Abstract
Spliceosomes remove introns from primary gene transcripts. They assemble de novo on each intron through a series of steps that involve the incorporation of five snRNP particles and multiple non-snRNP proteins. In mammals, all the intermediate complexes have been characterized on one transcript (MINX), with the exception of the very first, complex E. We have purified this complex by two independent procedures using antibodies to either U1-A or PRPF40A proteins, which are known to associate at an early stage of assembly. We demonstrate that the purified complexes are functional in splicing using commitment assays. These complexes contain components expected to be in the E complex and a number of previously unrecognized factors, including survival of motor neurons (SMN) and proteins of the SMN-associated complex. Depletion of the SMN complex proteins from nuclear extracts inhibits formation of the E complex and causes non-productive complexes to accumulate. This suggests that the SMN complex stabilizes the association of U1 and U2 snRNPs with pre-mRNA. In addition, the antibody to PRPF40A precipitated U2 snRNPs from nuclear extracts, indicating that PRPF40A associates with U2 snRNPs.
INTRODUCTION
In eukaryotes, the majority of primary gene transcripts (pre-mRNA) undergo splicing, a process that removes introns and joins exons to produce messenger RNA (mRNA). Splicing is catalysed by the spliceosomes, which contain five small ribonucleoprotein particles (U1, U2, U4, U5, and U6 snRNPs) and many non-snRNP proteins (1,2). The assembly of spliceosomes has been studied in most detail on transcripts containing a minimal functional unit (exon–intron–exon). Spliceosomes assemble in a series of consecutive steps that produce complexes E, A, B and C. First, in the E complex, the 5′- and 3′-splice sites (SS) of an intron are recognized by the specific binding of the U1 snRNP and the proteins U2 auxiliary factor (U2AF), respectively. Significantly, the pre-mRNA substrate is committed to the splicing pathway and the splice sites are in close proximity (3,4). The complex contains the U2 snRNP particle as a component, which is essential for its formation (5–7). At this stage, association of the U2 snRNP with the complex is weak but the underlying mechanism is not currently understood in detail. The next complex to form is complex A, which requires ATP. In this complex, the U2 snRNP is bound stably by base pairing to the branchpoint sequence, and U2-associated proteins of the SF3A/B complexes are bound to the anchoring site upstream of the branchpoint (8). This conformation serves as a binding platform for the U4/U6.U5 tri-snRNP, which culminates in the formation of complex B. The fully assembled spliceosome contains all five snRNPs and becomes competent for splicing through a series of rearrangements. These rearrangements result in the dissociation of U1 and U4 snRNPs and the formation of the catalytic centre for the first transesterification reaction, in which the 5′-exon is displaced and the lariat intron is formed. This produces complex C. The second transesterification reaction results in intron removal and the joining of exons (1,9).
The components of complexes A, B and C have been characterized in greatest detail on a transcript named MINX, which is derived from adenovirus sequences (10–19). However, the first complex in this series, E, has not been purified and characterized. The only E complexes characterized in any detail were assembled on substrates containing a neuron-specific exon, the N1 exon of c-src pre-mRNA (20,21). The protein composition of complexes formed on these transcripts provided important insights into the mechanism by which the exon is repressed, but it is not clear whether these complexes fit into the constitutive assembly pathway defined by MINX. For example, it is not clear whether the process of assembling complex A involves the same steps for c-src in WERI extracts as for MINX in HeLa. The progression from E complex to A complex can be understood only by determining the composition of both complexes on a common pre-mRNA. For this reason, we have purified and characterized complex E formed on MINX RNA in HeLa nuclear extracts.
The E complex we purified contains some factors in common with the A complex. In addition, we identified novel components that are specific for the E complex. These include the proteins of the survival of motor neurons (SMN) complex. Our data suggest that the SMN complex proteins are required for stabilizing the interactions between U1 and U2 snRNP with pre-mRNA in the E complex. Moreover, using a PRPF40A-specific antibody, we purified U2 snRNP complexes that contained PRPF40A and the SWI/SNF chromatin remodelling complex proteins. The E complex appears to be assembled from three principal sub-complexes: the U1 snRNP, the U2 snRNP and the SMN-associated complex.
MATERIALS AND METHODS
Antibodies
Rabbit polyclonal antibodies were raised (by Eurogentec) against peptides of U1A (amino acid 1–14), PRPF40A (amino acid 380–394), SF3A2 (amino acid 444–458) and purified using a SulfoLink column (Pierce) containing the antigenic peptides according to the manufacture procedure. The antibodies to SMN1 and GEMIN3 were purchased from BD Bioscience (manufacturer number 610647 and 612152, respectively). The antibodies to SIP1 (MANSIP1A), GEMIN5 (GEM5M), GEMIN6 (GEM6B) and GEMIN7 (GEM7B) were kindly provided by the MDA Monoclonal Antibody Resource (22). The antibodies to ACTL6A [BAF53 (N-19): sc-47808] and DDX15 [DDX15 (T-20): sc-67550 and (C-16): sc-67547] were purchased from Santa Cruz Biotechnology.
In vitro transcription and splicing
MINX pre-mRNA was synthesized from a PCR product template of pMINX plasmid (23) using MEGAshortscript kit (Ambion). [32P]-labelled MINX pre-mRNA was synthesized from the same template as described before (specific activity 315 000 cpm/pm) (24) and mixed with unlabelled MINX for easier monitoring of complexes. HeLa nuclear extract was prepared according to Dignam et al. (25). Splicing reactions were assembled either with the isolated E complex or an equivalent amount of naked pre-mRNA in the presence of 33% nuclear extract, treated or untreated with oligonucleotides/antibodies, 2.5 mM MgCl2, 2 mM ATP and 10 mM creatine phosphate. Reactions were incubated at 30°C for the times indicated. RNA was extracted and analysed by 14% PAGE followed by radiography.
Purification of the E complex with the antibody to U1A
To assemble E complex, nuclear extract was first dialysed against 20 mM HEPES (pH 7.9), 20% glycerol (26). A mix (2.4 ml) containing 40% nuclear extract and 65 mM KCl was depleted of endogenous ATP by incubating at 30°C for 20 min and then 15 nM MINX pre-mRNA was added. After assembly at 30°C for 10 min, the reaction was loaded on six 4 ml 10–50% glycerol gradients containing 20 mM HEPES (pH 7.9), 65 mM KCl and centrifuged in a Sorvall TH660 rotor at 29 000 rpm for 14 h. Gradients were fractionated manually into 175 µl aliquots and counted by Cherenkov. The major pre-mRNA peak, fractions 14–19, were combined and dialysed against 2 l of IP150 (20 mM HEPES, (pH 7.9), 150 mM NaCl, 0.05% NP-40, 0.5 mM DTT) for 3 h. For IPs, 400 µl of Protein A Sepharose (PAS, GE Healthcare) were charged with 150 µg of an antibody to U1A and pre-blocked with 0.3 mg/ml BSA and 50 µg/ml yeast tRNA. To each 30 µl aliquot of beads 1.5 ml of dialysed gradient fractions were added and incubated with rotation at 4°C for 3 h. Following six washes with IP150, complexes were eluted into 800 µl of the same buffer containing 0.6 mg/ml antigenic peptide. For analysis of complexes, the eluted material was loaded on two 4 ml 10–50% glycerol gradients containing 20 mM HEPES (pH 7.9), 65 mM KCl and centrifuged in a Sorvall TH660 rotor at 26 300 rpm for 13 h.
RNA was extracted from each gradient fraction with phenol/chloroform/isoamylalcohol (PCA, 25:24:1) and precipitated with ethanol. RNA was separated by 10% denaturing PAGE and visualized by silver staining. For identification of the protein composition by MS, corresponding gradient fractions were combined, diluted two-fold with 20 mM HEPES (pH 7.9), 150 mM NaCl and complexes were pelleted in a Sorvall TH660 rotor at 60 000 rpm for 6 h. The pellet was re-suspended in the remaining 60 µl of solution and the proteins were reduced in the presence of 1% SDS and 100 mM DTT at 37°C for 45 min.
For western blotting, eluted complexes were loaded on one 4 ml 10–30% glycerol gradient and 90 µl of each fraction were separated by 10% SDS–PAGE, transferred to a nitrocellulose membrane and probed with antibodies.
Purification of the E complex with the antibody to PRPF40A
Four hundred microlitres of the pre-blocked PAS were charged with 170 µg of affinity purified antibodies. IP was carried out from the E complex-assembly reaction (2.4 ml) diluted with 1 ml of IP150 buffer as described before. Complexes were eluted in 1.6 ml of IP150 containing 0.6 mg/ml antigenic peptide. The eluate was loaded on four 4 ml 10–50% glycerol gradients containing 20 mM HEPES (pH 7.9), 65 mM KCl and centrifuged in a Sorvall TH660 rotor at 27 400 rpm for 12 h. Gradient fractions were measured by Cherenkov counting and RNA was analysed by denaturing PAGE. For western blotting, 90 µl of each fraction were loaded directly on 10% SDS–PAGE and processed as described above.
Mass spectrometry of the E complex
Of each complex, 1.26 pmoles isolated with antibodies to PRPF40A and U1A were separated by 8/14% SDS–PAGE and stained with colloidal Coomassie G250. Each gel lane was cut into 54 slices and an in-gel trypsin digest was carried out. LC–MS/MS was performed upon each sample using a 4000 Q-Trap mass spectrometer (Applied Biosystems, Warrington, UK). Peptides resulting from in-gel digestion were loaded at high flow rate onto a reverse-phase trapping column (0.3 mm i.d. × 1 mm), containing 5 µm C18 300 Å Acclaim PepMap media (Dionex, UK) and eluted through a reverse-phase capillary column (75 µm i.d. × 150 mm) containing Waters Symmetry C18 100 Å media (Waters, UK) that was self-packed using a high pressure packing device (Proxeon Biosystems, Odense, Denmark). The output from the column was sprayed directly into the nanospray ion source of the 4000 Q-Trap mass spectrometer. Fragment ion spectra generated by LC–MS/MS were searched using the MASCOT (27) search tool against the National Center for Biotechnology Information non-redundant protein database using appropriate parameters: peptide mass tolerance = 0.4 Da; fragment mass tolerance = 0.6 Da; the enzyme was selected as trypsin with one missed cleavage site considered; carbamidomethyl (C) was selected as a fixed modification and oxidation (M) as a variable modification. The criteria for protein identification were based on the manufacturer's definitions (Matrix Science Ltd.). Basically candidate peptides with probability-based Mowse scores exceeding threshold (P < 0.05), and thus indicating a significant or extensive homology were referred to as ‘hits’. The highest scoring fragment ion spectrum from each protein hit was manually inspected to ensure its validity and thus minimize potential false positives in the data set.
Depletion of HeLa nuclear extracts
To deplete U1 and U2 snRNPs, 100 µl of nuclear extract in 20 mM HEPES (pH7.9), 65 mM KCl, 2.2 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, 0.1 mM DTT, 20% glycerol were incubated in the presence of 1 µg each oligonucleotide against U1 and U2 snRNAs (U2-E15 and U1-5′C) and 2 U of RNsae H (Promega) for 1.5 h as described previously (28). To analyse digestion of the targeted snRNP particles, the RNA was extracted from 10 µl aliquots and analysed by 10% PAGE followed by silver staining. For SMN depletion, 200 µl of PAS were charged with 40 µg of an antibody to SMN and pre-blocked with 0.3 mg/ml BSA and 50 µg/ml yeast tRNA. Two hundred microlitres of Hela nuclear extracts at either 200 or 700 mM NaCl were treated twice with PAS–SMN antibody-conjugate or PAS only as a control. Ten microlitres of extract were used for western blot analysis. The extracts were dialysed against buffer containing 20 mM HEPES (pH 7.9), 100 mM KCl, 0.5 mM DTT, 15% glycerol and used at concentrations of 40% for splicing and the E complex formation.
Analysis of E complex formation in the SMN-depleted extract by glycerol gradient centrifugation
The E complexes were assembled in a reaction containing 40% nuclear extract, 65 mM KCl, 10 nM MINX. The reactions were diluted two-fold with chilled buffer containing 20 mM HEPES (pH 7.9), 65 mM KCl, loaded on the 10–50% glycerol gradient and centrifuged in TH660 rotor at 24 500 rpm for 15 h.
Isolation of U2 snRNPs with the antibody to PRPF40A
The IP was carried out using 150 µl of pre-blocked PAS charged with 90 µg of affinity purified antibody to PRPF40A from the reaction (300 µl) containing 40% nuclear extract, 20 mM HEPES (pH 7.9), 65 mM KCl and depleted of endogenous ATP by incubating at 30°C for 20 min. IP was carried out in the IP150 buffer at 4°C for 2.5 h. The bound material was eluted in 400 µl of IP150 containing 0.6 mg/ml of antigenic peptide at 4°C for 1 h and loaded on the 4 ml 10–30% glycerol gradient containing 150 mM NaCl. Gradients were centrifuged in a Sorvall TH660 rotor at 24 800 rpm for 15 h. RNA from the gradient fractions was extracted and analysed by 10% PAGE followed by silver staining. Proteins from fractions 8–11 were precipitated with acetone, re-suspended in SDS-loading buffer and separated by 10/13% SDS–PAGE for MS analysis.
RESULTS
Isolation of the E complex
Spliceosomes are dynamic molecular machines that undergo multiple rearrangements during the splicing cycle. Their heterogeneity is a well-documented problem for structural studies (29,30). Therefore, to purify the E complex, we used antibodies to proteins that associate with the spliceosome transiently (16). We selected the U1 snRNP as it is a major factor that triggers spliceosome assembly and then dissociates from the complex before the catalytic spliceosome is formed. We raised an antibody to the U1 snRNP-associated protein U1A (encoded by the SNRPA gene), for which few interacting partners are known and the chances of being accessible to an antibody are higher. We also targeted PRPF40A (FBP11), a homologue of the essential yeast Prp40p protein, which has been proposed to bridge the intron's splice sites. The peptide antibodies raised against these proteins specifically recognized the corresponding polypeptides on western blots of the HeLa nuclear extract and efficiently precipitated pre-mRNA from splicing reactions (data not shown).
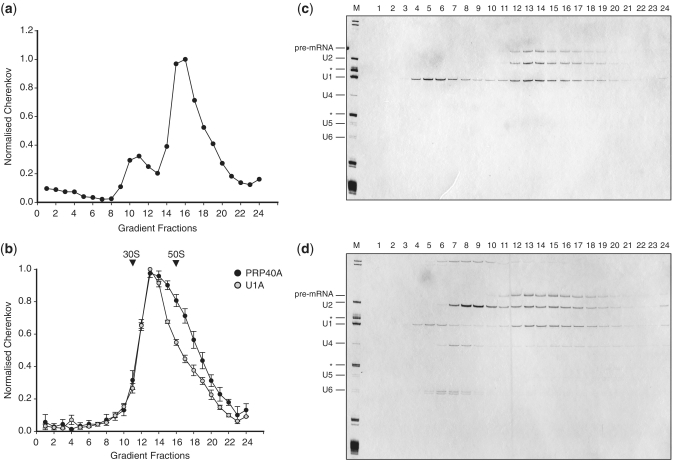
The E complex was assembled using MINX pre-mRNA in HeLa nuclear extracts in the absence of ATP (5,31). Before immunoprecipitation (IP) of complexes with the U1A-specific antibody, the E complex-assembly reaction was fractionated by 10–50% glycerol gradient centrifugation to separate spliceosomal complexes from free U1 snRNP particles. The concentration of U1 snRNPs in HeLa nuclear extracts is ∼200 nM, at least an order of magnitude greater than the concentration of the pre-mRNA (32). The position of complexes in the gradient (fractions 14–19) was determined by monitoring the distribution of radioactively labelled pre-mRNA by Cherenkov counting (Figure 1a). The sedimentation value of complexes was determined by comparison with the 30S and 50S ribosomal subunits and corresponds to 35–40 S, which is much larger than the 10S of U1 snRNPs. Complexes were isolated from fractions 14–19 using U1A-specific antibody immobilized on PAS and eluted from the column with the antigenic peptide to minimize any unspecific background. To prove the integrity of the isolated complexes, the eluted material was analysed again by 10–50% glycerol gradients. The material migrated as a single peak (Figure 1b; fractions 12–17), indicating that the isolated complexes are stable and homogeneous. The RNA was extracted from the gradient fractions and analysed by denaturing PAGE followed by silver staining (Figure 1c). Fractions 12–17 contained pre-mRNA and stoichiometric amounts of U1 and U2 snRNAs, which are characteristic of the E complex. Some U1 snRNPs were detected on the top of the gradient (Figure 1c; fractions 3–6). It is possible that some free U1 snRNPs were present in the E complex fractions and were precipitated independently of the spliceosomal complexes (33).
Figure 1.
Isolation and characterization of the E complex. (a) The E complex was assembled on MINX pre-mRNA in HeLa nuclear extract and the reaction was fractionated on a 10–50% glycerol gradient. The gradient fractions were counted by Cherenkov and the position of the labelled pre-mRNA is indicated. The major peak, fractions 14–19, was used in IPs with the U1A-specific antibody. (b) Complexes isolated with the U1A-specific antibody (grey circles) and with the PRPF40A-specific antibodies (black circles) were analysed by 10–50% glycerol gradients. Both purifications yielded complexes with similar 35–40S values. The positions of the E. coli 50S and 30S ribosomes are indicated. (c) RNA was recovered from the gradient fractions of the U1A-specific isolation, analysed by the 10% denaturing PAGE and visualized by silver staining. (d) The RNA profile from the gradient fractions of complexes isolated with the antibody to PRPF40A. Total RNA extracted from the nuclear extract was used as a marker (M). The identity of RNAs is indicated on the left. The asterisks correspond to the 5S and 5.8S ribosomal RNAs.
An alternative method for purification of the E complex began with direct addition of the assembly reaction to PAS beads charged with an antibody to PRPF40A. Complexes eluted from the beads using the antigenic peptide were further purified by glycerol gradient centrifugation under the same conditions as before. Significantly, both purifications resulted in the isolation of complexes with similar migration abilities (Figure 1b). RNA analysis of the gradient fractions showed that the isolated complexes contained pre-mRNA and U1 and U2 snRNPs (Figure 1d; fractions 12–17). In addition, a significant amount of U2 snRNA was found in fractions 7–10 (Figure 1d). This peak was absent from the U1A-associated complexes (Figure 1c) and is therefore unlikely to have arisen from dissociation during centrifugation. Indeed, we infer that these U2 snRNPs were in distinct complexes that were also selected by the PRPF40A-specific antibody. The relative migration of these U2-containing complexes corresponded to ∼20S (peak fractions 8–9), which is higher than the previously characterized 17S U2 snRNPs (34,35).
PRPF40A associates with U2 snRNPs
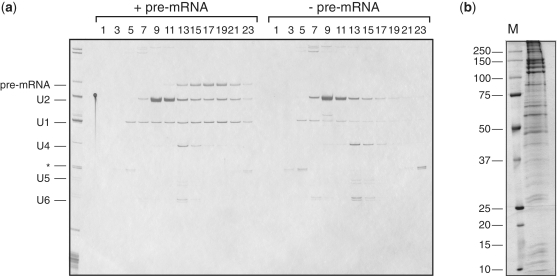
PRPF40A was not identified as a U2-associated protein in previous isolations of the U2 snRNPs (34,35). Moreover, its yeast homologue, Prp40p, is a component of the U1 snRNP and not U2 (36,37). To investigate whether there is an alternative form of U2 snRNPs present in HeLa nuclear extract, we carried out IPs with the PRPF40A-specific antibody from reactions in the absence of pre-mRNA. The analysis of an eluate by glycerol gradient centrifugation showed that U2 particles were selected again (Figure 2a; fractions 9–11), indicating the presence of such complexes in HeLa extract.
Figure 2.
Characterization of the PRPF40A–U2 snRNP complex. (a) The PRPF40A-specific antibodies precipitate U2 snRNPs from HeLa nuclear extract. The IPs were carried out from reactions containing pre-mRNA (left) or without pre-mRNA (right). Eluates from both IPs were fractionated by 10–30% glycerol gradients. RNA from the odd fractions was analysed by the 10% denaturing PAGE and visualized by silver staining. The identity of the RNAs is indicated on the left. (b) Identification of U2 snRNP-associated proteins. Proteins from the peak fractions of the U2 snRNP gradient (fractions 9–11) were recovered, fractionated by SDS–PAGE on the 10/13% polyacrylamide gel and stained with Coomassie Blue. The identity of proteins was determined by MS.
Next, we determined the full protein composition of the novel PRPF40A–U2 complex. Proteins were recovered from gradient fractions 9–11 containing U2 snRNAs, fractionated by SDS–PAGE, and stained with Coomassie Blue (Figure 2b). The identities of proteins were determined after in-gel digestion with trypsin using a Q-Trap 4000 mass spectrometer and are listed in Table 1 and Supplementary Table S1. Consistent with the presence of the U2 snRNA, the complex contained all the U2-specific proteins identified in previous isolations (34,35). In addition, we identified a number of novel components in association with the PRPF40A–U2 complex, including the SWI/SNF chromatin remodelling complex proteins such as ARID1A/1B/2, SMARCA1/2/4, PBRM1, SMARCB1, SMARCC1/2, SMARCD1, SMARCE1, ACTL6A and ACTBL2. In addition to these, we identified several transcription factors that have been identified as spliceosomal components in previous purifications of splicing complexes. These include RUVBL2/1 (TIP48/TIP49), RBM25 (RED120/S164), SART3 (TIP110), ARS2, BAT1 (UAP56), HTATSF1 (Tat-SF1), YEATS2 and CCNK. Other transcription factors produced insignificant scores, indicating that most of these proteins are more likely to be present in sub-stoichiometric amounts. The presence of additional proteins correlates with the increased S-value of the complex. The common trait shared by these proteins is their involvement in the regulation of transcriptional elongation by RNAP II, which is coupled to histone modification and chromatin remodelling. Therefore, our data strongly support the hypothesis that there is co-transcriptional recruitment of the U2 snRNP to nascent transcripts.
Table 1.
Proteins of the E complex and novel U2 snRNPs
| Gene name | Mol Mass | E complex Ab to PRPF40A |
E complex Ab to U1A |
U2 snRNP Ab to PRPF40A |
|||
|---|---|---|---|---|---|---|---|
| pept | emPAI | pept | emPAI | pept | emPAI | ||
| Sm proteins | |||||||
| SNRPB | 24 765 | 5 | 0.33 | 6 | 0.77 | 6 | N/A |
| SNRPD1 | 13 273 | 1 | N/A | 1 | 0.3 | 2 | N/A |
| SNRPD2 | 13 632 | 9 | 4.87 | 11 | 6.56 | ||
| SNRPD3 | 14 021 | 2 | 0.64 | 5 | 0.64 | 1 | N/A |
| SNRPE | 10 854 | 2 | 0.87 | 2 | 0.87 | 1 | N/A |
| SNRNPF | 9 776 | 1 | N/A | 5 | N/A | ||
| SNRPG | 8 547 | 3 | N/A | ||||
| U1 snRNP | |||||||
| SNRP70 | 51 583 | 4 | 0.32 | 12 | 0.52 | ||
| SNRPA | 31 259 | 4 | 0.41 | 7 | 0.77 | 1 | N/A |
| SNRPC | 17 552 | 3 | 3 | 0.82 | |||
| U2 snRNP | |||||||
| SNRPA1 | 28 512 | 7 | 1.11 | 6 | 1.11 | 11 | N/A |
| SNRNPB2 | 25 470 | 4 | 0.75 | 3 | 0.52 | 5 | N/A |
| SF3A1 | 88 888 | 19 | 0.92 | 38 | 2.13 | 40 | 2.4 |
| SF3A2 | 49 338 | 8 | 0.44 | 10 | 0.67 | 8 | 0.44 |
| SF3A3 | 59 154 | 22 | 0.73 | 15 | 1.08 | 20 | 1.65 |
| SF3B1 | 146 464 | 55 | 1.39 | 45 | 1.06 | 58 | 1.84 |
| SF3B2 | 97 710 | 29 | 1.18 | 15 | 0.4 | 33 | 1.35 |
| SF3B3 | 136 575 | 23 | 0.62 | 25 | 0.62 | 31 | 0.9 |
| SF3B4 | 44 414 | 2 | 0.18 | 2 | 0.08 | 3 | 0.27 |
| SF3B14 | 14 690 | 8 | 3.13 | 8 | 3.13 | 6 | N/A |
| PHF5A | 13 138 | 1 | N/A | 2 | N/A | ||
| U2 related | |||||||
| PRP40A | 109 022 | 9 | 0.26 | 9 | 0.31 | 5 | 0.14 |
| PAF60 | 60 009 | 13 | 0.94 | 29 | 2.99 | 3 | 0.06 |
| DDX46 | 117 803 | 17 | 0.59 | 5 | 0.17 | ||
| DHX15 | 91 673 | 11 | 0.49 | 1 | 30 | 1.21 | |
| SR140 | 118 675 | 2 | 0.03 | 13 | 0.24 | ||
| CHERP | 104 078 | 14 | 0.47 | ||||
| U4/U6.U5 tri-snRNP | |||||||
| PRPF8 | 274 738 | 52 | 0.7 | 2 | 0.03 | ||
| SNRNP200 | 246 006 | 36 | 0.54 | 3 | 0.03 | ||
| EFTUD2 | 110 336 | 13 | 0.48 | 8 | 0.22 | 4 | 0.07 |
| PRPF6 | 107 656 | 8 | 0.27 | 1 | 0.03 | ||
| DDX23 | 95 866 | 2 | 0.04 | 2 | 0.08 | ||
| PRPF3 | 77 652 | 14 | 0.52 | ||||
| PRPF31 | 55 649 | 4 | 0.21 | ||||
| PRPF4 | 59 097 | 4 | 0.28 | 2 | 0.06 | ||
| PPIH | 19 481 | 4 | N/A | ||||
| LSM4 | 15 511 | 1 | 0.25 | ||||
| SART1 | 90 371 | 2 | 0.08 | ||||
| USP39 | 65 739 | 2 | 0.12 | ||||
| SNRNP40 | 39 742 | 1 | N/A | ||||
| SMN associated | |||||||
| SMN1 | 32 285 | 1 | 0.12 | 1 | 0.12 | ||
| SIP1 | 32 021 | 2 | 0.25 | ||||
| DDX20 | 92 981 | 3 | 0.08 | 3 | 0.08 | ||
| GEMIN4 | 121 681 | 9 | 0.27 | 23 | 0.57 | ||
| GEMIN5 | 170 793 | 2 | 0.04 | 9 | 0.21 | ||
| GEMIN6 | 18 983 | 1 | 0.2 | 3 | 0.74 | ||
| GEMIN8 | 28 904 | 1 | 0.13 | ||||
| STRAP | 38 756 | 6 | 0.32 | ||||
| Splicing related | |||||||
| NCBP1 | 92 864 | 24 | 1.36 | 32 | 1.65 | ||
| NCBP2 | 18 161 | 1 | 0.21 | 4 | 0.21 | ||
| U2AF1 | 28 368 | 4 | 0.46 | 2 | 0.21 | ||
| U2AF2 | 53 809 | 14 | 1.39 | 7 | 0.49 | ||
| SF1 | 8 | 0.45 | |||||
| SRRM2 | 300 179 | 8 | 0.08 | 7 | 0.06 | ||
| SFRS1 | 27 842 | 10 | 1.78 | 7 | 0.89 | 1 | N/A |
| SFRS2 | 25 461 | 2 | 0.32 | ||||
| SFRS2B | 32 382 | 3 | 0.25 | 1 | 0.12 | ||
| SFRS3 | 19 546 | 2 | 0.43 | ||||
| SFRS4 | 56 759 | 2 | 0.07 | ||||
| SFRS5 | 31 359 | 5 | 0.57 | ||||
| SFRS6 | 39 677 | 2 | 0.2 | 3 | 0.31 | ||
| SFRS7 | 27 578 | 4 | 0.29 | 5 | 0.68 | ||
| SFRS8 | 105 190 | 1 | 0.04 | ||||
| SFRS11 | 53 624 | 2 | 0.07 | ||||
| SFRS12 | 59 402 | 2 | 0.06 | ||||
| SFRS15 | 126 132 | 2 | 0.03 | ||||
| HNRNPA0 | 30 993 | 2 | 0.26 | ||||
| HNRPA1L3 | 34 373 | 2 | 0.11 | 2 | 0.23 | 2 | N/A |
| HNRNPA2B1 | 37 464 | 1 | 0.1 | 1 | 0.1 | 6 | N/A |
| HNRNPC | 33 707 | 1 | 0.11 | 2 | 0.24 | ||
| HNRNPD | 38 581 | 1 | 0.1 | ||||
| HNRNPF | 45 985 | 2 | 0.08 | ||||
| HNRNPH1 | 49 484 | 1 | 0.08 | ||||
| HNRNPK | 51 230 | 1 | 0.07 | ||||
| HNRNPL | 64 720 | 2 | 0.12 | ||||
| HNRNPM | 77 749 | 6 | 0.2 | ||||
| SYNCRIP | 69 788 | 14 | 0.59 | 3 | 0.11 | 6 | 0.23 |
| HNRNPR | 71 184 | 6 | 0.16 | 8 | 0.36 | 1 | 0.05 |
| HNRNPU | 91 198 | 4 | 0.13 | 3 | 0.08 | 7 | 0.13 |
| RBM7 | 30 485 | 1 | 0.12 | 3 | 0.42 | ||
| RBM10 | 103 811 | 3 | 0.07 | ||||
| RBM17 | 45 162 | 1 | 0.08 | 8 | 0.75 | ||
| RBM25 | 94 407 | 34 | 1.42 | 12 | 0.31 | 8 | 0.21 |
| RBM39 | 59 628 | 7 | 0.35 | 6 | 0.27 | ||
| RBM42 | 50 496 | 1 | 0.07 | ||||
| RBMXL2 | 42 929 | 4 | 0.29 | ||||
| ARS2 | 101 060 | 18 | 0.65 | 9 | 0.24 | 7 | 0.11 |
| LUC7L3 (CROP) | 51 834 | 6 | 0.42 | 4 | 0.32 | ||
| LUC7L2 | 46 942 | 3 | 0.08 | ||||
| PCBP1 | 37 987 | 2 | 0.21 | 3 | 0.33 | ||
| PCBP2 | 38 955 | 3 | 0.32 | ||||
| SKIV2L2 | 118 756 | 5 | 0.13 | 3 | 0.1 | 3 | 0.1 |
| KHSRP | 73 443 | 2 | 0.1 | ||||
| FUS | 53 622 | 2 | 0.14 | ||||
| DDX5 | 69 618 | 3 | 0.17 | 6 | 0.37 | ||
| DDX17 | 72 953 | 3 | 0.1 | 2 | 0.05 | ||
| DHX9 | 142 181 | 4 | 0.05 | ||||
| DDX39 | 49 611 | 1 | |||||
| PRPF19 | 55 603 | 7 | 0.57 | 3 | 0.21 | ||
| SART3 | 110 721 | 3 | 0.1 | 6 | 0.18 | ||
| BCAS2 | 26 229 | 1 | 0.15 | ||||
| ZCCHC8 | 79 156 | 1 | 0.05 | ||||
| DNAJC8 | 29 823 | 4 | N/A | ||||
| THOC4 | 26 872 | 1 | 0.14 | ||||
| BAT1 | 49 416 | 1 | 0.08 | 4 | 0.16 | ||
| YBX1 | 35 903 | 1 | 0.1 | ||||
| RUVBL2 | 51 296 | 2 | 0.15 | 2 | 0.15 | 14 | 1.02 |
| RUVBL1 | 50 538 | 6 | 0.53 | ||||
| CPSF1 | 162 036 | 1 | 0.02 | ||||
| NUDT21 | 26 268 | 2 | 0.14 | ||||
| CPSF6 | 59 344 | 1 | 0.06 | 7 | 0.28 | ||
| CPSF7 | 52 189 | 1 | 0.07 | ||||
| HTATSF1 | 86 371 | 13 | 0.46 | 6 | 0.18 | 28 | 1.52 |
| Chromatin remodeling | |||||||
| ARID1A | 242 805 | 5 | 0.03 | ||||
| ARID1B | 237 057 | 17 | 0.22 | ||||
| ARID2 | 198 921 | 10 | 0.18 | 14 | 0.25 | ||
| SMARCA1 | 123 211 | 1 | 0.03 | ||||
| SMARCA2 | 181 794 | 15 | 0.22 | ||||
| SMARCA4 | 185 100 | 27 | 0.48 | 5 | 0.06 | 25 | 0.45 |
| PBRM1 | 194 080 | 39 | 0.76 | 8 | 0.08 | ||
| SMARCC2 | 133 196 | 25 | 0.73 | 21 | 0.43 | ||
| SMARCC1 | 123 303 | 3 | 0.09 | 10 | 0.27 | ||
| SMARCD1 | 55 195 | 2 | 0.14 | 11 | 0.8 | ||
| SMARCE1 | 46 678 | 5 | 0.26 | 9 | 0.59 | ||
| ACTL6A | 47 944 | 6 | 0.46 | 3 | 0.25 | 8 | 0.57 |
| SMARCB1 | 44 398 | 2 | 0.18 | 6 | 0.38 | ||
| ACTBL2 | 42 318 | 2 | 0.09 | ||||
| CHD4 | 219 393 | 20 | 0.26 | ||||
| CHD8 | 263 977 | 3 | 0.03 | 30 | 0.39 | ||
The E complex was assembled on MINX pre-mRNA in HeLa nuclear extract and purified using either U1A- or PRPF40A-specific antibodies. In addition, the antibody to PRPF40A precipitated novel U2 snRNPs. Proteins from purified complexes were fractionated by SDS–PAGE and subjected to MS analysis. The presence of a protein in a complex is shown by the number of unique peptides and the emPAI factor (60)—where available (note: N/A stands for not available). Calculated molecular masses for proteins and their corresponding gene names are taken from the UniProt database. Proteins are grouped according to their known function. Additional proteins identified in purifications which are not common components for all complexes are presented in the Supplementary Table S1.
The E complex is a functional stable spliceosomal intermediate
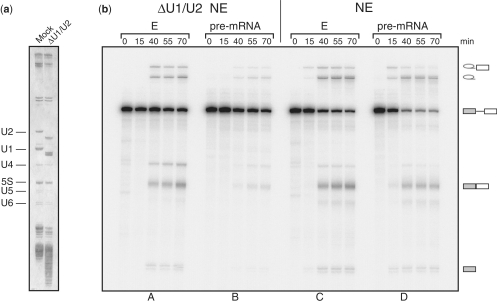
It was important to test whether the E complex that we isolated represents a functional spliceosomal intermediate. For this purpose, we treated an extract with RNase H in the presence of oligonucleotides complementary to U1 and U2 snRNAs (28). The U1 and U2 snRNPs were digested (Figure 3a), leading to a reduction in splicing activity. Then, we tested whether the isolated E complex would support splicing of the substrate pre-mRNA in this extract. Splicing in the E complex was unaffected by the depletion of U1 and U2 snRNPs (Figure 3b; panels A and B). This supports the view that the E complex we isolate is splicing competent and does not require the association of either U1 or U2 snRNPs, which indicates the integrity of the complexes. Notably, the E complex generated even more spliced product than naked pre-mRNA added to intact mock-treated extract (Figure 3b, panels C and D). This shows that pre-mRNA was protected from degradation in the E complex as compared to the naked pre-mRNA. On the other hand, the E complex had no kinetic advantage in the course of reaction, suggesting that later steps are limiting.
Figure 3.
The E complex represents a functional spliceosomal intermediate. (a) HeLa extract was depleted of U1 and U2 snRNPs using RNase H digestion with complementary oligonucleotides (ΔU1/U2). Total RNA from the corresponding extracts was analysed by the 10% denaturing PAGE and visualized by silver staining. The identity of RNAs is indicated on the left. (b) The time course of splicing in depleted (ΔU1/U2) and mock (NE) extracts. Equal amounts of the E complex isolated using the PRPF40A-specific antibody or naked pre-mRNA were supplemented with the mock (NE) or ΔU1/U2 nuclear extracts and allowed to splice. The aliquots of the reactions were taken at times indicated, RNA extracted, analysed by denaturing gel and visualized by exposure to a phosphoimager. The ΔU1/U2 extract is deficient in splicing naked pre-mRNA but pre-mRNA in the E complex is efficiently processed in this extract (compare panels A and B). In the mock extract, pre-mRNA is protected from degradation in the E complex as compared to the naked pre-mRNA (panels C and D). The identities of the precursor, splicing intermediates and the product are indicated on the right.
The protein composition of the E complex
The proteins in the E complex must be responsible for the initial steps in spliceosome formation and the pairing of splice sites. Having proved that the isolated E complexes represent a functional intermediate step in the spliceosome assembly pathway, we analysed their composition. Equal amounts of complexes purified by the PRPF40A- or U1A-specific antibodies were fractionated by SDS–PAGE and stained with Coomassie Blue (Figure 4a). Proteins identified by MS analysis are listed in Table 1 and Supplementary Table S1.
Figure 4.
Protein composition of the E complex. (a) Proteins from both preparations of the E complex isolated using either U1A- or PRPF40A-specific antibodies were fractionated by 8%/14% SDS–PAGE and stained with Coomassie Blue. The identities of proteins were determined by MS. (b) The U1A preparation of the E complex was analysed by 10–30% glycerol gradient and the gradient fractions were counted by Cherenkov to follow pre-mRNA distribution. (c) RNA and protein composition of U1A gradient fractions. RNA was extracted from the gradient fractions, separated by 10% denaturing PAGE and visualized by silver staining. Proteins from the gradient fractions were separated by 10% SDS–PAGE and western blotted with antibodies to SMN1, SIP1 and GEMIN6. (d) Complexes isolated with the antibody to PRPF40A were fractionated by 10–30% glycerol gradient and pre-mRNA was detected by Cherenkov counting. (e) RNA and protein analysis of gradient fractions from the PRPF40A-E complex isolation. The identities of RNA and antibodies are indicated on the left.
The comparison of protein compositions revealed that almost all proteins identified in the complex purified with the U1A-specific antibody were present in the complex isolated with the antibody to PRPF40A. Overall, more proteins were identified in the preparation with the PRPF40A-specific antibody and some common proteins were detected by a larger number of peptides. These observations are consistent with the fact that the purification procedure used to isolate complexes with the antibody to U1A was more rigorous and included fractionation of the complexes by glycerol gradient prior to affinity purification. This could result in the loss of more weakly associated proteins. For example, the U4/U6.U5 tri-snRNP proteins were better represented in the PRPF40A preparation, indicating the intrinsic affinity of the tri-snRNP particle for the pre-spliceosome, but at this stage of assembly this interaction is unlikely to be stable enough to withstand the conditions used for U1A isolation. Therefore, we believe that most of the common proteins in both isolations represent the more tightly bound components of the E complex.
Consistent with its RNA composition, the E complex contained U1 and U2 snRNP-associated proteins. Most of them were identified by a similar number of peptides in both preparations, confirming that equal amounts of complexes were subjected to the analysis. The E complex also contained proteins that associate with pre-mRNA directly. These include the cap-binding complex proteins and hnRNP proteins A1, A2/B1, C, Q, R and U together with poly(rC)-binding protein (PCBP1), SR proteins (SFRS1, 2B, 6, 7), RNA-binding motif proteins (RBM25, 39, 7), PUF60 and SR-related protein SRRM2 (SRp300) and CROP (Luc7A). The DEAD box family ATP-dependent RNA helicases DDX5 (p68) and DDX46 (hPRP5) were identified in both preparations. In addition, proteins that affect alternative splicing and therefore might influence spliceosome assembly were found in association with the E complexes. All these proteins were also identified as components of E complexes assembled on c-src pre-mRNA (20,21). Their presence on both MINX and the neuron-specific substrate suggests that they have constitutive functions in splicing. One difference between the complexes purified by the antibodies to PRPF40A and U1A is that the former lacked the proteins SF1 and DDX17 (p72). This did not prevent the complex functioning in a commitment assay (Figure 3b).
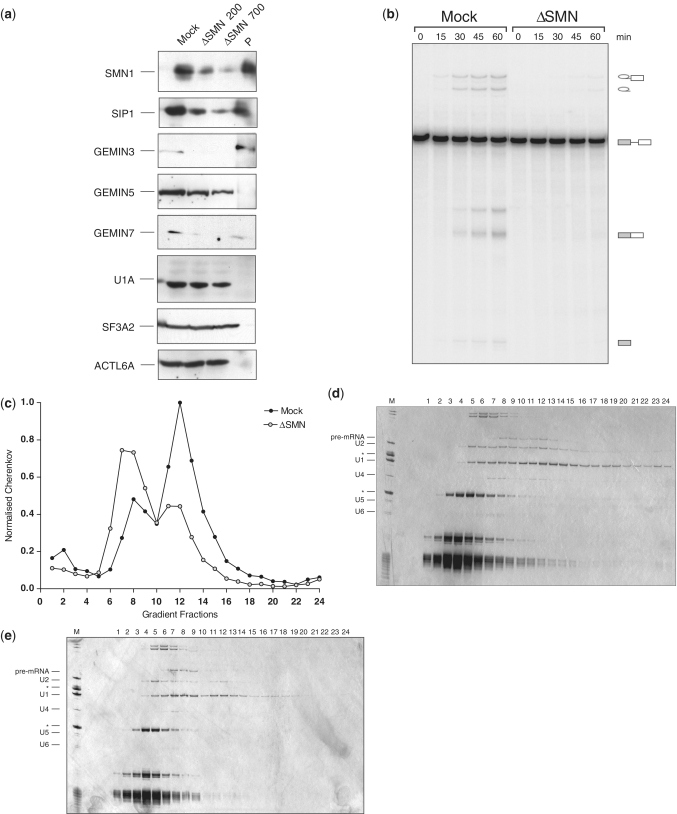
Significantly, the purified E complexes contain proteins of the SMN-associated complex. Although an indirect role for SMN in splicing has been suggested previously (39–43), this is the first time that the SMN complex has been identified as a possible spliceosomal component. The complex comprises nine proteins: SMN, GEMINS2-8 and STRAP (UNR-IP) (44–49). We identified all of them except GEMIN7, although GEMIN2 (SIP1) and GEMIN8 were detected only in the PRPF40A-specific preparation. Thus, it is likely that all proteins of the SMN complex co-purify with the E complex. Significantly, neither antibody, to U1A or PRPF40A, precipitated U1 or U2 snRNPs in association with the SMN complex. This is consistent with the idea that the presence of the SMN complex in the spliceosomal complex E is independent of snRNPs. To verify the MS data, the E complex preparations were analysed by western blotting. Complexes were purified as previously described and analysed by 10–30% glycerol gradients (Figure 4b and d). Antibodies against proteins SMN (SMN1), GEMIN2 (SIP1) and GEMIN6-7 were used for detecting proteins of the SMN complex across the entire gradients. The designated proteins were detected in fractions corresponding to the E complex with material purified via both U1A and PRPF40A antibodies (Figure 4c and e). This provides a strong evidence for the SMN complex proteins as components of the E complex.
Interestingly, analysis of the complexes purified with the antibody to PRPF40A showed two peaks for SMN (fractions 11, 12 and 14–16). The major peak (fractions 14–16) corresponds to the E complex, whereas fractions 11 and 12 do not coincide with any RNA species. It suggests that the antibody to PRPF40A also precipitated an SMN-containing complex in addition to the E complex and U2 snRNP. We infer that PRPF40A–SMN complexes might exist in HeLa nuclear extracts.
In addition, we tested some of the proteins identified in the PRPF40A–U2 snRNP complex (Figure 4e). The signal produced by an antibody against the U2 snRNP-specific protein SF3A2 corresponds to the distribution of U2 snRNA across the gradient. We also used an antibody to one of the SWI/SNF complex proteins, ACTL6A, and detected it in the PRPF40A–U2 snRNP complex.
Depletion of the SMN protein
To test whether SMN plays a role in the formation of the E complex, it was depleted from HeLa nuclear extract using an antibody to SMN. Depletion was done at two different salt concentrations, 200 and 700 mM NaCl. Depletion at the higher salt concentration resulted in more efficient removal of the proteins associated with SMN and their presence was detected in the eluate from the beads. Importantly, neither U1A nor SF3A proteins were affected, indicating that the SMN complex was removed without the U1 or U2 snRNPs (Figure 5a). Depletion inactivated the extract (Figure 5b).
Figure 5.
Depletions of the SMN and SMN-associated proteins affect E complex formation. (a) Western blot analysis of the mock and SMN-depleted extracts with different antibodies. HeLa nuclear extracts were depleted with the antibody to SMN at 200 and 700 mM NaCl and analysed by western blot together with the material removed. Mock—mock-treated extract, ΔSMN—depleted of SMN at 200 and 700 mM salt, correspondingly, P—precipitate. The identities of proteins detected by antibodies are shown on the left. (b) Depletion of SMN from the nuclear extract resulted in splicing inhibition as compared to the mock-treated extract. The pre-mRNA was incubated under splicing conditions in mock and SMN-depleted extracts. The aliquots of the reactions were taken at times indicated, RNA extracted, analysed by denaturing gel and visualized by exposure to a phosphoimager. Identities of the RNA are indicated on the right. (c) The E complex is not formed in the SMN-depleted extract as opposed to the mock-treated extract (fractions 11–13). Complexes assembled on pre-mRNA in mock and SMN-depleted extracts were fractionated by 10–50% glycerol gradient centrifugation. Gradient fractions were collected manually from the top to the bottom and counted by Cherenkov. (d) The RNA analysis of the E complex formed in mock-depleted extracts. RNA extracted from the gradient fractions was separated by the 10% denaturing PAGE and visualized by silver staining. Fractions 10-13 correspond to the E complex. (e) The RNA analysis of complexes assembled in the SMN-depleted extracts. The peak corresponding to the E complex is missing and pre-mRNA is found in non-productive complexes with amounts of the U2 snRNA greatly reduced. The identity of RNAs is indicated on the left. The asterisks correspond to the 5S and 5.8S ribosomal RNAs.
The ability of the SMN-depleted extract to form complexes was analysed by glycerol gradient centrifugation. The reactions were loaded on the gradient and the distribution of pre-mRNA across the gradients was monitored by Cherenkov counting. The comparison of pre-mRNA distributions displayed a striking difference between the two extracts (Figure 5c). In the mock-depleted extract, the major peak had the expected 35–40S value and formation of the E complex was unaffected. In contrast, complexes formed in the SMN-depleted extract produced a much smaller peak corresponding to the E complex (fractions 11–13) and the majority of pre-mRNA was found in slower-migrating fractions (7–9) (Figure 5c). To analyse complex formation in more detail, RNA was extracted from gradient fractions, separated by denaturing PAGE and visualized by silver staining (Figure 5d and e). In addition to the reduced amount of E complex, the distribution of U2 snRNA was significantly different in the SMN-depleted extract. The majority of the U2 snRNA was found in fractions 4–6, not associated with the pre-mRNA. This suggests that association of U2 snRNP in the E complex might require SMN and its associated proteins.
DISCUSSION
The E complex represents the first definite complex in the spliceosome assembly process. Previous studies identified some of its components but a full characterization of complex E on a standard substrate had not been done. Here, our objective was to isolate the E complex and characterize its components in detail. We carried out two independent immunopurifications with antibodies to U1A and PRPF40A and identified their protein composition using MS. A comparison of the isolates shows that most proteins are common to both purifications and therefore are likely to represent true components of the E complex. In addition to the known factors, we have identified several novel components. One of these is the SMN complex, which we show might be required for assembly. Analysis of the complexes formed in the SMN-depleted extracts showed that U2 snRNP is no longer associated with pre-mRNA. Since U2 snRNP is essential for the E complex formation, our data suggest that the SMN complex plays a role in U2 snRNP interaction within the complex. To confirm this it would be necessary to reconstitute the depleted extract with the SMN complex. However, reconstitution is not yet possible because recombinant proteins of the SMN complex are insoluble. For this reason, we cannot establish whether the entire SMN complex is necessary for spliceosome assembly.
At present the interactions between U2 snRNPs and other factors in the E complex are poorly characterized. Much is known about its interactions in the A complex where, upon conformational changes induced by ATP hydrolysis, the U2 snRNA becomes base paired to the branchpoint sequence and the U2-associated proteins of the SF3A/B complex form an anchoring site on the pre-mRNA. These rearrangements result in stable association of U2 snRNPs in the A complex. The SMN complex is not a component of the A complex and presumably dissociated during the transition. It will be of interest to elucidate the precise interactions of SMN in the E complex in greater detail.
Deficiency in the SMN protein causes spinal muscular atrophy (SMA). SMN and its associated proteins play a major role in snRNP biogenesis, and a recent study of an SMA mouse model detected widespread alterations in the stoichiometry of snRNAs in different tissues (43). The splicing of specific pre-mRNAs was inhibited in the SMA mice, but it was not established whether this arose from the changes in snRNP stoichiometry (43). Similar findings were reported for S. pombe when the expression of SMN was downregulated (39). The possible role of SMN in spliceosome formation may explain such defects in pre-mRNA processing observed in SMN-deficient cells.
Along with the E complex, a novel U2 snRNP-containing complex was selected by the antibody to PRPF40A from HeLa extract. These particles have a sedimentation coefficient of ∼20S. The earliest purification procedure for human U2 snRNPs involved several chromatographic steps and resulted in isolation of 12S and 17S U2 snRNPs, which contain only the most stably bound components. A more recent procedure used an antibody to SF3A2, raised against a peptide (amino acid 444–458). In this case a number of additional proteins were identified as components of U2, including DDX46 (PRP5), but not PRPF40A (35). The antibody to SF3A2 was raised against a peptide (amino acid 444–458) located in a proline-rich region of the protein that forms the interface with PRPF40A (56). Thus, it is possible that the use of this epitope in SF3A2 selects against co-purification with PRPF40A.
The major difference between the new and previously described U2 snRNPs is the association of the SWI/SNF complex proteins with the particles. The SWI/SNF complex interacts with chromatin (50), suggesting a model in which it might serve as an adaptor for U2 snRNP association with chromatin. The same SWI/SNF proteins were identified as components of the E complex in this study. Their recruitment into the spliceosome is likely to occur in a complex with the U2 snRNP. Interestingly, the BRM protein, a component of the SWI/SNF complex, was previously shown to associate with nascent transcripts and affect alternative splicing (51). This suggests that the SWI/SNF complex might be involved in differential recruitment of the U2 snRNP to nascent transcripts.
One of the most conspicuous features of the E complex is the proximity between the 5′- and 3′-SS and the branch sequence (3,52). Therefore, the identification of factors responsible for attaining this conformation is important for understanding E complex formation. PRPF40A has been proposed to mediate interactions between the 5′-SS and branch sequence (53). In S. cerevisiae, Prp40p is a component of the U1 snRNP (36,37,54) and interacts with branchpoint binding protein Msl5 (53). The human homologue of Prp40p, PRPF40A, has also been shown to interact with SF1, a homologue of Msl5 (53,55,56,57). Here, we identified PRPF40A as a component of the U2 snRNPs, but its interacting partners at the 5′-SS are currently unknown. It is possible that the use of peptide antibodies prevented the recovery of U1-PRPF40A complexes. Another proposed candidate for the bridging of SSs is a DEAD box family ATP-dependent RNA helicase, DDX46 (PRP5), which has been proposed to bridge the U1 and U2 snRNPs in the A complex (58). We identified DDX46 in the E complex, where it might also be involved in bridging interactions. Alternatively, SR proteins have been proposed to link the U1 snRNP bound at the 5′-SS with U2AF bound at the 3′-SS (59). Thus, it is plausible that many interactions between components of the E complex contribute to the link between the ends of the intron. The least understood are the interactions of the U2 snRNP in the E complex. Base pairing between the U2 snRNA and pre-mRNA has not been detected in the E complex and moreover, its association does not require a branchpoint sequence (5). Therefore, elucidating the interactions of SMN-associated proteins in the E complex might reveal the means by which U2 snRNPs associated with the complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1 and Supplementary Reference [60].
FUNDING
The Biotechnology and Biological Sciences Research Council (BB/C004272/1). Funding for open access charge: Department of Biochemistry, University of Leicester.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Glenn Morris for most generous advises during revision of the article and the MDA Muscular Dystrophy Association (U.S.A.) Monoclonal Antibody Resource for supplying antibodies to the proteins of the SMN complex. We are most grateful to Ian Eperon and John Schwabe for providing helpful discussions and comments. We thank other colleagues within Department of Biochemistry for advice and suggestions.
REFERENCES
- 1.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Will C, Luhrmann R. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 369–400. [Google Scholar]
- 3.Kent OA, MacMillan AM. Early organization of pre-mRNA during spliceosome assembly. Nat. Struct. Biol. 2002;9:576–581. doi: 10.1038/nsb822. [DOI] [PubMed] [Google Scholar]
- 4.Michaud S, Reed R. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 1993;7:1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- 5.Das R, Zhou Z, Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- 6.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donmez G, Hartmuth K, Luhrmann R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 9.Brow DA. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 10.Behzadnia N, Hartmuth K, Will CL, Luhrmann R. Functional spliceosomal A complexes can be assembled in vitro in the absence of a penta-snRNP. RNA. 2006;12:1738–1746. doi: 10.1261/rna.120606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl Acad. Sci. USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn AN, van Santen MA, Schwienhorst A, Urlaub H, Luhrmann R. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA. 2009;15:153–175. doi: 10.1261/rna.1332609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, Luhrmann R. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 2004;23:2381–2391. doi: 10.1038/sj.emboj.7600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf E, Kastner B, Deckert J, Merz C, Stark H, Luhrmann R. Exon, intron and splice site locations in the spliceosomal B complex. EMBO J. 2009;28:2283–2292. doi: 10.1038/emboj.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 20.Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 2008;15:183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young PJ, Le TT, thi Man N, Burghes AH, Morris GE. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- 23.Zillmann M, Zapp ML, Berget SM. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol. 1988;8:814–821. doi: 10.1128/mcb.8.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarova OV, Makarov EM, Luhrmann R. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J. 2001;20:2553–2563. doi: 10.1093/emboj/20.10.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konarska MM, Sharp PA. Association of U2, U4, U5, and U6 small nuclear ribonucleoproteins in a spliceosome-type complex in absence of precursor RNA. Proc. Natl Acad. Sci. USA. 1988;85:5459–5462. doi: 10.1073/pnas.85.15.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Black DL, Chabot B, Steitz JA. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985;42:737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- 29.Luhrmann R, Stark H. Structural mapping of spliceosomes by electron microscopy. Curr. Opin. Struct. Biol. 2009;19:96–102. doi: 10.1016/j.sbi.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Stark H, Luhrmann R. Cryo-electron microscopy of spliceosomal components. Annu. Rev. Biophys. Biomol. Struct. 2006;35:435–457. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- 31.Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 32.Hall KB, Konarska MM. The 5′ splice site consensus RNA oligonucleotide induces assembly of U2/U4/U5/U6 small nuclear ribonucleoprotein complexes. Proc. Natl Acad. Sci. USA. 1992;89:10969–10973. doi: 10.1073/pnas.89.22.10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez H, Makarova OV, Makarov EM, Morgner N, Muto Y, Krummel DP, Robinson CV. Isoforms of U1-70k control subunit dynamics in the human spliceosomal U1 snRNP. PLoS ONE. 2009;4:e7202. doi: 10.1371/journal.pone.0007202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Behrens SE, Tyc K, Kastner B, Reichelt J, Luhrmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol. Cell. Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Luhrmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kao HY, Siliciano PG. Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol. Cell. Biol. 1996;16:960–967. doi: 10.1128/mcb.16.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neubauer G, Gottschalk A, Fabrizio P, Seraphin B, Luhrmann R, Mann M. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl Acad. Sci. USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanackovic G, Kramer A. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol. Biol. Cell. 2005;16:1366–1377. doi: 10.1091/mbc.E04-11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campion Y, Neel H, Gostan T, Soret J, Bordonne R. Specific splicing defects in S. pombe carrying a degron allele of the Survival of Motor Neuron gene. EMBO J. 2010:1817–1829. doi: 10.1038/emboj.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meister G, Buhler D, Laggerbauer B, Zobawa M, Lottspeich F, Fischer U. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet. 2000;9:1977–1986. doi: 10.1093/hmg/9.13.1977. [DOI] [PubMed] [Google Scholar]
- 41.Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 2001;20:5443–5452. doi: 10.1093/emboj/20.19.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baccon J, Pellizzoni L, Rappsilber J, Mann M, Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 2002;277:31957–31962. doi: 10.1074/jbc.M203478200. [DOI] [PubMed] [Google Scholar]
- 45.Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- 48.Otter S, Grimmler M, Neuenkirchen N, Chari A, Sickmann A, Fischer U. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 2007;282:5825–5833. doi: 10.1074/jbc.M608528200. [DOI] [PubMed] [Google Scholar]
- 49.Pellizzoni L, Baccon J, Rappsilber J, Mann M, Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- 50.Kofler M, Schuemann M, Merz C, Kosslick D, Schlundt A, Tannert A, Schaefer M, Luhrmann R, Krause E, Freund C. Proline-rich sequence recognition I: marking GYF and WW domain assembly sites in early spliceosomal complexes. Mol. Cell. Proteomics. 2009;8:2461–2473. doi: 10.1074/mcp.M900191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 53.Donmez G, Hartmuth K, Kastner B, Will CL, Luhrmann R. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol. Cell. 2007;25:399–411. doi: 10.1016/j.molcel.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 54.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 55.Ester C, Uetz P. The FF domains of yeast U1 snRNP protein Prp40 mediate interactions with Luc7 and Snu71. BMC Biochem. 2008;9:29. doi: 10.1186/1471-2091-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bedford MT, Chan DC, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin KT, Lu RM, Tarn WY. The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol. Cell. Biol. 2004;24:9176–9185. doi: 10.1128/MCB.24.20.9176-9185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu YZ, Newnham CM, Kameoka S, Huang T, Konarska MM, Query CC. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 2004;23:376–385. doi: 10.1038/sj.emboj.7600050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 60.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.