D, Moncorge O, Ohlmann T. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007;35:e121. doi: 10.1093/nar/gkm682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarrosson-Wuilleme L, Goujon C, Bernaud J, Rigal D, Darlix JL, Cimarelli A. Transduction of nondividing human macrophages with gammaretrovirus-derived vectors. J. Virol. 2006;80:1152–1159. doi: 10.1128/JVI.80.3.1152-1159.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cimarelli A, Sandin S, Hoglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kent KA, Gritz L, Stallard G, Cranage MP, Collignon C, Thiriart C, Corcoran T, Silvera P, Stott EJ. Production and of monoclonal antibodies to simian immunodeficiency virus envelope glycoproteins. AIDS. 1991;5:829–836. doi: 10.1097/00002030-199107000-00006. [DOI] [PubMed] [Google Scholar]
- 44.Ricci EP, Mure F, Gruffat H, Decimo D, Medina-Palazon C, Ohlmann T, Manet E. Translation of intronless RNAs is strongly stimulated by the Epstein-Barr virus mRNA export factor EB2. Nucleic Acids Res. 2009;37:4932–4943. doi: 10.1093/nar/gkp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 47.Tritel M, Resh MD. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 2000;74:5845–5855. doi: 10.1128/jvi.74.13.5845-5855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blaak H, van der Ende ME, Boers PH, Schuitemaker H, Osterhaus AD. In vitro replication capacity of HIV-2 variants from long-term aviremic individuals. Virology. 2006;353:144–154. doi: 10.1016/j.virol.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 49.MacNeil A, Sarr AD, Sankale JL, Meloni ST, Mboup S, Kanki P. Direct evidence of lower viral replication rates in vivo in human immunodeficiency virus type 2 (HIV-2) infection than in HIV-1 infection. J. Virol. 2007;81:5325–5330. doi: 10.1128/JVI.02625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hizi A, Tal R, Shaharabany M, Loya S. Catalytic properties of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2. J. Biol. Chem. 1991;266:6230–6239. [PubMed] [Google Scholar]
- 51.Bolinger C, Boris-Lawrie K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology. 2009;6:8. doi: 10.1186/1742-4690-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dorman N, Lever A. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J. Virol. 2000;74:11413–11417. doi: 10.1128/jvi.74.23.11413-11417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueno F, Shiota H, Miyaura M, Yoshida A, Sakurai A, Tatsuki J, Koyama AH, Akari H, Adachi A, Fujita M. Vpx and Vpr proteins of HIV-2 up-regulate the viral infectivity by a distinct mechanism in lymphocytic cells. Microb. Infect. 2003;5:387–395. doi: 10.1016/s1286-4579(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 54.Owen SM, Ellenberger D, Rayfield M, Wiktor S, Michel P, Grieco MH, Gao F, Hahn BH, Lal RB. Genetically divergent strains of human immunodeficiency virus type 2 use multiple coreceptors for viral entry. J. Virol. 1998;72:5425–5432. doi: 10.1128/jvi.72.7.5425-5432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Babendure JR, Babendure JL, Ding JH, Tsien RY. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl Acad. Sci. USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muesing MA, Smith DH, Capon DJ. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 59.Biegalke BJ, Geballe AP. Sequence requirements for activation of the HIV-1 LTR by human cytomegalovirus. Virology. 1991;183:381–385. doi: 10.1016/0042-6822(91)90151-z. [DOI] [PubMed] [Google Scholar]
- 60.Chin DJ, Selby MJ, Peterlin BM. Human immunodeficiency virus type 1 Tat does not transactivate mature trans-acting responsive region RNA species in the nucleus or cytoplasm of primate cells. J. Virol. 1991;65:1758–1764. doi: 10.1128/jvi.65.4.1758-1764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dorin D, Bonnet MC, Bannwarth S, Gatignol A, Meurs EF, Vaquero C. The TAR RNA-binding protein, TRBP, stimulates the expression of TAR-containing RNAs in vitro and in vivo independently of its ability to inhibit the dsRNA-dependent kinase PKR. J. Biol. Chem. 2003;278:4440–4448. doi: 10.1074/jbc.M208954200. [DOI] [PubMed] [Google Scholar]
- 62.Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Svitkin YV, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 1994;68:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.SenGupta DN, Silverman RH. Activation of interferon-regulated, dsRNA-dependent enzymes by human immunodeficiency virus-1 leader RNA. Nucleic Acids Res. 1989;17:969–978. doi: 10.1093/nar/17.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maitra RK, McMillan NA, Desai S, McSwiggen J, Hovanessian AG, Sen G, Williams BR, Silverman RH. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 66.Chatterjee P, Garzino-Demo A, Swinney P, Arya SK. Human immunodeficiency virus type 2 multiply spliced transcripts. AIDS Res. Hum. Retroviruses. 1993;9:331–335. doi: 10.1089/aid.1993.9.331. [DOI] [PubMed] [Google Scholar]
- 67.Strong CL, Lanchy JM, Dieng-Sarr A, Kanki PJ, Lodmell JS. A 5′UTR-spliced mRNA isoform is specialized for enhanced HIV-2 gag translation. J. Mol. Biol. 2009;391:426–437. doi: 10.1016/j.jmb.2009.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yilmaz A, Bolinger C, Boris-Lawrie K. Retrovirus translation initiation: Issues and hypotheses derived from study of HIV-1. Curr. HIV Res. 2006;4:131–139. doi: 10.2174/157016206776055039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miele G, Mouland A, Harrison GP, Cohen E, Lever AM. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 1996;70:944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwartz S, Felber BK, Pavlakis GN. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol. Cell Biol. 1992;12:207–219. doi: 10.1128/mcb.12.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Charnay N, Ivanyi-Nagy R, Soto-Rifo R, Ohlmann T, Lopez-Lastra M, Darlix JL. Mechanism of HIV-1 Tat RNA translation and its activation by the Tat protein. Retrovirology. 2009;6:74. doi: 10.1186/1742-4690-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rivas-Aravena A, Ramdohr P, Vallejos M, Valiente-Echeverria F, Dormoy-Raclet V, Rodriguez F, Pino K, Holzmann C, Huidobro-Toro JP, Gallouzi IE, et al. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites. Virology. 2009;392:178–185. doi: 10.1016/j.virol.2009.06.050. [DOI] [PubMed] [Google Scholar]
- 73.Griffin SD, Allen JF, Lever AM. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 2001;75:12058–12069. doi: 10.1128/JVI.75.24.12058-12069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaye JF, Lever AM. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 1999;73:3023–3031. doi: 10.1128/jvi.73.4.3023-3031.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Centlivre M, Klaver B, Berkhout B, Das AT. Functional analysis of the complex trans-activating response element RNA structure in simian immunodeficiency virus. J. Virol. 2008;82:9171–9178. doi: 10.1128/JVI.00530-08. [DOI] [PMC free article] [PubMed] [Google Scholar]