Abstract
Targeted therapies have often given disappointing results when used as single agents in solid tumors, suggesting the importance of devising rational combinations of targeted drugs. We hypothesized that construction of such combinations could be guided by identification of growth and survival pathways whose activity or expression become upregulated in response to single-agent drug treatment. We mapped alterations in signaling pathways assessed by gene array and protein phosphorylation to identify compensatory signal transduction pathways in prostate cancer xenografts treated with a MEK inhibitor, PD325901. In addition to numerous components of the ERK signaling pathway, components of the IKK, Hedgehog, and PI3 Kinase/Akt/mTOR pathways were up regulated following treatment with PD325901. Combinations of PD325901 with inhibitors of any one of these upregulated pathways provided synergistically greater growth inhibition of in vitro cell growth and survival than the individual drugs alone. Thus, the identification of compensatory signal transduction pathways paves the way for rational combinatorial therapies for the effective treatment of prostate cancer.
Keywords: combination therapy, signal transduction, prostate cancer, reverse phase protein array
Introduction
Inhibitors targeting signaling molecules that are overexpressed and activated in cancer have shown only modest clinical benefit when used as single agents (1, 2). One explanation for this may rest in recent data demonstrating that extracellular signals are transmitted through a network of proteins rather than through hierarchical signaling pathways (3–5). This network model explains why inhibition of a single component of a canonical pathway is usually insufficient to have dramatic effects on the treatment of cancer: the biological outcome of signals propagated through a network is inherently more robust and resistant to inhibition of a single network component (6). Thus, it is becoming increasingly clear that for many cancers the most effective use of molecular targeted therapies for cancer will require a combination of several agents that inhibit key nodes or fragile points in the network.
However, the difficulty of developing combinations of targeted agents is geometrically more difficult than developing a single agent. Ideally, one would be able to identify pathways that compensate for or blunt the cytotoxic effects of single agents, and then utilize a second agent that targets this compensatory pathway. One potentially effective paradigm for identifying these pro-growth and survival compensatory signaling events is to identify changes in activity or expression of signaling components that are induced by specific targeted therapeutics. Here we demonstrate that these induced signaling changes do, in fact, reveal functionally significant compensatory changes and hence therapeutic targets that can guide the construction of rational therapies.
In prostate cancer, prior work by us and others suggests that activation of Ras signaling plays an important role in progression of prostate cancer to advanced, castration resistant disease (7–9). In earlier work we showed that activation of MAP kinase, an effector of Ras activation, correlates with prostate cancer progression in patient samples (9). Moreover, activation of Ras signaling was sufficient to reduce androgen dependence of growth in cultured cells and xenografts (8). Additionally, Ras-related signaling was necessary for castration resistant growth, as inhibition of Ras activity with dominant negative Ras restored androgen dependence both in vitro and in vivo (7). Taken together these results suggest that inhibition of Ras and/or its effectors such as MEK could be an effective therapy for advanced prostate cancer.
In this study we show, contrary to expectations, that inhibition of MEK was only partially effective at blocking castration-resistant growth of prostate cancer xenografts, suggesting that other pathways besides the MAP kinase pathway need to be co-targeted to achieve full therapeutic benefit in vivo. We identified several pro-growth and survival compensatory signaling pathways whose activity or expression were induced by inhibiting MEK in prostate cancer xenografts. We found that combining inhibitors of these compensatory responses with MEK inhibition effectively blocked cell growth.
Materials and Methods
Cell Culture and reagents
CWR22Rv1 cells were a kind gift from Steven Balk, Harvard University, and grown in DMEM (GIBCO-BRL, Grand Island, NY) with 10% fetal calf serum (Life Technologies, Inc, Grand Island, NY). LAPC4 cells (a gift from Dr. Charles Sawyers, Memorial Sloan-Kettering Cancer Center) were grown in DMEM/F12 (GIBCO-BRL, Grand Island, NY) supplemented with 10% fetal calf serum (Life Technologies, Inc, Grand Island, NY). The cell lines were verified by comparison to published 1) morphologic features, 2) growth properties in vitro and in vivo, 3) expression of the androgen receptor (AR), and 4) transcriptional response of a subset of genes to androgen stimulation. Cultures were maintained in a humidified chamber at 37°C with 5% CO2. PD325901 was a gift from Pfizer. SANT-1, SC-514, UO126, and Rapamycin were from EMD Biosciences (Darmstadt, Germany). See Supplemental Figure 1 for structures. PARP antibody was from Cell Signaling Technology (Danvers, MASS) and the anti-tubulin antibody from EMD Biosciences. Western blots were performed as previously described (10).
Animal Studies
5–6 week old athymic NCr-nu/nu mice (NCI) were bilaterally inoculated subcutaneously with 2×106 cells in Matrigel™ (BD Biosciences) at 50:50 by volume in a total of 100 μl/site. Tumor burden was measured weekly with calipers in two dimensions and volume was calculated using the following formula: [length x width2] × 0.5 = volume (11). In experiment one, tumor bearing animals were divided into 6 groups, 10 mice per group: untreated, vehicle alone, and 3.125, 6.25, 12.5, or 25 mg/kg/day PD325901 in 0.5% HPMT-0.2% Tween80. In experiment two, tumor bearing animals were divided into four groups, 12 mice per group: vehicle or 25mg/kg/day PD325901 in 0.5% HPMT-0.2% Tween80 with surgical or sham castration. Repeated measures models were used to compare overall tumor growth among dose groups and controls. F-tests based on contrasts were used to make specific comparisons between pairs of groups. Mice were sacrificed, placed on ice, tumors excised, divided into thirds, with 1/3 fixed in Zn++ buffered formalin and paraffin embedded for immunohistochemistry, 1/3 snap frozen in liquid nitrogen for protein analysis, and 1/3 placed in RNAlater (Ambion, Foster City, CA).
Microarray analysis
Sample preparation, cRNA labeling, hybridization to Affymetrix HG-U133 expression arrays and scanning was performed at the UVA Biomolecular Research Facility using the Affymetrix GeneChip System. The .cel files were quantile normalized and expression values estimated using GC-RMA (12). We applied a modified t-test using the limma package in Bioconductor to drug treated versus control to identify differentially expressed genes (12). To arrive at lists of genes for every comparison, we first corrected for multiple hypothesis testing by applying a False Discovery Rate (FDR) correction to the p-values and used a 5% FDR cutoff. We identified pathways impacted by the differentially expressed genes using Pathway Express (13).
Reverse Phase Protein Microarrays
Protein was generated by pulverizing tumor with mortar and pestle while frozen in liquid N2, resuspended in 1:1 T-PER (Pierce, Rockford, IL) and Laemmli sample buffer containing protease and phosphatase inhibitors, sonicated on ice and cleared. Lysates were then analyzed by reverse phase protein array as described (14). Briefly, approximately 40nL of lysate was printed in duplicate onto nitrocellulose-coated glass slides (FAST Slides, Whatman, Keene, NH) with an Aushon 2470 solid pin microarrayer (Aushon Biosystems, Billerica, MA) equipped with 350 μm pins. Samples were printed in 5 point, 1:2 serial dilution curves and 50 slides were printed for each group. Slides were stored desiccated at −20°C prior to staining with antibody.
For estimation of total protein amounts, selected arrays were stained with Sypro Ruby Protein Blot Stain (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and visualized on a Kodak ImageStation (Eastman Kodak, Rochester, NY). Printed slides were prepared for staining by treating with 1× Reblot (Chemicon, Temecula, CA) for 15 min, followed by 2×5 min washes with PBS. Slides were treated overnight with blocking solution (1g I-block (Applied Biosystems, Bedford, MA), 0.5% Tween-20 in 500mL PBS) with constant rocking at 4°C. Staining used an automated slide stainer (DAKO, Carpinteria, CA) using a biontinyl-linked peroxidase catalyzed signal amplification system as per the manufacturer’s recommendation and components from the manufacturer’s kit, unless otherwise indicated as previously described (15). Slides were stained with a set of 39 antibodies against phosphorylated or total forms of proteins involved in cell proliferation, survival, motility and apoptosis signaling. A complete list of antibodies, sources and dilutions used for these experiments is available in Supplementary Table 1. Stained slides were scanned individually on a UMAX PowerLook III scanner (UMAX, Dallas, TX) at 600 dpi and saved as TIF files in Photoshop 7.0 (Adobe, San Jose, CA). The TIF images for antibody-stained slides and Sypro-stained slide images were analyzed using MicroVigene v2.8.1.0 (VigeneTech, Carlisle, MA). Briefly, final data values for each sample were calculated using the factor average mode, and all values were negative control subtracted and normalized to total protein.
Growth assays
For three dimensional cell cultures, 500 CWR22Rv1 cells were seeded in 50% BD Matrigel™ Basement Membrane Matrix (BD Biosciences, Franklin Lakes, NJ) in 24wells and covered with DMEM/10%FBS. Drug or vehicle (DMSO) was administered daily with medium change. After 16 days of culture the 3D cultures were photographed and prepared for cell recovery. The gels were washed twice with PBS and then the cells were extracted from the gels with BD™ Cell Recovery Solution (BD Biosciences). Cell numbers were determined with the Quantos™ cell proliferation assay (STRATAGENE, Santa Clara, CA).
For two dimensional cell cultures, 15,000 cells were plated per well in a 24 well dish and treated with a single dose of drugs. After seven days crystal violet (Sigma) staining was used as a surrogate for cell number (16). Quantitation was performed on a BioTek Synergy 2 plate reader at 595nm absorbance following addition of 0.25ml 20% methanol, 30 minutes. Bliss independence is determined by the equation: Bliss independent effect = 1 - (Effect Drug 1 × Effect Drug 2) for specific dose of each drug (17; 18). When the observed experimental data match the predictions of Bliss independence, the inhibitors are additive whereas greater than predicted potency indicates synergism and lower than predicted potency indicates antagonism.
Results
MEK inhibition decreased CWR22Rv1 in vitro growth in 3D Matrigels
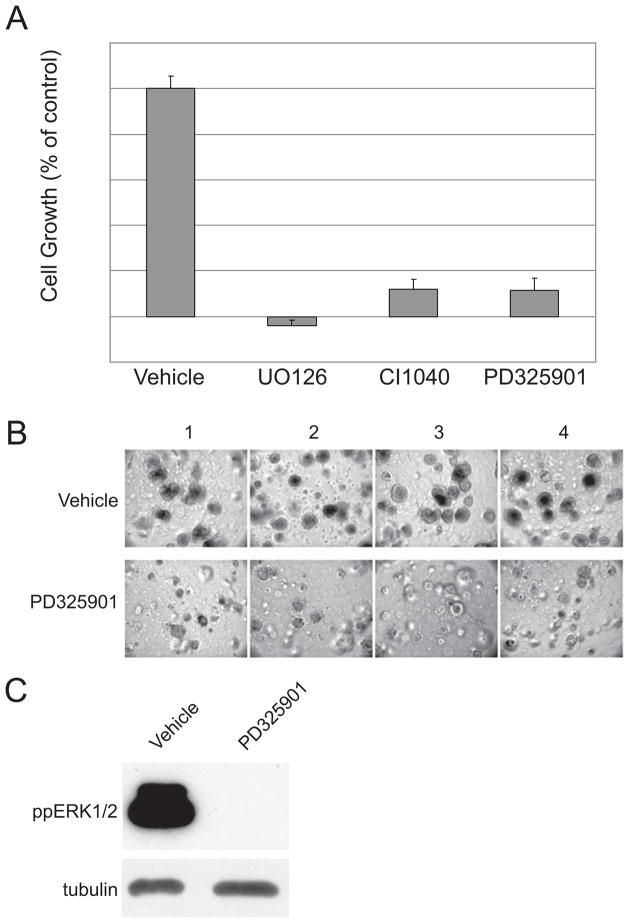
We selected CWR22Rv1 cells to test the efficacy of MEK inhibitors since CWR22Rv1 cells have detectable basal levels of MAPK activity (19) as occurs in advanced disease in humans (9). To reflect the in vivo situation in xenografts as close as possible, CWR22Rv1 cells were grown in 3 dimensional (3D) matrigel plugs and treated with different MEK inhibitors for a period of 2 weeks (Figure 1). Indirect cell counting was used to investigate the growth of cells in 3D following two weeks of treatment with UO126, CI1040, and PD325901 (Figure 1A). All three MEK inhibitors revealed strong growth inhibition. We focused on PD325901 in the following experiments as a representative of a family of allosteric MEK inhibitors in clinical development. Microscopy confirmed the cell counts; smaller and less numerous spheres developed following PD325901 treatment (Figure 1B). PD325901 effectively blocked ERK phosphorylation of CWR22Rv1 cells cultured in 3D (Figure 1C).
Figure 1.
MEK inhibitors effectively reduce growth of 3D in vitro growth. (A) Quantitation of cell number for CWR22Rv1 cells treated with vehicle or 1μM PD325901, 10μM UO126 or 1μM CI1040; n=4. (B) Images of four independent fields captured from separate experiments of CWR22Rv1 cells treated with either vehicle or 1μM PD325901. (C) Representative western blot of phospho-ERK levels of CWR22Rv1 cells isolated from 3D cultures.
PD325901 is highly selective in vivo
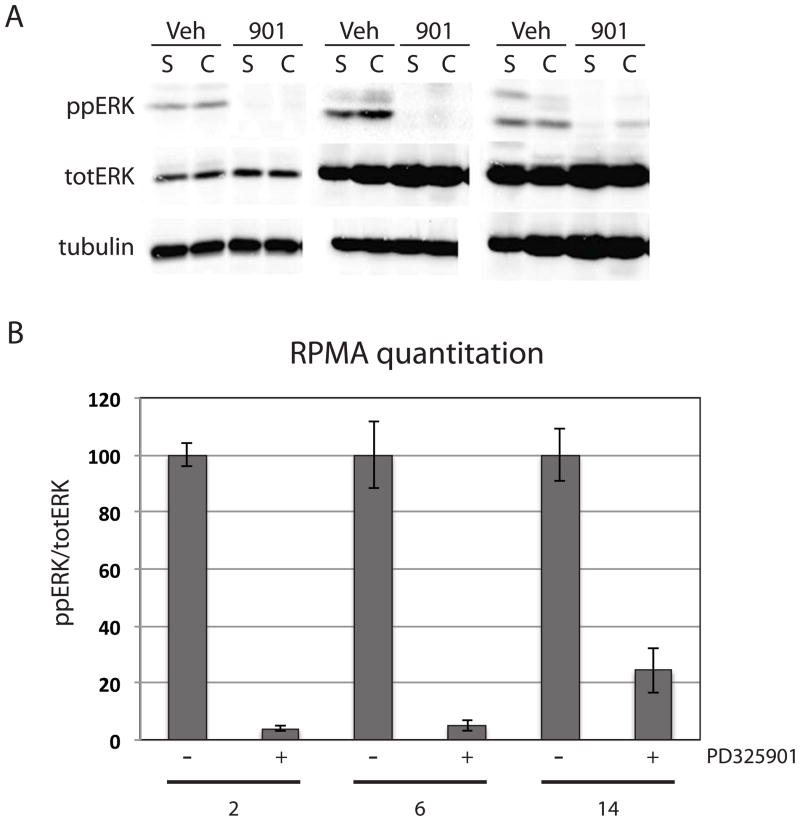
In order to assess the selectivity of PD325901 in vivo, castrated and sham operated nude mice carrying CWR22Rv1 tumors were treated with a single 25mg/kg dose of PD325901. Tumors were harvested at 2, 6, and 14 hours post-treatment. Western blot analysis showed effective inhibition of ERK phosphorylation at all times, although phospho-ERK levels were inhibited to a greater extent at 2 and 6 hours when compared to 14 hours post PD325901 treatment (Figure 2A). Analyzing xenografts over time by reverse phase protein array (RPMA) following a single dose of PD325901 showed a rapid decrease in only ERK phosphorylation (Figure 2B). Again, phospho-ERK levels are maximally suppressed at 2 and 6 hours. By 14 hours post PD325901 treatment phospho-ERK levels are still below control but have begun to rise. Thirty-eight different phosphorylation sites in tumor lysates were examined by RPMA (Supplemental Table 1). Only ERK and p90RSK, a kinase directly downstream and activated by ERK that showed a modest reduction in phosphorylation levels, changed during the first 14 hours following a single dose of PD325901. No other changes, either up or down, were observed in any of the other 36 phospho-proteins analyzed (data not shown). This is consistent with PD325901 being a highly selective small molecule MEK inhibitor in vivo, and provides important information complementary to the more usual kinase panel screening for drug specificity (20).
Figure 2.
PD325901 is selective for MEK inhibition in vivo. (A) Western blot analysis of CWR22Rv1 xenografts euthanized at 2, 6, and 14 hours following the first 25mg/kg dose of PD325901. Dually phosphorylated ERK, total ERK, and tubulin levels are shown. (B) Quantitation of RPMA phospho-ERK data normalized to total ERK levels. Two tumors for each treatment were independently analyzed in two separate RPMAs.
MEK inhibition decreased CWR22Rv1 subcutaneous tumor growth
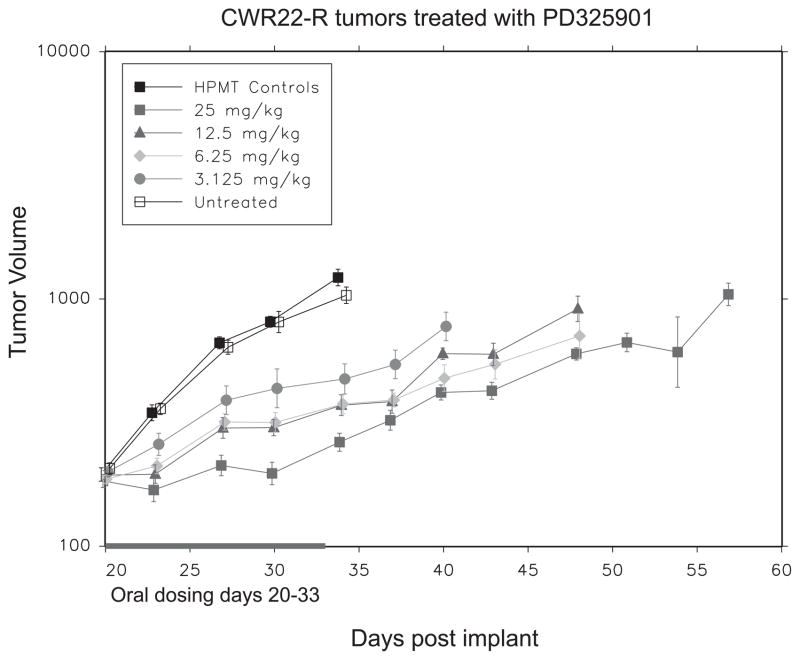
Nude mice carrying CWR22Rv1 tumors were randomized and treated daily for fourteen days with different doses of PD325901. We observed a dose dependent decrease in tumor growth during drug administration, with nearly complete tumor growth suppression observed at the highest dose (p<0.0001) (Figure 3). When drug was withdrawn tumor growth resumed at a slope comparable to vehicle treated animals. Previously we found that combining androgen ablation with blockade of Ras signaling (with dominant negative Ras) resulted in dramatic, rapid and complete tumor regression (7). Thus, we wanted to test if inhibition of MEK with PD325901 combined with androgen ablation would lead to tumor regression. Nude mice carrying CWR22Rv1 tumors were randomized and half were castrated. One week later half of each group received 25mg/kg PD325901 or vehicle daily for three weeks. Again, PD325901 inhibited tumor growth (p<0.0001; data not shown). However, there was no cooperative effect on inhibiting tumor growth when MEK inhibition was combined with androgen ablation (p=0.96; data not shown). MEK signaling was effectively inhibited following 3 weeks of 25mg/kg/day PD325901 (see below).
Figure 3.
PD325901 inhibits tumor growth. Plot of mean tumor volume over time for CWR22Rv1 xenografts in athymic nude mice either untreated, treated with vehicle alone, or 3.125, 6.25, 12.5, and 25 mg/kg/day of PD325901. The effect of PD325901 on growth within the sham group was p=<0.001.
The inability of MEK inhibition to mimic the growth-inhibitory effects of dominant negative Ras suggests that cells are able to compensate for the single-target inhibition by the small molecule more effectively than they can compensate for the multiple effects of a Ras blockade. In order to identify the compensatory signaling pathways induced by MEK inhibition as a paradigm for rationally selecting combinatorial therapeutics we analyzed the tumor transcriptome by gene array and functional protein signaling activation by RPMA. Three groups of xenografts were analyzed: control vehicle treated animals, PD325901 treated animals, and animals treated with PD325901 for three weeks and then allowed to recover for two weeks. Tumors were excised and divided, with representative tumor pieces from comparably sized tumors processed for protein or RNA analysis.
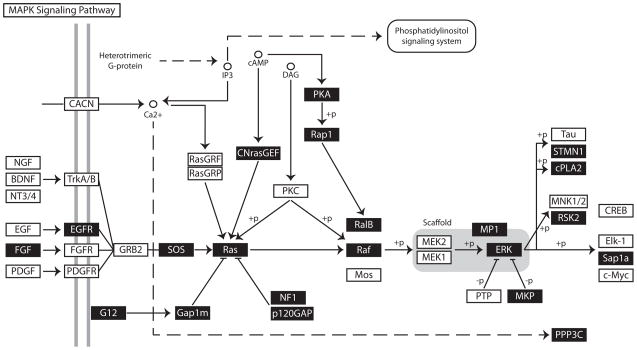
For gene array studies, RNA was isolated and analyzed using Affymetrix HgU133_Plus 2 gene chips. The treated and untreated samples clustered separately as expected. 5294 genes were scored as “upregulated” at least 1.5 fold P<0.05, and 833 genes were “downregulated” following treatment with PD325901. We found that approximately half the genes whose expression changed following treatment remained at least partially upregulated or downregulated even after drug was withdrawn, suggesting that MEK inhibition in prostate tumors led to persistent changes in the transcriptome. We used the Pathway Express tool to identify pathways affected by the differentially expressed genes (13). This revealed that numerous components of the ERK signaling pathway were upregulated in response to PD325901 treatment, including SOS, K-Ras, Rap1a, c-Raf, B-Raf, ERK1, ERK2, and MP1, as if massive overexpression of the pathway was a mechanism for overcoming the signaling blockade (Figure 4A). Furthermore, we found transcription of additional pathways were substantially upregulated following treatment with PD325901 (Figure 4B). Some were expected, such as PI3 Kinase and Akt. Others were more surprising, such as genes from the Sonic Hedgehog pathway, including Patched, DYRK1, and FU as well as genes of the Wnt, and Notch signaling pathways. It is striking that the pathways that change in response to MEK inhibition have been shown to be important in prostate development and/or cancer (21–24).
Figure 4.
Pathway Analysis of Gene Array Data from CWR22RV1 xenografts treated with PD325901 for three weeks. Three tumors per treatment group were analyzed by Affymetrix HgU133_Plus 2 gene chips. Shown is a schematic of the canonical MAPK pathway as described by Pathway Express. A black box with white letters denotes upregulated gene products.
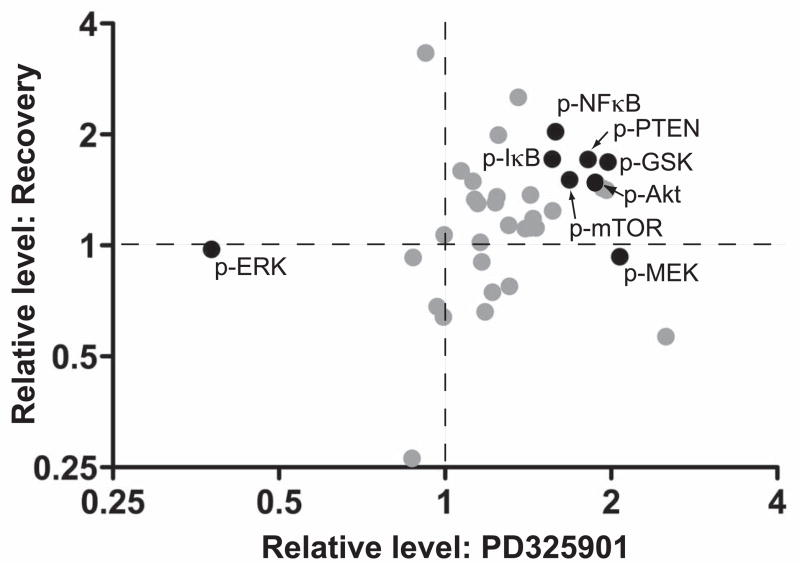
To determine the functional activation state of the signaling pathways, phosphorylation levels of key signaling proteins within these xenografts was analyzed using RPMA. ERK phosphorylation was inhibited for the three weeks of drug treatment and then returned to normal when drug was removed, as expected (Figure 5). Phosphorylation of MEK increased upon inhibition, presumably due to release of feedback inhibition due to phosphorylations by ERK on SOS and Raf (25). Thus, inhibition of MEK results in hyperactivation of the upstream components of the pathway, an important consideration in understanding how the compensatory survival pathways are activated. Regulatory phosphorylations of Akt, PTEN, and mTOR increased upon PD325901 treatment, consistent with an anti-apoptotic response perhaps due to the loss of feedback control at the level of Ras. Interestingly, these phosphorylations persisted even after withdrawal of the drug, suggesting that there was a selective advantage for maintaining this anti-apoptotic response. Additional phospho-proteins that increased and persisted following PD325901 withdrawal included SMAD2, GKS3b, IκB, and NFκB.
Figure 5.
Dynamic changes in the protein signaling architecture of CWR22RV1 xenografts treated with the MEK inhibitor. Three tumors for each treatment group was analyzed by reverse phase protein microarray were: 1) control, 2) PD325901 for 3 weeks, and 3) PD325901 for 3 weeks and then untreated for 3 weeks. Plotted are the relative change in phosphorylation of PD325901 vs control (x-axis) and drug withdrawal vs control (y-axis).
This constellation of changes in protein phosphorylation and gene transcription reflects changes in the cell signaling network triggered by MEK inhibition. We hypothesized that inhibition of one or more of these compensatory pathways will be necessary to complement MEK inhibition in prostate cancer therapy. In order to test if inhibition of the compensatory survival pathways cooperates with MEK inhibition to more effectively block prostate cancer cell growth we treated CWR22Rv1 cells with PD325901 in combination with inhibitors either of IKK, Hedgehog, or mTOR (Figure 6). These three protein targets were chosen based on 1) the magnitude and persistence of the change in phosphorylation following MEK inhibition; 2) the known role of these signaling pathways in prostate cancer (26–29); 3) the fact that these targets are down stream effectors of signaling pathways that had multiple proteins elevated – for example, in the PI3K signaling pathway PTEN, Akt, and mTOR were all elevated and in NFκB signaling IκB and NFκB were both elevated (Figure 6); 4) the occurrence of alterations detected at both the mRNA and protein levels (e.g. PI3K and NFκB; Figures 5, 6, and data not shown); 5) the existence of pathway cross-talk (e.g. PI3K signaling cross-talks with and can activate NFkB (30)); and 6) clinically relevant inhibitors for these targets exist (31–33). Thus, we chose inhibitors of mTOR, IKK, and Hedgehog for further analysis.
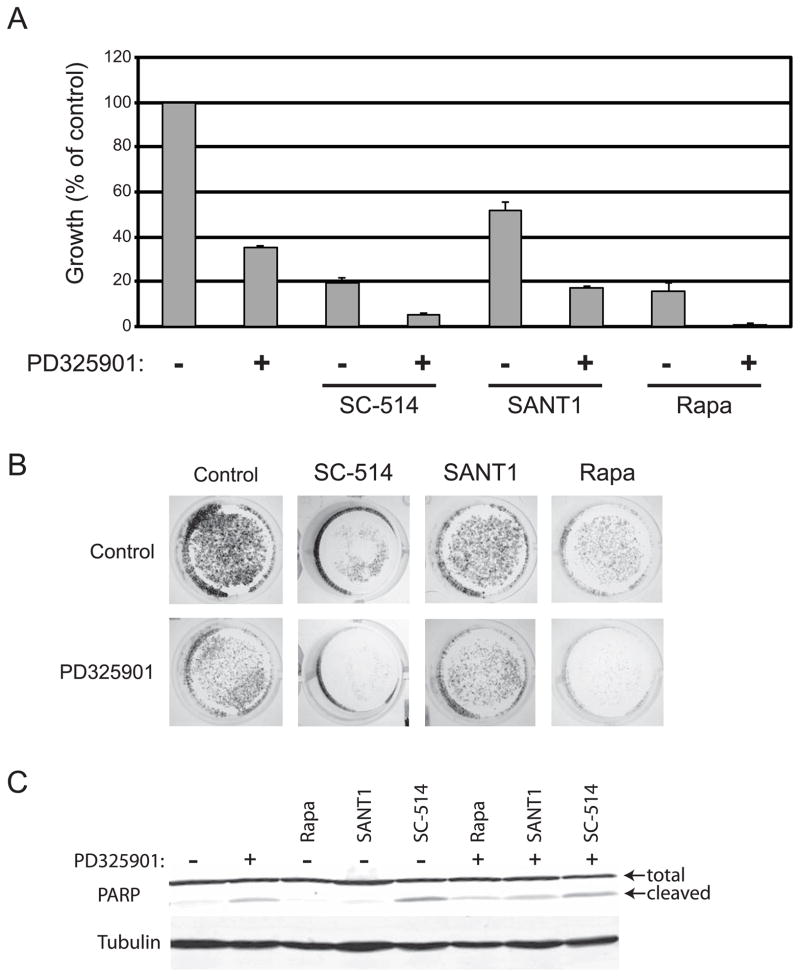
Figure 6.
Predicted drug combinations effectively inhibit in vitro prostate cancer cell growth. CWR22Rv1 cells were grown in the presence and absence of 10nM PD325901 and the inhibitors shown: 100μM SC-514 inhibits IKK, 10μM SANT1 inhibits hedgehog, and 100nM Rapamycin inhibits mTOR. (A) Quantitation of growth plotted as a percent of control, n=3. (B) Representative images of wells stained with crystal violet. (C) Western blot analysis of CRW22Rv1 cells treated as in (A) for total and cleaved PARP and tubulin. Shown is a representative blot on n=3.
CWR22Rv1 cells grown for seven days in the presence of 10nM PD325901 were inhibited nearly 70%. Figure 6 demonstrates that enhanced cytotoxicity can be achieved by combining PD325901 treatment with inhibitors either of IKK (NFκB pathway), Hedgehog, or mTOR (PI3K pathway). For each drug combination tested, the cytotoxicity observed was greater than the cytotoxicity of the single drugs. Additionally, the drug combinations of PD325901 with the IKK or mTOR inhibitors showed synergy as determined by the Bliss independence model (17). These experiments suggest that it is possible to enhance the therapeutic effectiveness of MAP kinase pathway inhibitors by combining with inhibitors of compensatory response pathways. While crystal violet staining is an effective measure of cell cytotoxicity (16), it does not provide any mechanistic insight. Therefore, we examined PARP cleavage to determine if the cytotoxic response we observed by crystal violet was due in part to apoptosis. We observed PARP cleavage when CWR22Rv1 cells were treated with PD325901 and with SC-514 alone as well as with all combinations of PD325901 with Rapamycin, SC-514, and SANT1 suggesting that the cytotoxic response is at least partially due to the induction of apoptosis.
These drug combinations were effective only in CWR22Rv1 cells. We tested combinations of PD325901 with IKK, Hedgehog, or mTOR inhibitors in three other AR positive prostate cancer cell lines, LNCaP, C4-2 and LAPC4. We did not observe enhanced cytotoxicity (data not shown). However, neither LNCaP, C4-2 or LAPC4 cells have elevated MAPK signaling in vitro nor are these cell lines sensitive to PD325901 (data not shown), thus making it difficult to effectively evaluate combinations involving MEK inhibition in additional prostate cancer cell lines.
Importantly, the cooperative growth inhibition observed is specific to inhibitors targeting the compensatory pathways elevated in response to MEK inhibition. Combinations of PD325901 with sixty other small molecule inhibitors targeting a range of signal transduction pathways uncovered only three drug combinations that showed a cooperative effect in CWR22Rv1 cells (data not shown). This contrasts with the success using combinations derived from observing the transcriptome and phosphoproteome changes in response to MEK inhibition.
Discussion
In our work, we have performed a functional protein signal pathway activation mapping and gene expression analysis of prostate tumor xenografts treated with a MEK inhibitor and found that MAP kinase inhibition caused hyperactivation of the upstream components of the canonical MAPK pathway as well as upregulation of other signaling events regulating cell growth and survival. This demonstrates the problem in basing a therapy on a simple catalogue of activated proteins in cancer cells, because an aberrantly activated protein could be part of a feedback or feed-forward control system. Without knowing its network role, inhibiting this protein could result in contributing to disease progression rather than curative therapy.
Previous studies have highlighted the complexity of selecting a targeted therapeutic agent based on the activation of a single network component (26, 27, 34). Elevated mTOR activity has been observed in multiple cancers and mTOR inhibitors have shown robust activity in model systems. However, the clinical trial results with mTOR inhibitors have been more modest than predicted (27). Studies have shown that while inhibiting mTOR activity in lung, breast, colon, and prostate cancer cells effectively suppressed the phosphorylation of downstream effectors such as p70S6K and 4EBP1, it increased the phosphorylation of AKT (26, 27). This increase in AKT activity attenuated the effect of mTOR inhibition and facilitated cancer cell growth and survival. Only upon discovery of this mTOR – AKT feedback control system could effective combinatorial treatments be determined; inhibition of IGFR-1 in breast and prostate cancer cell lines, and of PI3K in lung cancer cell lines, sensitized cells to mTOR inhibition (26, 27).
We utilized global analysis of protein activation and gene expression to identify compensatory events and facilitate the design of effective drug combinations. The usefulness of global analysis for identifying drug combinations was recently demonstrated using the KrasG12V/Lkb1−/− mouse model for NSCLC (35). Analysis of gene expression and phosphoproteome profiles between primary KrasG12V tumors, primary KrasG12V/Lkb1−/−tumors and metastatic KrasG12V/Lkb1−/− tumors showed an increase in genes associated with the FAK/Src and PI3K/AKT pathways. Targeting the PI3K/AKT, MAPK, and Src pathways in combination significantly reduced tumor burden in the KrasG12V/Lkb1−/− mice compared to targeting either Src alone or PI3K/AKT and MAPK together. These experiments conceptually overlap with our own results showing that identification of compensatory signaling pathways can be used to rationally develop drug combinations.
When we combined inhibitors of IKK (NFκB pathway), or mTOR (Phosphatidyl inositol pathway) with MEK inhibition we observed synergistic cytotoxicity in CWR22Rv1 cells and we observed additivity when we combined MEK and Hedgehog inhibition according to Bliss Independence (17, 18). Not yet determined is the precise mechanism of synergy with these drug combinations. An increase in NFκB signaling has been associated with prostate cancer (36). Moreover, a recent study has found that inflammatory infiltration and activation of IKK-alpha in tumor cells is associated with prostate cancer progression (28). The activation of IKK-alpha in tumor cells following castration was dependent upon IKK-beta in infiltrating immune cells and the release of lymphotoxin. Inhibition of any component of this signaling resulted in a significant delay in the appearance of castration-resistant prostate cancer. Inhibition of MEK may trigger up regulation of NFκB signaling since NFκB activation can lead to an increase in Bcl-X(L) in some systems (37). Such an up regulation could blunt the effectiveness of therapies by facilitating cell survival and castration-resistance.
mTOR is a protein kinase downstream of PTEN/PI3K/Akt signaling that regulates protein translation, cell growth, and apoptosis (38). The implication of inhibiting mTOR in isolation is described above. Our data suggest that inhibiting MEK in vivo leads to an increase in Akt and mTOR activity. This observation is consistent with previous work demonstrating that blockade of EGFR to MAPK signaling conferred a decrease in IRS-1 serine phosphorylation thereby promoting IGFR to Akt signaling (39). MAPK signaling can affect IRS-1 serine phosphorylation either through direct phosphorylation by ERK or through the ability of ERK to transactivate p70S6K (39; 40). The inhibition of MEK in prostate xenografts appears to trigger a similar response and the combination of MEK and mTOR inhibition may counteract the effect of MEK inhibition on IRS-1 phosphorylation.
Hedgehog signaling is a major regulator of cellular differentiation and proliferation that is elevated in prostate cancer (29). Previous studies have suggested cross talk between Hedgehog and MAPK signaling; specifically ERK involvement in Gli regulation (41–43). In pancreatic cancer Gli is required for KRas mediated tumorigenesis (42; 44). Recently, direct evidence for ERK and JNK binding and phosphorylation of Gli transcription factors was reported (45). Loss of ERK signaling in prostate cancer may trigger an increase in canonical Hedgehog signaling. The combination of MEK and Hedgehog inhibition then leads to additive growth inhibition.
One implication of these observations is that a combination therapy targeting MEK along with inhibiting IKK, mTOR, or Hedgehog may be efficacious for the treatment of prostate cancer, although further work is necessary testing these combinations in preclinical models. Previously we showed that in vivo Ras blockade could restore androgen sensitivity to a castration resistant prostate cancer xenograft, C4-2 cells (7). This suggests that combining MEK inhibition with IKK, mTOR, or Hedgehog inhibition may be effective with androgen ablation. Also, since multiple signaling pathways are elevated in response to MEK inhibition it may be more efficacious in the clinic to use a cocktail of drugs targeting the compensatory pathways. One fundamental question remaining is if the compensatory pathways elevated in response to MEK inhibition observed in this study will be observed clinically. In our hands, CWR22Rv1 cells are the only AR positive prostate cancer cell line with active MAPK in vitro. We did not observe any additive or synergistic effect on cell cytotoxicity when testing the above combinations on LNCaP, C4-2, and LAPC4 cells. This is likely due to the lack of active MAPK in vitro, however, it is possible that the compensatory effects and subsequent effective derived drug combinations may be unique to a given cell line or individual. The broader implication of the data presented herein suggests that the conceptual paradigm of a global analysis to identify the compensatory signal transduction pathways in response to a molecular targeted agent can be used to determine effective drug combinations for the treatment of cancer, especially in the context of personalized medicine.
Supplementary Material
Table 1.
Table lists pathways and percent of gene products altered in response to PD325901 treatment identified using Pathway Express.
| Pathway Name | % Pathway Genes Altered |
|---|---|
| mTOR | 49% |
| Phosphatidylinositol | 32% |
| VEGF | 32% |
| Wnt | 31% |
| Insulin | 31% |
| TGF-beta | 30% |
| Notch | 30% |
| MAPK | 29% |
| Apoptosis | 29% |
| Jak-STAT | 14% |
| Hedgehog | 16% |
Acknowledgments
Financial support: Prostate Cancer Foundation, NIH, NCI (P01 CA76465, R01 CA105402) to MJW and NIH, NCI (R01 CA124706) to DG.
References
- 1.Eastman A, Perez RP. New targets and challenges in the molecular therapeutics of cancer. Br J Clin Pharmacol. 2006;62:5–14. doi: 10.1111/j.1365-2125.2006.02720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saijo N. What are the reasons for negative phase III trials of molecular-target-based drugs? Cancer Sci. 2004;95:772–6. doi: 10.1111/j.1349-7006.2004.tb02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krogan NJ, Hughes TR. Signals and systems. Genome Biol. 2006;7:313. doi: 10.1186/gb-2006-7-4-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natarajan M, Lin KM, Hsueh RC, Sternweis PC, Ranganathan R. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8:571–80. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 5.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 6.Kitano H. Cancer as a robust system: implications for anticancer therapy. Nature Reviews Cancer. 2004;4:227–235. doi: 10.1038/nrc1300. [DOI] [PubMed] [Google Scholar]
- 7.Bakin RE, Gioeli D, Bissonette EA, Weber MJ. Attenuation of Ras signaling restores androgen sensitivity to hormone-refractory C4-2 prostate cancer cells. Cancer Res. 2003;63:1975–80. [PubMed] [Google Scholar]
- 8.Bakin RE, Gioeli D, Sikes RA, Bissonette EA, Weber MJ. Constitutive activation of the Ras/mitogen-activated protein kinase signaling pathway promotes androgen hypersensitivity in LNCaP prostate cancer cells. Cancer Res. 2003;63:1981–9. [PubMed] [Google Scholar]
- 9.Gioeli D, Mandell JW, Petroni GR, Frierson HF, Weber MJ. Activation of mitogen-activated protein kinase associated with prostate cancer progression. Cancer Res. 1999;59:279–84. [PubMed] [Google Scholar]
- 10.Gordon V, Bhadel S, Wunderlich W, Zhang J, Ficarro SB, Mollah SA, et al. CDK9 regulates AR promoter selectivity and cell growth through serine 81 phosphorylation. Mol Endocrinol. 2010;24:2267–80. doi: 10.1210/me.2010-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986 Apr;31(4):229–234. doi: 10.1002/jso.2930310402. [cited 2011 Feb 22 ] [DOI] [PubMed] [Google Scholar]
- 12.Gentleman R. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer Science+Business Media; 2005. [Google Scholar]
- 13.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–45. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grubb RL, Deng J, Pinto PA, Mohler JL, Chinnaiyan A, Rubin M, et al. Pathway biomarker profiling of localized and metastatic human prostate cancer reveal metastatic and prognostic signatures. J Proteome Res. 2009 Jun;8(6):3044–3054. doi: 10.1021/pr8009337. [cited 2011 Apr 16 ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wulfkuhle JD, Paweletz CP, Steeg PS, Petricoin EF, Liotta L. Proteomic approaches to the diagnosis, treatment, and monitoring of cancer. Adv Exp Med Biol. 2003;532:59–68. doi: 10.1007/978-1-4615-0081-0_7. [DOI] [PubMed] [Google Scholar]
- 16.Saotome K, Morita H, Umeda M. Cytotoxicity test with simplified crystal violet staining method using microtitre plates and its application to injection drugs. Toxicol In Vitro. 1989;3(4):317–321. doi: 10.1016/0887-2333(89)90039-8. [cited 2011 Apr 16 ] [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nature Chemical Biology. 2006;2:458–466. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 18.Bliss C. The toxicity of poisons applied jointly. Annals of Applied Biology. 1939;26:585–615. [Google Scholar]
- 19.Gregory CW, Whang YE, McCall W, Fei X, Liu Y, Ponguta LA, et al. Heregulin-induced activation of HER2 and HER3 increases androgen receptor transactivation and CWR-R1 human recurrent prostate cancer cell growth. Clin Cancer Res. 2005;11:1704–12. doi: 10.1158/1078-0432.CCR-04-1158. [DOI] [PubMed] [Google Scholar]
- 20.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villaronga M, Bevan C, Belandia B. Notch signaling: a potential therapeutic target in prostate cancer. Curr Cancer Drug Targets. 2008;8:566–580. doi: 10.2174/156800908786241096. [DOI] [PubMed] [Google Scholar]
- 22.Sarker D, Reid A, Yap T, de Bono J. Targeting the PI3K/AKT pathway for the treatment of prostate cancer. Clinical Cancer Research. 2009;15:4799–805. doi: 10.1158/1078-0432.CCR-08-0125. [DOI] [PubMed] [Google Scholar]
- 23.Vezina C, Bushman A. Hedgehog signaling in prostate growth and benign prostate hyperplasia. Current Urology Reports. 2007;8:275–80. doi: 10.1007/s11934-007-0073-x. [DOI] [PubMed] [Google Scholar]
- 24.Robinson D, Zylstra C, Williams B. Wnt signaling and prostate cancer. Current Drug Targets. 2008;9:571–80. doi: 10.2174/138945008784911831. [DOI] [PubMed] [Google Scholar]
- 25.Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell. 2005;17:215–24. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 26.Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–8. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 27.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ammirante M, Luo JL, Grivennikov S, Nedospasov S, Karin M. B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature. 2010;464:302–5. doi: 10.1038/nature08782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–12. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 30.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest. 2001 Feb;107(3):241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee D-F, Hung M-C. Advances in targeting IKK and IKK-related kinases for cancer therapy. Clin Cancer Res. 2008 Sep;14(18):5656–5662. doi: 10.1158/1078-0432.CCR-08-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maira S-M, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008 Jul;7(7):1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 33.Dierks C. GDC-0449--targeting the hedgehog signaling pathway. Recent Results Cancer Res. 2010;184:235–238. doi: 10.1007/978-3-642-01222-8_17. [DOI] [PubMed] [Google Scholar]
- 34.Petricoin EF, Espina V, Araujo RP, Midura B, Yeung C, Wan X, et al. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007;67:3431–40. doi: 10.1158/0008-5472.CAN-06-1344. [DOI] [PubMed] [Google Scholar]
- 35.Carretero J, Shimamura T, Rikova K, Jackson A, Wilkerson M, Borgman C, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17:547–59. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasparian AV, Yao YJ, Kowalczyk D, Lyakh LA, Karseladze A, Slaga TJ, et al. The role of IKK in constitutive activation of NF-kappaB transcription factor in prostate carcinoma cells. J Cell Sci. 2002;115:141–51. doi: 10.1242/jcs.115.1.141. [DOI] [PubMed] [Google Scholar]
- 37.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–96. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–41. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- 39.Buck E, Eyzaguirre A, Rosenfeld-Franklin M, Thomson S, Mulvihill M, Barr S, et al. Feedback mechanisms promote cooperativity for small molecule inhibitors of epidermal and insulin-like growth factor receptors. Cancer Res. 2008;68:8322–32. doi: 10.1158/0008-5472.CAN-07-6720. [DOI] [PubMed] [Google Scholar]
- 40.De Fea K, Roth RA. Modulation of insulin receptor substrate-1 tyrosine phosphorylation and function by mitogen-activated protein kinase. J Biol Chem. 1997;272:31400–6. doi: 10.1074/jbc.272.50.31400. [DOI] [PubMed] [Google Scholar]
- 41.Schnidar H, Eberl M, Klingler S, Mangelberger D, Kasper M, Hauser-Kronberger C, et al. Epidermal growth factor receptor signaling synergizes with Hedgehog/GLI in oncogenic transformation via activation of the MEK/ERK/JUN pathway. Cancer Res. 2009;69:1284–92. doi: 10.1158/0008-5472.CAN-08-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernandez-Zapico ME, et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23:24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kessaris N, Jamen F, Rubin LL, Richardson WD. Cooperation between sonic hedgehog and fibroblast growth factor/MAPK signalling pathways in neocortical precursors. Development. 2004;131:1289–98. doi: 10.1242/dev.01027. [DOI] [PubMed] [Google Scholar]
- 44.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–55. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 45.Whisenant TC, Ho DT, Benz RW, Rogers JS, Kaake RM, Gordon EA, et al. Computational prediction and experimental verification of new MAP kinase docking sites and substrates including Gli transcription factors. PLoS Comput Biol. 2010:6. doi: 10.1371/journal.pcbi.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.