Abstract
TRF1 and TRF2 are key proteins in human telomeres, which, despite their similarities, have different behaviors upon DNA binding. Previous work has shown that unlike TRF1, TRF2 condenses telomeric, thus creating consequential negative torsion on the adjacent DNA, a property that is thought to lead to the stimulation of single-strand invasion and was proposed to favor telomeric DNA looping. In this report, we show that these activities, originating from the central TRFH domain of TRF2, are also displayed by the TRFH domain of TRF1 but are repressed in the full-length protein by the presence of an acidic domain at the N-terminus. Strikingly, a similar repression is observed on TRF2 through the binding of a TERRA-like RNA molecule to the N-terminus of TRF2. Phylogenetic and biochemical studies suggest that the N-terminal domains of TRF proteins originate from a gradual extension of the coding sequences of a duplicated ancestral gene with a consequential progressive alteration of the biochemical properties of these proteins. Overall, these data suggest that the N-termini of TRF1 and TRF2 have evolved to finely regulate their ability to condense DNA.
INTRODUCTION
Telomeres are specialized structures protecting the natural termini of linear chromosomes from degradation and illicit repair (1). They are assembled through associations between telomeric DNA, a TTAGGG repeat containing sequence that ends with a single stranded 3′ overhang and telomere-specific proteins. The transcription of telomeric DNA produces a UUAGGG repeat containing RNA, TERRA, which is anticipated to play fundamental roles in telomere biology (2).
Several protein complexes associate with telomeric DNA. Among these, the shelterin complex in mammals involves six proteins (TRF1, TRF2, RAP1, TIN2, TPP1 and POT1) (3,4). Binding of this complex to DNA is mediated by the two double-stranded DNA (dsDNA)-binding proteins TRF1 (5,6) and TRF2 (7,8) and the G-tail binding protein POT1 (9). The other three proteins do not interact directly with DNA (10–14). Other complexes containing fewer members of the shelterin complex have also been described (15). Telomeres have the capacity to fold into t-loops, lasso-like structures that have been observed on purified telomeres of diverse origins (16–20). In vitro, formation of t-loops depends on TRF2 and has been proposed to involve the cis-oriented invasion of the telomeric dsDNA by the G tail (17,21). One interesting concept that emerged from recent biochemical studies on TRF2 is that its biological functions seem closely driven by its intrinsic properties. Its capacity to bind telomeric DNA ends (22,23) has been implicated in the inhibition of the non-homologous end-joining pathway (24). Its N-terminal domain binds the center of Holliday junctions, protecting them from resolvase cleavage (25), a property possibly explaining TRF2's ability to protect the t-loops from resolution. TRF2 has also been shown to stimulate the invasion of a telomeric single-strand inside a homologous duplex in free DNA (26) and in the context of the nucleosomal fiber (27), a process thought to participate in the formation of t-loops and chromatin loops. This stimulation was proposed to result from a change in topology generated by the condensation of the DNA. This latter property was shown to primarily involve the C-terminal Telobox/Myb-like and the homodimerization TRFH domains of the protein (26). Despite the close resemblance between these domains in TRF1 and TRF2, TRF1 exhibits a very different behavior towards DNA. In this study, we investigated this paradox and show that TRF1 and TRF2 are not as different as hitherto assumed. Through studies of different mutants and chimeras of these proteins, we uncover the important regulatory role played by the N-termini in the biochemical properties of these proteins.
MATERIALS AND METHODS
Plasmids and oligonucleotides
pTelo2 and PLTelo are both pUC19-based plasmid containing 650 bp of human telomeric repeats.
Proteins
Cloning of protein genes are described in Supplementary Data. TRF proteins and mutants were: TRF2 (3-500), TRF2ΔB (47-500), TRF2ΔD (3-44, 244-500),TRF2ΔBΔD (244-500), TRF2ΔL (3-248, 445-500), TRF2ΔDTRF1D (TRF2 3-44 TRF1 65-264 TRF2 244-500), TRF1 (2-439), TRF2hAΔB (hTRF1 2-67 TRF2 47-500), TRF2mAΔB (mTRF12-63 TRF2 47-500), TRF2cAΔB (cTRF1 2-27 TRF2 47-500) and TRF1ΔA (65-248). All proteins were fused to a 6-histidines tag and produced as published, a gel filtration chromatography step was added when necessary (26). A Coomassie blue-stained sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) gel of the wild type and mutant proteins is shown in Supplementary Figure S1A. Un-tagged TRF2 protein was also used in a control topology experiment (see below). In that case the tagged protein was cleaved using Tabacco Etch virus Protease I, the Histidine tag removed by Ni-column chromatography followed by two chromatographic steps (heparin and gel filtration).
Strand Invasion assay, electrophoretic mobility shift assay and topology assays
Strand invasion assay, electrophoretic mobility shift assay (EMSA) and topology assays were performed as published (26). A control topology using un-tagged TRF2 protein was performed to control the absence of effect of the Histidine Tag (Supplementary Figure S1B).
Surface plasmon resonance (SPR)-based invasion assay
The experiments were performed on a Biacore T100 (GE Healthcare) using C1 sensorchips (GE Healthcare). Binding of the T15G probe and the R100 control (a non-telomeric random 100 bases oligonucleotide, Supplementary Data) was performed via a streptavidin–biotin interaction in conditions recommended. A signal of 150 RU was achieved for both the captured telomeric (T15G) and control (R100) oligonucleotides. pTelo2 (50 nM) and proteins (200 nM) were pre-incubated in HBS-ET buffer (10 mM HEPES pH 7.4, 300 mM NaCl, 3 mM EDTA, 0.05% Tween 20) during 15 min at room temperature. Samples were injected at 10 µl/min and the washing step performed using HBS-ET buffer. Surfaces were regenerated by sequential injections of water, 1 M NaCl, and 0.1% SDS during 30 s. Each sensorgram was corrected by subtraction of the signal obtained from the control flow cell (R100).
Atomic force microscopy imaging
DNA was incubated with proteins for 20 min at 25°C in 5 mM HEPES pH 7.4, 150 mM KCl. The protein/DNA molar ratios used: 10/10 for TRF1, TRF2, TRF2hAΔB and TRF2ΔL; 3/6 for TRF1ΔA and TRF2ΔB; 6/9 for TRF2mAΔB; 10/9 for TRF2cAΔB. Samples were crosslinked with glutaraldehyde (0.1% final) for 30 min on ice and applied on freshly cleaved mica surfaces treated with 10 mM MgCl2. After 2 min, mica was washed with deionized water and dried. Imaging was performed on a Nanoscope IIIa equipped with E-scanner (Digital Instruments Inc., Santa Barbara, CA, USA), in air under Tapping Mode using silicon tips. Images were recorded at 1.5–2.0 Hz over scan areas 1 μm wide (512 × 512 pixels). Raw scanning force microscopy (SFM) images were flattened using the manufacturer's software and converted into TIF files. Contour lengths (CLs) were measured by the read-through length method using SigmaScan Pro software (SPSS Inc., Chicago, IL, USA). Volumes calculated as half-ellipsoids as published (26). Between 150 and 300 objects were scored for each condition. Detailed information on the construction of the 2D probability density maps are given in Supplementary Data.
Phylogenetic studies
Pblast
The Telobox sequence from the human TRF1 and TRF2 was blasted against the NCBI protein database. The resulting alignment was used to generate the PhyML tree. We downloaded the protein families defined in the Ensembl database version 56 (as of September 2009) (www.ensembl.org/) ENSFM00250000004074 and ENSFM00250000007334 and also added sequences from the NCBI database (www.ncbi.nlm.nih.gov/). All alignments were manually verified using SeaView to exclude redundant and improperly annotated sequences (28). We downloaded cDNAs from NCBI and also checked ESTs data. The best alignment carried out using ClustalW and Muscle (29,30) was considered and manually refined. All nucleotidic sequences retrieved were translated before our phylogenetic analyses.
RESULTS
The acidic domain of TRF1 inhibits its ability to condense DNA
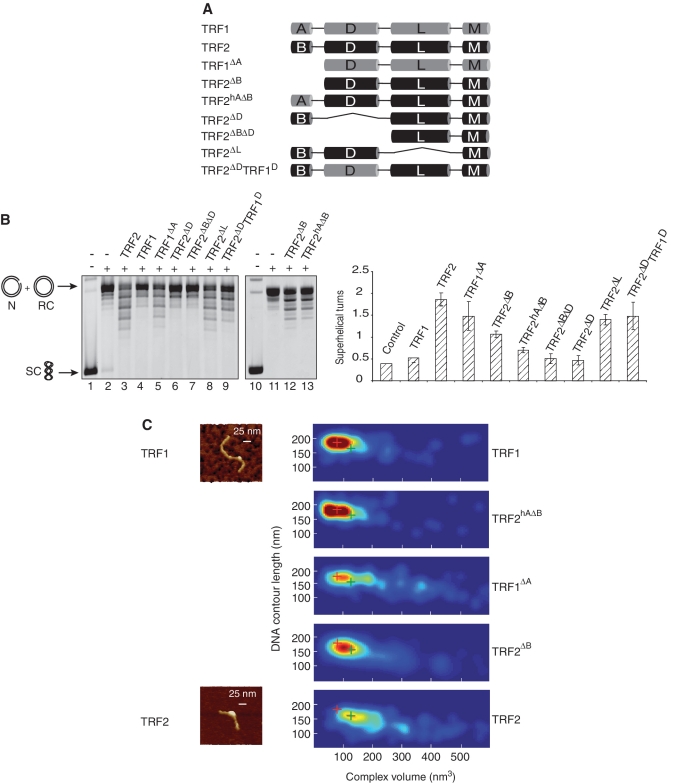
The TRFH domain of TRF2 (here called D domain, Figure 1A) plays a critical role in the ability of TRF2 to condense DNA and to stimulate telomeric invasion (26). In view of the structural homology between these D domains in TRF1 and TRF2, it was therefore surprising to observe that TRF1 inefficiently condensed DNA. Two hypotheses could explain this difference: (i) the TRF1 and TRF2 D domains could be functionally different or (ii) the D domain of TRF1 could also be capable of DNA condensation but this property is inhibited in the full-length protein. To investigate this question, we have constructed several mutants of TRF2 (Figure 1A, Supplementary Figure S1A) and analyzed their ability to condense DNA using a topology assay and atomic force microscopy (AFM) (26). The former assay is based on the analysis of the topology of DNA by gel electrophoresis after incubation with the protein in the presence of wheat germ topoisomerase I. This enzyme can remove the supercoils, located in the unbound part of a closed DNA molecule, generated by the binding on this DNA of a topologically active protein. Modification of DNA topology caused by this type of proteins can be visualized through the appearance of topoisomers on a gel (Figure 1B). This experiment allows the measurement of the average number of supercoils created (Figure 1B, right) and these turns can be characterized as being positive or negative by using chloroquine in the experiment. Indeed, this drug increases the rate of migration of positively supercoiled DNA and conversely decreases this rate for negatively supercoiled DNA compared to controls (Supplementary Figure S1C). AFM allows the direct visualization of the DNA–protein complexes and the measure of both the contour length (CL) of the DNA and the volume of the complexes (Figure 1C). From these numbers, color-coded 2D-probability density maps were drawn showing the probability (p(x,y)) of a given complex to have a volume x and a y DNA CL. As shown in Figure 1C, most of the TRF1 complexes exhibit a small volume and a long DNA CL, implying a lack of DNA condensation. This is confirmed by the topology assay showing that TRF1 inefficiently creates supercoils in DNA (Figure 1B). In contrast, TRF2 causes a significant decrease in DNA CL (Figure 1C), thus showing DNA condensation. TRF2 creates positive supercoils (Figure 1B and Supplementary Figure S1) that can also be observed by AFM imaging of the DNA resulting from the topology assay (Supplementary Figure S2). Comparison of the different mutants schematically shown in Figure 1A reveals several key points: (i) deleting the acidic domain of TRF1 (TRF1ΔA) greatly increases TRF1's ability to condense DNA, thus transforming it into a TRF2ΔB-like protein; (ii) adding the A domain of TRF1 on TRF2ΔB (TRF2hAΔB) does the reverse, transforming TRF2 in a TRF1-like protein; (iii) the linker or hinge domain that separates the D domain and the Myb-like domains has little role in this function (TRF2ΔL); (iv) the B domain of TRF2 seems to stimulate TRF2 ability to modify DNA topology (TRF2ΔB); (v) the TRFH domain of TRF2 is required (TRF2ΔD, TRF2ΔBΔD); and (vi) the TRFH domains of TRF1 and TRF2 seem interchangeable, since the chimeric protein TRF2ΔDTRF1D is very efficient in modifying DNA topology.
Figure 1.
The acidic domain of TRF1 prevents DNA condensation and also inhibits the modification of DNA topology. (A) Schematic representation of the TRF1 and TRF2 wild type and mutant proteins. (B) Topology assay performed with the telomeric plasmid pLTelo and 1 µM of each protein using wheat germ topoisomerase I (WG Topo I). Samples were analyzed by TBE agarose gel electrophoresis. SC stands for supercoiled plasmid, RC for relaxed circular, N for nicked. The plot on the right panel shows the average number of supercoils created by the proteins calculated from gels such as the one shown in (B). Error bars show standard deviation. (C) 2D-probability density maps of volumes and DNA contour length (CL) measured by AFM for the complexes formed with TRF1, TRF2hAΔB, TRF1ΔA, TRF2ΔB and TRF2. The red and green crosses indicate the position in the graph of the majority of TRF1 and TRF2 complexes, respectively.
In summary, both TRF1 and TRF2 have the ability to condense DNA, but the N-terminal acidic domain of TRF1 prevents this condensation.
TRF2 dramatically increases the rate of telomeric strand invasion
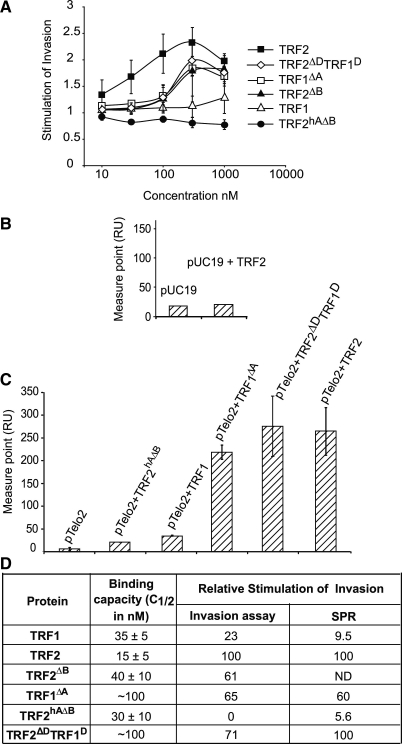
As a consequence of this change of DNA topology, TRF2 was shown to increase the invasion of telomeric double strand by an homologous single-stranded probe (26). In order to verify that the acidic domain could also be responsible for the lower efficiency of TRF1 in telomeric invasion, we analyzed the invasion activity using the pTelo2 telomeric plasmid and a single-stranded probe containing 15 TTAGGG telomeric repeats (T15G) with two different assays: an invasion assay based on gel electrophoresis thus measuring the association at steady state; an assay using SPR technology to monitor the same association in real-time. In the latter experiments, we measured the changes in refractive index (in arbitrary units RU) due to the binding of the pTelo2 plasmid on the T15G single-stranded probe immobilized on the chip (31). An SPR sensorgram presents two phases (Supplementary Figure S3A): injection, when the analyte is continuously injected and washing to follow dissociation. First, the samples were injected until the signal reached a plateau. With pTelo2 alone the response was very weak (RU max around 10) and reached a plateau in about 11 h. However, in the presence of TRF2, the signal reached the plateau in about 15 min for a response 60 times higher. This suggests that TRF2 dramatically increases the association rate. Then, we performed comparative analysis between TRF1 and TRF2 on different substrates. Samples were injected for only 3 min and the measure performed 1 min after the injection stop (measure point in Supplementary Figure S3B). Results are presented as histograms of the corresponding measure points. The response obtained in the presence of TRF2 was 44 times higher than the one obtained with pTelo2 alone and therefore corresponding to a stimulation of invasion of 44-fold. This increase could not be due to the binding of TRF2 on T15G since no change in SPR response could be seen when using pUC19 (Figure 2B). The dissociation phase was slow, suggesting that the major effect of the protein was on the association rate. TRF1 had a much lower effect than TRF2 (<10%, Supplementary Figure S3C and G). Gel-based assays performed with the same proteins (Supplementary Figure S3D, E and G) give the same trend. As expected, the effect measured for TRF2 was higher in SPR since, in this experiment, even unstable complexes would be scored. As seen in the invasion assay (26), the TRF2 effect was more potent on a closed supercoiled plasmid than on a linear molecule (Supplementary Figure S3F), showing the importance of topological constraints. Overall, data obtained with the two methods are in good agreement which suggests that both experiments monitor the same phenomenon. Furthermore, we can say that the main effect of TRF2 on telomeric invasion is to greatly increase the association rate of the double-stranded target with the invading single strand.
Figure 2.
TRF1 can stimulate invasion but is prevented to do so by its N-terminal acidic domain. (A) Invasion assays performed with supercoiled pTelo2 and increasing concentrations (10, 30, 100, 300 nM and 1 µM) of TRF1, TRF2, TRF2ΔDTRF1D, TRF1ΔA, TRF2ΔB and TRF2hAΔB. (B) Histogram of the SPR values obtained after injection of a control plasmid without telomeric repeats (pUC19, 50 nM) with or without pre-incubation with TRF2 (200 nM). (C) Histogram of the SPR values obtained after injection of supercoiled pTelo2 (50 nM) with or without pre-incubation with TRF1, TRF2, TRF2ΔDTRF1D, TRF1ΔA and TRF2hAΔB (200 nM). Error bars show standard deviation. (D) Summary table showing, for each protein, the concentration necessary for binding half the quantity of a double-stranded DNA in a standard EMSA (binding capacity), the relative stimulation of invasion calculated at maximum effect and the relative increase in SPR response both using TRF2 as reference.
The acidic domain of TRF1 inhibits its ability to promote strand invasion
Data obtained for a large panel of mutants clearly show a remarkable correlation between the ability to condense and to promote strand invasion (Figure 2). Proteins inefficient in condensing DNA (TRF1, TRF2hAΔB) were also poor in stimulating invasion in both gel-based and SPR assays. Conversely, efficient proteins in DNA condensation (TRF2, TRF1ΔA, TRF2ΔB and TRF2ΔDTRF1D) were found to be active in strand invasion. In summary, we clearly establish that TRF proteins have the inherent capacity of condensing DNA, modifying DNA topology and stimulating invasion but this intrinsic property is nearly lost in TRF1 through the presence of an acidic domain at its N-terminus while in TRF2 it is stimulated by the presence of a basic domain. This prompted us to investigate how these domains evolved.
Evolution of the N-terminal domains of TRF1 and TRF2 in the vertebrate's lineage
As telomeric DNA is highly conserved, we began our analysis by blasting (Pblast) the human Telobox DNA-binding domain against Metazoans. As expected (5,7), we found that TRF Telobox sequences form a distinct monophyletic group (Supplementary Figure S4). Moreover, it is possible to distinguish a signature differentiating TRF1 from TRF2 (Supplementary Figure S5). Studies of the available genomes of Prochordates, the Urochordates Ciona intestinalis and Ciona savignyi, and the Cephalochordate Amphioxus, Branchiostoma floridae, reveal a single copy of the telobox with sequences highly similar to that of the two human TRFs. Of note, the genome of the lamprey (Petromyzon marinus), one of the most basal organisms that split off from the Gnathostomes about 540 MYA ago, shows the two forms of telobox. Altogether, these results are in favor of a duplication of a unique ancestral gene during one of the two rounds of genome duplications (32) that occurred early in the Chordate lineage.
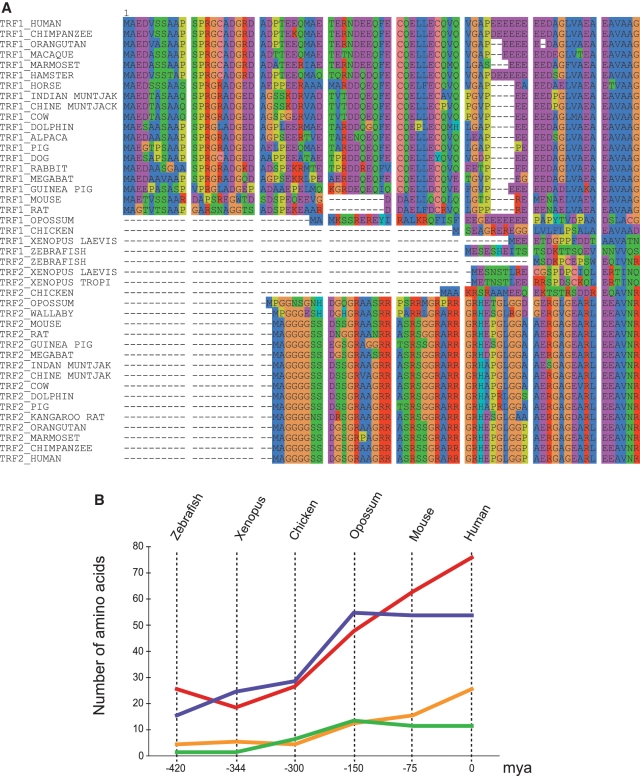
Outside Eutherians, only a few N-terminal sequences are available (Figure 3A, accession numbers in Supplementary Figure S6). However, some key species allow the plotting of a plausible scenario for the evolution of these domains. The TRF2 B domain is very short in zebrafish (16 residues), and increases in Xenopus, and even further in chicken. The gray short-tailed opossum has a B domain very similar to that of the placental mammals. For TRF1, the Xenopus sequences are shorter than the zebrafish domain (19 residues), but show a high proportion of acidic residues. The chicken has a longer domain (27 residues), with several acidic residues. The opossum has also a long domain (48 residues) but the Eutherians have the longest acidic domains (from 63 to 76 residues) which are also the richest in acidic residues. Thus, the evolution leading to the Eutherians seems to correlate with a gradual increase in both length and acidic/basic residues composition of the N-terminus of TRF proteins (Figure 3B).
Figure 3.
Evolutionary analyses of TRF genes. (A) Alignment of the N-terminal sequences of TRF proteins in different species. The N-terminal domains were defined by alignment of the protein sequences through their TRFH domain. (B) Curves showing the variations of the length in amino acids of the N-terminal domain of TRF1 (red) or TRF2 (blue) and of the number of acidic and basic residues in these domains (TRF1 in orange and TRF2 in green) as a function of the time since speciation on the lineage leading to the Eutherians.
Acquisition of these domains does not seem to result from exon addition but rather an extension of the first TRF exon by a continuous gain of the 5′ non-coding sequence, since: (i) we observe a gradual increase in the length and (ii) base composition of the 5′UTR is strikingly very similar to that of the corresponding A/B domains. In the vicinity of the human terf1 and terf2 genes, we calculated a striking GC content of 0.639 (5′UTR) and 0.677 (A domain) in TRF1, and 0.708 (5′UTR) and 0.793 (B domain) in TRF2. This suggests that the GC content of the 5′UTR is correlated to that of the A/B domain. Moreover, GC-rich nucleotidic sequences, when incorporated into a coding sequence, tend to yield larger numbers of charged residues (particularly D, E, R, S and G, which are hallmarks of the N-terminal domains of TRF1 and TRF2).
Altogether, pending on sequences available so far, the A/B domains of TRF proteins seem to originate from a gradual gain of amino acids by successive and iterative incorporation of the 5′UTR in the coding sequence all along the lineage leading to the Eutherians.
Importance of the length of the N-terminal acidic domain of TRF1
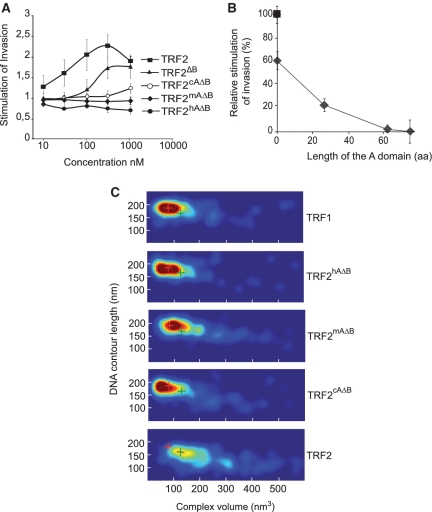
To test the consequences of the evolution of the N-terminal domains on the biochemical properties of TRF1 and TRF2, we analyzed the capacity of the acidic domain from mouse and chicken to inhibit strand invasion and DNA condensation (Figure 4). We fused these N-termini to TRF2ΔB (TRF2mAΔB and TRF2cAΔB for mouse and chicken sequences, respectively) and characterized these chimeric proteins. Mouse (63 residues) and chicken domains (27 residues) are shorter than the human one (76 residues), but have similar isoelectric points (3.65, 3.73 and 4.4 for human, mouse and chicken, respectively).
Figure 4.
Importance of the length of the N-terminal acidic domain of TRF1. (A) Invasion assays performed with supercoiled pTelo2 and increasing concentrations (10, 30, 100, 300 nM and 1 µM) of TRF2, TRF2ΔB, TRF2hAΔB, TRF2mAΔB and TRF2cAΔB. (B) Variations of the relative stimulation of invasion at maximum as a function of the length of the acidic N-terminal domain. (C) 2D-probability density maps of volumes and DNA CLs measured by AFM for the complexes formed with TRF1, TRF2, TRF2cAΔB, TRF2mAΔB and TRF2hAΔB. The red and green crosses indicate the position in the graph of the majority of TRF1 and TRF2 complexes, respectively.
Invasion assays (Figure 4A and B) and AFM (Figure 4C) experiments show a similar trend and suggest a correlation between the length of the acidic domain and the ability to inhibit DNA condensation and to stimulate invasion. The longer the A domain, the less strand invasion and DNA condensation was observed. Our results indicate that evolution of TRF1 proteins might have been associated with a progressive inhibition of their capacity to condense DNA and to stimulate telomeric invasion.
Telomeric RNA negatively regulates TRF2-mediated DNA condensation
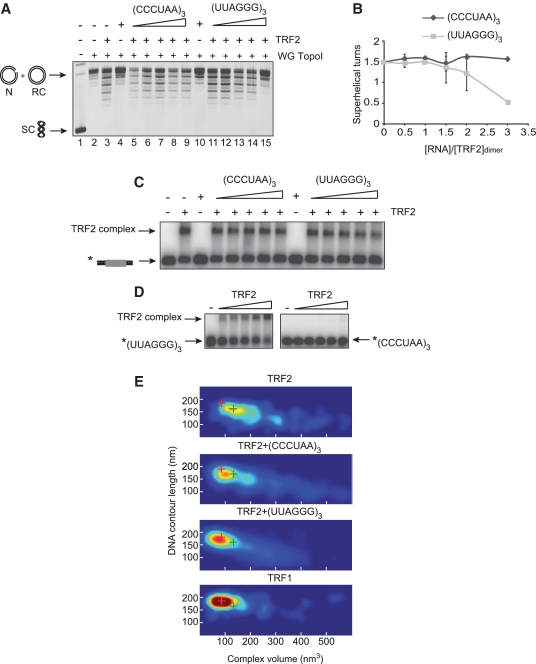
The binding of molecules to the N-termini of TRF1 and TRF2 is expected to have an effect on their behavior regarding strand invasion and DNA condensation. For TRF2, one possible candidate is TERRA, the telomeric RNA that was recently proposed to interact with the N-terminal basic domain of TRF2 (33). To study this, we performed topoisomerase I assays and AFM experiments in the presence of a G-RNA containing three (UUAGGG) repeats and a C-RNA bearing the (CCCUAA)3 sequence. As seen in Figure 5A and B, while the C-RNA had little effect on TRF2-mediated modification of topology, the G-RNA greatly reduced the ability of TRF2 to condense DNA. This was not due to a reduction in the binding of duplex DNA since no important changes in the amount of DNA bound by EMSA were observed in the presence of either RNA (Figure 5C). In accordance with published results (33), direct binding of TRF2 on these RNAs (Figure 5D) showed that TRF2 strongly prefers the G-RNA to the C-RNA, which could explain their differential effect. Of note, a faint band of C-RNA-TRF2 complex is visible at the highest concentration of protein, suggesting that recognition of the C-RNA by TRF2 is possible. AFM experiments show that adding G-RNA to TRF2 prior to DNA binding decreases its capacity to form condensed complexes. TRF2 complexes strikingly resemble those obtained for TRF1 (Figure 5E). The C-RNA has a much weaker but not negligible effect probably due to the residual binding mentioned above.
Figure 5.
Telomeric RNA negatively regulates TRF2-mediated DNA condensation. (A) Topology assay performed using the telomeric plasmid pLTelo and 1 µM of TRF2 in the presence of wheat germ topoisomerase I (WG Topo I) and increasing amounts of either G- or C-RNA (0.5, 1, 1.5, 2 and 3 µM). SC stands for supercoiled plasmid, RC for relaxed circular, N for nicked. (B) Plot of data in (A) showing the variations of the average number of supercoils as a function of the ratio between the concentration of dimers of TRF2 and the concentration of G-RNA (light gray curve) or C-RNA (dark gray curve). Error bars correspond to standard deviation. (C) EMSA using 5 nM of a double-stranded telomeric probe and 20 nM of TRF2 in the presence of increasing amounts of G- and C- RNA (10, 20, 30, 40 and 60 nM). (D) EMSA using 5 nM of labeled G- and C-RNA and increasing amounts of TRF2 (10, 30, 50, 100 and 300 nM). (E) 2D-probability density maps of volumes and DNA CLs measured by AFM for complexes formed with TRF1 and TRF2 in the presence of a ratio of 3 G- and C-RNA per TRF2 dimer. The red and green crosses indicate the position in the graph of the majority of TRF1 and TRF2 complexes, respectively.
Collectively, these experiments show that the binding of G-RNA on TRF2 severely hinders its capacity to condense DNA and to modify DNA topology.
DISCUSSION
Our phylogenetic studies on the terf1 and terf2 genes suggest that they originated from the duplication of an ancestral gene and that their N-termini gradually increased in size through iterative incorporation of sequences from their 5′UTR. These additions had profound consequences on the behavior of these proteins. They created binding sites for several telomere-interacting molecules (3) and conferred to TRF2 the ability to bind Holliday junctions and to protect them from resolvase cleavage (25). Here, we reveal that these domains also make important contributions to the ability of TRF1 and TRF2 to condense telomeric DNA and to stimulate telomeric invasion. Indeed, we show that TRF1 inefficiency in modifying topology and stimulating invasion can be attributed to the acidic nature of its N-terminus. Conversely, the basic nature of the N-terminus of TRF2 causes an increase in these capacities for TRF2. Analyzing the acidic domains of TRF1 proteins from chicken, mouse and human, we have found that the length rather than the pI seems important, indicating that the functional divergence between TRF1 and TRF2 may have gradually increased all along the Chordate lineage.
TRF2-mediated DNA condensation leads to the untwisting/unwrithing of the surrounding constrained DNA which is thought to stimulate telomeric invasion, a reaction involved in the folding of telomeric DNA into t-loop (26). Our data strongly reinforce this view since we observe a striking correlation between the capacity of TRF2 and its mutants to condense DNA, to modify DNA topology and to stimulate invasion. Furthermore, we have uncovered that stimulation of telomeric invasion is mediated by a striking increase in the kinetics of the invasion reaction (steady state is reached in 15 min in the presence of TRF2, 11 h in its absence).
Overall, these data suggest a role for TRF2 in the regulation of DNA topology on telomeres. In accordance, a recent study (34) shows that TRF2 acts together with topoisomerase 2 to protect telomeric DNA from replicative DNA damage. One major future challenge will be to elucidate the mechanism and the precise role of TRF2 in this process.
One interesting idea raised by our work is that modification(s) of the N-terminal domains of TRF1 and TRF2 might result in a TRF1 protein exhibiting TRF2-like behavior or vice versa. To our knowledge, modifications of TRF1 N-terminal domain have not been reported so far. However, one cannot exclude the possibility of a regulation through the binding of a protein partner. A tankyrase-binding motif exists in most of the TRF1 N-termini from mammals but tankyrases stand as poor candidates since mouse and rat TRF1s lack this motif and their ultimate role is the inhibition of TRF1 DNA binding (3). In contrast, the N-terminal basic domain of TRF2 is the target of several binding partners and, at least, one post-translational modification. Arginines 17 and 18 in this domain were shown to be methylated (35). Proteins such as the Werner protein as well as ORC1 were shown to recognize this domain (36,37). Similarly, the telomeric RNA, TERRA, was recently proposed to interact with the N-terminus of TRF2 (33). TERRA is a very attractive candidate for regulating the functions of the N-terminus of TRF2 since it could both mask the positive charges of this domain and repulse DNA. To investigate this hypothesis we performed AFM experiments and topology assays in the presence of small RNA molecules containing telomeric repeats. The effect of the G-rich small RNA was striking with a total abrogation of the capacity of TRF2 to condense DNA (Figure 5). Although these RNA molecules are far shorter than the natural TERRA, which can reach 9 kb, and lack the sub-telomeric sequences present in TERRA, it is tempting to speculate that TERRA may impact TRF2 behavior concerning DNA topology and even t-loop formation. One could imagine that TERRA could guide TRF2 complexes from a DNA-condensation/t-loop proficient type of complex to a more TRF1/shelterin proficient type. It follows that TERRA could impede t-loop formation, an event that could be deleterious to the cell but may also be a necessary step during replication. In accordance, TERRA has been proposed to regulate some aspects of telomere replication (33). TERRA could even be involved in the removal of the t-loop. Indeed, the N-terminal domain of TRF2 has been implicated in the protection of the t-loop (38) and Holliday junctions (25) against resolution events. One could speculate that TERRA binding could alleviate this protection and cause t-loop HR. In accordance with this hypothesis, it has been observed that depletion in Nonsense-mediated decay (NMD) proteins that increase the number of TERRA foci in human cells causes sudden telomere loss and a marked increase in telomeric fragments (39). This would make TERRA a crucial factor in regulating telomeric folding and state.
This work also raises the question of why different N-termini have evolved for TRF1 and TRF2 during Chordate evolution. We show here that these domains regulate the ability of TRF to condense telomeric DNA. Strikingly, the N-terminal part of TRF2 might facilitate heterochromatin formation by binding both ORC1 and TERRA (33), suggesting that the N-terminal parts of the TRF proteins are crucial to regulate telomeric chromatin condensation both through intrinsic properties and interactions with chromatin components. This is expected to be crucial for proper organization and dynamics of telomeric chromatin during the cell cycle, but it could also lead to long-range chromatin interactions between telomeres and non-telomeric chromosomal loci. Recent studies from our group and that of Zhou Songyang (40,41) have revealed the presence of TRF2 outside telomeres, specifically, on centromeric/pericentromeric satellite DNA and insterstitial telomeric sequences. Often located in the proximity of genes or within introns, these TRF2 binding sites may be involved in the regulation of the corresponding genes and thus participate in the cell transcriptional program. In view of the present results, it is tempting to speculate that the acquisition of specialized N-termini by TRF proteins during the evolution of chordates may have thus contributed to the establishment of new transcriptional regulatory programs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Information, Supplementary Figures 1–6.
FUNDING
Ligue Nationale contre le Cancer (équipe labellisée) and an ANR grant (Teloloop ANR 2010 BLAN 1512 01). A. P. and S. P. were funded by fellowships from the Ligue Contre le Cancer de Haute Savoie and the Cenci Bolognetti Pasteur Institute Foundation from Rome, Italy, respectively. Funding for open access charge: INCA program TELOFUN (number CB 000000/029661).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
A.P., S.P., C.F.-M., Z.H.-T., F.B., B. P., Y.-V. L. B., M.-H. L. D. and S.A. performed research. Y.T., F.M., N.H., F.A. and A.C. contributed reagents or analytic tools. E.G. and M.-J.G.-P. designed experiments and wrote the article.
REFERENCES
- 1.Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 2.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 4.Xin H, Liu D, Songyang Z. The telosome/shelterin complex and its functions. Genome Biol. 2008;9:232. doi: 10.1186/gb-2008-9-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996;24:1294–1303. doi: 10.1093/nar/24.7.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 7.Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 8.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 9.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 10.Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 11.Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 13.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 14.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc. Natl Acad. Sci. USA. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesare AJ, Quinney N, Willcox S, Subramanian D, Griffith JD. Telomere looping in P. sativum (common garden pea) Plant J. 2003;36:271–279. doi: 10.1046/j.1365-313x.2003.01882.x. [DOI] [PubMed] [Google Scholar]
- 17.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 18.Munoz-Jordan JL, Cross GA, de Lange T, Griffith JD. t-loops at trypanosome telomeres. EMBO J. 2001;20:579–588. doi: 10.1093/emboj/20.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikitina T, Woodcock CL. Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 2004;166:161–165. doi: 10.1083/jcb.200403118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomaska L, Makhov AM, Griffith JD, Nosek J. t-Loops in yeast mitochondria. Mitochondrion. 2002;1:455–459. doi: 10.1016/s1567-7249(02)00009-0. [DOI] [PubMed] [Google Scholar]
- 21.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3' telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanez GH, Khan SJ, Locovei AM, Pedroso IM, Fletcher TM. DNA structure-dependent recruitment of telomeric proteins to single-stranded/double-stranded DNA junctions. Biochem. Biophys. Res. Commun. 2005;328:49–56. doi: 10.1016/j.bbrc.2004.12.134. [DOI] [PubMed] [Google Scholar]
- 23.Yoshimura SH, Maruyama H, Ishikawa F, Ohki R, Takeyasu K. Molecular mechanisms of DNA end-loop formation by TRF2. Genes Cells. 2004;9:205–218. doi: 10.1111/j.1356-9597.2004.00719.x. [DOI] [PubMed] [Google Scholar]
- 24.Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 25.Poulet A, Buisson R, Faivre-Moskalenko C, Koelblen M, Amiard S, Montel F, Cuesta-Lopez S, Bornet O, Guerlesquin F, Godet T, et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, et al. A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 27.Baker AM, Fu Q, Hayward W, Victoria S, Pedroso IM, Lindsay SM, Fletcher TM. The telomere binding protein TRF2 induces chromatin compaction. PLoS One. 2011;6:e19124. doi: 10.1371/journal.pone.0019124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 31.Piliarik M, Vaisocherova H, Homola J. Surface plasmon resonance biosensing. Methods Mol. Biol. 2009;503:65–88. doi: 10.1007/978-1-60327-567-5_5. [DOI] [PubMed] [Google Scholar]
- 32.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Léger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, et al. TRF2 and Apollo cooperate with Topoisomerase 2alpha to protect human telomere from replicative damage. Cell. 2010;142:230–242. doi: 10.1016/j.cell.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell TR, Glenfield K, Jeyanthan K, Zhu XD. Arginine methylation regulates telomere length and stability. Mol. Cell. Biol. 2009;29:4918–4934. doi: 10.1128/MCB.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng Z, Dheekollu J, Broccoli D, Dutta A, Lieberman PM. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr. Biol. 2007;17:1989–1995. doi: 10.1016/j.cub.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 37.Li B, Jog SP, Reddy S, Comai L. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2DeltaB-mediated telomere shortening. Mol. Cell. Biol. 2008;28:1892–1904. doi: 10.1128/MCB.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 40.Simonet T, Zaragosi LE, Philippe C, Lebrigand K, Schouteden C, Augereau A, Bauwens S, Ye J, Santagostino M, Giulotto E, et al. The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res. 2011;21:1028–1038. doi: 10.1038/cr.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011;21:1013–1027. doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.