Abstract
Background
The tyramine producer Enterococcus durans IPLA655 contains all the necessary genes for tyramine biosynthesis, grouped in the TDC cluster. This cluster includes tyrS, an aminoacyl-tRNA synthetase like gene.
Results
This work shows that tyrS was maximally transcribed in absence of tyrosine at acidic pH, showing a greater than 10-fold induction in mRNA levels over levels occurring in presence of tyrosine. Mapping of the tyrS transcriptional start site revealed an unusually long untranslated leader region of 322 bp, which displays the typical features of the T box transcriptional attenuation mechanism. The tyrosine concentration regulation of tyrS was found to be mediated by a transcription antitermination system, whereas the specific induction at acidic pH was regulated at transcription initiation level.
Conclusions
The expression of the tyrS gene present in the TDC cluster of E. durans is transcriptionally regulated by tyrosine concentration and extracelular pH. The regulation is mediated by both an antitermination system and the promoter itself.
Background
Biogenic amines (BA) are low molecular weight organic bases present in a wide range of food products where they become organoleptically undesirable [1]. It is also worth noting that several toxicological problems resulting from the ingestion of food containing large amounts of BA have been described [2]. Although there is no specific legislation regarding BA content in many food products, it is generally assumed that they should not be allowed to accumulate [3-5].
Fermented foods are likely to contain high levels of BA, mainly due to the decarboxylase activity of some lactic acid bacteria (LAB). BA are produced by the decarboxylation of a precursor amino acid by the enzymatic action of an amino acid decarboxylase [6,7]. In these foods, the main BA are tyramine, histamine, cadaverine and putrescine, which are produced by decarboxylation of tyrosine, histidine, lysine and ornithine, respectively [8].
The presence of the genes encoding the amino acid decarboxylase and the amino acid-amine antiporter is a general feature observed in all the gene clusters involved in the biosynthesis of tyramine, histamine, putrescine and cadaverine [9-12]. We have found an open reading frame coding for a protein of 418 amino acids with a molar mass of 47.38 kDa located next to the tyrosine decarboxylase (tdcA) and the tyrosine-tyramine antiporter (tyrP) genes of Enterococcus durans IPLA655. The predicted amino acid sequence shares strong similarity to the tyrosyl-tRNA synthetase genes (tyrS) of gram positive bacteria. The aminoacyl-tRNA synthetases catalyze the covalent attachment of amino acids to their cognate tRNAs, a crucial reaction for the accuracy of protein synthesis. These enzymes are encoded by genes regulated strictly by antitermination systems; when the corresponding amino acid, tyrosine in this case, is at low concentration, it is not linked to the tRNA, and this uncharged tRNA interact with the antiterminator located between the promoter and the start codon, stabilizing it and allowing transcription. By contrast, when tyrosine is at high concentration, it is linked to the corresponding tRNA (charged tRNA) that cannot stabilize the antiterminator, and consequently the transcription stops [13]. Genes with high similarity to those encoding the corresponding aminoacyl-tRNA-synthetases are generally associated to the genes involved in BA biosynthesis in other species, in the case of tyramine (Enterococcus faecalis JH2-2 [14], Enterococcus faecalis V583 [15], Enterococcus faecium MV5 (GenBank HM921050), Enterococcus hirae (GenBank AY303667), Carnobacterium divergens 508 [16]), Lactobacillus brevis IOEB9809 [17], Lactobacillus brevis ATCC367 (GenBank NC_008497), and the histamine (Lactobacillus buchneri B301 [10], Lactobacillus hilgardii 0006 [18]).
The number of tyrS genes is heterogeneous within the genera Enterococcus. Some tyramine-producing species as E. faecium have a unique tyrS, whereas E. faecalis has two different tyrS genes [15]. So far, this aspect remains unknown for E. durans (no genomic data are available). Concerning the two different genes encoding tyrosyl-tRNA-sinthetases in E. faecalis V583, tyrS-2 is homologous to the tyrS of other bacteria with a unique tyrS, whereas tyrS-1 is located in the tyramine cluster. Database search revealed that tyrS-1 has higher similarity (> 80%) to other tyrS genes associated to tyramine biosynthesis clusters than to its own tyrS-2 gene located elsewhere in the genome (52%). The presence of two tyrS genes in the genome of a tyramine producing strain suggests that one (tyrS-2) would be implicated in protein biosynthesis, whereas the one linked to TDC cluster (tyrS-1) could be a sensor of the intracellular tyrosine pool to regulate tyrosine decarboxylation [9]. In addition, phylogenetic analyses of TyrS proteins associated to tyramine clusters, supported the hypothesis that these proteins made tight clusters and were clearly separated from their relatives encoded elsewhere in bacterial genomes. These results suggested a co-evolution of tyrS together with tdcA and tyrP. This fact prompted us to exhaustively investigate the transcriptional regulation mechanism of this gene and its putative role on the regulation of the tyramine operon.
Results
tyrS expression depends on the tyrosine concentration and extracellular pH
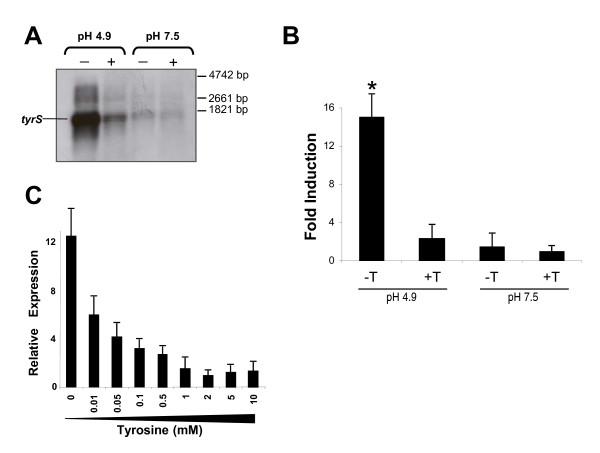
RNA from E. durans IPLA655 cultures grown in presence (10 mM) or absence of tyrosine at two different pH conditions (4.9 and 7.5), was analyzed by Northern blot hybridization with a tyrS specific probe (Figure 1A). Very low expression was detected in cells grown at pH 7.5 independently of the presence or absence of tyrosine. Noteworthy, at pH 4.9, an intense band corresponding to a transcript of 1.6 kb was observed. A policystronic mRNA including tyrS-tdcA was never detected [19]. The bigger band present in all lines with a low intensity would correspond to unspecific hybridization to the extremely abundant 23S rRNA molecules. This tyrS up-regulation was specially enhanced in absence of tyrosine, suggesting that the initiation of transcription or mRNA stability is controlled by pH and tyrosine.
Figure 1.
Transcriptional analysis of the tyrS gene. A: Northern blot analysis with a tyrS-specific probe and B: RT-qPCR quantification of tyrS mRNA levels in presence (+Y) and absence (-Y) of 10 mM tyrosine and different pH conditions (pH 4.9 and pH 7.5). The asterisk indicates statistically significant difference (p ≤ 0.005; Student's t-test) in comparison to the other conditions. C: Effect of different tyrosine concentrations (0, 0.01, 0.05, 0.1, 0.5, 1, 2, 5 and 10 mM) on tyrS expression at pH 4.9
The strength of these environmental conditions on tyrS expression was quantified by RT-qPCR. Data in Figure 1B confirmed that tyrS is maximally transcribed in absence of tyrosine and at pH 4.9, showing a greater than 10-fold induction in mRNA levels over levels occurring in presence of tyrosine. Even when tyrosine was not added to the media, no induction was detected at pH 7.5. These results confirmed that both conditions (acidic pH and absence of tyrosine) are needed for expression of tyrS gene.
Next, we examined whether intermediate tyrosine concentrations have an effect on tyrS expression. Therefore, we investigated at optimal pH 4.9, the effect that different tyrosine concentrations in the media (0, 0.01, 0.05, 0.1, 0.5, 1, 2, 5 and 10 mM) exert on gene expression by comparing RT-qPCR results obtained in each condition. As indicated in Figure 1C, tyrS expression showed an inverse correlation with the increased tyrosine concentration and exhibited a great sensitivity to very low tyrosine levels, since the maximal expression level was reached in absence of tyrosine and an increase of 0.01 mM tyrosine in the media was enough to reduce this level to the half. Not significant changes in transcription were observed above 2 mM tyrosine, probably because of saturating concentrations of tyrosine. Such concentrations were assayed because tyrosine can reach very high concentrations in some cheeses and even precipitate forming crystals [20].
Mapping of the tyrS transcription initiation site
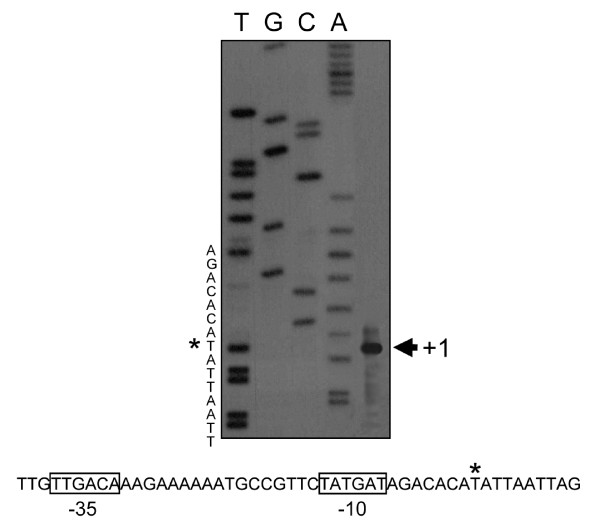
To map the precise start point of the transcription of tyrS, primer extension was performed using RNA samples extracted under optimal conditions of expression (pH 4.9 and absence of tyrosine).
A single band of 322 bp was observed, indicating that the position +1 of the mRNA corresponds to a T residue located 322 nucleotides upstream of the ATG codon (Figure 2). Seven nucleotides upstream this point, it was localized the -10 sequence TATGAT spaced 17 nucleotides downstream of the -35 sequence TTGACA, that nearly matched the consensus sequence for LAB promoters [21]. In a position 9-14 nucleotides upstream the ATG codon of this gene, it was identified the Shine-Dalgarno region (CGGAGG) (bases fitting with the consensus sequences are underlined).
Figure 2.
Primer extension identification of transcription start site (*) of tyrS and transcriptional regions -10 and -35 (boxes). Lines T, G, C and A correspond to DNA sequence reaction (note that the nucleotide sequences, 5' at the top and 3' at the bottom, represent the complementary DNA strand, so that it may be read directly)
Structural features of the tyrS leader region
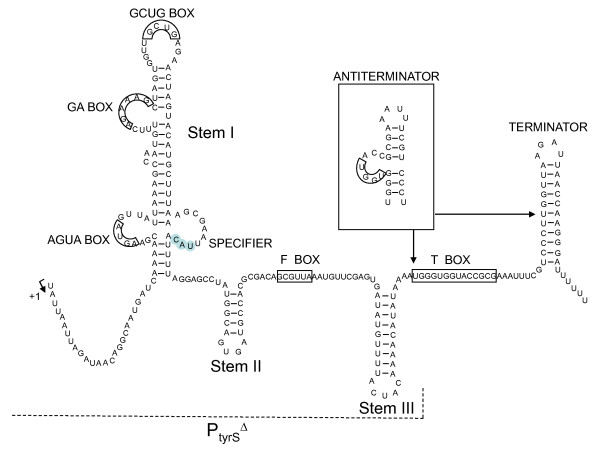
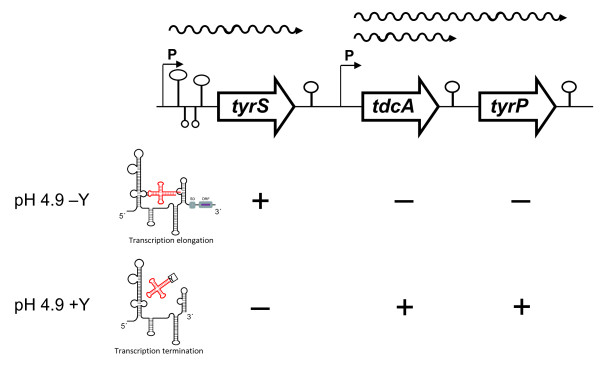
In E. durans IPLA655, the region upstream of the start codon of tyrS showed a 322 bp noncoding sequence that was named the tyrS leader region. A hypothetical representation of the secondary structure of the tyrS leader region is plotted on Figure 3. This region exhibits the sequence features of the tRNA-mediated antitermination systems described by Grundy et al. [22]. It contains the typical T box sequence UGGGUGGUACCGCG (nucleotides 187-200) (bases fitting with the consensus are underlined), a tyrosine specifier UAC (nucleotides 104-106), and most of the other less conserved boxes (AGUA-I box [AGUA, nucleotides 34-37], GA box [AGAAAG, nucleotides 58-63], GNUG box [GCUG, nucleotides 73-76], and F box [GCGUUA, nucleotides 142-147]). In addition to these conserved sequences, the tyrS leader region may be folded into three stem-loop structures (I, II and III) preceding a factor-independent transcriptional terminator/antiterminator. However, the AGUA-II and GAAC boxes that can be found in similar antitermination systems are not present.
Figure 3.
Primary-sequence and structural model of the E. durans IPLA655 tyrS mRNA leader region upstream the start of the coding region. The specifier (UAC), the Tbox sequence, and other highly-conserved motifs typical of genes regulated by tRNA-mediated antitermination appear highlighted in boxes. Sequence between arrows can adopt two alternative mutually exclusive structure conformations: terminator and antiterminator (stabilized by the cognate tRNA in absence of tyrosine). A transcriptional fusion of the tyrS promoter and the leader region with a deletion of the TBox-Terminator region (PtyrSΔ) was made (dashed line) to probe the role of the Tbox in the mechanism of tyrosine sensing
Tyrosine concentration sensing is mediated by an antitermination system
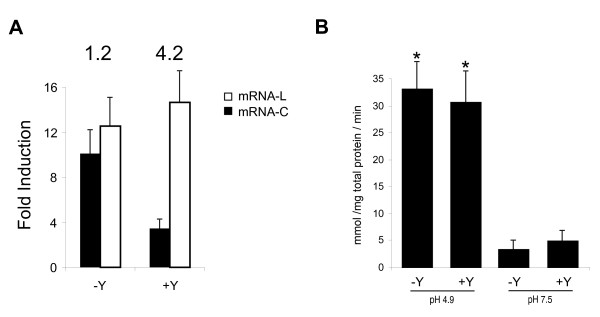
We investigated whether the conserved primary sequence and structural motifs located upstream the start of the coding sequence play a role in the regulation of tyrS expression by a transcription antitermination system. For this purpose we compared the amount of mRNA specific of the leader region (mRNA-L) and the amount of mRNA corresponding to the coding part of the gene (mRNA-C) under optimal expression condition (pH 4.9), and in presence or absence of tyrosine. This region-specific transcriptional quantification was performed by RT-qPCR using specific primer pairs for each region (see Methods).
As shown in Figure 4A, level of mRNA-L was not affected by tyrosine concentration, whereas mRNA-C level did not follow the same profile. In presence of tyrosine, the ratio mRNA-L/mRNA-C was 4.2, whereas this value decreased to 1.2 in absence of tyrosine (optimal conditions for tyrS expression). The ratio close to 1 observed in absence of tyrosine indicates no transcription termination and consequently the expression of tyrS. The 4.2 ratio indicates higher levels of mRNA-L than mRNA-C, suggesting that the transcription in the presence of tyrosine was under the control of a termination system. These data are coherent with a tyrosine concentration regulation of tyrS mediated by a transcription antitermination system.
Figure 4.
Regulatory effect of the Tbox on tyrS expression. A: Quantification of tyrS mRNA-C (in black) and mRNA-L (in white) levels at pH 4.9 in presence (+Y) and absence (-Y) of 10 mM tyrosine. Numbers above indicate the ratio mRNA-L/mRNA-C in the corresponding condition. B: Effect of Tbox deletion on β-Galactosidase activity of PtyrSΔ-lacZ fusions at different conditions of pH and presence/absence of 10 mM tyrosine (Y). Data represent the average of three independent experiments. The higher activity observed at pH 4.9 (asterisks) was statistically significant (p < 0.005; Student's t-test) in comparison to that at pH 7.5
Assessment of PtyrSΔ activity
The role of the T box in the mechanism of tyrosine sensing by tyrS was analyzed using a transcriptional fusion of lacZ reporter gene with the tyrS promoter and the leader region, but with a deletion of the T box-Terminator motif (PtyrSΔ) (Figure 4B). The lacZ activities under the control of PtyrSΔ at pH 4.9 were similar in the absence (33.8 mmol/mg total protein/min) and presence (31.5 mmol/mg total protein/min) of tyrosine, confirming that tyrosine regulation is located on the T box region. On the other hand, independently of the presence of tyrosine, promoter activities at neutral pH were lower than 5 mmol/mg total protein/min, showing an 8-fold higher strength of PtyrSΔ under acidic pH than at neutral pH. These data indicate that the induction of tyrS expression by pH is transcriptionally regulated by the promoter.
Putative role of tyrS in tyramine cluster
To test the hypothesis that TyrS plays a physiological role on tyramine biosynthesis and/or in the regulation of the related genes (tdcA and tyrP), tyrS was over-expressed under the control of the nisin promoter. In all cases, the concentration of tyrS transcripts (assessed by RT-qPCR) was 80-fold over the physiological expression level. The presence of soluble translated TyrS was tested by Anti-HIS immunodetection. An intense band of expected size was observed under induction conditions. Next, we analyzed the in vivo effect of the over-expression of tyrS in cells grown on the aforementioned conditions, (pH 4.9 in GM17-Y and GM17 + Y media). Negative controls of uninduced cultures were carried in parallel. Under these experimental conditions, level of tdcA-specific mRNA (quantified by RT-qPCR) was not affected by the overexpression of tyrS (data not shown). In addition, the concentration of tyramine in supernatants was examined by HPLC. Only the expected differences depending on the tyrosine concentration in the media were observed (260 ± 40 μM and 3100 ± 80 μM in GM17-Y and GM17 + Y cultures, respectively), but no significant differences between tyrS-induced cultures and the negative control were observed.
Discussion
The E. durans IPLA655 tyrS gene present in the tyramine cluster, which encodes a protein with high similarity to tyrosyl-tRNA-synthetases, is controlled by the intracellular concentration of tyrosine in a similar way than aminoacyl-tRNA synthetases are regulated by their substrate amino acid. The analysis of TyrS sequence revealed the typical HIGH and KMSKS domains of class I aminoacyl tRNA synthetases, being the HIGH motif perfectly conserved, and the KMSKS motif is represented by the KFGKT sequence, as in E. coli [23], Bacillus subtilis [24], and E. faecalis [14]. Mapping of the transcriptional start site revealed a long untranslated leader region of 322 bp with a highly conserved set of primary-sequence and secondary structure elements. These elements include three stem-loop structures, a highly conserved 14-bp sequence designated the T box, and a factor-independent transcriptional terminator (Figure 3). These features are also present in other genes of gram positive bacteria, mainly genes encoding aminoacyl-tRNA synthetases, but also amino acid biosynthetic genes and transporters [25-27]. Several studies have revealed a crucial role for conserved leader region motifs in regulation of gene expression at the level of premature termination of transcription [28]. In order to test whether this mechanism regulates the tyrS gene of E. durans TDC cluster, the levels of mRNA were quantified using specific primers for the leader and coding region of tyrS. When E. durans was starved for tyrosine, the predominant transcript was a 1.6 kb mRNA fragment, which is the expected size for full-length mRNA (mRNA-C). Interestingly, when tyrosine was present in excess, full-length mRNA was dramatically depleted, whereas the truncated mRNA-L species kept almost constant. Thus, tyrosine had no effect on the total number of mRNA-L molecules but caused a stoichiometric replacement of full-length mRNA by truncated RNA molecules. These data are consistent with the idea that tyrosine controls tyrS expression by promoting the premature termination of transcription rather than by inhibiting the initiation of transcription.
Experiments involving transcriptional fusions of the tyrS promoter with ß-galactosidase provided evidence for this mechanism. We showed that deletion of the T box-Terminator domain of the leader region originates a complete lost of regulation by tyrosine. Early termination at pH 4.9 in presence of tyrosine observed in vivo in the leader tyrS mRNA (which shows that this sequence promotes terminator formation specifically in presence of tyrosine) was not observed for the PtyrSΔ promoter. This effect can be expected because the T box sequence is present in a side bulge of the antiterminator overlapping the terminator-antiterminator structures.
In addition to the tyrosine regulation, transcription of tyrS is under strict pH control in E. durans, being expressed mostly at acidic growth conditions. The aminoacyl-tRNA synthetases catalyze the covalent attachment of amino acids to their cognate tRNAs. Incorporation of tyrosine to proteins is an essential function for cell life, which should not be restricted to acidic conditions. One explanation for this would be the presence of two different tyrosyl-tRNA synthetases, one of which would be induced under stress conditions (acidic pH and extremely low tyrosine concentration). In general, there is only one aminoacyl-tRNA-synthetase for each amino acid in most bacteria, however, several exceptions are known. Indeed, two very similar lysyl-tRNA synthetases, lysS (constitutive) and lysU (heat inducible) have been described in Escherichia coli [29]. In gram-positives, in addition to the aforementioned case of the two tyrS of E. faecalis, there are two distinct histidyl-tRNA synthetase genes in Lactococcus lactis [30], and two tyrosyl-tRNA synthetase genes (tyrS and tyrZ) and two threonyl-tRNA synthetase genes (thrS and thrZ) in Bacillus subtilis [31,32]. In this last case, the normally silent thrZ gene is induced during threonine starvation or by reducing the intracellular concentration of ThrS, which is the housekeeping threonyl-tRNA synthetase sufficient for normal cell growth [33].
The location of genes encoding an aminoacyl-tRNA-synthetase associated to the gene clusters involved in tyramine and histamine biosynthesis is a general feature [9,10,14,16-18,34]. One of the reasons to study the expression of tyrS in E. durans is to find out whether this protein could have a role on the genetic regulation of the tyramine cluster, being activated under limiting levels of tyrosine to prevent massive decarboxylation of this amino acid, ensuring its availability for protein synthesis. Consistent with this idea would be 1) the common location of genes encoding aminoacyl-tRNA sinthetases next to the operon of decarboxylation (BA-biosynthesis) of the corresponding aminoacid [9,10,14,16-18,34], 2) the expression of this gene only under acidic pH, which is the condition regulating positively the biosynthesis and accumulation of tyramine [19,35] and 3) the fact that tyrS and the genes of the tyramine biosynthesis pathway (tdcA and tyrP) require opposite conditions of tyrosine concentration for optimal expression (Figure 5) [19]. Altogether, these data raise the question whether TyrS could act as a negative regulator. However, overexpression of tyrS on multicopy plasmid during growth of the E. durans strain carrying the wild-type allele had no observable effect on the expression profile of the decarboxylating gene tdcA or on the tyramine concentration observed in the supernatant.
Figure 5.
Genetic organization and transcriptional profile of the TDC cluster in E. durans IPLA655. Promoters (P) and termination regions are indicated. The different mRNA are represented by wavy lines. Numbers indicate the size of the corresponding gene in base pairs (bp). Regulation of the genes by tyrosine and pH is indicated below. Acidic pH is required for optimal expression of the three genes. In addition, tdcA and tyrP are positively regulated by 10 mM tyrosine (+Y), whereas expression of tyrS is negatively affected by tyrosine (+: positively regulated; -: negatively regulated; tyrS: tyrosyl-tRNA synthetase; tdcA: tyrosine decarboxylase; tyrP: tyrosine-tyramine antiporter). Structures upstream tyrS represent the stems I, II, III and terminator of the leader region. The terminator/antiterminator mechanism that regulates the tyrS gene is also indicated: readthrough of the leader region is induced by limitation of tyrosine. Uncharged tyrtRNA stabilize formation of antiterminator structure in the mRNA, which prevents terminator formation (SD: Shine-Dalgarno; ORF: open reading frame of tyrS)
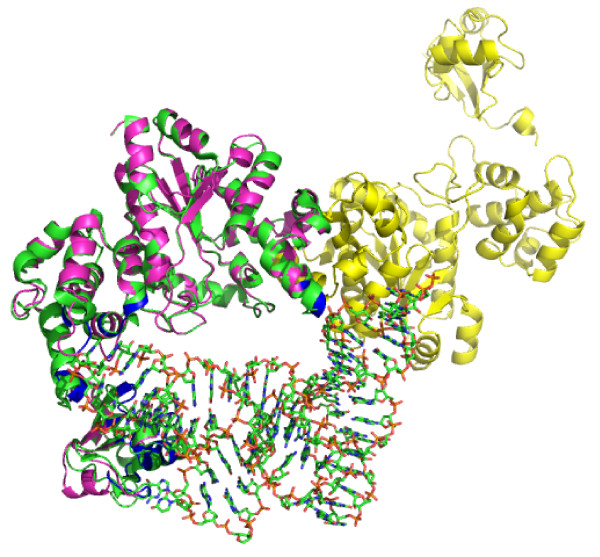
Computational three-dimensional modelling of E. durans TyrS protein revealed nucleic acids-binding domains that might suggest a role as transcriptional regulator. However, the same domains have been identified in the highly similar TyrS structure of Thermus thermophilus (Protein Data Bank: 1H3E), and predicted to interact with tRNA (Figure 6). This data is consistent with the electrophoretic mobility shift (EMSA) assays carried to test TyrS binding to the promoters of the TDC operon. Under the wide range of conditions studied (different pH, salt concentration, presence or absence of tyrosine...) no specific binding of TyrS was observed (data not shown). These data, together with the finding of tyramine clusters without a tyrS gene in Tetragenococcus halophilus (GenBank AB059363) and histamine biosynthesis clusters without a hisS gene [36], would suggest a non critical biological function of these genes in the modulation of the contiguous decarboxylation operon. In any case, it can not be discarded that tyrS could exert a post-transcriptional regulation of tyramine biosynthesis. In fact, both enzymes -TyrS and TdcA- share tyrosine as substrate.
Figure 6.
TyrS structural model achieved using Swiss-Pdb Viewer v. 4.04 software and structure superposition onto the highly similar Thermus terhmophilus tyrosyl-tRNA synthetase. (Protein Data Bank: 1H3E). 1H3E is shown in green, and TyrS model is shown in magenta and yellow. Analysis of the two aligned structures indicates that all of the DNA/RNA binding sites are in regions that interact with tRNA in the 1H3E structure (shown in blue).
Consequently, two are the possibilities that can be considered: i) there are two tyrS genes in E. durans -as described for E. faecalis- and the one ligated to TDC would be a stress-related gene to ensure sufficient charged TyrtRNA for protein biosynthesis in those conditions that tyrosine is being decarboxylated, or ii) this is the unique tyrS gene and the low expression levels observed under neutral pH conditions are enough to assure protein synthesis for general metabolism and the increased expression at acidic pH would guarantee protein biosynthesis when tyrosine is being decarboxylated. The presence of a second tyrS gene was investigated by Southern hybridizations of E. durans IPLA655 total DNA using as probes the tyrS gene of its own tyramine cluster and also tyrS2 from E. faecalis V583 (the one that is not linked to the tyramine cluster), and no additional tyrS genes were found (data not shown). Consistent with this would be the impossibility to knockout the tyrS gene, which would support the hypothesis of a single tyrS, therefore essential. Then, the most plausible model is the presence of a unique tyrS which exhibits a severe induction under acidic pH and low expression levels under neutral pH. It is important to highlight that the transcription quantification is relative, all the expression values were standardized to the reference condition, in this case pH 7.5 (Figure 1B). In fact, the signal observed by Northern blot (Figure 1A) is not zero in any of tested conditions. The tyrS basal expression that takes place at neutral pH would be enough to assure protein synthesis.
Conclusions
In this paper, we provide evidence that transcription of the E. durans IPLA655 tyrS gene is controlled at two levels, initiation and elongation. The initiation of transcription was shown to be enhanced in acidic environments, whereas elongation of transcription is subject to a tyrosine-dependent attenuation mechanism related to the T box system. This dual mechanism has thus never been described, since previously reported genes induced by tRNA-mediated antitermination, were not found to be pH dependent.
Methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table 1. Escherichia coli TOP10 (Invitrogen A/S, Taastrup, Denmark) and Lactococcus lactis NZ9000 were used for plasmid propagation. E. coli cells were grown in aeration in Luria-Bertani medium at 37°C. L. lactis and E. durans strains were grown at 30°C on M17 medium (Oxoid, Hampshire, United Kingdom) supplemented with 0.5% glucose (GM17) containing 10 mM tyrosine (GM17 + Y), or on GM17 without tyrosine (GM17-Y). The media was previously adjusted to the corresponding pH condition (pH 4.9 or pH 7.5). When needed, erythromycin (5 μg ml-1 for E. durans and L. lactis), ampicillin (100 μg ml-1 for E. coli) or chloramphenicol (5 μg ml-1 for E. durans and L. lactis) were added to the culture media.
Table 1.
Strains and plasmids used in this study
| Strain/Plasmid | Characteristics * | Source |
|---|---|---|
| STRAINS | ||
| E. coli TOP10 | Invitrogen | |
| L. lactis NZ9000 | Plasmid-free strain | [37,38] |
| E. durans IPLA655 | Isolated from artisanal cheese. Tyramine producer | IPLA Collection |
| PLASMIDS | ||
| pUC18 | AmpR | [39] |
| pNZ9530 | EryR, nisR-nisK | [40] |
| pILORI4 | EryR, lacZ promoterless gene | [41] |
| pNZcLIC | CmR, expression vector | [42] |
| pDA12 | AmpR, pUC18 with PtyrS cloned in SmaI | This work |
| pDA15 | AmpR, pUC18 with PtyrSΔ cloned in SmaI | This work |
| pDA16 | EryR, pILORI4 including PtyrSΔ -lacZ fusion | This work |
| pNZcTyrS | CmR, pNZcLIC including tyrS | This work |
* AmpR, ampicillin resistant; EryR, erythromycin resistant; CmR, chloramphenicol resistant
DNA manipulation procedures
Procedures for DNA manipulation, transformation of E. coli cells, and recombinant techniques used were essentially those described by Sambrook et al. [43]. Large-scale isolation of E. coli plasmids for nucleotide sequence analysis was performed with the Plasmid Midi Kit (Qiagen Ltd., Crawley, United Kingdom) according to the manufacturer's instructions. Constructions for E. durans were achieved using L. lactis NZ9000 as intermediate host. Plasmid and chromosomal DNA of E. durans and L. lactis were isolated and transformed as described previously [44]. All enzymes for DNA technology were used according to the manufacturer's specifications. DNA hybridizations were performed using the non-radioactive DNA Labelling and Detection Kit (Roche Molecular Biochemicals) following the manufacturer's instructions.
RNA manipulation and northern blot analysis of tyrS transcripts
Total RNA was isolated from cells of E. durans IPLA655 grown in GM17-Y and GM17 + Y at pH 4.9 and pH 7.5 to exponential phase (optical density at 600 nm [OD600] of 0.6). Purified RNAs were resuspended in DEPC 0.1% (diethyl pyrocarbonate) treated water, and total concentration and yield were determined by UV spectrophotometry by measuring absorbance at 260 nm using a BioPhotometer (Eppendorf, NY).
After extraction, RNA samples were treated with DNase (Fermentas, Vilnius, Lithuania), as described by the manufacturer, to eliminate any genomic contaminations. 20 μg of each sample were subjected to electrophoresis through a 1.5% agarose gel containing 5% formaldehyde and 1X MOPS buffer [20 mM 3-N-morfolino-propanesulfonic acid (MOPS), 1 mM EDTA, 5 mM sodium acetate; pH 7.0]. Transfers and hybridizations were performed as described by Sambrook et al. [43]. DNA probes were labeled with [α-32P]dATP by nick translation with the DNA polimerase/DNase I (Invitrogen A/S, Taastrup, Denmark). Primers used in the PCR-amplification of the probes are summarized in Table 2.
Table 2.
Oligonucleotides used in this study
| Primer | Function* | Sequence (5' to 3') |
|---|---|---|
|
TDC11 RT1 |
tyrS probe amplification (F) tyrS probe amplification (R) |
TCAATTACAGATCGGTGGGG ACTTACCATCGAATGCATCAAATG |
|
TRNA2B(2) TRNA(P) TyrS prom (F) tyrS prom (R) |
mRNA-C quantification (F) mRNA-C quantification (R) mRNA-L quantification (F) mRNA-L quantification (R) |
CGTAAATTAGAAGGGCCAGAGGCAG GATCAAGCCAGATTGCGCCACCTGCAG AACAGGCAATGATCAAAACGAAGTA CATAGGCTCCTAAAATGTAATTCGC |
| TYR2 | PtyrS mapping (R) | ACTTCGTTTTGATCATTGCCTG |
|
TDC123 TDC130 |
PtyrSΔ cloning and sequencing (F) PtyrSΔ cloning and sequencing (R) |
AAAACTTCCCATATGCATTGTAACG CTGCAGCATTTTATATGTTTTGTAGTAA |
|
TYS (F) TYS(R) |
tyrS overexpression (F) tyrS overexpression (R) |
ATGGGTGGTGGATTTGCTAATATTATCGATGAATTAACTT TTGGAAGTATAAATTTTCATCAACTACTTTGGCCAAAAAG |
*F, forward; R, reverse
Quantification of tyrS expression by reverse transcription quantitative PCR (RT-qPCR)
Gene expression analysis was carried out by RT-q PCR on a 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA) using SYBR® Green PCR Master Mix (Applied Biosystems, Carlsbad, CA). cDNA samples were synthesized from total RNA using the iScript™ cDNA Synthesis kit (Bio-Rad, Hercules, CA). After dilution of cDNA, 5 μl were added to 20 μl of PCR mixture (12.5 μl of SYBR Green Supermix, 1 μL of each primer at 7 μM, 5.5 μl of RNAse free water). Amplifications were performed with specific primers (Table 2) designed using Primer Express software (Applied Biosystems, Carlsbad, CA), using primers for 16S ribosomal RNA as an internal control to normalize RNA concentration [19]. Cycling settings were those default-established by Applied Biosystems, Carlsbad, CA. For each condition, RT-qPCR analysis was performed on RNA purified from three independently grown cultures.
Primer extension and DNA sequencing
Total RNA isolated from E. durans IPLA655 grown in GM17-Y pH 4.9 was used as template for primer extension. The reaction was done mixing 15 μg of RNA with 1 pmol of oligonucleotide 5'end-labeled with [γ-32P]-dATP using T4 polynucleotide kinase (New England Biolabs Inc. Ipswich, MA) and a mixture containing deoxynucleotides triphosphate (1 mM each), RNase inhibitor (Gibco, BRL), and 20 U of Avian Myeloblastosis Virus reverse transcriptase (Promega, Madison, USA). The control sequence used as size standard was determined with the same reverse primer using as template 2 μg of the double-stranded recombinant plasmid pDA12 (Table 1). Each sequence reaction was performed according to the dideoxynucleotide chain termination sequencing method using 5 μCi of [α-32P]dCTP and the T7 Sequencing™ Mixes Kit (Pharmacia Biotech Inc. Piscataway) under manufacturer instructions. Finally, the samples were resolved in parallel on a denaturing 8% polyacrylamide gel containing 7 M urea to determine the endpoints of the extension products.
Construction of PtyrSΔ-lacZ transcriptional fusion
A fragment including the tyrS promoter and leader region with a deletion of the T-box-Terminator domain (PtyrSΔ) (Figure 3), was amplified using primers TDC123 and TDC130 (Table 2) and cloned in the SmaI site of pUC18, resulting pDA15 plasmid. A 450 bp EcoRI-PstI fragment including PtyrSΔ was cloned into the low copy number vector pILORI4 [41] for fusion to a promoterless lacZ reporter gene, yielding the pDA16 plasmid. All clones were sequenced.
Plasmid pDA16 was firstly constructed in L. lactis NZ9000 strain, and subsequently transformed into E. durans IPLA655, and lacZ activity was tested under specific pH and in presence or absence of tyrosine in the culture media. For promoter assay, a negative control with the pILORI4 plasmid was carried.
Determination of β-galactosidase activity
In order to determine β-galactosidase activity, cells grown in the corresponding conditions of pH and tyrosine concentration at OD600 = 0.6 were harvested, resuspended in 1 ml of 10 mM Na2HPO4, pH 7.0, and then disrupted with 0.2 g of glass beads (Sigma). After centrifugation at 12000 g for 10 min at 4°C, different dilutions of the resulting supernatants (CFE) were immediately assayed in a final volume of 1 ml, which included 750 μl of 10 mM sodium phosphate buffer pH 7.0, 200 μl of CFE and 50 μl of 20 mM o-nitrophenilgalactopiranoside (ONPG). The mixture was immediately incubated at 37°C and absorbance was measured (λ = 420 nm). Each condition was assayed independently by triplicate and the values were standardized to protein contents of cell extracts, determined by using the BCA Protein Assay Reagent Kit (Pierce, Rockford, Ill.).
Overexpression of tyrS and immunodetection
The gene encoding for TyrS was amplified using primers TYSF and TYSR (Table 2) and cloned into a pNZcLIC expression vector using the VBEx system [45], yielding the corresponding derivative pNZcTyrS. For detection purposes, a decaHis-tag was added to the C-terminal of the target protein.
tyrS expression was carried using the NICE system [46]. The genes encoding nisR and nisK were introduced in E. durans IPLA655 in the low copy number plasmid pNZ9530 [40]. After induction with 2 μg L-1 nisin, expression of the protein was confirmed by Western blotting analysis of cell lysates by 10% SDS-PAGE electrophoresis gels, subsequently electroblotted and immunodetected with an anti-His-tag antibody (Amersham Pharmacia Biotech Inc. Piscataway). Chemiluminescence detection was done using the Western-Light kit (Tropix Inc. Bedford, MA) and quantified using the Fujifilm LAS-3000 imaging system (Fuji Photo Film Co. Ltd; Tokyo).
Analysis of tyramine by HPLC
The quantitative analysis of tyramine production was undertaken by reverse-phase high performance liquid chromatography (RP-HPLC) using a Waters liquid chromatograph controlled by Millenium 32 Software (Waters, Milford, MA, USA). The samples were prepared by centrifugation at 8,000 × g for 10 min. The resulting supernatants were filtered using Millipore 0.2 μm filters and derivatized using dabsyl chloride, as described by Krause et al. [47]. Separations were performed using a Waters Nova-pack C18 column (150 × 3.9 mm). Usually, 10 μl of the derivatized sample was injected and detection performed at 436 nm. The solvent gradient and detection conditions were similar to those described by Krause et al. [47].
Authors' contributions
DML designed and performed the experiments, and drafted the manuscript. MF and MAA designed experimental procedures and helped to write the manuscript. All authors read and approved the manuscript.
Contributor Information
Daniel M Linares, Email: daniml@ipla.csic.es.
Maria Fernández, Email: mfernandez@ipla.csic.es.
Beatriz Del-Río, Email: beadelrio@ipla.csic.es.
Victor Ladero, Email: ladero@ipla.csic.es.
Maria Cruz Martin, Email: mcm@ipla.csic.es.
Miguel A Alvarez, Email: maag@ipla.csic.es.
Acknowledgements
This research was performed with financial support from the Ministry of Science and Innovation, Spain (AGL2010-18430) and the European Community's Seventh Framework Programme (BIAMFOOD-211441). We are grateful to Paloma López for technical assistance with Primer Extension experiments, and Begoña Redruello for experienced support provided for protein modelling and structure alignment. Strain L. lactis NZ9000 and plasmid pNZ9530 were kindly provided by NIZO food research, and plasmids pILORI4 and pNZcLIC were kindly provided by Oscar Kuipers and Bert Poolman, respectively. D. M. Linares is the recipient of a contract from Gobierno del Principado de Asturias. B. del Río is beneficiary of a JAE DOC contract (CSIC).
References
- Fernández M, Linares DM, Del-Río B, Ladero V, Álvarez MA. HPLC quantification of biogenic amines in cheeses: correlation with PCR-detection of tyramine-producing microorganisms. J Dairy Res. 2007;74(Suppl 3):276–282. doi: 10.1017/S0022029907002488. [DOI] [PubMed] [Google Scholar]
- Ladero V, Calles-Enríquez M, Fernández M, Álvarez MA. Toxicological effects of dietary biogenic amines. Curr Nutr Food Sci. 2010;6:145–156. [Google Scholar]
- Maintz L, Novak N. Histamine and histamine intolerance. Am J Clin Nutr. 2007;85:1185–1196. doi: 10.1093/ajcn/85.5.1185. [DOI] [PubMed] [Google Scholar]
- Ten-Brink B, Damink C, Joosten HM, Huis In't Veld JH. Occurrence and formation of biologically active amines in foods. Int J Food Microbiol. 1990;11:73–84. doi: 10.1016/0168-1605(90)90040-c. [DOI] [PubMed] [Google Scholar]
- Shalaby AR. Significance of biogenic amines to food safety and human health. Food Res Int. 1996;29:675–690. [Google Scholar]
- Spano G, Russo P, Lonvaud-Funel A, Lucas P, Alexandre H, Grandvalet C, Coton E, Coton M, Barnavon L, Bach B, Rattray F, Bunte A, Magni C, Ladero V, Álvarez M, Fernández M, Lopez P, De-Palencia PF, Corbi A, Trip H, Lolkema JS. Biogenic amines in fermented foods. Eur J Clin Nutr. 2010;64(Suppl 3):S95–S100. doi: 10.1038/ejcn.2010.218. [DOI] [PubMed] [Google Scholar]
- Linares DM, Cruz Martín M, Ladero V, Álvarez MA, Fernández M. Biogenic amines in dairy products. Critical Rev Food Sci Nutr. 2011;51:691–703. doi: 10.1080/10408398.2011.582813. [DOI] [PubMed] [Google Scholar]
- Coton M, Romano A, Spano G, Ziegler K, Vetrana C, Desmarais C, Lonvaud-Funel A, Lucas P, Coton E. Occurrence of biogenic amine-forming lactic acid bacteria in wine and cider. Food Microbiol. 2010;27(Suppl 8):1078–1085. doi: 10.1016/j.fm.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Fernández M, Linares DM, Álvarez MA. Sequencing of the tyrosine decarboxylase cluster of Lactococcus lactis IPLA 655 and the development of a PCR method for detecting tyrosine decarboxylating lactic acid bacteria. J Food Prot. 2004;67(Suppl 11):2521–2529. doi: 10.4315/0362-028x-67.11.2521. [DOI] [PubMed] [Google Scholar]
- Martín MC, Fernández M, Linares DM, Álvarez MA. Sequencing, characterization and transcriptional analysis of the histidine decarboxylase operon of Lactobacillus buchneri. Microbiology. 2005;151:1219–1228. doi: 10.1099/mic.0.27459-0. [DOI] [PubMed] [Google Scholar]
- Marcobal A, De Las-Rivas B, Moreno-Arribas MV, Muñoz R. Identification of the ornithine decarboxylase gene in the putrescine producer Oenococcus oeni BIFI- 83. FEMS Microbiol Lett. 2004;239:213–220. doi: 10.1016/j.femsle.2004.08.036. [DOI] [PubMed] [Google Scholar]
- Soksawatmaekhin W, Kuraishi A, Sakata K, Kashiwagi K, Igarashi K. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol Microbiol. 2004;51:1401–1412. doi: 10.1046/j.1365-2958.2003.03913.x. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Rollins SM, Henkin TM. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J Bacteriol. 1994;176(Suppl 15):4518–4526. doi: 10.1128/jb.176.15.4518-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connil N, Le-Breton Y, Dousset X, Auffray Y, Rince A, Prevost H. Identification of the Enterococcus faecalis tyrosine decarboxylase operon involved in tyramine production. Appl Environ Microbiol. 2002;68:3537–3544. doi: 10.1128/AEM.68.7.3537-3544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, Deboy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Coton M, Coton E, Lucas P, Lonvaud-funel A. Identification of the gene encoding a putative tyrosine decarboxylase of Carnobacterium divergens 508 Development of molecular tools for the detection of tyramine producing bacteria. Food Microbiol. 2004;21:125–130. [Google Scholar]
- Lucas P, Landete J, Coton M, Coton E, Lonvaud-funel A. The tyrosine decarboxylase operon of Lactobacillus brevis IOEB 9809: characterization and conservation in tyramine-producing bacteria. FEMS Microbiol Lett. 2003;229:65–71. doi: 10.1016/S0378-1097(03)00787-0. [DOI] [PubMed] [Google Scholar]
- Lucas P, Wolken WAM, Claisse O, Lolkema JS, Lonvaud-funel A. Histamine producing pathway encoded on an unstable plasmid in Lactobacillus hilgardii 0006. Appl Environ Microbiol. 2005;71:1417–1424. doi: 10.1128/AEM.71.3.1417-1424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares DM, Fernández M, Martín MC, Alvarez MA. Tyramine biosynthesis in Enterococcus durans is transcriptionally regulated by the extracellular pH and tyrosine concentration. Microb Biotechnol. 2009;2(Suppl 6):625–633. doi: 10.1111/j.1751-7915.2009.00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dox AW. The occurrence of tyrosine crystals in Roquefort cheese. J Am Chem Soc. 1911;33:423–425. [Google Scholar]
- Gasson MJ, De-Vos WM. Genetics and biotechnology of lactic acid bacteria. 74. Glasgow, England: Blackie Academic & Professional; 1994. [Google Scholar]
- Grundy FJ, Moir TR, Haldeman MT, Henkin TM. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res. 2002;30:1646–1655. doi: 10.1093/nar/30.7.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker A, Bruton D, Winter G. The tyrosyl-tRNA synthetase from Escherichia coli: Complete nucleotide sequence of the structural gene. FEBS Lett. 1982;150:419–423. doi: 10.1016/0014-5793(82)80781-3. [DOI] [PubMed] [Google Scholar]
- Henkin TM, Glass BL, Grundy FJ. Analysis of the Bacillus subtilis tyrS gene: conservation of a regulatory sequence in multiple tRNA synthetase genes. J Bacteriol. 1992;174:1299–1306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochhar S, Paulus H. Lysine-induced premature transcription termination in the lysC operon of Bacillus subtilis. Microbiology. 1996;142(Suppl 7):1635–1639. doi: 10.1099/13500872-142-7-1635. [DOI] [PubMed] [Google Scholar]
- Delorme C, Ehrlich SD, Renault P. Regulation of expression of the Lactococcus lactis histidine operon. J Bacteriol. 1999;181(Suppl 7):2026–2037. doi: 10.1128/jb.181.7.2026-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584(Suppl 2):318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveque F, Plateau P, Dessen P, Blanquet S. Homology of lysS and lysU, the two E. coli genes encoding distinct lysyl-tRNA synthetase species. Nucleic Acids Res. 1990;18:305–312. doi: 10.1093/nar/18.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault P, Godon JJ, Goupil N, Delorme C, Corthier G, Ehrlich SD. Metabolic operons in Lactococci. Dev Biol Stand. 1995;85:431–441. [PubMed] [Google Scholar]
- Glaser P, Danchin A, Kunst F, Debarbouille M, Vertes A, Dedonder R. A gene encoding a tyrosine-tRNA synthetase is located near sac in Bacillus subtilis. J DNA Mapping Sequencing. 1990;1:251–261. doi: 10.3109/10425179109020780. [DOI] [PubMed] [Google Scholar]
- Putzer H, Brackhage AA, Grunberg-Manago M. Independent genes for two threonyl-tRNA synthetases in Bacillus subtilis. J Bacteriol. 1990;172:4593–4602. doi: 10.1128/jb.172.8.4593-4602.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzer H, Gendron N, Grunberg-Manago M. Co-ordinate expression of the two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J. 1992;11:3117–3127. doi: 10.1002/j.1460-2075.1992.tb05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coton M, Fernández M, Trip H, Ladero V, Mulder NL, Lolkema JS, Álvarez MA, Coton E. Characterization of the tyramine-producing pathway in Sporolactobacillus sp. P3J. Microbiology. 2011;157:1841–1849. doi: 10.1099/mic.0.046367-0. [DOI] [PubMed] [Google Scholar]
- Fernández M, Linares DM, Rodríguez A, Álvarez MA. Factors affecting tyramine production in Enterococcus duran IPLA 655. Appl Microbiol Biotechnol. 2007;73(Suppl 6):1400–1406. doi: 10.1007/s00253-006-0596-y. [DOI] [PubMed] [Google Scholar]
- Calles-Enríquez M, Eriksen BH, Andersen PS, Rattray FP, Johansen AH, Fernández M, Ladero V, Álvarez MA. Sequencing and transcriptional analysis of the Streptococcus thermophilus histamine biosynthesis gene cluster: factors that affect differential hdcA expression. Appl Environ Microbiol. 2010;76(Suppl 18):6231–6231. doi: 10.1128/AEM.00827-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers OP, De-Ruyter PG, Kleerebezem M, De-Vos WM. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64:15–21. [Google Scholar]
- Linares DM, Kok J, Poolman B. Genome sequences of Lactococcus lactis MG1363 (revised) and NZ9000 and comparative physiological studies. J Bacteriol. 2010;192:5806–5812. doi: 10.1128/JB.00533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M, Beerthuyzen MM, Vaughan EE, De-Vos WM, Kuipers OP. Controlled gene expression systems for lactic acid bacteria: Transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol. 1997;63:4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen R, Buist G, Kuipers OP, Kok J. ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. J Bacteriol. 2004;186:1147–1157. doi: 10.1128/JB.186.4.1147-1157.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares DM, Geertsma ER, Poolman B. Evolved Lactococcus lactis strains for enhanced expression of recombinant membrane proteins. J Mol Biol. 2010;401:45–55. doi: 10.1016/j.jmb.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Sambrook JD, Russell D. Molecular Cloning a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- De-Vos WM, Vos P, Dehaard H, Boerritger I. Cloning and expression of the Lactococcus lactis ssp cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989;85:169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- Geertsma ER, Groeneveld M, Slotboom DJ, Poolman B. Quality control of overexpressed membrane proteins. Proc Natl Acad Sci USA. 2008;105(Suppl 15):5722–5727. doi: 10.1073/pnas.0802190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers OP, De-Ruyter PG, Kleerebezem M, De-Vos WM. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol. 1997;15:135–140. doi: 10.1016/S0167-7799(97)01029-9. [DOI] [PubMed] [Google Scholar]
- Krause I, Bockhardt A, Neckermann H, Henle T, Klostermeyer H. Simultaneous determination of amino acids and biogenic amines by reversed-phase high performance liquid chromatography of the dabsyl derivatives. J Chromatogr A. 1995;715:67–79. [Google Scholar]