Abstract
Peptides derived from cytosolic, mitochondrial, and nuclear proteins have been detected in extracts of animal tissues and cell lines. To test whether the proteasome is involved in their formation, HEK293T cells were treated with epoxomicin (0.2 μM or 2 μM) for 1 hour and quantitative peptidomics analysis was performed. Altogether, 147 unique peptides were identified by mass spectrometry sequence analysis. Epoxomicin treatment decreased the levels of the majority of intracellular peptides, consistent with inhibition of the proteasome beta-2 and beta-5 subunits. Treatment with the higher concentration of epoxomicin elevated the levels of some peptides. Most of the elevated peptides resulted from cleavages at acidic residues, suggesting that epoxomicin increased the processing of proteins through the beta-1 subunit. Interestingly, some of the peptides that were elevated by the epoxomicin treatment had hydrophobic residues in P1 cleavage sites. Taken together, these findings suggest that while the proteasome is the major source of intracellular peptides, other peptide-generating mechanisms exist. Because intracellular peptides are likely to perform intracellular functions, studies using proteasome inhibitors need to be interpreted with caution as it is possible that the effects of these inhibitors are due to a change in the peptide levels rather than inhibition of protein degradation.
Keywords: Peptides, Proteasome, Protease, Peptidase
INTRODUCTION
Many previous peptidomics studies have been performed in a variety of organisms, ranging from invertebrates to humans.1-6 Although most peptidomics studies have been focused on neuropeptides, which are secreted from cells and function in cell-cell communication, the peptidomics techniques have detected a number of additional peptides which are derived from cytosolic, mitochondrial, and nuclear proteins.7, 8 There is growing interest in cytosolic peptides and the possibility that they may serve multiple functions within the cell.9, 10 It is well established that a fraction of the peptides produced in the cytosol are translocated into the endoplasmic reticulum, some of which then bind to major histocompatability complex class I molecules and traffic to the cell surface for presentation to T cells. In addition, cytosolic peptides may have other functions within the cell. It has been proposed that cytosolic peptides perform a regulatory role by modulating protein-protein interactions.7, 9 Relatively small peptides of 10-20 amino acids can mimic or block protein function and have been used in many systems to produce physiological changes in cellular function.10-12 Peptides generated from mitochondrial proteins by the matrix-localized protease ClpP are able to signal the stress of protein misfolding and activate nuclear-encoded mitochondrial chaperone genes in C. elegans.13 In Drosophila, peptides of 11 to 32 amino acids encoded by small open reading frame genes are able to control epidermal differentiation by modifying the transcription factor Shavenbaby.14 Together, these data suggest that intracellular peptides can play an important physiological role controlling a variety of cellular functions.
The ubiquitin-proteasome system is considered to be the major mechanism for the turnover of cytosolic proteins.15 The proteasome consists of multiple subunits, of which three are active proteases: the beta-1 subunit cleaves peptide bonds with acidic residues in the P1 position, the beta-2 subunit cleaves peptide bonds with basic residues in the P1 position, and the beta-5 subunit cleaves peptide bonds with hydrophobic residues in the P1 position.16, 17 The proteasome complex degrades proteins into short peptides of 3-30 amino acids, with an average size around 10 amino acids.18 Many inhibitors of proteasomes have been reported, each with a different specificity for the various active subunits. Crystal structure of the epoxomicin-20S proteasome complex revealed that at lower concentrations epoxomicin displays a high degree of selectivity for covalent binding to beta-5 subunits, while at higher concentrations this inhibitor can covalently bind to the substrate binding pocket of all catalytic subunits.19 In the HeLa cell line, epoxomicin has been shown to inhibit the proteasome with greatest potency for the beta-5 subunit and lower potency for the beta-2 subunit without affecting protein degradation by the beta-1 subunit.20
In addition to the proteasome, there are three other families of endopeptidases that are common to all cells and which can cleave cytosolic proteins: calpains, caspases, and lysosomal enzymes.21-23 None of these other protease families are known to be inhibited by epoxomicin concentrations up to 50 μM.24 In general, calpains and caspases are considered to perform limited proteolysis of proteins, playing a key role in activation or inactivation of protein function, rather than a direct role in protein turnover. Lysosomes are thought of as degradative organelles that convert proteins into amino acids by a series of endo- and exopeptidase activities. There are also several cytosolic oligopeptidases that can cleave peptides; examples include endopeptidase 24.15 (also known as thimet oligopeptidase), endopeptidase 24.16 (also known as neurolysin), post-prolyl oligopeptidase, insulin degrading enzyme, and others.25-30 These oligopeptidases do not cleave most cellular proteins, and instead are selective for a subset of peptides.25-31 Peptides produced by the proteasome and other intracellular peptidases are subsequently cleaved into amino acids by a series of aminopeptidases and other enzymes.32, 33
The focus of the present study was to test the hypothesis that cytosolic peptides detected in cells are produced by the proteasome. For this, HEK293T cells were treated with 0.2 or 2 μM epoxomicin for 1 hour; these concentrations and length of treatment were based on a previous study on protein turnover in cultured cells.20 Then, the levels of peptides in epoxomicin-treated cells were compared to levels in untreated cells using a quantitative peptidomics approach that employs amine-labeling reagents that are chemically identical except for the ratio of stable isotopes in each compound.3, 34 Because the peptides labeled with the different isotopic tags coelute from HPLC, the relative levels of peptide in the original extract can be determined from the peak intensity of each isotopic form detected by mass spectroscopy.34 Using this technique, a number of peptides were previously detected in the HEK293T cells.8, 31 In the present study, a large number of these peptides were again detected, as well as new peptides. The levels of many peptides were reduced by the treatment with epoxomicin, although other peptides were elevated by the treatment, presumably reflecting the switch in the cleavage patterns due to the inhibition of the beta-2 and beta-5 subunits. This finding demonstrates that proteasome inhibitors do not only affect protein levels but also affect the cellular peptidome by changing the processing pathways. If these cellular peptides have physiological functions, this global change in peptide pattern caused by proteasome inhibitors may contribute to some of the physiological effects of these compounds.
MATERIALS AND METHODS
Reagents
Acetonitrile was purchased from Fisher. Hydrochloridric acid and trifluoroacetic acid mass spectroscopy grade were from Pierce Thermo Scientific. Hydroxylamine, glycine, sodium hydroxide, sodium phosphate, and dimethyl sulfoxide (DMSO), were obtained from Sigma. Epoxomicin was obtained from Calbiochem. The 4-trimethylammoniumbutyryl-N-hydroxysuccinimide (TMAB-NHS) stable isotopic labeling reagents, containing either 0, 3, or 9 atoms of deuterium (D0-, D3-, and D9-, respectively) or 9 atoms of deuterium and three 13C atoms (D12-) were synthesized as described.35
Large scale cell culture, epoxomicin treatment, and peptide extraction
HEK293T cells were grown to 90% confluence in 15 cm cell culture plates in high glucose Dulbecco’s Modified Eagle’s Medium (D-MEM, Invitrogen), supplemented with 10% fetal bovine serum and 1% pen/strep antibiotic. Two plates of cells were used per group. At the start of the experiment, media were removed from all plates and media with or without epoxomicin were placed into the plates incubated at 37°C for 1 hr. Two concentrations of epoxomicin were used: 0.2 μM and 2 μM, prepared by a 1:2500 dilution of 0.5 and 5 mM solutions of epoxomicin in DMSO, respectively. For each concentration of drug, two groups of cells were treated with the drug and two groups of cells were untreated controls.
Following incubation, cells were washed three times with Dulbecco’s phosphate-buffered saline (Invitrogen) and centrifuged at 800 × g for 5 min. The pellet was resuspended in 1 mL of 80°C water and incubated in an 80°C water bath for 20 min. The cell lysate was centrifuged at 13,000 rpm in a microfuge for 15 minutes at 4°C, and stored at −80°C overnight. The samples were thawed and centrifuged again at 13,000 rpm for 30 minutes at 4°C and the supernatant collected for peptide extraction. To extract peptides, samples were first concentrated in a vacuum centrifuge to ~750 μL, cooled on ice, and acidified with 75 μL of ice-cold 0.1 M HCl to a final concentration of 10 mM HCl. The samples were incubated 15 minutes on ice and centrifuged at 13,000 x g for 40 min at 4°C. The supernatant was transferred to a new 2 mL low retention tube and stored at −70°C until labeling.
Isotopic labeling and mass spectrometry
The labeling procedure has been previously described in detail.35 In brief, each group of cells within an experiment was labeled with one of four isotopic TMAB-NHS labels. The two biological replicates of the 0.2 μM epoxomicin-treated cells were labeled with D0- and D12-TMAB-NHS while the two biological replicates of the control groups were labeled with D3- and D9-TMAB-NHS. This labeling was altered between experiments; for the other concentration of epoxomicin (2 μM), the inhibitor-treated samples were labeled with D3- and D9-TMAB-NHS while the untreated controls groups were labeled with D0- and D12-TMAB-NHS.
The peptide extracts were combined with 250 μL of 0.4 M phosphate buffer, pH 9.5. The pH of the samples was adjusted to 9.5 with 1 M NaOH. A total of 15 mg of TMAB-NHS label per group (i.e. two 15 cm plates of cells) was used for the labeling procedure. Each TMAB-NHS was dissolved in DMSO at a concentration of 0.4 mg/μL and 1/7th of the volume (5.4 μL) was added at a time to the peptide extracts. After 10 min at room temperature, an appropriate volume of 1.0 M NaOH was added to the reaction mixture to adjust the pH back to 9.5, and the reaction was further incubated for 10 min. This procedure was repeated for a total of 7 full cycles of TMAB-NHS addition and pH adjustment. After the final addition of TMAB-NHS, the mixture was incubated at room temperature an additional 60 min. To quench any remaining labeling reagent, 30 μL of 2.5 M glycine was added to the reaction. After 40 min at room temperature, the samples labeled with the different isotopic label were combined and concentrated to 2 mL in a vacuum centrifuge. The concentrated samples were pooled and filtered through a Microcon YM-10 unit (Millipore). The pH was adjusted to 9.0, and 2.0 M hydroxylamine (a total of 5 μL per 15 mg label) was added to remove TMAB labels from Tyr residues. The addition of hydroxylamine was split into three cycles, with the adjustment of the pH to 9.0 before each addition of hydroxylamine. The samples were desalted with a C18 spin columns (Pierce-Thermo Sci) the resin from two columns were combined into one, and the activation, washing and elution conditions were as described by the manufacturer. The peptides were eluted from the C18 column with 160 μL of 0.5% trifluoroacetic acid in 70% acetonitrile. The eluate was dried in a vacuum centrifuge and analyzed by liquid chromatography/mass spectrometry (LC-MS/MS) on a Synapt G1 mass spectrometer with a nanoACQUITY UltraPerformance LC System (Waters Co., EUA). The peptide mixture was desalted online for 15 min using a Symmetry C18 trapping column (5 μm particles, 180 μm i.d.× 20 mm, Waters, USA) and the trapped peptides were then separated by elution with a water/acetonitrile 0.1% formic acid gradient through a BEH 130 -C18 column (1.7 μm particles, 100 μm i.d. × 100 mm, Waters, USA), as previously described in more detail.31 Data were acquired in data-dependent mode and selected peptides dissociated by collisions with argon. The liquid chromatography and electrospray ionization conditions included a flow rate of 600 nL/min, nanoflow capillary voltage of 3.5 kV, block temperature of 100°C, and cone voltage of 100 V.
The MS spectra were analyzed using the MassLynx software (Waters) to identify groups of peaks representing peptides labeled with the different isotope tags. Quantification was performed by determining the relative intensity of each isotopic peak, considering both the mono-isotopic and the peak containing one atom of 13C and subtracting the baseline due to overlapping lower-mass peaks.36 The intensity of each of the four isotopic TMAB-labeled peptides was compared to the average of the two control replicates in each group. For these analyses, a single spectrum corresponding to the highest ion current was analyzed if the relative levels of the different isotopic-labeled peptides were representative of the other spectra for the peptide. In some cases, several spectra were averaged together prior to quantification if this provided a more representative view of the relative levels among spectra.
To identify the peptides, the MS/MS data was analyzed using the Mascot search engine (Matrix Science Ltd, UK) with variable modifications of N-terminal acetylation, methionine oxidation, and the isotopic D0-, D3-, and D9-TMAB tags used in our study (listed on Mascot as GIST-Quat). The D12-TMAB tag used for one of the replicates in our study is not available as a search option in Mascot. All results were manually interpreted to eliminate false positives, using criteria previously described.31, 35-39 In brief, these criteria include nine points: (i) the isotopic form of TMAB matched by Mascot corresponds to the isotopic form based on analysis of the peak set. Mascot does not consider the peak set and recognize which of the individual peaks correspond to the D0, D3, D9, or D12 forms. Therefore, correlation of the isotopic TMAB form in the observed peak set with the predicted Mascot match is a simple and necessary step, and most false positives (~75%) will fail this test. (ii) The number of tags incorporated into the peptide matches the number of free amines (i.e. the N-terminus, if not blocked by modification, and side chain amines of Lys). If multiple tags are incorporated, all should be the same isotopic form on a particular peptide. False positives will rarely have consistent forms of the tags, while real positives are always the same isotopic form. (iii) The Mascot score is either the top score of all potential peptides, or the other peptides with comparable scores can be excluded by the above criteria. (iv) The vast majority of the major MS/MS fragment ions match predicted a, b, or y ions, internal ions, or precursor ions with loss of trimethylamine. (v) A minimum of five fragment ions match b- or y-series ions. (vi) The mass accuracy of the fragment ions is within the accepted specification for the q-TOF instrument used for the analysis, usually within 20 parts per million. (vii) The charge state should be reasonable based on the peptide sequence, usually corresponding to the number of isotopic tags (which are positively charged) plus the number of Arg and His residues, although His residues are not always positively charged and often exist with two different charge states. Some peptides pick up an additional proton to give a charge state one higher than the maximum predicted from the number of amines, and some lose a proton (presumably from –COOH groups on Glu, Asp, and the C-terminus) to give a charge state lower than expected. However, in both cases, these forms are usually minor, relative to the ion with the correct charge state. (viii) The fragment ions match the expected ions based on the peptide sequence. Fragmentation of Xaa-Pro bonds is favored, while Pro-Xaa bonds is unfavored, as are cleavages near Gly residues. The b2 (and a2) ions are usually very strong if the N-terminus has a TMAB-label attached (i.e. not acetylated), unless there is an N-terminal Pro or a basic residue in positions 1 or 2. (ix) For some peptides, MS/MS fragmentation was not obtained in any of the four LC/MS runs performed for the samples in the present study, but the peptide was previously identified by MS/MS analysis of another HEK 293 cell sample, using the above criteria to validate the Mascot results. In these cases, the peptide detected in the present study was considered to be the same as the previously identified peptides if the observed mass was within 40 ppm of the theoretical value and the number of TMAB tags, charge state, and elute time from the LC column were comparable to the previously identified peptide (elute time within 1-2 minutes). Mascot results or annotated MS/MS spectra of representative peptides are included in a supplemental file. To reduce the size of this file, some of the peptides previously identified in HEK 293 cell extracts and previously published8, 31 are excluded from the attached supplement.
RESULTS
Epoxomicin, a natural product isolated from Actinomycetes, is a cell permeable, potent, highly selective and irreversible proteasome inhibitor that inhibits the beta-5 site at low concentrations (0.15 μM) and also inhibits the beta-2 site when incubated with cells at higher concentrations such as 2 μM.20, 24 Therefore, we incubated HEK293 cells for 1 hour with either medium alone, or medium containing 0.2 or 2 μM epoxomicin and examined the effect on the cellular peptidome using a quantitative peptidomics approach. Quantification of the relative levels of peptides in the control and epoxomicin-treated cells was performed by measurement of peak intensity for each of the four isotopic-labeled peptides. A total of 147 distinct peptides were identified by MS/MS sequence analysis and another 135 peptides could not be identified from the sequence analysis. The 147 peptides represent naturally-occurring fragments of 43 different proteins: fragments of all but 6 of these proteins were found in previous peptidomics studies of HEK293, MCF7 and SH-SY5Y cells.8, 31
The entire data set is shown in Table S1 (supplement). This table contains 478 entries, each line representing a peak set of a peptide found in one of the LC/MS runs. If the peptide was found in two different charge states, it appears in two lines of this table, reflecting the different m/z values of the peaks that were detected. For those peptides found in all four of the samples that were pooled for the LC/MS run (i.e. control replicates and epoxomicin-treated replicates), the table shows the ratio of each of these four samples relative to the average of the two control replicates of this sample. The comparison of the individual control samples to the average of the two controls provides an indication of the variability of this peptide among replicates. The comparison of the epoxomicin-treated samples to the average controls indicates whether the peptide was present at lower levels after epoxomicin treatment (ratio below 1.0), or elevated by the treatment (ratio above 1.0). For those peptides detected only in the control samples, and not in the epoxomicin-treated samples, the epoxomicin/control ratio was listed as <0.2 to reflect the detection limit of a typical peptide. Conversely, those peptides detected only in the epoxomicin-treated samples and not in the controls were listed with an epoxomicin/control ratio of >5.
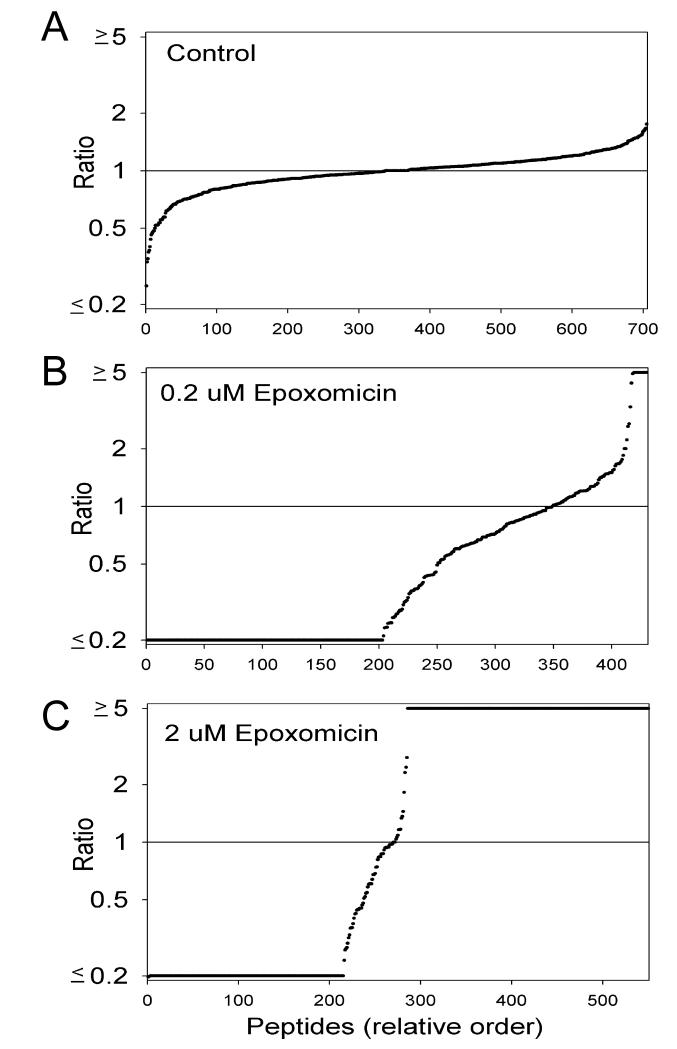
A variety of analyses were performed on the data. To visualize the variation of each peptide among replicates, the individual ratios for all the control groups were placed into a spreadsheet and sorted from low to high. Then, these values were plotted (Figure 1A). The y-axis shows the ratio of each peptide, relative to the average of the two controls in each run, and the x-axis is the relative rank order of the peptide after sorting. Nearly all of the peptides in the control replicates were within 2-fold of the average control value (i.e. ratio between 0.5 and 2.0). In contrast, when the ratio of the epoxomicin-treated samples relative to the average control values were plotted, only a small number of peptides fell within the 0.5-2.0 range (Figure 1B, C). After treatment of cells with 0.2 μM of epoxomicin for 1h, approximately 1/2 of the peptides were not detected in the cells (ratio <0.2), and many more were decreased with ratios of 0.2-0.8 (Figure 1B). A small number of peptides showed elevated epoxomicin/control ratios after treatment with 0.2 μm epoxomicin (Figure 1B). When cells were treated with the higher concentration of epoxomicin, ~200 peptides showed a ratio of <0.2 (Figure 1C); this is similar to the number that decreased substantially with the lower concentration of epoxomicin. However, many additional peptides showed a ratio of >5; these were detected only in the epoxomicin-treated cells and not in the control replicates (Figure 1C). Thus, treatment of HEK293 cells with 0.2 or 2 μM epoxomicin alters the dynamic equilibrium of the intracellular peptides, with virtually all the identified peptides affected by the higher dose of 2 μM.
Figure 1. Analysis of the peptidome of HEK293 cells.
The relative level of every peptide, both identified and unidentified by MS/MS analysis, was calculated for each of the replicates and expressed relative to the average level of the peptide in the two replicates of untreated cells. Then, the relative level was sorted from low to high and plotted on the y-axis; for these plots, the x-axis represents the rank order of each peptide when sorted by the relative level. Peptides only present in the epoxomicin-treated cells and not detected in the control cells were listed with a ratio of ≥5, and those peptides only present in the control cells and not detected in the epoxomicin-treated cells were listed with a ratio of ≤0.2. For this analysis, if a peptide was detected with two different charge states, each value is indicated separately. A: The ratio of each control replicate was expressed versus the average of the controls. B: The ratio of cells treated for 1 hour with 0.2 μM epoxomicin, relative to the average of the untreated control cells. C: The ratio of cells treated for 1 hour with 2 μM epoxomicin, relative to the average of the untreated control cells.
For the summary plots shown in Figure 1, all peptides were included, both identified and unidentified. In order to provide information on the variability of a peptide within the replicates of each experiment and between experiments, we further analyzed the data using a heat map plot (Figure 2). For this plot, only the 147 known peptides were considered, and each of the rows represents a different peptide. For those peptides found multiple times with different charge states, the ratios were averaged such that a single value is indicated for each replicate. In general, the multiple charge states showed similar ratios within a particular replicate (see Table S1). In the heat map, each column represents a separate group of plates within each experiment (i.e. each of the two replicates) for either the epoxomicin-treated samples (left columns) or corresponding control samples (right columns). The color scheme used in the heat map is indicated on the bottom of Figure 2; peptides showing a very large decrease relative to the average control (ratio <0.2) are in bright green; peptides that greatly increased (>5) are in bright red, and peptides showing smaller changes are in gradated shades, with grey representing peptides that did not substantially change and white representing peptides either not detected or for which peak overlap precluded analysis. The data for this plot (i.e. information on each peptide, ratio values, and statistical calculations) are included in Supplemental Table S2. For the majority of peptides, the relative level of peptide in the two replicates of epoxomicin-treated cells was fairly close and was either the same for both replicates, or reflected minor variability (Figure 2, Table S2). Similarly, the peptides in the control samples did not show large variability among replicates (Figure 2, Table S2). Most of the peptides that were decreased by the treatment with 0.2 μM of epoxomicin were also decreased by treatment with 2 μM epoxomicin. In contrast, many of the peptides greatly elevated by treatment with 2 μM epoxomicin (bright red squares) were not detected in the control samples or in the 0.2 μM epoxomicin-treated samples (white squares). Most of the peptides which greatly increased upon treatment with 2 μM epoxomicin required cleavage to the C-terminal side of an Asp or Glu residue, which is due to the beta-1 subunit of the proteasome. The majority of the peptides which were produced by cleavage at Lys or Arg residues, catalyzed by the beta-2 subunit of the proteasome, were decreased with 2 μM epoxomicin but were not greatly influenced by 0.2 μM epoxomicin (Figure 2). Most of the peptides that required cleavage at hydrophobic residues, which represents the beta-5 proteasome subunit activity, were decreased by treatment with either concentration of epoxomicin. Peptides that required other P1 cleavage site residues (Pro, Gly, Ala, Ser, Thr, Cys, Asn, Gln, and His), which may be substrates of the beta-5 proteasome subunit, were also generally decreased by treatment with either concentration of epoxomicin (Figure 2). The two small proteins detected in our analysis, thymosin beta-4 and beta-10, were unaltered by the epoxomicin treatment (Figure 2). While most of the observed changes fit the expected results, there were a number of exceptions (Figure 2), and so further analysis was performed.
Figure 2. Heat map analysis of identified peptides.
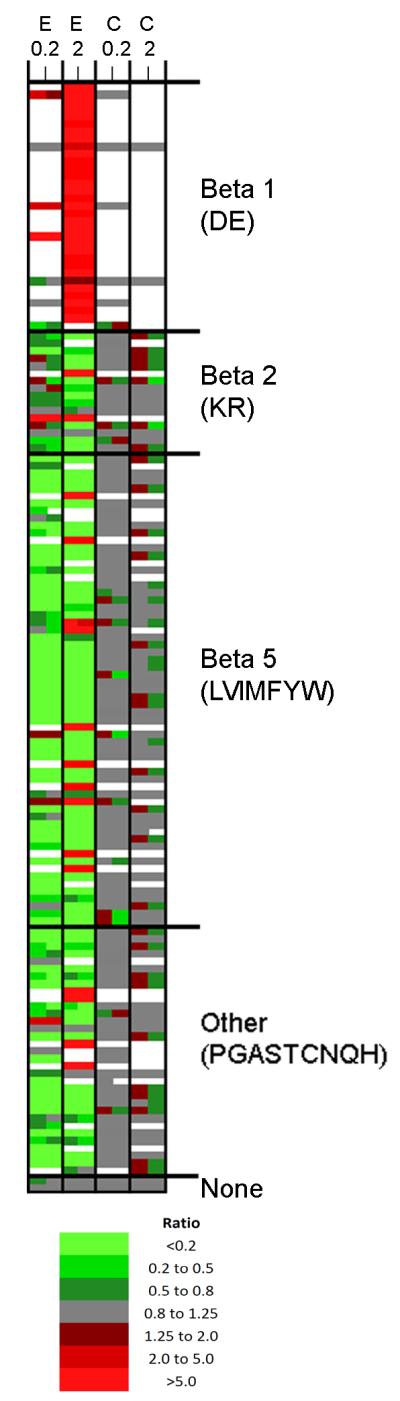
The 147 different peptides identified by MS/MS sequence analysis were plotted as separate rows; for this analysis, peptides found in two or more different charge states were averaged together. The individual replicates for each experiment are indicated in separate columns. E, epoxomicin; C, control. The controls for the 0.2 and 2 μM epoxomicin experiments are indicated separately; all four of these replicates represent similar controls. The ratio was color coded using the scheme shown in the bottom of the figure, with green representing decreases and red representing increases, relative to the average of the untreated control samples. White represents peptides that were not detected, or which were detected but showed peak overlap with another peptide that prevented accurate quantification of the relative level. The various peptides were sorted by cleavage site required to generate the observed peptide; beta-1 sites (with D or E in the P1 position), beta-2 (K or R in this position), beta-5 (L, V, I, M, F, Y, or W in this position), or other amino acids that are neither acidic, basic, or hydrophobic (P, G, A, S, T, C, N, Q, or H), and which may be cleaved by the beta-5 subunit. Two peptides corresponded to small proteins (thymosin beta-4 and beta-10) that did not require any cleavage, other than removal of the N-terminal initiation methionine (which was not considered in this analysis as a cleavage). For this analysis, only half of the peptides represented N- or C-terminal protein fragments, which required a single cleavage to generate the peptide. For the internal peptides that required two cleavages, if either side was an acidic residue, it was grouped in the beta-1 (DE) group. For the remaining peptides, if either side was a basic residue it was placed in the beta-2 (KR) group. Of the remaining peptides, only those with a hydrophobic group on both sides were placed in the beta-5 (LVIMFYW) group, and the remainder were placed in the “other” group. Names of proteins, peptide sequences, and other information including the values of the ratios and statistical calculations are included in supplemental Table S2.
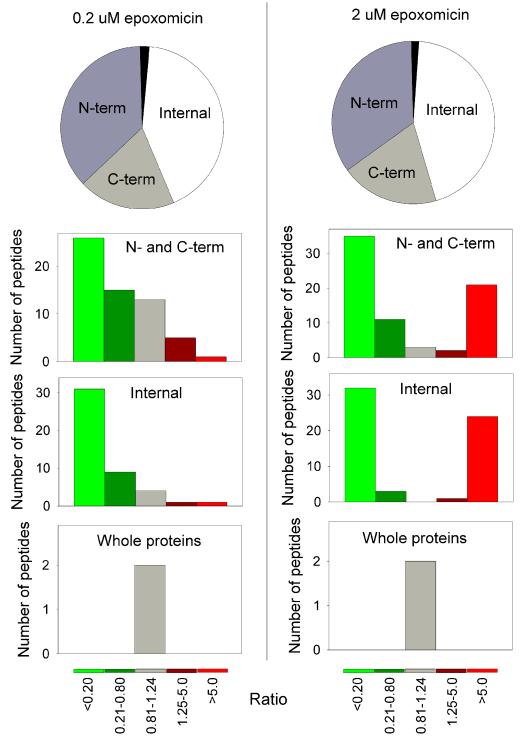
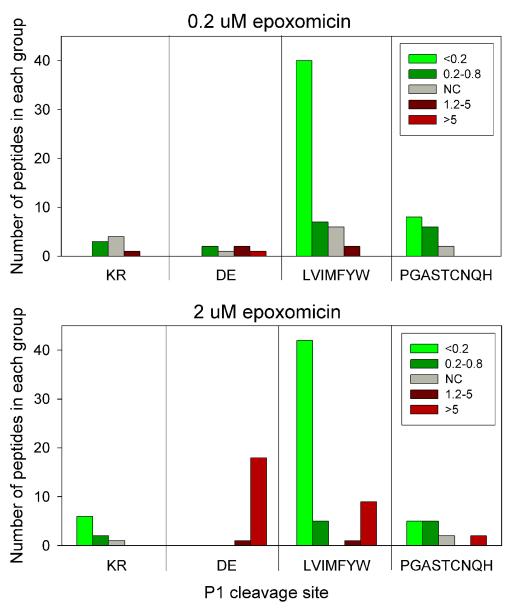
In previous peptidomic studies on mouse brain and human cell lines, approximately half of the protein fragments corresponded to either an N- or C-terminal piece of the protein.7, 8 A similar result was found for the 147 peptides identified in the present study (Figure 3). In general, the peptides corresponding to the N- or C-terminal fragments were affected by epoxomicin to the same degree as the internal protein fragments (Figure 3). This result indicates that the N- and C-terminal protein fragments are generated by the proteasome, as are the internal fragments. Therefore, for the subsequent analysis we focused mainly on the N- and C-terminal fragments because the cleavage site required to generate these peptides is clear; a single cleavage liberates the peptide from the protein. In contrast, the internal peptides require two cleavages to produce the peptide, and if not the same (i.e. beta 1 site on one side, beta 2 on the other), it would not be easy to predict whether the peptide should increase or decrease after epoxomicin treatment. Therefore, only those internal peptides that contained the same type of residue in the P1 position on both sides were included in the further analysis along with all of the N- and C-terminal protein fragments. The majority of peptides that decreased upon treatment with either concentration of epoxomicin contain hydrophobic residues in the P1 position of the cleavage sites, representing the beta-5 activity (Figure 4). Peptides with Lys or Arg residues in the P1 positions of the cleavage sites were not greatly affected by the low concentration of epoxomicin but were greatly decreased by the high concentration of epoxomicin (Figure 4). Only a small number of peptides resulting from cleavages at acidic residues were detected in the 0.2 μM epoxomicin study, and many more were detected only in the 2 μM epoxomicin-treated samples (Figure 4). Furthermore, the levels of the peptides that required cleavage at acidic residues were greatly increased by treatment with the high concentration of epoxomicin, consistent with the expectation that inhibition of the beta-2 and beta-5 subunits would result in elevated beta-1 cleavages. However, a number of peptides requiring cleavage at hydrophobic sites were also greatly elevated by treatment with 2 μM epoxomicin (Figure 4); this does not fit with the expected inhibition of the beta-5 subunit of the proteasome.
Figure 3. Location of the peptide within the protein, and influence of epoxomicin on the levels of each subgroup of peptides.
The pie graphs show the fraction of peptides representing N-terminal (N-term), C-terminal (C-term), or internal fragments of the precursor protein. The black slice represents the two small proteins detected in this analysis (thymosins) that do not require cleavages. For each group, the effect of epoxomicin on peptide levels is indicated, using the coloring scheme shown at the bottom of the figure. The y-axes show the number of peptides detected for each of these groups, and with the indicated ratio for the epoxomicin versus untreated cells.
Figure 4. Effect of epoxomicin treatment on peptide levels; correlation with cleavage site.
For this analysis, only peptides with a single cleavage (i.e. N- or C-terminal peptides) or internal peptides that required cleavage at P1 sites which fell into the same group were considered. Peptides that reflected internal fragments requiring distinct cleavages were excluded from this analysis. The number of peptides found in each group is indicated.
The exceptions to the expected change in peptide levels were further examined. Seven of these peptides represent N- and C-terminal protein fragments, and in all cases there were additional N- and C-terminal fragments detected. For example, the 1712.88 Da C-terminal fragment of cytochrome c oxidase 5a is formed by cleavage of a Leu-Asn bond, and this peptide increased >5 fold when cells were treated with 2 μM epoxomicin (Table 1). Four other C-terminal fragments of this protein were detected, two of which decreased while the others increased. The two decreased peptides represented cleavages after Leu, and the two elevated peptides represented cleavages after Glu; this fits the general observation for other peptides. Similarly, the other peptides that do not fit the expectation that cleavages at hydrophobic residues would decrease upon epoxomicin treatment also have related peptides that do fit with the expected changes (Table 1). In several cases, there is a larger peptide that is greatly elevated by the epoxomicin treatment, raising the possibility that the shorter “anomalous” peptide is cleaved from the larger peptide by intracellular peptidase(s).
Table 1.
| Protein | Observed Mass |
N term | Peptide sequence | C term | Ratio 0.2 μM |
Ratio 2 μM |
|---|---|---|---|---|---|---|
| Cytochrome c oxidase subunit 5a | 1356.71 | l | GISTPEELGLDKV | * | <0.2 | 0.41 |
| Cytochrome c oxidase subunit 5a | 1469.80 | e | LGISTPEELGLDKV | * | nd | >5 |
| Cytochrome c oxidase subunit 5a | 1712.88 | l | NELGISTPEELGLDKV | * | nd | >5 |
| Cytochrome c oxidase subunit 5a | 2180.17 | l | RPTLNELGISTPEELGLDKV | * | <0.2 | <0.2 |
| Cytochrome c oxidase subunit 5a | 2293.25 | e | LRPTLNELGISTPEELGLDKV | * | 1.05 | >5 |
| Heat shock 10kDa protein 1 (chaperonin 10) | 1249.63 | f | RDGDILGKYVD | * | <0.2 | <0.2 |
| Heat shock 10kDa protein 1 (chaperonin 10) | 2063.04 | d | KDYFLFRDGDILGKYVD | * | nd | >5 |
| Heat shock 10kDa protein 1 (chaperonin 10) | 2293.09 | l | DDKDYFLFRDGDILGKYVD | * | nd | >5 |
| Triosephosphate isomerase 1 | 1400.77 | l | KPEFVDIINAKQ | * | 0.71 | >5 |
| Triosephosphate isomerase 1 | 2914.51 | l | ASQPDVDGFLVGGASLKPEFVDIINAKQ | * | 1.17 | <0.2 |
| Triosephosphate isomerase 1 | 3284.73 | c | KELASQPDVDGFLVGGASLKPEFVDIINAKQ | * | <0.2 | 0.77 |
| Eukaryotic translation elongation factor 1 beta | 1060.56 | *m | GFGDLKSPAGL | q | nd | >5 |
| Eukaryotic translation elongation factor 1 beta | 1287.68 | *m | GFGDLKSPAGLQV | l | 0.47 | <0.2 |
| Eukaryotic translation elongation factor 1 beta | 1400.77 | *m | GFGDLKSPAGLQVL | n | <0.2 | 0.61 |
| Eukaryotic translation elongation factor 1 beta | 1629.84 | *m | GFGDLKSPAGLQVLND | y | nd | >5 |
| Eukaryotic translation elongation factor 1 beta | 1905.98 | *m | GFGDLKSPAGLQVLNDYL | a | <0.2 | <0.2 |
| Eukaryotic translation elongation factor 1 beta | 2092.05 | *m | GFGDLKSPAGLQVLNDYLAD | k | nd | >5 |
| Eukaryotic translation elongation factor 1 beta | 2583.32 | *m | GFGDLKSPAGLQVLNDYLADKSYI | e | nd | >5 |
| Macrophage migration inhibitory factor | 2073.07 | *m | PMFIVNTNVPRASVPDGFL | s | 1.61 | >5 |
| Macrophage migration inhibitory factor | 2872.48 | *m | PMFIVNTNVPRASVPDGFLSELTQQL | a | 0.38 | 0.51 |
| Phosphatidylethanolamine-binding protein 1 | 1397.76 | *m | PVDLSKWSGPLSL | q | 0.54 | 4.92 |
| Phosphatidylethanolamine-binding protein 1 | 1654.86 | *m | PVDLSKWSGPLSLQE | v | >5 | >5 |
| Phosphatidylethanolamine-binding protein 1 | 1868.95 | *m | PVDLSKWSGPLSLQEVD | e | nd | >5 |
| Phosphatidylethanolamine-binding protein 1 | 3198.60 | *m | PVDLSKWSGPLSLQEVDEQPQHPLHVTY | a | 0.33 | <0.2 |
DISCUSSION
The initial purpose of the present study was to identify the enzymatic source of the cellular peptides found in previous peptidomics studies using human cell lines, many of which have also been detected in extracts of mouse brain.7, 35, 37, 38, 40-42 Although some of the protein precursors of these peptides are common cytosolic, mitochondrial, or nuclear proteins, only a few correspond to the most abundant cellular proteins.8, 43 The protein precursors of the peptides detected in the human cell lines do not have especially high turnover rates based on a proteomic analysis of protein turnover in a human adenocarcinoma cell line.8, 44 A recent study on the ubiquitin-modified peptides in HEK293 cells found only a small number of the peptide precursor proteins detected in the present study,45 further suggesting that these proteins are not the most unstable proteins in a cell. One possible explanation is that the peptides detected in the human cell lines represent selective cleavages of the proteins rather than degradation fragments. Consistent with this idea is the finding that 50% of the peptides detected in mouse brain and human cell lines represent the N- or C-terminus of the protein, raising the possibility that limited proteolysis within the N- or C-terminal region generates the peptide without complete protein degradation.7, 8 Intracellular proteases such as caspases and calpains perform limited cleavage of proteins.21 However, the specificity of caspases for cleavage at Asp residues doesn’t fit with the observed intracellular peptides, most of which are produced by cleavages at hydrophobic residues. The possibility that calpains contribute to the production of the intracellular peptides is unlikely because a previous study found that activation of calpains (by treatment of cells with a calcium ionophore) did not alter the levels of most intracellular peptides.8 The present finding that most peptides detected in HEK293T cells are either reduced or increased by treatment with epoxomicin, a highly specific irreversible proteasome inhibitor, suggests that the proteasome is the main enzyme responsible for intracellular peptide production.
Peptides whose levels decrease upon treatment with the low concentration of epoxomicin generally contain hydrophobic residues in the P1 sites, consistent with the known specificity of epoxomicin for the beta-5 proteasome subunit and the preference of this subunit for hydrophobic residues in the P1 site.20, 24, 46 Epoxomicin has been tested in a variety of cell lines, and typically has shown its proteasome inhibitory effects in concentrations that vary from nM to low μM.20, 24, 47 At the higher concentration of epoxomicin used in the present study (2 μM), most of the peptides that required cleavage at sites with basic residues in the P1 position were decreased. This is consistent with the lower potency of epoxomicin at the beta-2 proteasome subunit, which cleaves at Lys and Arg residues, compared to the beta-5 proteasome subunit.20, 24, 46 The increase in the levels of peptides in response to the epoxomicin treatment is mainly found for peptides requiring cleavage at acidic residues, which is catalyzed by the proteasome beta-1 subunit; this activity is not inhibited by 2 μM epoxomicin. The most likely explanation of the increase in these peptides is that the inhibitor alters the processing pattern of the protein by blocking the major processing activities and allowing for minor pathways to contribute to a greater extent. Alternatively, it is possible that the beta-1 proteasome subunit is activated upon epoxomicin treatment. Previous studies report the existence of a non-catalytic modifier site of peptide hydrolysis by the 20S proteasome in which effectors of proteasome activity (inhibitors or substrates) activate specific subunits by binding to non-catalytic sites.48 The existence of a mutual regulation of beta-5 and the beta-1 activities of the proteasome by their substrates has also been suggested as the substrates of the beta-5 subunit stimulate the beta-1-like activity and also activate the beta-5 site.49 The beta-5-like activity of the yeast 20S proteasome is enhanced in the presence of the beta-2-like substrates but reduced in the presence of beta-1-like substrates.50 Thus, the regulation of the various subunits of the proteasome is complex and the increase in beta-1-like products upon epoxomicin treatment may reflect activation of their cleavage.
Some of the peptides found to greatly increase when cells were treated with 2 μM epoxomicin arise from cleavage at hydrophobic residues, which is not consistent with inhibition of the beta-5 subunit. It is possible that some of the effects of epoxomicin reflect an off-target effect, although epoxomicin is reported to be highly specific for the proteasome.19, 20, 24 Alternatively, the peptides found to increase upon epoxomicin treatment may be generated by a distinct pathway, and not by the beta-5 proteasome subunit. A number of intracellular enzymes are known to process proteins and peptides inside the cells in addition to the proteasome. Tripeptidyl peptidase II has been suggested to be involved in the processing of proteins into peptides and although it primarily cleaves three residues from the N-terminus of peptides/proteins, it can also function as an endopeptidase.33, 51, 52 Several endopeptidases are known to be present in the cytosol, although they are selective for relatively small peptides and therefore unlikely to be responsible for the initial cleavage of the proteins. Examples include insulin degrading enzyme,30 endopeptidase 24.15,27, 28, 31 endopeptidase 24.16,26, 27 prolyl endopeptidases,29 and other enzymes.53 Although they are unlikely to initiate the cleavage of the proteins due to their substrate size restriction, these oligopeptidases may contribute to the production of the observed peptides from larger intermediates. For example, the 1712 Da peptide derived from the C-terminal region of cytochrome C oxidase subunit 5a (NELGISTPEELGLDKV) requires cleavage of a Leu-Asn bond, but was greatly elevated when cells were treated with epoxomicin (Table 1). A longer C-terminal peptide of 2293 Da (LRPTLNELGISTPEELGLDKV) was also greatly elevated by the epoxomicin treatment, which fits with the Glu-Leu cleavage necessary to generate this peptide (Table 1). If the 2293 Da peptide was converted into the 1712 Da peptide by an intracellular peptidase, then this would explain the large increase observed for the 1712 Da peptide upon epoxomicin treatment. Similarly, fragments of eukaryotic translation elongation factor 1 beta (GFGDLKSPAGL) and phosphatidylethanolamine-binding protein 1 (PVDLSKWSGPLSL) are elevated upon treatment of cells with epoxomicin despite the hydrophobic residues in the P1 cleavage site; longer forms of both peptides with acidic residues in the P1 site are detected and these longer peptides are elevated upon epoxomicin treatment (Table 1). Therefore, if these longer peptides are cleaved by intracellular peptidases into the shorter forms, this would explain the apparent paradox in the increased levels of the shorter form upon treatment of cells with epoxomicin. However, not all of the peptides which contain hydrophobic residues in the P1 position of the cleavage site and which are greatly elevated by the epoxomicin treatment correspond to smaller forms of peptides which are likely to be produced by the beta-1 proteasome subunit (Table 1).
The finding that the proteasome is responsible for the production of the majority of intracellular peptides raises the question as to why ~50% of these peptides represent the N- or C-terminal protein fragment. In theory, only a small fraction of the potential proteasome products should correspond to the N- or C-terminus of the protein. In support of this, analysis of peptides derived from secretory pathway proteins showed that only ~10% corresponded to N- or C-terminal fragments.7 Thus, selective detection of the N- or C-terminal fragments in the peptidomics analysis is unlikely to account for the large number of cytosolic and mitochondrial protein termini found in our analyses. Another possibility is selective production of these N- and C-terminal fragments. In addition to the classical destructive action, the proteasome is known to function in the limited proteolysis of several proteins. For example, to generate the active nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), transcriptional dimeric complexes (NF-κB1 and NF-κB2) undergo limited proteolytic processing by the proteasome to yield the respective shorter active subunits p50 and p52 that represent the N-terminal domains of their precursors.54 In addition, Drosophila Cubitus interruptus (Ci), and its vertebrate homologs Gli2 and Gli3, as well as the homologous yeast proteins Spt23 and Mga2, are only partially digested by the proteasome resulting in smaller protein fragments with new biological functions.55, 56 Although only a few examples are known of proteins that are selectively processed by the proteasome, a large number of cytosolic proteins undergo selective processing; a study examining proteins isolated from human Jurkat cells found that ~50% of the protein N-termini did not correspond to that predicted from the gene sequence, including signal peptide or pro-peptide removal.57 Thus, it appears that protein processing is much more common than previously thought, and some of this processing may be due to selective cleavage by the proteasome.
An alternative explanation for the large fraction of N- and C-terminal protein fragments in the cellular peptidome observed in this study, as well as previous studies,7, 8 is that these peptides are selectively preserved while other fragments are degraded. A previous study reported that the half-life of peptides within cells was less than 10 seconds, although this study examined a single peptide that was modified by a bulky fluorescent group and therefore may not reflect the turnover of most cellular peptides.58 It is possible that a subset of peptides (i.e. those observed in the various peptidomics studies) are bound to cellular proteins and therefore protected from further degradation, while the unbound peptides are degraded by cellular peptidases. A study on peptides that associate with major histocompatibility complex class I molecules found that a cytosolic pool of certain peptides was detectable hours after the production of the peptides was inhibited, and this cytosolic pool required heat-shock protein 90.59 The peptides observed in the present study may also bind to heat-shock protein 90, or to a variety of other cellular proteins, and this binding may potentially affect protein function. Previous studies have found that synthetic peptides of 10-20 amino acids can bind to proteins, thus affecting protein-protein or protein-substrate interactions.11, 12 Furthermore, synthetic peptides that correspond to peptides found in the cytosol of rat brain have been found to alter various cellular processes such as G protein-coupled receptor signal transduction when introduced into cell lines.10 Moreover, endopeptidase 24.15 overexpression itself changed both angiotensin II and isoproterenol signal transduction, suggesting a physiological function for its intracellular substrates/products.10 Subsequently, endopeptidase 24.15 overexpression was shown to affect only a limited set of specific peptides, despite the existence of a large number of intracellular peptides in HEK293T cells.31 Together, these data suggest that intracellular peptide metabolism can play an important physiological role controlling signal transduction. Because intracellular peptides can have widespread effects on many cellular processes, it is possible that the effects of proteasome inhibitors are due in part to the changes in the intracellular peptidome, and not just on the changes of the cellular proteome as previously considered. In general, the effect of proteasome inhibitors such as epoxomicin on cellular levels of proteins is rather small, whereas the effect of epoxomicin on levels of peptides is much more dramatic. If these peptides are functional, as proposed,7, 9 the altered peptidome may contribute to some of the biological effects of epoxomicin and other proteasome inhibitors.
Supplementary Material
Supplemental Data
File 1: Supplemental tables
Table S1. Data for all identified and unidentified peptides
Table S2. Data for the peptides shown in the heat map (Figure 2)
File 2: MS/MS data (Mascot results and/or annotated MS/MS spectra)
ACKNOWLEDGMENTS
This work was primarily supported by National Institutes of Health grant DA-004494, (L.D.F.), and by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) through the Rede Genoprot (559698/2009-7), and partially funded by the University of São Paulo (Grant#2011.1.9333.1.3, NAPNA). L.M.C., F.C.G., and E.S.F are fellowship recipients from CNPq.
References
- 1.Hummon AB, Amare A, Sweedler JV. Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom.Rev. 2006;25:77–98. doi: 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- 2.Baggerman G, Verleyen P, Clynen E, Huybrechts J, De Loof A, Schoofs L. Peptidomics. J.Chromatogr.B Analyt.Technol.Biomed.Life Sci. 2004;803:3–16. doi: 10.1016/j.jchromb.2003.07.019. [DOI] [PubMed] [Google Scholar]
- 3.Fricker LD, Lim J, Pan H, Che FY. Peptidomics: Identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom.Rev. 2006;25:327–344. doi: 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt JJ, McIlwain S, Page D, Christie AE, Li L. Combining MALDI-FTMS and bioinformatics for rapid peptidomic comparisons. JProteome Res. 2008;7:887–896. doi: 10.1021/pr070390p. [DOI] [PubMed] [Google Scholar]
- 5.Svensson M, Skold K, Nilsson A, Falth M, Svenningsson P, Andren PE. Neuropeptidomics: expanding proteomics downwards. Biochem.Soc.Trans. 2007;35:588–593. doi: 10.1042/BST0350588. [DOI] [PubMed] [Google Scholar]
- 6.Sasaki K, Takahashi N, Satoh M, Yamasaki M, Minamino N. A peptidomics strategy for discovering endogenous bioactive peptides. Journal of proteome research. 2010;9:5047–5052. doi: 10.1021/pr1003455. [DOI] [PubMed] [Google Scholar]
- 7.Fricker LD. Analysis of mouse brain peptides using mass spectrometry-based peptidomics: implications for novel functions ranging from non-classical neuropeptides to microproteins. Mol.Biosyst. 2010;6:1355–1365. doi: 10.1039/c003317k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelman JS, Sironi J, Castro LM, Ferro ES, Fricker LD. Peptidomic analysis of human cell lines. J.Proteome.Res. 2011;10:1583–1592. doi: 10.1021/pr100952f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferro ES, Hyslop S, Camargo AC. Intracellullar peptides as putative natural regulators of protein interactions. J.Neurochem. 2004;91:769–777. doi: 10.1111/j.1471-4159.2004.02757.x. [DOI] [PubMed] [Google Scholar]
- 10.Cunha FM, Berti DA, Ferreira ZS, Klitzke CF, Markus RP, Ferro ES. Intracellular peptides as natural regulators of cell signaling. J.Biol.Chem. 2008;283:24448–24459. doi: 10.1074/jbc.M801252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubinstein M, Niv MY. Peptidic modulators of protein-protein interactions: progress and challenges in computational design. Biopolymers. 2009;91:505–513. doi: 10.1002/bip.21164. [DOI] [PubMed] [Google Scholar]
- 12.Arkin MR, Whitty A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr.Opin.Chem.Biol. 2009;13:284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 13.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol.Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 16.Navon A, Ciechanover A. The 26 S proteasome: from basic mechanisms to drug targeting. The Journal of biological chemistry. 2009;284:33713–33718. doi: 10.1074/jbc.R109.018481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groll M, Nazif T, Huber R, Bogyo M. Probing structural determinants distal to the site of hydrolysis that control substrate specificity of the 20S proteasome. Chemistry & biology. 2002;9:655–662. doi: 10.1016/s1074-5521(02)00144-8. [DOI] [PubMed] [Google Scholar]
- 18.Kisselev AF, Akopian TN, Goldberg AL. Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J.Biol.Chem. 1998;273:1982–1989. doi: 10.1074/jbc.273.4.1982. [DOI] [PubMed] [Google Scholar]
- 19.Groll M, Kim KB, Kairies N, Huber R, Crews CM. Crystal structure of epoxomicin: 20S proteasome reveals a molecular basis for selectivity of alpha’,beta ’-epoxyketone proteasome inhibitors. J Am Chem Soc. 2000;122:1237–1238. [Google Scholar]
- 20.Kisselev AF, Callard A, Goldberg AL. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J.Biol.Chem. 2006;281:8582–8590. doi: 10.1074/jbc.M509043200. [DOI] [PubMed] [Google Scholar]
- 21.Vosler PS, Brennan CS, Chen J. Calpain-mediated signaling mechanisms in neuronal injury and neurodegeneration. Mol.Neurobiol. 2008;38:78–100. doi: 10.1007/s12035-008-8036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timmer JC, Salvesen GS. Caspase substrates. Cell Death.Differ. 2007;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- 23.Turk V, Turk B, Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orlowski M, Reznik S, Ayala J, Pierotti AR. Endopeptidase 24.15 from rat testes. Isolation of the enzyme and its specificity toward synthetic and natural peptides, including enkephalin-containing peptides. Biochem.J. 1989;261:951–958. doi: 10.1042/bj2610951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rioli V, Kato A, Portaro FC, Cury GK, te KK, Vincent B, Checler F, Camargo AC, Glucksman MJ, Roberts JL, Hirose S, Ferro ES. Neuropeptide specificity and inhibition of recombinant isoforms of the endopeptidase 3.4.24.16 family: comparison with the related recombinant endopeptidase 3.4.24.15. Biochem.Biophys.Res.Commun. 1998;250:5–11. doi: 10.1006/bbrc.1998.8941. [DOI] [PubMed] [Google Scholar]
- 27.Barrett AJ, Brown MA, Dando PM, Knight CG, McKie N, Rawlings ND, Serizawa A. Thimet oligopeptidase and oligopeptidase M or neurolysin. Methods Enzymol. 1995;248:529–556. doi: 10.1016/0076-6879(95)48034-x. [DOI] [PubMed] [Google Scholar]
- 28.Saric T, Graef CI, Goldberg AL. Pathway for degradation of peptides generated by proteasomes: a key role for thimet oligopeptidase and other metallopeptidases. J.Biol.Chem. 2004;279:46723–46732. doi: 10.1074/jbc.M406537200. [DOI] [PubMed] [Google Scholar]
- 29.Gass J, Khosla C. Prolyl endopeptidases. Cell Mol.Life Sci. 2007;64:345–355. doi: 10.1007/s00018-006-6317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grasso G, Rizzarelli E, Spoto G. The proteolytic activity of insulin-degrading enzyme: a mass spectrometry study. J.Mass Spectrom. 2009 doi: 10.1002/jms.1550. [DOI] [PubMed] [Google Scholar]
- 31.Berti DA, Morano C, Russo LC, Castro LM, Cunha FM, Zhang X, Sironi J, Klitzke CF, Ferro ES, Fricker LD. Analysis of intracellular substrates and products of thimet oligopeptidase (EC 3.4.24.15) in human embryonic kidney 293 cells. J.Biol.Chem. 2009;284:14105–14116. doi: 10.1074/jbc.M807916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsui M, Fowler JH, Walling LL. Leucine aminopeptidases: diversity in structure and function. Biol.Chem. 2006;387:1535–1544. doi: 10.1515/BC.2006.191. [DOI] [PubMed] [Google Scholar]
- 33.Reits E, Neijssen J, Herberts C, Benckhuijsen W, Janssen L, Drijfhout JW, Neefjes J. A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity. 2004;20:495–506. doi: 10.1016/s1074-7613(04)00074-3. [DOI] [PubMed] [Google Scholar]
- 34.Che FY, Fricker LD. Quantitative peptidomics of mouse pituitary: Comparison of different stable isotopic tags. J.Mass Spectrom. 2005;40:238–249. doi: 10.1002/jms.743. [DOI] [PubMed] [Google Scholar]
- 35.Morano C, Zhang X, Fricker LD. Multiple Isotopic Labels for Quantitative Mass Spectrometry. Anal.Chem. 2008;80:9298–9309. doi: 10.1021/ac801654h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wardman J, Fricker LD. Quantitative peptidomics of mice lacking Peptide-processing enzymes. Methods Mol.Biol. 2011;768:307–323. doi: 10.1007/978-1-61779-204-5_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Che FY, Zhang X, Berezniuk I, Callaway M, Lim J, Fricker LD. Optimization of neuropeptide extraction from the mouse hypothalamus. J.Proteome.Res. 2007;6:4667–4676. doi: 10.1021/pr060690r. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Che FY, Berezniuk I, Sonmez K, Toll L, Fricker LD. Peptidomics of Cpe(fat/fat) mouse brain regions: implications for neuropeptide processing. J.Neurochem. 2008;107:1596–1613. doi: 10.1111/j.1471-4159.2008.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gelman JS, Wardman JH, Bhat VB, Gozzo FC, Fricker LD. Quantitative peptidomics to measure neuropeptide levels in animal models relevant to psychiatric disorders. Methods Mol.Biol. 2012;829:487–503. doi: 10.1007/978-1-61779-458-2_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Che FY, Lim J, Biswas R, Pan H, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol.Cell.Proteomics. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Che FY, Vathy I, Fricker LD. Quantitative peptidomics in mice: Effect of cocaine treatment. J.Mol.Neurosci. 2006;28:265–275. doi: 10.1385/JMN:28:3:265. [DOI] [PubMed] [Google Scholar]
- 42.Castro LM, Berti DA, Russo LC, Coelho V, Gozzo FC, Oliveira V, Ferro ES. Similar Intracellular Peptide Profile of TAP1/beta2 Microglobulin Double-Knockout Mice and C57BL/6 Wild-Type Mice as Revealed by Peptidomic Analysis. AAPS.J. 2010;12:608–616. doi: 10.1208/s12248-010-9224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schirle M, Heurtier MA, Kuster B. Profiling core proteomes of human cell lines by one-dimensional PAGE and liquid chromatography-tandem mass spectrometry. Mol.Cell Proteomics. 2003;2:1297–1305. doi: 10.1074/mcp.M300087-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Doherty MK, Hammond DE, Clague MJ, Gaskell SJ, Beynon RJ. Turnover of the Human Proteome: Determination of Protein Intracellular Stability by Dynamic SILAC. J.Proteome.Res. 2009;8:104–112. doi: 10.1021/pr800641v. [DOI] [PubMed] [Google Scholar]
- 45.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Molecular cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris JL, Alper PB, Li J, Rechsteiner M, Backes BJ. Substrate specificity of the human proteasome. Chem.Biol. 2001;8:1131–1141. doi: 10.1016/s1074-5521(01)00080-1. [DOI] [PubMed] [Google Scholar]
- 47.Cheng B, Maffi SK, Martinez AA, Acosta YP, Morales LD, Roberts JL. Insulin-like growth factor-I mediates neuroprotection in proteasome inhibition-induced cytotoxicity in SH-SY5Y cells. Molecular and cellular neurosciences. 2011;47:181–190. doi: 10.1016/j.mcn.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidtke G, Emch S, Groettrup M, Holzhutter HG. Evidence for the existence of a non-catalytic modifier site of peptide hydrolysis by the 20 S proteasome. The Journal of biological chemistry. 2000;275:22056–22063. doi: 10.1074/jbc.M002513200. [DOI] [PubMed] [Google Scholar]
- 49.Kisselev AF, Akopian TN, Castillo V, Goldberg AL. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Molecular cell. 1999;4:395–402. doi: 10.1016/s1097-2765(00)80341-x. [DOI] [PubMed] [Google Scholar]
- 50.Wakata A, Lee HM, Rommel P, Toutchkine A, Schmidt M, Lawrence DS. Simultaneous fluorescent monitoring of proteasomal subunit catalysis. J Am Chem Soc. 2010;132:1578–1582. doi: 10.1021/ja907226n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.York IA, Bhutani N, Zendzian S, Goldberg AL, Rock KL. Tripeptidyl peptidase II is the major peptidase needed to trim long antigenic precursors, but is not required for most MHC class I antigen presentation. J.Immunol. 2006;177:1434–1443. doi: 10.4049/jimmunol.177.3.1434. [DOI] [PubMed] [Google Scholar]
- 52.Marcilla M, Villasevil EM, de Castro JA. Tripeptidyl peptidase II is dispensable for the generation of both proteasome-dependent and proteasome-independent ligands of HLA-B27 and other class I molecules. Eur.J.Immunol. 2008;38:631–639. doi: 10.1002/eji.200737444. [DOI] [PubMed] [Google Scholar]
- 53.Turner AJ, Nalivaeva NN. New insights into the roles of metalloproteinases in neurodegeneration and neuroprotection. Int.Rev.Neurobiol. 2007;82:113–135. doi: 10.1016/S0074-7742(07)82006-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kravtsova-Ivantsiv Y, Cohen S, Ciechanover A. Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Mol.Cell. 2009;33:496–504. doi: 10.1016/j.molcel.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 55.Tian L, Matouschek A. Where to start and when to stop. Nature structural & molecular biology. 2006;13:668–670. doi: 10.1038/nsmb0806-668. [DOI] [PubMed] [Google Scholar]
- 56.Schrader EK, Harstad KG, Holmgren RA, Matouschek A. A Three-part Signal Governs Differential Processing of Gli1 and Gli3 Proteins by the Proteasome. The Journal of biological chemistry. 2011;286:39051–39058. doi: 10.1074/jbc.M111.274993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity. 2003;18:97–108. doi: 10.1016/s1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 59.Lev A, Takeda K, Zanker D, Maynard JC, Dimberu P, Waffarn E, et al. The exception that reinforces the rule: crosspriming by cytosolic peptides that escape degradation. Immunity. 2008;28:787–798. doi: 10.1016/j.immuni.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data
File 1: Supplemental tables
Table S1. Data for all identified and unidentified peptides
Table S2. Data for the peptides shown in the heat map (Figure 2)
File 2: MS/MS data (Mascot results and/or annotated MS/MS spectra)