Abstract
Signaling through the mammalian target of rapamycin, complex 1 (mTORC1), positively regulates the transcription of ribosomal RNA (rRNA) and the synthesis of ribosomal proteins, thereby promoting the complex process of ribosome biogenesis. The major rRNAs are transcribed as a single precursor, which must be processed to create the 5.8S, 18S and 28S rRNAs. We used a new non-radioactive labeling approach to study the effects of rapamycin, an inhibitor of mTORC1, on rRNA synthesis. Rapamycin not only impaired synthesis of new 18S, 28S or 5S rRNA but also induced their decay. This prompted us to examine the effects of rapamycin on rRNA processing. We show that rapamycin also interferes with the processing events that generate 18S and 28S rRNA. rRNA transcription and processing occur in regions of the nucleus known as nucleoli. We find that the mTORC1 components raptor and mTOR are both present in nucleoli, where they may regulate rRNA maturation events. While rapamycin has no effect on overall nucleolar morphology or its proteome, it does induce loss of mTOR and raptor from them. These data show that mTORC1 is located in nucleoli where it acts to regulate events involved in ribosome biogenesis including the maturation of rRNA molecules.
INTRODUCTION
Signaling through the mammalian target of rapamycin (complex 1), mTORC1, regulates many diverse cellular processes, especially those contributing to cell growth and proliferation (1), including the production of ribosomes (2). This process, termed ribosome biogenesis, is a complex one that involves the synthesis and subsequent processing of 4 different ribosomal RNAs (rRNAs) and around 80 proteins, which are assembled into ribosomes in the nucleolus, aided by many additional proteins and small RNAs. mTORC1 signaling promotes the transcription of the 47S precursor by positively regulating Pol I [which makes three of the rRNAs, (3)] and the translation of the mRNAs encoding ribosomal proteins (4).
Ribosome biogenesis is critical for maintaining cells’ capacity for protein synthesis, especially during cell growth (hypertrophy) or proliferation. Indeed, increases in the size and number of nucleoli have long been recognized as a key feature of cancer cells (5). Furthermore, deregulation of ribosome biogenesis may pre-dispose toward cancer and other conditions (6,7). Defective ribosome biogenesis can also activate the tumor suppressor p53 and/or induce apoptosis (8,9).
Three of the rRNAs, 5.8S, 18S and 28S, are transcribed as a single precursor (47S) by RNA polymerase I (Pol I) in the nucleolus, which is then processed through several steps to generate the mature rRNAs. Here, we show for the first time that rapamycin interferes with the processing of rRNA in human cells indicating that mTORC1 signaling controls pre-rRNA processing in addition to the synthesis of rRNA and ribosomal proteins. Furthermore, we show that mTORC1 is present in the nucleolus and this localization is disrupted by rapamycin. This likely allows mTORC1, which is recognized as a positive regulator of anabolic cellular functions, to promote and coordinate the complex multiplicity of steps involved in manufacturing new ribosomes.
MATERIALS AND METHODS
Cell culture and reagents
Human cervical carcinoma HeLa cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (v/v) fetal calf serum (FCS), 100 U/ml of penicillin and 100 µg/ml streptomycin in 5% CO2 at 37°C.
4-Thiouridine (4-TU) was used at 100 µM (Sigma T4509), actinomycin D (Sigma, A1410) was used at the concentrations given in the text. Rapamycin (Calbiochem 553210) was used at 100 nM (unless stated otherwise) and for the times indicated in the legends. AZD8055 was kindly provided by AstraZeneca UK.
RNA preparation and biotinylation of 4-TU-labeled RNA
The procedure for labeling newly synthesized RNA within cells using modified uridine was adapted from those described earlier (10,11). Total RNA was extracted from HeLa cells using Trizol (Invitrogen, 15596-018) following the manufacturer's protocol.
To biotinylate 4-TU-labeled RNA, total RNA was incubated with EZ-Link Biotin-HPDP (Pierce 21341). Biotin-HPDP was suspended in dimethylformamide at a concentration of 1 mg/ml. One microliter of this reagent was used per 1 µg of RNA extracted from the labeling reaction. Biotinylation of 4-TU-labeled RNA was performed in 10 mM Tris–HCl (pH 7.5) and 1 mM EDTA in a final volume five times greater than that of biotin-HPDP in the reaction. RNA was then incubated with biotin-HPDP at room temperature for 2 h in the dark. To precipitate RNA, 1/10 of the reaction volume of 5 M NaCl and equal reaction volume of isopropanol were added and the sample was centrifuged at 14 500 rpm for 20 min. The pellet was rinsed with 75% (v/v) ethanol and resuspended in RNase-free water. The samples were stored at −80°C until required.
Isolation of 4-TU-labeled RNA from total RNA
To purify 4-TU-labeled RNA, Magnetic Porous Glass (MPG) streptavidin beads (PUREBiotech MSTR0510) were used to capture the biotinylated RNA. One microliter of MPG streptavidin beads was used per microgram of total RNA. The beads were incubated with carrier yeast tRNA (1 µg tRNA per 5 µl beads) for 20 min at ambient temperature to prevent non-specific binding and washed three times in 500 µl MPG Buffer (1 M NaCl, 10 mM EDTA, 100 mM Tris–HCl, pH 7.5) before adding RNA. MPG beads were collected in a magnetic stand and resuspended in MPG buffer to the original volume of beads. The volume of biotinylated RNA sample was adjusted with RNase-free water to the same volume as the beads. The biotinylated RNA was then added to the beads and incubated at ambient temperature for 1 h with rotation. The beads were collected on the magnetic stand for more than 1 min and followed by washing three times in 500 µl MPG buffer, the first two times at ambient temperature and the last one at 65°C. Two more washes were performed at ambient temperature to make sure that all the non-4-TU-labeled RNA was washed away. Bound RNA that was labeled by 4-TU was eluted by suspending the MPG beads in freshly diluted 5% β-mercaptoethanol volume equal to that of the original beads. The mixture was incubated at room temperature for at least 20 min with rotation. The beads were collected on the magnetic stand for about 1 min and the supernatant was saved. One microgram of glycogen (Roche #10 901 393 001) were added as carrier and RNA was precipitated using isopropanol and NaCl. The pellet was rinsed with 75% (v/v) ethanol and resuspended in RNase-free water. The resuspended RNA sample was then placed in magnetic stand for >1 min to collect any excess beads from earlier steps. The supernatant was transferred to a new tube; this sample contains purified 4-TU-labeled RNA.
RT–PCR amplification analysis
Total purified 4-TU-labeled RNA was treated with RNase-free DNAse (optional Roche Cat. No. 04 716 728 001) and then subjected to SuperScript® III reverse transcription (Invitrogen #18080-044) with an oligo(dT)15 primer (Invitrogen #18418-012) and random primers (Invitrogen #48190-011) following the manufacturer's protocol. Subsequently, PCR using specific primers (PrimerDesign HY-SY-hu-600) was employed to amplify 18S rRNA and other house-keeping genes according to the product protocol. The comparative Ct method was employed to measure amplification of the new rRNAs from control and treated samples. The quantity of new (labeled) rRNA was normalized to the total amount of 18S rRNA in this sample before the pulldown. These data were compared with the corresponding data for untreated cells and the results are expressed as, e.g., ‘new 18S rRNA/control’.
Uridine uptake
Uridine transport was monitored in cultured HeLa cells in medium either containing or lacking Na+ ions, using a method based on those used in earlier studies (12,13). The medium contained 5.4 mM KCl, 1.8 mM CaCl2, 1.2 mM MgSO4, 10 mM HEPES (pH 7.4) and 137 mM Choline chloride and [3H]uridine (1 µCi/nmol). After 20-s incubation, uptake was stopped by washing three times in 2 ml of ice-cold transport buffer containing 1 mM unlabeled competitor uridine and two inhibitors of nucleoside uptake, 1 µM S-(4-Nitrobenzyl)-6-thioinosine (Sigma N2255) and dipyridamole (Sigma D9766). Cells were lysed in 500 µl of a solution containing 0.5% Triton X-100 and 100 mM NaOH before quantification of radioactivity by scintillation counting, data being corrected to disintegrations per minute.
Immunofluorescence microscopy
HeLa cells were grown on coverslips in complete DMEM/FCS for 15 h, and then incubated with rapamycin or actinomycin D for 2 h. Cells were washed twice in phosphate-buffered saline (PBS), fixed with formaldehyde (4% in PBS) for 15 min at ambient temperature, permeabilized with PBS/Triton X-100 0.1% for 10 min and blocked with 1% bovine serum albumin in PBS for 1 h.
Antibodies for nucleophosmin/B23 were from Sigma (FC82291) and for block of proliferation1 (BOP1) from BioVaria. Primary antibodies were incubated for 2 h at room temperature. After washing in PBS, labeled secondary antibodies (rhodamine-conjugated AffiniPure donkey anti-mouse IgG, Jackson ImmunoResearch) were incubated at room temperature for 2 h in the dark at room temperature. Cells were washed with PBS and nuclei were counterstained with TO-PRO-3 (Invitrogen, T3605) 1:1000 for 15 min at room temperature in dark. Coverslips were mounted with 50 µl of ProLong Gold anti-fade (Invitrogen, P36930) overnight in the dark at room temperature.
RNA preparation, northern blot and pre-rRNA species
Total RNA was extracted by the proteinase K method (14). For the northern blot analysis, RNA (5 µg) was fractionated on agarose-formaldehyde gels 1.5% and transferred to GeneScreen Plus membrane (PerkinElmer Life Sciences) for 15 h. Northern blotting was performed essentially as recommended by the manufacturer. Oligonucleotides were end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (BioLabs M0201S) for 30 min at 37°C. Membranes were hybridized overnight in hybridization buffer [6X SSPE, 5X Denhardt's solution, 0.1 mg/ml fish sperm DNA and 1% sodium dodecyl sulfate (SDS)]. Washing was performed in 2X SSPE, 1% SDS at 65°C for 30 min. Membranes were exposed to storage phosphor screen (GE-Healthcare) and analyzed using a Phosphorimager (STORM, GE Healthcare). Quantitation of northern blots was performed using the Image-Quant software (Amersham Biosciences).
Polysome analysis
HeLa cells were lysed with 300 µl of lysis buffer [10 mM NaCl, 10 mM MgCl2, 10 mM Tris–HCl, pH 7.5, 1% (v/v) Triton X-100, 1% sodium deoxycholate, 36 U/ml RNase inhibitor, 1 mM dithiothreitol] and layered in a 20–50% sucrose gradient containing 30 mM Tris–HCl, pH 7.5, 100 mM NaCl and 10 mM MgCl2, and centrifuged in a Beckman SW41 rotor for 150 min at 37 000 rpm (234,000 rcf). Fractions were collected while monitoring absorbance at 254 nm. RNA was extracted from each fraction and northern blot were performed as previously described (15). Radioactive probes were prepared by the random primer technique. Quantitation of northern blots was performed using a PhosphorImager and Image-Quant software (GE Healthcare).
Oligonucleotides
The oligonucleotide sequences complementary to the human rDNA used were:
Subcellular fractionation
To prepare total cell extracts, HeLa cells were washed twice with cold PBS and then lysed in high salt buffer [50 mM Tris HCl pH 7.5, 350 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 0.1 mM EGTA and 1% (v/v) Triton X-100] supplemented with protease inhibitor cocktail tablet (mini EDTA-free complete protease inhibitor cocktail tablet, Roche 11873580001). The cell lysate was incubated on ice for 30 min and centrifuged at 14 500 rpm for 10 min at 4°C.
For the fractionation of nuclei and cytoplasm, HeLa cells were washed twice with cold PBS and resuspended in cold buffer A comprising 10 mM Tris–HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.05% of Nonidet P-40 and protease inhibitor. The cell lysates were centrifuged at 700 rpm for 5 min at 4°C. After removal of the supernatant (the cytoplasmic fraction), the resulting pellet (the nuclear fraction) was washed five to six times with buffer A and resuspended in cold buffer B containing 20 mM Tris–HCl pH 7.5, 420 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 20% (v/v) glycerol, 1% (v/v) Nonidet P-40 and protease inhibitors. The nuclear extract was incubated for 1 h at 4°C on a rotating tube mixer. It was then centrifuged for 10 min at 13 000 rpm (rcf 15 700) at 4°C and the supernatant was carefully removed and stored at −20°C. Nucleoli were isolated as described (18).
Western blotting and antibodies
Protein concentrations were determined by Bradford reagent. Cell lysate was heated at 95°C for 5 min in sample buffer [62.5 mM Tris–HCl, 7% (w/v) SDS, 20% (w/v) sucrose and 0.01% (w/v) bromophenol blue] and subjected to polyacrylamide gel electrophoresis and electrophoretic transfer to nitrocellulose membranes. Membranes were then blocked in PBS–Tween containing 5% (w/v) skimmed milk powder for 30 min at room temperature. The membranes were probed with the indicated primary antibody overnight at 4°C. Antibodies specific for the following proteins were used: mTOR (Cell Signaling Technology, #2972), raptor (Cell Signaling Technology #4978), Lamin B (Santa Cruz sc-6216), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Abcam ab9485), histone H3 (Millipore, 06.755), phospho-S6 ribosomal protein Ser235/236 and Ser240/244 (Cell Signaling Technology, #5316 and #2215), ribosomal protein (Rp)S6 (Cell Signaling Technology, #2317), Pes1 (Abnova B02), p70 S6 kinase (Cell Signaling Technology, #9202) and UBF (Santa Cruz, sc-9131). The antibody for S6K2 was a kind gift from Professor Ivan Gout (University College, London). After incubation with fluorescently tagged secondary antibody, the signals were scanned using a Licor Odyssey® imaging system.
RESULTS
Rapamycin induces the decay of new rRNA molecules
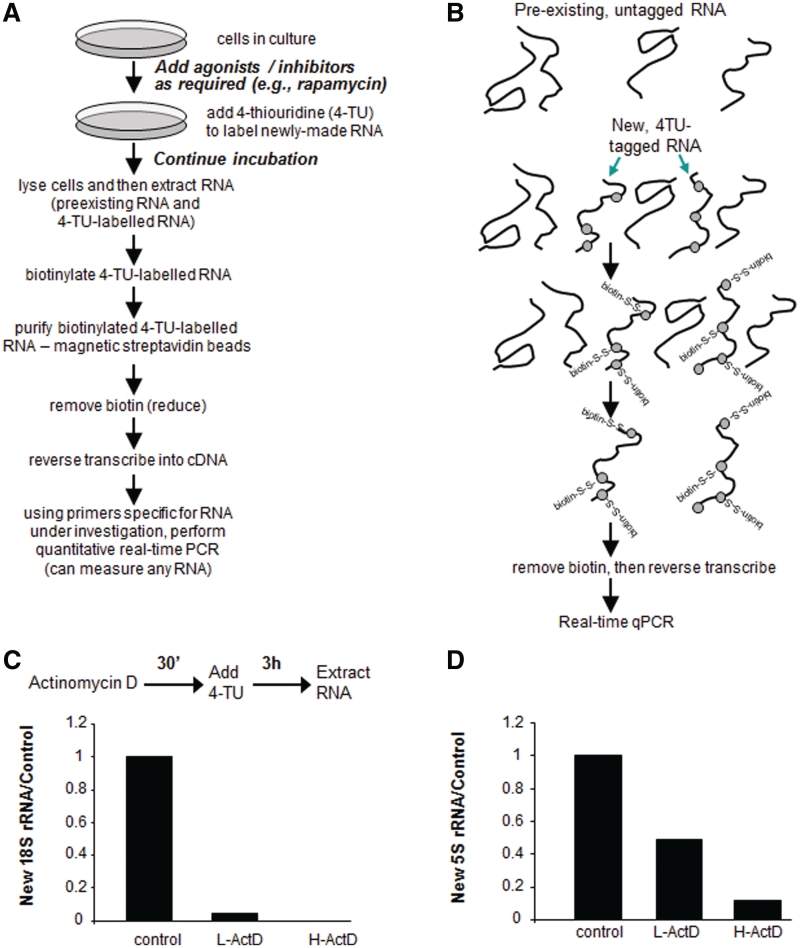
We used a novel approach to study the synthesis of new rRNA molecules, in which they are labeled, in vivo, with 4-thiouridine (4-TU), allowing them subsequently to be biotinylated, specifically purified and subjected to reverse transcription (10,11). The resulting cDNA can then be used as the template for quantitative real-time PCR to determine levels of any specific RNA (Figure 1A and B illustrates the overall procedure). This new approach offers several important advantages over earlier methods using, e.g. [14C]orotic acid to radiolabel new rRNA. In addition to avoiding the use of radioactivity, it allows one easily to quantify specific RNA components by using appropriate qPCR primers, thus obviating the need to run gels to resolve individual rRNA or pre-rRNA species and can readily be configured as a pulse-chase experiment (see below) without complications due to loss of label due to excision of rRNA fragments during processing.
Figure 1.
Procedure used to label newly synthesized RNA molecules with 4-TU. (A) Flow chart and (B) scheme showing the labeling of new RNAs with 4-TU, followed by derivatization with biotin and purification of tagged RNAs. After reverse transcription, any individual cDNA (RNA) species can be quantitated by qPCR. (C and D) HeLa cells were pre-cultured with actinomycin D for 30 min at different concentrations, as indicated (high: 2 μg/ml or low: 20 ng/ml), and then the cells were incubated with 4-TU 100 nM for 3 h. 4-TU-labeled RNAs were extracted and purified as described in the ‘Materials and Methods’ section. The amounts of cDNAs derived from specific RNAs were quantitated by RT–qPCR and are expressed as described in the ‘Materials and Methods’ section: (C) Data for 18S rRNA (also measures its precursors); (B) Data for 5S rRNA.
Figure 1C shows that 4-TU is incorporated into 18S rRNA in HeLa cells, and that, as expected, this is almost totally inhibited by low concentrations of actinomycin D. Label was also incorporated into 5S rRNA (Figure 1D) and this was also partially inhibited by actinomycin D. Consistent with the lower sensitivity of RNA polymerase III to actinomycin D, higher concentrations were needed to achieve strong inhibition of labeling of 5S rRNA. Taken together, the sensitivity of the labeling to actinomycin D indicates that labeling is dependent upon ongoing transcription, i.e. reflects new rRNA synthesis.
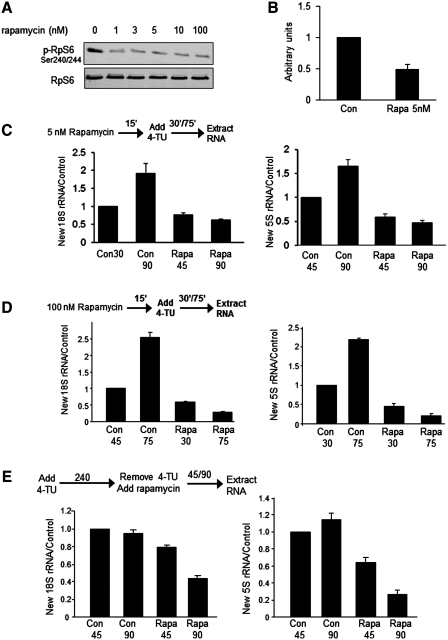
One potential concern was that, as with any labeling technique applied to intact cells, the signaling inhibitors to be used might interfere with the uptake of the label into the cells in addition to affecting, in this case, RNA synthesis by impairing relevant signaling events. Rapamycin is a highly specific inhibitor of mTORC1 when used at high concentrations (19), but has been reported to impair the transport of nucleotides into mammalian cells (20). To try to minimize such effects, we tested the ability of a range of concentrations to inhibit mTORC1 signaling in HeLa cells, monitoring the phosphorylation of ribosomal protein (RpS6) at Ser240/244 as a readout [these residues are specific targets of the S6 kinases which lie downstream of mTORC1 (21)]. As shown in Figure 2A, concentrations as low as 1 nM rapamycin could inhibit RpS6 phosphorylation, although maximal inhibition was only consistently seen at 5 nM. Even at 5 nM, rapamycin inhibited the uptake of radio-labeled [3H]uridine into HeLa cells (Figure 2B); higher concentrations of rapamycin inhibited to a similar extent (data not shown). This effect needs to be taken into account when measuring RNA synthesis using 4-TU or other labeled nucleoside precursors. On the other hand, for pulse-chase experiments, of the type used here to study RNA stability, this effect of rapamycin on nucleoside uptake is not an issue (as discussed below). It should be noted that for the uridine uptake experiments, we took care to pre-treat the cells with rapamycin for a period corresponding to the longest time of exposure for the 4-TU incorporation experiments as described below.
Figure 2.
Rapamycin inhibits the accumulation of new rRNA and promotes its decay. (A) HeLa cells maintained in medium containing serum were treated with or without rapamycin at the indicated concentrations for 90 min. Levels of phosphorylated and total RpS6 were analyzed by western blot; data are typical of three separate experiments. (B) Uridine uptake was measured after 20 s incubation with [3H]uridine in the absence or presence of 5 nM rapamycin (added 90 min prior to the uptake assay). The radioactivity was determined by scintillation counting, normalized to the total protein level for that sample and plotted as a percentage of the uptake into non-treated HeLa cells. (C and D) HeLa cells were pre-incubated with rapamycin (5 or 100 nM, as indicated) for 15 min before the addition of 4-TU. Total RNA was extracted 30 min or 75 min after addition of 4-TU and processed to measure levels of labeled rRNA species as described in the ‘Materials and Methods’ section. Levels of purified labeled rRNA were compared with those in untreated controls that had been labelled for 45 min, data being normalized to the total level of 18 S rRNA. Data are presented as mean ± SEM (n = 3). The times given below each column indicate the period of 4-TU labeling. (E) HeLa cells were pre-incubated with 4-TU for 4 h. After washing with 1 × PBS to remove excess 4-TU and transfer to fresh medium, rapamycin (100 nM) was added. RNA was extracted and samples were processed as for Panels C and D. Data are presented and expressed as described in the ‘Materials and Methods’ section, in this case being normalized to the 45 min control and given as mean ± SEM (n = 3).
We then studied the effects of rapamycin on the synthesis and stability of rRNA using the 4-TU-labeling procedure. It should be noted that these short times of rapamycin treatment (2–6 h) are insufficient to affect cell-cycle progression. Rapamycin inhibited the labeling of newly synthesized 18S or 5S rRNA at 45 min and completely inhibited further incorporation up to 90 min of labeling (Figure 2C and D), which appears consistent with several lines of evidence showing that signaling through mTORC1, or equivalent complexes in other species, promotes rRNA synthesis [reviewed in (2)]. While the reduced level of labeling at 45 min almost certainly reflects, at least in part, impaired uptake of label into the cells, the absence of a further increase in labeled rRNA at 90 min indicates a strong block in new rRNA synthesis over and beyond any impairment of uptake. In fact, and surprisingly, in rapamycin-treated cells, the labeling of all three rRNAs was actually lower at the later time-point than at the earlier one (Figure 2C), suggesting that rapamycin may induce the degradation of new rRNA. Similar data were obtained using rapamycin at 5 nM or 100 nM (Figure 2C and D), the latter concentration being the one we have employed previously to study mTORC1 signaling [see e.g. (22,23)].
One advantage of the 4-TU-labeling approach is that it can readily be configured as a pulse-chase experiment (Figure 2E), where new RNA is labeled with 4-TU and the medium is then changed to one lacking 4-TU, adding e.g. rapamycin and the incubation is continued. In this format, the effect of, e.g. rapamycin on nucleoside uptake is not relevant, as rapamycin is only added after switching to label-free medium. Using this method, no loss of new 18 S or 5S rRNA was seen after the end of the pulse in control cells, while, in contrast, rapamycin-induced rapid loss of 4-TU-labeled 18S or 5S rRNA (Figure 2E), indicating that rapamycin induces the decay of newly made rRNA.
Since 18S rRNA is derived from a single precursor that also contains the 28S rRNA, it seemed likely that rapamycin might also induce decay of new 28S rRNA. Using the same approach, we did indeed observe that rapamycin caused the loss of newly made (4-TU-labeled) 28S rRNA (data not shown). Taken together, these data suggest that rapamycin might interfere with the downstream processing of new rRNA molecules resulting in their degradation.
Interestingly, rapamycin also induced the decay of newly made 5S rRNA (Figure 2D and E), although this rRNA does not require processing. This may reflect the inhibition by rapamycin of the overall process of ribosome assembly, indicating that impaired ribosome biogenesis affects 5S rRNA stability. This type of effect may also explain why low concentrations of actinomycin D partially impair the accumulation of new 5S rRNA (Figure 1D), even though at this concentration it has little direct inhibitory effect on Pol III.
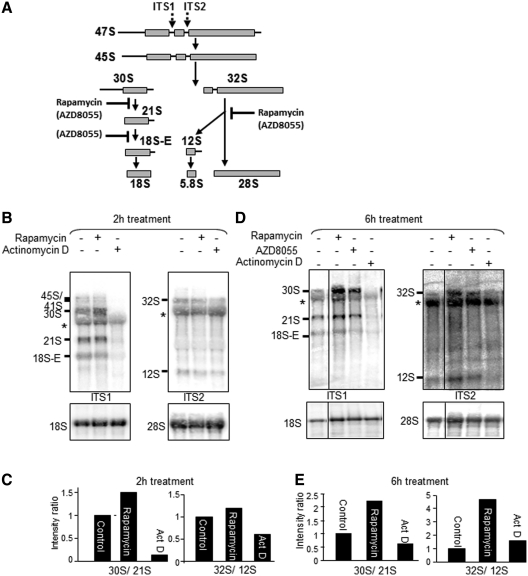
Rapamycin also affects rRNA processing
To study rRNA processing, we used northern blot analysis employing two different probes (ITS1/2) that recognize distinct regions of the 47S rRNA precursor (for details, see Figure 3A). Using the ITS1 probe, it is clear that rapamycin causes a marked increase in the amount of the 30S pre-rRNA, which is a precursor of the 18S rRNA (Figure 3B and C). This is especially apparent after 6-h rapamycin treatment, but is evident as early as 2 h. A modest increase in the 21S species was also observed in response to treatment with rapamycin indicating that mTORC1 signaling likely affects the conversion of the 30S to the 21S pre-rRNA. Similar analysis with the ITS2 probe revealed that treatment with rapamycin for 6 h also caused an accumulation of the 32S pre-rRNA species, the precursor of 5.8S and 28S rRNA (Figure 3D and E), indicating that mTORC1 signaling impacts on both major ‘arms’ of 47S pre-rRNA processing. Thus, inhibition of mTORC1 impairs the processing of 30S to 21S rRNA and also the conversion of 32S to 12S pre-rRNA and 28S rRNA (Figure 3B and C), with the former being affected more quickly by rapamycin. The accumulation of the 32S and 30S pre-rRNA species was more pronounced after 6 h rapamycin treatment than after 2 h (Figure 3D and E and cf. Figure 3B and C).
Figure 3.
Rapamycin affects pre-rRNA processing in HeLa cells. (A) Schematic diagram of pre-rRNA processing pathways in HeLa cells. Structure of the primary 47S rRNA transcript containing two internal transcriber spacers (ITS1 and ITS2) and two external transcribed spacers. The 47S pre-rRNA is processed through intermediate precursors to mature 18S, 28S and 5.8S rRNAs. The locations of the probes used are shown (ITS1 and ITS2). (B and D) effects of rapamycin and AZD8055 (each used at 100 nM) on rRNA processing in serum-fed cells. Northern hybridization of total RNA from HeLa cells after treatment with rapamycin or actinomycin D (20 ng/ml). After 2–6 h treatment (as indicated), total RNA was extracted and subjected to northern blotting analysis for pre-rRNA species. Membranes were hybridized with the ITS1 and ITS2 probes and probes for 18S and 28S rRNAs. The asterisk indicates cross-reaction with 28S rRNA and the vertical line denotes that these are non-adjacent lanes from the same gel/northern blot. (C and E) Ratios of different pre-rRNA species. Bar graphs show signal quantification using a phosphorimager and the bars indicate the ratios of 30S to 21S and 32S to 12S after rapamycin or actinomycin D treatment.
Since inhibition of rRNA transcription would result in the loss of early intermediates in the pathway of rRNA processing, whereas we actually observe an increase in their levels (Figure 3B and D), the effects of rapamycin on processing cannot merely be a consequence of inhibition of rRNA transcription. This is consistent with the effects of low concentrations of actinomycin D (which potently inhibit Pol I; Figure 1C and D) on the processing intermediates that are quite different from those of rapamycin (Figure 3B and C): as expected, actinomycin D caused a loss of processing intermediates, reflecting blocked Pol I transcription. Thus, mTORC1 signaling regulates the processing of pre-rRNA, affecting at least two steps in this process, the conversion of 30S to 21S and the processing of 32S rRNA, in the pathways that lead to 18S and 28S+5.8S rRNAs, respectively (as indicated in Figure 3A).
It was also of interest to assess whether a different type of mTOR inhibitor also impaired the processing of rRNA. AZD8055 inhibits mTOR by acting as a competitive inhibitor of its kinase activity (24), unlike rapamycin, which is an allosteric inhibitor. As shown in Figure 3D, AZD8055 treatment also led to the accumulation of the 30S and 32S species, although the build up of the 32S species was less pronounced than for rapamycin. Interestingly, AZD8055 also caused a modest but reproducible decrease in the amount of the 18S-E species, indicating that it also blocks further processing of the 21S intermediate, which rapamycin does not appear to do. These data indicate that inhibition of mTOR kinase activity, like rapamycin, impairs pre-rRNA processing, but also suggest qualitative differences. This is also clear from the fact that AZD8055 decreased the levels of the 12S intermediate while rapamycin did not (Figure 3D). These differences between the effects of rapamycin and AZD8055 may reflect that fact that the latter compound inhibits mTORC2 as well as mTORC1 and also blocks functions of mTORC1 which are insensitive to rapamycin (25). Similar data were obtained with a different (and slightly less specific) mTOR-kinase inhibitor, PP242 (25; data not shown).
Effects of rapamycin on translation of Rp mRNAs
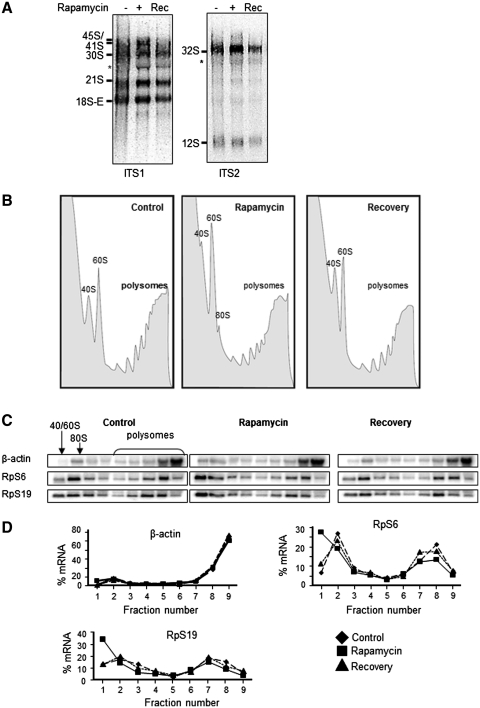
To assess whether the effect of rapamycin on pre-rRNA processing was readily reversible, we treated HeLa cells with rapamycin for 4 h and then, in some cases, changed the medium to one lacking rapamycin, and continued the cell culture for a further 2 h. RNA was extracted and analysed by northern blot. As seen in Figure 4A, removing rapamycin led to a loss of the build up of processing intermediates.
Figure 4.
Effect of rapamycin and its removal on ribosome distribution. HeLa cells were incubated with 100 nM rapamycin for 4 h and then the medium was replaced with fresh medium without the inhibitor for 2 h before the lysis. (A) Total RNA was extracted and subjected to northern blot analysis for pre-rRNA species. The membrane was hybridized with probes ITS1, ITS2, 18S and 28S as previously described. (B) Polysomal profile after rapamycin removal of HeLa cells. The polysomal and non-polysomal fractions are indicated. (C) Northern blot analysis of cytoplasmic RNA extracts from each fraction of sucrose gradient and analyzed with indicated probes (RpS6, RpS19 and β-actin). (D) Quantification of the signals is reported as a linear plot of percentage of mRNAs in each fraction.
Many Rps are involved in the processing of rRNA (26) and their synthesis is modulated by mTORC1 signaling: they are encoded by mRNAs that contain a 5′ tract of pyrimidines (5′ TOP), which modulates their translational efficiency, such that rapamycin impairs their recruitment into polysomes when serum-starved cells are stimulated with serum (27). Thus, it was possible that inhibiting mTORC1 signaling affected rRNA processing by depleting the levels of Rps, especially in the nucleolus where rRNA in processed. We first tested whether rapamycin's effect on pre-rRNA processing was readily reversible, by removing rapamycin from cells after 4 h treatment and allowing them to ‘recover’ for a further 2 h.
We also assessed the effects of rapamycin treatment and ‘recovery’ on the protein synthesis machinery and on translation of selected 5′ TOP mRNAs encoding Rps. As shown in Figure 4B, rapamycin treatment of HeLa cells grown in serum caused only a modest decrease in the levels of active polyribosomes (and a rise in the levels of ‘free’ subunits; especially clear for 60S subunits). Removal of rapamycin led to a substantial, but not quite complete, recovery. Gradient fractions were analyzed by northern blot for the RpS6 and RpS19 mRNAs and also the β-actin mRNA which is not a 5′ TOP message and therefore acts as a ‘negative control’. Neither addition nor removal of rapamycin affected the distribution of the β-actin mRNA between polysomal and non-polysomal fractions (Figure 4C; quantified in Figure 4D). In contrast, rapamycin caused a clear, but only partial, shift of the Rp mRNAs from large polysomes into smaller polysomes and non-polysomal material. Overall, the amount of the Rp mRNAs associated with polysomes decreased only slightly (from 50% of total being associated with polysomes to about 40%; Figure 4D). Removal of rapamycin allowed a recovery in this, although the effect was incomplete (Figure 4C and D). The fact that the changes in Rp mRNA translation caused by rapamycin are small in serum-fed cells, and that recovery is only partial, seems to contrast with the strong inhibition of rRNA processing caused by rapamycin, and the complete recovery seen after removal of rapamycin (Figure 4A).
While these data do not allow us to rule out that changes in Rp levels underlie the effects on rapamycin on rRNA processing, they suggest that they are unlikely to play a major role in them. It is perhaps more likely that alterations in the activity (and phosphorylation) of nucleolar components involved in rRNA processing account for this.
Inhibiting mTORC1 does not induce disruption of the nucleolus
The nucleolus is the site of rRNA transcription, rRNA processing and ribosome assembly. Thus, rapamycin might interfere with rRNA processing simply by inducing the disruption of nucleoli. To assess this, we first used immunofluorescence to study the localization of B23 (nucleophosmin) and BOP1 (block of proliferation 1), both of which are nucleolar proteins involved in rRNA processing (28,29). TOPRO (4-[3-(3-methyl-2(3H)-benzothiazolylidene)]-1-[3-(trimethylamminio)propyl]-, diiodide) was used to stain DNA and thus the nuclei; nucleoli are visible as darker areas within nuclei (yellow arrowheads; Figure 5). Actinomycin D, which at low concentrations only inhibits Pol I, does not cause complete loss of nucleoli as they are still visible against the background nuclear staining. However, it did cause marked disaggregation or redistribution of BOP1 and B23 away from nucleoli so they appear spread across the nucleoplasm (Figure 5). In contrast, rapamycin had little effect on the distribution of B23 and BOP1 (Figure 5), although they did appear slightly more diffuse than in untreated cells. Overall it seems that unlike actinomycin D, rapamycin has, at most, only a modest effect on overall nucleolar integrity.
Figure 5.
Rapamycin treatment induces a partial redistribution of B23 but not of BOP1 into the nucleoplasm. HeLa cells were treated for 2 h with rapamycin or actinomycin D, and then fixed, permeabilized and analyzed by immunofluorescence with monoclonal anti-B23 (red) or anti-BOP1 (green). Cell nuclei were visualized by TO-PRO3 staining. Yellow arrowheads indicate the darker intranuclear regions corresponding to nucleoli.
mTORC1 is localized to the nucleolus
Given that mTORC1 signaling affects the transcription and the processing of rRNA, both of which occur in nucleoli, it was important to ascertain whether components of this pathway were present in the nucleolus. We studied this by subcellular fractionation followed by western blot, rather than by ectopic (over)expression of, e.g. GFP-tagged proteins, as this would likely perturb the stoichiometry and likely the dynamics/distribution of, e.g. mTOR complexes. We detected mTOR and the mTORC1-specific component raptor in the nucleolar fraction as well as the cytoplasmic and nuclear ones (Figure 6A and B). This shows that mTORC1 is present in the cellular locales where mTORC1-dependent transcription and processing of rRNA occur. This is consistent with the findings that (30,31) mTOR is associated with the (nucleolar) Pol I-transcribed genes.
Figure 6.
Cytoplasmic, nuclear and nucleolar distribution of mTORC1 and other proteins in HeLa cells. (A and B) Subcellular localization of endogenous mTOR components or targets and other proteins in HeLa cells. Where indicated, cells were treated with rapamycin (100 nM). The total, cytoplasmic, nuclear and nucleolar fractions were isolated after 2 h treatment and analyzed by western blot. GAPDH (cytoplasmic), lamin B (nuclear) and B23 and UBF (nucleolar) were used as fractionation markers. (C) Bar graph shows signal quantification of nucleolar on the total fraction of mTOR, B23 and UBF after 2 h treatment with rapamycin. (D and E) Localization of endogenous S6K1 and S6K2 in HeLa cells. Since the signal for S6K2 is weak, a longer exposure is also shown for the nucleolar fraction.
We also asked whether rapamycin altered the nucleolar levels of mTOR and other proteins. Rapamycin completely blocked the phosphorylation of nucleolar RpS6, confirming that it inhibits mTORC1 signaling in this compartment (Figure 6B). It caused a modest decrease in the levels of B23 and UBF in the nucleolus. We normalized the data to levels of histone H3 (Figure 6C). Although rapamycin also affected them (Figure 6B), the change was small; it may reflect the alterations in nucleolar morphology seen in Figure 5. Figure 6C reveals that the effect of rapamycin on the amount of nucleolar mTOR was much greater that its effect on either UBF or B23, and thus that the effect of rapamycin on the nucleolar localization is rather selective, not a general effect on all nucleolar proteins.
The S6 kinases (S6Ks 1 and 2) are key downstream targets of mTORC1 that are implicated in ribosome biogenesis [reviewed in (32)]. We were unable to detect S6K1 within the nuclei or nucleoli of HeLa cells (Figure 6D), although a trace of S6K2 was evident on long exposure (Figure 6E). Our data for S6K1 apparently contrast with the recent findings that S6K1 is found in the nucleus when phosphorylated at the mTORC1 site, Thr389 (33,34), which were performed in a different cell type. It may be that nuclear levels of S6K1 are simply too low to detect. The ribosomal protein RpS6 in HeLa cells was also found in the nucleolus (as expected since this is the site of ribosome assembly): rapamycin decreased this, likely due to inhibition of new RpS6 synthesis, reflecting the fact that the translation of its mRNA is inhibited by rapamycin (Figure 4).
DISCUSSION
Our findings demonstrate that, in addition to controlling the transcription of rRNA and the translation of the mRNAs that encode ribosomal proteins, mTORC1 signaling also regulates the processing of pre-rRNAs in human cells. It thus exerts concerted control over multiple steps in ribosome biogenesis—rRNA transcription, pre-rRNA processing and the synthesis of ribosomal proteins. Such coordinated control over ribosome production makes clear physiological sense and is also consistent with the importance of ribosome production for cell growth and proliferation, cellular processes that are positively regulated by mTORC1 signaling.
We show that inhibition of mTORC1 by rapamycin interferes with at least distinct stages in pre-rRNA processes, the conversion of 30S pre-rRNA to the 21S intermediate and the processing of 32S rRNA, the precursor of the 28S and 5.8S rRNAs. AZD8055, a selective mTOR kinase inhibitor, appears to have additional effects pointing to roles for rapamycin-insensitive outputs from mTORC1 (25) or perhaps a role for mTORC2 in pre-rRNA processing. It remains to be established how mTORC1 promotes pre-rRNA processing. Interestingly, rapamycin has also been shown to impair pre-rRNA processing in budding yeast (35), indicating that control of this process by (m)TORC1 emerged early during eukaryotic evolution.
Since some Rps participate in the processing of rRNA (36) and rapamycin inhibits the synthesis [reviewed in (4)] of Rps, it was possible the changes in the levels of newly made r-proteins might be responsible for the effects of rapamycin on rRNA processing, as recently described in yeast (37). Since there are about 80 different ribosomal proteins, it is not feasible to examine the potential involvement of each of them in the effects of mTORC1 inhibition on pre-rRNA processing all of them. Effects of rapamycin on the translation of the 5′-TOP mRNAs were quite modest. This contrasts with data from budding yeast where even very brief treatment with rapamycin (15 min) drastically decreased Rp synthesis (37), perhaps reflecting the steep fall in Rp mRNA levels caused by rapamycin in this species [which probably arises because TORC1 promotes transcription of Rp genes in yeast, whereas it does not do so in mammals (35)]. We also studied a possible role for RpS19, a protein that is mutated in Diamond-Blackfan anaemia, a condition which is associated with impaired ribosome biogenesis including the accumulation of early rRNA processing intermediates (36). This indicates a key role for RpS19 in rRNA processing. However, overexpression of RpS19 did not overcome the defects in rRNA processing caused by rapamycin, indicating that the effects of mTOR inhibition cannot be attributed solely to possible changes in levels of this Rp (data not shown). Thus additional events are important in the control of rRNA processing downstream of mTORC1.
Our finding that mTORC1 is located in the nucleolus suggested a second potential mechanism by which rapamycin might affect rRNA processing; i.e. that affects the amount of mTORC1 associated with nucleoli. To study this, cells were pre-treated with rapamycin prior to fractionation. Rapamycin completely blocked the phosphorylation of RpS6 at Ser235/236 and Ser240/244 [the latter sites being specific targets for the S6 kinases (21)], showing that it is effective in inhibiting mTORC1 signaling in the cellular compartments studied here (Figure 6A). Rapamycin did not affect the distribution of GAPDH or lamin B (cytoplasmic and nuclear markers, respectively), but did cause a marked decrease in the amount of mTOR associated with nucleolar fraction, but not the nuclear fraction (Figure 6A and B). This suggests that rapamycin causes the relocation of mTORC1 away from sites of active pre-rRNA processing, which may provide a mechanism by which it can regulate this process. The decrease in nucleolar mTOR levels may also play a role in the inhibitory effects of rapamycin on Pol I-mediated transcription (2). It was clearly possible that the mTORC1 substrates S6K1 or (since we find it in the nucleus, Figure 6E, albeit at low levels) S6K2 might play a role in rRNA processing. However, overexpressing S6K1 or S6K2 did not protect rRNA processing from inhibition by rapamycin, although it did eliminate its effect on RpS6 phosphorylation (data not shown). Clearly, further work is required to dissect the molecular events that underlie the control of pre-rRNA processing by mTORC1.
The observation that low levels of rapamycin (the minimum required to effectively inhibit mTORC1 signaling) impair uridine uptake suggests that mTORC1 signaling may promote the transport of nucleosides into mammalian cells; this would make good physiological sense as they are precursors for RNA and thus ribosome production, but further work is required to establish whether this effect genuinely reflects a role for mTORC1 signaling (as suggested by the fact that the inhibition caused by 5 nM rapamycin was almost maximal) or is an indirect or ‘off-target’ effect of this compound.
FUNDING
Ajinomoto Amino Acid Research Program (Japan); AstraZeneca UK and the British Heart Foundation 08/099/26124 (to C.G.P.); Canadian Institutes of Health Research MOP-81244 (to E.J.). Funding for open access charge: Research support from the Ajinomoto Amino Acid Research Program.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank Dr David Johnston of the Biomedical Imaging Unit at Southampton General Hospital for his invaluable help with immunofluorescence microscopy and Professor Ivan Gout for anti-S6K2. We are grateful to Dr Harry de Koning (University of Glasgow) for helpful advice on measuring nucleoside transport. We are grateful to AstraZeneca for providing AZD8055.
REFERENCES
- 1.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 3.White RJ. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 4.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 5.Pianese G. Beitraege zur Histologie und Aetiologie der Carconoms [Contributions to the histology and etiology of cancer]. Beitr. Pathol. Anat. Allgem. Pathol. 1896;142:1–193. [Google Scholar]
- 6.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 7.Montanaro L, Trere D, Derenzini M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 9.Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27:5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- 10.Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 2005;23:232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- 11.Woodford TA, Schlegel R, Pardee AB. Selective isolation of newly synthesized mammalian mRNA after in vivo labeling with 4-thiouridine or 6-thioguanosine. Anal. Biochem. 1988;171:166–172. doi: 10.1016/0003-2697(88)90138-8. [DOI] [PubMed] [Google Scholar]
- 12.del SB, Valdes R, Mata J, Felipe A, Casado FJ, Pastor-Anglada M. Differential expression and regulation of nucleoside transport systems in rat liver parenchymal and hepatoma cells. Hepatology. 1998;28:1504–1511. doi: 10.1002/hep.510280609. [DOI] [PubMed] [Google Scholar]
- 13.Duflot S, Riera B, Fernandez-Veledo S, Casado V, Norman RI, Casado FJ, Lluis C, Franco R, Pastor-Anglada M. ATP-sensitive K(+) channels regulate the concentrative adenosine transporter CNT2 following activation by A(1) adenosine receptors. Mol. Cell. Biol. 2004;24:2710–2719. doi: 10.1128/MCB.24.7.2710-2719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA. 2008;14:1730–1736. doi: 10.1261/rna.1037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadjiolova KV, Nicoloso M, Mazan S, Hadjiolov AA, Bachellerie JP. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 1993;212:211–215. doi: 10.1111/j.1432-1033.1993.tb17652.x. [DOI] [PubMed] [Google Scholar]
- 18.Lam Y, Lamond AI. Isolation of nucleoli. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. Vol. 2. Burlington, MA: Elsevier Academic Press; 2006. pp. 103–108. [Google Scholar]
- 19.Lorberg A, Hall MN. TOR: the first 10 years. In: Thomas G, Sabatini DM, Hall MN, editors. TOR Target of rapamycin. Berlin: Springer; 2004. pp. 1–18. [Google Scholar]
- 20.Huang M, Wang Y, Cogut SB, Mitchell BS, Graves LM. Inhibition of nucleoside transport by protein kinase inhibitors. J. Pharmacol. Exp. Ther. 2003;304:753–760. doi: 10.1124/jpet.102.044214. [DOI] [PubMed] [Google Scholar]
- 21.Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang BPH, Wang Y, Wang X, Proud CG. Formation of eIF4F complexes is required for translation of structured mRNAs but not for the mTORC1-dependent activation of general protein synthesis in cardiomyocytes. Am. J. Physiol. Heart. Circ. Physiol. 2009;296:H505–H514. doi: 10.1152/ajpheart.01105.2008. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Fonseca BD, Tang H, Liu R, Elia A, Clemens MJ, Bommer UA, Proud CG. Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J. Biol. Chem. 2008;283:30482–30492. doi: 10.1074/jbc.M803348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, Vincent JP, Ellston R, Jones D, Sini P, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 25.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS. Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14:1918–1929. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jefferies HBJ, Reinhard G, Kozma SC, Thomas G. Rapamycin selectively represses translation of the ‘polypyrimidine tract’ mRNA family. Proc. Natl Acad. Sci. USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strezoska Z, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 2000;20:5516–5528. doi: 10.1128/mcb.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savkur RS, Olson MO. Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res. 1998;26:4508–4515. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang CK, Liu H, Zheng XF. mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle. 2010;9:953–957. doi: 10.4161/cc.9.5.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei Y, Zheng XF. TORC1 association with rDNA chromatin as a mechanism to co-regulate Pol I and Pol III. Cell Cycle. 2009;8:3802–3803. doi: 10.4161/cc.8.23.10039. [DOI] [PubMed] [Google Scholar]
- 32.Jastrzebski K, Hannan KM, Tchoubrieva EB, Hannan RD, Pearson RB. Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors. 2007;25:209–226. doi: 10.1080/08977190701779101. [DOI] [PubMed] [Google Scholar]
- 33.Rosner M, Schipany K, Hengstschlager M. p70 S6K1 nuclear localization depends on its mTOR-mediated phosphorylation at T389, but not on its kinase activity towards S6. Amino Acids. 2011 doi: 10.1007/s00726-011-0965-4. (in press). doi: 10.1007/s00726-011-0965-4. [DOI] [PubMed] [Google Scholar]
- 34.Rosner M, Hengstschlager M. Nucleocytoplasmic localization of p70 S6K1, but not of its isoforms p85 and p31, is regulated by TSC2/mTOR. Oncogene. 2011;30:4509–4522. doi: 10.1038/onc.2011.165. [DOI] [PubMed] [Google Scholar]
- 35.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da CL, Gleizes PE. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiter A, Steinbauer R, Philippi A, Gerber J, Tschochner H, Milkereit P, Griesenbeck J. Reduction in ribosomal protein synthesis is sufficient to explain major effects on ribosome production after short-term TOR inactivation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2011;31:803–817. doi: 10.1128/MCB.01227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]