Abstract
Chronic kidney disease (CKD) is an important public health problem with a genetic component. We performed genome-wide association studies in up to 130,600 European ancestry participants overall, and stratified for key CKD risk factors. We uncovered 6 new loci in association with estimated glomerular filtration rate (eGFR), the primary clinical measure of CKD, in or near MPPED2, DDX1, SLC47A1, CDK12, CASP9, and INO80. Morpholino knockdown of mpped2 and casp9 in zebrafish embryos revealed podocyte and tubular abnormalities with altered dextran clearance, suggesting a role for these genes in renal function. By providing new insights into genes that regulate renal function, these results could further our understanding of the pathogenesis of CKD.
Author Summary
Chronic kidney disease (CKD) is an important public health problem with a hereditary component. We performed a new genome-wide association study in up to 130,600 European ancestry individuals to identify genes that may influence kidney function, specifically genes that may influence kidney function differently depending on sex, age, hypertension, and diabetes status of individuals. We uncovered 6 new loci associated with estimated glomerular filtration rate (eGFR), the primary measure of renal function, in or near MPPED2, DDX1, SLC47A1, CDK12, CASP9, and INO80. CDK12 effect was stronger in younger and absent in older individuals. MPPED2, DDX1, SLC47A1, and CDK12 loci were associated with eGFR in African ancestry samples as well, highlighting the cross-ethnicity validity of our findings. Using the zebrafish model, we performed morpholino knockdown of mpped2 and casp9 in zebrafish embryos and revealed podocyte and tubular abnormalities with altered dextran clearance, suggesting a role for these genes in renal function. These results further our understanding of the pathogenesis of CKD and provide insights into potential novel mechanisms of disease.
Introduction
Chronic kidney disease (CKD) affects nearly 10% of the global population [1], [2], and its prevalence continues to increase [3]. Reduced estimated glomerular filtration rate (eGFR), the primary measure used to define CKD (eGFR<60 ml/min/1.73 m2) [4], is associated with an increased risk of cardiovascular morbidity and mortality [5], acute kidney injury [6], and end stage renal disease (ESRD) [6], [7].
Using genome-wide association studies (GWAS) in predominantly population-based cohorts, we and others have previously identified more than 20 genetic loci associated with eGFR and CKD [8]–[11]. Although most of these genetic effects seem largely robust across strata of diabetes or hypertension status [9], evidence suggests that some of the loci such as the UMOD locus may have heterogeneous effects across these strata [11]. We thus hypothesized that GWAS in study populations stratified by four key CKD risk factors - age, sex, diabetes or hypertension status - may permit the identification of novel eGFR and CKD loci. We carried this out by extending our previous work [9] to a larger discovery sample of 74,354 individuals with independent replication in additional 56,246 individuals, resulting in a total of 130,600 individuals of European ancestry. To assess for potential heterogeneity, we performed separate genome-wide association analyses across strata of CKD risk factors, as well as in a more extreme CKD phenotype.
Results
Meta-analyses of GWAS on the 22 autosomes were performed for: 1) eGFR based on serum creatinine (eGFRcrea) and CKD (6,271 cases) in the overall sample, 2) eGFRcrea and CKD stratified by the four risk factors, and 3) CKD45, a more severe CKD phenotype defined as eGFRcrea <45 ml/min/1.73 m2 in the overall sample (2,181 cases). For the stratified analyses, in addition to identifying loci that were significant within each stratum, we performed a genome-wide comparison of the effect estimates between strata of the four risk factors. A complete overview of the analysis workflow is given in Figure S1. All studies participating in the stage 1 discovery and stage 2 replication phases are listed in Tables S1 and S2. The characteristics of all stage 1 discovery samples by study are reported in Table S3, and information on study design and genotyping are reported in Table S4. Results of the eGFRcrea analyses are summarized in the Manhattan and quantile-quantile plots reported in Figures S2 and S3. A total of 21 SNPs from the discovery stage were carried forward for replication in an independent set of 56,246 individuals (Tables S5 and S6). These SNPs were selected for replication for the following (Figure S1): 5 reached genome-wide significance in either eGFRcrea overall or stratified analyses, 1 based on a test of direction-consistency of SNP-eGFR associations across the discovery cohorts for eGFRcrea overall, 4 demonstrated a P value≤10−6 and high between-study homogeneity (I2<25%) in the CKD45 analysis (Table S7), and 11 demonstrated between-strata P value≤5×10−5 along with a P value≤5×10−5 for association with eGFRcrea in at least one of the two strata (Table S8).
While none of the loci identified for CKD45 or the test for between-strata difference analyses replicated, all 6 loci identified from the eGFRcrea overall analysis, stratified analyses, and the direction test did (Table 1). These 6 loci were identified and replicated in the overall analysis (rs3925584, located upstream of the MPPED2 gene; rs6431731 near the DDX1 gene), in the diabetes-free sub-group (rs2453580 in an intron of the SLC47A1 gene), in the younger age stratum (rs11078903 in an intron of the CDK12 gene; rs12124078 located near the CASP9 gene), and the direction test (rs2928148, located in the INO80 gene, see Methods for details). In the combined meta-analysis of all 45 studies used in the discovery and replication stages, all six SNPs met the genome-wide significance threshold of 5×10−8, with individual P values ranging from 4.3×10−8 to 8.4×10−18 (Table 1). The imputation quality of these SNPs is reported in Table S9, and Figure S4 shows the regional association plots for each of the 6 loci. We also confirmed all previously identified renal function loci in the current data (Table S10). Brief descriptions of the genes included within the 6 new loci uncovered can be found in Table S11. Forest plots for the associations between the index SNP at each of the 6 novel loci and eGFR across all discovery studies and all strata are presented in Figures S5 and S6. Most of the 6 new loci had similar associations across strata of CKD risk factors except for the CDK12 locus, which revealed stronger association in the younger (≤65 years of age) as compared to the older age group (>65 years of age).
Table 1. Novel loci associated with eGFRcrea.
| Locus description | Discovery analysis | Replication analysis | Combined analysis† | ||||||||||
| Analysis subgroup | SNP ID | Chr | Position (bp)‡ | Genes nearby‡ | Ref./Non-Ref. alleles (RAF) | Effect(SE)§ | P value§ | Effect(SE) | 1-sided P value | Q value | Effect(SE) | P value | I2 |
| Overall | rs3925584 | 11 | 30,716,911 | MPPED2 | T/C(0.54) | −0.0077(0.0013) | 1.0×10−09 | −0.0073(0.0013) | 4.0×10−9 | 1.1×10−08 | −0.0075(0.0009) | 8.4×10−18 | 21% |
| Overall | rs6431731 | 2 | 15,780,453 | DDX1 | T/C(0.94) | −0.0181(0.0033) | 4.6×10−08 | −0.0065(0.0034) | 0.0277 | 0.0195 | −0.0127(0.0023) | 4.3×10−08 | 11% |
| No Diabetes | rs2453580 | 17 | 19,378,913 | SLC47A1 | T/C(0.59) | 0.0076(0.0014) | 4.6×10−08 | 0.0038(0.0014) | 0.0037 | 0.0039 | 0.0059(0.0010) | 2.1×10−09 | 21% |
| Age≤65 yrs* | rs12124078 | 1 | 15,742,486 | DNAJC16, CASP9, AGMAT | A/G(0.70) | 0.0096(0.0015) | 9.8×10−10 | 0.0098(0.0017) | 5.0×10−9 | 1.1×10−08 | 0.0097(0.0011) | 1.5×10−17 | 20% |
| Age≤65 yrs | rs11078903 | 17 | 34,885,450 | CDK12, MED1, FBXL20 | A/G(0.76) | −0.0103(0.0017) | 2.4×10−09 | −0.0083(0.0023) | 1.4×10−4 | 2.0×10−04 | −0.0096(0.0013) | 9.0×10−13 | 0% |
| Direction Test (Overall)** | rs2928148 | 15 | 39,188,842 | INO80, EXD1, CHAC1 | A/G(0.52) | 0.0064(0.0012) | 1.2×10−07 | 0.0033(0.0015) | 0.0145 | 0.0122 | 0.0051(0.0009) | 4.0×10−08 | 0% |
SNPs are listed in the stratum where the smallest P value in the discovery analysis was observed. Sample size/number of studies in the discovery phase: 74,354/26 (overall, direction test), 66,931/24 (No Diabetes), 46,435/23 (age ≤65 years); replication phase: 56,246/19 (overall, direction test), 41,218/17 (No Diabetes), 28,631/16 (age ≤65 years); combined analysis: 130,600/45 (overall, direction test), 108,149/41 (No Diabetes), 75,066/39 (age ≤65 years).
Chr.: chromosome; bp: base-pairs; Ref./Non-Ref. All.: reference/non-reference alleles; RAF: reference allele frequency; SE: standard error.
Genes nearby were based on RefSeq genes (build 36). The gene closest to the SNP is listed first and is in boldface if the SNP is located within the gene.
Effects on log(eGFRcrea); post GWAS meta-analysis genomic control correction applied to P values and SEs.
While being uncovered in the younger samples, this locus showed consistent results in the non-diabetic group (combined-analysis P value 5.7×10−16) and in the overall population (P value 9.5×10−22) - see Tables S16 and S10 for additional details.
The direction test was performed in the overall dataset; the genomic control corrected P value from the direction test for the SNP rs2928148 was 4.0×10−7. In the combined analysis, the largest effect size (0.0054 on log eGFR in ml/min/1.73 m2) and the smallest P value (3.7×10−8) were observed in the non-diabetic group.
All results were confirmed by random-effect meta-analysis.
We further examined our findings in 8,110 African ancestry participants from the CARe consortium [12] (Table 2). Not surprisingly, given linkage disequilibrium (LD) differences between Europeans and African Americans, none of the 6 lead SNPs uncovered in CKDGen achieved significance in the African American samples. Next, we interrogated the 250 kb flanking regions from the lead SNP at each locus, and showed that 4 of the 6 regions (MPPED2, DDX1, SLC47A1, and CDK12) harbored SNPs that achieved statistical significance after correcting for multiple comparisons based on the genetic structure of each region (see Methods for details). Figure 1 presents the regional association plots for MPPED2, and Figure S7 presents the plots of the remaining loci in the African American sample. Imputation scores for the lead SNPs can be found in Table S12. We observed that rs12278026, upstream of MPPED2, was associated with eGFRcrea in African Americans (P value = 5×10−5, threshold for statistical significance: P value = 0.001). While rs12278026 is monomorphic in the CEU population in HapMap, rs3925584 and rs12278026 have a D′ of 1 (r2 = 0.005) in the YRI population, suggesting that these SNPs may have arisen from the same ancestral haplotype.
Table 2. Interrogation of the six novel loci uncovered in the European ancestry (EA) individuals (CKDGen consortium) in individuals of African ancestry (AA) from the CARe consortium for the trait eGFRcrea.
| Results for the lead SNPs in the CARe AA individuals | Best SNP in region in the CARe AA individuals | |||||||||||
| SNP ID* | Nearby genes§ | Ref./Non-Ref. alleles (RAF) | Effect(SE) | P value | SNP ID | Position (build 36) | LD (R2) with lead SNP | RAF (Ref./Non-Ref. alleles) | Effect(SE) | P value | S** | Bonferroni P value threshold (0.05/S) |
| rs3925584 | MPPED2 | T/C (0.88) | −0.0005(0.0066) | 0.9349 | rs12278026 | 30,744,460 | 0.005 | 0.89 (A/G) | 0.0342(0.0084) | 4.6×10−5 | 46 | 0.0011 |
| rs6431731 | DDX1 | T/C (0.99) | −0.0181(0.0213) | 0.3948 | rs4669002 | 15,874,859 | NA† | 0.56 (T/C) | −0.0196(0.0047) | 2.6×10−5 | 78 | 6.4×10−4 |
| rs12124078 | SLC47A1 | A/G (0.69) | −0.0024(0.0045) | 0.5956 | rs1472554 | 15,987,920 | 0.004 | 0.50 (C/G) | −0.0120(0.0041) | 0.0035 | 44 | 0.0011 |
| rs2453580 | DNAJC16, CASP9, AGMAT | T/C (0.59) | 0.0056(0.0049) | 0.2524 | rs1800869 | 19,505,226 | 0.011 | 0.93 (C/G) | −0.0294(0.0082) | 3.6×10−4 | 33 | 0.0015 |
| rs11078903‡ | CDK12, MED1, FBXL20 | A/G (NA‡) | NA‡ | NA‡ | rs1874226 | 34,982,557 | 0.112 | 0.34 (T/C) | 0.0157(0.0045) | 4.2×10−4 | 15 | 0.0033 |
| rs2928148 | INO80, EXD1, CHAC1 | A/G (0.22) | −0.0003(0.0053) | 0.9497 | rs8039934 | 39,284,719 | 0.105 | 0.50 (T/C) | −0.0086(0.0042) | 0.0412 | 22 | 0.0023 |
Ref./Non-Ref. All.: reference/non-reference alleles; RAF: reference allele frequency; SE: standard error.
Characteristics of the six lead SNPs in the EA individuals from the CKDGen consortium can be found in Table 1.
The gene closest to the SNP is listed first and is in boldface if the SNP is located within the gene.
S = number of independent, typed SNPs interrogated.
No LD information available in the HapMap database between the target SNP and the best SNP in the DDX1 region.
The SNP rs11078903 was not present in the CARe consortium database.
Figure 1. Genetic association and LD distribution of the MPPED2 gene locus in European and African ancestry populations.
Regional association plots in the CKDGen European ancestry discovery analysis (N = 74,354) (A) and in the CARe African ancestry discovery analysis (N = 8,110) (B). LD structure: comparison between the HapMap release II – CEU and YRI samples in the region included within +/−100 kb from the target SNP rs3925584 identified in the CKDGen GWAS. The green circle highlights a stream of high LD connecting the two blocks, indicating the presence of common haplotypes (C).
We also performed eQTL analyses of our 6 newly identified loci using known databases and a newly created renal eSNP database (see Methods) and found that rs12124078 was associated with cis expression of the nearby CASP9 gene in myocytes, which encodes caspase-9, the third apoptotic activation factor involved in the activation of cell apoptosis, necrosis and inflammation (P value for the monocyte eSNP of interest = 3.7×10−13). In the kidney, caspase-9 may play an important role in the medulla response to hyperosmotic stress [13] and in cadmium-induced toxicity [14]. The other 5 SNPs were not associated with any investigated eQTL. Additional eQTL analyses of 81 kidney biopsies (Table S13) did not reveal further evidence of association with eQTLs (Table S14).
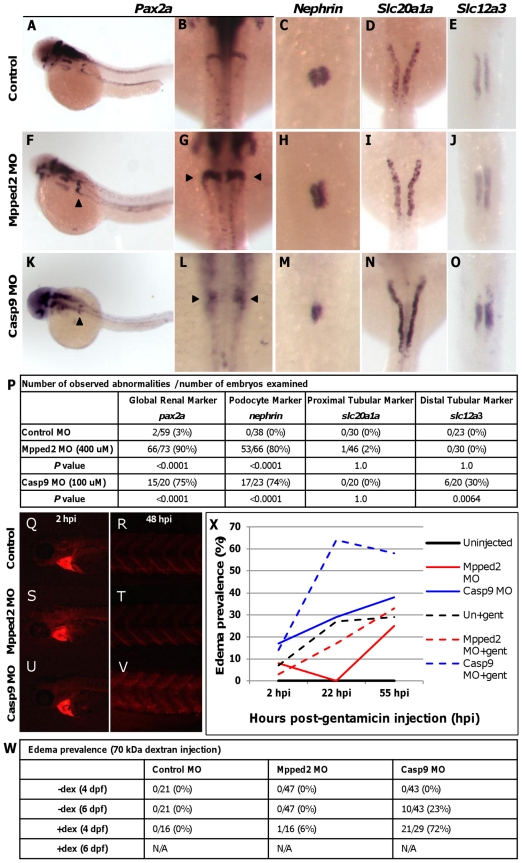
Of the 6 novel loci identified, 2 (MPPED2 and DDX1) were in regions containing only a single gene, and 1 (CASP9) had its expression associated with the locus lead SNP. Thus, to determine the potential involvement of these three genes during zebrafish kidney development, we independently assessed the expression of 4 well-characterized renal markers following morpholino knockdown: pax2a (global kidney) [15], nephrin (podocyte) [16], slc20a1a (proximal tubule) [17], and slc12a3 (distal tubule) [17]. While we observed no abnormalities in ddx1 morphants (Figure S8), mpped2 and casp9 knockdown resulted in expanded pax2a expression in the glomerular region in 90% and 75% of morphant embryos, respectively, compared to 0% in controls (P value<0.0001 for both genes; Figure 2A versus 2F and 2K; 2B versus 2G and 2L; and 2P). Significant differences were also observed in expression of the podocyte marker nephrin (Figure 2C versus 2H and 2M; 80% and 74% abnormalities for mpped2 and casp9, respectively, versus 0% in controls, P value<0.0001 for both genes). For mpped2, no differences were observed in expression of the proximal or distal tubular markers slc20a1a and slc12a3 (P value = 1.0; Figure 2D versus 2I and 2E versus 2J). Casp9 morphants and controls showed no differences in proximal tubular marker expression (Figure 2D versus 2N), but abnormalities were observed in distal tubular marker expression in casp9 knockdown embryos (30% versus 0%; Figure 2E versus 2O; P value = 0.0064).
Figure 2. Mpped2 and casp9 knockdowns result in defective kidney development.
(A–E) Whole mount in situ hybridization in control embryos demonstrates normal expression of the global kidney marker pax2a (A: lateral view; B: dorsal view), the glomerular marker nephrin (C), and the tubular markers slc20a1a (proximal tubule, D), and slc12a3 (distal tubule, E) at 48 hours post fertilization (hpf). (F–J) Mpped2 morpholino (MO) knockdown embryos develop glomerular gene expression defects (F–H, arrowheads), but tubular marker expression is normal (I, J). (K–O) Casp9 MO knockdown embryos demonstrate reduced glomerular gene expression (K–M, arrowheads) and shortened distal tubules (O). (P) Quantification of observed abnormalities per number of embryos reveal significant differences in expression of pax2a and nephrin in response to knockdown of both mpped2 and casp9 (Fisher's exact test). (Q–V) Embryos were injected with control, mpped2, or casp9 MO at the one-cell stage and subsequently injected with 70,000 MW fluorescent rhodamine dextran at 80 hpf. Dextran fluorescence was monitored over the next 48 hours. All dextran-injected embryos show equal loading into the cardiac sinus venosus at 2 hours post-injection (2 hpi/82 hpf; Q, S, U). Compared to control MO-injected embryos (R) and mpped2 knockdown embryos (T), knockdown of casp9 resulted in reduced dextran clearance at 48 hpi as shown by increased trunk fluorescence (V). (W) Casp9 knockdown results in increased susceptibility to edema formation both spontaneously (−dex) (P value = 0.0234, Fisher's exact test) and after dextran challenge (+dex) (P value<0.0001). Embryos injected with both MO and dextran did not survive to 6 dpf (N/A). (X) Edema develops earlier and with higher frequency in casp9 morphants following injection of the nephrotoxin gentamicin.
Casp9 morphants displayed diminished clearance of 70,000 MW fluorescent dextran 48 hours after injection into the sinus venosus compared to controls, revealing significant functional consequences of casp9 knockdown (Figure 2Q–2V). No clearance abnormalities were observed in mpped2 morphants. The occurrence of abdominal edema is a non-specific finding that is frequently observed in zebrafish embryos with kidney defects. We examined the occurrence of edema in mpped2 and casp9 knockdown embryos at 4 and 6 days post fertilization (dpf), both in the absence and presence of dextran, and observed a significant increase in edema prevalence in casp9 with (P value<0.0001) and without (P value = 0.0234) dextran challenge but not in mpped2 morphants (Figure 2W).
In order to further demonstrate differences in kidney function in response to knockdown of mpped2 and casp9, we injected the nephrotoxin gentamicin which predictably causes edema in a subset of embryos. Casp9 morphants were more susceptible to developing edema compared to both controls and mpped2 morphants (Figure 2X). In addition, edema developed earlier and was more severe, encompassing a greater area of the entire embryo (Figure S9). Together, these findings suggest that casp9 and mpped2 knockdowns result in altered kidney gene expression and function. Specifically, abnormal expression of pax2a and nephrin in casp9 morphants in addition to dextran retention and edema formation suggest loss of casp9 impacts glomerular development and function.
The lead SNP at the MPPED2 locus is located approximately 100 kb upstream of the gene metallophosphoesterase domain containing 2 (MPPED2), which is highly evolutionary conserved and encodes a protein with metallophosphoesterase activity [18]. It has been recognized for a role in brain development and tumorigenesis [19] but thus far not for kidney function.
To determine whether the association at our newly identified eGFRcrea loci was primarily due to creatinine metabolism or renal function, we compared the relative associations between eGFRcrea and eGFR estimated using cystatin C (eGFRcys) (Figure S10, File S1). The new loci showed similar effect sizes and consistent effect directions for eGFRcrea and eGFRcys, suggesting a relation to renal function rather than to creatinine metabolism. Placing the results of these 6 loci in context with our previously identified loci [8], [9] (23 known and 6 novel), 18 were associated with CKD at a 0.05 significance level (odds ratio, OR, from 1.05 to 1.26; P values from 3.7×10−16 to 0.01) and 11 with CKD45 (OR from 1.08 to 1.34; P values from 1.1×10−5 to 0.047; Figure S11 and Table S15).
When we examined these 29 renal function loci by age group, sex, diabetes and hypertension status (Tables S16, S17, S18, and S19), we observed consistent associations with eGFRcrea for most loci across all strata, with only two exceptions: UMOD had a stronger association in older individuals (P value for difference 8.4×10−13) and in those with hypertension (P value for difference 0.002), and CDK12 was stronger in younger subjects (P value for difference 0.0008). We tested the interaction between age and rs11078903 in one of our largest studies, the ARIC study. The interaction was significant (P value = 0.0047) and direction consistent with the observed between-strata difference.
Finally, we tested for associations between our 6 new loci and CKD related traits. The new loci were not associated with urinary albumin-to-creatinine ratio (UACR) or microalbuminuria [20] (Tables S20 and S21), with blood pressure from the ICBP Consortium [21] (Table S22) or with myocardial infarction from the CARDIoGRAM Consortium [22] (Table S23).
Discussion
We have extended prior knowledge of common genetic variants for kidney function [8]–[11], [23] by performing genome-wide association tests within strata of key CKD risk factors, including age, sex, diabetes, and hypertension, thus uncovering 6 loci not previously known to be associated with renal function in population-based studies (MPPED2, DDX1, CASP9, SLC47A1, CDK12, INO80). In contrast to our prior genome-wide analysis [8], [9], the majority of the new loci uncovered in the present analysis have little known prior associations with renal function. This highlights a continued benefit of the GWAS approach by using large sample sizes to infer new biology.
Despite our hypothesis that genetic effects are modified by CKD risk factors, most of the identified variants did not exhibit strong cross-strata differences. This highlights that many genetic associations with kidney function may be shared across risk factor strata. The association of several of these loci with kidney function in African Americans underscores the generalizability of identified renal loci across ethnicities. Zebrafish knockdown of mpped2 resulted in abnormal podocyte anatomy as assessed by expression of glomerular markers, and loss of casp9 led to altered podocyte and distal tubular marker expression, decreased dextran clearance, edema, and enhanced susceptibility to gentamicin-induced kidney damage. These findings demonstrate the potential importance of these genes with respect to renal function and illustrate that zebrafish are a useful in vivo model to explore the functional consequences of GWAS-identified genes.
Despite these strengths, there are some limitations of our study that warrant discussion. Although we used cystatin C to separate creatinine metabolism from true filtration loci, SNPs within the cystatin C gene cluster have been shown to be associated with cystatin C levels [8], which might result in some degree of misclassification in absolute levels. While we used standard definitions of diabetes and hypertension in the setting of population-based studies, these may differ from those definitions used in clinical practice. In addition, we were unable to differentiate the use of anti-hypertension medications from other clinical indications of these agents or type 1 from type 2 diabetes. The absence of association between our six newly discovered SNPs and the urinary albumin to creatinine ratio, blood pressure, and cardiovascular disease may have resulted from disparate genetic underpinnings of these traits, the overall small effect sizes, or the cross-sectional nature of our explorations; and we were unable to differentiate between these potential issues. Finally, power was modest to detect between-strata heterogeneity.
With increased sample size and stratified analyses, we have identified additional loci for kidney function that continue to have novel biological implications. Our primary findings suggest that there is substantial generalizability of SNPs associations across strata of important CKD risk factors, specifically with hypertension and diabetes.
Materials and Methods
Phenotype definition
Serum creatinine and cystatin C were measured as detailed in Tables S1 and S2. To account for between-laboratory variation, serum creatinine was calibrated to the US nationally representative National Health and Nutrition Examination Study (NHANES) standards in all discovery and replication studies as described previously [8], [24], [25]. GFR based on serum creatinine (eGFRcrea) was estimated using the four-variable MDRD Study equation [26]. GFR based on cystatin C (eGFRcys) was estimated as eGFRcys = 76.7×(serum cystatin C)−1.19 [27]. eGFRcrea and eGFRcys values<15 ml/min/1.73 m2 were set to 15, and those >200 were set to 200 ml/min/1.73 m2. CKD was defined as eGFRcrea <60 ml/min/1.73 m2 according to the National Kidney Foundation guidelines [28]. A more severe CKD phenotype, CKD45, was defined as eGFRcrea <45 ml/min/1.73 m2. Control individuals for both CKD and CKD45 analyses were defined as those with eGFRcrea >60 ml/min/1.73 m2.
Covariate definitions
In discovery and replication cohorts, diabetes was defined as fasting glucose ≥126 mg/dl, pharmacologic treatment for diabetes, or by self-report. Hypertension was defined as systolic blood pressure ≥140 mmHg or diastolic blood pressure ≥90 mmHg or pharmacologic treatment for hypertension.
Discovery analyses
Genotyping was conducted as specified in Table S4. After applying quality-control filters to exclude low-quality SNPs or samples, each study imputed up to ∼2.5 million HapMap-II SNPs, based on the CEU reference samples. Imputed genotypes were coded as the estimated number of copies of a specified allele (allelic dosage). Additional, study-specific details can be found in Table S1.
Primary association analysis
A schematic view of our complete analysis workflow is presented in Figure S1. Using data from 26 population-based studies of individuals of European ancestry, we performed GWA analyses of the following phenotypes: 1) loge(eGFRcrea), loge(eGFRcys), CKD, and CKD45 overall and 2) loge(eGFRcrea) and CKD stratified by diabetes status, hypertension status, age group (≤/>65 years), and sex. GWAS of loge(eGFRcrea) and loge(eGFRcys) were based on linear regression. GWAS of CKD and CKD45 were performed in studies with at least 25 cases (i.e. all 26 studies for CKD and 11 studies for CKD45) and were based on logistic regression. Additive genetic effects were assumed and models were adjusted for age and, where applicable, for sex, study site and principal components. Imputation uncertainty was accounted for by including allelic dosages in the model. Where necessary, relatedness was modeled with appropriate methods (see Table S1 for study-specific details). Before including in the meta-analysis, all GWA data files underwent to a careful quality control, performed using the GWAtoolbox package in R (www.eurac.edu/GWAtoolbox.html) [29].
Meta-analyses of study-specific SNP-association results, assuming fixed effects and using inverse-variance weighting, i.e.: the pooled effect  is estimated as
is estimated as  , where
, where  is the effect of the SNP on the outcome in the i
th study, K is the number of studies, and
is the effect of the SNP on the outcome in the i
th study, K is the number of studies, and  is the weight given to the i
th study. The meta-analyses were performed using METAL [30], with genomic control correction applied across all imputed SNPs [31] if the inflation factor λ>1 at both the individual study level and after the meta-analysis. SNPs with minor allele frequency (MAF)<1% were excluded. All SNPs with a meta-analysis P value≤5×10−8 for any trait or any stratum were deemed genome-wide significant [32].
is the weight given to the i
th study. The meta-analyses were performed using METAL [30], with genomic control correction applied across all imputed SNPs [31] if the inflation factor λ>1 at both the individual study level and after the meta-analysis. SNPs with minor allele frequency (MAF)<1% were excluded. All SNPs with a meta-analysis P value≤5×10−8 for any trait or any stratum were deemed genome-wide significant [32].
In the eGFRcrea analyses, after excluding loci that were previously reported [8], [9], we selected for replication all SNPs with P value<5×10−8 in any trait or stratum that were independent (defined by pairwise r 2<0.2), in the primary association analysis. This yielded five SNPs in five independent loci. The same criterion was applied to the CKD analysis, where no SNPs passed the selection threshold. Given the smaller number of cases with severe CKD resulting in less statistical power, a different selection strategy was adopted for the CKD45 analysis: selected for replication were SNPs with discovery P value≤5×10−6, MAF≥5%, and homogeneous effect size across studies (I2≤25%). Four additional SNPs were thereby selected for replication from the CKD45 analysis.
Direction test to identify SNPs for replication
In addition to identifying SNPs for replication based on the genome-wide significance threshold from a fixed effect model meta-analysis, we performed a “direction test” to identify additional SNPs for which between-study heterogeneity in effect size might have obscured the overall association that was nevertheless highly consistent in the direction of allelic effects. Under the null hypothesis of no association, the a priori probability that a given effect allele of a SNP has either a positive or negative association with eGFRcrea is 0.5. Because the meta-analysis includes independent studies, the number of concordant effect directions follows a binomial distribution. Therefore, we tested whether the number of discovery cohorts with the same sign of association (i.e. direction of effect) was greater than expected by chance given the binomial distribution and a null expectation of equal numbers of associations with positive and negative sign. The test was only applied for eGFRcrea in the overall analysis. Multiple testing was controlled by applying the same P value threshold of 5×10−8 as in the overall GWAS. Given that no SNP met this criterion, we selected for replication one novel SNP with the lowest P value of 4.0×10−7.
Genome-wide between-strata difference test to identify SNPs for replication
Based on the results of the stratified GWAS of eGFRcrea and CKD, for each SNP we tested the hypothesis whether the effect of a SNP on eGFRcrea or CKD was the same between strata (null hypothesis), i.e. diabetes versus non-diabetes subjects, hypertensive versus normotensive, younger versus older, females versus males. We used a two-sample test defined as Z = (b1−b2)/(SE(b1)2+SE(b2)2)0.5, with b1 and b2 indicating the effect estimates in the two strata and SE(b1) and SE(b2) their standard errors [33]. For large samples, the test statistic follows a standard normal distribution. SNPs were selected for replication if they had a between-stratum difference P value≤5×10−5, an association P value≤5×10−5 in one of the two strata, and MAF≥10%. Independent loci were defined using the same criteria as described above. Eleven further SNPs, one per locus, were selected for replication from the between-strata difference test.
Replication analysis
Replication was performed for a total of 21 SNPs including 5 from the overall and stratified eGFRcrea analyses, 1 from the direction test on eGFRcrea, 4 from the overall CKD45 analysis, and 11 from the between-strata difference test. Replication studies used the same phenotype definition, and had available genotypes from imputed in silico genome-wide SNP data or de novo genotyping. The same association analyses including the identical stratifications were performed as in discovery studies. Details can be found in the Tables S2, S5 and S6. Study-specific replication results for the selected SNPs were combined using the same meta-analysis approach and software as in the discovery stage. One-sided P values were derived with regard to the effect direction found in the discovery stage. Based on the P value distribution of all SNPs submitted for replication (the 10 from eGFRcrea and CKD45 and the 11 from the between strata difference test), we estimated the False Discovery Rate as a q-value using the QVALUE [34] package in R. SNPs with q-value<0.05 were called significantly replicating, thus specifying a list of associations expected to include not more than 5% false positives.
Finally, study-specific results from both the discovery and replication stage were combined in a joint inverse-variance weighted fixed-effect meta-analysis and the two-sided P values were compared to the genome-wide significance threshold of 5×10−8 to test whether a SNP was genome-wide significant. Between-study heterogeneity of replicated SNPs was quantified by the I2 statistic [35].
Replication genotyping
For de novo genotyping in 10,446 samples from KORA F3, KORA F4, SAPHIR and SAPALDIA, the MassARRAY system at the Helmholtz Zentrum (München, Germany) was used, using Assay Design v3.1.2 and the iPLEX chemistry (Sequenom, San Diego, USA). Assay design failed for rs1322199 and genotyping was not performed. Ten percent of the spectra were checked by two independent, trained persons, and 100% concordance between investigators was obtained. SNPs with a P value<0.001 when testing for Hardy-Weinberg equilibrium (rs10490130, rs10068737, rs11078903), SNPs with call rate <90% (rs500456 in KORA F4 only) or monomorphic SNPs (rs2928148) were excluded from analyses without attempting further genotyping. The call rates of rs4149333 and rs752805 were near 0% on the MassARRAY system. These SNPs were thus genotyped on a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, USA). Mean call rate across all studies and SNPs ranged from 96.8% (KORA F4) to 99% (SAPHIR). Duplicate genotyping was performed in at least 14% of the subjects in each study with a concordance of 95–100% (median 100%). In the Ogliastra Genetic Park Replication Study (n = 3000) de novo genotyping was conducted on a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, USA), with a mean call rate of 99.4% and 100% concordance of SNPs genotyped in duplicate.
Between-strata analyses for candidate SNPs in replication samples
Twenty-nine SNPs, including the 6 novel loci reported in the current manuscript along with 23 previously confirmed to be associated with renal function [9], were tested for differential effects between the strata. The same Z statistics as described for discovery (above) was used and the Bonferroni-adjusted significance level was set to 0.10/29 = 0.003.
SNP-by-age interaction, for the one SNP showing significantly different effects between strata of age, was tested in the ARIC study by fitting a linear model on log(eGFRcrea) adjusted for sex, recruitment site, the first and the seventh genetic principal components (only these two were associated with the outcome at P value<0.05). Both the interaction term and the terms for the main effects of age and the SNP were included in the model.
Power to assess between-strata effect difference
To assess genome-wide between-strata differences, with alpha = 5×10−8 and power = 80%, the maximum detectable difference was 0.025 when comparing nonDM versus DM and 0.015 when comparing nonHTN versus HTN. Similarly, when testing for between-strata differences the 29 known and new loci (Bonferroni-corrected alpha = 0.003) in the combined sample (n = ∼125,000 in nonDM and n = ∼13,000 in DM) we had 80% power to detect differences as large as 0.035.
Look-up in African Americans (CARe)
For each of the 6 lead SNPs identified in our European ancestry samples, we extracted eGFR association statistics from a genome-wide study in the CARe African ancestry consortium [12]. We further investigated potential allelic heterogeneity across ethnicities by examining the 250 kb flanking region surrounding each lead SNP to determine whether other SNPs with stronger associations exist in each region. A SNP with the smallest association P value with MAF>0.03 was considered the top SNP in the African ancestry sample. We defined statistical significance of the identified lead SNP in African ancestry individuals based on a region-specific Bonferroni correction. The number of independent SNPs was determined based on the variance inflation factor (VIF) with a recursive calculation within a sliding window of 50 SNPs and pairwise r2 of 0.2. These analyses were performed using PLINK.
Analyses of related phenotypes
For each replicating SNP, we obtained association results for urinary albumin-to-creatinine ratio and microalbuminuria from our previous genome-wide association analysis [20], and for blood pressure and myocardial infarction from genome-wide association analysis from the ICBP [21] and CARDIoGRAM [22] consortia, respectively.
eSNP analysis
Significant renal SNPs were searched against a database of expression SNPs (eSNP) including the following tissues: fresh lymphocytes [36], fresh leukocytes [37], leukocyte samples in individuals with Celiac disease [38], lymphoblastoid cell lines (LCL) derived from asthmatic children [39], HapMap LCL from 3 populations [40], a separate study on HapMap CEU LCL [41], peripheral blood monocytes [42], [43], adipose [44], [45] and blood samples [44], 2 studies on brain cortex [42], [46], 3 large studies of brain regions including prefrontal cortex, visual cortex and cerebellum (Emilsson, personal communication), liver [45], [47], osteoblasts [48], skin [49] and additional fibroblast, T cell and LCL samples [50]. The collected eSNP results met criteria for statistical significance for association with gene transcript levels as described in the original papers.
A second expression analysis of 81 biopsies from normal kidney cortex samples was performed as described previously [51], [52]. Genotyping was performed using Affymetrix 6.0 Genome-wide chip and called with GTC Software (Affymetrix). For eQTL analyses, expression probes (Affymetrix U133set) were linked to SNP probes with >90% call-rate using RefSeq annotation (Affymetrix build a30). P values for eQTLs were calculated using linear multivariable regression in both cohorts and then combined using Fisher's combined probability test (see also [52]). Pairwise LD was calculated using SNAP [53] on the CEU HapMap release 22.
Zebrafish functional experiments
Zebrafish were maintained according to established IACUC protocols. Briefly, we injected zebrafish embryos with newly designed (mpped2, ddx1) or previously validated (casp9 [54]) morpholino antisense oligonucleotides (MO, GeneTools, Philomath OR) at the one-cell stage at various doses. We fixed embryos in 4% PFA at the appropriate stages for in situ hybridization (http://zfin.org/ZFIN/Methods/ThisseProtocol.html). Different anatomic regions of the kidney were visualized using a panel of 4 established markers: pax2a (global kidney marker) [15], nephrin (podocyte marker) [16], slc20a1a (proximal tubule) [17], and slc12a3 (distal tubule marker) [17]. Abnormalities in gene expression were independently scored by two investigators. We compared the number of abnormal morphant embryos to control embryos, injected with a standard control MO designed by GeneTools, with the Fisher's exact test, at the Bonferroni-corrected significance level of 0.0125, i.e.: 0.05/4 markers. We documented the development of gross edema at 4 and 6 days post-fertilization in live embryos.
We performed dextran clearance experiments following previously described protocols [55]. Briefly, 80 hours after MO injection, we anesthetized embryos in 4 mg/ml Tricaine in embryo water (1∶20 dilution), then positioned embryos on their back in a 1% agarose injection mold. We injected an equal volume of tetramethylrhodamine dextran (70,000 MW; Invitrogen) into the cardiac sinus venosus of each embryo. We then returned the embryos to fresh embryo water. Using fluorescence microscopy, we imaged the embryos at 2 hours post-injection (82 hpf) to demonstrate equal loading, then at 48 hours post-injection (128 hpf) to evaluate dextran clearance.
Embryos were injected with control, mpped2, or casp9 MOs at the one-cell stage. At 48 hpf, embryos were manually dechorionated, anesthetized in a 1∶20 dilution of 4 mg/ml Tricaine in embryo water, and oriented on a 1% agarose injection mold. As previously described [56], embryos were injected with equal volumes of 10 mg/ml gentamicin (Sigma) in the cardiac sinus venosus, returned to fresh embryo water, and subsequently scored for edema (prevalence, time of onset) over the next 3 days.
Supporting Information
Flowchart of the project.
(TIF)
Genome-wide −log10 P values plot from stage 1 discovery meta-analysis. Plots show the discovery analysis of eGFRcrea in the overall group, with known loci [8], [9] highlighted in orange and novel loci highlighted in blue (A), and in strata of the main CKD risk factors (B, C, D, and E), with complementary groups being contrasted each other. The dotted line indicates the genome-wide significance threshold at P value = 5×10−8. The unmarked locus is RNASEH2C on chromosome 11, colored in gray despite genome-wide significance. The P value for the current stage 1 discovery for rs4014195 was 2.7×10−9. This locus previously did not replicate [9]; when we additionally considered our prior non-overlapping in silico and de novo replication data, the current stage 2 P value was 0.8832, yielding a combined stage 1+stage 2 P value of 2.6×10−7. Therefore, we did not submit this SNP for further replication.
(PDF)
Quantile-quantile plots of observed versus expected −log10 P values from the discovery analysis of eGFRcrea overall (A) and by strata of the main CKD risk factors (B). The orange line and its 95% confidence interval (shaded area) represent the null hypothesis of no association. In panel (A), results are compared when considering all SNPs (black dots) and when removing SNPs from loci that were already reported in previous GWAS [8], [9] (orange dots). The meta-analysis inflation factor λ is reported along with the discovery sample size. Individual-study minimum, maximum and median λs are also reported for comparison. Genomic-control correction was applied twice: on individual study results, before the meta-analysis, and on the meta-analysis results.
(PDF)
Regional association plots for the six new loci in the European ancestry discovery samples: (A) MPPED2; (B) DDX1; (C) SLC47A1; (D) CASP9; (E) CDK12; (F) INO80. −log10 P values are plotted versus genomic position(build 36). The lead SNP in each region is labeled. Other SNPs in each region are color-coded based on their LD to the lead SNP(LD based on the HapMap CEU, see color legend). Gene annotations are based on UCSC Genome Browser(RefSeq Genes, build 36) and arrows indicate direction of transcription. Graphs were generated using the stand-alone version of LocusZoom [57], version 1.1.
(PDF)
Forest plots of the six novel loci in the discovery phase.
(TIF)
Results from discovery meta-analysis of eGFRcrea for the six new loci: overall sample and all strata are considered. Reported is the effect size on log(eGFRcrea) and its 95% confidence interval. The stratum where the SNP was discovered is marked with a triangle for discovery based on meta-analysis P value or with a circle for discovery based on direction test.
(TIF)
Regional association plots for the six new loci in the African ancestry CARe samples: (A) MPPED2; (B) DDX1; (C) SLC47A1; (D) CASP9; (E) CDK12; (F) INO80. −log10 P values are plotted versus genomic position (build 36). The lead SNP in each region is labeled and identified by a blue arrow and blue P value. The SNP with the smallest P value in the region is indicated by a red arrow. Other SNPs in each region are color-coded based on their LD to the lead SNP (based on the HapMap YRI, see color legend). Gene annotation is based on UCSC Genome Browser (RefSeq Genes, build 36) and arrows indicate direction of transcription. Graphs were generated using the stand-alone version of LocusZoom [57], version 1.1.
(PDF)
Ddx1 knockdown does not affect kidney gene expression. (A–E) Uninjected control embryos show normal kidney development as demonstrated by in situ hybridization for the renal markers pax2a (A, B), nephrin (C), slc20a1a (D) and slc12a3 (E). (F–J) Ddx1 morpholino(MO)-injected embryos do not show significant changes in renal marker expression. (K) Number of observed abnormalities per number of embryos examined at 400 uM MO injection for renal gene expression analysis.
(TIF)
Casp9 and mpped2 knockdown embryos are more susceptible to gentamicin-induced kidney injury. Compared to control embryos (A), casp9 and mpped2 knockdown embryos develop edema at 103 hpf (C, E), suggestive of a renal defect. When injected with gentamicin, a nephrotoxin that reproducibly induces edema in control embryos (B), mpped2 and casp9 knockdown embryos develop edema earlier, more frequently, and in a more severe fashion (D, F). Whereas control embryos primarily develop cardiac edema, mpped2 and casp9 knockdown embryos display cardiac (arrowhead), ocular (black arrow), and visceral (white arrow) edema, demonstrating that mpped2 and casp9 knockdown predisposes embryos to kidney injury. (G) Quantification of edema prevalence in control, mpped2, and casp9 knockdown embryos 2, 22, and 55 hours post-injection (hpi) of gentamicin. These numbers are presented graphically in Figure 2X.
(TIF)
Comparison of the effect size on eGFRcrea and on eGFRcys for the lead SNPs of known and new loci. Results are based on the largest sample size available for each locus, i.e. the combined discovery and replication sample for the novel loci (N = 130,600), the discovery sample only for the known loci (N = 74,354). Sign of effect estimates has been changed to reflect the effects of the eGFRcrea lowering alleles. Original beta coefficients and their standard errors for the two traits can be downloaded from the File S1.
(TIF)
Odds ratios (ORs) and 95% confidence intervals of CKD and CKD45 for the lead SNPs of all known and new loci, sorted by decreasing OR of CKD.
(TIF)
Effect size on eGFRcrea and on eGFRcys for the lead SNPs of known and new loci.
(XLSX)
Study-specific methods and full acknowledgments—discovery studies.
(DOC)
Study-specific methods and full acknowledgments—replication studies and functional follow-up studies.
(DOC)
Characteristics of stage 1 discovery studies.
(DOC)
Study-specific genotyping information for stage 1 discovery studies.
(DOC)
Characteristics of stage 2 replication studies.
(DOC)
Study-specific genotyping information for stage 2 in silico replication studies.
(DOC)
Top four SNPs from the CKD45 analysis.
(DOC)
Loci identified by the test for differential effects between strata in the GWAS. Results are sorted by trait, group and chromosome. For each SNP, the P value of the test for difference between strata is reported.
(DOC)
Imputation quality of replicated SNPs in all discovery and replication studies: median MACH-Rsq and interquartile range (IQR) are reported.
(DOC)
Effects of novel and known loci on log(eGFRcrea) in the overall population.
(DOC)
Genes nearest to loci associated with renal traits.
(DOC)
Imputation Quality (MACH-Rsq) for the best SNPs in the African ancestry samples of the CARe consortium (1.00 refers to genotyped data).
(DOC)
Baseline characteristics of the kidney biopsies for the eQTL analysis.
(DOC)
Analysis of the new loci for eQTL status in meta-analysis of two cohorts of kidney biopsies.
(DOC)
Association of novel and known loci with CKD and CKD45: Odds Ratios (OR), 95% confidence intervals (95%CI) and P values.
(DOC)
Association between novel and known loci and log(eGFRcrea) in individuals without and with diabetes and test for difference between strata.
(DOC)
Association between novel and known loci and log(eGFRcrea) in individuals without and with hypertension and test for difference between strata.
(DOC)
Association between novel and known loci and log(eGFRcrea) in individuals younger and older than 65 years and test for difference between strata.
(DOC)
Association between novel and known loci and log(eGFRcrea) in females and in males and test for difference between strata.
(DOC)
Effects of novel loci on the logarithm of urinary albumin-to-creatinine ratio (log(UACR)) in the overall sample and by diabetes and hypertension status.
(DOC)
Effects (log odds ratios) of novel loci on microalbuminuria (MA) in the overall sample and by diabetes and hypertension status.
(DOC)
Association of novel loci with diastolic and systolic blood pressure in the ICBP consortium.
(DOC)
Association of novel loci with myocardial infarction in the CARDIoGRAM consortium.
(DOC)
Footnotes
The authors have declared that no competing interests exist.
The AGES study has been funded by NIH contract N01-AG-1-2100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). The Amish study was supported by grants and contracts from the NIH including R01 AG18728 (Amish Longevity Study), R01 HL088119 (Amish Calcification Study), U01 GM074518-04 (PAPI Study), U01 HL072515-06 (HAPI Study), U01 HL084756 and NIH K12RR023250 (University of Maryland MCRDP), the University of Maryland General Clinical Research Center, grant M01 RR 16500, the Baltimore Veterans Administration Medical Center Geriatrics Research and Education Clinical Center, and the Paul Beeson Physician Faculty Scholars in Aging Program. The ASPS research reported in this article was funded by the Austrian Science Fund (FWF) grant number P20545-P05 and P13180. The Medical University of Graz supported the databank of the ASPS. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. A Köttgen and C Hundertmark were supported by the grant KO3598/2-1 (Emmy Noether Programme) of the German Research Foundation. The BLSA was supported in part by the Intramural Research Program of the NIH (National Institute on Aging). The CHS research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, and grant numbers U01 HL080295 and R01 HL087652 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center for Research Resources grant M01RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The ERF study was supported by grants from the Netherlands Organization for Scientific Research (NWO; Pioneergrant), Erasmus Medical Center, the Centre for Medical Systems Biology (CMSB), and the Netherlands Kidney Foundation. The Family Heart Study (FHS) work was supported in part by NIH grants 5R01HL08770003, 5R01HL08821502 (M Province) from the NHLBI and 5R01DK07568102, 5R01DK06833603 from the NIDDK (I Borecki). The Framingham Heart Study research reported in this paper was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by the National Heart, Lung, and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The GENOA research was partially supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health R01 HL-87660. The Health Aging and Body Composition Study (Health ABC) was funded by the National Institutes of Aging. This research was supported by NIA contracts N01AG62101, N01AG62103, and N01AG62106. The GWAS was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. For the KORA F3 and F4 studies, the genetic epidemiological work was funded by the NIH subcontract from the Children's Hospital, Boston, US, (HE Wichmann, IM Heid, prime grant 1 R01 DK075787-01A1), the German National Genome Research Net NGFN2 and NGFNplus (H.E.Wichmann 01GS0823; WK project A3, number 01GS0834), the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ, and by the Else Kröner-Fresenius-Stiftung (P48/08//A11/08; CA Böger, BK Krämer). The kidney parameter measurements in F3 were funded by the Else Kröner-Fresenius-Stiftung (CA Böger, BK Krämer) and the Regensburg University Medical Center, Germany; in F4 by the University of Ulm, Germany (W Koenig). Genome-wide genotyping costs in F3 and F4 was in part funded by the Else Kröner-Fresenius-Stiftung (CA Böger, BK Krämer). De novo genotyping in F3 and F4 was funded by the Else Kröner-Fresenius-Stiftung (CA Böger, BK Krämer). The KORA research platform and the MONICA Augsburg studies were initiated and financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, by the German Federal Ministry of Education and Research, and by the State of Bavaria. Genotyping was performed in the Genome Analysis Center (GAC) of the Helmholtz Zentrum München. The LINUX platform for computation was funded by the University of Regensburg for the Department of Epidemiology and Preventive Medicine at the Regensburg University Medical Center. The NHS/HPFS type 2 diabetes GWAS (U01HG004399) is a component of a collaborative project that includes 13 other GWAS (U01HG004738, U01HG004422, U01HG004402, U01HG004729, U01HG004726, U01HG004735, U01HG004415, U01HG004436, U01HG004423, U01HG004728, RFAHG006033; National Institute of Dental and Craniofacial Research: U01DE018993, U01DE018903) funded as part of the Gene Environment-Association Studies (GENEVA) under the NIH Genes, Environment and Health Initiative (GEI). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Genotyping was performed at the Broad Institute of MIT and Harvard, with funding support from the NIH GEI (U01HG04424), and Johns Hopkins University Center for Inherited Disease Research, with support from the NIH GEI (U01HG004438) and the NIH contract “High-throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). Additional funding for the current research was provided by the National Cancer Institute (P01CA087969, P01CA055075) and the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK058845). We thank the staff and participants of the NHS and HPFS for their dedication and commitment. The Korcula study was supported through the grants from the Medical Research Council UK to H Campbell, AF Wright, and I Rudan and by Ministry of Science, Education, and Sport of the Republic of Croatia to I Rudan (number 108-1080315-0302). The MICROS study was supported by the Ministry of Health and Department of Educational Assistance, University and Research of the Autonomous Province of Bolzano, the South Tyrolean Sparkasse Foundation, and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). The Northern Swedish Population Health Study was supported by grants from the Swedish Natural Sciences Research Council, the European Union through the EUROSPAN project (contract no. LSHG-CT-2006-018947), the Foundation for Strategic Research (SSF), and the Linneaus Centre for Bioinformatics (LCB). The NHS renal function and albuminuria work was supported by DK66574. Additional funding for the current research was provided by the National Cancer Institute (P01CA087969, P01CA055075) and the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK058845). ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh. The popgen study was supported by the German Ministry of Education and Research (BMBF) through the National Genome Research Network (NGFN) and the Ministry of Science, Commerce, and Transportation of the State of Schleswig-Holstein. The project has also received infrastructure support through the DFG excellence cluster “Inflammation at Interfaces.” The Sorbs study was funded by grants from the German Research Council KFO-152 (to M Stumvoll) and the IFB (Integrated Research and Treatment Center) AdiposityDiseases (K7-37 to M Stumvoll and A Tönjes). We also thank Dr. Knut Krohn (Microarray Core Facility of the Interdisciplinary Centre for Clinical Research, University of Leipzig, Germany) for providing the genotyping platform. The research of Inga Prokopenko is funded in part through the European Community's Seventh Framework Programme (FP7/2007–2013), ENGAGE project, grant agreement HEALTH-F4-2007- 201413. R Mägi acknowledges financial support from the European Commission under a Marie Curie Intra-European Fellowship. For the Rotterdam Study-I and Rotterdam Study-II, the GWAS was funded by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Consortium for Healthy Aging (NCHA) project nr. 050-060-810. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, The Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture, and Science, the Ministry for Health, Welfare, and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The Erasmus Computing Grid, Rotterdam (The Netherlands) and the national German MediGRID and Services@MediGRID part of the German D-Grid were both funded by the German Bundesministerium fuer Forschung und Technology under grants #01 AK 803 A-H and # 01 IG 07015 G, for access to their grid resources. A Dehghan is supported by NWO grant (vici, 918-76-619). The Study of Health in Pomerania (SHIP) is part of the Community Medicine Research net of the University of Greifswald, Germany, funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania. Genome-wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany, and the Federal State of Mecklenburg-West Pomerania. The University of Greifswald is a member of the ‘Center of Knowledge Interchange’ program of the Siemens AG. The Vis study was supported through the grants from the Medical Research Council UK to H Campbell, AF Wright, and I Rudan; and Ministry of Science, Education, and Sport of the Republic of Croatia to I Rudan (number 108-1080315-0302) and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). The WGHS is supported by HL 043851 and HL69757 from the National Heart, Lung, and Blood Institute and CA 047988 from the National Cancer Institute, the Donald W. Reynolds Foundation and the Fondation Leducq, with collaborative scientific support and funding for genotyping provided by Amgen. The 3 City Study was supported by the National Foundation for Alzheimer's disease and related disorders, the Institut Pasteur de Lille and the Centre National de Génotypage. The 3 City Study was performed as part of a collaboration between the Institut National de la Santé et de la Recherche Médicale (Inserm), the Victor Segalen Bordeaux II University and Sanofi-Synthélabo. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study was also funded by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, MGEN, Institut de la Longévité, Agence Française de Sécurité Sanitaire des Produits de Santé, the Aquitaine and Bourgogne Regional Councils, Fondation de France and the joint French Ministry of Research/INSERM “Cohortes et collections de données biologiques” programme. Lille Génopôle received an unconditional grant from Eisai. The Blue Mountains Eye Study (BMES) has been supported by the Australian RADGAC grant (1992–94) and Australian National Health and Medical Research Council, Canberra Australia (Grant Nos: 974159, 211069, 991407, 457349). The GWAS studies of BMES population are supported by the Australian National Health and Medical Research Council (Grant Nos: 512423, 475604, 529912) and the Wellcome Trust, UK (2008), as part of Wellcome Trust Case Control Consortium 2 (A Viswanathan, P McGuffin, P Mitchell, F Topouzis, P Foster, grant numbers 085475/B/08/Z and 085475/08/Z). EG Holliday and JJ Wang are funded by the Australian National Health and Medical Research Council Fellowship Schemes. The CoLaus study received financial contributions from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, and the Swiss National Science Foundation (33CSCO-122661). M Bochud is supported by the Swiss School of Public Health Plus (SSPH+). The Cardiovascular Risk in Young Finns study (YFS) is supported by the Academy of Finland (grant no. 117797, 121584, and 126925), the Social Insurance Institution of Finland, University Hospital Medical funds to Tampere and Turku University Hospitals, and the Finnish Foundation of Cardiovascular Research. The Emil Aaaltonen Foundation (T Lehtimäki). EGCUT received support from FP7 grants ((201413 ENGAGE, 212111 BBMRI, 205419 ECOGENE, 245536 OPENGENE) and also received targeted financing from Estonian Government SF0180142s08 and from the European Union through the European Regional Development Fund, in the frame of Centre of Excellence in Genomics. The research of the FamHS-II was conducted in part using data and resources from the NHLBI Family Heart Study supported in part by NIH grant 5R01HL08770002. For the GoDARTs study, the Wellcome Trust provides support for Wellcome Trust United Kingdom Type 2 Diabetes Case Control Collection and the informatics support is provided by the Chief Scientist Office, and the Wellcome Trust funded Scottish Health Informatics Programme (SHIP). The INGI-Carlantino and INGI-FVG studies were supported by grants from Telethon, FVG region, and Fondo Trieste. The INGI-Cilento study was supported by grants from the EU (Vasoplus-037254), the Italian Ministry of Universities (FIRB -RBIN064YAT), the Assessorato Ricerca Regione Campania, the Ente Parco Nazionale del Cilento e Vallo di Diano, and the Fondazione Banco di Napoli to M Ciullo. The INGI – Val Borbera Study was supported from Compagnia di San Paolo, Torino, Italy, the Cariplo Fundation, Milano, Italy, and Italian Ministry of Health Progetto Finalizzato 2007 and 2009. The JUPITER trial and the genotyping were supported by AstraZeneca. The Ogliastra Genetic Park (OGP) - Replication Study and OGP - Talana study were supported by grants from the Italian Ministry of Education, University, and Research (MIUR) no. 5571/DSPAR/2002 and (FIRB) D. M. no. 718/Ric/2005. The Prospective Study of Pravastatin in the Elderly at Risk (PROSPER) trial was supported by an investigator initiated grant from Bristol-Myers Squibb, USA. The study was conducted, analyzed, and reported independently of the company. The SAPALDIA study was supported by the Swiss National Science Foundation (grants no 33CSCO-108796, 3247BO-104283, 3247BO-104288, 3247BO-104284, 3247-065896, 3100-059302, 3200-052720, 3200-042532, 4026-028099), the Federal Office for Forest, Environment, and Landscape, the Federal Office of Public Health, the Federal Office of Roads and Transport, the canton's government of Aargau, Basel-Stadt, Basel-Land, Geneva, Luzern, Ticino, Zurich, the Swiss Lung League, the canton's Lung League of Basel Stadt/Basel Landschaft, Geneva, Ticino, and Zurich. The SAPHIR-study was partially supported by a grant from the Kamillo Eisner Stiftung to B Paulweber and by grants from the “Genomics of Lipid-associated Disorders – GOLD” of the “Austrian Genome Research Programme GEN-AU” to F Kronenberg. eQTL analysis: HJ Gierman received support from the AFAR/EMF postdoctoral fellowship and the Stanford Dean's postdoctoral fellowship. HE Wheeler and SK Kim were supported by grants from the NIA, NHGRI and NIGMS.
References
- 1.Meguid El Nahas A, Bello AK. Chronic kidney disease: The global challenge. Lancet. 2005;365(9456):331–340. doi: 10.1016/S0140-6736(05)17789-7. [DOI] [PubMed] [Google Scholar]
- 2.Imai E, Matsuo S. Chronic kidney disease in asia. Lancet. 2008;371(9631):2147–2148. doi: 10.1016/S0140-6736(08)60928-9. [DOI] [PubMed] [Google Scholar]
- 3.Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, et al. Prevalence of chronic kidney disease in the united states. JAMA. 2007;298(17):2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 4.Levey AS, de Jong PE, Coresh J, El Nahas M, Astor BC, et al. The definition, classification, and prognosis of chronic kidney disease: A KDIGO controversies conference report. Kidney Int. 2011;80(1):17–28. doi: 10.1038/ki.2010.483. [DOI] [PubMed] [Google Scholar]
- 5.van der Velde M, Matsushita K, Coresh J, Astor BC, Woodward M, et al. Lower estimated glomerular filtration rate and higher albuminuria are associated with all-cause and cardiovascular mortality. A collaborative meta-analysis of high-risk population cohorts. Kidney Int. 2011;79(12):1341–1352. doi: 10.1038/ki.2010.536. [DOI] [PubMed] [Google Scholar]
- 6.Gansevoort RT, Matsushita K, van der Velde M, Astor BC, Woodward M, et al. Lower estimated GFR and higher albuminuria are associated with adverse kidney outcomes. A collaborative meta-analysis of general and high-risk population cohorts. Kidney Int. 2011;80(1):93–104. doi: 10.1038/ki.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Astor BC, Matsushita K, Gansevoort RT, van der Velde M, Woodward M, et al. Lower estimated glomerular filtration rate and higher albuminuria are associated with mortality and end-stage renal disease. A collaborative meta-analysis of kidney disease population cohorts. Kidney Int. 2011;79(12):1331–1340. doi: 10.1038/ki.2010.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kottgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41(6):712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42(5):376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers JC, Zhang W, Lord GM, van der Harst P, Lawlor DA, et al. Genetic loci influencing kidney function and chronic kidney disease. Nat Genet. 2010;42(5):373–375. doi: 10.1038/ng.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudbjartsson DF, Holm H, Indridason OS, Thorleifsson G, Edvardsson V, et al. Association of variants at UMOD with chronic kidney disease and kidney stones-role of age and comorbid diseases. PLoS Genet. 2010;6:e1001039. doi: 10.1371/journal.pgen.1001039. doi: 10.1371/journal.pgen.1001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu CT, Garnaas MK, Tin A, Kottgen A, Franceschini N, et al. Genetic association for renal traits among participants of african ancestry reveals new loci for renal function. PLoS Genet. 2011;7:e1002264. doi: 10.1371/journal.pgen.1002264. doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allan LA, Clarke PR. Apoptosis and autophagy: Regulation of caspase-9 by phosphorylation. FEBS J. 2009;276(21):6063–6073. doi: 10.1111/j.1742-4658.2009.07330.x. [DOI] [PubMed] [Google Scholar]
- 14.Gobe G, Crane D. Mitochondria, reactive oxygen species and cadmium toxicity in the kidney. Toxicol Lett. 2010;198(1):49–55. doi: 10.1016/j.toxlet.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 15.Drummond IA, Majumdar A, Hentschel H, Elger M, Solnica-Krezel L, et al. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125(23):4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- 16.Kramer-Zucker AG, Wiessner S, Jensen AM, Drummond IA. Organization of the pronephric filtration apparatus in zebrafish requires nephrin, podocin and the FERM domain protein mosaic eyes. Dev Biol. 2005;285(2):316–329. doi: 10.1016/j.ydbio.2005.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wingert RA, Selleck R, Yu J, Song HD, Chen Z, et al. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3:e189. doi: 10.1371/journal.pgen.0030189. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyagi R, Shenoy AR, Visweswariah SS. Characterization of an evolutionarily conserved metallophosphoesterase that is expressed in the fetal brain and associated with the WAGR syndrome. J Biol Chem. 2009;284(8):5217–5228. doi: 10.1074/jbc.M805996200. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz F, Eisenman R, Knoll J, Gessler M, Bruns G. cDNA sequence, genomic organization, and evolutionary conservation of a novel gene from the WAGR region. Genomics. 1995;29(2):526–532. doi: 10.1006/geno.1995.9973. [DOI] [PubMed] [Google Scholar]
- 20.Boger CA, Chen MH, Tin A, Olden M, Kottgen A, et al. CUBN is a gene locus for albuminuria. J Am Soc Nephrol. 2011;22(3):555–570. doi: 10.1681/ASN.2010060598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehret GB, Munroe PB, Rice KM, Bochud M, et al. The International Consortium for Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43(4):333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattaro C, De Grandi A, Vitart V, Hayward C, Franke A, et al. A meta-analysis of genome-wide data from five european isolates reveals an association of COL22A1, SYT1, and GABRR2 with serum creatinine level. BMC Med Genet. 2010;11:41. doi: 10.1186/1471-2350-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fox CS, Larson MG, Leip EP, Culleton B, Wilson PW, et al. Predictors of new-onset kidney disease in a community-based population. JAMA. 2004;291(7):844–850. doi: 10.1001/jama.291.7.844. [DOI] [PubMed] [Google Scholar]
- 25.Coresh J, Astor BC, McQuillan G, Kusek J, Greene T, et al. Calibration and random variation of the serum creatinine assay as critical elements of using equations to estimate glomerular filtration rate. Am J Kidney Dis. 2002;39(5):920–929. doi: 10.1053/ajkd.2002.32765. [DOI] [PubMed] [Google Scholar]
- 26.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, et al. A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. modification of diet in renal disease study group. Ann Intern Med. 1999;130(6):461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 27.Stevens LA, Coresh J, Schmid CH, Feldman HI, Froissart M, et al. Estimating GFR using serum cystatin C alone and in combination with serum creatinine: A pooled analysis of 3,418 individuals with CKD. Am J Kidney Dis. 2008;51(3):395–406. doi: 10.1053/j.ajkd.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Kidney Foundation. K/DOQI clinical practice guidelines for chronic kidney disease: Evaluation, classification, and stratification. Am J Kidney Dis. 2002;39(2 Suppl 1):S1–266. [PubMed] [Google Scholar]
- 29.Fuchsberger C, Taliun D, Pramstaller PP, Pattaro C on behalf of the CKDGen consortium. GWAtoolbox: An R package for fast quality control and handling of GWAS meta-analysis data. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr679. 10.1093/bioinformatics/btr679. [DOI] [PubMed] [Google Scholar]
- 30.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 32.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 33.Cohen A. Comparing regression coefficients across subsamples. Sociol Methods Res. 1983;12:77–94. [Google Scholar]
- 34.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39(10):1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 37.Idaghdour Y, Czika W, Shianna KV, Lee SH, Visscher PM, et al. Geographical genomics of human leukocyte gene expression variation in southern morocco. Nat Genet. 2010;42(1):62–67. doi: 10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heap GA, Trynka G, Jansen RC, Bruinenberg M, Swertz MA, et al. Complex nature of SNP genotype effects on gene expression in primary human leucocytes. BMC Med Genomics. 2009;2:1. doi: 10.1186/1755-8794-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39(10):1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 40.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, et al. Population genomics of human gene expression. Nat Genet. 2007;39(10):1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40(2):225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 42.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, et al. Tissue-specific genetic control of splicing: Implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, et al. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS ONE. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 45.Greenawalt DM, Dobrin R, Chudin E, Hatoum IJ, Suver C, et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21(7):1008–1016. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, et al. Genetic control of human brain transcript expression in alzheimer disease. Am J Hum Genet. 2009;84(4):445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schadt EE, Molony C, Chudin E, Hao K, Yang X, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grundberg E, Kwan T, Ge B, Lam KC, Koka V, et al. Population genomics in a disease targeted primary cell model. Genome Res. 2009;19(11):1942–1952. doi: 10.1101/gr.095224.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, et al. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet. 2010;87(6):779–789. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325(5945):1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wheeler HE, Metter EJ, Tanaka T, Absher D, Higgins J, et al. Sequential use of transcriptional profiling, expression quantitative trait mapping, and gene association implicates MMP20 in human kidney aging. PLoS Genet. 2009;5:e1000685. doi: 10.1371/journal.pgen.1000685. doi: 10.1371/journal.pgen.1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, et al. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, bcl-2, and caspase-3. Cell. 2008;133(5):864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hentschel DM, Mengel M, Boehme L, Liebsch F, Albertin C, et al. Rapid screening of glomerular slit diaphragm integrity in larval zebrafish. Am J Physiol Renal Physiol. 2007;293(5):F1746–50. doi: 10.1152/ajprenal.00009.2007. [DOI] [PubMed] [Google Scholar]
- 56.Hentschel DM, Park KM, Cilenti L, Zervos AS, Drummond I, et al. Acute renal failure in zebrafish: A novel system to study a complex disease. Am J Physiol Renal Physiol. 2005;288(5):F923–9. doi: 10.1152/ajprenal.00386.2004. [DOI] [PubMed] [Google Scholar]
- 57.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flowchart of the project.
(TIF)
Genome-wide −log10 P values plot from stage 1 discovery meta-analysis. Plots show the discovery analysis of eGFRcrea in the overall group, with known loci [8], [9] highlighted in orange and novel loci highlighted in blue (A), and in strata of the main CKD risk factors (B, C, D, and E), with complementary groups being contrasted each other. The dotted line indicates the genome-wide significance threshold at P value = 5×10−8. The unmarked locus is RNASEH2C on chromosome 11, colored in gray despite genome-wide significance. The P value for the current stage 1 discovery for rs4014195 was 2.7×10−9. This locus previously did not replicate [9]; when we additionally considered our prior non-overlapping in silico and de novo replication data, the current stage 2 P value was 0.8832, yielding a combined stage 1+stage 2 P value of 2.6×10−7. Therefore, we did not submit this SNP for further replication.
(PDF)
Quantile-quantile plots of observed versus expected −log10 P values from the discovery analysis of eGFRcrea overall (A) and by strata of the main CKD risk factors (B). The orange line and its 95% confidence interval (shaded area) represent the null hypothesis of no association. In panel (A), results are compared when considering all SNPs (black dots) and when removing SNPs from loci that were already reported in previous GWAS [8], [9] (orange dots). The meta-analysis inflation factor λ is reported along with the discovery sample size. Individual-study minimum, maximum and median λs are also reported for comparison. Genomic-control correction was applied twice: on individual study results, before the meta-analysis, and on the meta-analysis results.
(PDF)
Regional association plots for the six new loci in the European ancestry discovery samples: (A) MPPED2; (B) DDX1; (C) SLC47A1; (D) CASP9; (E) CDK12; (F) INO80. −log10 P values are plotted versus genomic position(build 36). The lead SNP in each region is labeled. Other SNPs in each region are color-coded based on their LD to the lead SNP(LD based on the HapMap CEU, see color legend). Gene annotations are based on UCSC Genome Browser(RefSeq Genes, build 36) and arrows indicate direction of transcription. Graphs were generated using the stand-alone version of LocusZoom [57], version 1.1.
(PDF)
Forest plots of the six novel loci in the discovery phase.
(TIF)
Results from discovery meta-analysis of eGFRcrea for the six new loci: overall sample and all strata are considered. Reported is the effect size on log(eGFRcrea) and its 95% confidence interval. The stratum where the SNP was discovered is marked with a triangle for discovery based on meta-analysis P value or with a circle for discovery based on direction test.
(TIF)
Regional association plots for the six new loci in the African ancestry CARe samples: (A) MPPED2; (B) DDX1; (C) SLC47A1; (D) CASP9; (E) CDK12; (F) INO80. −log10 P values are plotted versus genomic position (build 36). The lead SNP in each region is labeled and identified by a blue arrow and blue P value. The SNP with the smallest P value in the region is indicated by a red arrow. Other SNPs in each region are color-coded based on their LD to the lead SNP (based on the HapMap YRI, see color legend). Gene annotation is based on UCSC Genome Browser (RefSeq Genes, build 36) and arrows indicate direction of transcription. Graphs were generated using the stand-alone version of LocusZoom [57], version 1.1.
(PDF)
Ddx1 knockdown does not affect kidney gene expression. (A–E) Uninjected control embryos show normal kidney development as demonstrated by in situ hybridization for the renal markers pax2a (A, B), nephrin (C), slc20a1a (D) and slc12a3 (E). (F–J) Ddx1 morpholino(MO)-injected embryos do not show significant changes in renal marker expression. (K) Number of observed abnormalities per number of embryos examined at 400 uM MO injection for renal gene expression analysis.
(TIF)
Casp9 and mpped2 knockdown embryos are more susceptible to gentamicin-induced kidney injury. Compared to control embryos (A), casp9 and mpped2 knockdown embryos develop edema at 103 hpf (C, E), suggestive of a renal defect. When injected with gentamicin, a nephrotoxin that reproducibly induces edema in control embryos (B), mpped2 and casp9 knockdown embryos develop edema earlier, more frequently, and in a more severe fashion (D, F). Whereas control embryos primarily develop cardiac edema, mpped2 and casp9 knockdown embryos display cardiac (arrowhead), ocular (black arrow), and visceral (white arrow) edema, demonstrating that mpped2 and casp9 knockdown predisposes embryos to kidney injury. (G) Quantification of edema prevalence in control, mpped2, and casp9 knockdown embryos 2, 22, and 55 hours post-injection (hpi) of gentamicin. These numbers are presented graphically in Figure 2X.
(TIF)
Comparison of the effect size on eGFRcrea and on eGFRcys for the lead SNPs of known and new loci. Results are based on the largest sample size available for each locus, i.e. the combined discovery and replication sample for the novel loci (N = 130,600), the discovery sample only for the known loci (N = 74,354). Sign of effect estimates has been changed to reflect the effects of the eGFRcrea lowering alleles. Original beta coefficients and their standard errors for the two traits can be downloaded from the File S1.
(TIF)
Odds ratios (ORs) and 95% confidence intervals of CKD and CKD45 for the lead SNPs of all known and new loci, sorted by decreasing OR of CKD.
(TIF)
Effect size on eGFRcrea and on eGFRcys for the lead SNPs of known and new loci.
(XLSX)
Study-specific methods and full acknowledgments—discovery studies.
(DOC)
Study-specific methods and full acknowledgments—replication studies and functional follow-up studies.
(DOC)
Characteristics of stage 1 discovery studies.
(DOC)
Study-specific genotyping information for stage 1 discovery studies.
(DOC)
Characteristics of stage 2 replication studies.
(DOC)
Study-specific genotyping information for stage 2 in silico replication studies.
(DOC)
Top four SNPs from the CKD45 analysis.
(DOC)
Loci identified by the test for differential effects between strata in the GWAS. Results are sorted by trait, group and chromosome. For each SNP, the P value of the test for difference between strata is reported.
(DOC)
Imputation quality of replicated SNPs in all discovery and replication studies: median MACH-Rsq and interquartile range (IQR) are reported.
(DOC)
Effects of novel and known loci on log(eGFRcrea) in the overall population.
(DOC)
Genes nearest to loci associated with renal traits.
(DOC)
Imputation Quality (MACH-Rsq) for the best SNPs in the African ancestry samples of the CARe consortium (1.00 refers to genotyped data).
(DOC)
Baseline characteristics of the kidney biopsies for the eQTL analysis.
(DOC)
Analysis of the new loci for eQTL status in meta-analysis of two cohorts of kidney biopsies.
(DOC)
Association of novel and known loci with CKD and CKD45: Odds Ratios (OR), 95% confidence intervals (95%CI) and P values.
(DOC)
Association between novel and known loci and log(eGFRcrea) in individuals without and with diabetes and test for difference between strata.
(DOC)
Association between novel and known loci and log(eGFRcrea) in individuals without and with hypertension and test for difference between strata.
(DOC)
Association between novel and known loci and log(eGFRcrea) in individuals younger and older than 65 years and test for difference between strata.
(DOC)
Association between novel and known loci and log(eGFRcrea) in females and in males and test for difference between strata.
(DOC)
Effects of novel loci on the logarithm of urinary albumin-to-creatinine ratio (log(UACR)) in the overall sample and by diabetes and hypertension status.
(DOC)
Effects (log odds ratios) of novel loci on microalbuminuria (MA) in the overall sample and by diabetes and hypertension status.
(DOC)
Association of novel loci with diastolic and systolic blood pressure in the ICBP consortium.
(DOC)
Association of novel loci with myocardial infarction in the CARDIoGRAM consortium.
(DOC)