Abstract
The expressions of chloroplast and mitochondria genes are tightly controlled by numerous nuclear-encoded proteins, mainly at the post-transcriptional level. Recent analyses have identified a large, plant-specific family of pentatricopeptide repeat (PPR) motif-containing proteins that are exclusively involved in RNA metabolism of organelle genes via sequence-specific RNA binding. A tandem array of PPR motifs within the protein is believed to facilitate the RNA interaction, although little is known of the mechanism. Here, we describe the RNA interacting framework of a PPR protein, Arabidopsis HCF152. First, we demonstrated that a Pfam model could be relevant to the PPR motif function. A series of proteins with two PPR motifs showed significant differences in their RNA binding affinities, indicating functional differences among PPR motifs. Mutagenesis and informatics analysis putatively identified five amino acids organizing its RNA binding surface [the 1st, 4th, 8th, 12th and ‘ii’(-2nd) amino acids] and their complex connections. SELEX (Systematic evolution of ligands by exponential enrichment) and nucleobase preference assays determined the nucleobases with high affinity for HCF152 and suggested several characteristic amino acids that may be involved in determining specificity and/or affinity of the PPR/RNA interaction.
INTRODUCTION
Chloroplasts and mitochondria originated from free-living bacterial ancestors (1,2). During evolution, the vast majority of the endosymbionts’ genes were transferred to the nucleus. Current chloroplasts and mitochondria genomes encode only a fraction of the genetic information. Therefore, numerous nuclear encoded factors are imported into the organelles to maintain organelle biogenesis. The nuclear encoded factors either originated from the symbiont, the host nucleus, or are novel factors acquired after endosymbiosis. Consequently, the biochemical and genetic features of plant organelles arose in the context of coordinated co-evolution between the organellar and nuclear genomes. Genome sequencing has revealed the presence of large families of proteins and/or motifs whose functions have not been assigned or validated. Genome sequencing of Arabidopsis thaliana identified one such group, the pentatricopeptide repeat (PPR) motif, comprising a degenerate motif of 35 amino acids (aa), similar to the tetratricopeptide (TPR) repeat (3). The PPR-containing proteins normally have a tandem array of PPR motifs and are found in all eukaryotes (4). All known PPR proteins are nuclear encoded, yet most are predicted to be localized in mitochondria or chloroplasts (5). Their origin is unknown; however, the PPR protein is likely to have been acquired for maintaining the symbiotic organelles.
PPR proteins are particularly expanded in vesicular plants: plant genomes encode nearly 500, whereas animal genomes encode few to several dozens, with the exception of 28 PPRs in trypanosoma (4,6). Many genetic studies found that PPR proteins have essential roles in diverse plant phenomena, such as maintenance of chloroplasts and mitochondria (4), organelle-to-nuclear signaling (7), embryogenesis (8), fertility restoration of cytoplasmic male sterility (9), abiotic stress response (10) and metabolite biosynthesis (11). These PPR proteins are proposed to interact with a single, or a small subset of, specific RNA molecule(s), and affect various aspects of RNA metabolism, including RNA editing (12), splicing (13), cleavage (14), RNA stability (15), translation or some combination of these functions (16). Several PPR proteins have been shown to interact with RNA by in vitro studies (17–20), or by co-immunoprecipitation (21,22), and it is suggested that the PPR motif itself does not catalyze any RNA processing. Alternatively, it is suggested that PPR proteins act as adapters, with the tandem array of PPR motifs facilitating binding to nucleic acids in a sequence-specific manner.
Recently, the structure of a mitochondrial RNA polymerase containing two PPR motifs has been solved (23). The 35-aa PPR motif forms a pair of anti-parallel α-helices. A protein with a long PPR tract is predicted to form consecutive helical hairpins to form a super helical structure. The helical-hairpin model has been experimentally confirmed by circular dichroism spectrum analysis and analytical ultracentrifugation using maize PPR5 (24). Structural prediction suggests that helix A of the PPR motif is located at the concave surface. The inner face of the protein is positively charged, which might provide an interface for interaction with nucleic acids (25). Several hypotheses have been proposed for the RNA interacting residues (3,26,27). However, little is understood about the molecular basis of PPR/RNA interaction, and experimental support is lacking.
Here, we present an initial biochemical analysis of the PPR motif involved in RNA interaction, using an Arabidopsis PPR protein, HCF152. First, we determined the functional criterion of the PPR motif, the definition of which is currently controversial among domain search programs. Experiments using a series of truncated proteins with two PPR motifs showed remarkable differences in RNA binding affinities among PPR motifs. Amino acid substitution and structural modeling identified five aa [the 1st, 4th, 8th 12th and ii (−2nd) aa] putatively forming the RNA interacting surface. We addressed the nucleobase specificity by a SELEX assay of the full-length protein and a binding assay using ribonucleotide homo-polymer and the truncated proteins. The results identified aa that are putatively involved in affinity for RNA and in recognizing specific nucleobases. We also revealed complex connections of the RNA interacting residues between intra- or inter motif(s).
MATERIALS AND METHODS
Production of mini-PPR proteins and mutagenized proteins
Mini-PPR proteins were produced by PCR amplification from the corresponding DNA sequence using the oligonucleotides shown in Supplementary Table S1. The PCR product was inserted into the pBAD/Thio-TOPO vector (Invitrogen, Carlsbad, CA, USA), allowing the protein to be expressed as an N-terminal thioredoxin fusion protein with six histidine residues at the C-terminus. Expression and purification of the mini-PPR proteins and the full-length HCF152 protein (HCF152/F) were performed as described previously (28), and their purities were verified (Supplementary Figure S1). The expression vectors for the mutagenized proteins were prepared as shown in Supplementary Table S1.
Preparation of RNA probes
Preparation of the 32P-labeled Dd120 RNA was performed as described previously (17). Briefly, a PCR fragment containing a T7 promoter sequence and a 120-mer Arabidopsis chloroplast DNA fragment (Dd120) was used to transcribe an (α-32P) UTP-labeled RNA probe. The non-radioactive RNAs for the competitive gel shift assay were produced by T7 Ribomax (Promega, Madison, WI, USA) and appropriate DNA fragments. The ribonucleotide homo-polymer RNA probes (N25; A25, U25, G25 and C25, Supplementary Table S1) were chemically synthesized (Dharmacon, Boulder, CO, USA). A linker sequence was attached at the 5′-end of the homo-nucleotide 25-mer, to normalize the radiolabeling efficiency. The 5′-end-32P-labeled N25 probe was prepared using (γ-32P) ATP and polynucleotide kinase.
Gel shift assay
The gel shift assay was performed as previously described (29). Briefly, various amounts of the recombinant protein were incubated with (α-32P)-labeled Dd120 RNA probe (250 pM) in 20 µl of 10 mM Tris-HCl (pH 8.0), 40 mM KCl, 6 mM MgCl2, 0.05 mM EDTA, 2 mM DTT and 8% glycerol (w/v) at 25°C for 15 min. Samples were then subjected to 10% native polyacrylamide gel electrophoresis (PAGE), using Tris-borate-EDTA (TBE) buffer. Gels were dried and imaged with a FLA-3000 (Fuji Photo Film, Tokyo, Japan). The overall apparent KD value was determined from the concentration of protein at which 50% of the RNA probe bound, as an indication of the RNA binding affinity. A competitive gel shift assay was performed using HCF152/F (100 nM) and the 32P-labeled Dd120 RNA probe (250 pM), with the addition of 160 mM KCl and 0.5 mg/ml heparin.
Structural modeling
The structure model for the Arabidopsis HCF152 protein was automatically constructed using the Phyre program and the full-length sequence of HCF152 protein as the query (30) (http://www.sbg.bio.ic.ac.uk/phyre/). From the built models, a 148-aa region (from aa 376 to 523; eight helixes including three PPR motifs and a PPR motif-like structure), was identified using a template of O-GlucNAc transferase, was used for the analysis. Visualization and evaluation of the structure model was carried out using the Mac Pymol software (http://pymol.org/).
Systematic evolution of ligands by exponential enrichment (SELEX)
We used a strategy based on the original SELEX method of Tuerk and Gold (31). The double-stranded DNA template for the initial RNA pool was obtained by annealing SELEX_B_lig and SELEX_B_25-F (Supplementary Table S1) and an extension oligonucleotide. The DNA was gel-purified and used for transcription of the initial RNA pool, using T7 Ribomax (Promega). The selection was performed by beads or gel selection. Prior to beads selection, the initial RNA pool (2000 pmol) was mixed with Ni-NTA resin (Promega) in the binding buffer [50 mM Tris-HCl (pH 8.0), 40 mM KCl, 10 mM MgCl2, 0.05 mM EDTA, 0.5 mM EGTA] containing 1 mg/ml BSA to remove the RNA species adsorbing to the Ni-NTA resin. Meanwhile, the HCF152/F protein (200 pmol, Protein: RNA = 1:10) (28) was immobilized on 20 µl of Ni-NTA resin. The pre-treated RNA was mixed with the protein-immobilized resin at room temperature for 20 min with gentle flicking, and then washed three times with 1 ml of the binding buffer. The remaining RNA was eluted, together with the protein, by binding buffer containing 200 mM imidazole. Alternatively, for gel-selection, the RNA pool was incubated with the protein in the binding buffer at room temperature for 20 min and subjected to 8% PAGE at 4°C for 45 min (200 mV, 20 mA). After electrophoresis, the gel was excised every 1 cm from the top of the gel, and RNA was extracted in 300 µl of RNA extraction buffer [300 mM NaAc (pH 5.5), 25 mM EDTA, 1% SDS]. RNA associated with the HCF152 protein was extracted from gel sections 1 and 2, whereas free RNA was found in sections 6 and 7 (Supplementary Figure S2C). The amounts of RNA pool, protein and competitor (yeast RNA) were varied in each round of the selection to increase the stringency (Supplementary Figure S2B). The recovered RNA from the beads- or gel-selection was reverse-transcribed to produce cDNA. The cDNA was PCR amplified using the SELEX_B_25-F and -R oligonucleotides. The obtained DNA fragments were used for subsequent selection or cloned into the vector of the Zero Blunt TOPO PCR cloning kit (Invitrogen) to determine the sequence. The consensus motif was analyzed by MEME (http://meme.sdsc.edu/meme/intro.html) with the 21-mer of putative HCF152 binding sequence (17). The sequence logo of the consensus motif was created by WebLogo (http://weblogo.berkeley.edu/). The thioredoxin protein, expressed using empty pBAD/Thio-TOPO vector (Invitrogen), was used as a control protein in the selection.
Filter binding assay (FBA)
The binding reactions of 32P-labeled N25 RNA (250 pM) and proteins (200 nM) were performed as described above in the gel shift assay with the addition of 160 mM KCl and 0.5 mg/ml heparin. Binding reactions were filtered through stacked nitrocellulose (PROTEIN BA85, 0.45 mm; Schleicher & Schuell, Keene, NH, USA) and nylon membranes (Hybond N+; GE Healthcare, Piscataway, NJ, USA) in a slot blot manifold. Slots were washed three times by vacuum filtration with 400 µl of wash buffer (445 mM Tris, 445 mM Boric acid and 1 mM EDTA). The protein–RNA complexes were retained on the nitrocellulose membrane. The RNA that passed through the nitrocellulose was trapped on the nylon membrane underlay. The membranes were dried and analyzed by autoradiography. The ratio of protein–RNA complexes was estimated from the signal intensity on the nitrocellulose membrane against that of both nitrocellulose and nylon membranes.
Statistical analysis
The aa sequences of 5669 Arabidopsis PPR motifs were obtained from Uniprot (IPR002885; http://www.uniprot.org/). According to the IDs of the sequences, the distances between PPR motifs were calculated from their starting positions, and sequences were eliminated that had a distance of >10 between motifs. As a result, 4614 sequences were selected. The intra- and inter-motif connections between the corresponding positions of the aa [1st, 4th, 8th, 12th aa and ‘ii’ (−2nd)] of the forward and behind motifs were estimated by a chi-squared test. This used the actual and theoretical values, which were classified from their aa properties, i.e. hydrophobic (G, A, V, L, I, P, M, F, W), hydrophilic and neutral (S, T, C, N, Q, Y), hydrophilic and acidic (D, E), and hydrophilic and basic (K, R, H).
RESULTS
Two pairs of PPR motifs in the Pfam model confer RNA binding activity
The PPR proteins typically comprise a tandem array of dozens of PPR motifs, which are assumed to provide a sequence-specific RNA binding capacity. A previous study using the HCF152 protein (12 PPR motifs) reported that the full-length protein had high affinity and robust specificity for the target RNA molecules. However, truncated proteins displayed partial RNA binding properties, and at least two PPR motifs were required to detect the RNA binding activity (17).
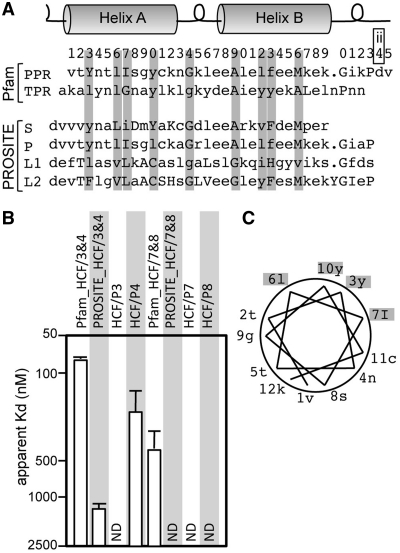
In this study, we aimed to simplify the analysis of the protein–RNA interaction using truncated proteins containing PPR motifs that were as short as possible. However, the definition of the length and start position of a PPR motif is currently controversial among the domain search programs. The PPR motif was originally identified as a 35-aa motif, and later the PPR motif was sub-divided into P (classical PPR; 35 aa), PPR-like S (short; 31 aa), PPR-like L1 (long; 35 aa) and L2 (36 aa) from their sequence characteristics (Figure 1A) (5). The Pfam model defines the 1st aa of the PPR motif as the beginning of helix A (http://Pfam.sanger.ac.uk/, PF01535; Val of PPR in Figure 1A). In contrast, the PROSITE model defines the 1st aa as the loop before helix A (http://expasy.org/prosite/, PS51375; 34th aa for the Pfam, Asp of S to L2 in Figure 1A), to compensate for the length differences among the PPR sub-types (5).
Figure 1.
Amino acid sequence of the PPR motif and its RNA binding activities. (A) The consensus aa sequences for PPR and TPR motifs in the Pfam models (PF01535 and PF00515, respectively). The sequences of PPR subtypes, S (PPR-like short), P (classical PPR), L1 (PPR-like long) and L2, are also shown, whose numbering are coincident with the PROSITE model (PS51375). The helix and loop structure is schematically represented. The number above the sequence indicates the first digit of the position of the PPR motif in the Pfam model. Positions containing conserved aa in either TPR and PPR motifs are shaded in gray. The 34th aa is re-designated as the ‘ii’ aa (see the text). (B) The RNA binding affinity of the HCF152 truncated proteins. The gel shift assay was performed with the Dd120 RNA probe and several dilutions of the indicated proteins; the proteins having two PPR motif (HCF/3&4 and 7&8), but conforming to different motif models (Pfam or PROSITE); or the proteins containing a single PPR motif (HCF/P3, P4, P7 and P8). The apparent KD values are graphically shown. The original gel image is shown in Supplementary Figure S4. (C) Helical wheel model and the position of aa. The aa forming helix A of the PPR motif in (A) are plotted on the model with the positions indicated.
We first addressed the RNA binding activity of truncated proteins containing two PPR motifs of two different PPR models. The gel shift assay was performed using the Dd120 RNA probe, which contains the putative target RNA sequence for HCF152 [coding region of chloroplast psbH and following untranslated region (UTR); Supplementary Figure S3] (17). Multiple species of protein–RNA complexes were observed in the gel shift assay (Supplementary Figure S4), assuming a non-cooperative binding model. This complexity allowed us to estimate the overall apparent KD value from the concentration of protein in which 50% of the RNA probe bound, as an indication of the RNA binding activity. From the apparent KD, the proteins of the Pfam model displayed definite RNA binding activities; however, the proteins in the PROSITE model were less active (or not detected; Figure 1B), indicating that the Pfam model is relevant to the functionality.
We also re-examined the RNA binding activity of proteins containing a single PPR motif, which showed no activity in a previous study (17). Consistent with the previous results, three out of four tested motifs demonstrated extremely low binding affinities (KD >2500 nM; Figure 1B), suggesting that it is hard to compare the RNA binding characteristics of single PPR motifs using our current experimental conditions. Thus, we decided to analyze the RNA binding properties of a series of truncated proteins consisting of two PPR motifs (mini-PPR proteins) derived from the HCF152 protein, following the Pfam criterion.
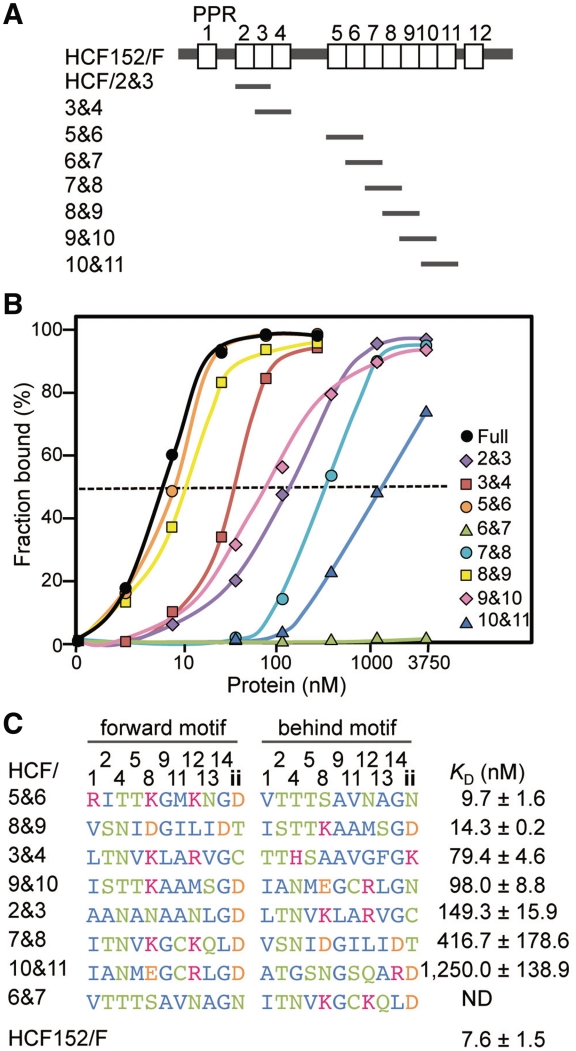
Characterization of RNA binding activities of mini-PPR proteins
A typical PPR protein consists of tandem and seamless arrays of PPR motifs. Therefore, eight mini-PPR proteins were constructed (2nd and 3 rd, 3 rd and 4th, 5th and 6th, 6th and 7th, 7th and 8th, 8th and 9th, 9th and 10th and 10th and 11th PPR motifs; Figure 2A). Several pairs of PPR motifs were not used (1st and 2nd, 4th and 5th and 11th and 12th), because they contain intervening aa (48, 77 and 19 aa, respectively). While the apparent KD for the full-length HCF152 protein was estimated as 7.6 nM, the RNA binding activities of the various mini-PPR proteins were significantly different (Figure 2B). From the apparent KD, the difference in RNA binding activity was >200-fold from the highest (9.7 nM; HCF/5&6) to the lowest (KD >2500 nM; HCF/6&7; Figure 2C), indicating that the pairs of PPR motifs had different RNA interacting natures.
Figure 2.
Mini-PPR proteins and their RNA binding activities. (A) Schematic representation of the full-length HCF152 protein (HCF152/F) and the mini-PPR proteins used in this study (HCF/2&3, 3&4, 5&6, 6&7, 7&8, 8&9, 9&10 and 10&11). Open boxes show PPR motifs. (B) The RNA binding activities of the full-length HCF152 and the mini-PPR proteins. The RNA binding activities were determined by gel shift assays, as described in Figure 1, and plotted. The original gel image is shown in Supplementary Figure S4. The symbols corresponding to the full-length and the mini-PPR proteins are shown at the right of panel. The dashed line indicates 50% of RNA probe bound. (C) Sequences of the mini-PPR proteins and their RNA binding activities. The unique aa of the PPR motif are shown sorted by their extent of RNA binding activities (the apparent KD) with their standard deviations (n≥3). The KD for HCF152/F is also shown. ND means not detected (the KD of >2500 nM). The 3 rd, 6th, 7th, 10th and later aa of PPR motif are not shown. Hydrophobic, hydrophilic & neutral, basic and acidic aa are colored in blue, green, red and orange, respectively.
Helix A has been predicted to be responsible for PPR's RNA interaction (3,25). The conserved residues, in either PPR or TPR motifs (3rd, 6th, 7th and 10th aa), are mostly hydrophobic (or Tyr; Figure 1A). When the residues are plotted on a conventional helical wheel model with a periodicity of 3.67 aa per turn, these aa are positioned on one side, presumably for helix formation, as indicated previously (Figure 1C). Therefore, the RNA binding surface would lie on the opposite side. The aa sequences of the mini-PPR proteins were sorted in order of their KD, i.e. the extent of RNA binding activity, with designation of the 1st and 2nd PPR motifs in the mini-PPR protein as the forward and behind motif, respectively (Figure 2C). The aa were highly diverse, and conservation of particular aa species was not found in the PPR motifs of either high or low activity proteins.
Survey of aa involved in the RNA interaction
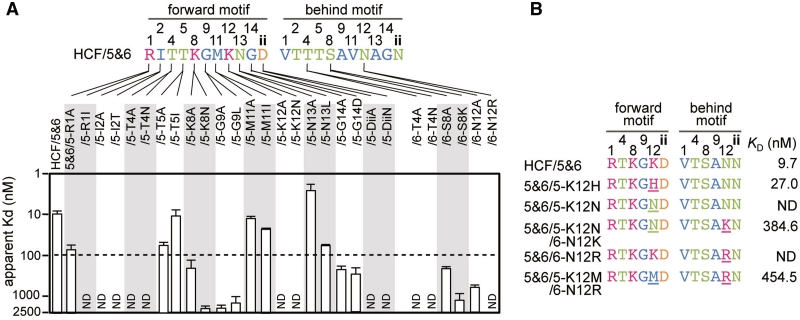
To identify the aa involved in the RNA interaction, the RNA binding activity was examined in mutants of HCF/5&6 with three regions of aa substitutions. First, the residues in helix A from the 2nd to 11th positions based on the helical wheel model (2nd, 9th, 5th, 12th, 1st, 8th, 4th and 11th aa; Figure 1C) were substituted. Second, the 13th and 14th aa of the loop region connecting helix A and B were substituted. Finally, the 34th aa located in the loop region in the front of helix A, which has been proposed to be involved in the RNA interaction [position 1 in the reference (27)], was substituted. The 34th aa was re-designated as ‘ii’ (-2nd) in this study, because the functionality of a PPR motif was retained in the Pfam criterion (Figure 1B), whereas the position should be designated as two aa before the 1st aa of next PPR motif, rather than as the 34th aa (see below).
The aa substitutions were performed by the introduction of alanine, or by an aa that could be found in other PPR motifs in HCF152, based on the hypothesis that the substitutions might reveal the different RNA binding activities of the mini-PPR proteins. Substitution was considered significant if a more than 10-fold reduction of the KD compared with the original HCF/5&6 protein was observed. By this criterion, substitutions into the 1st, 2nd, 4th, 8th, 9th, 12th, 14th and ‘ii’ aa caused reduction in RNA binding, whereas those in the 5th, 11th and 13th aa did not (Figure 3A). When the residues located in the loop between helix A and B were substituted, substitutions of the 14th aa reduced RNA binding, whereas those at the 13th aa had no effect. The 14th aa contains a conserved glycine in both PPR and TPR motifs (Figure 1A), suggesting a common contribution to both motifs. Thus, the 14th aa was not analyzed further. Notably, introduction of Asn into the 4th aa (5&6/5-T4N) and Lys into the 8th aa (5&6/6-S8K) of HCF/5&6 caused reductions. These aa are frequently observed in other mini-PPR protein with high RNA binding affinity (ex. 4th Asn in the HCF/8&9 and 3&4, Figure 2C). This suggests that the RNA interaction may be dependent on the combination of a plurality of aa, as well as the individual aa characteristics.
Figure 3.
RNA binding affinities of the mini-PPR proteins carrying aa substitution(s). The gel shift assay was performed as described in Figure 1. The apparent KD was estimated from Supplementary Figure S4 and shown. (A) RNA binding affinity for the derivatives of HCF/5&6. The motif and position of the substituted aa is denoted in the protein name. The dashed line indicates the 10-fold reduction of RNA binding affinity from that of HCF/5&6. (B) Coordinated action of 12th aa for the RNA interaction. The residues involved in the RNA interaction are shown with the substituted aa (underlined). The RNA binding activities (KD) are shown at the right. ND indicates a KD of >2500 nM. The aa color scheme follows that of Figure 2.
One such combination of aa could be found at the 12th aa, a conserved basic residue of which has been postulated as a generalized RNA anchor in the PPR–RNA interaction (Figure 1A) (3,25). The substitution of the 12th Lys to His, another basic residue, resulted in similar RNA binding activity to that of the original protein (5&6/5-K12H; Figure 3B). The introduction of Asn at the same position reduced the RNA binding activity (5&6/5-K12N), and the reduction was partially recovered by introduction of Lys in the 12th aa of another PPR motif (5&6/5-K12N/6-N12K). The results suggested that the 12th basic residue promotes the RNA interaction. However, the aa substituted protein containing a basic aa at the 12th position of both PPR motifs displayed a reduced RNA binding activity (5&6/6-N12R). The reduction was partially recovered by the removal of the basic 12th aa from one motif (Met-Arg; 5&6/5K12M/6-N12R). These results suggest a pair of basic and neutral (hydrophilic or hydrophobic) 12th aa are involved in the RNA interaction, and that there is an interaction of the 12th aa with adjoining PPR motifs in the mini-PPR protein.
Putative RNA interacting surface in the predicted PPR structure
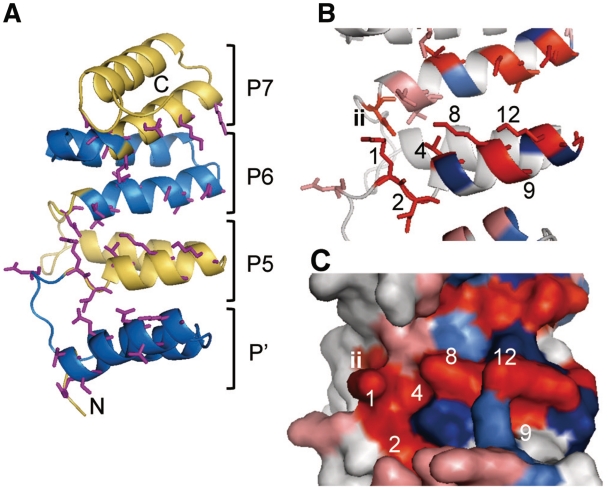
A series of substitutions suggested that seven aa (1st, 2nd, 4th, 8th, 9th, 12th and ‘ii’) could be involved in RNA interaction. The 1st, 4th, 8th and 12th aa are located side by side, whereas the 2nd and 9th aa are separate on helix A (Figure 1C), and the ‘ii’ aa is predicted to be positioned in the loop before helix A. To gain some structural insight, we constructed a structural model of HCF152. The Phyre software automatically presented several structural models for the full-length HCF152 protein using several TPR proteins as templates, with an E-value of <2× 10−6, which has been considered reliable (32,33). Modeling using the 148-aa region (376th to 523rd aa; eight helixes, including three PPR motifs and a PPR-like structure) displayed highly similar models using several templates (Supplementary Figure S5) and was also highly similar to previous structural models for PPR proteins (25,27). Structural prediction using other PPR proteins as queries also resulted in similar models. Therefore, we considered that the principal structure of above packed helixes would be reliable and further analyzed the model using O-GlucNAc transferase. The structural model suggested that the 1st, 4th, 8th and 12th aa are on the solvent-exposed surface and form a line, supporting the hypothesis that they act as the RNA binding surface of the PPR motif (Figure 4).
Figure 4.
Structural model of HCF152. (A) Cartoon diagram of the model of the 148 aa from HCF152 containing a PPR-like helical structure (P′) and three PPR motifs (5th, 6th and 7th PPR motifs; P5-P7). The helical repeats are colored alternately in blue or yellow. The side chains of aa involved in the RNA interaction are shown by sticks (1st, 2nd, 4th, 8th, 9th, 12th and ‘ii’ aa). N and C indicate the N- and C-terminus, respectively. (B) Magnification of the model containing the 5th and 6th PPR motif. The numbers of residues in the 5th motif are shown. Residue for which mutations had reduced RNA binding activity are colored in red (1st, 2nd, 4th, 8th, 9th, 12th, 14th and ‘ii’ aa; or salmon pink for the corresponding position, but not experimentally determined). Residues for which mutations had no effect (5th, 11th and 13th aa) are displayed in blue (or light blue for the corresponding position), respectively. (C) Surface representation of the model. The numbers and colors of the residues are the same as in (B).
The positions of the ‘ii’ aa are disordered in this model; it faced the 1st aa of the same motif or behind motif, or occasionally in another direction, depending on the template structure (Figure 4A and B). The result shown in Figure 1B indicates that the position of ‘ii’ aa is relevant at the end of a PPR motif (34th aa of the Pfam criterion). The model suggests the ‘ii’ aa acts with the residues in the last helix A (Figure 4B); however, the PPR subtypes have length differences in this loop region (Figure 1A). Therefore, the position could be relevant to define the two aa before the 1st aa of the next motif, rather than the 34th aa. The 2nd and 9th aa might be important in maintaining the overall structure. The aa are predicted to be on the facing surface of helix A and B. Based on biochemical analysis and structural modeling, we proposed that five aa (1st, 4th, 8th, 12th and ‘ii’) organize the RNA binding surface of the PPR motif.
Nucleobase specificity of PPR motifs
We next addressed the most intriguing issue of PPR function, namely how each member recognizes the distinct RNA target, and how the above five RNA interacting residues are involved in RNA recognition. A previous study identified that the HCF152 protein interacts with RNAs, including the 21-mer of the UTR between chloroplast psbH and petB, in vitro (17). To gain further insight, we adapted a SELEX assay to the full-length HCF152 protein of 12 PPR motifs. An RNA pool containing a random 30-mer window was mixed with the recombinant full-length HCF152 protein containing a histidine tag. The bound RNA was enriched by purification on nickel affinity beads (beads selection), or excision of slowly migrating bands, i.e. the protein–RNA complexes, from gels after PAGE (gel selection; Supplementary Figure S2C). The bound RNA was reverse transcribed and PCR amplified to produce the RNA pool for subsequent rounds of selection.
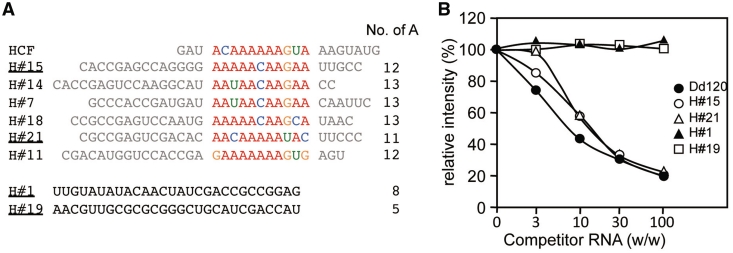
After seven rounds of selection, we sequenced 24 independent cDNA clones to obtain the information on the RNA consensus sequence for the binding of HCF152 (Supplementary Table S2). G/C-rich sequences were frequently acquired in the selected RNAs, using either the HCF152 or control thioredoxin protein as bait, suggesting the G/C-rich sequences might be aptamers that are selected depending our selection procedure. The motif search by MEME found a consensus motif between six representative clones in the 24 HCF152 selected RNA molecules and the previously identified 21-mer of HCF152 target sequence (17). The consensus motif contains adenine-rich sequences, which were interrupted by guanine or other nucleotides (Figure 5A). The consensus motif was not found either in the RNAs selected using the control thioredoxin protein or from the initial RNA pool. In addition, the SELEX assay using other two PPR proteins resulted in the selection of different contexts of RNA sequences (data not shown), strengthening the identification of the high affinity binding of HCF152 to the RNA molecules containing the consensus motif. The competitive gel shift assay verified that several positively selected sequences specifically bind to the HCF152 protein (Figure 5B). No significant difference was observed in the binding by the selected RNA molecules in the presence or absence of guanine in the consensus motif (H#15 and H#21, Supplementary Figure S2D).
Figure 5.
Sequences of HCF152-selected RNA molecules. (A) SELEX assay was performed and the sequences of random window of eight RNA molecules are shown with the putative target sequence for HCF152 (HCF, 21-mer). The alignment was performed by MEME. The number of adenine in the 30-mer window and the negatively selected sequences are also shown (H#1 and H#19), i.e. those contained in the HCF152 selected RNA pool, but not aligned with Dd120 RNA in the MEME analysis. The RNA species used in competitive gel shift assay are underlined. Nucleobases are colored in red (A), green (U), orange (G) or blue (C). (B) Competitive gel shift assay for the selected RNA molecules. The gel shift assay was conducted using the full-length HCF152 and the Dd120 RNA probe with the competitors of non-labeled selected RNA molecules (89-mer, H#15 & H#21), as well as Dd120 RNA probe and negatively selected RNA molecules (H#1 & H#19). The non-labeled RNA was added at 3- to 100-fold excess over the radiolabeled RNA (w/w). The intensities for protein-RNA complexes were estimated from the intensity of the complex in the absence of competitor RNA, which was set at 100% and the averages (n=3) were plotted. The symbols corresponding to the competitor RNAs are shown at the right of the panel. The gel images are shown in Supplementary Figure S2D.
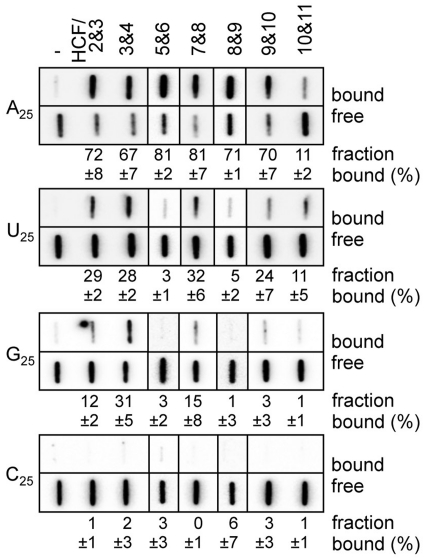
To address the correspondence of the nucleobase versus each PPR motif in HCF152, the nucleobase specificity of the mini PPR protein was analyzed using a synthesized ribonucleotide homo-polymer (N25). The binding experiment was performed with a filter-binding assay (FBA), because the G25 RNA probe migrates heterogeneously, and the retarded band was stacked at the edge of gel in the gel shift assay. The result was similar to the SELEX assay and the putative target sequence for HCF152: many mini-PPR proteins displayed a high preference for the A25 (67–81%; HCF/2&3, 3&4, 5&6, 7&8, 8&9 and 9&10), with a weak preference for the U25 (Figure 6). Notably, HCF/3&4 displayed significant affinity to the G25 (31 %) in addition to the A25. HCF/10&11 displayed a preference for the A25 and U25 with low affinity (11%), probably because of its low RNA binding affinity (the KD = 1250 nM, Figure 2C). The nucleobase preferences of the mini-PPR proteins are graphically represented in Figure 7B.
Figure 6.
Nucleobase preference of the mini-PPR proteins. The filter binding assay was conducted with ribonucleotide homo-polymer (A25, U25, G25 and C25; 250 pM) and the indicated mini-PPR protein (200 nM). Samples were filtered through the nitrocellulose and nylon membranes layer. The protein–RNA complexes were captured on the nitrocellulose membrane (bound). RNA that passed through the nitrocellulose was retained on the underlay of nylon membrane (free). Averages of the ratio of protein–RNA complexes (fraction bound, %) and the standard deviation (n ≥ 3) are shown.
Figure 7.
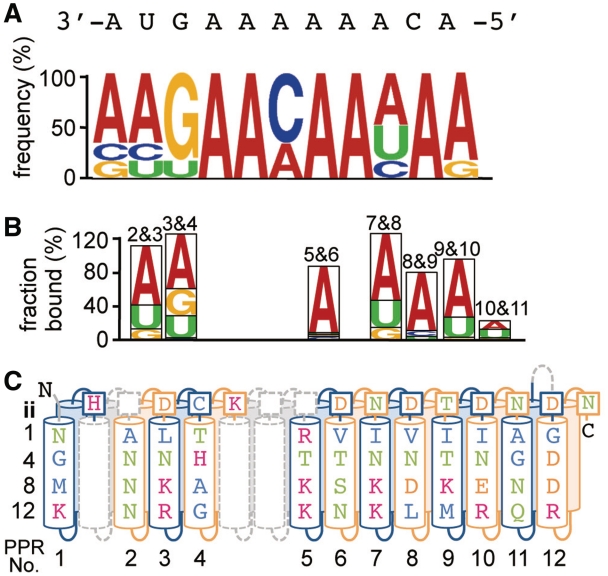
Model for the RNA recognition of the HCF152 protein. The examined nucleobase specificity and the residues of HCF152 protein were aligned. (A) The sequence logo for the consensus HCF152 binding motif derived from the SELEX assay (Figure 5A). The putative target RNA sequence for HCF152 in chloroplasts is shown above the logo. The sequences are arranged in a 3′ to 5′ orientation, with the position of guanine as an index. (B) The result for the nucleobase specificity of mini-PPR protein in Figure 6 is graphically shown by the order of preferred nucleobase. (C) The putative RNA interacting residues (1st, 4th, 8th, 12th and ‘ii’ aa) in individual PPR motif are shown in the schematic HCF152 structure. The PPR motifs are colored alternately in blue or orange. The position and predicted structure of intervening aa between PPR motifs are shown as dashed gray lines. The colors of the aa and nucleotides are the same as in Figures 2 and 5, respectively.
To interpret the results of SELEX and FBA against the putative RNA binding residues of HCF152, we proposed a model where the consensus motif of SELEX assay was arranged in 3′ to 5′ orientation with fitting of a guanine residue to the middle of the 3rd and 4th PPR motif. This was because the HCF3&4 displayed significant preference for guanine and adenine, and the mini-PPR proteins containing 5th to 11th PPR motifs displayed high preferences for adenine (Figure 7).
The 1st, 4th and ‘ii’ residues were recently suggested to be involved in the determination of nucleobase specificity (27). By focusing on these residues, the latter half (5th to 12th motif) may be responsible for the recognition of adenine and is rich Val/Ile at the 1st aa (Figure 7C). The Asp/Asn at the ‘ii’ aa and Asn/Thr at the 4th aa also frequently appeared in the mini-PPR protein, displaying a high preference for adenine. In contrast, the HCF/3&4 displayed a preference for both adenine and guanine and contains several characteristic residues, including 1st (Leu) and ‘ii’ (Cys), in the 3rd motif, and unique residues at all positions in the 4th motif. Some of the characteristic aa described above might be involved in the preference for adenine and purine (adenine and guanine), respectively.
When the FBA was performed to validate their involvement, the mini-PPR proteins with a single aa substitution at the 1st, 4th or ‘ii’ residue displayed significant reductions in binding affinities to specific nucleobase(s), as well-reduced affinities to Dd120 RNA (e.g. 3&4/3-L1I; Supplementary Figures S4 and S6). The mini-PPR protein containing a substitution at the 12th residue (5&6/5-K12H) also showed reduced binding affinities to both poly(A) and Dd120 RNA (Figure 3 and Supplementary Figure S6C). This suggested that single aa are not sufficient to provide the affinities to specific nucleobases. We did not identify aa-substituted proteins displaying altered nucleobase preferences without reductions in RNA binding affinities.
Statistical analysis for intra-, inter-motif connections for PPR-RNA interaction
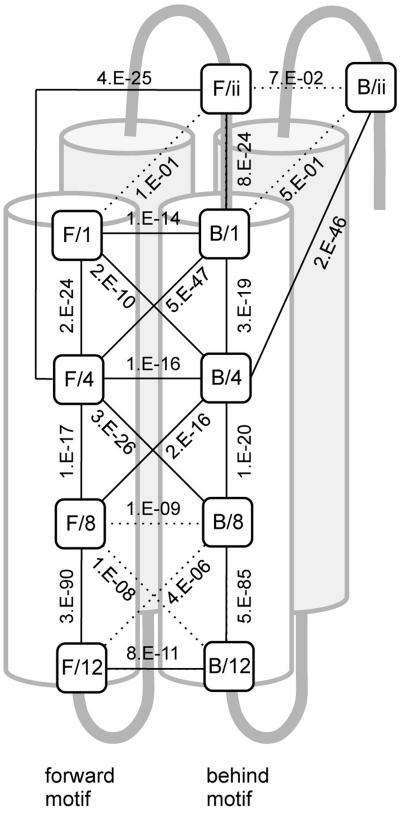
The above results suggested complex connections between the RNA interacting residues in the intra- and inter- motif(s). We therefore statistically examined the intra- and inter-motif connections between the adjoining residues of the 1st, 4th, 8th, 12th and ‘ii’ aa using 4614 Arabidopsis PPR motifs that had fewer than 10 aa between their motifs. The connections were estimated by the deflection between the actual and expected aa distribution at the two positions (Figure 8). The inter-motif connection between the 12th aa was experimentally suggested (Figure 3B). Therefore, the statistics indicated that P < E-10 was a significant connection. Intra-motif connections were observed through the motif from the 1st to 12th aa, with the exception of that between the ‘ii’ and 1st aa. The strongest intra-motif connection was found for the 8th to 12th interaction. Complex inter-motif connections were also found for several positions. The 4th residue had the most complex intra- (to the 1st, 8th and ‘ii’ aa) and inter- (to the 1st, 4th and 8th aa of the adjoining motif) connections. Taken together, the RNA interaction of the PPR protein is suggested to be achieved by an RNA binding surface containing a complex connection of residues between neighboring motifs, as well as within a motif.
Figure 8.
Statistical analysis for the intra- and inter-motif connections. The connections between the adjoining putative RNA interacting residues (1st, 4th, 8th, 12th and ‘ii’ aa) in the forward (F) and behind (B) motifs were statistically examined using 4614 PPR motifs. The difference between the actual and expected aa distribution was analyzed by a chi-squared test, and the P-value is shown. The residues are plotted on a schematic structure of two PPR motifs (forward and behind motif). Helix B is shaded in gray. The connection showing a significant P-value (> E-10) is drawn as a solid line. The original analytical data can be found in Supplementary Table S3.1-3.22.
DISCUSSION
In the present study, we identified and/or characterized putative RNA interacting residues in the PPR motif in vitro. Proteins with multi-repeat RNA binding domains are observed within other classes of RNA binding protein, e.g. RRM, KH and Zinc binding domains. Characterizations of these domains have been performed by dividing the domain into minimum functional units and/or by extensive mutagenesis (34,35). Accordingly, the present study was mostly conducted using recombinant proteins carrying two PPR motifs.
Characterization of the PPR motif
Initially, we demonstrated that the Pfam model could be relevant to the PPR motif function, if the motif is defined as a functional unit (Figure 1B). This also suggests the importance of ‘ii’ aa and its position for PPR function. Whereas a single PPR motif might correspond to a single nucleotide, many proteins carrying a single PPR motif displayed very weak RNA binding affinities. Furthermore, proteins carrying two PPR motifs displayed various RNA binding affinities, e.g. the HCF/5&6 and 7&8 displayed affinity to the RNA, although the overlapping protein of HCF/6&7 did not. The 6th and 7th PPR motif were assigned as PPR motifs with high E-values (1.6 × 10−5 and 1.5 × 10−6, respectively) compared with other PPR motifs in HCF152 (0.19–1.2 × 10−6) by the Pfam program, suggesting that conservation as a PPR motif may not guarantee the RNA binding activity of a protein with two PPR motifs. The high binding activities of several mini-PPR proteins, in contrast to the low activity of HCF6&7 and of the proteins carrying a single PPR motif, suggest that the observed RNA-binding activities may result from a cooperative effect between two motifs, rather than the simple sum of individual motif activities. This might be analogous to the combination of two or more RNA (or DNA) binding domains, such as RRM and the zinc finger domain, that often drastically increase the affinity for the ligand(s) (36,37).
The apparent KD for the full-length HCF152 was estimated as 7.6 nM, which is comparable to those of other characterized PPR proteins [CRR4 and PpPPR_38 (1.6 and 13.4 nM, respectively)] (18,38), and less than those of Rf1 and PPR10 (0.17 and 0.1 nM, respectively) (20,29). Significant elevation of the binding affinity was not observed between the protein of two PPR motifs (e.g. HCF/5&6, 9.7 nM; Figure 2B) and the full-length protein containing 12 PPR motifs, in contrast to that observed between the proteins of single and two PPR motif(s). This suggests that at least one repetition of the PPR motif might be significant for the RNA binding capacity in vitro.
The significant differences in RNA binding activities of the proteins having two PPR motifs may also suggest the presence of PPR motifs of low and high RNA binding affinities, i.e. different contributions of the motifs to the whole protein function. The ‘low’ motif might be involved in the recognition of specific nucleobases or function as a wobble for adaptation to variant RNA sequences. Thus, RNA binding capacity of respective motifs should be studied using the mutagenized full-length protein both in vitro and in vivo.
Putative RNA interacting residues in the PPR motif
The hypothetical RNA binding surface of the PPR motif was initially proposed by the discovery of the motif [2nd, 4th, 5th, 8th, 12th and 32nd aa, (3)]. Later, another hypothesis was proposed involving the 4th, 8th and 12th aa on the surface of the PPR motif (26). Recently, Fujii et al. suggested the 1st, 4th and ‘ii’ aa as the specificity-determining residues, which have been proposed because of their high diversifying rates in restorer-like PPR proteins and a constrain modeling of PPR–RNA complex [the position 1, 3 and 6 in the reference (27)]. Complementation tests have indicated the significance of several residues (8th, 12th and 14th aa) for protein functions (39,40).
The present mutagenesis study demonstrated the involvement of various aa positions in the overall RNA binding capacity of the PPR motif in vitro. It is also possible that aa at other positions may be involve in the RNA binding capacity, because the mutagenesis, in this study, was conducted by introductions of few aa species at limited positions. However, this study, combining the mutagenesis and the structural modeling, identified five residues (1st, 4th, 8th, 12th and ‘ii’) that form a putative RNA interacting surface of the PPR motif. The five residues are exposed on the solvent surface in the determined structure of the PPR motifs in the human mitochondrial RNA polymerase, although the structure did not imply the mechanism of PPR-RNA interaction (23). The mutagenesis and statistical analyses also suggested that the RNA binding capacity of a PPR motif could involve complex cooperation of the RNA-interacting residues, as well as their individual characteristics, which might be in addition to their hydropathy or charge. We cannot discuss the details of the intra- and inter-connections suggested by statistical analysis here. The aa are highly diverged and are thus less informative for interpreting their functional relevance. For example, when a single substitution was introduced (e.g. the 12th residue) to analyze a connection (e.g. the 8–12 intra-connection), the substitution could involve other connections (e.g. the 12–12 inter connection). This experimental verification requires a systematic, large scale of RNA binding analyses.
Subsequent analyses showed the high preference of the HCF152 protein, and the PPR motifs within, for adenine and purine (adenine and guanine; Figure 7). The nucleobase specificity is consistent with a former study suggesting that the editing factor can distinguish purine/pyrimidine and, at some positions, recognize specific bases (41). This analyses also indicated the aa signature at 1st, 4th and ‘ii’ aa, which might be responsible for determining the nucleobase specificity, in agreement with a recent informatics suggestion (27). However, their significance in nucleobase discrimination is still inconclusive because of the reduction of RNA binding capacity of mini-PPR proteins by aa substitutions. The 4th residue might be particularly important for the PPR function; the substitutions of the 4th residues resulted in severe reductions in RNA binding affinity (Figure 3A), and the 4th residue contains intra- and inter-connections with all adjoining residues (Figure 8). To elucidate a set of aa for nucleobase correspondence, PPR motifs displaying a preference for other nucleobases, such as pyrimidine, must be distinguished in other PPR protein(s) and the residues at the corresponding positions (1st, 4th and ‘ii’ aa) characterized.
We could not find a correlation between the aa species at the 8th and 12th residues and the nucleobase preferences for the mini-PPR proteins. The importance of the positively charged 12th aa was suggested by the general preference of a basic residue for the phosphate of a nucleic acid (25), as also shown in Figure 3B. The 12th aa might facilitate the RNA binding capacity of the PPR motif, together with the 8th aa, which contains the highest intra-motif connection with 12th aa at the statistical level (Figure 8). Further analyses will be required to elucidate the characteristics and significances of the putative RNA interacting residues (1st, 4th, 8th, 12th and ‘ii’) in vivo.
This study merely attempted to characterize the RNA binding capacity of PPR motifs in the HCF152 protein, but also presented the sequence context in which the HCF152 protein can bind with high affinity and specificity in vitro. All experiments performed here and in a previous study suggested the interaction of HCF152 with an adenine-rich region (17). However, it should be mentioned that the binding does not necessarily imply the interaction in vivo. A previous study of an hcf152-deficient strains suggested pleiotropic functions of HCF152, with a pronounced effect in the formation and/or stability of the psbH 3′ termini and petB 5′ termini. In addition, the HCF152 protein has also been shown to interact with other RNA(s) in vitro (17). A recent report proposed a direct binding of HCF152 to the psbH 3′ and petB 5′ over-lapping region (Supplementary Figure S3) (15). It is also possible that the HCF152 protein interacts with other sequence(s), because of competition for binding to multiple proteins, or the folding of RNA into alternate structures, in chloroplasts. Determination of the in vivo effect of the binding of HCF152 to the proposed region and the mode of action of HCF152 in RNA processing(s) requires substantial analyses, combining in vitro RNA binding assays and complementation tests, using mutagenized proteins. The results presented here could facilitate the understanding of the molecular actions of PPR proteins, including HCF152, further the elucidation of the set of aa responsible for nucleobase discrimination.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–6.
FUNDING
The Precursory Research for Embryonic Science and Technology (PREST) from the Japan Science and Technology Agency (JST), the Kyushu University Research Superstar Program (SSP, based on the budget of Kyushu University allocated under the President's initiative) from Kyushu University, the Program for Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN) from Bio-oriented Technology Research Advancement Institution, and the Ministry of Education, Culture, Sports, Science and Technology (Grant-in-Aid 22681028 and 22380008 to T.N.). Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology (Grant-in-Aid 22681028 to T.N.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank T. Shikanai (Kyoto University, Kyoto, Japan) for valuable discussions and critical reading of the text, and R. Nakamura (Kyushu University, Fukuoka, Japan) and M. Ide (Nagoya University, Nagoya, Japan) for technical assistance.
REFERENCES
- 1.Kurland CG, Andersson SG. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 2000;64:786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharya D, Archibald JM, Weber AP, Reyes-Prieto A. How do endosymbionts become organelles? Understanding early events in plastid evolution. BioEssays. 2007;29:1239–1246. doi: 10.1002/bies.20671. [DOI] [PubMed] [Google Scholar]
- 3.Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 4.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mingler MK, Hingst AM, Clement SL, Yu LE, Reifur L, Koslowsky DJ. Identification of pentatricopeptide repeat proteins in Trypanosoma brucei. Mol. Biochem. Parasit. 2006;150:37–45. doi: 10.1016/j.molbiopara.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- 8.Cushing DA, Forsthoefel NR, Gestaut DR, Vernon DM. Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta. 2005;221:424–436. doi: 10.1007/s00425-004-1452-x. [DOI] [PubMed] [Google Scholar]
- 9.Chase CD. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2007;23:81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Zsigmond L, Rigo G, Szarka A, Szekely G, Otvos K, Darula Z, Medzihradszky KF, Koncz C, Koncz Z, Szabados L. Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol. 2008;146:1721–1737. doi: 10.1104/pp.107.111260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi K, Suzuki M, Tang J, Nagata N, Ohyama K, Seki H, Kiuchi R, Kaneko Y, Nakazawa M, Matsui M, et al. LOVASTATIN INSENSITIVE 1, a novel pentatricopeptide repeat protein, is a potential regulatory factor of isoprenoid biosynthesis in Arabidopsis. Plant Cell Physiol. 2007;48:322–331. doi: 10.1093/pcp/pcm005. [DOI] [PubMed] [Google Scholar]
- 12.Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433:326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- 13.Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gobert A, Gutmann B, Taschner A, Gossringer M, Holzmann J, Hartmann RK, Rossmanith W, Giege P. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 2010;17:740–744. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 15.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5' and 3' mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamazaki H, Tasaka M, Shikanai T. PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 2004;38:152–163. doi: 10.1111/j.1365-313X.2004.02035.x. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura T, Meierhoff K, Westhoff P, Schuster G. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 2003;270:4070–4081. doi: 10.1046/j.1432-1033.2003.03796.x. [DOI] [PubMed] [Google Scholar]
- 18.Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T. A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 2006;281:37661–37667. doi: 10.1074/jbc.M608184200. [DOI] [PubMed] [Google Scholar]
- 19.Hammani K, Colas des Francs-Small C, Takenaka M, Tanz SK, Okuda K, Shikanai T, Brennicke A, Small I. The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. J. Biol. Chem. 2011;286:21361–21371. doi: 10.1074/jbc.M111.230516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5' region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cel. Biol. 2008;28:5337–5347. doi: 10.1128/MCB.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 24.Williams-Carrier R, Kroeger T, Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008;14:1930–1941. doi: 10.1261/rna.1077708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 26.Stern DB, Hanson MR, Barkan A. Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci. 2004;9:293–301. doi: 10.1016/j.tplants.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Fujii S, Bond CS, Small ID. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. USA. 2011;108:1723–1728. doi: 10.1073/pnas.1007667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazama T, Nakamura T, Watanabe M, Sugita M, Toriyama K. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 2008;55:619–628. doi: 10.1111/j.1365-313X.2008.03529.x. [DOI] [PubMed] [Google Scholar]
- 30.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 31.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 32.Bujnicki JM, Elofsson A, Fischer D, Rychlewski L. Structure prediction meta server. Bioinformatics. 2001;17:750–751. doi: 10.1093/bioinformatics/17.8.750. [DOI] [PubMed] [Google Scholar]
- 33.Cristobal S, Zemla A, Fischer D, Rychlewski L, Elofsson A. A study of quality measures for protein threading models. BMC Bioinformatics. 2001;2:5. doi: 10.1186/1471-2105-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serin G, Joseph G, Ghisolfi L, Bauzan M, Erard M, Amalric F, Bouvet P. Two RNA-binding domains determine the RNA-binding specificity of nucleolin. J. Biol. Chem. 1997;272:13109–13116. doi: 10.1074/jbc.272.20.13109. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Mayoral MF, Hollingworth D, Masino L, Diaz-Moreno I, Kelly G, Gherzi R, Chou CF, Chen CY, Ramos A. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure. 2007;15:485–498. doi: 10.1016/j.str.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 38.Hattori M, Sugita M. A moss pentatricopeptide repeat protein binds to the 3′-end of plastid clpP pre-mRNA and assists with mRNA maturation. FEBS J. 2009;276:5860–5869. doi: 10.1111/j.1742-4658.2009.07267.x. [DOI] [PubMed] [Google Scholar]
- 39.Tavares-Carreon F, Camacho-Villasana Y, Zamudio-Ochoa A, Shingu-Vazquez M, Torres-Larios A, Perez-Martinez X. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 2008;283:1472–1479. doi: 10.1074/jbc.M708437200. [DOI] [PubMed] [Google Scholar]
- 40.Lipinski KA, Puchta O, Surandranath V, Kudla M, Golik P. Revisiting the Yeast PPR Proteins - Application of an Iterative Hidden Markov Model Algorithm Reveals New Members of the Rapidly Evolving Family. Mol. Biol. Evol. 2011;10:2935–2948. doi: 10.1093/molbev/msr120. [DOI] [PubMed] [Google Scholar]
- 41.Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell. 2009;21:3686–3699. doi: 10.1105/tpc.109.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.