Abstract
For more than half a century, researchers have studied the basic biology of Adenovirus (Ad), unraveling the subtle, yet profound, interactions between the virus and the host. These studies have uncovered previously unknown proteins and pathways crucial for normal cell function that the virus manipulates to achieve optimal virus replication and gene expression. In the infecting virion, the viral DNA is tightly condensed in a virally encoded protamine-like protein which must be remodeled within the first few hours of infection to allow for efficient expression of virus-encoded genes and subsequent viral DNA replication. This review discusses our current knowledge of Ad DNA–protein complex within the infected cell nucleus, the cellular proteins the virus utilizes to achieve chromatinization, and how this event contributes to efficient gene expression and progression of the virus life cycle.
INTRODUCTION
Human Adenovirus (Ad) was first isolated from adenoid tissue in the 1950s as novel viral agents associated with respiratory infections (1,2). Over 100 Ad family members have been identified and characterized in a wide range of host organisms, from a variety of mammals and birds, to reptiles and amphibians (3). In the early 1960s, researchers showed that some human Ads can cause tumours in rodents (4,5), which led to a surge in studies of the molecular biology, genetics and physiology of Ads which continues to this day. Since Ads must manipulate the host cell to promote a microenvironment conducive to virus replication, studies of basic Ad biology have contributed a great deal of novel insight into all fields of cellular biology, including DNA replication, tumourigenesis and control of gene expression in the host cell.
While the pool of knowledge regarding the Ad lifecycle is immense, few studies have investigated the structure and protein association of Ad DNA within the infected cell nucleus. Considering the fundamental importance of chromatin in regulating gene expression in host cells, it is surprising that, until recently, it remained unclear whether Ad DNA interacted with cellular histones or assembled into chromatin. This review summarizes our current knowledge of the nucleoprotein structure of the Ad genome within the infected cell.
ADENOVIRUS BIOLOGY
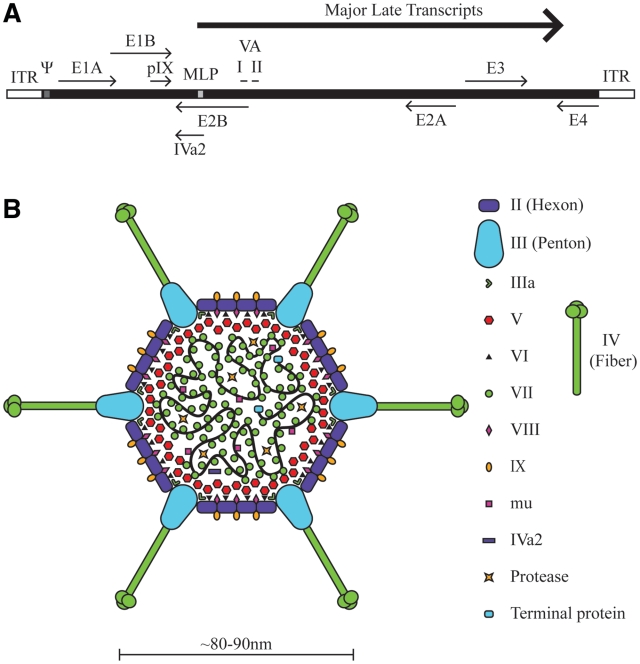
All Ads have the same general structural characteristics. The virion is a non-enveloped icosahedral capsid with a diameter of ∼80–90 nm, containing a linear double stranded DNA genome of ∼30–40 kb (Figure 1) (3). Of the human Ads, serotype 2 (Ad2) and 5 (Ad5), both of subclass C, are the most extensively characterized. The Ad5 genome is ∼36 kb in size and encodes ∼39 genes, which are classified as either early or late depending on whether they are expressed before or after DNA replication (Figure 1A) (6). Four early transcription units (E1a, E1b, E3 and E4) encode proteins that are required for transactivating other viral regions, modifying the host cellular environment or altering the immune response. E2 encodes proteins directly involved in viral DNA replication. All major late proteins, organized in the transcription units L1 to L5, are expressed from a common major late promoter and are generated by alternative splicing of a single transcript. However, recent work has shown that the L4-22K and L4-33K proteins, which are themselves involved in regulation of the major late promoter, are initially expressed from a novel promoter (7). The late transcripts generally encode virion structural proteins. Four other small late transcripts are also produced: protein IX (pIX, encoding a minor structural protein), IVa2 (encodes a protein involved in encapsidating the viral DNA into the immature virion) and VA RNA I and II (the RNA itself blocks activation of the interferon response). Inverted terminal repeats (ITR) of ∼100 bp flank both ends of the viral DNA and contain the origins of replication. Directly adjacent to the left ITR is the viral packaging sequence (∼150 bp). The genome organization is relatively conserved through all Ad species.
Figure 1.
Schematic of the adenovirus genome and virion. (A) A simplified map of the Ad5 genome showing the early genes (E1–E4) and the region from which the major late transcript is produced (the extensively spliced L1–L5 transcripts produced from alternative splicing of the major late transcript are not shown). The relative position of pIX, VA RNA I and II and IVa2 are indicated. Also shown are the viral inverted terminal repeats (ITR) located at each end of the genome, the viral packaging element (Ψ) located adjacent to the left ITR, and the position of the major late promoter (MLP). Please note that these features are not drawn to scale. (B) Model of the Ad5 virion, adapted from (9), with modifications based on additional data provided by (8,10,11).
The Ad5 capsid is composed of three major (II, III and IV) and five minor (IIIa, IVa2, VI, VIII and IX) polypeptides (Figure 1B) (8–11). The facets are composed primarily of hexons (trimers of protein II) with pentons (five molecules of protein III) capping each vertex. The latter is the base from which extends fibre (trimer of protein IV), the distinctive projections at the Ad capsid vertices. Within the capsid, the viral DNA is associated with three highly basic proteins, core proteins VII, V and Mu (µ) (12–14). Protein VII is a protamine-like protein and is responsible for wrapping and condensing the viral DNA (15). The protein VII-DNA nucleoprotein complex is organized into a central dense core with 12 large spherical nucleoprotein projections, termed adenosomes, which extend into each vertex (16,17). A shell of protein V is thought to coat the protein VII-DNA complex (16,18). Protein V is believed to make contact with the outer capsid in several different ways; protein V interacts directly with penton, and indirectly with peripentonal hexon and the remainder of hexon bridged through IIIa and protein VI, respectively (10,11,19–22). Mu is synthesized as a 79 amino acid precursor protein, pre-Mu, which is cleaved by the Ad-encoded proteinase to its final 19 amino acid, highly basic mature form (23). Pre-Mu is speculated to interact with and aid in tightly condensing the viral DNA within the capsid, and cleavage of pre-mu may serve to partially relax this structure prior to its entry into the nucleus (24). Although the viral DNA does not interact directly with the major capsid proteins (10,25,26), the DNA still appears to contribute to the physical stability to the virion; packaging of subgenomic sized DNA [<90% of the wild-type (wt) genome length] results in virions that are less stable than wtAd (27,28).
Many of the details of Ad5 infection of cells have been elucidated. Initially, the Ad fibre protein binds to the Coxsackie-Adenovirus receptor (CAR), which is the primary receptor for both Ad5 and Coxsackie B virus (29,30). This binding is followed by a secondary interaction between Ad penton and αvβ3 or αvβ5 integrins (31). Recent studies have shown that Ad5 can enter cells using heparin sulfate proteoglycans as an alternative receptor, either through direct binding to the Ad fibre shaft (32), or bridged through interaction of Ad with blood factors such as factor IX, factor X or complement component C4-binding protein (33–35). Ad is internalized by receptor-mediated endocytosis and evades degradation by escaping from the early endosome (36). The virion is transported through the cytoplasm to the nucleus along the microtubule network (36), and the capsid is slowly disassembled en route (37). Upon reaching the nuclear pore complex, the protein VII-wrapped Ad DNA enters the nucleus (14,37,38), while the rest of the capsid remains at the nuclear membrane and is subsequently degraded. Viral DNA replication and assembly of progeny virions occur entirely within the nucleus of infected cells. The life cycle takes 24–36 h, although the time for completion of the lifecycle is slightly extended in primary cells. A single cell infected with a single virus can produce ∼104 daughter virions.
EARLY EVENTS WITHIN THE INFECTED CELL NUCLEUS
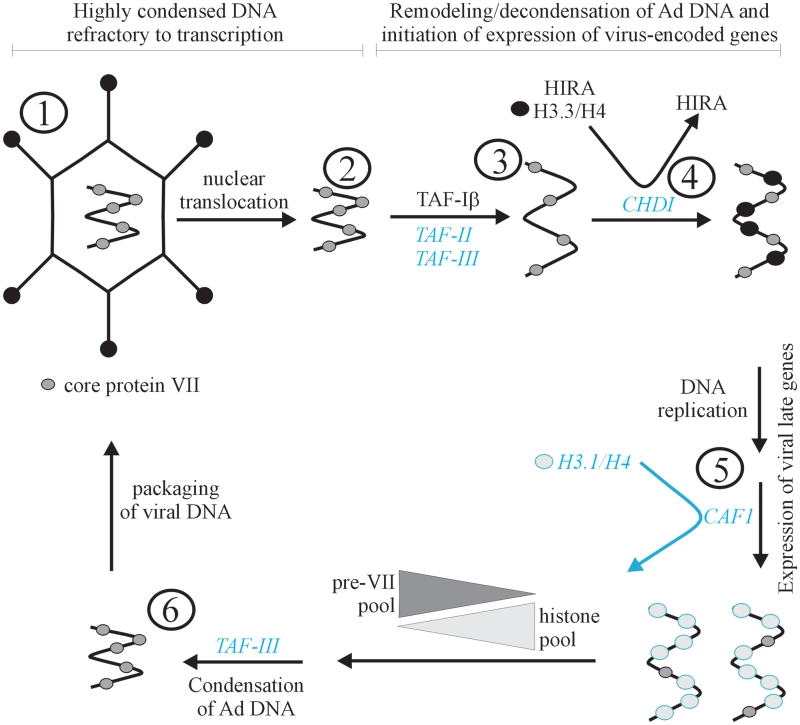
An overview of our current understanding of Ad DNA chromatin state in the infected cell is shown in Figure 2. Although a number of Ad capsid proteins reach the nucleus, it is only the protein VII-wrapped DNA that enters the nucleus (14). Histone H1 (H1) escorts the Ad DNA–protein complex through the nuclear pore (39); however, this function appears to be independent of any structural role for H1 on the viral DNA. During this phase of infection, protein VII protects the viral DNA from activating the DNA damage response (40). The ultimate fate of protein VII after entry into the nucleus is currently a topic of debate. Some studies suggest that protein VII stably associates with Ad DNA throughout the early phase of infection (14,41), while other groups suggest that VII is removed gradually from at least certain regions of the genome during this same time period (42,43). Other reports have shown that the overall level of VII within the infected cell declines rapidly within the first few hours of infection with a concomitant decline in VII association with viral genomes (44). Whether the eviction of protein VII requires active transcription of the Ad genome is also in dispute (41,44,45).
Figure 2.
Adenovirus DNA chromatin structure throughout infection. In black—elements that we know. In blue and italics—speculation/areas of future research. DNA in the Ad capsid is highly condensed with core protein VII (1), along with protein V and µ (not shown). The protein VII-DNA complex transits to the nucleus (2), and undergoes remodeling to decondense the core before transcription of early genes can begin (3). Remodeling may involve loss of at least some VII. At the time when viral gene expression is first detected, histones can be found bound to the viral DNA along with VII. Histone variant H3.3 is preferentially deposited on the viral DNA, through the action of HIRA and CHD1 (4). Onset of viral DNA replication may be accompanied by a shift to deposition of H3.1 by the CAF1 complex on the DNA, as is observed for HSV1 (5). As the histone pool is depleted and the intracellular levels of newly synthesized pre-protein VII increase, there is a transition of Ad DNA association from nucleosomes to pre-protein VII, possibly mediated by TAF-III (6). The viral DNA condensed with pre-protein VII is packaged into the Ad capsid (1).
Cell-free systems developed to study Ad DNA replication have shown that the compacted nature of the VII-wrapped viral DNA allows for only limited transcription and DNA replication (46,47). This observation suggests that the core/DNA structure must be remodeled to allow these processes to proceed with greater efficiency. Three cellular proteins have been identified that can remodel the Ad core in these cell-free systems: template activating factor Iβ (TAF-Iβ) [also known as SET (46)], TAF-II [NAP-1 (48)] and TAF-III [B23/nucleophosmin (49)]. Using the cell-free system, all three TAF's were shown to stimulate replication from the Ad core, while TAF-Iβ and TAF-II were also shown to enhance transcription. TAF-1β, the best characterized TAF in the context of Ad core remodeling, forms a tertiary complex with the VII-wrapped DNA (41,50,51), which results in increased accessibility of the viral DNA to nucleases and restriction enzymes and, presumably, transcriptional activators (47). It is not clear if increased accessibility was due to actual removal or only shifting of VII on the DNA template. siRNA-mediated knockdown of TAF-Iβ in infected cells delayed virus gene expression, DNA replication and virus yield (42), although the effect was relatively modest. Knockdown of TAF-Iβ did not affect the binding level of protein VII on viral DNA as assessed by chromatin immunoprecipitation (ChIP), at least at 4 hpi (43). Thus, additional proteins are likely involved in preparing the Ad core for efficient gene expression and DNA replication within the infected cell nucleus.
Several lines of evidence suggest that VII remains associated with Ad DNA during the early stage of infection. First, VII can be cross-linked to the viral DNA at virtually all stages of infection (52). Second, based on immunofluorescence analysis, foci of VII can be observed in the nucleus of infected cells, which represent the VII-wrapped viral DNA (40,41,45,53). Third, ChIP studies have shown directly that VII is bound to the viral DNA up to at least ∼10 hpi (41,43,44,51,54). There is some disagreement in these studies regarding the level of VII association over time; some studies indicate that VII association is constant and does not change throughout the early stage of infection (41,45,52), while others suggest a gradual (or more rapid) decline in VII association with the viral DNA during this time period (40,43,44,51). Based on ChIP experiments by Komatsu et al. (43), it appears that the degree and timing of VII association with the viral genome during the early phase of infection can vary depending on the region of the genome that is analysed. For example, between 1 and 10 hpi, VII remains stably associated with the late-gene hexon coding region, but shows declining association over time with the major late promoter (43). This observation somewhat rationalizes the previous disparate studies. In plasmid-based in vitro assays, addition of small amounts of protein VII with the DNA actually enhanced transcription over naked DNA, suggesting that small quantities of protein VII may function in part to keep repressive histone/chromatin features from forming on certain promoter regulatory elements (43). Taken together, these results suggest that dynamic regulation of protein VII is necessary for optimal viral growth; sufficient protein VII must be removed or remodeled to decondense the viral DNA–nucleoprotein complex to allow access to the transcription machinery, but some protein VII must remain to stimulate transcription.
It is unclear whether transcription through the Ad DNA template is required for removal or remodeling of VII. Inhibition of transcription has been correlated with prolonged retention of VII on the Ad genome (40,45), although in other studies inhibition of transcription or transcription elongation did not affect loss of VII (43,44). Although it has been suggested that de novo expressed E1A (the first viral gene product expressed within the infected cell) associates with protein VII and is involved in stimulating transcription on the viral genome which subsequently strips VII from the viral DNA (45,54), this function is likely not completely necessary since VII is still removed in the absence of E1A or active transcription (43,44). It is possible that other proteins within the cell can perform this function in the absence of E1A; indeed, E1-deleted Ad can complete a full replication cycle in certain cell types, although the time required to complete the replication cycle is extended (55), suggesting that compensating proteins may exist.
AD DNA ASSOCIATES WITH CELLULAR HISTONES IN THE INFECTED CELL NUCLEUS
In eukaryotic cells, the basic unit of chromatin is the nucleosome, with 147 bp of DNA wrapped around a histone octamer, composed of two sets of H2A–H2B and H3–H4 dimers. The notion that nucleosomes are simply ‘beads on a string’ has been challenged by the realization that histones and nucleosomes play key roles in gene regulation (56). The post-translationally modified N-terminal tails of histones serve as docking/recognition sites for other regulatory proteins (57), providing the epigenetic information governing gene expression, as dictated by the ‘histone code’ (58).
Conflicting data from the 1980s suggested that Ad DNA is or is not associated with cellular histones within the infected cell (59–63). With the development of more sensitive techniques, the subject of the Ad nucleoprotein structure within the infected cell has been revisited recently. Based on ChIP analysis, Ad and its derivative vectors (either E1-deleted, replication defective Ad or helper-dependent Ad [hdAd—devoid of all Ad protein coding sequences (64)]) do interact with cellular histones within a few hours of infection (43,44,65). Histones can be detected on the Ad DNA as early as 1-h post-infection, and ChIP/re-ChIP experiments show that both protein VII and histones can be found associated with the same DNA molecule in the cell (43). Since the histones almost certainly bind directly to the Ad DNA, at least some VII must be removed from the viral DNA at these time points to allow for binding of histones. The mechanism by which VII is removed, and the cellular protein(s) involved in this process, have yet to be determined.
Within the cell, deposition of cellular histones can occur either through a replication-coupled or replication-independent mechanism, and there are specific histone variants and chaperones associated with each mechanism (66). Histone variant H3.1 is expressed exclusively during S-phase and is deposited on de novo synthesized DNA by the Chromatin Assembly Factor I (CAF1) complex in what is considered a replication-coupled mechanism (67). In contrast, the replacement histone variant H3.3, which differs from H3.1 by only five amino acids, is expressed at all phases of the cell cycle, and is deposited through a replication-independent mechanism (66). H3.3 is deposited on actively transcribed genes by the histone chaperone HIRA, or on specific regions of the chromosome [such as pericentric DNA repeats and on telomeres (68–70)] by the H3.3 chaperone DAXX (68). The H3.3 variant is also deposited on incoming male pro-nuclear DNA shortly after fertilization utilizing the histone chaperone HIRA (71). Although TAF-Iβ can act as a chaperone to transfer histones to DNA templates (47), it does not appear to perform this function during Ad infection (43).
As Ad can infect both dividing and non-dividing cells (and only induces cell cycle progression after viral gene expression has initiated), it suggests that Ad DNA is likely to be chromatinized by exploiting a mechanism independent of DNA replication. Chromatin immunoprecipitation (ChIP) experiments have recently demonstrated hdAd and E1-deleted Ad (44), and wtAd (our unpublished data) DNA preferentially associates with H3 variant H3.3 as early as 4 hpi, suggesting that chromatinization does indeed occur by a replication-independent mechanism. A preferential association with H3.3 was also found with Herpes Simplex Virus 1 (HSV1) DNA during the early phase of infection (72). siRNA-mediated knockdown of HIRA reduced the total association of H3 with the hdAd and HSV1 DNA, as well as reducing expression of virally encoded genes for both viruses, suggesting that chromatinization was necessary for efficient expression. The involvement of the H3.3 chaperone HIRA, and not DAXX, is consistent with the observation that DAXX is actively degraded during normal Ad infection (73). As deposition of H3.3 on Ad (44) and on HSV1 (72) was dependent on HIRA, it suggests a common mechanism for deposition of histones on the genomes of invading dsDNA nuclear viruses. Moreover, the similarity between the chromatinization of sperm DNA and nuclear virus DNA suggests that both use a similar pathway to achieve chromatinization in the absence of cellular DNA replication. In vitro observations suggest that histone chaperones, such as HIRA, either do not assemble nucleosomes or assemble them at a greatly reduced rate in the absence of ATP-utilizing factors (74). In the male pronucleus, HIRA is necessary for delivery of H3.3 to the site of nucleosome formation (71,75), but it is the ATP-dependent chromatin remodeling complex CHD1 that is required for H3.3 deposition (74). In HeLa extracts, HIRA interacts directly with CHD1 (74). Thus, it is likely CHD1 that is directly involved in deposition of histones on the Ad DNA, although this has yet to be formally proven.
Work by Komatsu et al. (43) showed that wtAd can be found associated with all members of the nucleosome, H2A–H2B and H3–H4, as early as 1 hpi which, together with studies showing Ad DNA in the nucleus is wrapped in a repeating ∼200 bp structure, suggests that the DNA may be wrapped in complete, physiologically spaced nucleosomes (44,59–62,76). The chaperone responsible for deposition of H2A/H2B is unknown. It has been estimated that up to 40% of infecting wtAd DNA is contained in nucleosomes at 3 hpi, and all regions of the genome are represented in micrococcal nuclease-protected fractions (59). The observation that both protein VII and histones can be found bound to the same viral DNA molecule at the same time suggests that the viral chromatin may not completely resemble that of the host cell (43). Interestingly, HSV1 genomic DNA associates with regularly spaced nucleosomes in a latent infection, but the spacing becomes ‘unstable’ during a lytic infection and generates heterogeneously sized fragments upon MNase digestion (77). Whether wtAd assembles into stable or unstable chromatin during a productive infection remains to be determined. Electron microscopy analysis of viral genomes isolated during late time points of infection (16–18 hpi) showed irregularly spaced nucleosome-like particles at approximately one-tenth the density of cellular chromatin in HeLa cells [3 versus 26 nucleosomes per µm of DNA, respectively (61)]. However, it is not clear whether this is due to ‘unstable’ chromatin or the limited quantities of histones that are available late in Ad infection. During the replicative phase of the HSV1 lifecycle, there is a switch from early association with H3.3 to deposition of H3.1 (72). Whether a similar phenomenon occurs with wtAd has yet to be determined. The observation that Ad-induced shut-off of host protein synthesis results in a reduction in histone gene expression (78,79) suggests that the virus may simply switch from association of nucleosomes containing H3.3 to re-deposition of pre-protein VII in preparation for DNA packaging into the viral capsid at the final stage of the virus lifecycle (discussed below).
THE IMPORTANCE OF AD DNA CHROMATINIZATION
Since Ad DNA is chromatinized, this suggests that epigenetic regulation may be as important for expression of Ad-encoded genes as is it for expression of genes encoded by the host cell. ChIP analysis showed that there was an increase over time in the level of association of acetylated H3 at all Ad promoters tested (43). Since acetylated histones are commonly associated with actively transcribed genes, it suggests that as these promoters become active, they adopt an epigenetic status similar to cellular genes, which may aid in recruiting appropriate co-factors for optimal gene expression. Interestingly, the cell also uses an epigenetic approach in an attempt to down-regulate expression from some foreign, invading DNAs, including Ad. Indeed, DAXX can act as an anti-Ad defense factor and down-regulate gene expression during wtAd infection (73,80). Thus, Ad-induced degradation of DAXX at late times during infection may be a mechanism that the virus uses to evade down-regulation of its expressed genes (73). A similar phenomenon occurs for vectors based on Ad (65), and this can affect vector function in vitro and in vivo (81). In these latter studies, the vector chromatin was preferentially associated with deacetylated histones, which is a marker of transcriptionally inactive chromatin (65); thus the DNA was ‘marked’ to reduce expression of vector-encoded genes. These observations clearly illustrate the ongoing battle between host and pathogen, and the importance of epigenetic regulation of viral DNA at the chromatin level.
LATE STAGE PROTEIN VII REPLACEMENT
During the final stage of virus replication, the viral DNA must be condensed once again into the compact structure required for packaging within the viral capsid. The histones must therefore be displaced from the Ad DNA and replaced with pre-protein VII, the precursor of the mature protein VII [the N-terminus of pre-protein VII is cleaved by the viral-encoded protease during virion maturation (3)]. Little is known about how this switch occurs.
In eukaryotic cells, expression and synthesis of new histones is tightly regulated to coincide with cellular DNA replication (82). Interestingly, however, there is a dramatic decline in histone synthesis at late times during Ad infection (78,79). This puts forth the hypothesis that at the late stage of infection, the decline of available histones relative to the increased levels of pre-protein VII leads, by default, to the deposition of pre-VII on the newly synthesized viral DNA (60,62,83). Experiments in cell free systems have shown that simply mixing Ad DNA and purified pre-protein VII leads to the formation of an insoluble complex, suggesting that a specific cellular chaperone(s) mediates the placement of pre-protein VII on Ad DNA (84). Based on co-immunoprecipitation studies using extracts from infected cells, TAF-III/nucleophosmin was shown to have a greater affinity for pre-protein VII than the mature protein VII, suggesting that TAF-III may be involved in placing pre-protein VII on the viral DNA (85). In a cell free system, TAF-III was able to transfer pre-VII onto DNA, suggesting that TAF-III is indeed a pre-VII chaperone. However, additional studies are required to further support the role for TAF-III as a chaperone during normal Ad infection of a cell.
CONCLUSION
Insight into the mechanism of Ad chromatinization has the potential to impact three specific areas of research. First, Ad is widely used as a gene delivery system for basic studies and gene therapy applications (64), and improved understanding of the parameters that aid in establishing gene expression within the host cell will improve vector efficacy and safety. Second, Ad is a significant and often overlooked human pathogen (86), and understanding early events in the cell that permit expression of viral genes may lead to the identification of new therapeutic targets to limit or prevent wtAd-induced morbidity and mortality. Finally, numerous studies of basic aspects of Ad biology have contributed significantly to our understanding of how the host cell works (3). Undoubtedly, delineation of the proteins and pathways involved in Ad DNA chromatinization will also improve our understanding of this process within the host cell.
FUNDING
Canadian Institutes of Health Research and Cancer Research Society (CIHR) (Canada) (grants for research in the Parks laboratory) and Ontario Graduate Scholarships in Science and Technology (OGSST) from the Ontario Government (to A.N.G. and A.R.D.). Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Drs F. Jeffrey Dilworth, David J. Picketts and Rashmi Kothary (Ottawa Hospital Research Institute) for critical evaluation of the manuscript and helpful discussion.
REFERENCES
- 1.Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953;84:570–573. doi: 10.3181/00379727-84-20714. [DOI] [PubMed] [Google Scholar]
- 2.Hilleman MR, Werner JH. Recovery of new agents from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 1954;85:183–188. doi: 10.3181/00379727-85-20825. [DOI] [PubMed] [Google Scholar]
- 3.Berk AJ. Adenoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th edn. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 2355–2394. [Google Scholar]
- 4.Trentin JJ, Yabe Y, Taylor G. The quest for human cancer viruses. Science. 1962;137:835–841. doi: 10.1126/science.137.3533.835. [DOI] [PubMed] [Google Scholar]
- 5.Yabe Y, Trentin JJ, Taylor G. Cancer induction in hamsters by human type 12 adenovirus. Effect of age and of virus dose. Proc. Soc. Exp. Biol. Med. 1962;111:343–344. doi: 10.3181/00379727-111-27786. [DOI] [PubMed] [Google Scholar]
- 6.Davison AJ, Benko M, Harrach B. Genetic content and evolution of adenoviruses. J. Gen. Virol. 2003;84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 7.Morris SJ, Scott GE, Leppard KN. Adenovirus late-phase infection is controlled by a novel L4 promoter. J. Virol. 2010;84:7096–7104. doi: 10.1128/JVI.00107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen JB, Byrd SA, Walker AK, Strahler JR, Andrews PC, Imperiale MJ. Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J. Virol. 2008;82:9086–9093. doi: 10.1128/JVI.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell WC. Adenoviruses: update on structure and function. J. Gen. Virol. 2009;90:1–20. doi: 10.1099/vir.0.003087-0. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. Crystal structure of human adenovirus at 3.5 A resolution. Science. 2010;329:1071–1075. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maizel JV, Jr, White DO, Scharff MD. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology. 1968;36:126–136. doi: 10.1016/0042-6822(68)90122-0. [DOI] [PubMed] [Google Scholar]
- 13.Russell WC, Laver WG, Sanderson PJ. Internal components of adenovirus. Nature. 1968;219:1127–1130. doi: 10.1038/2191127a0. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee PK, Vayda ME, Flint SJ. Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J. Mol. Biol. 1986;188:23–37. doi: 10.1016/0022-2836(86)90477-8. [DOI] [PubMed] [Google Scholar]
- 15.Mirza MA, Weber J. Structure of adenovirus chromatin. Biochim. Biophys. Acta. 1982;696:76–86. doi: 10.1016/0167-4781(82)90012-4. [DOI] [PubMed] [Google Scholar]
- 16.Brown DT, Westphal M, Burlingham BT, Winterhoff U, Doerfler W. Structure and composition of the adenovirus type 2 core. J. Virol. 1975;16:366–387. doi: 10.1128/jvi.16.2.366-387.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newcomb WW, Boring JW, Brown JC. Ion etching of human adenovirus 2: structure of the core. J. Virol. 1984;51:52–56. doi: 10.1128/jvi.51.1.52-56.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everitt E, Sundquist B, Pettersson U, Philipson L. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology. 1973;52:130–147. doi: 10.1016/0042-6822(73)90404-2. [DOI] [PubMed] [Google Scholar]
- 19.Everitt E, Lutter L, Philipson L. Structural proteins of adenoviruses. XII. Location and neighbor relationship among proteins of adenovirion type 2 as revealed by enzymatic iodination, immunoprecipitation and chemical cross-linking. Virology. 1975;67:197–208. doi: 10.1016/0042-6822(75)90417-1. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee PK, Vayda ME, Flint SJ. Interactions among the three adenovirus core proteins. J. Virol. 1985;55:379–386. doi: 10.1128/jvi.55.2.379-386.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart PL, Fuller SD, Burnett RM. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews DA, Russell WC. Adenovirus core protein V is delivered by the invading virus to the nucleus of the infected cell and later in infection is associated with nucleoli. J. Gen. Virol. 1998;79(Pt 7):1671–1675. doi: 10.1099/0022-1317-79-7-1671. [DOI] [PubMed] [Google Scholar]
- 23.Anderson CW, Young ME, Flint SJ. Characterization of the adenovirus 2 virion protein, mu. Virology. 1989;172:506–512. doi: 10.1016/0042-6822(89)90193-1. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Berna AJ, Marabini R, Scheres SH, Menendez-Conejero R, Dmitriev IP, Curiel DT, Mangel WF, Flint SJ, San Martin C. Structure and uncoating of immature adenovirus. J. Mol. Biol. 2009;392:547–557. doi: 10.1016/j.jmb.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabry CM, Rosa-Calatrava M, Conway JF, Zubieta C, Cusack S, Ruigrok RW, Schoehn G. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 2005;24:1645–1654. doi: 10.1038/sj.emboj.7600653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silvestry M, Lindert S, Smith JG, Maier O, Wiethoff CM, Nemerow GR, Stewart PL. Cryo-electron microscopy structure of adenovirus type 2 temperature-sensitive mutant 1 reveals insight into the cell entry defect. J. Virol. 2009;83:7375–7383. doi: 10.1128/JVI.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith AC, Poulin KL, Parks RJ. DNA genome size affects the stability of the adenovirus virion. J. Virol. 2009;83:2025–2028. doi: 10.1128/JVI.01644-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy MA, Parks RJ. Adenovirus virion stability and the viral genome: size matters. Mol. Ther. 2009;17:1664–1666. doi: 10.1038/mt.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 30.Tomko RP, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl Acad. Sci. USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 32.Smith TA, Idamakanti N, Rollence ML, Marshall-Neff J, Kim J, Mulgrew K, Nemerow GR, Kaleko M, Stevenson SC. Adenovirus serotype 5 fiber shaft influences in vivo gene transfer in mice. Hum. Gene Ther. 2003;14:777–787. doi: 10.1089/104303403765255165. [DOI] [PubMed] [Google Scholar]
- 33.Shayakhmetov DM, Gaggar A, Ni S, Li ZY, Lieber A. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J. Virol. 2005;79:7478–7491. doi: 10.1128/JVI.79.12.7478-7491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waddington SN, McVey JH, Bhella D, Parker AL, Barker K, Atoda H, Pink R, Buckley SMK, Greig JA, Denby L, et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Kalyuzhniy O, Di Paolo NC, Silvestry M, Hofherr SE, Barry MA, Stewart PL, Shayakhmetov DM. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl Acad. Sci. USA. 2008;105:5483–5488. doi: 10.1073/pnas.0711757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leopold PL, Ferris B, Grinberg I, Worgall S, Hackett NR, Crystal RG. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum. Gene Ther. 1998;9:367–378. doi: 10.1089/hum.1998.9.3-367. [DOI] [PubMed] [Google Scholar]
- 37.Greber UF, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 38.Strunze S, Engelke MF, Wang IH, Puntener D, Boucke K, Schleich S, Way M, Schoenenberger P, Burckhardt CJ, Greber UF. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell. Host Microbe. 2011;10:210–223. doi: 10.1016/j.chom.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 39.Trotman LC, Mosberger N, Fornerod M, Stidwill RP, Greber UF. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell. Biol. 2001;3:1092–1100. doi: 10.1038/ncb1201-1092. [DOI] [PubMed] [Google Scholar]
- 40.Karen KA, Hearing P. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J. Virol. 2011;85:4135–4142. doi: 10.1128/JVI.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue Y, Johnson JS, Ornelles DA, Lieberman J, Engel DA. Adenovirus protein VII functions throughout early phase and interacts with cellular proteins SET and pp32. J. Virol. 2005;79:2474–2483. doi: 10.1128/JVI.79.4.2474-2483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haruki H, Okuwaki M, Miyagishi M, Taira K, Nagata K. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J. Virol. 2006;80:794–801. doi: 10.1128/JVI.80.2.794-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komatsu T, Haruki H, Nagata K. Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res. 2011;39:889–901. doi: 10.1093/nar/gkq783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross PJ, Kennedy MA, Christou C, Risco Quiroz M, Poulin KL, Parks RJ. Assembly of helper-dependent adenovirus DNA into chromatin promotes efficient gene expression. J. Virol. 2011;85:3950–3958. doi: 10.1128/JVI.01787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Morral N, Engel DA. Transcription releases protein VII from adenovirus chromatin. Virology. 2007;369:411–422. doi: 10.1016/j.virol.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 46.Matsumoto K, Nagata K, Ui M, Hanaoka F. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem. 1993;268:10582–10587. [PubMed] [Google Scholar]
- 47.Okuwaki M, Nagata K. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J. Biol. Chem. 1998;273:34511–34518. doi: 10.1074/jbc.273.51.34511. [DOI] [PubMed] [Google Scholar]
- 48.Kawase H, Okuwaki M, Miyaji M, Ohba R, Handa H, Ishimi Y, Fujii-Nakata T, Kikuchi A, Nagata K. NAP-I is a functional homologue of TAF-I that is required for replication and transcription of the adenovirus genome in a chromatin-like structure. Genes Cells. 1996;1:1045–1056. doi: 10.1046/j.1365-2443.1996.d01-223.x. [DOI] [PubMed] [Google Scholar]
- 49.Okuwaki M, Iwamatsu A, Tsujimoto M, Nagata K. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 2001;311:41–55. doi: 10.1006/jmbi.2001.4812. [DOI] [PubMed] [Google Scholar]
- 50.Gyurcsik B, Haruki H, Takahashi T, Mihara H, Nagata K. Binding modes of the precursor of adenovirus major core protein VII to DNA and template activating factor I: implication for the mechanism of remodeling of the adenovirus chromatin. Biochemistry. 2006;45:303–313. doi: 10.1021/bi051248+. [DOI] [PubMed] [Google Scholar]
- 51.Haruki H, Gyurcsik B, Okuwaki M, Nagata K. Ternary complex formation between DNA-adenovirus core protein VII and TAF-Ibeta/SET, an acidic molecular chaperone. FEBS Lett. 2003;555:521–527. doi: 10.1016/s0014-5793(03)01336-x. [DOI] [PubMed] [Google Scholar]
- 52.Chatterjee PK, Vayda ME, Flint SJ. Adenoviral protein VII packages intracellular viral DNA throughout the early phase of infection. EMBO J. 1986;5:1633–1644. doi: 10.1002/j.1460-2075.1986.tb04406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walkiewicz MP, Morral N, Engel DA. Accurate single-day titration of adenovirus vectors based on equivalence of protein VII nuclear dots and infectious particles. J. Virol. Methods. 2009;159:251–258. doi: 10.1016/j.jviromet.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson JS, Osheim YN, Xue Y, Emanuel MR, Lewis PW, Bankovich A, Beyer AL, Engel DA. Adenovirus protein VII condenses DNA, represses transcription, and associates with transcriptional activator E1A. J. Virol. 2004;78:6459–6468. doi: 10.1128/JVI.78.12.6459-6468.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson JE, Kay MA. Persistence of recombinant adenovirus in vivo is not dependent on vector DNA replication. J. Virol. 1997;71:8902–8907. doi: 10.1128/jvi.71.11.8902-8907.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- 57.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 58.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 59.Sergeant A, Tigges MA, Raskas HJ. Nucleosome-like structural subunits of intranuclear parental adenovirus type 2 DNA. J. Virol. 1979;29:888–898. doi: 10.1128/jvi.29.3.888-898.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daniell E, Groff DE, Fedor MJ. Adenovirus chromatin structure at different stages of infection. Mol. Cell. Biol. 1981;1:1094–1105. doi: 10.1128/mcb.1.12.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beyer AL, Bouton AH, Hodge LD, Miller OL., Jr Visualization of the major late R strand transcription unit of adenovirus serotype 2. J. Mol. Biol. 1981;147:269–295. doi: 10.1016/0022-2836(81)90441-1. [DOI] [PubMed] [Google Scholar]
- 62.Dery CV, Toth M, Brown M, Horvath J, Allaire S, Weber JM. The structure of adenovirus chromatin in infected cells. J. Gen. Virol. 1985;66(Pt 12):2671–2684. doi: 10.1099/0022-1317-66-12-2671. [DOI] [PubMed] [Google Scholar]
- 63.Wong ML, Hsu MT. Psoralen-cross-linking study of the organization of intracellular adenovirus nucleoprotein complexes. J. Virol. 1988;62:1227–1234. doi: 10.1128/jvi.62.4.1227-1234.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amalfitano A, Parks RJ. Separating fact from fiction: assessing the potential of modified adenovirus vectors for use in human gene therapy. Curr. Gene Ther. 2002;2:111–133. doi: 10.2174/1566523024605618. [DOI] [PubMed] [Google Scholar]
- 65.Ross PJ, Kennedy MA, Parks RJ. Host cell detection of noncoding stuffer DNA contained in helper-dependent adenovirus vectors leads to epigenetic repression of transgene expression. J. Virol. 2009;83:8409–8417. doi: 10.1128/JVI.00796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 67.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 68.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 72.Placek BJ, Huang J, Kent JR, Dorsey J, Rice L, Fraser NW, Berger SL. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J. Virol. 2009;83:1416–1421. doi: 10.1128/JVI.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schreiner S, Wimmer P, Sirma H, Everett RD, Blanchette P, Groitl P, Dobner T. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 2010;84:7029–7038. doi: 10.1128/JVI.00074-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonnefoy E, Orsi GA, Couble P, Loppin B. The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization. PLoS Genet. 2007;3:1991–2006. doi: 10.1371/journal.pgen.0030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tate VE, Philipson L. Parental adenovirus DNA accumulates in nucleosome-like structures in infected cells. Nucleic Acids Res. 1979;6:2769–2785. doi: 10.1093/nar/6.8.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lacasse JJ, Schang LM. During lytic infections, herpes simplex virus type 1 DNA is in complexes with the properties of unstable nucleosomes. J. Virol. 2009;84:1920–1933. doi: 10.1128/JVI.01934-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hodge LD, Scharff MD. Effect of adenovirus on host cell DNA synthesis in synchronized cells. Virology. 1969;37:554–564. doi: 10.1016/0042-6822(69)90273-6. [DOI] [PubMed] [Google Scholar]
- 79.Tallman G, Akers JE, Burlingham BT, Reeck GR. Histone synthesis is not coupled to the replication of adenovirus DNA. Biochem. Biophys. Res. Commun. 1977;79:815–822. doi: 10.1016/0006-291x(77)91184-6. [DOI] [PubMed] [Google Scholar]
- 80.Ullman AJ, Hearing P. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol. 2008;82:7325–7335. doi: 10.1128/JVI.00723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parks RJ, Bramson JL, Wan Y, Addison CL, Graham FL. Effects of stuffer DNA on transgene expression from helper-dependent adenovirus vectors. J. Virol. 1999;73:8027–8034. doi: 10.1128/jvi.73.10.8027-8034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robbins E, Borun TW. The cytoplasmic synthesis of histones in hela cells and its temporal relationship to DNA replication. Proc. Natl Acad. Sci. USA. 1967;57:409–416. doi: 10.1073/pnas.57.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brown M, Weber J. Virion core-like organization of intranuclear adenovirus chromatin late in infection. Virology. 1980;107:306–310. doi: 10.1016/0042-6822(80)90297-4. [DOI] [PubMed] [Google Scholar]
- 84.Burg JL, Schweitzer J, Daniell E. Introduction of superhelical turns into DNA by adenoviral core proteins and chromatin assembly factors. J. Virol. 1983;46:749–755. doi: 10.1128/jvi.46.3.749-755.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Samad MA, Okuwaki M, Haruki H, Nagata K. Physical and functional interaction between a nucleolar protein nucleophosmin/B23 and adenovirus basic core proteins. FEBS Lett. 2007;581:3283–3288. doi: 10.1016/j.febslet.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 86.Wold WS, Horwitz MS. Adenoviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th edn. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 2396–2436. [Google Scholar]