Abstract
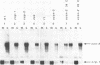
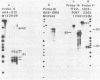
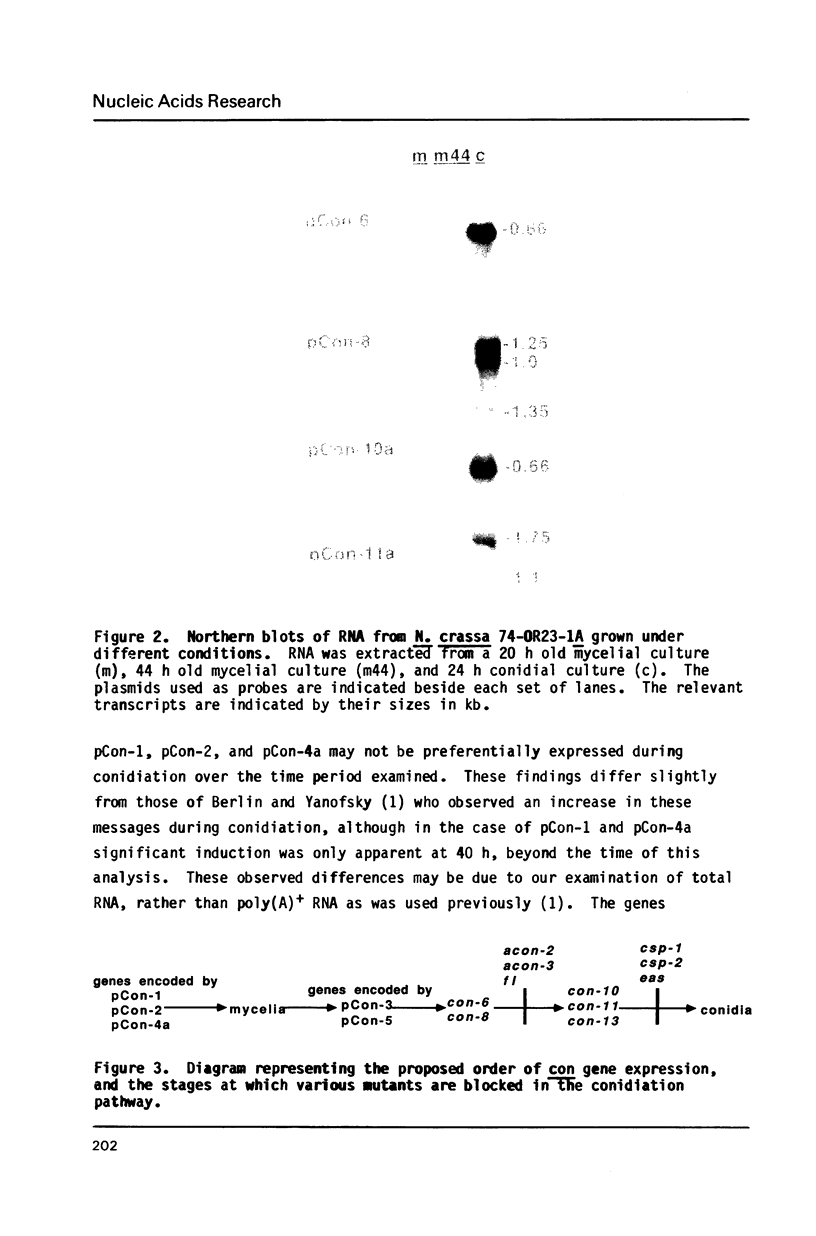
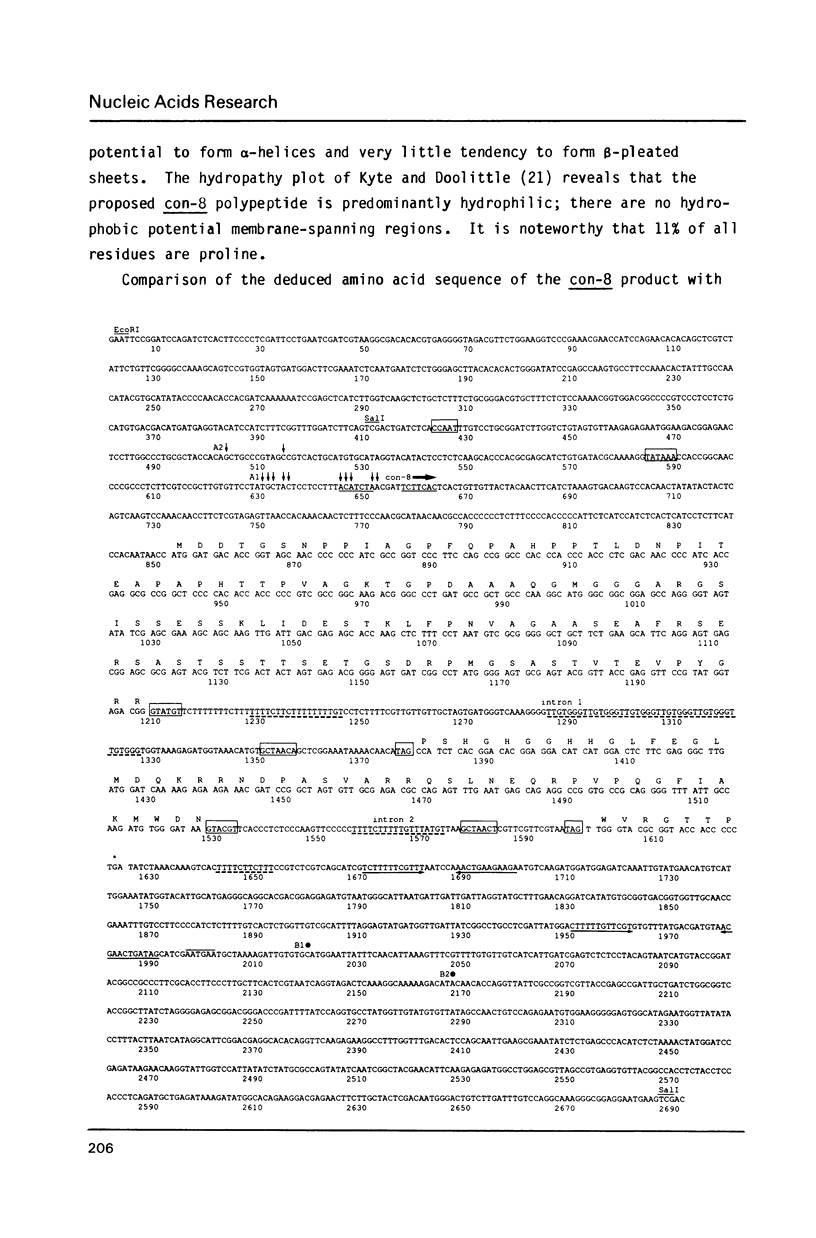
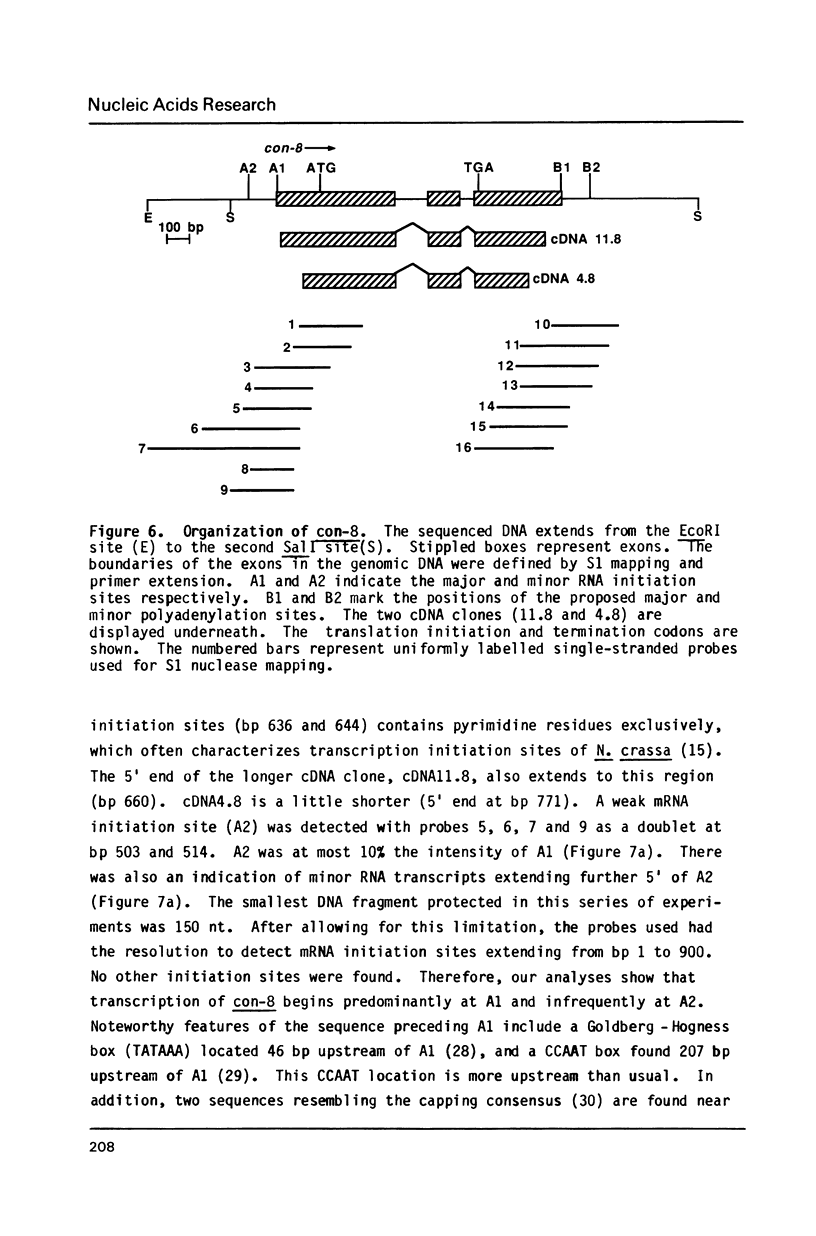
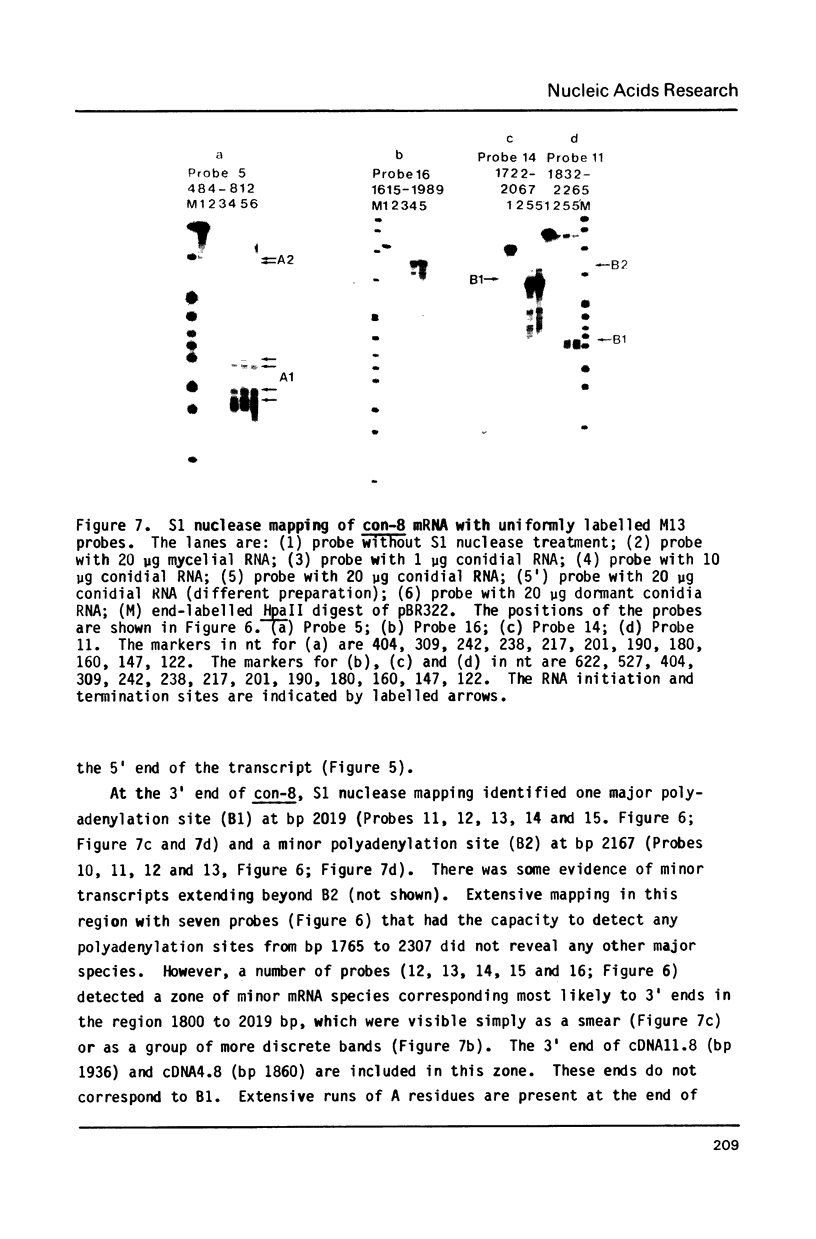
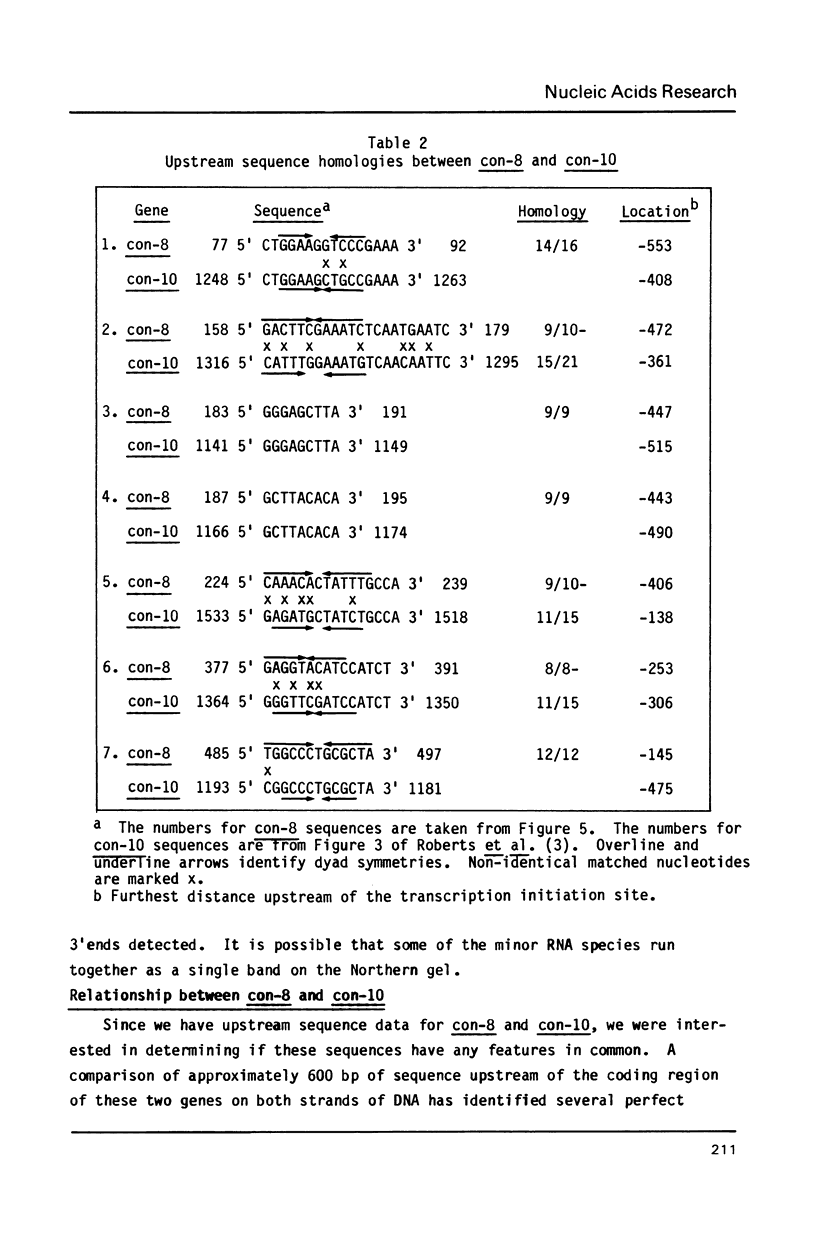
The filamentous fungus Neurospora crassa, by a series of defined changes, differentiates from a mycelium composed of branching hyphae to form dormant spores, called conidia. Several genes of unknown function (con genes) that are preferentially expressed during this period have been cloned. Transcription of these genes has been examined in conidiation-defective mutants, and the results obtained revealed that con-6, con-8, con-10, con-11 and con-13 are most likely to play a unique role during conidiation, con-8 is expressed early during conidial differentiation. Genomic and cDNA sequence analyses with con-8 clones identified one open reading frame, interrupted by two introns, which encodes a weakly acidic 18.4 kDa polypeptide containing 176 amino acid residues, con-8 is unusual in that it is transcribed as two mRNA species, 1.0 and 1.25 kb in length. S1 nuclease mapping and primer extension analyses identify one major initiation site, one major polyadenylation site, and demonstrated the existence of heterogeneity at the messenger's 5' and 3' ends.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballance D. J. Sequences important for gene expression in filamentous fungi. Yeast. 1986 Dec;2(4):229–236. doi: 10.1002/yea.320020404. [DOI] [PubMed] [Google Scholar]
- Berlin V., Yanofsky C. Isolation and characterization of genes differentially expressed during conidiation of Neurospora crassa. Mol Cell Biol. 1985 Apr;5(4):849–855. doi: 10.1128/mcb.5.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin V., Yanofsky C. Protein changes during the asexual cycle of Neurospora crassa. Mol Cell Biol. 1985 Apr;5(4):839–848. doi: 10.1128/mcb.5.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Lue N. F., Kornberg R. D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986 Mar;5(3):603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkema R., Dekker B. M., van Ormondt H. Gene organization of the transforming region of adenovirus type 7 DNA. Gene. 1982 May;18(2):143–156. doi: 10.1016/0378-1119(82)90112-3. [DOI] [PubMed] [Google Scholar]
- Dorn A., Bollekens J., Staub A., Benoist C., Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987 Sep 11;50(6):863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- Gallwitz D., Sures I. Structure of a split yeast gene: complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 May;77(5):2546–2550. doi: 10.1073/pnas.77.5.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1986 Jan 10;14(1):11–15. doi: 10.1093/nar/14.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager K. M., Mandala S. M., Davenport J. W., Speicher D. W., Benz E. J., Jr, Slayman C. W. Amino acid sequence of the plasma membrane ATPase of Neurospora crassa: deduction from genomic and cDNA sequences. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7693–7697. doi: 10.1073/pnas.83.20.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G. S., Davidson B. E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol. 1984 Dec 25;180(4):1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kreader C. A., Heckman J. E. Isolation and characterization of a Neurospora crassa ribosomal protein gene homologous to CYH2 of yeast. Nucleic Acids Res. 1987 Nov 11;15(21):9027–9042. doi: 10.1093/nar/15.21.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986 Jul;6(7):2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh J. L., Orbach M. J., Legerton T. L., Yanofsky C. The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3728–3732. doi: 10.1073/pnas.85.11.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Roberts A. N., Berlin V., Hager K. M., Yanofsky C. Molecular analysis of a Neurospora crassa gene expressed during conidiation. Mol Cell Biol. 1988 Jun;8(6):2411–2418. doi: 10.1128/mcb.8.6.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romans P., Firtel R. A. Organization of the actin multigene family of Dictyostelium discoideum and analysis of variability in the protein coding regions. J Mol Biol. 1985 Nov 20;186(2):321–335. doi: 10.1016/0022-2836(85)90108-1. [DOI] [PubMed] [Google Scholar]
- Rutledge B. J. Molecular characterization of the qa-4 gene of Neurospora crassa. Gene. 1984 Dec;32(3):275–287. doi: 10.1016/0378-1119(84)90003-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schechtman M. G., Yanofsky C. Structure of the trifunctional trp-1 gene from Neurospora crassa and its aberrant expression in Escherichia coli. J Mol Appl Genet. 1983;2(1):83–99. [PubMed] [Google Scholar]
- Staden R. A computer program to enter DNA gel reading data into a computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):499–503. doi: 10.1093/nar/12.1part2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Villa V. D., Storck R. Nucleotide composition of nuclear and mitochondrial deoxyribonucleic acid of fungi. J Bacteriol. 1968 Jul;96(1):184–190. doi: 10.1128/jb.96.1.184-190.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer S. J., Yanofsky C. Efficient cloning of genes of Neurospora crassa. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]