Abstract
Several subunits of the multifunctional eukaryotic translation initiation factor 3 (eIF3) contain well-defined domains. Among them is the conserved bipartite PCI domain, typically serving as the principal scaffold for multisubunit 26S proteasome lid, CSN and eIF3 complexes, which constitutes most of the C-terminal region of the c/NIP1 subunit. Interestingly, the c/NIP1-PCI domain is exceptional in that its deletion, despite being lethal, does not affect eIF3 integrity. Here, we show that a short C-terminal truncation and two clustered mutations directly disturbing the PCI domain produce lethal or slow growth phenotypes and significantly reduce amounts of 40S-bound eIF3 and eIF5 in vivo. The extreme C-terminus directly interacts with blades 1–3 of the small ribosomal protein RACK1/ASC1, which is a part of the 40S head, and, consistently, deletion of the ASC1 coding region likewise affects eIF3 association with ribosomes. The PCI domain per se shows strong but unspecific binding to RNA, for the first time implicating this typical protein–protein binding domain in mediating protein–RNA interactions also. Importantly, as our clustered mutations severely reduce RNA binding, we conclude that the c/NIP1 C-terminal region forms an important intermolecular bridge between eIF3 and the 40S head region by contacting RACK1/ASC1 and most probably 18S rRNA.
INTRODUCTION
Translation initiation is a multistep process ensuring formation of the 80S elongation–competent complex composed of both ribosomal subunits with the P-site occupied by the methionyl initiator tRNA (Met-tRNAiMet) base-paired with the mRNA's AUG start codon. In the first step, Met-tRNAiMet is bound by eukaryotic initiation factor 2 (eIF2) in its GTP form to produce the ternary complex (TC). eIF3 together with eIFs 1, 1A and 5 then promotes TC recruitment to the small ribosomal subunit (40S) producing the 43S preinitiation complex (PIC). Subsequently, the 43S PIC interacts with the 5′-end of capped mRNA in a reaction promoted by eIF4F, eIF4B, PABP and eIF3 [reviewed in (1)]. Thus formed 48S PIC then scans the mRNA leader until the AUG start codon is recognized. This step is controlled by GTP-hydrolysis on eIF2 stimulated by eIF5 and by the subsequent release of free Pi from the 48S PIC triggered by eIF1. The scanning-arrested 48S PIC can now join the large ribosomal subunit with the help of GTP-bound eIF5B, upon which most eIFs are ejected with the exception of eIF1A and eIF3 (2–4). Finally, GTP hydrolysis on eIF5B triggers the release of eIF1A and eIF5B producing an active 80S ribosome poised for elongation.
One of the key facilitators of the eukaryotic initiation pathway is the multifunctional, multiprotein complex eIF3. Yeast S. cerevisiae eIF3 is composed of 6 subunits (a/TIF32, b/PRT1, c/NIP1, i/TIF34, g/TIF35 and j/HCR1), all of which have corresponding orthologs in the 13-subunit mammalian eIF3 complex. Yeast eIF3, as part of a higher order ribosome-free structure called the multifactor complex (MFC) composed also of the TC and eIFs 1 and 5 (5), was shown to enhance the efficiency of the 43S and 48S PIC assembly processes and also to stimulate the postassembly steps such as scanning and AUG recognition (6–15). The importance of eIF3 as a pivotal coordinator of the PIC assembly most probably lies in the fact that whereas its body resides on the solvent-exposed side of the small ribosomal subunit, several of its flexible protein interaction domains are thought to reach out to the interface side (16–18). Moreover, at least on the yeast 40S, the locations of both the mRNA entry and exit pores are thought to overlap with the eIF3 position. Thus, eIF3 is ideally suited for spatial distribution of other eIF3-interacting eIFs over the 40S surface as well as for mRNA loading.
To better understand the molecular details of the eIF3 role in the PIC assembly process, we have begun a systematic mapping of the positions of specific domains of various eIF3 subunits on the 40S (16). We found that the N-terminal domain (NTD) of a/TIF32 forms a crucial intermolecular bridge between eIF3 and the 40S by interacting with small ribosomal protein RPS0 in the vicinity of the mRNA exit pore (3). In addition, we observed that deleting the C-terminal domain (CTD) of a/TIF32 reduced the MFC association with the 40S when the connection between eIF3 and eIF5/TIF5 in the MFC was impaired by the tif5-7A mutation (16). Interestingly, the a/TIF32-CTD was found to interact with helices 16–18 of 18S rRNA (16) and RPS2 and RPS3 (14), all constituents of the mRNA entry channel (19). Consistently, RPS2 also interacts with the CTD of j/HCR1; i.e. the direct binding partner of the a/TIF32-CTD (12,14). The g/TIF35 subunit interacts with the 40S beak proteins RPS3 and RPS20 (13); however, its contribution to the overall 40S-binding affinity of eIF3 is currently unknown. Conversely, the RNA recognition motif (RRM) of b/PRT1 plays a direct role in anchoring eIF3 to the ribosome (9,12); however, its binding partner(s) remains to be identified. Finally, deletion of the C-terminal 240 residues of c/NIP1 completely eliminated binding of the mutant form of the complex to the 40S in vivo when competing with the wild-type eIF3. This result suggested that the c/NIP1-CTD also represents an important intermolecular bridge between eIF3 and some component(s) of the small ribosome (16).
Interestingly, the c/NIP1-CTD is formed by the bipartite PCI domain that is known to serve as the principal scaffold for the 26S proteasome lid, COP9 signalosome (CSN) and eIF3 [reviewed in (20)]. The PCI domain is defined by a conserved arrangement of curved bihelical tetratricopeptide-like repeats followed by a globular winged helix (WH) subdomain (21). Besides c/NIP1, yeast eIF3 contains only one additional PCI subunit in a/TIF32, whereas mammalian eIF3 is composed of a ‘canonical number’ of 6 PCI subunits (a, c, e, k, l, m). The PCI subunits are believed to regulate proper complex assembly via interactions between each other as well as with other protein partners. Despite the recent findings that several PCI proteins occur in complexes related to nucleic acid regulation (22), they are not known to be capable of direct RNA binding.
Among all 33 small ribosomal proteins, there is one that deserves a special attention—the yeast ribosomal protein ASC1 and its mammalian ortholog RACK1. They are both members of the WD40 (Trp-Asp) repeat scaffold protein family that adopts a seven-bladed β-propeller structure. RACK1/ASC1 (designated as ASC1 thereafter) is located on the head of the 40S ribosomal subunit near the mRNA exit tunnel and makes extensive contacts with helices h39 and h40 of 18S rRNA and ribosomal proteins RPS16, 17, and 3 (19,23). Importantly, ASC1 was also shown to interact with a number of signaling molecules on and off the ribosome and thus it is believed to play a crucial role in a multitude of biological processes and serve as a regulatory link between signaling and translation [reviewed in (24)]. For example, RACK1 recruits activated protein kinase C to the ribosome, which leads to the stimulation of translation through the phosphorylation of initiation factor 6 (25). In a ribosome-free form RACK1 associates with membrane-bound receptors (26), indicating that it can promote the docking of ribosomes at sites where local translation is required, such as focal adhesions.
Despite being implicated in all these cellular processes, yeast ASC1 is not essential for growth. The asc1Δ null strains were shown to impair cellular growth, to produce halfmer polysomes that could arise either from a defective subunit joining step or from a defect in 60S biogenesis, to impact translational rates in a transcript-specific manner, to deregulate GCN4 translational control under amino acid starvation and to increase sensitivity to drugs affecting cell wall biosynthesis (27–31). The breadth of the observed phenotypes may seem consistent with the documented multitasking by RACK1; however, there is one caveat. The ASC1 gene contains an intron encoding a small nucleolar RNA U24 (SNR24) (27) that plays a critical role in biogenesis of the large ribosomal subunit (32). Because all aforementioned asc1Δ null strains were also deleted for SNR24, it is unclear what phenotypes derive from the lack of what functional molecule and so what is the true contribution of the ASC1 protein to general translation.
In this study, we have focused on the role of the C-terminal PCI domain of the c/NIP1 subunit of eIF3 in promoting association of eIF3 and other MFC components with the 40S. We demonstrate that a short C-terminal deletion in nip1-Δ60 and a specific clustered alanine-scanning mutation (CAM) nip1–743A752 impinging into the WH subdomain of the PCI domain produce slow growth (Slg−) phenotypes and significantly reduce the amounts of 40S-bound MFC components in vivo, consistent with the idea that the c/NIP1-PCI forms an important intermolecular bridge between eIF3 and the 40S. Whereas the extreme C-terminal region of c/NIP1 was found to interact directly with blades 1–3 of ASC1, the PCI domain, the 3D structure of which was modeled in silico, shows strong but unspecific binding to RNA that is severely reduced by two of our CAM mutants. To our knowledge, this is the first report showing that the protein–protein interacting PCI domain is also capable of direct RNA binding. Finally, we demonstrate that the halfmer phenotype previously associated with the ASC1 deletion is in fact caused by deletion of its intron encoding U24, which has been previously accidentally overlooked. Importantly, however, the deletion of the only ASC1 coding region produced the Slg− phenotype and also reduced the eIF3-binding affinity toward the 40S subunit in vivo, ascribing RACK1/ASC1 the eIF3-docking role in general translation initiation. In addition, 40S-binding of eIF5 and the TC was also reduced, and, accordingly, the asc1Δ strain derepressed GCN4 translational control under non-starvation conditions producing the Gcd− phenotype suppressible by overexpressing TC. Together, we conclude that the c/NIP1-PCI domain promotes the PIC assembly process by linking eIF3 with the head region of the small ribosomal subunit.
MATERIALS AND METHODS
Yeast strains, plasmids and biochemical methods
Lists of strains, plasmids and oligonucleotides used in this study (Supplementary Tables S1–S3), details of their construction, as well as description of all well-established biochemical assays used throughout the study can be found in the Supplementary Data.
40S-binding assay
The 40S ribosomal subunits were purified as described previously (33). GST pull-down assays with purified 40S subunits were performed according to (34) with minor modifications. Briefly, the glutathione-Sepharose beads adsorbed with wt GST-c/NIP1 fusion proteins were first washed with the binding buffer [20 mM HEPES (pH 7.5), 2.5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA and 1 mM DTT] and subsequently incubated with purified ribosomal 40S subunits at final concentration of 0.2 μM in the same buffer supplemented with 1% dry milk for 30 min at 22°C. After three washes with the binding buffer, the resulting complexes were subjected to SDS–PAGE electrophoresis followed by immunoblotting with anti-Rps0 and anti-ASC1 antibodies.
RNA-binding assays
The [32P]-labeled mRNA species were prepared in vitro using the MAXIscriptTM SP6 or T7 transcription kit (Ambion), [α-32P]UTP (10 mCiml−1), and enzymatically linearized RNA-encoding vectors (see Supplementary Table S1 for their description) according to the vendor's instructions. The transcripts were purified using a size exclusion column (NucAway Spin Column, Ambion).
GST pull downs were carried out as follows: WT and mutant c/NIP1-CTD proteins fused to the GST moiety and immobilized on glutathione-Sepharose beads were incubated with 100 ng of [32P]-labeled RNA species in 250 μl of the binding buffer (10 mM HEPES [pH 7.6], 3 mM MgCl2, 40 mM KCl, 5% glycerol, 1 mM DTT, 1.5% 2 -β-mercaptoethanol) for 30 min at 26°C. [To increase specificity of binding, 200 ng of yeast total tRNA (Sigma) were added to each reaction as a competitor RNA.] The beads were then washed three times with 1 ml of binding buffer, and bound RNAs were separated by electrophoresis on 5.5% denaturing (8 M urea) polyacrylamide gel and subjected to autoradiography. For control experiments, the same procedure was carried out using beads containing only the GST moiety or beads preincubated with bacterial extracts derived from plain E. coli BL21 cells.
For the RNA electrophoretic mobility shift assay we used the optimized procedure originally described by (35). GST-c/NIP1-CTD fusion protein and GST alone demobilized from the glutathione-Sepharose beads at final concentration of ∼0.2 μM were incubated with [32P]-labeled RNA species at final concentration of ∼5 nM in 10 μl of binding buffer [10 mM HEPES (pH 7.5), 3 mM MgCl2, 14 mM KCl, 5% glycerol, 1 mM DTT, 0.2% IGEPAL CA-630] for 30 min at 22°C. RNA loading dye was added to each sample and the RNA–protein complexes were resolved on 3.5% polyacrylamide mini-gel (acrylamide:bis acrylamide of 37.5:1) at 4°C and subjected to autoradiography.
RESULTS
The 3D architecture of the C-terminal PCI domain of the c/NIP1 subunit of eIF3 predicted in silico
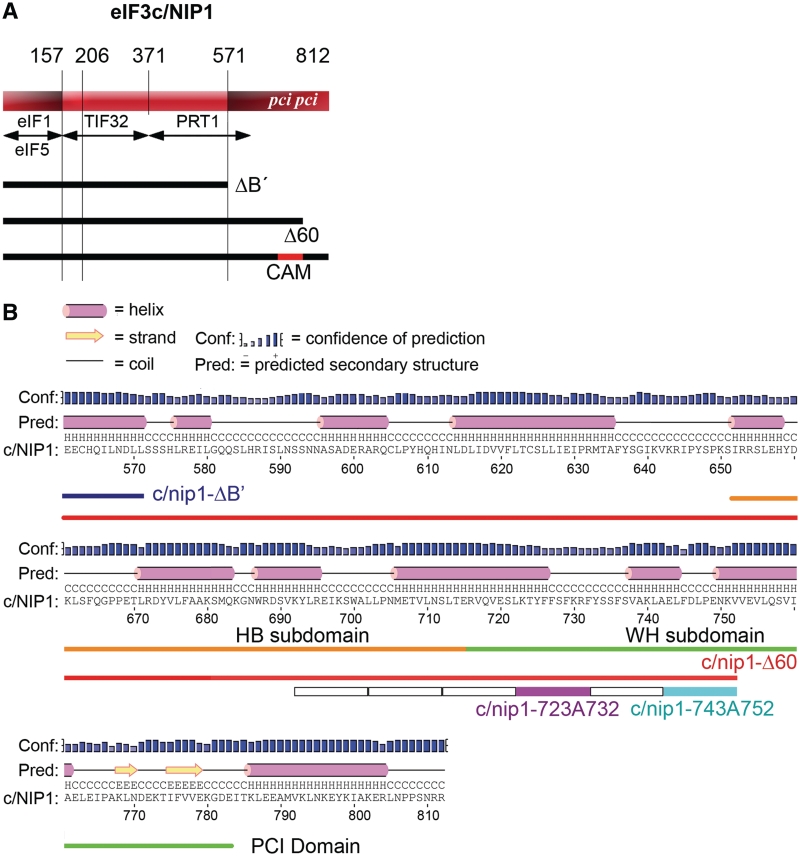
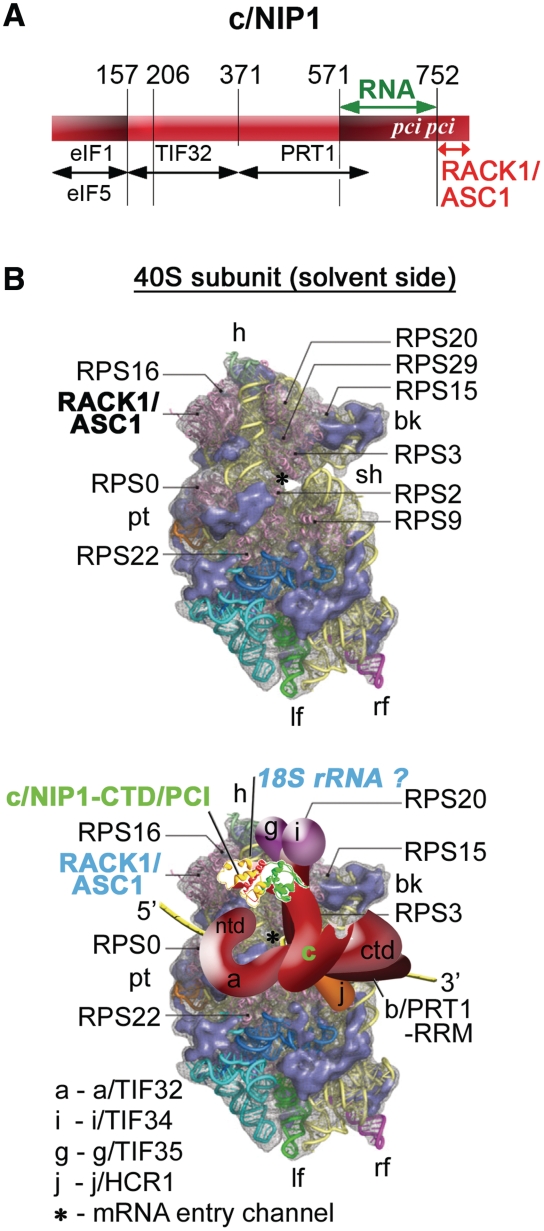
We previously identified and characterized several important domains of eIF3 subunits that mediate binding of the MFC components with the 40S subunit, which allowed us to propose a model of a spatial organization of the 43S PIC (3,12–14,16). Among the candidate domains to directly contact the ribosome was also the C-terminal one-third (in particular the last 240 amino acid residues) of eIF3c/NIP1 (16). The c/NIP1-CTD is not involved in the interaction network linking together eIF3 subunits or all other MFC components and as such, its otherwise lethal truncation in nip1-ΔB′ (Figure 1A) had no impact on the overall MFC integrity (7). However, when expressed in the background of the wild type NIP1 gene, the mutant form of eIF3 containing c/nip1-ΔB′ could not compete with the wild-type eIF3 and failed to associate with 40S ribosomes in vivo (16). To eliminate the effect of competition and examine the true importance of the c/NIP1-CTD for the 43–48S PIC assembly and cell viability, we first computationally predicted its secondary and tertiary structure and subsequently subjected this region to two mutagenic approaches to generate viable mutants that would enable us to study the role of the c/NIP1-CTD directly in the absence of the wt allele.
Figure 1.
The amino acid sequence of the c/NIP1-CTD with predicted secondary structure by PSIPRED. (A) Schematic of c/NIP1 with arrows delimiting the minimal binding domains for the indicated proteins identified previously. The lines beneath the schematic depict two C-terminally truncated mutants (ΔB′ and Δ60) and indicate the segment of c/NIP1-CTD that was subjected to clustered alanine mutagenesis (CAM) herein. The location of the PCI homology domain is indicated in the colored rectangle. (B) Secondary structure prediction of the c/NIP1-CTD (residues 560–812) by PSIPRED. The C-terminal truncations ΔB′ and Δ60 are marked by lines in blue and red, respectively. The 10-Ala substitutions in six consecutive blocks between residues 693 and 752 are indicated by open boxes with the nip1-723A732 and nip1-743A752 mutants highlighted in magenta and sky-blue, respectively. The WH and BH subdomains of the PCI domain are shown in green and orange, respectively.
The putative secondary structure of the last 240 amino acid residues was analyzed by the PSIPRED secondary structure prediction method (36). As shown in Figure 1B, this stretch of residues consists of a series of α-helices distributed over the entire region and two C-terminal β-sheets. Importantly, a major part of this stretch is formed by a well characterized canonical PCI domain (between residues 651 and 783) (20) (Figure 1A and B). Typically, PCI domains mediate protein–protein interactions to built large multiprotein assemblies. They are not well conserved in their primary sequence but based on the biochemical and structural studies, a classical PCI domain comprises two subdomains, the N-terminal helical bundle (HB) subdomain and the C-terminal WH that are tightly connected through an interface helix to build a complete and autonomous domain (21).
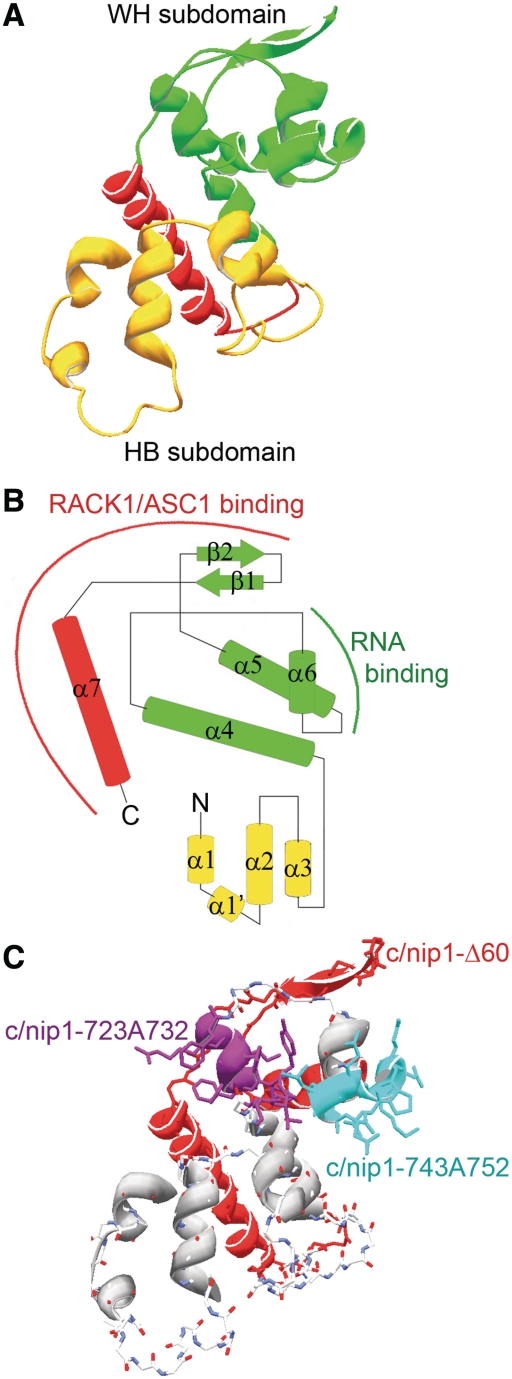
To compute a 3D model of the c/NIP1-PCI region spanning residues 651–812, we used the 3D threading engine on the Phyre2 protein fold recognition server (Figure 2A and B) (37,38). To examine the concordance of our predicted model, we compared it with two experimentally determined PCI structures: (i) a crystal structure of the PCI domain of Arabidopsis thaliana CSN7, where both WH and HB structural subdomains are distinguished (21); and (ii) an NMR solution structure of the C-terminal WH subdomain from the Mus musculus COP9 subunit SGN4 (Suzuki, S., Yokoyama, S; protein data bank accession code 1UFM). Despite the fact that the c/NIP1-PCI predictably shares low sequence homology with both experimental structures, they all share significant structural homology through the WH subdomain—backbone atoms RMSD is <1.7 Å [measured by DaliLite online structure alignment server (39)]. Based on this homology, it was possible to align the 3D model of the c/NIP1-PCI to the structures of SGN4 and to CSN7 by Swiss-PDBViewer 4.0.1. (40) (Supplementary Figure S1A). Despite the apparent similarities of the overall fold, both termini of our c/NIP1-PCI model slightly vary from the experimental structures. Specifically, the N-terminus of the c/NIP1-PCI model (Figure 2B, in yellow; Supplementary Figure S1A, in green) differs from the corresponding part of CSN7 (Supplementary Figure S1A, in yellow) in the number of pairs of the anti-parallel helices; whereas two pairs are found at the CSN7 HB subdomain structure (αN1-αN2 and α1/α1′-α2), the c/NIP1-PCI contains only the α1/α1′-α2 pair. However, since all our attempts to compute a meaningful structural prediction of the extreme N-terminal part of the c/NIP1-HB subdomain unexpectedly failed, it is important to note that this difference may have only a ‘technical’ character and does not necessarily mean that the first more N-terminal pair is missing in real c/NIP1-PCI structure. In the C-terminus of the c/NIP1-PCI model, the WH subdomain is extended by an extra α-helix that turns backward to the HB subdomain (Figure 2A and B, in red).
Figure 2.
The 3D threading model of the c/NIP1-PCI domain from residues 651–812 computed by Phyre2 (A) with a topology depiction of the secondary structure (B). The WH subdomain is shown in green, the HB subdomain in yellow and the flanking C-terminal helix in red. Binding regions of the c/NIP1-CTD-PCI domain for ASC1 and RNA determined in this study are indicated by red and green arches. (C) The 3D model of the c/NIP1-PCI domain as in (A) depicting positions of individual mutations; the nip1-723A732 and nip1-743A752 10-Ala substitutions are shown in magenta and sky-blue, respectively, and the truncated segment in nip1-Δ60 in red.
Interestingly, the electrostatic surface of the c/NIP1-PCI bears a few well distinguished patches of charged residues (Supplementary Figure S1B). Most of the HB subdomain's surface is dominated by positive charge that further extends into C-terminal end of helix α4 of the WH subdomain, whereas the rest of the WH subdomain is predominantly negative with a few positively charged residues sticking out into the solvent. Remarkably, there is also a positively charged large groove at the base of the predicted structure formed partially by the extended helix α7.
The c/NIP1-PCI serves as an important intermolecular bridge between eIF3 and the 40S subunit
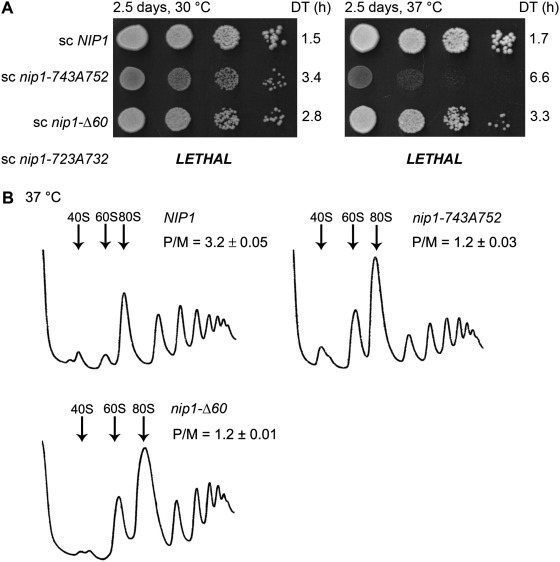
As the next step toward our goal to characterize the role of the c/NIP1-CTD in translation, we generated a battery of progressive C-terminal truncation mutants taking into account our 3D structural model. All mutations were generated in a plasmid-borne NIP1 allele with a C-terminal Myc tag and introduced into the nip1Δ strain by plasmid shuffling. The shortest c/NIP1-CTD truncation of the last 60 amino acid residues in nip1-Δ60 (Figures 1A and B, 2C—in red), which removes the last α-helix and both β-sheets of the PCI domain, was not lethal and produced a desired slow growth (Slg−) phenotype with a doubling time (DT) of 2.8 h (compare to 1.5 h of the wt strain) that is slightly exacerbated at higher temperature (Figure 3A). All other larger truncations were lethal (data not shown), strongly suggesting that the integrity of the PCI is vital for the c/NIP1 role in cell proliferation. To further support the latter conclusion and uncover the molecular details of the essential function of the c/NIP1-PCI, we introduced Ala substitutions in six consecutive blocks of 10 residues between residues 693 and 752 just upstream of the nip1-Δ60 truncation (Figure 1A and B). The most C-terminal substitution of residues 743 through 752 in nip1-743A752 (Figures 1B and 2C—in blue) impinging into a turn between helices 5 and 6 of the WH subdomain (Figure 2B) produced a temperature-dependent Slg− with DT of 3.4 h at 30°C and 6.6 at 37°C (Figure 3A). In addition, the more N-terminal substitution between residues 723 and 732 in nip1-723A732 (Figures 1B and 2C in magenta) interfering with the C-terminal end of the helix 4 of the WH subdomain (Figure 2B) was lethal even when overexpressed. Steady-state protein levels of all mutants were found comparable with those of the wt cells (Supplementary Figure S2A and S2B). None of the four remaining substitutions produced any strong growth defects (data not shown).
Figure 3.
The intact PCI domain of c/NIP1 is required to promote optimal translation initiation rates and thus to ensure wild-type cell growth. (A) nip1-743A752 and nip1-Δ60 produce temperature-dependent Slg− phenotypes. The HMJ08 (nip1Δ) strain was transformed with YCpNIP1-Myc-L (row 1), YCpNIP1-743A752-Myc-L (row 2), YCpNIP1-Δ60-Myc-L (row 3), and YCpNIP1-723A732-Myc-L (row 4), and the resident YCpNIP1-His-U plasmid was evicted from the resulting transformants on SD medium containing 5-FOA (with the exception of YCpNIP1-723A732-Myc-L encoding a lethal mutation). Thus generated mutant strains were then spotted in four serial 10-fold dilutions on YPD medium and incubated at 30 and 37°C for 2.5 days. Doubling times (DT) are given on the right-hand side of the panels. (B) Polysome profiles of the c/NIP1-CTD mutants reveal ∼2.6-fold decrease in the polysome to monosome ratio. The c/NIP1 strains as in panel A were cultured in YPD medium at 37°C to an exponential growth phase and treated with cycloheximide 5 min prior to harvesting. WCEs were prepared and subsequently separated by high velocity sedimentation (at 39 000 rpm for 2.5 h) through 5–45% sucrose gradients. The resulting gradients were collected and scanned at 254 nm to visualize the ribosomal species. Positions of 40S, 60S and 80S species are indicated by arrows, and P/M ratios are given above the profiles.
We next wished to demonstrate that the observed growth phenotypes of the nip1-743A752 and nip1-Δ60 cells are associated with a defect in translation initiation by analyzing their polysome content. Cells were grown into exponential phase and the translating ribosomes were locked on mRNA by the cycloheximide treatment. Thus pretreated cells were lysed and the whole-cell extracts (WCEs) were resolved by velocity sedimentation through 5–45% sucrose gradients (41). In both nip1-743A752 and nip1-Δ60 WCEs an increased amount of 80S monosomes (Figure 3B) was observed leading to a similar ∼2.6-fold decrease in the polysome to monosome (P/M) ratio indicating a reduced rate of general translation initiation.
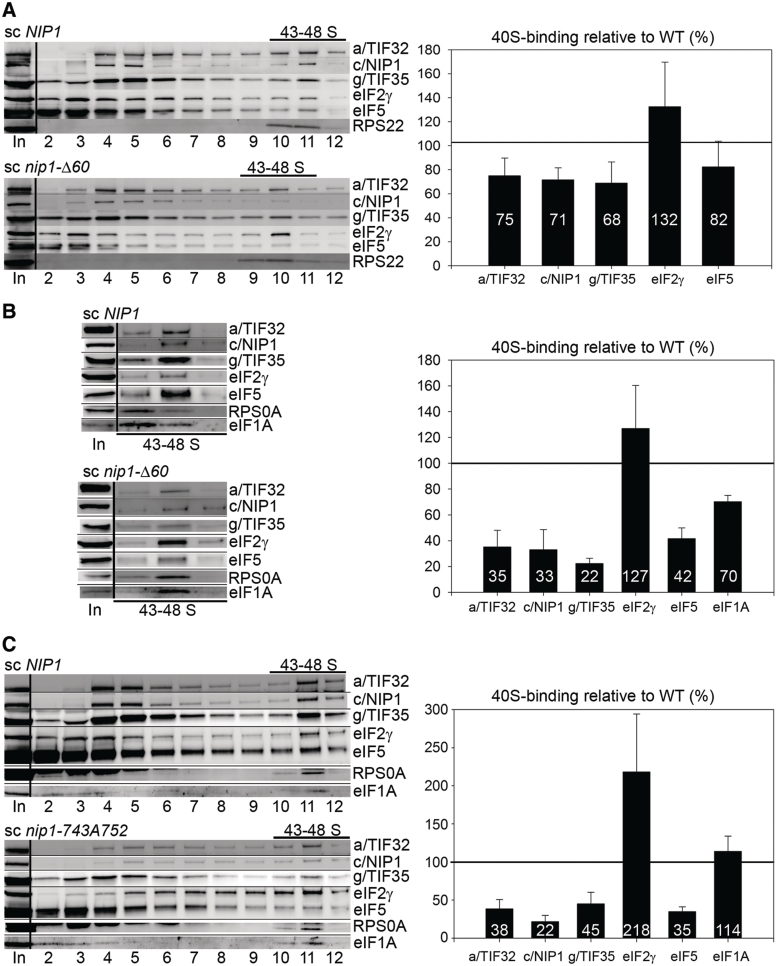
To test our prediction that the c/NIP1-CTD represents an important molecular link between the MFC and the 40S ribosomes, we measured binding of individual eIF3 subunits and other MFC components to 40S subunits in WCEs of mutant nip1-Δ60 and nip1-743A752 cells treated with 2% formaldehyde. This treatment cross-links eIFs to 40S ribosomes in vivo and thus provides the best approximation of the native 43S/48S PICs composition (41). HCHO-pretreated WCEs were resolved by velocity sedimentation through 7–30% sucrose gradients and the collected gradient fractions were subjected to western blot analysis with antibodies against MFC components and the 40S subunit proteins RPS0A or RPS22. Combined western blot signals in the fractions containing 43–48S PICs were quantified and normalized to RPS0A or RPS22 levels, and the mean eIF/40S ratio for each MFC component was determined from at least three replicate experiments. The values obtained for mutant cells were then plotted in percentages relative to wt. In the case of nip1-Δ60 (Figure 4A), we observed ∼30% reductions in the amounts of selected eIF3 subunits sedimenting in the 40S-containing fractions. Binding of 40S-associated eIF5 was also reproducibly reduced by ∼20%. The eIF2 amounts were found on average slightly accumulated, although the variability of this result was somewhat higher than desirable for an unknown reason (see ‘Discussion’ section). To further support these findings, we applied the resedimentation protocol (41), where the 40S-containing fractions collected from the first gradient are pooled and resolved by the same sedimentation procedure on a second sucrose gradient. This helps to discriminate between eIFs cross-linked to 40S subunits and non-cross-linked factors present non-specifically in 40S fractions and hence it ensures a better resolution of the 43S/48S PICs composition. Application of the resedimentation protocol confirmed a defect in eIF3 and eIF5 binding to 40S subunits and also indicated a higher amount of 40S-associated eIF2 (Figure 4B). Importantly, binding of eIF1A, which associates with the 40S ribosomes in the MFC-independent manner, was affected only modestly clearly suggesting that this c/NIP1 mutation selectively impacts 40S binding of the eIF3-associated factors. The fact that the overall reductions in binding seem higher (∼60–65%) than those observed with the single-gradient protocol (compare quantification diagrams in Figure 4A and B) can be explained by the following two, mutually compatible possibilities. First, a certain proportion of the western blotting signals detected in 40S factions in Figure 4A could arise from trailing of free factors not cross-linked to PICs in vivo into heavier sucrose fractions during sedimentation. Second, more pronounced loss of signals from 40S fractions in Figure 4B can be caused by partially reversed cross-linking during the in vitro manipulations. A similar phenomenon has been also previously observed by others (42).
Figure 4.
The c/NIP1-PCI serves as an important intermolecular bridge between eIF3 and the 40S subunit. The nip1-Δ60 (A and B) and nip1-743A752 (C) mutants and the corresponding wt NIP1 strain (all as in Figure 3A) were grown in YPD medium at 37°C to an OD600 of ∼1.5 and cross-linked with 2% HCHO prior to harvesting. WCEs were prepared and subsequently separated on 7.5–30% sucrose gradients by centrifugation at 41 000 rpm for 5 h. Proteins from the collected fractions were subjected to western blot analysis using antibodies against the indicated proteins. For nip1-Δ60 (B), the resedimentation protocol was applied as follows. Bottom fractions containing the 43–48S PICs were collected, pooled, concentrated and resolved on a second 7.5–30% gradient—the resulting 43–48S fractions were then subjected to western blotting. The Myc-tagged c/nip1-Δ60 mutant protein was visualized by anti-Myc antibodies. An aliquot of each WCE was analyzed in parallel (In, input) showing that the steady-state levels of the c/NIP1 and other analyzed proteins were are not affected by the c/NIP1 mutations. The 43-48S PICs containing fractions (43–48S) are indicated. Proportions of the 40S-bound proteins relative to the amount of 40S subunits in each mutant were calculated by fluorescence imaging (Quantity One—Bio-Rad) from at least three independent experiments. The resulting values obtained with the wt strain were set to 100% and those obtained with mutant strains were expressed as percentages of the wt and are shown as bar diagrams to the right of the western blots (SDs are given).
The nip1-743A752 led to larger (∼60%) reductions in the eIF/40S ratios for all tested eIF3 subunits and eIF5 when compared to nip1-Δ60 subjected to a single-gradient protocol (compare Figures 4A and C). These results clearly indicate that nip1-743A752 has a more dramatic impact on 40S binding of eIF3 and eIF5 than nip1-Δ60, which is consistent with the severity of their growth phenotypes at 37°C (Figure 3A). In analogy with nip1-Δ60, eIF2 amounts in 40S fractions of nip1-743A752 were also found increased (by ∼2-fold) and binding of eIF1A remained virtually unaffected. The lack of an increase in abundance of unbound factors in the gradients with this mutant, which might have been expected given a more severe loss of these proteins from the PICs, is attributed to increased instability of free eIFs relative to those stably associated in PICs during the course of the sedimentation experiment, as seen before (6,43,44). Quantification of input lanes ruled out a possibility of a significantly increased proteolysis of eIF3 and eIF5 in living cells (data not shown). Taken together, we propose that the initiation defect in both of these mutants is by a large degree caused by a reduced rate of the 43S PIC formation and/or by its decreased stability.
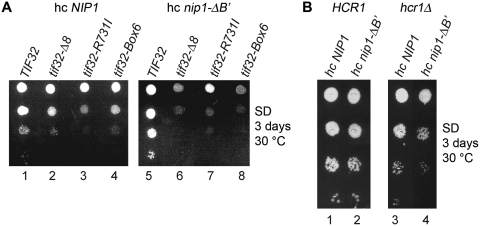
Further genetic support for the latter conclusion comes from our findings that overexpressing the original nip1-ΔB′ truncation results in synthetic exacerbation of growth defects of two mutants in the a/TIF32 subunit of eIF3, namely tif32-Δ8 (Figure 5A, compare lanes 2 versus 6) and tif32-box6 (Figure 5A, compare lanes 4 versus 8), that were previously shown to severely impair 40S-association of eIF3 as well as the other MFC-components (3,14). In contrast, high dosage of nip1-ΔB′ in the background of the tif32-R731I mutation, which does not interfere with formation of 43S PICs (14) had no further impact on its Slg− phenotype (Figure 5A, compare lanes 3 versus 7). Finally, deletion of the HCR1 subunit of eIF3 is also believed to affect binding of eIF3 to 40S subunits to a certain extent (9,12) and, analogously, overexpression of nip1-ΔB′ also worsened the growth phenotype of the hcr1Δ cells (Figure 5B, compare lanes 4 versus 2). Hence we conclude that the C-terminal ∼1/3 of c/NIP1 formed for the most part by the canonical PCI domain serves as an important intermolecular bridge between eIF3 and the 40S subunit.
Figure 5.
Overexpression of the c/NIP1 truncated form lacking the C-terminal 240 amino acid residues exacerbates growth defects of several eIF3 mutants previously shown to impair 40S-binding of eIF3. (A) The tif32Δ strains harboring TIF32 (lanes 1 and 5), tif32Δ8 (lanes 2 and 6), tif32-R731I (lanes 3 and 7), and tif32-Box6 (lane 4 and 8) alleles on a single copy plasmid were transformed with YEpNIP1-His-U (hc NIP1, lanes 1–4) or YEpNIP1-ΔB′-His-U (hc nip1-ΔB′, lanes 5–8) and the resulting transformants were spotted in four serial dilutions on the SD plates and incubated at 30°C for 3 days. (B) The hcr1Δ strains harboring YEpLVHCR1-L (HCR1, lanes 1 and 2) or YEplac181 (hcr1Δ, lanes 3 and 4) were transformed with YEpNIP1-His-U (hc NIP1, lanes 1–2) or YEpNIP1-ΔB′-His-U (hc nip1-ΔB′, lanes 3–4) and the resulting transformants were spotted in four serial dilutions on the SD plates and incubated at 30°C for 3 days.
The extreme CTD of c/NIP1 interacts with the small ribosomal protein ASC1 and the preceding PCI domain binds RNA
Having established that the c/NIP1-CTD anchors eIF3 to the 40S binding in vivo, we next wished to identify its potential interacting partner(s) among the constituents of the small ribosomal subunit. We began by testing yeast two-hybrid interactions among two C-terminal fragments of c/NIP1 (residues 571–812 and 701–812) and all 33 small ribosomal proteins (RPSs) fused to either the GAL4-activation or DNA-binding domains, respectively. From all pairwise combinations both c/NIP1 fragments interacted only with ASC1. ASC1 is a protein comprised of 7 WD40 repeats that forms the head of the 40S ribosomal subunit near the mRNA exit tunnel (19,23).
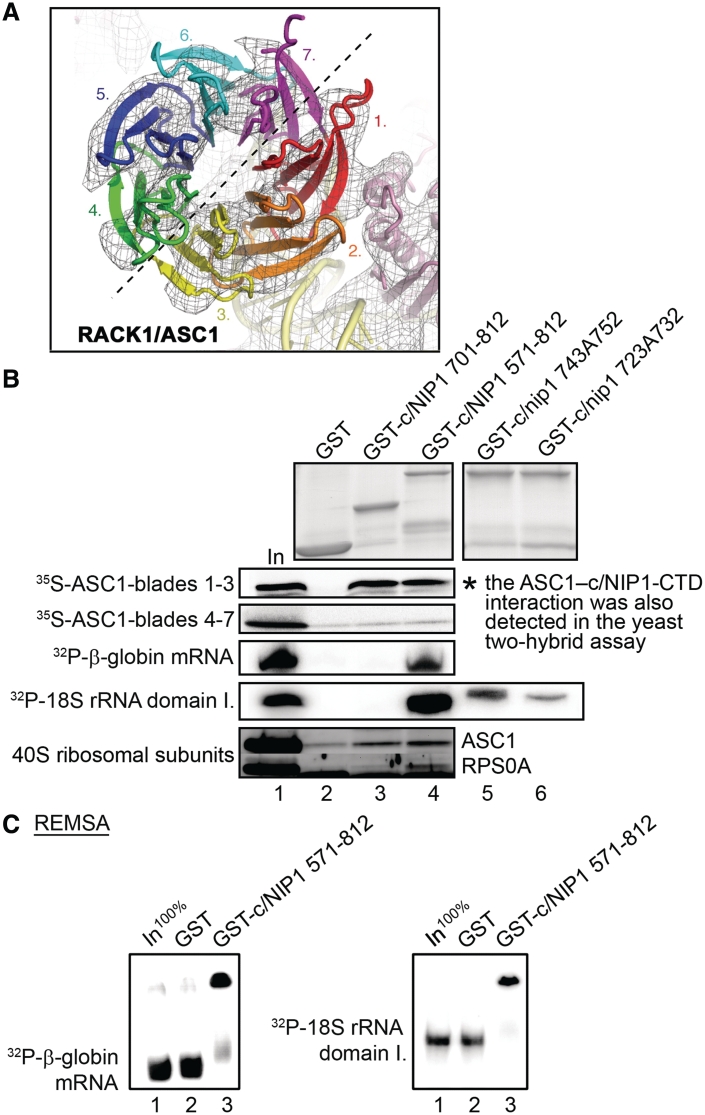
To confirm these two-hybrid interactions and determine what part of the ASC1 protein mediates them, the c/NIP1 fragments were fused with the GST moiety and tested for binding to [35S]-labeled segments of ASC1 synthesized in vitro. ASC1 adopts a seven-bladed β-propeller structure and, according to localization of the ASC1 density on the 40S ribosome, was previously subdivided into two distinct halves corresponding to blades 1–3 and 4–7 (Figures 6A and 7B, upper panel) (31,45). As shown in Figure 6B, whereas the blades 1–3 of ASC1 interacted strongly and specifically with both GST-c/NIP1-CTD fusions but not with the GST alone, the fragment carrying the blades 4–7 showed no binding. Hence the extreme C-terminal 112 amino acid residues are sufficient for direct c/NIP1 binding to ASC1. In effort to demonstrate that these residues are also necessary for the interaction, we attempted to make several GST fusions with the C-terminal c/NIP1 fragment (571–812) lacking the extreme 112, 80, 60 or 50 residues. However, all these attempts failed due to instability of the resulting fusion proteins. Nevertheless, our observations that full-length GST-c/NIP1 bound blades 1–3 with a similar intensity to these seen with both CTD fusions (data not shown) and that the nip1-723A732 and nip1-743A752 mutations had no impact on ASC1 binding (Supplementary Figure S2C, lanes 5 and 6) strongly indicates that the c/NIP1 interaction site for the ASC1 blades 1–3 resides in the terminal 60 amino acid residues formed by the last two β-sheets of the PCI-WH subdomain and the extended α-helix (Figures 7A and 2C in red). It is important to note that we also tested binding of our GST-c/NIP1-CTD fusions to all remaining ribosomal proteins including RPS0A, RPS2, RPS3 and RPS20 that interact with other eIF3 segments (3,12–14,16) and observed no additional interactions (data not shown). This fact plus our two hybrid results significantly underscore the specificity of the c/NIP1 contact with ASC1. Taking all these data together with our aforementioned findings showing the 40S-binding defect of nip1-Δ60, we conclude that the extreme c/NIP1-CTD anchors eIF3 to the small ribosomal subunit via its WD40-repeat scaffold protein ASC1. This result is in perfect agreement with our recently revised model of eIF3 placement on the upper half of the solvent side of the 40S subunit (Figure 7B, lower panel) (13).
Figure 6.
The intact c/NIP1-PCI domain is capable of stable RNA binding in vitro and the extreme CTD of c/NIP1 interacts with the small ribosomal protein RACK1/ASC1; the c/NIP1-CTD also pulls down purified 40S ribosomes. (A) Top view of RACK1/ASC1 with the seven propellers color-coded [reproduced from (31)]. The EM density indicates that primary interactions with the 40S subunit are coordinated via blade 1 of ASC1. Density of the ASC1 structure further indicates the molecule to have two distinct hemicycles with one half formed by tight interactions between blades 1–3 (roughly encoded by the ASC1 exon 1) and a second half from blades 4–7 (roughly encoded by the ASC1 exon 2). (B) GST fusions of two segments of the c/NIP1-CTD [residues 701–812 (lane 3) or 571–812 (lane 4)] or GST alone (lane 2) were tested for binding to either 35S-labeled fragments of ASC1 corresponding to its exon 1 or 2, or 32P-labeled β-globin mRNA and domain I. of 18S rRNA, or purified wt 40S ribosomal subunits. Lane 1 (In) contains 10, 2.5, and 20% of input amounts of proteins, RNAs and 40S subunits, respectively, added to each reaction mixture. The GST fusions were visualized by Coomassie blue staining (top) and the radiolabeled molecules were detected by autoradiography. In addition, the 10-Ala substitutions 743A752 (lane 5) and 723A732 (lane 6) were introduced into the GST-c/NIP1 571-812 fusion and tested for binding to domain I. of 18S rRNA (right-handed parts of the very top and 18S rRNA panels). Binding to 40S ribosomes was detected by western blotting with antibodies to ASC1 and RPS0A. (C) RNA electrophoretic mobility shift assay carried out with 32P-labeled β-globin mRNA and 18S rRNA (domain I.) and GST fusion proteins described in panel B that were demobilized from glutathione-Sepharose beads. Lane 1 (In) contains 100% of an input amount the RNA.
Figure 7.
Model of the hypothetical location of eIF3 on the S. cerevisiae small ribosomal subunit. (A) Schematic as in Figure 1A with additional arrows delimiting newly identified c/NIP1-binding domains for RNA (in green) and for RACK1/ASC1 (in red). (B) (Upper panel) The Cryo-EM reconstruction of the 40S subunit is shown from the solvent side with ribosomal RNA represented as tubes. Ribosomal proteins, with known homologs and placement, are shown as pink cartoons and labeled [reproduced from (45), with permission from Elsevier]. The position of RACK1/ASC1 is highlighted in bold. The mRNA entry channel is designated by an asterisk. (Lower panel) Hypothetical location of S. cerevisiae eIF3 on the back side of the 40S subunit based on the data presented in this study and elsewhere, including the interactions between c/NIP1-CTD and ASC1 (and potentially also with 18S rRNA), RPS0 and a/TIF32-NTD, RPS2 and j/HCR1, RPS2 and 3 and a/TIF32-CTD, helices 16–18 of 18S rRNA and a/TIF32-CTD, and RPS3 and 20 and g/TIF35 (see text for further details). The 3D structural model of the c/NIP1-PCI domain determined in this study was used to replace the original schematic representation of the same molecule. The yellow lines represent mRNA.
The fact that the nip1-743A752 mutation did not affect binding to ASC1 in vitro (Supplementary Figure S2C) yet it showed even stronger 40S-binding defect in vivo than nip1-Δ60 (Figure 4) indicated to us that there must be an additional anchor point on the 40S ribosomal surface contacting the c/NIP1-PCI domain from the residue 752 upstream. Interestingly, it was proposed but not examined experimentally that the PCI domain's molecular function might, besides the protein–protein interactions, also include nucleic acid binding (46). This potential interaction was even modeled (21). These ideas prompted us to investigate whether the c/NIP1-PCI domain may specifically interact with ribosomal 18S rRNA in vitro. Toward this end, we incubated the aforementioned GST-c/NIP1-CTD fusions with radiolabeled 18S rRNA [either full length or subdivided into three distinct domains (16,47)], 25S rRNA and β-globin mRNA. Strikingly, the longer but not shorter GST-c/NIP1 fusion protein strongly interacted with all mentioned RNA moieties (Figure 6B and data not shown), clearly indicating that the PCI domain of c/NIP1 as a whole is indeed capable of stable but unspecific binding to RNA. Importantly, in both 10-alanine substitutions (in GST-c/nip1-743A752 and GST-c/nip1-723A732) this PCI RNA-binding ability was nearly completely abolished or strongly reduced (Figure 6B). Notably, the first mutation severely impairs eIF3 association with the 40S subunits (Figure 4C) and produces the Slg− phenotype (Figure 3A) while the second, more deleterious mutation with respect to RNA binding, is lethal.
To further support these novel findings, we also performed a RNA electrophoretic mobility shift assay (REMSA) with RNAs and the PCI-containing c/NIP1 fragment fused to GST but demobilized from the beads. As can be seen in Figure 6C, the c/NIP1-571–812 fusion showed robust RNA-binding affinity as it band shifted majority of the β-globin mRNA as well as 18S rRNA species in the non-denaturing gel. In contrast, GST alone showed no binding whatsoever.
Finally, we examined whether the c/NIP1-CTD, carrying two potential anchor sites for the small subunit—in ASC1 and 18S rRNA, interacts with purified 40S ribosomes in vitro as a whole. To do that, we performed GST pull down assays with both GST-c/NIP1-CTD fusions and 40S species isolated from the wt H503 strain. Even though this type of an in vitro assay does not seem very efficient, we did observe a weak but specific interaction between 40S subunits and the shorter GST-c/NIP1 construct when compared to the GST alone control (Figure 6B, bottom panel). In accord with our in vivo data, more ribosomes (by ∼2–3-fold) were pulled down when the entire CTD including the RNA-binding PCI domain was used in this assay.
On the basis of all these results, we established a linkage map between the C-terminal ∼1/3 of the c/NIP1 subunit of eIF3 and the 40S components, where the established binding site of the c/NIP-PCI to RNA is marked in green and binding of the extreme CTD to ASC1 is in red (Figure 7A). We propose that whereas the extreme c/NIP1-CTD contacts ASC1, the PCI domain including the WH helices 5 and 6 most probably mediates the eIF3-40S interaction by binding to 18S rRNA (Figure 2B). To our knowledge, this is the first report unambiguously showing the RNA-binding capability of the canonical PCI domain.
ASC1 deletion destabilizes formation of the 43S PICs and derepresses GCN4 translation under non-starvation conditions
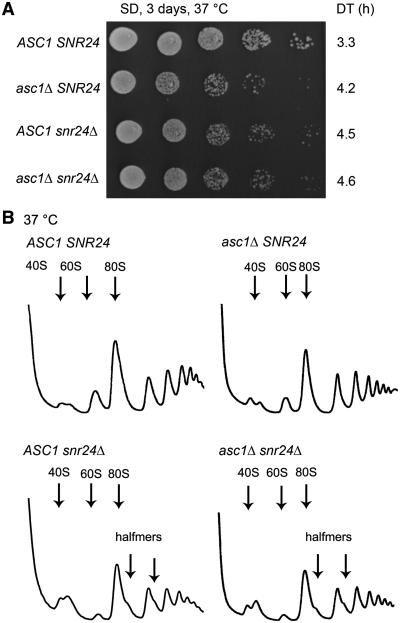
We next wished to support this proposal by showing that the deletion of the ASC1 gene, which is non-essential, has an impact on formation of the 43S PICs as could be predicted. Previously, asc1Δ was suggested to impair growth, produce halfmer polysomes at 37°C, affect translational rates and interfere with cell wall biosynthesis (27–29,31). (Halfmers are formed by mRNAs containing elongating 80S ribosomes and the 48S PICs stuck in the mRNA leader.) However, the ASC1 gene also contains an intron carrying SNR24 encoding the C/D box U24 snoRNA that is required to guide 2′-O-methylation of the large subunit rRNA during 60S biogenesis (32,48). To our knowledge, all previous studies worked with strains deleted for both ASC1 and SNR24 and overlooking this fact, ascribed the aforementioned phenotypes solely to the loss of the ASC1 gene.
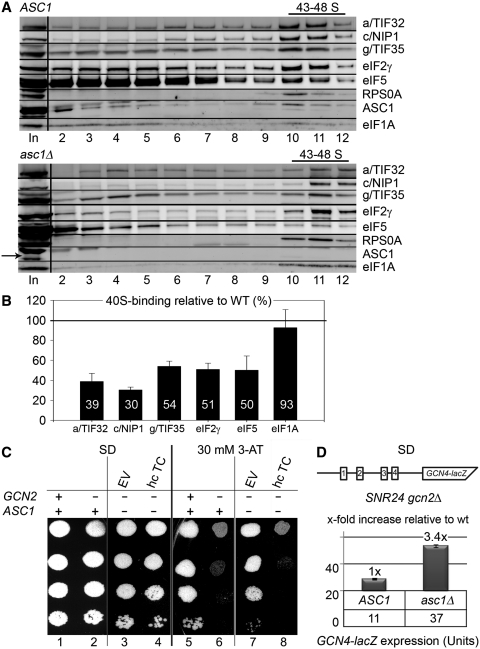
In keeping with our goal, we started by testing the effects of the ASC1 or SNR24 single deletions on formation of halfmer polysomes. To do that we constructed the asc1Δ snr24Δ double deletion strain expressing ASC1 and SNR24 on two different shuttling vectors. First, we noticed that single deletions of ASC1 and SNR24 each produced equally strong slow growth phenotype that had a non-additive character when both deletions were combined (Figure 8A). Importantly, in contrast to what was previously published [see for example (27)], halfmer polysomes formed in snr24Δ to the same extent as in the double deletion strain but were absent in asc1Δ (Figure 8B). Hence we propose that the insufficient amount of mature 60S ribosomal species conferred by the loss of U24 but not a subunit joining defect is the prime cause of the halfmer phenotype in the original ‘asc1-deleted’ strains. This implies that the true ASC1 role in translation initiation should be revised to account for these novel facts. Hence we next performed sucrose gradient analysis of the WCEs derived from HCHO-pre-treated wt and asc1Δ cells, carried out essentially the same as described above. We observed ∼50% reduction in the 40S-associated amounts of all selected eIF3 subunits and other MFC-associated factors like eIF2 and eIF5 (Figure 9A and B). In contrast, binding of eIF1A was almost unaffected. These results nicely correlate with those obtained with nip1-Δ60 (Figure 4) and provide further support for our conclusion that ASC1 anchors eIF3 to the 40S ribosome by binding to the extreme CTD of c/NIP1, which in turn stabilizes 40S-binding of other eIF3-associated components of the MFC such as TC.
Figure 8.
Single deletions of ASC1 and SNR24 impair cell growth but only snr24Δ produces polysomal halfmers. (A) The ED43 (tif32Δ asc1Δ snr24Δ) strain harboring pRSTIF32-His-L was transformed with the following combinations of vectors: pRSASC1-intron-less-U and pFL45s/ACTU24 (ASC1 SNR24; row 1), pRS416 and pFL45s/ACTU24 (asc1Δ SNR24; row 2), pRSASC1-intron-less-U and pFL45S (ASC1 snr24Δ; row 3), and pRS416 and pFL45S (asc1Δ snr24Δ; row 4). The resulting double transformants were spotted in five serial 10-fold dilutions on SD medium and incubated at 37°C for 3 days. Doubling times (DT) are given on the right-hand side of the panel. (B) The strains as in (A) were then subjected to polysome profiling as described in Figure 3B.
Figure 9.
The ASC1 deletion destabilizes formation of the 43–48S PICs. (A) The wt ASC1 SNR24 and asc1Δ SNR24 strains were grown in SD medium at 37°C to an OD600 of ∼1 and subjected to formaldehyde cross-linking as described in Figure 4A. (B) Histogram showing proportions of the 40S-bound proteins in asc1Δ relative to the amounts in the wt ASC1 strain. (C) The ASC1 deletion impairs GCN4 translational control (produces the Gcd− phenotype) in the canonical manner suppressible by overexpressing the TC. The gcn2Δ SNR24 asc1Δ strain was transformed with either empty vector or all three subunits of eIF2 + IMT4 on a high copy plasmid (hc TC). The resulting transformants and isogenic strains H2880 (GCN2 SNR24 ASC1) and H2881 (gcn2Δ SNR24 ASC1) transformed with empty vector were spotted in four serial dilutions on SD media with or without 30 mM 3-AT and incubated at 30°C for 3 and 5 days, respectively. (D) Quantification of the Gcd− phenotype. The gcn2Δ SNR24 ASC1 and gcn2Δ SNR24 asc1Δ strains were transformed with p180 containing the GCN4–lacZ fusion with all four uORFs present (shown as schematic above the plot), grown in minimal media for 6 h, and the β-galactosidase activities were measured in the WCEs and expressed in units of nmol of o-nitrophenyl-b-d-galactopyranoside (ONPG) hydrolyzed per min per mg of protein. The mean values and standard deviations obtained from at least 3 independent measurements with three independent transformants, and the x-fold increases in activities of the mutant strains relative to the corresponding wt, are given in the histogram.
The defect in the TC recruitment to 40S ribosomes can be also examined genetically with help of a specific translational control mechanism that governs expression of the yeast transcription factor GCN4, and has been extensively used in the past as a molecular tool to monitor various steps of translation initiation [reviewed in (49)]. Translation of GCN4 is regulated mainly in response to amino acids availability and relies on the presence of four upstream open reading frames (uORFs 1–4) in its mRNA leader. After translation of the first uORF1, sizeable number of small ribosomal subunits does not dissociate from the GCN4 mRNA and instead resumes scanning downstream. To reinitiate on any of the downstream uORFs or on the main GCN4 ORF, these rescanning subunits have to first reacquire the TC. When cells are grown in rich media, intracellular levels of the TC are high so most of rescanning ribosomes pick it up before reaching the AUG start codon of the inhibitory uORF4 at which they reinitiate. This uORF does not allow resumption of scanning of post-termination 40S ribosomes and thus blocks further reinitiation. However, when cells are starved for amino acids, the GCN2 kinase induces dramatic decrease in the TC levels, which enables many of the re-scanning ribosomes to skip the trap of uORF4 by picking up the TC after scanning through it. As a result, the AUG of GCN4 is reached and its ORF gets translated. Mutants that reduce 40S-binding affinity of eIF2 deregulate this system by allowing (derepressing) GCN4 translation even under non-starvation condition. This phenomenon is called the Gcd- phenotype and can be monitored by growing the mutant cells with GCN2 deleted in the presence of a histidine biosynthesis inhibitor 3-aminotriazol (3-AT). Whereas wt cells cannot cope with this drug, mutant cells with defects in the TC binding become resistant to it. Overexpressing the TC then cancels out the 3-AT resistance. Accordingly, whereas the gcn2Δ SNR24 ASC1 wt cells failed to grow on SD plates supplemented with 30 mM 3-AT (Figure 9C, lane 6), the gcn2Δ SNR24 asc1Δ mutant displayed significant resistance that was fully abolished by high copy TC (lanes 7 versus 8). Depression of GCN4 expression under non-starvation conditions was further confirmed and quantified by assaying the β-galactosidase activity of a GCN4-lacZ reporter containing all four uORFs in the mRNA leader (Figure 9D). The wt gcn2Δ SNR24 ASC1 strain showed constitutively low GCN4-lacZ expression, however, the gcn2Δ SNR24 asc1Δ mutant increased β-galactosidase activity by ∼3.4-fold (after normalizing for its effect on the expression of an incontrollable GCN4-lacZ construct lacking all four uORFs). Taken together, we propose that the ASC1 contributes to translation initiation by promoting assembly of the entire 43S preinitiation complexes.
DISCUSSION
In this article, we (i) predicted a 3D structure of the C-terminal ∼1/3 of the c/NIP1 protein containing a well-defined PCI domain by computer modeling; (ii) showed that the PCI domain is capable of strong RNA binding and that the extreme CTD binds RACK1/ASC1; (iii) generated two specific substitution mutants in the PCI domain and a truncation of the extreme CTD having a significant impact on formation of the PICs and cellular growth; and (iv) demonstrated that deletion of ASC1 only without its SNR24-coding intron also disturbs PICs assembly but does not produce the halfmer phenotype as previously believed. Together, we propose that the C-terminal PCI domain of c/NIP1 stimulates docking of eIF3 and its associated eIFs to the small ribosomal subunit by contacting its head region where ASC1 resides.
The c/NIP1-PCI domain binds RNA
In silico modeling of the prospective 3D structure of the last 240 residues of the c/NIP1-CTD revealed a typical fold for the PCI domain with two groups of specifically aligned α-helices each representing a different PCI subdomain and two antiparallel β-sheets at the end of the WH subdomain that is in the case of c/NIP1 followed by an extra α-helix (Figure 2). Importantly, this structure prediction model was tested for its congruence by superimposing it with the experimentally determined structures of two PCI-containing proteins in CSN7 from A. thaliana and SGN4 from M. musculus (Supplementary Figure S1A, left panel). Overall, only minor differences were found indicating a high structural homology between the tested molecules. One of the differences was the extra α-helix (#7) flanking the WH subdomain of the c/NIP1-PCI at its very C-terminus. Interestingly, a similar structural element was observed also in the crystal of the PCI-containing ‘k’ subunit of human eIF3 (50). There this flanking C-terminal α-helix winds up backward to the helical part of the HB subdomain, forming an encapsulated structure. In our model the α-helix 7 also turns backward, reaching the rear part of the HB subdomain (Figure 2A, in red). Because the extreme CTD of c/NIP1 specifically interacts with ASC1 (Figure 6, see also below), this extended α-helix 7 is one of the foremost candidates for mediating this contact. If so, it could mean that the c/NIP1-PCI domain docks onto the ribosomal surface through its back side, exposing the WH subdomain for further interactions, for example with RNA as observed here (Figure 6). Alternatively, the ASC1 contact could be bridged by the two antiparallel β-sheets of the WH subdomain.
Our observation that the c/NIP1-PCI domain binds RNA is rather intriguing, given the fact that so far a PCI domain in itself was considered to organize large multiprotein complexes via strictly protein–protein interactions. Chamovitz (46) noted that the WH subdomain comprises a canonical helix-turn-helix that has a structure and electrostatic potential similar to canonical WH nucleic acid-binding proteins, which raised the question of whether the presence of this structural motif could indicate a capability of a PCI domain to bind nucleic acids. However, even though theoretical modeling supported this possibility by showing that the WH domain of CSN7 can dock with nucleic acids (21); until this work no evidence of direct nucleotide binding has been reported. In striking accord with this notion, both CAM mutations that were generated and shown to severely compromise RNA binding of the c/NIP1-PCI domain in vitro are in fact predicted to disturb the helix-turn-helix motif (Figure 6). In particular, the region mutated in nip1-723A732 with the most deleterious effect on RNA binding and growth is rich in evolutionarily conserved aromatic and basic residues, which are known to mediate protein–RNA contacts (Figures S3). Indeed, the electrostatic potential of its surface is dominated by positive charge (Supplementary Figure S1B). In the case of nip1-743A752, there is only a conserved Phe745 residue and Lys751 that could be responsible for the RNA interaction. In fact, Lys751 markedly sticks out into the solvent from the otherwise negatively charged surface (Supplementary Figure S1B). Alternatively, either of these regions could disturb RNA binding indirectly, when mutated, by altering the WB subdomain's fold. Actually, the importance of a proper fold is reflected in the fact that the WH subdomain failed to interact with RNA when expressed on its own (Figure 6). This suggests that the HB subdomain is also required for the RNA binding and its most likely contribution is to provide a structural support for proper assembly of the entire PCI domain with the WH helix-turn-helix exposed to the surface to be able to interact with RNA. The fact that the PCI domain binds RNA in a non-specific manner could perhaps mean that it requires some ribosome-associated interacting partner (for example ASC1) with an allosteric effect on its RNA-binding ability to make it more specific. A real structural analysis of the c/NIP1-PCI domain bound to RNA and perhaps also ASC1 is indeed needed to address the precise molecular details of its nucleic acid-interaction.
The c/NIP1-PCI domain is one of the key eIF3 contributors to the process of the 43-48S PIC assembly
In our long-standing pursuit of mapping the binding site of yeast eIF3 on the 40S ribosomal surface, we identified several key contacts between specific domains of eIF3 subunits and either protein or RNA components of the 40S ribosome that placed the major body of eIF3 to its solvent-exposed head, beak and shoulder regions (Figure 7B). Among them are the interactions between the NTD of a/TIF32 and the small ribosomal protein RPS0A, the a/TIF32-CTD and helices 16–18 of 18S rRNA and RPS2 and RPS3, g/TIF35 and RPS3 and RPS20, and finally the CTD of j/HCR1 and RPS2 (3,12–14,16). Here, the c/NIP1-CTD has been added to the list of the important eIF3-40S intermolecular bridges with ASC1 and most likely 18S rRNA serving as its binding partners. Given the ASC1 location on the 40S head, the c/NIP1-PCI domain could interact with some of the surface-exposed RNA loops of the major domain III of 18S rRNA (19,47).
In support of the c/NIP1-CTD role in formation of 43-48S PICs, we showed that the nip1-743A752 10-Ala substitution and the very C-terminal truncation nip1-Δ60 impaired cell growth producing polysome profiles typical for translation initiation mutants, and, most importantly, significantly impaired composition of the PICs (Figures 3 and 4). The 40S-associated amounts of eIF3 and its direct interacting partner eIF5 (51) were markedly reduced in both mutants with nip1-743A752 having a more deleterious impact, which could have been anticipated giving its more severe growth phenotype. Importantly, the levels of eIF1A, which is not a component of the MFC but an important constituent of the PICs, remained virtually unchanged. Interestingly, the amounts of eIF2 were found to be increased in nip1-743A752 and to a certain extent also in the nip1-Δ60 mutant. eIF2 makes two contacts with eIF3 in the MFC—indirect via c/NIP1 and eIF5 and direct via a/TIF32 (7), and in contrast to eIF3 is believed to interact with the interface side of the 40S ribosome to direct Met-tRNAiMet to the ribosomal P-site. This implies that upon eIF3-stimulated ternary complex recruitment to the 40S ribosome, eIF2 has to subsequently relocate to or near the P-site. Hence it is conceivable that the increased amounts of the PICs containing specifically eIF2 could arise from simultaneous binding of all MFC-associated eIFs to the ribosome, followed by rapid dissociation of unstably bound eIF3 and eIF5 during the initial conformational changes of the 40S ribosome. These are required to start scanning along the mRNA's 5′-UTR (52) and should also include the transfer/accommodation of the TC to the P-site area. It is understood that these events would not influence stability of the MFC-independent eIF1A on the ribosome. The physiological importance of the c/NIP1-PCI for the PCI assembly was also supported genetically. Combining the c/NIP1-CTD mutations with mutations in other eIF3 subunits such as a/TIF32 and j/HCR1 that are known to affect 40S binding of eIF3 had an additive effect, resulting in synthetic exacerbation of growth defects of these mutants (Figure 5).
ASC1 promotes formation of PICs by anchoring eIF3 to the head region of the 40S subunit
As mentioned above, over the years, we have identified numerous contacts between eIF3 subunits and the 40S ribosomal components, the physiological importance of which was verified by mutagenic approaches targeted against the relevant eIF3 subunits. However, the true importance of the ribosomal components in these bridging interactions has not been directly examined as yet. Here we have initiated this effort by deleting the ASC1 coding region and demonstrating that the asc1Δ cells impair 40S-binding of all MFC components, including eIF2, to a similar extent as nip1-Δ60, which lacks the ASC1-binding site. Why did we not detect accumulated amounts of eIF2 in the 40S-containing sucrose fractions as in the case of nip1-Δ60? The ASC1 protein is a dominant component of the 40S head region and thus it is conceivable that the loss of ASC1 might change the overall conformation of the 40S head relative to its body in such a way that indirectly weakens eIF2/TC binding affinity. In support, cryo-electron microscopy images of yeast ribosomes showed that the structure of the entire 80S subunit is affected by ASC1 depletion suggesting that binding of ASC1 to the 40S subunit stabilizes the proper 80S conformation (23). Consistently, asc1Δ but not nip1-Δ60 cells impart a severe Gcd− phenotype suppressible by overexpression of all three eIF2 subunits and IMT4-encoding Met-tRNAiMet (Figure 9C and D), which is an established indicator of a defect in the TC recruitment (49).
Owing to the fact that ASC1 interacts with a number of signaling and regulatory molecules, it is thought to serve as a link between signaling and translational control (24,53,54). Interestingly, a recent cryo-EM analysis subdivided ASC1 density into two distinct halves corresponding to blades 1–3 and 4–7. To accommodate a large spectrum of proteins proposed to interact with ASC1, it was suggested that the two halves behave as independent docking stations for separate proteins to bind simultaneously (45). Our findings that the extreme CTD of c/NIP1 specifically interacted only with the half corresponding to blades 1–3, but not to blades 4–7, supports this interesting concept. It is thus conceivable that the signaling molecules bound to ASC1 next to eIF3 could directly influence the translational rates by acting upon eIF3. Taking these ideas into account, our findings not only uncover another important element required for stable binding and perhaps a proper organization of eIFs on the 40S ribosome, but also provide deeper insight into the proposed connection between translation initiation and signaling pathways linked to the ASC1 scaffold protein.
Last but not least, contrary to what was previously believed, our study clearly shows that deletion of the ASC1 intron encoding small nucleolar RNA U24, but not of the coding region of ASC1 per se, is the primary cause of the halfmer phenotype. Because U24 guides 2′-O-methylation of 25S rRNA at several positions during the 60S subunit maturation (32), it is highly likely that the halfmer phenotype associated with the deletion of SRN24 arises from defective subunit joining due to insufficient amounts of mature 60S subunits. While this manuscript was under preparation, other group came to the same conclusion by showing that the intron-encoded U24 snoRNA, but not the coding sequence of ASC1, affects 60S biogenesis (55). We recommend taking these findings into account when contemplating the various functions ascribed to ASC1 in the studies that worked with a complete deletion of the ASC1 intron-containing gene. Hence, to our knowledge, stimulation of the 43S PIC assembly described here is so far the first and only unambiguously documented function of ASC1 in general translation in yeast.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Tables 1–3, Supplementary Figures 1–3 and Supplementary References (3,6–9,12,14,16,28,32,40–41,56–58,60–62).
FUNDING
The Wellcome Trust (076456/Z/05/Z, 090812/B/09/Z); and Inst. Research Concept (AV0Z50200510). Funding for open access charge: The Wellcome Trust (090812/B/09/Z).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Alan G. Hinnebusch and Monica Liu for critical reading of the manuscript, Jinsheng Dong, Sarah Mitchell and Petr Šimandl for pilot experiments, the members of the Valášek and Krásný laboratories for helpful suggestions, and Olga Krýdová for technical and administrative assistance.
REFERENCES
- 1.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szamecz B, Rutkai E, Cuchalova L, Munzarova V, Herrmannova A, Nielsen KH, Burela L, Hinnebusch AG, Valášek L. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 2008;22:2414–2425. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munzarová V, Pánek J, Gunišová S, Dányi I, Szamecz B, Valášek LS. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet. 2011;7:e1002137. doi: 10.1371/journal.pgen.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valášek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol. 2004;24:9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valášek L, Nielsen KH, Hinnebusch AG. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002;21:5886–5898. doi: 10.1093/emboj/cdf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen KH, Szamecz B, Valášek LJA, Shin BS, Hinnebusch AG. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen KH, Valášek L, Sykes C, Jivotovskaya A, Hinnebusch AG. Interaction of the RNP1 motif in PRT1 with HCR1 promotes 40S binding of eukaryotic initiation factor 3 in yeast. Mol. Cell. Biol. 2006;26:2984–2998. doi: 10.1128/MCB.26.8.2984-2998.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jivotovskaya A, Valášek L, Hinnebusch AG, Nielsen KH. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto Y, Singh CR, Marintchev A, Hall NS, Hannig EM, Wagner G, Asano K. The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3, and eIF4G. Proc. Natl Acad. Sci. USA. 2005;102:16164–16169. doi: 10.1073/pnas.0507960102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ElAntak L, Wagner S, Herrmannová A, Karásková M, Rutkai E, Lukavsky PJ, Valášek L. The indispensable N-terminal half of eIF3j co-operates with its structurally conserved binding partner eIF3b-RRM and eIF1A in stringent AUG selection. J. Mol. Biol. 2010;396:1097–1116. doi: 10.1016/j.jmb.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuchalová L, Kouba T, Herrmannová A, Danyi I, Chiu W-L, Valášek L. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol. 2010;30:4671–4686. doi: 10.1128/MCB.00430-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu W-L, Wagner S, Herrmannova A, Burela L, Zhang F, Saini AK, Valášek L, Hinnebusch AG. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell. Biol. 2010;30:4415–4434. doi: 10.1128/MCB.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell SF, Walker SE, Algire MA, Park E-H, Hinnebusch AG, Lorsch JR. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol. Cell. 2010;39:950–962. doi: 10.1016/j.molcel.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valášek L, Mathew A, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG. The yeast eIF3 subunits TIF32/a and NIP1/c and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 2003;17:786–799. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava S, Verschoor A, Frank J. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit-subunit interface. J. Mol. Biol. 1992;220:301–304. doi: 10.1016/0022-2836(92)90946-h. [DOI] [PubMed] [Google Scholar]
- 18.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 19.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 20.Pick E, Hofmann K, Glickman MH. PCI complexes: beyond the proteasome, CSN, and eIF3 troika. Mol. Cell. 2009;35:260–264. doi: 10.1016/j.molcel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Dessau M, Halimi Y, Erez T, Chomsky-Hecht O, Chamovitz DA, Hirsch JA. The arabidopsis COP9 signalosome subunit 7 is a model PCI domain protein with subdomains involved in COP9 signalosome assembly. Plant Cell Online. 2008;20:2815–2834. doi: 10.1105/tpc.107.053801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sengupta J, Nilsson J, Gursky R, Spahn CMT, Nissen P, Frank J. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol. 2004;11:957–962. doi: 10.1038/nsmb822. [DOI] [PubMed] [Google Scholar]
- 24.Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Carlo Marchisio P, Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 26.Liliental J, Chang DD. Rack1, a receptor for activated protein kinase C, interacts with integrin β subunit. J. Biol. Chem. 1998;273:2379–2383. doi: 10.1074/jbc.273.4.2379. [DOI] [PubMed] [Google Scholar]
- 27.Chantrel Y, Gaisne M, Lions C, Verdiere J. The transcriptional regulator Hap1p (Cyp1p) is essential for anaerobic or heme-deficient growth of Saccharomyces cerevisiae: genetic and molecular characterization of an extragenic suppressor that encodes a WD repeat protein. Genetics. 1998;148:559–570. doi: 10.1093/genetics/148.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerbasi VR, Weaver CM, Hill S, Friedman DB, Link AJ. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell. Biol. 2004;24:8276–8287. doi: 10.1128/MCB.24.18.8276-8287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shor B, Calaycay J, Rushbrook J, McLeod M. Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J. Biol. Chem. 2003;278:49119–49128. doi: 10.1074/jbc.M303968200. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann B, Mosch HU, Sattlegger E, Barthelmess IB, Hinnebusch A, Braus GH. The WD protein Cpc2p is required for repression of Gcn4 protein activity in yeast in the absence of amino-acid starvation. Mol. Microbiol. 1999;31:807–822. doi: 10.1046/j.1365-2958.1999.01219.x. [DOI] [PubMed] [Google Scholar]
- 31.Coyle SM, Gilbert WV, Doudna JA. Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol. 2009;29:1626–1634. doi: 10.1128/MCB.01718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 33.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR, Jon L. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 34.Nemoto N, Singh CR, Udagawa T, Wang S, Thorson E, Winter Z, Ohira T, Ii M, Valášek L, Brown SJ, et al. Yeast 18S rRNA is directly involved in the ribosomal response to stringent AUG selection during translation initiation. J. Biol. Chem. 2010;285:32200–32212. doi: 10.1074/jbc.M110.146662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomson AM, Rogers JT, Walker CE, Staton JM, Leedman PJ. Optimized RNA gel-shift and UV cross-linking assays for characterization of cytoplasmic RNA-protein interactions. Biotechniques. 1999;27:1032–1039, 1042. doi: 10.2144/99275rr03. [DOI] [PubMed] [Google Scholar]
- 36.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 37.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 38.Jefferys BR, Kelley LA, Sternberg MJE. Protein folding requires crowd control in a simulated cell. J. Mol. Biol. 2010;397:1329–1338. doi: 10.1016/j.jmb.2010.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 40.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 41.Valášek L, Szamecz B, Hinnebusch AG, Nielsen KH. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 2007;429:163–183. doi: 10.1016/S0076-6879(07)29008-1. [DOI] [PubMed] [Google Scholar]
- 42.Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nanda JS, Cheung Y-N, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 2009;394:268–285. doi: 10.1016/j.jmb.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, Harvey SC, Frank J. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chamovitz DA. Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep. 2009;10:352–358. doi: 10.1038/embor.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 48.Samarsky DA, Fournier MJ. A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:161–164. doi: 10.1093/nar/27.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 50.Wei Z, Zhang P, Zhou Z, Cheng Z, Wan M, Gong W. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J. Biol. Chem. 2004;279:34983–34990. doi: 10.1074/jbc.M405158200. [DOI] [PubMed] [Google Scholar]
- 51.Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 53.Guo J, Wang S, Valerius O, Hall H, Zeng Q, Li JF, Weston DJ, Ellis BE, Chen JG. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 2011;155:370–383. doi: 10.1104/pp.110.160663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jannot G, Bajan S, Giguere NJ, Bouasker S, Banville IH, Piquet S, Hutvagner G, Simard MJ. The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 2011;12:581–586. doi: 10.1038/embor.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z, Lee I, Moradi E, Hung N-J, Johnson AW, Marcotte EM. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009;7:e1000213. doi: 10.1371/journal.pbio.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.Algire MA, Maag D, Savio P, Acker MG, Tarun SZ, Jr, Sachs AB, Asano K, Nielsen KH, Olsen DS, Phan L, et al. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–397. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gietz RD, Sugino A. New yeast—Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 59.Valášek L, Hašek J, Nielsen KH, Hinnebusch AG. Dual function of eIF3j/Hcr1p in processing 20 S Pre-rRNA and translation initiation. J. Biol. Chem. 2001;276:43351–43360. doi: 10.1074/jbc.M106887200. [DOI] [PubMed] [Google Scholar]
- 60.Asano K, Vornlocher H-P, Richter-Cook NJ, Merrick WC, Hinnebusch AG, Hershey JWB. Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits: possible roles in RNA binding and macromolecular assembly. J. Biol. Chem. 1997;272:27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- 61.Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. Conserved bipartite motifs in yeast eIF5 and eIF2Be, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mueller PP, Hinnebusch AG. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45:201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.