Abstract
In this work, we examine regulation of DNA methyltransferase 1 (DNMT1) by the DNA damage inducible protein, GADD45α. We used a system to induce homologous recombination (HR) at a unique double-strand DNA break in a GFP reporter in mammalian cells. After HR, the repaired DNA is hypermethylated in recombinant clones showing low GFP expression (HR-L expressor class), while in high expressor recombinants (HR-H clones) previous methylation patterns are erased. GADD45α, which is transiently induced by double-strand breaks, binds to chromatin undergoing HR repair. Ectopic overexpression of GADD45α during repair increases the HR-H fraction of cells (hypomethylated repaired DNA), without altering the recombination frequency. Conversely, silencing of GADD45α increases methylation of the recombined segment and amplifies the HR-L expressor (hypermethylated) population. GADD45α specifically interacts with the catalytic site of DNMT1 and inhibits methylation activity in vitro. We propose that double-strand DNA damage and the resulting HR process involves precise, strand selected DNA methylation by DNMT1 that is regulated by GADD45α. Since GADD45α binds with high avidity to hemimethylated DNA intermediates, it may also provide a barrier to spreading of methylation during or after HR repair.
INTRODUCTION
In mammals, methylation is directed primarily at the cytosine residues of CpG dinucleotides. DNA methylation induces transcriptional repression by preventing binding of basal transcription machinery or other transcription factors that require contact with cytosine residues (1). There are two types of DNA methylation, stable and metastable. Stable methylation is inherited through generations in a male- or female-specific fashion. In contrast, metastable methylation is variable and generates different methylation patterns among individual cells and cell types. It is modified by environment and changes during the lifetime of individual somatic cells. Defective metastable methylation is inherently dangerous and can lead to loss of somatic cell growth regulation and cellular transformation. The root cause of inappropriate methylation is not understood, although it may be related to DNA damage pathways (2–4).
DNA is continually being exposed to cellular metabolites and exogenous DNA-damaging agents, leading to cell death and/or changes in gene expression, which attend loss of growth control. Of the various forms of DNA damage, the most dangerous are DNA double-strand breaks (DSBs), which may create serious problems arising from inappropriate recombination such as chromosomal translocations (5–8). To deal with the threats posed by DSBs, cells have developed multiple mechanisms to detect, signal and repair the regions. Two main pathways, homologous recombination (HR) and non-homologous end-joining (NHEJ), are involved in the repair of DSBs (6). In prokaryotes, HR has been known to be a major pathway for the repair of DSBs, while in eukaryotes, NHEJ was thought to be preferred. More recently, HR has also been shown to be a major pathway in mammals. These pathways are largely distinct from one another and function in complementary ways (9–11). NHEJ involves the ligation of two DNA ends without homology and is highly error prone while HR is essentially error free. In this process, often called gene conversion, a donor DNA sequence with homology to both sides of the DSB supplies genetic information to repair the DSB (12,13). The homologous sequence is copied into the broken locus, making the repaired locus an exact copy of donor sequence, without altering the donor sequence. We previously examined the epigenetic status of DNA repaired by HR and gene conversion to determine whether the repaired DNA region was silenced by DNA methylation (3). This work demonstrated that repaired sites were subject to methylation by DNMT1, which directs methylation to one strand primarily 3′ of the double strand (DS) break. This leaves open the question of how DNMT1 action might be directed to act as a de novo methylase yielding a hemimethylated DNA product that does not convert to full methylation (see model, Supplementary Figure S2).
In order to identify DNMT1 regulatory partners in HR, we evaluated DNA damage inducible factors that have also been implicated in methylation events. One of these is the GADD45α (growth arrest and DNA damage-inducible 45 alpha), which is a small, 18.4 kDa acidic protein originally isolated from cells treated with UV irradiation (14). Subsequently, it was found to be induced by a variety of DNA-damage agents, including ionizing radiation (IR), methyl methansulfonate (MMS) and medium depletion (15,16). Three GADD45-like proteins, GADD45α (GADD45), GADD45β (MyD118) and GADD45γ (CR6), have been identified, sharing 55–60% sequence identity (17). All three genes are inducible by cellular stress, but their expression profile is distinct in various tissues (18). GADD45α (G45a) has a strong p53-binding site in the third intron (19). Its induction by IR is dependent on p53, but the induction by UV, MMS and medium starvation is not (19,20). G45a is also regulated by BRCA1 in a p53 dependent manner (21,22), which suggests a role in HR. The GADD45 family of proteins interact with multiple intracellular proteins including proliferating cell nuclear antigen (PCNA), p21 protein, Cdc2-cyclinB1 complex, core histones and MTK1/MEKK4 in a JNK pathway (18,23–26). These various interactions suggest that the GADD45 pathway has multiple and important roles in signaling of DNA damage in both p53 dependent and independent modes (27). Some of these roles are common to all three isoforms while others may appear to be isoform specific. A known role for G45a is in growth arrest following genotoxic stress. GADD45 proteins bind Cdc2, displace cyclinB1 from Cdc2 and induce cell cycle arrest (21,28). In addition, the suppression of GADD45α or GADD45γ by siRNA abrogates growth arrest supporting the importance of their up-regulation after cellular stress (29). Another biological role of G45a is in DNA repair. Specifically, G45a-like DNMT1 interacts with PCNA and may play a role in nucleotide excision repair (NER) (23). Antisense RNA experiments revealed that depleting G45a levels sensitize cells to UV or cisplatin (30) and G45a null mice exhibit genomic instability and are highly susceptible to carcinogenesis induced by IR or UV (26,31). In addition, it has been reported that G45a interacts directly with core histones to destabilize histone-DNA complexes following UV irradiation (25). These findings imply that G45a binds damaged DNA in a chromatin setting.
A possible role of G45a in DNA demethylation has been reported (32) that G45a promotes DNA demethylation and erases epigenetic marks. G45a over-expression activated a methylation silenced reporter plasmid and interacts with repair endonuclease XPG to direct DNA demethylation suggesting a potential role in active demethylation; however, this finding was challenged by others (33). Recent findings implicate G45a in active demethylation associated with base excision repair (34) and in plants, genomic methylation patterns can be changed through active demethylation involving a family of methyl-cytosine glycosylases (35–37).
Here we investigate the role of G45a in DNA methylation and HR repair. We report that G45a interacts with the conserved catalytic domain of DNMT1 and inhibits methylation. Since G45a is transiently induced during HR, binds HR chromatin and negatively regulates methylases, we hypothesize that it may inhibit DNA methylation of one strand of HR repaired DNA. Using an HR GFP recombination system as a reporter (38), we found that G45a increases the expression of repaired DNA suggesting that G45a provides the choreography associated with strand specific methylation.
MATERIALS AND METHODS
Plasmids
The cDNAs encoding human GADD45α (G45a) from HeLa cells were cloned into a pCMV-MYC1 (39) vector. Expression vectors for V5 epitope tagged full length DNMT1 and deletion mutants (1–419, 412–1113 and 1114–1616 mutants) have been described (39). DNMT1 deletion mutants and intein fused G45a for GST pull-down experiments were generated by PCR using DNMT1-V5 and pCMV-MYC1-G45a as templates, respectively. DNMT1 deletion mutants were cloned into pGEX-5X-1 (GE Healthcare Life Sciences) and G45a was cloned into pTXB1 (NEB). To clone a V5-G45a, pCMV-nV5-His6 plasmid was constructed by inserting a V5-His6 linker into pcDNA3.1(+) (Invitrogen) using HindIII and BamHI sites. G45a PCR products were cloned into pCMV-nV5-His6 using EcoRV and XbaI sites. Using Myc-G45a or V5-G45a as a template, point mutants of G45a, RT34AA, LC56AA, ED63AA and CE83AA, were generated by PCR using site-directed mutagenesis kit (Stratagene). Primers used in this study are listed in Supplementary Table S1.
Cell culture and transfection
HeLa-DR-GFP cells, containing a single integrated copy of the HR reporter (also known as HO-1 cells) (3) were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (Gibco). In some experiments (where indicated), the HeLa DR-GFP cells were analyzed as a pool of DR-GFP cells and were an uncloned population representing all integration events. In other experiments, it was necessary to use cloned HO-1 cells. Both cell systems gave concordant results. HEK293FT cells (Invitrogen) were grown in DMEM with 10% fetal bovine serum. Transfection of plasmids was performed using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. Transfection efficiency was measured by assaying β-galactosidase activity of an included pSVβGal vector (Promega).
Viral production and infection
Lentiviral vectors (shRNA#49 and shRNA#50) for G45a were purchased from Open Biosystems (clone numbers: TRCN0000062349 and TRCN0000062350). Scramble shRNA lentiviral vector for use as a negative control was obtained from Addgene. We transfected HEK293FT cells cultured on 6-cm dishes with the shRNA lentiviral vectors (2 µg) together with the packaging plasmids psPAX2 (1.5 µg) and pMD2G (0.5 µg) using Lipofectamine 2000 reagents. Viral supernatants were harvested on three consecutive days starting 24 h after transfection, yielding a total of ∼15 ml of supernatant per virus. HeLa-DRGFP cells were infected with 150 ∼ 400 µl of viral supernatant at a density of 200 000 cells/well (in 6-well plates) in the presence of polybrene (9 µg/ml).
Co-immunoprecipitations
Whole-cell extracts were prepared by lysing cells in RIPA buffer (1× PBS pH 7.5, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 10% glycerol) containing 1× protease inhibitor cocktail (Roche), 1 mM DTT, 1 mM MgCl2 and 2 mM PMSF. For co-immunoprecipitation, the whole-cell extracts were diluted four times with 0.5× PBS containing 1× protease inhibitor cocktail, 1 mM DTT, 1 mM EDTA and 2 mM PMSF. Antibody (1 µg) was added to samples for 3 h, followed by 30 µl of protein A Sepharose 4B beads (Zymed) at 4°C for 18 h. Beads were washed three times with 1× PBS, 0.5% NP-40 and bound proteins eluted with sample buffer (60 mM Tris/HCl pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue) and analyzed by western blotting. The antibodies and their commercial sources were as follows: anti-V5 (Invitrogen), anti-DNMT1 (NEB), anti-Flag (Sigma) and anti-Myc (Upstate).
GST pull downs
Escherichia coli strain BL21 (DE3) was transformed with plasmid harboring GST fused DNMT1 deletion mutant or intein fused G45a. Proteins were induced by adding IPTG to final concentration of 0.4 mM for 4 h at 37°C. Intein fused G45a was purified according to manufacturer's instructions (NEB). To purify GST fused DNMT1 deletion mutants, cell pellets were resuspended in PBS containing protease inhibitor cocktail (Roche), 1 mM PMSF, 10% glycerol and lysozyme (Sigma, 100 µg/ml) and incubated for 20 min on ice. Cells were sonicated and lysates cleared by centrifugation. Proteins were purified using GST-Sepharose 4B (GE Healthcare Life Sciences). To determine the purification yield of each deletion mutant, small aliquots of each bead preparation were mixed with sample buffer and analyzed by SDS–PAGE. Equal amounts of each deletion mutants were mixed with 100 ng of purified G45a and incubated overnight. Beads were extensively washed and bound proteins were eluted using sample buffer. The eluted proteins were analyzed by western blot using anti-G45a antibody (Santa Cruz, C-4).
Chromatin immunoprecipitation
Cells were transfected with either empty vector or I-SceI plasmid (5 µg) along with V5-GADD45α (1.25 µg) in 150-mm dishes. After 48 h, cells were fixed with 1% formaldehyde. Chip-IT Express kit (Active Motif) was used for chromatin immunoprecipitation (ChIP) experiments. ChIP reactions were set up according to the manufacturer's instructions. Briefly, the sheared chromatin (corresponding to 18 µg of DNA) was mixed with protein G magnetic beads and 2 µg of each antibody or normal mouse IgG (Santa Cruz). Antibodies used are: anti-V5 (Invitrogen), anti-G45a (H-165, Santa Cruz) and anti-DNMT1 (N-16, Santa Cruz). The reaction mixtures were incubated at 4°C for 48 h. Beads were washed and immunoprecipitated DNA was recovered. The enriched DNAs were analyzed by PCR using Rec and 3′-common primers (see Supplementary Figure S1) (3). PCR was performed in a 20 -µl reaction mixture containing 5 µl of recovered DNA (or 100 ng of input DNA), 0.2 mM dNTP, 1.25 U of HotStarTaq polymerase (Qaigen) and 5 µM of each primer. Amplifications were performed using the following conditions: 95°C/5 min× 1 cycle; 95°C/45 s, 66°C/40 s, and 72°C/40 s × 40 cycles; and 72°C/10 min× 1 cycle.
Methylated-dependent immunoprecipitation (MeDIP) and methyl-sensitive restriction analysis (MSRA)
For MeDIP analysis, HeLa cells were transfected with 4 µg of I-SceI expression vector and with 100 nM G45a or EZH2 siRNAs (Dharmacon). Six days later, the genomic DNA was prepared with QIAamp DNA Mini Kit (Qiagen) according with the manufacturer instructions, and 10 µg were digested with EcoRI, HindIII and HpaII (Roche) overnight at 37°C. The digested genomic DNA was then extracted with phenol-chloroform-isoamyl alcohol (25:24:1), ethanol precipitated, resuspended in TE (10 mM Tris pH 8.0, 1 mM EDTA) and quantified by absorbance at 260 nm. Next, 2.5 µg DNA was diluted in 500 µl of IP buffer (0.15% SDS, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 10 mM Tris pH 8.0, 0.1% BSA, 7 mM NaOH) and incubated at 95°C for 10 min before the immunoprecipitation with 5 µg of 5-methyl cytosine antibody (Abcam). As a control, 5 µg of normal mouse IgG (Santa Cruz) was included. The samples were incubated (overnight at 4°C on a rotating platform) followed by addition of 20 µl of Protein A/G Plus-Agarose (Sigma) and a 2 h (4°C). Beads were washed as follows: 2× Low Salt buffer (0.1% SDS, 0.1% DOC, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 10 mM Tris pH 8.0); 1× High Salt buffer (0.1% SDS, 0.1% DOC, 1% Triton X-100, 500 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris, pH 8.0); 1× LiCl buffer (0.25 M LiCl, 0.5% DOC, 0.5% NP-40, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris, pH 8.0); 2× TEE buffer (1 mM EDTA, 0.5 mM EGTA, 10 mM Tris, pH 8.0). The immunocomplexes were resuspended in 200 µl of Proteinase K solution [50 mM Tris, (pH 8.0) 10 mM EDTA, 0.5% SDS] and digested with 200 µg/ml Proteinase K (Roche) for 4 h at 55°C. The samples were then extracted with phenol-chloroform-isoamyl alcohol and ethanol precipitated, resuspended in 10 mM Tris pH 8.0 and 1 mM EDTA pH 8.0. For the PCR analysis, 2 µl of immunoprecipitated DNA and 2 µl of digested genomic DNA were used for the PCR reaction. The primers used are in Supplementary Table S2. In this analysis, two control genes (H19, an imprinted gene and UBE2B, an unmethylated DNA control) were included to validate that methyl-DNA was being selectively precipitated by the method. PCR was performed in a 20 -µl reaction mixture containing 0.2 µM of each primer and 10 µl of QuantiFast SYBR Green PCR Kit 2× (Qiagen). Amplifications were performed using the following conditions: 95°C/15 min× 1 cycle; 95°C/45 s, 65°C/30 s, and 72°C/1 min× 40 cycles; and 72°C/10 min× 1 cycle for the Rec2-DRGFP ChIP LOWER R primers. PCR conditions for H19ICR and UBE2B were: 95°C/10 min× 1 cycle; 95°C/45 s, 59°C/30 s, and 72°C/30 s × 5 cycles; 95°C/45 s, 56.5°C/30 s and 72°C/30 s × 35 cycles; 72°C/10 min× 1 cycle.
For MSRA, genomic DNA was isolated from cells with DNeasy Blood and Tissue kit (Qiagen). To remove unrepaired DNA from repaired DNA, total genomic DNA was digested with I-SceI endonuclease (Fermentas) prior to bisulfite modification. The genomic DNA (5 µg) was digested for 18 h at 37°C with 10 U of I-SceI enzyme. The digested DNA was cleaned up using QIAprep Spin Miniprep kit (Qiagen). To monitor methylation of repaired sites after HR, 1 µg of genomic DNA was digested with 5 U of HpaII before amplification with Rec and 3′-common primers. The methylation was quantified using a GeneTools software (SynGene, Cambridge, UK).
In vitro DNA methylation assays
DNA methylation activity assays were performed as described (39) with slight modifications. Briefly, DNMT1 activity was measured in the presence or absence of purified GADD45α by the incorporation of labeled Ado-Met S-adenosyl-l-[methyl-3H] methionine (PerkinElmer) into a 30-bp oligonucleotide substrate containing a single CpG site in hemimethylated form (D1subHMF: 5′-GAA GCT GGG ACT TCM GGC AGG AGA GTG CAA-3′, D1subUMF: 5′-TTG CAC TCT CCT GCC GGA AGT CCC AGC TTC-3′, where M denotes 5-methylcytosine) or the same oligo as an unmethylated substrate. (oligonucleotides from Integrated DNA Technologies). Double-stranded oligonucleotides were annealed by mixing equal amounts of complimentary oligonucleotides, heating to 95°C for 5 min, 65°C for 10 min and cooling down to 20°C. Under these conditions, reconstruction experiments confirmed that annealing is essentially complete (>99%). The methylation reaction was carried out at a concentration of 0.5 µM DNA, 0.5 µCi Ado-Met and 400 ng of DNMT1 (purified from baculovirus) in methylation buffer (20 mM Tris/HCl pH 7.5, 5 mM EDTA, 5 mM DTT, 1 mM PMSF and 10% glycerol) at 37°C. At defined times, the reactions were stopped by adding phenol/chloroform and DNA was precipitated by adding the same volume of isopropyl alcohol. The DNA pellet was dissolved in TE buffer and transferred to Whatman filter paper. Radioactivity was determined using a LS6500 scintillation counter (Beckman Coulter).
Analysis of homologous recombination
Homologous recombination (HR) assays were performed as previously described (3). HeLa-DRGFP cells or cell transduced with lentivirus expressing shRNAs were transfected with I-SceI plasmid (sometimes along with Myc-G45a plasmid) as well as pSVβGal. After transfection, cells were harvested at defined time points and GFP positive cells were analyzed by flow cytometry using FACSCalibur (BD Biosciences). Statistical analyses of FACS data and expression classification (HR-L and HR-H) was carried out using previously published methods (see Supplementary Figure S8) (3).
RNA extraction and RT–PCR
Total RNA was isolated from cells with RNeasy Minikit (Qiagen). cDNA synthesis and amplification was performed with OneStep RT-PCR kit (Qiagen) using gene specific primers. Primer sequences are listed in Supplementary Table S1.
Surface plasmon resonance
DNA–protein interaction analysis was performed by Surface Plasmon Resonance (SPR) experiments using a Reichert SR7000 SPR refractometer (Reichert Inc., Depew, NY, USA). Two types of binding experiments were performed. First, duplex biotinylated DNA oligomers (either unmethylated, hemimethylated or fully methylated DNA oligomers described above for DNMT1 activity assays) were immobilized to a neutravidin coated sensor slide (Reichert Inc., Depew, NY) at a flow rate 6.67 µl/min. Second, purified His-GADD45α was bound to the sensor slide (through amide chemistry) at 6.67 µl/min (in this case binding orientation of the protein was random). Unbound material was washed off with PBS containing 0.2% Tween-20. HBS buffer (10 mM HEPES pH 7.4, 150 mM NaCl and 3 mM EDTA) was used as a running buffer. To determine equilibrium dissociation constants (KD) for DNA–protein interactions, various concentrations of either oligo or GADD45α were applied at a flow rate of 13.3 µl/min at 37°C in HBS buffer. Reichert Labview software was used for data collection. Biologic Scrubber 2 software (Campbell, Australia) was used for curve fitting and data analysis. The sensogram raw data were subjected to curve fitting with regression values in excess of 97%.
Statistical analyses
Data are presented as the means ± SE of at least three experiments in triplicates (n ≥ 9). Statistical significance between groups was determined using Student's t-test (matched pairs test or unmatched test were used as indicated in the figure legends). Statistical analysis was performed using the JMP 6.0.3 software by S.A.S (www.sas.com).
RESULTS
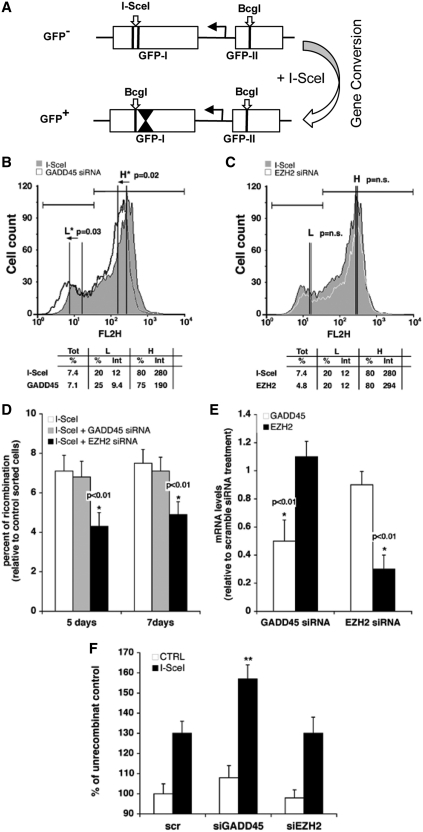
We previously showed that HR results in methylation revisions of the post-HR templates using a DR-GFP reporter (Supplementary Figure S1) (3). These methylation events localized to a region 3′ of the I-SceI cleavage site, indicated by inverted triangles in Figure 1A. Following HR repair, hemimethylated intermediates resolve into two populations of GFP positive cells (corresponding to a high expression class or HR-H, which are hypomethylated GFP molecules and a low expression class or HR-L, which are hypermethylated molecules, see Supplementary Figures S1 and S2). The HR-H and L populations derive from templating events off the methylated and unmethylated strands as the cells divide (3). These two populations were FACS sorted and the methylation status confirmed by bisulfite sequencing analysis; therefore, we validated that FACS analysis represents a precise, albeit independent, measurement of methylation of repaired segments post-HR. We selectively silenced G45a in HeLa DR-GFP pools of cells (representing a wide range of integration events) (3) following I-SceI expression and examined the frequency of recombination at 5- and 7-day post-HR (Figure 1D). In addition, HR-L and H expression classes were examined as a readout for repair induced methylation (Figure 1B and C), and methylation of the 3′ GFP segment was directly evaluated over a 437-bp region 3′ of the I-SceI site (Figure 1F). The data show that G45a depletion (Figure 1E) during repair stimulates formation of hypermethylated recombinant GFP products (Figure 1B). This was confirmed by showing an increase in DNA methylation levels at the 3′ end of the I-SceI site by methyl-DNA immunoprecipitation (Figure 1F). Methyl-DNA immunoprecipitation (MeDIP) experiments were replicated for statistical analysis (Supplementary Figure S9). Two external control genes (H19, imprinted and UBE2B, unmethylated) were included in the experiment to validate antibody quality and specificity. In addition, methyl-C DNA recovery was unchanged before and after HR at these two control genes, demonstrating that changes in DNA methylation selectively target the 3′ GFP region after I-SceI cleavage (data not shown). The PRC2 repressor component EzH2, responsible for H3K27me3 marks that silence genes during differentiation (40), was compared with the G45a results (Figure 1C and E). Since EzH2 is also recruited to DS DNA breaks (4), we evaluated the impact of its depletion on post-repair silencing and recombination frequency. Depletion of EzH2 alters HR frequency which introduces a bias in analyzing GFP expression. For this reason, we asked whether G45a siRNA might similarly affect the frequency of HR. In contrast to EzH2, HR frequency was not influenced by G45a siRNA tested at 5- and 7-day post-I-SceI. As additional evidence, we show that HR frequency was not affected by depleting G45 using two different shRNAs and the DR-GFP reporter cell system (HO-1 cells, containing a single integrated copy of DR-GFP reporter, Supplementary Figure S7E). The overall trend is consistent with the view that when G45a is depleted, greater methylation of GFP molecules is a direct or indirect result. Using an independent method (MSRA, methylation sensitive restriction analyses), we confirmed that two different shRNA G45a knockdowns in HO-1 cells showed this same trend (enhanced DNMT1 methylation 3′ of the I-SceI break) without any change in HR frequency, as noted (Supplementary Figure S7). The HO-1 cell system is a more tractable system for analysis of the kinetics of the HR repair silencing, compared to experiments that use pools of transfected cells (see next section).
Figure 1.
Silencing of G45a modifies targeted methylation and GFP expression in recombinant cells. (A) Map of the DR-GFP reporter (13). HeLa cells containing the DR-GFP reporter shown as two cassettes (5′ GFP-I and 3′ GFP-II) are transfected by an I-SceI expression plasmid. This results in HR and gene conversion whereby wild-type cassette II serves as donor template to convert the I-SceI site in cassette in GFP-I to a BcgI site. The inverted triangles in GFP-I of wild-type GFP+ cells mark the 437-bp region of methylation reprogramming that attends this process. A more detailed map of the process, including the PCR primers used to unambiguously distinguish recombinant (5′ Rec) molecules is given in Supplementary Figure S1. (B and C) Analysis of GFP expression classes. HeLa DR-GFP cells (as a pool of DR-GFP transfected HeLa cells) were transiently transfected with G45a, EzH2 or scrambled control siRNAs along with the I-SceI expression vector. After 5 days the cells were subjected to FACS analysis to quantify HR-H (H) and HR-L (L) cell populations. The arrows indicate the shift of the cells following the transfection with siRNA. A paired t-test to compare mean GFP intensity was used; P < 0.05 is indicated by the asterisk (n.s., ‘not significant’). (D) HR recombination frequency in silenced cells. Genomic DNA was extracted from HeLa DR-GFP cells (silenced by siRNA G45a or EzH2) at 5- or 7-day post-I-SceI transfection. Specific GFP primers (Rec) were used to determine the frequency of HR relative to control cells (I-SceI only) using RT-PCR as described previously (3). The matched statistical analysis used to derive P-values is shown in Supplementary Figure S9. (E) RNA analysis in G45a and EzH2 silenced cells. The total RNA was extracted from HeLa DR-GFP cells 48 h after the transfection with I-SceI. In all transfections, G45a and EzH2 mRNA levels were determined by RT-PCR (data normalized to the GAPDH gene). The statistical analysis is presented in Supplementary Figure S8. (F) Methyl-C DNA content (MeDIP) in G45a silenced cells. Genomic DNA was extracted from HeLa DR-GFP cells 5 days after I-SceI transfection, restriction digested and immunoprecipitated with anti-methyl-C specific antibodies. The 5′ Rec and Unrec primers (Supplementary Figure S1) were used to amplify DNA recovered from the immunoprecipitate. The data were normalized for the percentage of immunoprecipitated unrecombinant control. Scr is the scrambled RNA negative control. Mean comparisons were calculated using Student’s t-test (**P = 0.0046, G45a and EZH2; P = 0.0047, G45a and Scr control; Scr and EZH2 no significant difference, P = 0.99) (Supplementary Figure S9). Control experiments with an H19 (a known imprinted gene) and UBE2B (unmethylated control DNA) were included to demonstrate specificity of the commercial antibody for methylated DNA; recovery of immunoprecipitated methyl-C DNA in the control genes was not affected by I-SceI transfection (data not shown).
Ectopic G45a relieves HR mediated repression of recombinants
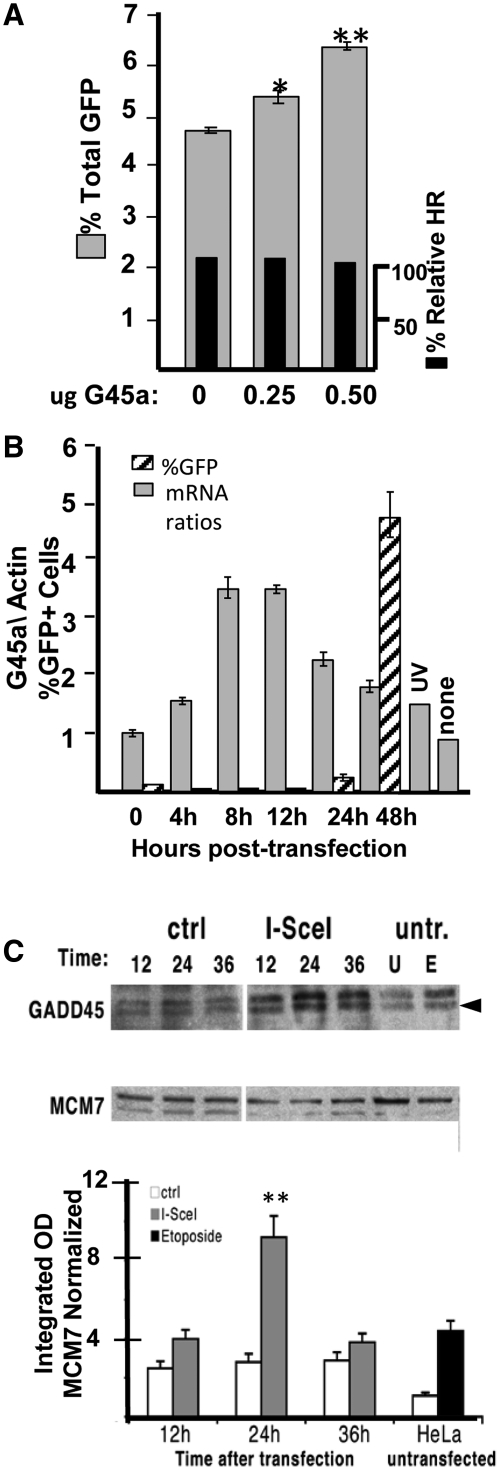
We next tested the influence of ectopic high level expression on total GFP response in the DR-GFP reporter cell system using a single clonal isolate in HO-1 cells (2). Increasing amounts of G45a increase total GFP expression without a detectable change in HR frequency (Figure 2A). The increase, while not large was highly reproducible and statistically significant (Supplementary Figure S10); therefore, we conclude that elevated G45a relieves transcriptional repression due to alterations in epigenetic reprogramming. We next tested the kinetics of expression of the G45a gene after I-SceI transfection of HO-1 cells (Figure 2B and C). G45a mRNA appears relatively soon after double strand DNA break formation (4 h), peaks between 8 and 12 h and is reduced to half by 48 h (and continues to decline at later times, not shown). Similar results were seen in UV treated cells (24 h induction shown in Figure 2B). The first detectable GFP positive cells appear between 24 and 48 h (Figure 2B, Supplementary Figure S4). This time period coincides with the appearance of G45a protein (Figure 2C). The peak amount of G45a protein following DSB (Figure 2C) was slightly delayed relative to the mRNA (which peaked between 8 and 24 h, Figure 2B). Overall, mRNA and endogenous protein levels increase by ∼2- to 4-fold after I-SceI exposure. Topoisomerase II induced DNA breaks due to etoposide treatment showed a strong induction of G45a protein in untransfected HeLa cells as expected (greater than UV). These data establish that G45a induction is early and transient in the reporter cells. Results with pools of DR-GFP Hela cells (Figure 1) and a single clone (HO-1 cells) were concordant.
Figure 2.
Influence of G45a on HR and kinetics of G45a expression in HO-1 cells. (A) Influence of ectopic G45a on GFP expression. HO-1 cells containing a single integrated copy of DR-GFP (see ‘Materials and Methods’) were transfected with I-Sce plus or minus a plasmid encoding G45a (both plasmids express constitutive levels using CMV enhancer). GFP percent was determined 48 h later concurrent with an analysis of HR frequency using suitable primer pairs [see Supplementary Figure S1 and (3) for details]. *P < 0.03 between 0 and 0.25 µg; P < 0.0002 between 0 and 0.5 µg (statistical significance determined using paired t-test and four experiments, see Supplementary Figure S10). (B) Time course of G45a mRNA induction after DSB. HeLa DR-GFP (HO-1) reporter cells were transfected by I-SceI and RT-PCR performed to measure G45a mRNA levels (shaded boxes) at various times post-transfection as described in the ‘Materials and Methods’ section. Concurrently, GFP expression was monitored by FACS analysis. The UV induction control to measure G45a mRNA was performed 24 h after UV exposure. (C) Time course of G45a protein induction by DSB. HeLa DR-GFP cells were transfected with I-SceI plasmid or with a control vector. Control cultures were untransfected cells untreated (U) or etoposide treated for 18 h (E). Cell extracts were prepared at the indicated intervals (12–24–36 h after the I-SceI transfection) for transfected and at 36 h for the etoposide samples (untr., no I-SceI transfection). A representative immunoblot with specific antibodies to G45a and a reference nuclear protein (MCM7) in cell extracts from HeLa DRGFP cells treated (E) or not (U) with etoposide or transfected with I-SceI or a control vector, and analyzed at the times indicated. The lower panel shows the statistical analysis of the G45a signals compared to a reference control (MCM7). The data are derived from three independent experimental determinations (P < 0.0001 comparing 24–36 h and 12 h points) (Supplementary Figure S10).
Endogenous G45a and DNMT1 found in recombinant GFP chromatin
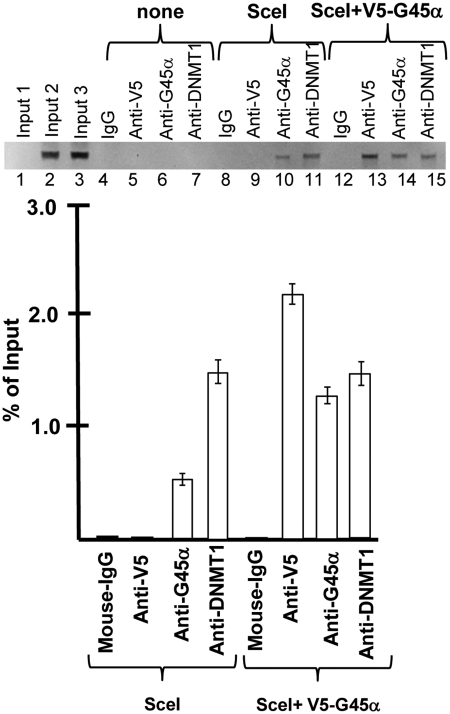
To test the idea that endogenous G45a binds to DNA and/or DNMT1 and regulates methylation during HR, we performed chromatin immunoprecipitation (ChIP) analysis. For this analysis, we transfected cells with I-SceI and performed a ChIP experiment using specific and control antibodies and the Rec primer (Supplementary Figure S1 map) to amplify recombinant DNA products specifically. In the absence of I-SceI, Rec DNA products were not recovered with any of the antibodies tested (Figure 3, Lanes 4–7). ChIPs were performed with endogenous G45a and ectopic V5-G45a. The data show that there is a selective recovery of Rec products with G45a (endogenous and V5 tag) as well as DNMT1 (3) (Figure 3, Lanes 8–15). The result is highly reproducible as seen with the DNMT1 ChIP control demonstrating nearly identical recovery (compare Lanes 11 and 15 and quantitative data in graph) for both experiments (comparing endogenous and V5-G45a). Recovery of endogenous G45a was reproducibly lower than that seen with V5-G45a, which is expected since the V5 promoter construct is constitutively expressed at higher levels.
Figure 3.
Chromatin immunoprecipitation during HR. ChIP assays before and after HR repair. ChIP assays were performed as described in the ‘Materials and Methods’ section in cells transfected with vector DNA (‘none’), I-SceI plasmid (‘SceI’) or co-transfected with V-5-G45a (N-terminal tag) plus I-SceI expression constructs at 48 h after I-SceI transfection. The precipitating antibodies used in each analysis are shown. The 5′ Rec primer amplifies only HR recombinants. The input Lanes 1–3 correspond to DNA inputs for none, SceI and SceI+V5-G45a samples.
G45a and DNMT1 interactions modulate methylase activity
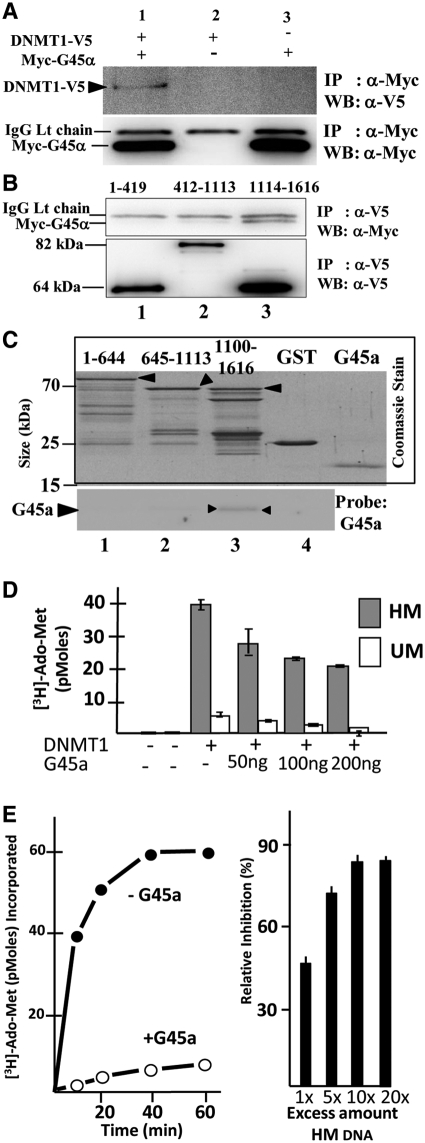
DNA methylation at HR segments is primarily mediated by DNMT1 (3) and we considered the possibility that G45a and DNMT1 might also physically interact (Figure 4A). G45a binds the C-terminal domain of DNMT1, the conserved catalytic center of the methylase (Figure 4C). In a chromatin context, many protein/protein interactions are likely involved in directing DNMT1 at HR templates; however, a fundamental question is whether additional bridging proteins are involved with G45a. Using GST tagged protein, G45a forms a complex with the C-terminal catalytic region of DNMT1 (Figure 4B) showing that the two bind directly in vitro. The result of this interaction was further examined in vitro by testing the influence of G45a on DNMT1 methylation of un- or hemi-methylated oligo targets (Figure 4D). G45a strongly inhibits DNMT1 activity on both substrates and the inhibition was clearly affecting the methylation rate (Figure 4E). Inhibition is partially but not totally overcome by addition of excess hemi-methylated DNA target; therefore G45a and DNMT1 also compete for DNA binding (Figure 4E, right panel). It is likely that G45a has some advantage over DNMT1 in terms of DNA binding since the KD value for G45a/hemi-methylated DNA is ∼0.28 µM compared to DNMT1/hemi-methylated DNA KD of 1.07 µM (Supplementary Figure S6). A binding preference for substrate, combined with occlusion of the DNMT1 catalytic center could explain the inhibitory effects of G45a on DNMT1 methylation (given that G45a also preferentially binds hemimethylated DNA). Recent protein structure studies on DNMT1 further demonstrate the importance of the catalytic domain interactions with a central region that makes close contact with each CpG target at the site of methylation (41). G45a would interfere with this interaction and disrupt the activity of DNMT1 as a result.
Figure 4.
DNMT1 and G45a binding. (A) Immunoprecipitation followed by western blotting show DNMT1 and G45a interact as binding partners. DR-GFP HeLa cells were transfected with tagged DNMT1 (C-terminal V5) and G45a (N-terminal Myc tag) genes. Extracts were subjected to immunoprecipitation with Myc IgG and western blot performed using a V5 IgG probe (upper panel) or a Myc probe (lower panel). The position of each polypeptide is marked along with the IgG light chain that is present in all samples (lower blot). (B) Domain mapping. The DNMT1 protein was divided into three domains corresponding to the N-terminus (residues 1-419), central domain (412–1113) and C-terminus (1114–1616) containing the conserved catalytic region of the methylase (39,41). The deletion mutants are all V5 tagged and differences in expression levels were normalized for each mutant and taken into account. HeLa cells were transfected with Myc-G45a and DNMT1 mutants. At 48-h post-transfection, extracts were immunoprecipitated with anti-V5 IgG and recovery products probed on western blots with anti-Myc IgG (upper blot). (C) Direct binding between DNMT1 domains and G45a. GST tagged, purified domain segments of DNMT1 (residues 1–644, 645–1113 or 1100–1616) were incubated with purified G45a and isolated by the GST tag. After extensive washing of the glutathione affinity resin, recovery products were run on an SDS–PAGE, which was stained with Coomassie Blue (boxed upper panel). The same samples were subjected to western blotting using an anti-G45a IgG probe as described in the ‘Materials and Methods’ section. (D) Effects of G45a binding on DNMT1 methylation activity in vitro. DNMT1 methylase assays were carried out for 60 min using 200 ng purified DNMT1 (from Baculovirus) and the indicated amount of purified G45a (E. coli). The substrate was a 30-bp duplex oligo containing a single CpG site that was either hemi- or unmethylated as indicated. Radioactive Ado-Met incorporation was measured as described in the ‘Materials and Methods’ section. The stoichiometric (molar) ratios of G45a:DNMT1 were calculated to range from 2.5:1 (50 ng G45a) to 7.5:1 (200 ng G45a) using a predicted molecular mass for the dimer form of G45a. (E) Time course of methylation and inhibition reversal. A time course of methylation of hemi-methylated (HM) DNA substrate is shown plus or minus G45a (left panel). The influence of excess HM DNA on inhibition is shown on the right panel (under conditions that gave 50% inhibition by G45a with a fixed amount of HM DNA substrate).
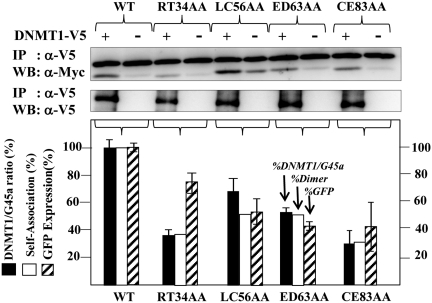
Alanine substitutions that alter dimerization potential affect DNMT1 methylation outcomes
Structural data suggest that G45a proteins function in vivo as multimers (homodimers or higher oligomers) to induce cell cycle checkpoints as a stress responder (8,42). Since we demonstrated that ectopic V5-G45a substitutes for the endogenous activity (Figures 1 and 2), we made alanine scanning mutations to examine dimerization potential in cells (Supplementary Figure S3), and tested for three parameters of G45a function in HR silencing (Figure 5): (i) ability to bind DNMT1; (ii) ability to form homodimers (or higher multimers) and (iii) influence on total GFP expression post-HR. DNMT1/G45a interaction data are shown above the histogram in Figure 5. These data (black boxes, Figure 5 histogram) reveal that DNMT1 binding correlates generally with its ability to dimerize, suggesting a multi-protein assembly in chromatin is likely to be strongly dependent on the ability of G45a to self-associate. Cross tag (V5-G45a, Myc-G45a) pull down analyses with different alanine substitutions (Supplementary Figure S5A) show that dimerization was impaired from 30% to 50% (Figure 5 open boxes, histogram). In addition, total GFP expression (stippled boxes) was reduced in three of the mutants in proportion to their ability to dimerize and interact with DNMT1; however, one mutant (RT34AA) had less impact on GFP expression possibly due to lower overall expression levels for this mutant (not shown). A direct comparison between wild-type G45a and the CE83AA mutant relative to cells that were influenced by endogenous G45a alone (no ectopic G45a) indicate that the CE83AA mutant is dominant negative (Supplementary Figure S5B).
Figure 5.
Analysis of G45a alanine dimerization mutants. Alanine substitution mutations in G45a predicted to disrupt dimer formation (Supplementary Figure S3) were used (8). The degree of self-association was assayed using cross tag immunoprecipitation (Myc and V5 tags) for each mutation (see Supplementary Figure S5). The ability of each mutant to interact with DNMT1 in HeLa cells was assessed by co-transfecting each V5 tagged DNMT1 with Myc-tagged G45a mutant and I-SceI, performing an immunoprecipitation with anti-V5 IgG followed by western probing with anti-Myc IgG (upper blots). The graph below shows the collective digitized data. The shade bars represent the efficiency of DNMT1/G45a binding. The band intensities of co-imunoprecipitated G45a mutants were quantified using the GeneTools program (SynGene, Cambridge, UK). The amount of co-immunoprecipitated G45a was normalized to the amount of immunoprecipitated DNMT1 and plotted as a relative amount against wild-type band intensity. The statistical data are based on three independent experiments. The white bars represent the efficiency of dimer formation (from Supplementary Figure S5) and the stippled bars the percent total GFP expression.
DISCUSSION
A role for G45a in HR silencing
In this work, we used the GFP reporter system, described by Jasin and co-workers (38), to examine mechanisms that control DNA methylation of repaired segments. We previously showed that this process involves DNMT1 mediated methylation of a region that is 3′ of the DSB (relative to the GFP transcription orientation) (3). The methylation status of the 3′ wild-type donor GFP (cassette II) does not direct the final epigenetic outcome, nor do the recombinant clones carry the methylation pattern of the 5′ cassette I. Instead, methylation patterns are either reprogrammed or overlaid with new patterns resulting in GFP+ clones that are either hypo-methylated or hyper-methylated. Thus, the HR repair product is a hemi-methylated intermediate (2,3) that resolves into two GFP expressor populations in daughter cells (hyper and hypomethylated) (see Supplementary Figure S2). The intermediate could arise by active demethylation, mediated by G45a (32,34,43,44), passive (new homologous strand synthesis) or de novo methylation mechanisms (3) but results in strand-specific templating that imprints the methylation pattern in a permanent fashion. Thus, the process that transpires during HR repair of the break becomes a permanent revision to the epigenetic landscape (Supplementary Figure S2). Previously, we demonstrated the presence of positive acting factors that act as co-repressors in HR silencing, such as the DNMT1 binding protein DMAP1 (2). In this article, we ask what mechanism(s) might prevent or limit re-methylation of the hemi-methylation intermediate by DNMT1 (an attractive substrate for the maintenance methylase). We searched for factors that have been implicated in active epigenetic reprogramming, that are rapidly induced by DNA damage and/or genotoxic stress, and might appear early and transiently in the recombination process (32,43,44). We find that the small DNA damage-inducible protein G45a fits these criteria. G45a is rapidly induced in response to genotoxic events such as DSBs induced by I-SceI damage (14,15,19). Rapid induction and turnover of the G45a gene is mediated both at the transcriptional and post-transcriptional levels (45–47) and in non-stressed (resting) cells, G45a transcripts have a short half-life (47). The following pieces of evidence suggest a role for G45a as a negative modulator of DNMT1 targeted methylation events at sites of double strand break repair by HR.
First, ectopic overexpression of G45a during HR repair results in hypomethylated GFP products as attested by overall increases in GFP expression. We found no evidence that G45a altered the efficiency or frequency of HR in either G45a overexpressing cells or specific knock downs. Although the overall increase in total GFP expression due to constitutive G45a is not large (20–40% increases), other data are consistent with this general picture. Specifically, in G45a knock down cells, GFP+ cells were reduced and the HR-L class (low expressors) elevated to similar extents. Therefore, manipulating G45a levels results in a mutually consistent outcome. DNMT1 is inhibited when G45a is in excess and activated when G45a is depleted, which results in hypomethylation and hypermethylation, respectively.
Second, DNMT1 and G45a are interactive partners and in vitro data show that the two bind directly. Thus, the mechanism underlying G45a inhibitory action in vivo appears to be at least partially due to its ability to bind DNMT1. Moreover, since G45a binds the catalytic center of DNMT1, G45a could block radial access to the DNA target. We cannot rule out the prospect that G45 also sequesters DNMT1 to reduce activity; however, this seems unlikely for a couple of reasons. For example, G45a binds hemimethylated DNA with a slightly higher avidity compared to DNMT1 and in chromatin G45a forms extremely stable complexes with genomic DNA (M. T. Muller and B. Lee, manuscript in preparation). In addition, G45a displays a significantly higher affinity for hemi-methylated DNA over non-methylated or fully methylated sites (50 - to 100-fold). A recent report showed that G45a binds to RNA and such complexes were partially dissociated by adding hemi-methylated competitor DNA (48). Moreover, these authors reported an RNase insensitive complex of G45a present at the nuclear periphery of nuclei (48). In our biochemical assays, G45a inhibition was partially relieved by adding back excess hemimethylated DNA substrate (Figure 4E). We propose that G45a is retained at hemi-methylated sites that appear somewhere along the DNA during (or soon after) formation of the 3′ overhang that attends HR synapse. High affinity binding of hemimethylated DNA also suggests a means whereby G45a is likely to occupy sites arising from de novo methylation by DNMT1. Subsequently, G45a restricts DNMT1 activity by blocking or occluding the opposing strand. Stable G45a complexes could provide an effective barrier to long range spreading of methylation during HR. Recent X-ray structure data with DNMT1 define the importance of the interaction between the target recognition domain (catalytic center) and regions surrounding the BAH domains that bind CpG. Interactions between these domains have been reported to be critical in regulating de novo versus maintenance methylation by DNMT1 (41). Directed binding of G45a at the catalytic center of DNMT1 would certainly inhibit both mechanisms, as shown here (Figure 4). G45a could limit DNMT1 and methylation spreading by a combinatorial mechanism of catalytic site binding and DNA target affinity. The process is of course dynamic and involves precise timing of appearance of each participating protein or subunit (see next point).
Third, based on ChIP data, G45a and DNMT1 are associated with HR recombinant chromatin which places endogenous reactants directly in contact with the 437-bp region that undergoes gene conversion 3′ of the break site. When we compared endogenous and ectopically expressed G45a, higher ChIP signals were seen when G45a was overexpressed (Figure 3, using the same anti-G45a antibody). DNMT1 signals were nearly identical in these two situations; therefore, ectopically expressed G45a enters the 437-bp segment of chromatin without attracting additional DNMT1. G45a acts by preventing the existing DNMT1 from gaining access to the DNA that is 3′ of the I-SceI site. We would further argue that G45a shields the opposing strand to limit methyltransferase action by a mechanism involving both hemimethylated DNA and DNMT1 binding. Finally, it is worth noting that DNMT1 and G45a interact by direct contact in vitro, although we cannot rule out important bridging factors that may operate to attract and retain DNMT1 at HR sites (including hemimethylated DNA, a good substrate).
Fourth, alanine scanning mutations that alter the ability of G45a to dimerize, adversely affect its ability to act as a regulator of HR methylation (Figure 5). Structural studies of Gadd45γ by X-ray crystallography showed the importance of dimerization ability for this class of proteins to interact with other binding partners in cell cycle checkpoints following genotoxic events (8). The alanine substitution mutants (Supplementary Figure S3) displaying poor dimerization ability also bound DNMT1 poorly. As a consequence, GFP expression after HR was reduced, presumably due to the fact that DNMT1 action was elevated in the absence of the G45a over ride. A consistent model emerges from these collective and diverse observations. G45a appears to negatively modulate the action of DNMT1 (and possibly other methylases that contain the conserved catalytic domain) on hemi and unmethylated DNA both in vitro and in a chromatin setting. Since G45a is transiently expressed, its influence is limited to situations where some kind of DNA damage has occurred (HR or base excision repair); therefore, G45a acts to modulate methylation under a strict set of physiological conditions. Once the damage is repaired and the cell cycle checkpoint reversed, G45a transcripts rapidly turnover as expression is extinguished.
Mechanism of G45a action early in HR silencing
We compared kinetics of G45a gene transcription with the appearance of the silenced population of GFP molecules. G45a transcription was detected as early as 4-h post-I-SceI, reached maximum levels by 8–12 h and began to decline around 24 h. During this period of transcriptional decline, G45a protein accumulates to highest levels and the first GFP positive cells begin to appear; however, most of these first GFP positive cells were HR-L (low expressors; Supplementary Figure S4). The HR-L fraction was elevated until 48 h; thus, it appears that the first GFP+ cells are silenced molecules. Since these HeLa cell reporters have a population doubling time in excess of 20 h, events that proceed during the first 24 h represent the original parental cell population undergoing HR (founder cells). Founder cells represent HR and silencing processes that are replete with hemi-methylated products and these do not express well. That G45a protein appears about the same time (or just prior to) HR-L founders, suggests an early role for G45a in establishing or regulating the epigenetic alterations that attend HR (3).
Reports from several other labs indicate that G45a is an important mediator of active demethylation (32,34,43,44) and our findings do not contradict these results but instead add new information regarding the mechanistic underpinnings for the process. Although we cannot distinguish active versus passive demethylation, and considering the rate of accumulation of HR-H cells, we suggest that during repair, G45a maintains the hemimethylated state of the repaired DNA by inhibiting DNMT1 locally (after repair). Before disappearing, G45a may support active demethylation coupled to transcription (M. Muller, M. Gottesman and E. Avvedimento, manuscript in preparation). Indeed, our data clearly demonstrate that G45a displays high avidity binding at sites of hemimethylation and this small protein has the unusual ability to inhibit DNMT1 methylation activity as a result of targeted binding to the catalytic domain. This places G45a at HR regions undergoing methylation re-programming in chromatin (confirmed by ChIP experiments). Catalytic inhibition of the methylase then blocks localized DNMT1 from acting on previously demethylated sites (arising by passive mechanisms such as HR strand synthesis or active G45a mediated events) (32,34,43,44). These data provide a mechanistic overview of the importance of Gadd45 isoforms in setting DNA methylation programming in cells and we would further argue that this family of proteins should be excellent anti-cancer targets, given the clinical importance of hypomethylation in therapy. Future studies will be critical in establishing this potential.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–10.
FUNDING
National Institutes of Health Grants (CA098214 and CA127416 to M.T.M, partial). Funding for open access charge: The University of Central Florida Burnett School of Biomedical Sciences.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge technical assistance from M. Taylor who assisted with SPR analysis. We thank E. Avvedimento and Max Gottesman for their input and critical evaluation of the work.
REFERENCES
- 1.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee GE, Kim JH, Taylor M, Muller MT. DNA methyltransferase 1 associated protein (DMAP1) is a co-repressor that stimulates DNA methylation globally and locally at sites of double strand break repair. J. Biol. Chem. 2010;285:37630–37640. doi: 10.1074/jbc.M110.148536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Di Pardo A, Messina S, Iuliano R, Fusco A, Santillo MR, et al. DNA damage, homology-directed repair, and DNA methylation. PLoS Genet. 2007;3:e110. doi: 10.1371/journal.pgen.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell. Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair. 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 8.Schrag JD, Jiralerspong S, Banville M, Jaramillo ML, O'Connor-McCourt MD. The crystal structure and dimerization interface of GADD45gamma. Proc. Natl Acad. Sci. USA. 2008;105:6566–6571. doi: 10.1073/pnas.0800086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Essers J, van Steeg H, de Wit J, Swagemakers SM, Vermeij M, Hoeijmakers JH, Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 12.Taghian DG, Nickoloff JA. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornace AJ, Jr, Alamo I, Jr, Hollander MC. DNA damage-inducible transcripts in mammalian cells. Proc. Natl Acad. Sci. USA. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fornace AJ, Jr, Nebert DW, Hollander MC, Luethy JD, Papathanasiou M, Fargnoli J, Holbrook NJ. Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol. Cell. Biol. 1989;9:4196–4203. doi: 10.1128/mcb.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papathanasiou MA, Kerr NC, Robbins JH, McBride OW, Alamo I, Jr, Barrett SF, Hickson ID, Fornace AJ., Jr Induction by ionizing radiation of the gadd45 gene in cultured human cells: lack of mediation by protein kinase C. Mol. Cell. Biol. 1991;11:1009–1016. doi: 10.1128/mcb.11.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Bae I, Krishnaraju K, Azam N, Fan W, Smith K, Hoffman B, Liebermann DA. CR6: A third member in the MyD118 and Gadd45 gene family which functions in negative growth control. Oncogene. 1999;18:4899–4907. doi: 10.1038/sj.onc.1202885. [DOI] [PubMed] [Google Scholar]
- 18.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 19.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhan Q, Chen IT, Antinore MJ, Fornace AJ., Jr Tumor suppressor p53 can participate in transcriptional induction of the GADD45 promoter in the absence of direct DNA binding. Mol. Cell. Biol. 1998;18:2768–2778. doi: 10.1128/mcb.18.5.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan W, Jin S, Tong T, Zhao H, Fan F, Antinore MJ, Rajasekaran B, Wu M, Zhan Q. BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs. J. Biol. Chem. 2002;277:8061–8067. doi: 10.1074/jbc.M110225200. [DOI] [PubMed] [Google Scholar]
- 22.Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD, et al. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 23.Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, Kastan MB, O'Connor PM, Fornace AJ., Jr Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 24.Kearsey JM, Coates PJ, Prescott AR, Warbrick E, Hall PA. Gadd45 is a nuclear cell cycle regulated protein which interacts with p21Cip1. Oncogene. 1995;11:1675–1683. [PubMed] [Google Scholar]
- 25.Carrier F, Georgel PT, Pourquier P, Blake M, Kontny HU, Antinore MJ, Gariboldi M, Myers TG, Weinstein JN, Pommier Y, et al. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol. Cell. Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollander MC, Sheikh MS, Bulavin DV, Lundgren K, Augeri-Henmueller L, Shehee R, Molinaro TA, Kim KE, Tolosa E, Ashwell JD, et al. Genomic instability in Gadd45a-deficient mice. Nat. Genet. 1999;23:176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 27.Reinhardt HC, Hasskamp P, Schmedding I, Morandell S, van Vugt MA, Wang X, Linding R, Ong SE, Weaver D, Carr SA, et al. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Mol. Cell. 40:34–49. doi: 10.1016/j.molcel.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vairapandi M, Balliet AG, Hoffman B, Liebermann DA. GADD45b and GADD45g are cdc2/cyclinB1 kinase inhibitors with a role in S and G2/M cell cycle checkpoints induced by genotoxic stress. J. Cell. Physiol. 2002;192:327–338. doi: 10.1002/jcp.10140. [DOI] [PubMed] [Google Scholar]
- 29.Zerbini LF, Wang Y, Czibere A, Correa RG, Cho JY, Ijiri K, Wei W, Joseph M, Gu X, Grall F, et al. NF-kappa B-mediated repression of growth arrest- and DNA-damage-inducible proteins 45alpha and gamma is essential for cancer cell survival. Proc. Natl Acad. Sci. USA. 2004;101:13618–13623. doi: 10.1073/pnas.0402069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith ML, Kontny HU, Zhan Q, Sreenath A, O'Connor PM, Fornace AJ., Jr Antisense GADD45 expression results in decreased DNA repair and sensitizes cells to u.v.-irradiation or cisplatin. Oncogene. 1996;13:2255–2263. [PubMed] [Google Scholar]
- 31.Hollander MC, Kovalsky O, Salvador JM, Kim KE, Patterson AD, Haines DC, Fornace AJ., Jr Dimethylbenzanthracene carcinogenesis in Gadd45a-null mice is associated with decreased DNA repair and increased mutation frequency. Cancer Res. 2001;61:2487–2491. [PubMed] [Google Scholar]
- 32.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 33.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano M, et al. Thymine DNA Glycosylase Is Essential for Active DNA Demethylation by Linked Deamination-Base Excision Repair. CELL. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl Acad. Sci. USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnaud P, Feil R. MEDEA takes control of its own imprinting. Cell. 2006;124:468–470. doi: 10.1016/j.cell.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 38.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee B, Muller MT. SUMOylation enhances DNA methyltransferase 1 activity. Biochem. J. 2009;421:449–461. doi: 10.1042/BJ20090142. [DOI] [PubMed] [Google Scholar]
- 40.Simon J, Lange C. Roles of EzH2 histone methyltransferase in cancer epigenetica. Mutat. Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 41.Song J, Rechkoblit O, Bestor T, Patel D. Structure of DNMT1-DNA Complex Reveals a Role for Autoinhibition in Maintenance DNA Methylation. Science. 2011;331:1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez R, Pantoja-Uceda D, Prieto J, Diercks T, Marcaida MJ, Montoya G, Campos-Olivas R, Blanco FJ. Solution structure of human growth arrest and DNA damage 45alpha (Gadd45alpha) and its interactions with proliferating cell nuclear antigen (PCNA) and Aurora A kinase. J. Biol. Chem. 285:22196–22201. doi: 10.1074/jbc.M109.069344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol. Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Jin S, Fan F, Fan W, Zhao H, Tong T, Blanck P, Alomo I, Rajasekaran B, Zhan Q. Transcription factors Oct-1 and NF-YA regulate the p53-independent induction of the GADD45 following DNA damage. Oncogene. 2001;20:2683–2690. doi: 10.1038/sj.onc.1204390. [DOI] [PubMed] [Google Scholar]
- 46.Maekawa T, Sano Y, Shinagawa T, Rahman Z, Sakuma T, Nomura S, Licht JD, Ishii S. ATF-2 controls transcription of Maspin and GADD45 alpha genes independently from p53 to suppress mammary tumors. Oncogene. 2008;27:1045–1054. doi: 10.1038/sj.onc.1210727. [DOI] [PubMed] [Google Scholar]
- 47.Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, Brewer G, Gorospe M. Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol. Cell. 2006;22:117–128. doi: 10.1016/j.molcel.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 48.Sytnikova Y, Kubarenko A, Schafer A, Weber A, Niehrs C. Gadd45a is an RNA binding protein and is localized in nuclear speckles. PLoS One. 2011;6:e14500. doi: 10.1371/journal.pone.0014500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.