Abstract
Alternative splicing (AS) coupled to nonsense-mediated decay (NMD) is a post-transcriptional mechanism for regulating gene expression. We have used a high-resolution AS RT–PCR panel to identify endogenous AS isoforms which increase in abundance when NMD is impaired in the Arabidopsis NMD factor mutants, upf1-5 and upf3-1. Of 270 AS genes (950 transcripts) on the panel, 102 transcripts from 97 genes (32%) were identified as NMD targets. Extrapolating from these data around 13% of intron-containing genes in the Arabidopsis genome are potentially regulated by AS/NMD. This cohort of naturally occurring NMD-sensitive AS transcripts also allowed the analysis of the signals for NMD in plants. We show the importance of AS in introns in 5′ or 3′UTRs in modulating NMD-sensitivity of mRNA transcripts. In particular, we identified upstream open reading frames overlapping the main start codon as a new trigger for NMD in plants and determined that NMD is induced if 3′-UTRs were >350 nt. Unexpectedly, although many intron retention transcripts possess NMD features, they are not sensitive to NMD. Finally, we have shown that AS/NMD regulates the abundance of transcripts of many genes important for plant development and adaptation including transcription factors, RNA processing factors and stress response genes.
INTRODUCTION
Alternative splicing (AS) is an important mechanism to control gene expression and increase the proteome complexity of higher eukaryotes (1–3). Regulated AS drives developmental pathways and responses to environmental pressures. Following transcription, splicing of the exons requires removal of introns by assembling a large RNP complex, the spliceosome, with five snRNPs and about 180 proteins (4). Splice site selection has to be precise but consensus sequences defining splice sites are degenerate and how a splice site is selected from many similar sites within a transcript remains a major question. In many cases, specific splice sites are used in all transcripts (constitutive splicing) while in alternative splicing, other splice sites are used to various extents giving rise to alternate transcripts with variable sequences. It is now well established that in addition to splice sites, sequence elements within exons and introns, termed either splicing enhancers or silencers are binding sites for splicing factors which either enhance or repress splicing depending on their activities (5,6). These splicing regulators are, for example, SR and hnRNP protein families, and other cell-, stage- or tissue-specific proteins involved in constitutive and alternative splicing which establish the splicing code and determine which splice site is selected (7–10). The regulation of alternative splicing is brought about by the relative levels of the RNA-binding proteins determining how efficiently different splice sites are used to generate more than one spliced mRNA from one gene.
Alternatively spliced mRNA variants can produce functionally different protein isoforms with altered amino acid sequences and protein domains resulting in changes in activity, localization, interaction partners or post-translational modifications (1,11). In addition, alternative splicing can regulate mRNA levels through the targeted degradation of specific AS isoforms by nonsense-mediated decay (NMD) (see below). In particular, alternative splicing can result in mRNAs with premature termination codons (PTCs) which could give rise to truncated proteins which are detrimental to cell survival and energy costly for the cell. RNA quality control mechanisms have evolved at all levels of gene expression to identify and remove aberrant RNA transcripts. One of the best investigated mRNA quality control mechanisms is NMD which degrades mRNAs which possess a premature termination codon (PTC+) and other physiological mRNAs without a PTC such as transcripts with long 3′-UTRs [reviewed in (12–18)]. Despite great advances in understanding of the NMD pathway, it is apparent that not every PTC triggers NMD and that this pathway controls the abundance of certain mRNAs which do not contain known NMD features, arguing that not all the factors inducing NMD have been identified yet.
Several features of NMD-sensitive, PTC+ transcripts have been elucidated and have led to models of how PTCs are recognized and degradation triggered. In the current model for mammals, NMD initiates the rapid decay of a transcript if translation termination is perturbed [reviewed in (12–18)]. Efficient translation termination of the ribosome is proposed to involve the interaction of the release factor, eRF3, and poly(A) binding proteins (PABP) on the poly(A) tail of the mRNA. If this interaction is prevented or impaired by, for example, an unusually long 3′-UTR, the eRF3 on the ribosome will bind UPF1 which then recruit UPF2 and UPF3, all core NMD proteins. This functional NMD complex (which includes many other proteins) then elicits the phosphorylation of UPF1 and rapid degradation of the transcript. This ‘long 3′UTR’ mechanism is characteristic for transcripts in invertebrates and yeast. In mammals, the NMD response triggered by a ribosome terminating at a PTC is stimulated by UPF3 associated with a downstream exon-junction complex (EJC) which is deposited on the mRNA 20–25 nt upstream of a spliced exon–exon junction (19,20). In the course of splicing the EJC complex binds the NMD factors UPF2/UPF3 which can then associate with a ribosome terminating at a PTC upstream of the EJC which has recruited UPF1 in the SURF complex (SMG1-UPF1-eRF1-eRF3) (21). On a normal, non-PTC-containing mRNA, the EJC is removed in the first round of translation (22) except when the EJC is located in the 3′-UTR. This is consistent with the observation that introns in the 3′-UTR may significantly enhance NMD. Thus, in mammals, both the length of the 3′-UTR and the presence of an EJC complex downstream of a PTC can trigger NMD. However, the recent demonstration in Drosophila that EJCs are not deposited at each splice junction such that only some introns are able to trigger intron-dependent NMD (23) suggests that NMD may rely more widely on long 3′UTR signals.
The NMD pathway in plants is as yet not well characterized (24,25). Plants possess orthologues of the key eukaryotic NMD proteins, UPF1, UPF2, UPF3 and SMG-7 (but not SMG-1, SMG-5 or SMG-6) and these have been shown to be involved in degrading mRNAs with PTCs (26–31). Efforts to determine the rules for NMD substrates suggest that like mammals, plants are able to recognize different types of PTC-containing transcripts (Figure 1c). Firstly, it was shown that both long 3′-UTRs and introns located in 3′-UTRs are signals for efficient NMD (30,32–34). This indicates that like in invertebrates and yeast, the distance between a stop codon and the PABP on the poly(A) sequence is important and that translation termination is also likely to require the interaction of the release factor-containing ribosome with PABP. Secondly, EJC components, which are required for the intron-based NMD mechanism proposed for mammalian NMD, have been demonstrated to be similarly important in plants (28). Furthermore, upstream open reading frames (uORFs) of more than 35 amino acids can trigger NMD in plants (35).
Figure 1.
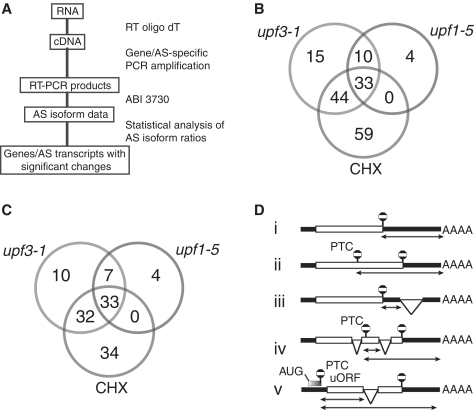
Analysis of alternatively spliced NMD substrates. (A) Schematic figure of RT–PCR panel analysis (see ‘Materials and Methods’ section). (B) Venn diagram of the number of transcripts which increase significantly in upf mutants and cycloheximide treatment. (C) Venn diagram of the number of genes with splice isoforms which increase significantly in upf mutants and cycloheximide treatment. (D) General features of transcripts which trigger NMD: (i) long 3′-UTR; (ii) PTC—long 3′-UTR; (iii) splice junction downstream of authentic stop codon (3′-UTR intron); (iv) PTC—downstream splice junctions and long 3′-UTR; (v) uORFs in 5′-UTR. Endogenous transcripts regulated by NMD contain long 3′-UTRs, introns in the 3′-UTR where the splice junction is >50–55 nt from the authentic stop or UORFs (i, iii and v, respectively). Transcripts which contain PTC in the coding region or 5′-UTR (uORF) also generate long 3′-UTRs with or without downstream splice junctions (ii, iv and v, respectively). Exons—open boxes; UTRs—black rectangles; thin lines—introns; diagonal lines—splicing events; stop sign—PTC or authentic termination codon.
Alternative splicing in plants is an important regulatory process for plant development and for the response of plants to environmental factors. However, its frequency of occurrence has been grossly underestimated largely due to low depth of sequencing and relatively few available ESTs. The most recent estimate based on next generation sequencing is that about 42% of intron-containing genes undergo alternative splicing (36) and this is still likely to be a significant underestimate. In humans, over 95% of genes undergo alternative splicing and more importantly around 20–30% of alternatively spliced transcripts contain PTCs and are potentially turned over by NMD (37–40). The importance of the link between AS and NMD has been highlighted by the regulation of functional transcript levels of key splicing factors such as SR proteins and PTB through alternative splicing via conserved splice sites (41,42). Conservation of alternative splice sites to produce PTC-containing transcripts has also been demonstrated for SR protein genes in lower and higher plants (43) and more generally, NMD seems to play a regulatory role in gene expression using alternative spliced transcripts (27). The best characterized examples of gene regulation by AS/NMD in plants are GRP7 and GRP8, components of a slave oscillator coupled to the circadian clock. These glycine-rich RNA-binding proteins bind their own pre-mRNAs inducing AS which produces NMD-sensitive transcripts thereby auto- and cross-regulating their mRNA levels (44,45). Other examples of such regulation are SR protein genes (46), polypyrimidine tract binding protein (PTB) (47), possibly SUPPRESSOR OF OVEREXPRESSION OF CO1 (48) and riboswitch-regulated alternative splicing controlling NMD (49).
The definition of the rules of NMD in plants have mainly relied on mutations in a small number of model transcripts and it is necessary to examine how these features correspond to those in NMD-sensitive endogenous transcripts. As about 78% of alternative transcripts in Arabidopsis introduced in-frame PTCs more than 55 nt upstream of exon junction, it was speculated that NMD is a widespread mechanism for regulating gene expression (36), however this has not been experimentally addressed. Two previous studies performed genome-wide transcriptome profiling of NMD-defective plants using tiling and expression microarrays (31,50) and found that only about 1% of plant protein-coding genes were up-regulated in NMD-deficient plants. These arrays have limited ability to distinguish AS transcripts in contrast to the splicing-sensitive microarrays successfully used in animals (51–53), and it is therefore necessary to thoroughly investigate the fate and characteristics of endogenous plant AS transcripts turned over by NMD.
In the absence of splicing-sensitive microarrays for plants, we have used a high-resolution RT–PCR system (54) which is able to detect multiple AS transcript isoforms simultaneously and obtained isoform-level measurements from strong, but still viable mutant alleles (26,27) of the NMD protein genes, UPF1 and UPF3, and cycloheximide (an inhibitor of translation and thus of NMD) treated plants. To address the link between AS and NMD we (i) investigated the effect of NMD impairment on a population (∼950) of endogenous alternatively spliced transcripts, (ii) identified NMD-sensitive AS isoforms from significant changes in the ratio of AS isoforms, (iii) identified the characteristics of AS transcripts which trigger NMD, and (iv) identified transcripts which contain NMD features but which are insensitive to NMD. Our results demonstrate that alternative splicing and NMD affect a broad range of different genes in Arabidopsis and regulate expression of these genes via targeted degradation of specific AS transcripts using different mechanisms depending on the position of the AS event in the gene.
MATERIALS AND METHODS
Plant material, growth conditions, treatments and RNA isolation
Wild-type (ecotype Col-0) and UPF mutant Arabidopsis plants were used for the analysis. UPF mutants, upf1-5 and upf3-1, (26) were a gift from Brendan Davies (Centre for Plant Sciences, University of Leeds, UK). Plants were grown in vitro on plates containing germination medium (55). Plants were maintained in 16-h light/8-h dark cycle at 22°C. Three week old plants were transferred into liquid half-strength Murashige and Skoog medium (56) and infiltrated with either 20 µM cycloheximide or the same volume of dimethylsulfoxide as a control. Samples of cycloheximide-treated plants were collected after 5 h (27). RNA was isolated using RNeasy Plant Mini Kit (Qiagen).
High-resolution alternative splicing RT–PCR panel and data analysis
The original panel (54) was expanded to 289 primer pairs by identifying alternative splicing events which were either published, annotated in The Arabidopsis Information Resource (TAIR8—http://www.arabidopsis.org/) or in the Alternative Splicing in Plants database (ASIP—http://www.plantgdb.org/ASIP/). Primer pairs where one primer is fluorescently labelled were designed as described previously (54). Primer pairs used are listed in Supplementary Table S1. RT–PCR analysis was performed as described earlier (54). In brief, the reverse transcription reaction was carried out with total RNA using oligo-dT primers and the first-strand cDNA was aliquoted into microtitre plates, and PCR with the gene/alternative splicing event-specific primers performed using 24 cycles. We have previously shown that 24 cycles was still in the linear amplification range for various splicing substrates using [32P]-labelling (57) and for a number of the AS primers used here to amplify transcripts of different abundance and size (54). The high-resolution RT–PCR system is capable of detecting multiple different AS transcripts from a gene, distinguishing alternative splicing events involving small size differences in transcripts (as few as 2–3 nt) and identifying small but significant changes in the ratios of alternatively spliced variants. The AS variants for each of the genes are amplified simultaneously by the same primers in the same reaction. The different AS isoforms usually have substantial common sequence which will reduce variation in amplification efficiency. In addition, if there are differences in amplification efficiency among particular AS isoforms, these differences will occur in the PCR reactions with wild-type, mutants and cycloheximide treatment. Electropherograms produced by the ABI 3730 genotyping software identified the exact size of the RT–PCR products for each primer pair. Peak areas for each RT–PCR product were extracted from the three reps, ratios of the different peaks were calculated generating a mean and standard error for each AS transcript as a percentage of the total transcript across the three reps.
Statistical analysis
The response to genotype, treatment and genotype by treatment interaction was assessed by analysis of variance (ANOVA). Each peak of each primer was analysed separately assuming a completely randomized design with three replicate values for each treatment combination. Response was measured as the percentage contribution of a particular isoform to the total transcripts measured and ANOVA was carried out after an angular transformation of the percentage values. In addition to assessing the significance of genotype and treatment main effects and their interaction, three specific comparisons (contrasts) were made: wild-type versus upf1-5, wild-type versus upf3-1 and wild-type versus cycloheximide treatment. Residual plots were used to monitor the ANOVA assumptions of approximate normality and equality of variance. For the small number of cases where these assumptions did not hold either the response levels were all very low (or all very high) or the differences between treatments were so large as to render the ANOVA redundant.
In the analysis of the direct comparisons (above and Table 1) P-values were determined. In the subsequent analysis, we focussed on those transcripts which showed a significant percentage increase or decrease with a 3% difference between the means of wild-type plants and mutants/cycloheximide-treated plants. This level of difference was selected because we previously determined that when comparing variation in technical reps in the AS RT–PCR system, the majority of transcripts showed a standard error of the mean of <3% (54).
Table 1.
Selected AS transcripts which increase significantly in upf mutants and/or cycloheximide treatment
| Primer pair | Gene ID | Name | Band size (bp) | AS event | NMD features | Mean ratio of transcripts |
|||
|---|---|---|---|---|---|---|---|---|---|
| wt | upf1-5 | upf3-1 | CHX | ||||||
| 7 | At1g55310 | At-SCL33 | 361 | E4 Alt3′ss(+162) | PTC+; ds SJ | 0.15 | 0.19 | 0.32 | 0.47 |
| 7 | At1g55310 | At-SCL33 | 364 | E4 Alt3′ss(+165) or AE (165) | PTC+; ds SJ | 0.02 | 0.03 | 0.05 | 0.08 |
| 7 | At1g55310 | At-SCL33 | 293 | Unknown | Unknown | 0.05 | 0.08 | 0.09 | 0.07 |
| 21 | At2g37340 | At-RS2Z33 | 341 | E3 Alt3′ss(+218) | PTC+; ds SJ | 0.04 | 0.07 | 0.13 | 0.20 |
| 86 | At4g16845 | VRN2 | 396 | IR2(170); E5 Alt3′ss(−4); E5 Alt5′ss(+22) | PTC+; ds SJ | 0.07 | 0.09 | 0.12 | 0.13 |
| 90 | At4g39260 | GRP8/CCR1 | 316 | E1 Alt5′ss(+158) | PTC+; ds SJ | 0.03 | 0.05 | 0.12 | 0.14 |
| 109 | At1g77080 | MAF1 | 118 | E4 Alt3′ss(−38) | PTC+; ds SJ | 0.14 | 0.22 | 0.29 | 0.19 |
| 118 | At2g02960 | Zinc finger (C3HC4) protein | 233 | E2 Alt3′ss(+37) | uORFs; uORF(26aa) | 0.03 | 0.05 | 0.07 | 0.07 |
| 118 | At2g02960 | Zinc finger (C3HC4) protein | 237 | E2 Alt3′ss(+41) | uORFs; uORF(26aa) | 0.04 | 0.05 | 0.07 | 0.07 |
| 118 | At2g02960 | Zinc finger (C3HC4) protein | 240 | E2 Alt3′ss(+44) | uORFs; uORF(26aa) | 0.02 | 0.02 | 0.03 | 0.02 |
| 118 | At2g02960 | Zinc finger (C3HC4) protein | 289 | E2 Alt3′ss(+92) | uORFs; uORF(26aa) | 0.25 | 0.30 | 0.33 | 0.51 |
| 125 | At2g46790 | APRR9/TL1 | 251 | E2 Alt5′ss(+8) | PTC+; ds SJ | 0.22 | 0.32 | 0.47 | 0.37 |
| 131 | At2g38880 | NF-YB1/HAP3a | 373 | E6 Alt5′ss(+62) | 3′UTR intron | 0.21 | 0.26 | 0.30 | 0.41 |
| 194 | At3g49430 | At-SR34a | 366 | I1 AE(224) | uORFs; uORF(61aa) | 0.07 | 0.21 | 0.35 | 0.30 |
| 195 | At3g01150 | At-PTB2a | 156 | E8 Alt5′ss(−47) | PTC+; no ds SJ - possible long 3′UTR | 0.10 | 0.12 | 0.16 | 0.19 |
| 196 | At3g01150 | At-PTB2a | 268 | I2 AE(102) | PTC+; ds SJ | 0.11 | 0.17 | 0.25 | 0.37 |
| 202 | At3g13570 | At-SCL30a | 351 | I3 AE(161) | PTC+; ds SJ | 0.18 | 0.30 | 0.48 | 0.52 |
| 204 | At3g53500 | At-RS2Z32 | 376 | E3 Alt3′ss(+218) | PTC+; ds SJ | 0.09 | 0.11 | 0.25 | 0.40 |
| 205 | At3g61860 | At-RS31 | 556 | I2 AE(393) | PTC+; ds SJ | 0.01 | 0.04 | 0.10 | 0.27 |
| 206 | At2g21660 | GRP7/CCR2 | 349 | E1Alt5′'ss(+166) | PTC+; long 3′UTR | 0.02 | 0.07 | 0.16 | 0.13 |
| 213 | At5g53180 | At-PTB2b | 198 | I3 AE(58) | PTC+; ds SJ | 0.08 | 0.12 | 0.19 | 0.31 |
| 213 | At5g53180 | At-PTB2b | 201 | I3 AE(61) | PTC+; ds SJ | 0.06 | 0.09 | 0.14 | 0.22 |
| 217 | At1g16610 | SR45 | 175 | FS | no reason for NMD | 0.68 | 0.71 | 0.72 | 0.77 |
| 218 | At2g30260 | U2B'' | 134 | E2 Alt5′'ss(−35) | PTC+; ds SJ | 0.05 | 0.09 | 0.19 | 0.17 |
| 219 | At4g25500 | At-RS40 | 382 | I2 AE(257) | PTC+; ds SJ | 0.14 | 0.33 | 0.54 | 0.34 |
| 220 | At3g55460 | At-SCL30 | 590 | I3 AE(449) | PTC+; ds SJ | 0.01 | 0.05 | 0.11 | 0.08 |
| 220 | At3g55460 | At-SCL30 | 673 | Unknown | Unknown | 0.01 | 0.02 | 0.05 | 0.13 |
| 223 | At2g29210 | Splicing factor PWI domain-containing protein | 202 | E6 Alt3′ss(+50) | PTC+; ds SJ | 0.36 | 0.36 | 0.59 | 0.63 |
| 237 | At1g07830 | RPL29 family | 123 | E2 Alt3′ss(−70) | uORFs; uORF(31aa) | 0.01 | 0.02 | 0.03 | 0.02 |
| 237 | At1g07830 | RPL29 family | 273 | E1 Alt5′ss, predicted | uORFs | 0.02 | 0.05 | 0.09 | 0.07 |
| 241 | At1g02090 | FUS5/CSN7/COP15 | 119 | E8 Alt3′ss(−17) | PTC+; ds SJ | 0.15 | 0.27 | 0.29 | 0.30 |
| 249 | At1g72560 | PSD/Exportin-t | 174 | E13 Alt5′ss(+25) | 3′UTR intron | 0.04 | 0.15 | 0.04 | 0.05 |
| 259 | At3g17090 | PP2C group D | 214 | E2 Alt3′ss(+17) | PTC+; ds SJ | 0.07 | 0.11 | 0.23 | 0.14 |
| 284 | At4g33060 | CYP57 | 290 | I5 AE(87) | PTC+; ds SJ | 0.26 | 0.34 | 0.49 | 0.71 |
| 284 | At4g33060 | CYP57 | 295 | I5 AE(92) | PTC+; ds SJ | 0.05 | 0.06 | 0.08 | 0.11 |
| 306 | At2g46830 | CCA1 | 218 | FS | No reason for NMD from AS event—other AS/NMD events known | 0.79 | 0.85 | 0.87 | 0.89 |
| 309 | At5g65060 | MAF3 | 223 | E4 Alt3′ss(−38) | PTC+; ds SJ | 0.19 | 0.35 | 0.42 | 0.38 |
| 309 | At5g65060 | MAF3 | 219 | E4 Alt3′ss(−38); E5 Alt3′ss(−4) | PTC+; ds SJ | 0.04 | 0.06 | 0.08 | 0.07 |
| 311 | At5g65080 | MAF5/AGL68/FCL1 | 462 | E5 Alt3′ss(−4) | PTC+; ds SJ | 0.76 | 0.82 | 0.80 | 0.75 |
| 343 | At3g29160 | AKIN11 | 159 | FS | No reason for NMD from AS event—other AS/NMD events known | 0.27 | 0.27 | 0.34 | 0.40 |
| 344 | At3g29160 | AKIN11 | 195 | E9 Alt3′ss(−5) | PTC+; ds SJ | 0.08 | 0.12 | 0.17 | 0.15 |
| 363 | At5g24270 | SOS3/CBL4 | 337 | E5 Alt5′ss(+29) | PTC+; ds SJ | 0.06 | 0.08 | 0.13 | 0.25 |
| 370 | At5g35410 | SOS2/CIPK24 | 120 | E9 Alt3′ss(+5) | PTC+; ds SJ | 0.42 | 0.49 | 0.59 | 0.63 |
| 374 | At4g36960 | RRM-containing protein | 399 | IR1(172) | uORFs; uORF(73aa) | 0.05 | 0.15 | 0.32 | 0.09 |
| 375 | At3g20270 | lipid-binding serum glycoprotein family protein | 195 | FS | uORFs; uORF(22aa) | 0.54 | 0.62 | 0.64 | 0.59 |
| 384 | At4g02200 | At-Di19-5 | 149 | E2 Alt5′ss(+32) | PTC+; ds SJ | 0.06 | 0.13 | 0.21 | 0.22 |
| 384 | At4g02200 | At-Di19-5 | 110 | Unknown | Unknown | 0.01 | 0.01 | 0.02 | 0.04 |
Values are mean values from three biological reps and significance is P < 0.1 (see text). En Alt5′ss and En Alt3′ss—alternative 5′ splice site and alternative 3′ splice site in exon, where n is the exon number. The number in brackets indicates the number of nucleotides added (+) or removed (−) from the exonic sequence. IRn—retention of intron, where n is intron number [number in brackets indicates the size (nt) of the intron]. In AE—alternative (cryptic) exon created in an intron, where n is the intron number [number in brackets indicates the size(s) of the alternative exon(s)]. Exons and introns are numbered with respect to the TAIR reference splice variant. FS—fully spliced; PTC+—transcript containing premature termination codon(s); ds SJ—presence of downstream splice junction(s). Numbers in bold—significant percentage difference >3%; numbers in bold italic—significant percentage difference <3%. Full data set is presented in the Supplementary Table S2. Note that Arabidopsis SR proteins are named according to the recently proposed nomenclature (77).
Sequencing analysis of AS RT–PCR products
Many RT–PCR products corresponded to unknown splice variants. To identify these products, RT–PCR reactions were purified using Agencourt AMPure beads (Beckman Coulter Genomics) and re-amplified prior to cloning into pGEM-T. Clones with differently sized inserts were identified by colony PCR and sequenced by standard procedures. Sequences were analysed either by using ClustalW or spliced alignments generated by GeneSeqer (http://www.plantgdb.org/tool/GeneSeqer/) (58). An in-house Perl script was used to parse the output from GeneSeqer for categorizing the annotations of alternative splicing events.
RESULTS
Analysis of alternative splicing coupled to nonsense-mediated decay using a high-resolution RT–PCR panel
To analyse the levels of alternatively spliced isoforms in mutants in the NMD protein genes, UPF1 and UPF3, and to investigate the link between AS and NMD in Arabidopsis thaliana, we have exploited an alternative splicing RT–PCR panel. This unique high-resolution system is very sensitive and capable of detecting small but significant changes in alternative splicing at single nucleotide resolution (54) (Figure 1A). It detects different AS variants from the same gene/region simultaneously, variants containing more than one AS event and novel AS transcripts (Supplementary Figure S1). The AS RT–PCR panel used here consists of 289 primer pairs covering different alternative splicing events in 270 genes, and 3 control primer pairs (Supplementary Table S1). The AS events were selected from publications or from plant alternative splicing databases without prior knowledge of NMD-sensitivity with the exception of GRP7 and GRP8 (Figure 2). The AS events were mainly from genes encoding transcription factors, RNA-interacting proteins (including splicing factors) and stress-related proteins and included many important regulatory genes. The relative levels of AS isoforms were compared between wild-type, the two upf mutants and cycloheximide treatment (see Figures 2 and 3 and Table 1 for examples). The upf1-5 and upf3-1 mutants are impaired in NMD and have severe growth phenotypes but are viable (26,27). Translation is required for NMD (22,59) and the translation inhibitor, cycloheximide, leads to accumulation of NMD-sensitive transcripts in plants (27). Therefore a 5 hour cycloheximide treatment was used to inhibit translation and thereby NMD (27). Consequently, AS isoforms which are targets of NMD are expected to increase in their levels in the upf mutants and cycloheximide-treated plants. Three biological replicates were analysed for each, and significant changes in alternative splicing ratios were determined by statistical analysis (see ‘Materials and Methods’ section).
Figure 2.
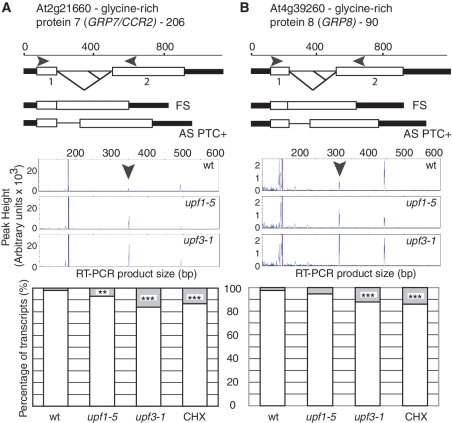
Regulation of GRP7 and GRP8 by alternative splicing and NMD. (A) GRP7 and (B) GRP8 are known to be regulated by AS/NMD. Figures show the GRP7 and GRP8 gene and transcript structures and the alternative splicing events in the introns: alternative 5′ splice sites generate AS isoforms which increase in abundance in the upf1-5 and upf3-1 mutants as illustrated on scans generated from the ABI 3730 data by GeneMapper (transcripts are arrowed). The significant increases in NMD-sensitive transcript abundance are shown in histograms of the ratio of normally spliced and alternatively spliced isoforms (shaded). Significance: ***P< 0.01; **0.01 > P < 0.05. For diagram key see legend to Figure 1.
Figure 3.
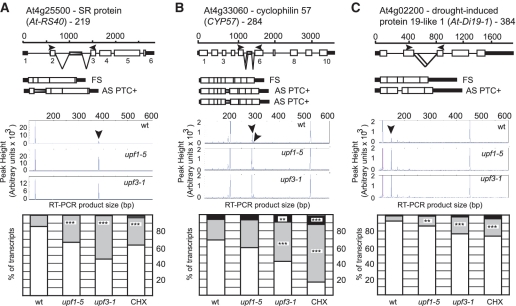
Genes with AS isoforms which increase in upf mutants. (A) At4g25500—SR protein gene, At-RS40. (B) At4g33060—cyclophilin 57, CYP57. (C) At4g02200—drought-induced protein gene, At-Di19-1. For all, the gene and transcript structures and relevant splicing events are shown. AS isoforms which increase in the upf mutants are labelled with arrows on ABI3730 scans and the ratios of transcripts are shown in histograms and significant increases are indicated. Significance: ***P < 0.01; **0.01 > P < 0.05. For diagram key see legend to Figure 1.
High frequency of novel alternatively spliced transcripts in regulatory plant genes
Based on publications or plant databases the majority of AS events were expected to generate two alternative transcripts. However, the number of observed RT–PCR products varied among the different amplified regions from a single product to as many as 15 different alternatively spliced products. Just over 950 RT–PCR products were observed using the 289 primer pairs and therefore approximately 350 new transcripts were discovered. To identify the nature of the novel AS transcripts, cloning and sequencing of RT–PCR products was carried out (for examples of analysis see Supplementary Figure S1). In addition, many were identified by RNA-Seq (our unpublished data). In general, our data shows an increase of AS frequency in our gene set by one third compared to presently annotated events. The identification of so many novel products illustrates that far more alternative splicing occurs in Arabidopsis than is currently known.
Identification of endogenous alternatively spliced targets of nonsense-mediated decay
Our high-resolution RT–PCR system allowed the determination of the ratio of AS transcript variants for each gene region amplified. Mean ratios of AS products obtained with the upf mutants and cycloheximide treatment were compared to the wild-type values to identify the particular AS isoforms which increased significantly when NMD is impaired. Significance was determined at the P < 0.1 level, although for the vast majority significant changes in AS isoform levels, the P-value was considerably smaller (Supplementary Table S2). Of the >950 transcripts in the study, 638 showed no significant change in the transcript ratios between wild-type, mutants or cycloheximide-treated plants. Of the 313 RT–PCR products that showed a significant change, 165 increased in amount in at least one of the upf mutants or cycloheximide treatments (Figure 1B; Supplementary Table S2). Thirty-three transcripts increased in both mutants and cycloheximide-treated plants (Figure 1B). A total of 106 transcripts were increased in one or other mutant while 59 showed a significant increase only in the cycloheximide-treated plants. Cycloheximide is used widely as an NMD inhibitor but as cycloheximide is a general translational inhibitor, other RNA degradation pathways or cellular processes might also be affected by this treatment and impact on transcript levels. Therefore, the 106 transcripts which increased in the upf mutants and the 165 which increased in mutants and cycloheximide treatment represent a range of naturally occurring alternatively spliced transcripts which are putatively turned over by NMD and make up 11–17% of the total transcripts analysed and 16–25% of the alternatively spliced transcripts analysed.
We found that 87 and 121 genes of the 270 AS genes on the panel (Figure1C; Supplementary Table S3) had at least one AS isoform with increased abundance in the upf mutants or in the mutants plus CHX treatment, respectively, suggesting that ∼32% and 45% of AS genes are regulated by NMD to some extent. At least 42% (9273 genes out of 22 302) of intron-containing genes in Arabidopsis are alternatively spliced (36). With the caveat that our gene set may contain some bias, we can extrapolate to suggest that around 13–18% of intron-containing genes may be regulated by AS and NMD in Arabidopsis.
GRP7 and GRP8 are genes encoding components of a slave oscillator and are known to be auto- and cross-regulated by alternative splicing and NMD (44,45) and were included as controls. For both genes, the AS isoform which is turned over by NMD increased significantly as expected. The GRP7 isoform increased significantly in both mutants and cycloheximide treatment (Figure 2A; Table 1, primer pair 206) and significant increases in the GRP8 isoform were observed in upf3-1 and cycloheximide treatment (Figure 2B; Table 1, primer pair 90). These results demonstrate that the AS RT–PCR panel is able to detect significant changes in AS isoforms due to NMD. Other examples of AS/NMD transcripts are shown in Figure 3. At-RS40, an SR protein gene, CYP57, a peptidyl-prolyl cis-trans isomerase gene, and the drought-induced protein 19-like 1 gene, At-Di19-5, have alternative splicing events which introduce PTCs and increase significantly in mutants and cycloheximide treated plants. In general, the increase in levels of the NMD-sensitive AS isoforms in the mutants and after cycloheximide treatment varied with different genes. Interestingly, the steady state levels of the AS transcripts turned over by NMD varied greatly in wild-type plants from being virtually undetectable to tens of percent of the transcripts from a gene (Figure 3B and C; Table 1 and Supplementary Table S2). Thus, AS/NMD transcripts from different genes are differentially abundant in wild-type plants which reflects the different efficiency of alternative splicing and rates of turnover by NMD. This analysis shows that coupled AS/NMD can influence gene expression significantly and that even low abundant alternative transcripts (at steady state level) might turn over a quite significant proportion of the RNA produced from a gene.
upf1-5, upf3-1 and cycloheximide treatment impair nonsense-mediated decay to different degrees
More transcripts (102 transcripts) increased significantly in the upf3-1 mutant than in upf1-5 (47 transcripts) while 136 transcripts increased significantly in the cycloheximide-treated plants (Figure 1B). This suggests that in terms of NMD impairment, the upf3-1 allele is stronger than the upf1-5 allele which conforms to the severity of the phenotype of the particular mutants which was described previously (26,27,31) and that UPF3 transcripts are up-regulated in upf1-5 (60). In addition to the described late flowering phenotype, we have observed that both upf1-5 and upf3-1 mutants have accelerated senescence and again upf3-1 showed the stronger phenotype (Supplementary Figure S2). Some transcripts only increased with cycloheximide treatment and were not visible in the wild-type or mutant plants. Thus, cycloheximide has a much stronger effect on the number of transcripts which increased in abundance and on the degree of increase than upf3-1 with the smallest increases being seen in upf1-5 (Supplementary Tables S4 and S5). Some transcripts did not follow this pattern suggesting perhaps differential or additional functions of the two different NMD factors in different mechanisms of NMD (28). There was substantial overlap between transcripts which increased in abundance in cycloheximide treatment and in the mutants (Figure 1B). In addition, we analysed 11 transcripts which increased only in the cycloheximide treatment in detail and the majority showed NMD characteristics (PTCs/downstream splice junctions or uORFs) (Supplementary Table S2). This suggests that AS transcripts which increase in the cycloheximide treatment are turned over by NMD with the caveat that some transcripts may be turned over by other degradation pathways.
Features of alternatively spliced transcripts which are sensitive to nonsense-mediated decay
Previously, 1% of plant protein-coding genes was shown to have increased transcript levels in NMD-deficient plants using tiling arrays but the features of the AS transcripts which were NMD targets were not specifically investigated (50). The 165 endogenous AS isoforms which show significant increases in levels in the mutants and cycloheximide-treated plants (Table 1; Supplementary Table S2) are expected to be turned over by NMD and therefore should contain NMD signals. To identify the NMD features, each transcript which increased significantly in at least one of the mutants and 11 of the transcripts which increased only in cycloheximide-treated plants (117 transcripts in total) was characterized in terms of whether they contained PTCs, had splice junctions downstream of the authentic stop codon or PTCs, had long 3′-UTR sequences or contained an upstream ORF (Figure 1D). In addition, other AS events in the same gene (annotated in TAIR8) and novel RT–PCR products for which sequence was generated were analysed. Of the 117 transcripts, the sequences of 8 remain elusive and could not be characterized. Of the remaining 109 transcripts, 94 (86%) clearly contained NMD features. This includes a major group of 74 transcripts containing PTCs more than 50–55 nt upstream of splice junctions and long 3′UTRs, classical features of NMD substrates (61). The NMD-sensitivity of a further nine transcripts could be explained on the basis of long 3′-UTRs and/or where the distance between the authentic stop codon and splice junction of an intron in the 3′-UTR was changed due to alternative splicing. In addition, the alternative splicing events of 12 genes (14 transcripts) involved introns in the 5′-UTR which affected the presence or absence, length and position of uORFs. In 11 of these transcripts, the presence of one or more uORFs correlated with NMD. Finally, for the remaining 12 transcripts, the AS event monitored did not explain the turnover by NMD but of these, four genes had known AS events elsewhere in the transcripts which could generate NMD. The remaining transcripts could either be genes/transcripts which have unknown NMD-inducing AS events elsewhere in the gene or may represent genes where changes in AS isoform levels are due to secondary effects on mRNA accumulation. Therefore, the vast majority of the endogenous NMD-sensitive transcripts analysed had characteristic features of NMD substrates in plants.
Alternative splicing in 3′-UTRs modulates nonsense-mediated decay
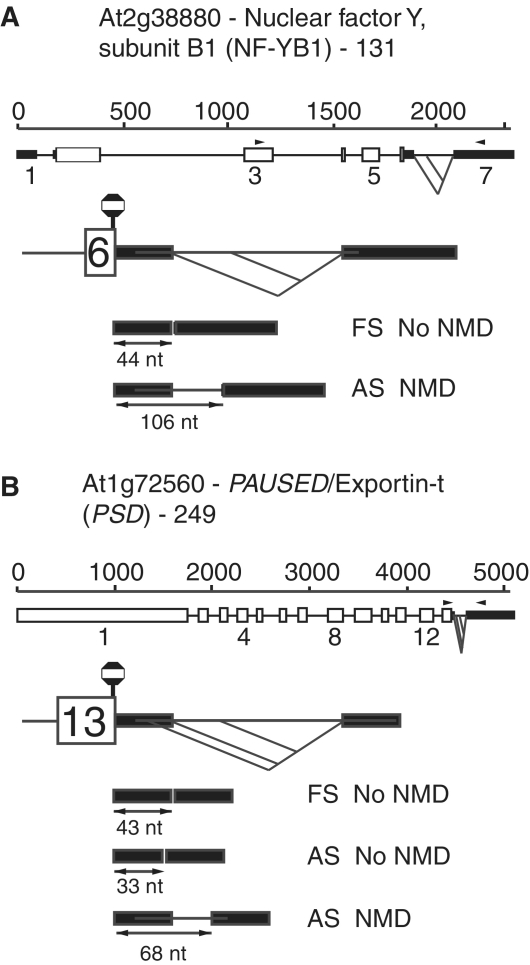
It is not widely appreciated that alternative splicing in the untranslated regions of a gene (which does not create a PTC) may induce NMD sensitivity and be a mechanism of fine regulation of transcript abundance. Also, little is known about the consequences of AS of such UTR introns. We have identified two genes in our panel where AS occurred in 3′-UTR introns and thus did not create a PTC, but at least one AS isoform in each gene was sensitive to NMD. Alternative splicing in the 3′-UTR of At2g38880 (NF-YB1/HAP3A transcription factor) and At1g72560 (PAUSED/Exportin-t) generated two and three isoforms, respectively, with different sensitivity to NMD (Figure 4). Only the isoforms with a distance >50–55 nt from the authentic stop codon were subjected to NMD (Table 1). Thus, alternative splicing can affect the distance between the authentic stop codon and downstream splice junction in 3′-UTRs and determine whether a transcript is turned over by NMD.
Figure 4.
Alternative splicing of introns in the 3′-UTR influences turnover of AS isoforms by NMD. Exon–intron structures of genes and transcripts of (A) At2g38880—NF-YB1 transcription factor, NF-YB1/HAP3a and (B) At1g72560—PSD/exportin-t. Alternative splicing of the 3′-UTR introns in these genes generate transcripts with different distances between the authentic stop codon and downstream splice junction consistent with the 50–55 nt rule where distances >50–55 nt trigger NMD. Stop codon to splice junction distances are indicated along with whether the AS isoform is turned over by NMD or not. For diagram key see legend to Figure 1.
The position of PTCs defines the length of 3′-UTRs which can trigger nonsense-mediated decay
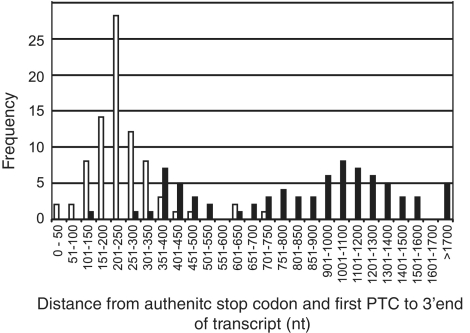
Current models for NMD suggest that the length of the 3′-UTR (distance between the stop codon or PTC and 3′-end of the transcript—long 3′-UTR) can be one of the triggers of NMD (Figure 1c). For all of the analysed AS transcripts containing PTCs, we analysed the distance between the first PTC and the 3′-end of the transcript (Figure 5; Supplementary Table S2). These distances appeared to show a bimodal distribution with the majority (58 transcripts) being longer than 600 nt and twenty transcripts (from 16 genes) shorter than 550 nt. When these latter transcripts were examined, two-thirds were from genes with relatively short coding sequences (500–720 bp) and all but five had splice junctions downstream of the PTC such that they conformed to expected features of NMD substrates. The five transcripts without a downstream splice junction had PTC to 3′-end distances of 366, 370, 440 nt and two transcripts had 441 nt and assuming that there are no other AS events in the genes causing NMD, these could represent ‘long 3′–UTR’ transcripts. We also determined the distance between the authentic stop codon and 3′-end of the gene in the normally spliced transcripts from the same genes. The mean distance was 242 nt and the majority of transcripts were in the range of 22–350 nt (Figure 5). Eight fully spliced transcripts (from 7 genes) had a 3′-UTR of >350 nt ranging from 354 to 718 nt. Taken together, these data suggest that long 3′-UTRs which trigger NMD in Arabidopsis mRNAs are in general >350 nt but there are exceptions where transcripts with longer 3′-UTRs do not show evidence of NMD.
Figure 5.
Distribution of stop codon to 3′-end distances. Distribution of frequency of distances in nucleotides (nt) between the first premature termination codon and the 3′-end of the transcript (shaded) compared to the distance between the authentic stop codon and 3′-end of the transcript of cognate genes (unshaded).
uORFs overlapping the main start codon induce nonsense-mediated decay
Previous studies in Arabidopsis suggest that uORF can trigger NMD (35,60). Preliminary features of such uORFs were defined using model constructs as being the first uORF in a transcript, at least 10 nt from the 5′-end and longer than 35–50 amino acids (35). However, the uORF in AtMHX was only 13 amino acids long and affected mRNA levels and translational efficiency (60). In addition, the link between AS in 5′UTRs and the presence, size, and positions of uORFs which may then trigger NMD has never been investigated in plants or in endogenous transcripts. In our study, we identified 12 genes with alternative splicing of 5′-UTR introns where transcripts increased significantly in upf mutants and/or cycloheximide treated plants. uORFs of between 3 and 123 amino acids were present in the fifteen different AS isoforms of these genes.
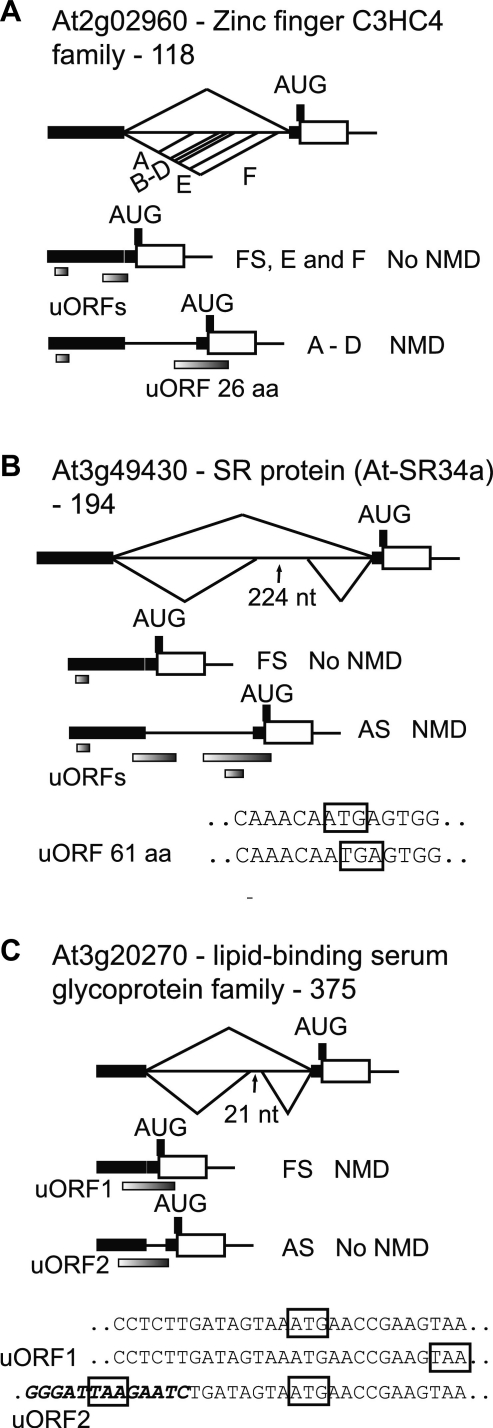
Seven genes had uORFs with the interesting unifying feature that the AS isoforms turned over by NMD contained an uORF which overlapped the translation start site of the main ORFs (Figure 6A–C; Supplementary Figure S3; Table 1). For example, in the zinc finger protein gene (At2g02960), alternative splicing produces six different AS transcripts through use of multiple alternative 3′ splice sites (Figure 6A). Four of these produce a 26 amino acid uORF which overlaps the AUG translation start site of the main ORF and all four are NMD sensitive. The fully spliced transcript and two shorter alternatively spliced isoforms contained short uORFs which do not overlap with the AUG. AS in the 5′UTR of At3g49430 (At-SR34a) generated three new uORFs of 13, 30 and 61 amino acids (Figure 6B). The stop codon of the 61 amino acid uORF overlapped the AUG of the main ORF and this transcript is NMD sensitive. Similarly, the fully spliced product of At3g20270 contains a uORF of 22 amino acids where the stop codon of the uORF lies downstream of the AUG of the main ORF (Figure 6C) and is NMD sensitive whereas the other AS transcripts without this feature are NMD resistant. Other examples of genes showing this phenomenon are shown in Supplementary Figure S3. Taken together, our analysis shows that many transcripts containing a uORF which overlaps the authentic start codon are subject to NMD and identifies a new feature of uORFs which is capable of inducing NMD in plants. The analysis also indicates that not all features of uORFs which trigger NMD have been resolved as we find examples where there is no correlation of the presence of uORFs of between 43 and 92 amino acids and NMD (see ‘Discussion’ section).
Figure 6.
Alternative splicing of introns in the 5′-UTR affects the presence, size and position of uORFs and influences turnover of AS isoforms by NMD. Exon–intron structures of genes and transcripts of (A) At2g02960—zinc finger transcription factor, (B) At3g49430—SR protein gene, At-SR34a, and (C) At3g20270—lipid-binding serum glycoprotein gene. These examples illustrate alternative splicing events in 5′-UTR introns which generate uORFs which overlap the main ORF and correlate with NMD. (A) four of the AS isoforms contain a 26 amino acid uORF (A–D) which overlaps the translation start site of the main ORF and correlates to NMD; (B) an uORF of 61 amino acids overlaps the authentic translation start codon in the AS product; and (C) uORF1 overlaps the translation start of the main coding sequence in the fully spliced transcript while in the alternatively spliced isoform the stop codon of uORF2 lies upstream of the main translation start site. Shaded rectangles below transcripts—uORFs; FS—fully spliced; AS—alternatively spliced. Sequences below the figures show the relationship between the stop codon of uORFs and the translational start AUG of the main ORF. For diagram key see legend to Figure 1.
Splice isoforms with retained introns are not sensitive to nonsense-mediated decay
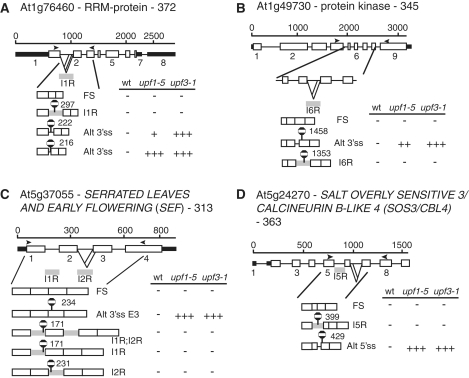
Intron retention is the most abundant type of alternative splicing in plants—41% (62). In general, due to the UA-richness of plant introns, most intron retention events create PTC transcripts. These transcripts are considered as potential targets of NMD as it has been shown in other organisms that transcripts with retained introns are turned over by NMD (63). Of the 90 characterized AS transcripts which increased in one or both of the upf mutants, only four had retained introns (Supplementary Table S2) suggesting that IR transcripts were under-represented. We therefore examined all of the readily detectable intron retention events on our panel where the IR transcripts made up at least 2% of the total. Of the 29 such IR transcripts, nineteen contained PTCs with downstream splice junctions and/or had long 3′-UTRs (all >400 nt) and therefore have features of typical NMD substrates (Table 2; Figures 1D and 7). Despite containing these NMD signals, these intron retention isoforms did not increase in abundance in the upf mutants and/or cycloheximide treatment suggesting that they are not turned over by the NMD pathway. Interestingly, in a number of these genes, other alternatively spliced transcripts were produced which contained PTCs and were subject to NMD. In two cases in particular, the other AS events involved the same intron as the retained intron (Figure 7A and B). In At5g37055 (SEF—SERRATED LEAVES AND EARLY FLOWERING), there are three different intron retention transcripts involving introns 1 and 2, all containing PTCs, none of which is a target of NMD (Figure 7C). However, use of an alternative 3′ splice site in exon 3 generates a PTC+ transcript which is turned over by NMD (Figure 7C and Table 2). A further example is At5g24270, coding for SOS3—SALT OVERLY SENSITIVE 3, a calcineurin-like protein, where retention of intron 5 was NMD resistant while use of an alternative 5′ splice site in the next intron, also creating a PTC, significantly up regulated the latter transcript in upf3-1 and cycloheximide-treated plants (Figure 7D and Table 2). The first PTC generated in these two transcripts are only 30 nt apart (Figure 7D) arguing against a position effect of the PTC. Thus, transcripts from the same gene which generate PTCs in very similar positions either through alternative splicing or intron retention events can be differentially sensitive to NMD. It appears that if a transcript is generated where an intron has not been spliced (retained intron) then the transcript is not NMD sensitive.
Table 2.
PTC+, NMD-insensitive intron retention transcripts and NMD-sensitive AS transcripts from the same genes
| Primer pair | GeneID | Name | Exon no. | Primer sites | AS event | Transcript | wt | upf1-5 | upf3-1 | NMD |
|---|---|---|---|---|---|---|---|---|---|---|
| 50 | At5g43910 | pfkB-type carbohydrate kinase family protein | 12 | Ex8-10 | FS | 0.58 | 0.46 | 0.39 | ||
| IR9 | PTC+; ds SJ | 0.05 | 0.05 | 0.04 | No NMD | |||||
| E10 Alt 3′ss(−19) | PTC+; ds SJ | 0.35 | 0.48 | 0.56 | NMD | |||||
| 346 | At4g23260 | CRK18 | 7 | Ex1–3 | FS | 0.09 | 0.09 | 0.17 | ||
| IR1 | PTC+; ds SJ | 0.90 | 0.90 | 0.82 | No NMD | |||||
| unknown (245 bp) | 0.00 | 0.00 | 0.01 | NMD | ||||||
| 316 | At2g28550 | TOE1 | 8 | Ex1–4 | FS | 0.93 | 0.94 | 0.96 | ||
| IR2 | PTC+; ds SJ | 0.07 | 0.06 | 0.04 | No NMD | |||||
| 312 | At5g13790 | AGL15 | 8 | Ex1–7 | FS | 0.45 | 0.47 | 0.47 | ||
| IR2 and IR3 | PTC+; ds SJ | 0.06 | 0.06 | 0.07 | No NMD | |||||
| 363 | At5g24270 | SOS3/CBL4 | 8 | Ex4–7 | FS | 0.83 | 0.82 | 0.77 | ||
| E5 Alt5′SS(+29) | PTC+; ds SJ | 0.06 | 0.08 | 0.13 | NMD | |||||
| IR4 | PTC+; ds SJ | 0.03 | 0.03 | 0.02 | No NMD | |||||
| unknown (299 bp) | 0.02 | 0.02 | 0.03 | NMD | ||||||
| unknown (330 bp) | 0.00 | 0.01 | 0.01 | NMD | ||||||
| 313 | At5g37055 | SEF | 4 | Ex1–4 | FS | 0.79 | 0.78 | 0.77 | ||
| IR1 and IR2 | PTC+; ds SJ | 0.03 | 0.03 | 0.03 | No NMD | |||||
| IR1 | PTC+; ds SJ | 0.13 | 0.13 | 0.12 | No NMD | |||||
| IR2 | PTC+; ds SJ | 0.01 | 0.01 | 0.01 | No NMD | |||||
| E3 Alt 3′ss(−11) | PTC+; ds SJ | 0.02 | 0.04 | 0.05 | NMD | |||||
| 372 | At1g76460 | RRM-containing protein | 7 | Ex1–3 | FS | 0.88 | 0.84 | 0.79 | ||
| IR1 | PTC+; ds SJ | 0.03 | 0.03 | 0.02 | No NMD | |||||
| E2 Alt 3′ss(+9) | PTC+; ds SJ | 0.04 | 0.05 | 0.07 | NMD | |||||
| E2 Alt 3′ss(+54) | PTC+; ds SJ | 0.05 | 0.08 | 0.12 | NMD | |||||
| 345 | At1g49730 | protein kinase | 9 | Ex4–7 | FS | 0.89 | 0.81 | 0.77 | ||
| IR6 | PTC+; ds SJ | 0.02 | 0.03 | 0.03 | No NMD | |||||
| E7 Alt 3′ss(+37) | PTC+; ds SJ | 0.08 | 0.16 | 0.21 | NMD | |||||
| 321 | At4g27410 | RD26 | 3 | Ex1–3 | FS | 0.93 | 0.92 | 0.93 | ||
| IR1 | PTC+; ds SJ | 0.02 | 0.02 | 0.02 | No NMD | |||||
| 325 | At2g47890 | Zn finger (B-box type) protein | 4 | Ex1–4 | FS | 0.92 | 0.93 | 0.88 | ||
| IR3 | C-terminal change | 0.06 | 0.06 | 0.10 | No NMD | |||||
| IR2 | PTC+; ds SJ | 0.01 | 0.01 | 0.01 | No NMD | |||||
| 373 | At3g13224 | RRM-containing protein | 6 | Ex4–5/6 | FS | 0.90 | 0.89 | 0.90 | ||
| IR5 | PTC+; long 3′ UTR (1395 nt) | 0.10 | 0.11 | 0.10 | No NMD | |||||
| 335 | At5g66210 | CPK28 | 13 | Ex11–13 | FS (I12 in 3′ UTR) | 0.01 | 0.01 | 0.01 | ||
| IR11 and IR12 | PTC+; long 3′UTR (686 nt) | 0.90 | 0.91 | 0.90 | No NMD | |||||
| ES12 | C-terminal change | 0.02 | 0.02 | 0.01 | No NMD | |||||
| 326 | At1g69250 | NTF2 | 8 | Ex6–8 | FS | 0.92 | 0.92 | 0.94 ** | ||
| IR6 | PTC+; long 3′UTR (415 nt) | 0.08 | 0.08 | 0.06 | No NMD | |||||
| 333 | At3g16800 | PP2C | 6 | Ex4–6 | FS | 0.97 | 0.97 | 0.97 | ||
| IR5 | PTC+; long 3′UTR (469 nt) | 0.03 | 0.03 | 0.03 | No NMD | |||||
| 317 | At2g28550 | TOE1 | 8 | Ex6–8 | FS | 0.97 | 0.98 | 0.98 | ||
| IR7 | PTC+; long 3′UTR (493 nt) | 0.02 | 0.02 | 0.01 | No NMD | |||||
| 366 | At5g25610 | RD22 | 4 | Ex1–3 | FS | 0.97 | 0.95 | 0.97 | ||
| IR2 | PTC+; dsSJ | 0.03 | 0.05 | 0.02 | No NMD | |||||
| 374 | At4g36960 | RRM-containing protein | 13 | Ex1–3 | FS | 0.84 | 0.74 | 0.61 | ||
| IR1 (5′ UTR) | PTC+; uORF in 5′ UTR | 0.05 | 0.15 | 0.32 | NMD | |||||
| IR2 | PTC+; ds SJ | 0.11 | 0.10 | 0.07 | No NMD |
Primers are positioned in the exons indicated and amplify across at least two introns. En Alt5′ss and En Alt3′ss—alternative 5′ splice site and alternative 3′ splice site in respective exon, where n is an exon number. The number in brackets indicates the number of nucleotides added (+) or removed (−) from the exonic sequence. IRn—retention of intron, where n is an intron number. The number in brackets indicates the size (nt) of the intron. Exons and introns are numbered with respect to the TAIR reference splice variant. FS—fully spliced; PTC+—transcript containing premature termination codon(s); ds SJ—presence of downstream splice junction(s); Numbers in bold—significant difference >3%; numbers in bold italic—significant difference <3%.
Figure 7.
Intron retention transcripts are not turned over by NMD. Schematic figures of genes which produce detectable intron retention transcripts and other alternatively spliced transcripts with different NMD phenotypes: (A) At1g76460; (B) At1g49730; (C) At5g37055 and (D) At5g24270. Below each gene, structures of transcripts in the amplified region are shown and display different alternative splicing events. Data for each transcript is shown alongside (−: no change; +, ++ and +++: transcript level increases significantly in upf mutants with P < 0.1; 0.01 > P < 0.05 and P < 0.01, respectively. For diagram key see legend to Figure 1. Grey lines below introns labelled IR—retained introns; FS—fully spliced; AS—alternatively spliced.
Besides the above PTC+ NMD-insensitive events, six other IR transcripts were effectively PTC- and were not targets of NMD (Supplementary Table S6). In these transcripts, the introns were either (i) in frame (no PTC), (ii) towards the end of the transcript such that the PTC was close to the authentic stop and would lead to a change in C-terminal sequence, (iii) in the 5′-UTR with uORFs which do not trigger NMD, or (iv) there was no evidence of an intron at the suggested position in the transcript.
Only four intron retention transcripts increased significantly in the mutants and/or cycloheximide treatment (Supplementary Tables S2 and S6). One of these (At4g36960) retained intron 1 in the 5′UTR which generated an uORF overlapping the authentic translation start site and therefore is expected to trigger NMD. In the other cases, potential NMD-causing AS events may occur elsewhere in the gene.
In conclusion, our analysis suggests that plant transcripts with retained introns are usually not targets for NMD provided there is no other alternative splicing event which produces features for NMD in the transcript.
DISCUSSION
Alternative splicing is a major determinant in the production of variant mRNA transcripts some of which contain PTCs and might be targeted by NMD. This pathway has a significant impact on the expression of genes involved in plant development and adaptation [reviewed (24,25)]. This raises the important questions of how frequently the expression of plant genes is regulated by coupled AS/NMD and what are the structural features of endogenous NMD substrates. Using a high-resolution RT–PCR system we have examined a large population of 950 endogenous transcripts from 270 genes and have characterized over 100 NMD-sensitive AS transcripts in detail. This represents the most extensive and accurate analysis of AS and AS/NMD in endogenous transcripts in plants. We demonstrate (i) a previously unknown high overall prevalence of AS and AS/NMD; (ii) that NMD-sensitive transcripts are readily detected in wild-type plants often representing substantial proportions of the total transcripts of a gene; (iii) that AS in 5′-UTRs and 3′-UTRs regulates transcript levels by rendering them NMD sensitive; (iv) that uORFs overlapping the start codon can trigger NMD; and (v) that transcripts with intron retention events in plants do not trigger NMD even though they possess classical features inducing NMD.
Coupling AS to NMD is a frequent event in plant gene expression
In the course of our analysis we have discovered an unexpectedly high number of novel AS transcripts. This follows from the sensitivity of the RT–PCR system which can detect transcripts of <1% of the total transcripts of a gene; on the other hand, many of the novel transcripts were abundant but not represented in databases. Thus, clearly much more AS is occurring in Arabidopsis than is currently estimated and annotated, especially considering that we have assessed AS/NMD only in plants at one developmental stage (3 week old) and have not taken into account many developmental stage-, tissue- or condition-specific AS events.
Genome-wide analyses in eukaryotes have shown that up to 20% of the transcriptome can be affected by NMD [reviewed in (12,53,64)] and that around 20–30% of alternatively spliced transcripts in humans contain PTCs and are potential targets of NMD (37). In plants, little is known about the contribution of NMD to regulation of gene expression. A recent genome-wide tiling array analysis in Arabidopsis found that only around 1% of plant protein-coding genes were affected (50) while an RNA-Seq analysis which showed that 42% of Arabidopsis genes underwent AS predicted that around 78% of AS isoforms could be putative targets of NMD (36). By comparing our AS/NMD gene set to those detected using expression/tiling arrays (31,50) we found only two genes in common (data not shown) and, in addition, known AS/NMD substrates such as GRP7 and GRP8 or SR genes (45,46) were not detected using expression or tiling arrays (31,50). Similarly, very little overlap in NMD-affected gene sets was found in Drosophila when comparing expression microarray and splicing-sensitive array results (53). This discrepancy is most likely due to the sensitivity and resolution of the AS RT–PCR panel which is able to detect significant changes in individual transcript levels which would not be detected in microarray experiments where the usual cut-off is >1.5–2-fold.
In this study, 11–17% of the total number of transcripts and 16–25% of the alternatively spliced transcripts analysed were potential NMD substrates suggesting that about 32–44.8% of AS genes are regulated by NMD. Extrapolating from these values and the estimate of the frequency of AS (36), about 13–18% of Arabidopsis intron-containing genes are potentially regulated by AS/NMD. This compares well to the 14% and 20% reported for Drosophila and Caenorhabiditis elegans (64,65).
Features of NMD-sensitive transcripts in plants
Rules for NMD in plants have been established based on the behaviour of a small number of genes or artificial constructs (28,32–35). While the general principles of intron-based and long 3′-UTR dependent NMD have been described (Figure 1C), little investigation of this behaviour in endogenous NMD-sensitive transcripts has been performed until now. Here, we determined the features of individual AS transcripts and found that the majority of AS/NMD transcripts (∼85%) contained PTCs with downstream splice junctions and/or long 3′-UTRs and therefore comply with existing NMD rules. In plants the average length of the 3′-UTR is 241 nt (66) and our results show that a ‘long 3′-UTR’ capable of triggering NMD in Arabidopsis is usually >350 nt. A similar estimate was obtained using NMD-test constructs where instead of the 3′-UTR length, the distance between a PTC and the authentic stop codon was defined previously as around 300 nt (34). In addition, however, we identified a number of exceptions where transcripts with 3′-UTRs >350 nt were not turned over by NMD suggesting that additional yet unidentified features are involved in triggering NMD.
Our results demonstrate that alternative splicing of introns in either the 3′-UTR or 5′-UTR can determine whether transcripts of endogenous genes are targets of NMD or not and thereby regulate transcript levels. We identified two genes where AS in 3′-UTR introns rendered AS transcripts NMD-sensitive by increasing the distance between the authentic stop codon and the splice junction to more than 50 nt. This agrees with the rules for EJC complexes trigging NMD (Figure 1D). Interestingly, one of the genes showing regulation by AS/NMD in a 3′-UTR intron is the NF-YB1 transcription factor subunit involved in photoperiod-regulated flowering and in drought stress responses, and whose over-expression leads to increased drought resistance (67).
Alternative splicing of introns in 5′-UTRs can change the length and sequence of the 5′-UTR or remove the authentic AUG (again altering the 5′-UTR) and thereby affect the presence/absence, number, size and position of uORFs. In human, polymorphisms or mutations which create or remove uORFs can suppress mRNA and protein levels and cause disease (68). uORFs in 5′-UTR regions can affect gene expression by different mechanisms: encoding an active peptide, affecting translational efficiency or reducing transcript levels by triggering NMD (68–70). In eukaryotes, ribosomes generally load onto mRNAs at the 5′-end and scan to the first AUG translation start codon. If an uORF is translated, the uORF stop codon might be recognized as a PTC (with the additional features of creating a long 3′-UTR and the high likelihood of downstream splice junctions) and thereby targeting the transcript to the NMD pathway. Around 20% of plant genes contain uORFs (35,71) but their fate in terms of whether they are translated or scanned through, trigger NMD or allow re-initiation of translation is not known. We found a strong correlation between presence of an uORF which overlapped the AUG of the main ORF and NMD most likely triggered by generating a long 3′-UTR and downstream splice junctions. We also found other genes where the presence of ‘fully upstream’ uORFs correlated with activation of NMD which indicates inefficient reinitiation of translation of their main ORFs. However, other AS transcripts contained short uORFs and/or ‘long’ uORFs (e.g. 43, 55 and 92 amino acids) which did not trigger NMD. Thus, the factors which determine whether or not particular uORFs activate NMD are clearly complex and poorly understood. With around 20% of Arabidopsis genes containing uORFs and the frequent occurrence of AS in 5′-UTRs, AS/NMD involving uORFs is likely to be important in regulation of expression of many plant genes.
Retained introns do not trigger NMD
Besides identifying NMD-sensitive AS transcripts, we also identified AS transcripts which contained NMD signals but which were immune to NMD. A comprehensive analysis of AS/NMD in mammalian tissues indicated that not all characteristics of NMD-targeted RNAs have been identified and that not all RNAs containing known NMD features are in fact turned over by NMD (52). Surprisingly, we found that the majority of intron retention transcripts which we analysed were not turned over by NMD despite containing PTCs, downstream splice junctions and long 3′-UTRs. However, transcripts from the same gene with other types of alternative splicing events in the same or nearby intron which generated PTCs in very similar positions were sensitive to NMD. This unexpected finding is in contrast to current assumptions that plant transcripts with retained introns and PTCs are subject to NMD as such transcripts have been found on ribosomes (72,73), a prerequisite for NMD. More importantly, in other organisms transcripts with retained introns and PTCs are subjected to NMD suggesting a different strategy in plants as intron retention transcripts avoid the NMD machinery and have a different fate [see below; (63,74)]. In addition, our clear demonstration that intron retention events which create a PTC are NMD insensitive may explain the high frequency of IR events identified in plants where it constitutes the major AS event.
We previously detected aberrant mRNAs in the nucleolus of Arabidopsis of which around 80% were intron retention events (75). We also found UPF2 and UPF3 to localize to the nucleolus and hypothesized that this may be the site of assembly of NMD factors onto aberrant mRNAs prior to NMD. From the data presented here, IR transcripts avoid NMD and their accumulation in the nucleolus may therefore have a different function. Although the current model of NMD in mammals is that PTCs are recognized in the pioneer round of translation as the mRNA exits the nuclear pore, it is not clear whether this model applies to plants. However, if translation is required for NMD, one possible explanation is that transcripts containing introns or intron fragments are recognized as aberrant prior to export by virtue of proteins binding to the UA-rich intron sequences (75) and therefore do not connect with the NMD machinery. Further research will be needed to determine the fate of different plant PTC-containing transcripts in terms of their intranuclear and intracellular localization, dynamics and transport and how and why intron retention transcripts escape NMD in plants.
Significance of AS/NMD for plant gene expression pattern
The endogenous NMD-sensitive transcripts showed great variation in their steady state levels (i.e. levels detectable in wild-type plants) and in the degree of increased abundance in the different mutants and cycloheximide treatment. This variation is likely to reflect gene-specific differences in transcription levels, frequency of AS producing the different isoforms, or tissue-specific AS occurring only in particular organs or cell types. In addition, the transcript-specific efficiency of NMD turnover could reflect features of the transcripts such as position of PTC and downstream splice junctions, length of 3′-UTR and RNA secondary structure. Importantly, for some genes, non-productive mRNAs (PTC-containing; unable to produce full-length protein) which includes transcripts targeted by NMD and transcripts which are NMD-insensitive such as intron retention transcripts, form a significant proportion of steady state levels of transcripts in wild-type plants. One consequence is that traditional expression microarrays are unable to distinguish between productive and non-productive mRNAs and therefore functional transcript abundance for these genes is over-estimated. Clearly, alternative splicing information must be integrated with transcriptional data to provide true measures of gene expression.
The coupling of AS and NMD is an important general mechanism in gene expression regulation. It modulates the relative levels of mRNA isoforms from a gene which are either productive (protein-coding) or unproductive AS variants and thereby regulates protein levels. Recent examples of plant genes regulated or putatively regulated by AS/NMD in plants are GRP7/8 and SOC1 (involved in the circadian clock and flowering control, respectively), SR and PTB protein splicing factors (involved in a range of developmental and stress response processes) and HSF2A (a heat shock factor) (43–48,76). Here, despite only around 270 genes being analysed, AS/NMD has been identified in 121 genes (Table1; Supplementary Table S3) playing central roles in cellular processes: transcription factors, splicing factors, RNA-binding proteins, RNA helicases, spliceosome and exon junction complex proteins, tRNA export, signal recognition particle and ribosomal proteins. Components of developmental pathways also show NMD-mediated turnover of AS transcripts, for example, different MAF genes and VRN2 (flowering time) and CCA1 and PRR9 (core circadian clock). Finally, a number of genes involved in signalling and stress response pathways undergo AS/NMD: the calcium-dependent salt stress signalling pathway protein genes SOS2 and SOS3, phosphatases and kinases (e.g. the SNF1-like protein kinase, AtKIN11) and various temperature, drought and salt response factors (e.g. SRF2, HSF2A). The identification of many genes in a wide range of processes and pathways suggests that AS/NMD is a widespread regulatory mechanism in plants.
ACCESSION NUMBERS
The Arabidopsis Genome Initiative numbers for UPF1 and UPF3 are At5g47010 and At1g33980. AGI locus identifiers of genes analyzed in this article are listed in Supplementary Table S1.
SUPPLEMETARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–6, Supplementary Figures S1–3.
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) [BB/G000212/1 and BB/G024979/1 (ERA-NET Plant Genomics (PASAS)]; Scottish Government Rural and Environment Research and Analysis Directorate (RERAD) WP114; EU FP6 Programme Network of Excellence on Alternative Splicing (EURASNET) LSHG-CT-2005-518238; Austrian Science Fund (FWF) [SFB 1710, 1711; DK W1207; ERA-NET Plant Genomics (PASAS) I254]; Austria Genomic Program (GENAU II, III; ncRNAs). H.Q.D. acknowledges support from the WWTF Science Chair to Arndt von Haeseler. Funding for open access charge: Austrian National Science Foundation (FWF).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Monika Maronova for help in preparing plant material, and TAIR and especially Philippe Lamesch for providing custom Arabidopsis datasets.
REFERENCES
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 4.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 7.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 8.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 9.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 12.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 13.Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 15.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol. Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 23.Sauliere J, Haque N, Harms S, Barbosa I, Blanchette M, Le Hir H. The exon junction complex differentially marks spliced junctions. Nat Struct Mol Biol. 2010;17:1269–1271. doi: 10.1038/nsmb.1890. [DOI] [PubMed] [Google Scholar]
- 24.Belostotsky DA, Sieburth LE. Kill the messenger: mRNA decay and plant development. Curr. Opin. Plant Biol. 2009;12:96–102. doi: 10.1016/j.pbi.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Chiba Y, Green P. mRNA degradation machinery in plants. J. Plant Biol. 2009;52:114–124. [Google Scholar]
- 26.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- 27.Hori K, Watanabe Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 2005;43:530–540. doi: 10.1111/j.1365-313X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- 28.Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 2008;27:1585–1595. doi: 10.1038/emboj.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riehs N, Akimcheva S, Puizina J, Bulankova P, Idol RA, Siroky J, Schleiffer A, Schweizer D, Shippen DE, Riha K. Arabidopsis SMG7 protein is required for exit from meiosis. J. Cell Sci. 2008;121:2208–2216. doi: 10.1242/jcs.027862. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Kang JH, Hettenhausen C, Baldwin IT. Nonsense-mediated mRNA decay (NMD) silences the accumulation of aberrant trypsin proteinase inhibitor mRNA in Nicotiana attenuata. Plant J. 2007;51:693–706. doi: 10.1111/j.1365-313X.2007.03173.x. [DOI] [PubMed] [Google Scholar]
- 31.Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 32.Hori K, Watanabe Y. Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol. 2007;48:1072–1078. doi: 10.1093/pcp/pcm075. [DOI] [PubMed] [Google Scholar]
- 33.Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D. Both introns and long 3'-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006;34:6147–6157. doi: 10.1093/nar/gkl737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz AM, Komarova TV, Skulachev MV, Zvereva AS, Dorokhov Iu L, Atabekov JG. Stability of plant mRNAs depends on the length of the 3′-untranslated region. Biochemistry. 2006;71:1377–1384. doi: 10.1134/s0006297906120145. [DOI] [PubMed] [Google Scholar]
- 35.Nyiko T, Sonkoly B, Merai Z, Benkovics AH, Silhavy D. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 2009;71:367–378. doi: 10.1007/s11103-009-9528-4. [DOI] [PubMed] [Google Scholar]
- 36.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hillman RT, Green RE, Brenner SE. An unappreciated role for RNA surveillance. Genome Biol. 2004;5:R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lareau LF, Green RE, Bhatnagar RS, Brenner SE. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Baek D, Green P. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc. Natl Acad. Sci. USA. 2005;102:12813–12818. doi: 10.1073/pnas.0506139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 42.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalyna M, Lopato S, Voronin V, Barta A. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 2006;34:4395–4405. doi: 10.1093/nar/gkl570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D. Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- 45.Schoning JC, Streitner C, Meyer IM, Gao Y, Staiger D. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 2008;36:6977–6987. doi: 10.1093/nar/gkn847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palusa SG, Reddy AS. Extensive coupling of alternative splicing of pre-mRNAs of serine/arginine (SR) genes with nonsense-mediated decay. New Phytol. 2010;185:83–89. doi: 10.1111/j.1469-8137.2009.03065.x. [DOI] [PubMed] [Google Scholar]
- 47.Stauffer E, Westermann A, Wagner G, Wachter A. Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J. 2010;64:243–255. doi: 10.1111/j.1365-313X.2010.04321.x. [DOI] [PubMed] [Google Scholar]
- 48.Song HR, Song JD, Cho JN, Amasino RM, Noh B, Noh YS. The RNA binding protein ELF9 directly reduces SUPPRESSOR OF OVEREXPRESSION OF CO1 transcript levels in arabidopsis, possibly via nonsense-mediated mRNA decay. Plant Cell. 2009;21:1195–1211. doi: 10.1105/tpc.108.064774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, et al. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl Acad. Sci. USA. 2009;106:2453–2458. doi: 10.1073/pnas.0808902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barberan-Soler S, Zahler AM. Alternative splicing regulation during C. elegans development: splicing factors as regulated targets. PLoS Genet. 2008;4:e1000001. doi: 10.1371/journal.pgen.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, Rehwinkel J, Gallusser FL, Izaurralde E, Rio DC, Dudoit S, et al. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simpson CG, Fuller J, Maronova M, Kalyna M, Davidson D, McNicol J, Barta A, Brown JW. Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J. 2008;53:1035–1048. doi: 10.1111/j.1365-313X.2007.03392.x. [DOI] [PubMed] [Google Scholar]
- 55.Valvekens D, Van Montague M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants using kanamycin selection. Proc. Natl Acad. Sci. USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 1962;15:473–497. [Google Scholar]
- 57.Simpson CG, Clark GP, Lyon JM, Watters J, McQuade C, Brown JWS. Interactions between introns via exon definition in plant pre-mRNA splicing. Plant J. 1999;18:293–302. [Google Scholar]
- 58.Usuka J, Zhu W, Brendel V. Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics. 2000;16:203–211. doi: 10.1093/bioinformatics/16.3.203. [DOI] [PubMed] [Google Scholar]
- 59.Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 60.Saul H, Elharrar E, Gaash R, Eliaz D, Valenci M, Akua T, Avramov M, Frankel N, Berezin I, Gottlieb D, et al. The upstream open reading frame of the Arabidopsis AtMHX gene has a strong impact on transcript accumulation through the nonsense-mediated mRNA decay pathway. Plant J. 2009;60:1031–1042. doi: 10.1111/j.1365-313X.2009.04021.x. [DOI] [PubMed] [Google Scholar]
- 61.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 62.Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 63.de Lima Morais DA, Harrison PM. Large-scale evidence for conservation of NMD candidature across mammals. PLoS ONE. 2010;5:e11695. doi: 10.1371/journal.pone.0011695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramani AK, Nelson AC, Kapranov P, Bell I, Gingeras TR, Fraser AG. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol. 2009;10:R101. doi: 10.1186/gb-2009-10-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pesole G, Liuni S, Grillo G, Saccone C. Structural and compositional features of untranslated regions of eukaryotic mRNAs. Gene. 1997;205:95–102. doi: 10.1016/s0378-1119(97)00407-1. [DOI] [PubMed] [Google Scholar]
- 67.Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl Acad. Sci. USA. 2007;104:16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl Acad. Sci. USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meijer HA, Thomas AA. Control of eukaryotic protein synthesis by upstream open reading frames in the 5'-untranslated region of an mRNA. Biochem J. 2002;367:1–11. doi: 10.1042/BJ20011706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kochetov AV, Sirnik OA, Rogosin IB, Glazko GV, Komarova ML, Shumny VK. Contextual features of higher plant mRNA 5’ untranslated regions. Mol. Biol. 2002;36:510–516. [PubMed] [Google Scholar]
- 72.Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 73.Jiao Y, Meyerowitz EM. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 2010;6:419. doi: 10.1038/msb.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol. Cell. 2008;31:360–370. doi: 10.1016/j.molcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JW. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell. 2009;21:2045–2057. doi: 10.1105/tpc.109.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sugio A, Dreos R, Aparicio F, Maule AJ. The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell. 2009;21:642–654. doi: 10.1105/tpc.108.062596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barta A, Kalyna M, Reddy AS. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell. 2010;22:2926–2929. doi: 10.1105/tpc.110.078352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.