Abstract
Anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase incorporated with Cdh1 and/or Cdc20 recognizes and interacts with specific substrates, and faithfully orchestrates the proper cell cycle events by targeting proteins for proteasomal degradation. Experimental identification of APC/C substrates is largely dependent on the discovery of APC/C recognition motifs, e.g., the D-box and KEN-box. Although a number of either stringent or loosely defined motifs proposed, these motif patterns are only of limited use due to their insufficient powers of prediction. We report the development of a novel GPS-ARM software package which is useful for the prediction of D-boxes and KEN-boxes in proteins. Using experimentally identified D-boxes and KEN-boxes as the training data sets, a previously developed GPS (Group-based Prediction System) algorithm was adopted. By extensive evaluation and comparison, the GPS-ARM performance was found to be much better than the one using simple motifs. With this powerful tool, we predicted 4,841 potential D-boxes in 3,832 proteins and 1,632 potential KEN-boxes in 1,403 proteins from H. sapiens, while further statistical analysis suggested that both the D-box and KEN-box proteins are involved in a broad spectrum of biological processes beyond the cell cycle. In addition, with the co-localization information, we predicted hundreds of mitosis-specific APC/C substrates with high confidence. As the first computational tool for the prediction of APC/C-mediated degradation, GPS-ARM is a useful tool for information to be used in further experimental investigations. The GPS-ARM is freely accessible for academic researchers at: http://arm.biocuckoo.org.
Introduction
The 2001 Noble Prize in Physiology or Medicine was awarded to Leland Hartwell, Paul Nurse and Timothy Hunt for their seminal discoveries of key regulators of the cyclin-dependent kinases (CDKs) which are active in the cell cycle and cellular proliferation [1], [2]. Besides CDK-mediated phosphorylation, cell cycle proteins are modulated by other mechanisms, such as ubiquitin-dependent degradation, which is mainly mediated by the Skp1-cullin-F box (SCF) and the APC/C [3]–[8]. As a high-molecular-mass complex composed of 13 core subunits [3], [5], APC/C was first identified as an E3 ligase for the degradation of mitotic cyclins [9]. Beyond mitosis, APC/C-mediated degradation also plays an important role in regulating Rho GTPase activity [10], [11], axon growth [12], cell adhesion [13] and glycolysis [14], [15]. In this regard, the identification of APC/C-specific degradation substrates is fundamental to understanding the molecular mechanisms and regulatory roles of APC/C.
In 1991, Glotzer et al. first characterized an ennea-peptide (9aa) located at the N-terminus of cyclin B which is responsible for its degradation during mitotic exit [16]. Further analyses revealed that the destruction box or D-box follows a minimal consensus of RXXL (where X is any amino acid), while two co-activators of APC/C, Cdh1 and Cdc20, directly target and interact with the D-box [17]–[19]. Recently, a structural analysis revealed that a core APC/C subunit of Apc10 can also interact with the D-box and contribute to recognition specificity together with Cdh1 [20]. A second APC/C degron, the KEN-box motif with a consensus sequence of KEN, is recognized by Cdh1 and Cdc20 [21], [22]. Although a number of non-canonical destruction signals were experimentally identified, such as the A-box (QRVL) of Aurora-B kinase [23], the GXEN motif in Xenopus chromokinesin Kid (Xkid) [24], the CRY-box in mammalian Cdc20 [25] and so on, the D-box and the KEN-box are still regarded as the major APC/C recognition motifs [3]–[6], [26].
Conventional experimental identification of APC/C targets using a site-directed mutagenesis strategy is time-consuming, labor-intensive and inefficient [16]–[18], [20]. Although many experimental efforts have been undertaken over the past two decades, the number of known APC/C substrates is still quite limited. In contrast with the experimental approaches, computational prediction and analysis of the D-box and the KEN-box proteins can generate useful information for further experimental manipulation. Recently, the SLiMSearch 2.0 web server was developed for identifying user-defined short linear Motif in a proteome, using evolutionary conservation and protein structural disorder context to score occurrences [27]. However, it is evident that that the prediction with the two loosely defined motifs of RXXL and KEN will most likely generate too many false positive hits, suggesting that more sophisticated approaches are needed. For example, Michael et al. predicted 25 KEN-box proteins as potentially new APC/C targets by means of a combination of the enrichment of the cell cycle Gene Ontology (GO) terms together with native disorder prediction and motif conservation information [28]. However, only four known APC/C substrates were included in their results (CycA, P14785; KIF22, Q14807; BUB1B, O60566; PDS1, P40316. See in Table S1). In this regard, the development of a general and efficient predictor for D-boxes and KEN-boxes is urgently needed.
In this work, we developed a novel GPS-ARM software package for the prediction of potentially functional D-boxes and KEN-boxes in APC/C substrates. The experimental data was collected from the scientific literature, while the previously developed GPS 2.2 algorithm was adopted for training and prediction. By extensive evaluations, the prediction performance of GPS-ARM determined to be promising and much better than using simple short motifs. With this powerful tool, we systematically analyzed the functional abundance and diversity of D-box and KEN-box proteins in H. sapiens. From the results, it is evident that KEN-box proteins are the ones more clearly implicated in cell cycle and mitosis, while both the D-box and KEN-box proteins regulate a variety of biological processes in addition to the cell cycle. Moreover, with additional co-localization information, we predicted hundreds of mitosis-specific D-box and KEN-box proteins in eukaryotes with high confidence. Taken together, the prediction and analysis results are helpful for further experimental consideration, and the GPS-ARM can serve as a useful program for experimentalists. The online service and local packages of GPS-ARM 1.0 were implemented in JAVA and could be freely accessed for academic research at: http://arm.biocuckoo.org.
Materials and Methods
Data preparation
We searched the PubMed database with the keywords of “D box” and “KEN box”, followed by a review of the scientific literature published before August 16th, 2011. The non-canonical motifs were discarded, while the collected D-boxes and KEN-boxes were required to follow the consensus motifs of RXXL and KEN, respectively. In total, we obtained a non-redundant dataset with 74 experimentally identified D-boxes in 68 unique proteins and 44 known KEN-boxes in 42 APC/C substrates (Table S1). The corresponding sequences of these proteins were retrieved from the UniProt database.
Here, we defined an APC/C recognition motif ARM(m, n) as a core motif of RXXL (for the D-box) or KEN (for the KEN-box) flanked by m amino acids upstream and n amino acids downstream. As previously described [29], all experimentally verified D-boxes or KEN-boxes were regarded as positive data(+), while all other ARM(m, n) peptides in the same proteins were taken as negative data(−). Ultimately 217 and 16 negative peptides were obtained for the D-box and the KEN-box, respectively.
For proteome-wide analysis, we also downloaded 6,620, 3,334, 3,124, 16,384 and 20,245 reviewed protein sequences of S. cerevisiae, C. elegans, D. melanogaster, M. musculus and H. sapiens, respectively, from the UniProt database.
Performance evaluation
As previously described [29], we used the five measurements of accuracy (Ac), precision (Pr), sensitivity (Sn), specificity (Sp) and Mathew's Correlation Coefficient (MCC) to evaluate the prediction performance. The measurements were defined as below:
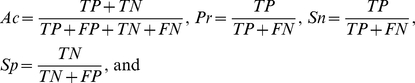 |
In this work, the leave-one-out (LOO) validation and 4-, 6-, 8- and 10-fold cross-validations were performed. The Receiver Operating Characteristic (ROC) curves were drawn, and AROC (area under ROC) values were calculated.
The algorithm
Recently, we developed the GPS 2.2 algorithm mainly for the prediction of protein pupylation sites in prokaryotes [29]. In this work, that algorithm was adopted and described as below.
The GPS 2.2 algorithm comprises two major parts, scoring strategy and performance improvement. In the former step, based on the hypothesis that similar short peptides exhibit similar biochemical properties and biological functions [29], it is possible to use an amino acid substitution matrix, e.g., BLOSUM62, to calculate the similarity between the two ARM(m, n) peptides of A and B as below:
The Score(A[i], B[i]) represents the substitution score of the two amino acids of A[i] and B[i] in the amino acid substitution matrix. If S(A, B)<0, we simply redefined it as S(A, B) = 0.
The second procedure comprises the three sequential steps of motif length selection (MLS), weight training (WT) and matrix mutation (MaM). To evaluate the performance improvement, we fixed the Sp at 90% and compared the Sn values of the LOO validation.
Motif length selection (MLS). In this step, the combinations of ARM(m, n) (m = 1, …, 30; n = 1, …, 30) were extensively tested, while the optimized combination of ARM(m, n) was determined to have the highest LOO performance.
- Weight training (WT). Since different positions can generate different contributions to recognition specificity, we can refine the substitution score between the two ARM(m, n) peptides A and B can be refined as:
Here wi is the weight of position i. Again, if S′(A, B)<0, we simply redefined it as S′(A, B) = 0. Initially, w was defined as 1 for each position. We randomly picked out the weight of any position for +1 or −1, and adopted the manipulation if the LOO performance was increased. The process was repeated until convergence was attained.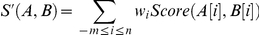
Matrix mutation (MaM). BLOSUM62 was chosen as the initial matrix and the leave-one-out performance was calculated. Subsequently, we improved the performance by randomly picking out an element of the matrix for +1 or −1. The procedure was terminated when the performance was not increased any further.
Implementation of the online service and local packages
The online service and local packages of GPS-ARM 1.0 were implemented in JAVA and are freely available at http://arm.biocuckoo.org/. For the online service, we tested GPS-ARM 1.0 on a variety of internet browsers, including Internet Explorer 8.0 and Mozilla Firefox 7.0.1 under the Windows XP Operating System (OS), Mozilla Firefox 7.0 under Fedora Core 6 OS (Linux), and Safari 3.0 under Apple Mac OS X 10.4 (Tiger) and 10.5 (Leopard). For the Windows and Linux systems, the latest version of Java Runtime Environment (JRE) package (JAVA 1.4.2 or later versions) should be pre-installed. However, for Mac OS, GPS-ARM 1.0 can be directly used without any additional packages. For convenience, we also developed local packages of GPS-ARM 1.0 which support the three major Operating Systems Windows, Linux and Mac.
Results
Development of GPS-ARM for the prediction of the D-boxes and KEN-boxes
Although not true in all cases, most of the APC/C substrates contain the D-box and/or the KEN-box, which can be recognized and interact with Cdh1 and/or Cdc20 as two major APC/C recognition motifs [3]–[6], [26]. Since the core motifs were too short and not stringent, extended consensus sequences were utilized, such as RXXLXX-I/V-XN (Motif-D1) [3], RXXLXXXXN (Motif-D2) [4], [5], [16], and RXXLXX-L/I/V/M (Motif-D3, from the Eukaryotic Linear Motif resource) [30] for the D-box, and KENXXX-N/D (Motif-KEN) [3], [22] for the KEN-box. However, in our dataset, only 9, 18, 41 and 7 boxes follow the patterns of Motif-D1, Motif-D2, Motif-D3 and Motif-KEN, respectively (Table S1). Thus, these motifs can not be used as predictive indicators due to a low sensitivity.
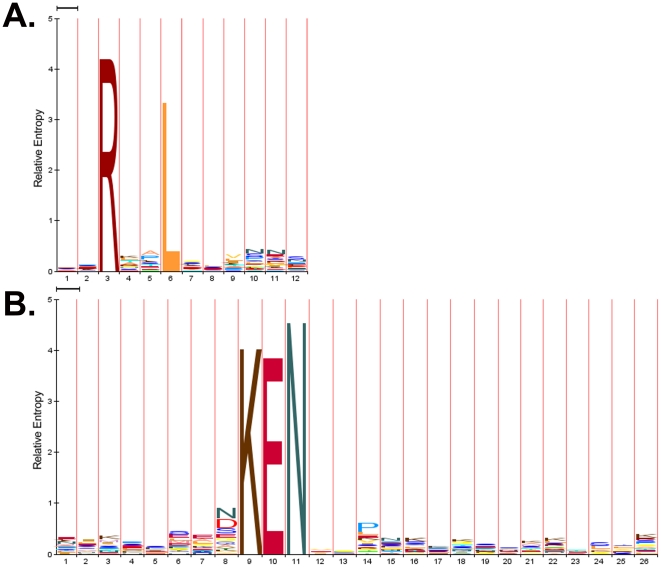
In this work, we hypothesized that flanking sequences around RXXL and KEN could contribute additional specificity for APC/C recognition. In this regard, a recently developed GPS 2.2 algorithm [29] was used for training and predicting. The ARM(2, 6) and ARM(8, 15) were determined as the optimal motifs of the D-box and KEN-box, respectively. To strengthen our hypothesis, the sequence logos of ARM(2, 6) and ARM(8, 15) were created by the HMM-Logo [31] for the D-box (Figure 1A) and KEN-box (Figure 1B), respectively. For the D-box, amino acid residues of V, N, and N preferentially appear at positions of +3, +4 and +5, although weakly (Figure 1A). For the KEN-box, the N residue is weakly informative at positions of +4 (Figure 1B). Thus, the features of known motifs were largely included in the computational models of GPS-ARM. Also, we observed that the amino acids located in RXXL are also weakly informative (Figure 1A), whereas residues of N/D and P are moderately informative at positions of −1 and +3 for the KEN-box (Figure 1B). In this regard, our models contained more useful information than known simple motifs.
Figure 1. The Sequence logos of ARM(2, 6) and ARM(8, 15) were generated by the HMM-Logo (LogoMat-M) [31] for the (A) D-box and (B) KEN-box, respectively.
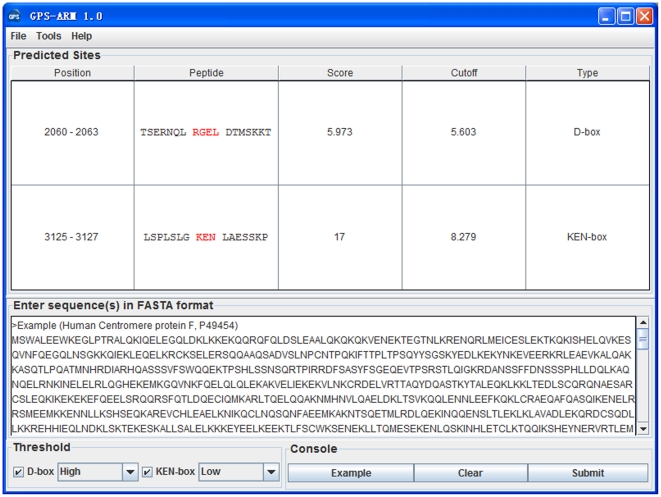
The software packages of GPS-ARM 1.0 were implemented, and the ARM(7, 7) is shown for convenience. As an example, the prediction results of human centromere protein F/CENP-F (UniProt ID: P49454) was shown (Figure 2). Although human CENP-F is a large protein (3210aa) with up to eight putative KEN-boxes, experimental analysis of its C-terminal fragment (630aa) revealed that disruption of a single KEN-box (3125–3127) is sufficient to inhibit degradation [21]. In these results, this motif was correctly predicted as the only positive hit, while an additionally predicted D-box (RGEL, 2060–2063) should prove useful for further experimental verification (Figure 2).
Figure 2. Screen snapshot of the GPS-ARM 1.0 software.
The default thresholds were chosen for the D-box (high) and KEN-box (low). As an example, the prediction results for the human centromere protein F/CENP-F (UniProt ID: P49454) are shown.
Performance evaluation and comparison
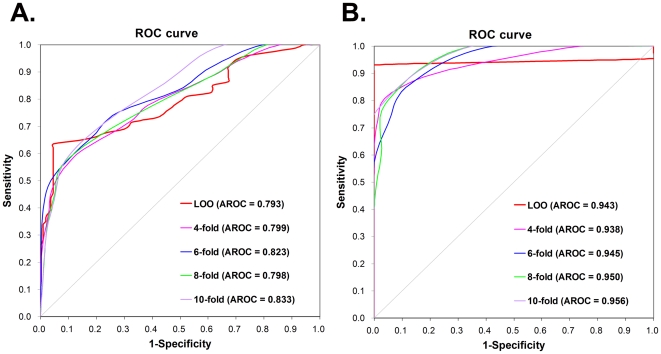
To evaluate the prediction performance and robustness of GPS-ARM, LOO validation and 4-, 6-, 8-, 10-fold cross-validations were performed (Figure 3). ROC curves were drawn, while the AROC values were 0.793 (LOO), 0.799 (4-fold), 0.823 (6-fold), 0.798 (8-fold) and 0.833 (10-fold) for the D-box (Figure 3A), and 0.943 (LOO), 0.938 (4-fold), 0.945 (6-fold), 0.950 (8-fold) and 0.956 (10-fold) for the KEN-box (Figure 3B). Since the results of the 4-, 6-, 8- and 10-fold cross-validations were similar to the LOO validation, GPS-ARM 1.0 is evidently a stable and robust predictor. The performance of the LOO validation was also used for the cut-off setting and further comparison, and the three thresholds of high, medium and low were selected (Table 1). In addition, given the highest MCC values, the high (0.6463) and low (0.8858) thresholds were chosen as the default thresholds of the D-box and KEN-box, respectively (Table 1).
Figure 3. The prediction performance of GPS-ARM 1.0.
The LOO validation and 4-, 6-, 8- and 10-fold cross-validations were performed for (A) the D-box and (B) the KEN-box.
Table 1. Performance evaluation and comparison of the GPS-ARM with known motifs.
| Method | Threshold | Ac(%) | Pr(%) | Sn(%) | Sp(%) | MCC |
| D-box (GPS-ARM) | High | 87.29 | 82.46 | 63.51 | 95.39 | 0.6463 |
| Medium | 80.76 | 61.54 | 64.86 | 86.18 | 0.5018 | |
| Low | 76.63 | 53.26 | 66.22 | 80.18 | 0.4346 | |
| e | 81.10 | 95.24 | 27.03 | 99.54 | 0.4471 | |
| KEN-box (GPS-ARM) | High | 86.67 | 100.00 | 81.82 | 100.00 | 0.7385 |
| Medium | 91.67 | 100.00 | 88.64 | 100.00 | 0.8218 | |
| Low | 95.00 | 100.00 | 93.18 | 100.00 | 0.8858 | |
| Motif-D1a | 77.32 | 90.00 | 12.16 | 99.54 | 0.2797 | |
| Motif-D2b | 75.95 | 56.25 | 24.32 | 93.55 | 0.2488 | |
| Motif-D3c | 76.29 | 53.25 | 55.41 | 83.41 | 0.3832 | |
| Motif-KENd | 38.33 | 100.00 | 15.91 | 100.00 | 0.2192 |
To clearly demonstrate the superiority of GPS-ARM, we also calculated the performances of Motif-D1 [3], Motif-D2 [4], [5], [16], Motif-D3 [30] and Motif-KEN [3], [22] (Table 1). For the D-box, we fixed the GPS-ARM Sp value to be identical with that of Motif-D1 (Table 1). By comparison, the Sn value of GPS-ARM is much larger than that of Motif-D1 (27.03% vs. 12.16%) (Table 1). Also, the performance of GPS-ARM is better than Motif-D2, since both the Sn and Sp scores are higher (Sn: 27.03% vs. 24.32%; Sp: 99.54% vs. 93.55%) (Table 1). Furthermore, both the Sn and Sp values of GPS-ARM are much better than Motif-D3 (Sn: 64.86% vs. 55.41%; Sp: 86.18% vs. 83.41%) (Table 1). For the KEN-box, although the Sp score of Motif-KEN can reach as high as 100%, its Sn value is much lower (Table 1). In addition, we compared the prediction results of GPS-ARM and various motifs for five eukaryotic proteomes (Table 2). In total, there are 143,972 RXXL and 6,443 KEN peptides in S. cerevisiae, C. elegans, D. melanogaster, M. musculus and H. sapiens, while GPS-ARM with the default thresholds predicted 11,417 (∼8%) and 3,932 (∼61%) positive hits for D-boxes and KEN-boxes, respectively (Table 2). Although Motif-D1, Motif-D2, and Motif-KEN generated fewer hits, it can be expected that a large proportion of the real boxes are missed due to their low sensitivity. Also, although Motif-D3 can generate more predicted hits (>3-fold) against GPS-ARM, the results will contain too many false positive hits due to its low specificity (Table 2). Taken together, the prediction performance of GPS-ARM 1.0 is much better than that of the simple short motifs.
Table 2. The predicted D-boxes and KEN-boxes in five eukaryotic organisms.
| Method | S. cerevisiae | C. elegans | D. melanogaster | M. musculus | H. sapiens | Total | |
| RXXL | Boxa | 12,814 | 7,555 | 9,520 | 51,065 | 63,018 | 143,972 |
| Pro.b | 4,723 | 2,519 | 2,569 | 13,503 | 16,477 | 39,791 | |
| Motif-D1 | Box | 118 | 51 | 39 | 216 | 247 | 671 |
| Pro. | 117 | 51 | 39 | 213 | 244 | 664 | |
| Motif-D2 | Box | 817 | 415 | 445 | 1,746 | 2,123 | 5,546 |
| Pro. | 732 | 374 | 395 | 1,581 | 1,916 | 4,998 | |
| Motif-D3 | Box | 3,154 | 1,849 | 2,442 | 12,478 | 15,138 | 35,061 |
| Pro. | 2,207 | 1,192 | 1,411 | 7,523 | 8,963 | 21,296 | |
| GPS 2.2 | Box | 1,104 | 635 | 815 | 4,022 | 4,841 | 11,417 |
| Pro. | 958 | 517 | 638 | 3,221 | 3,832 | 9,166 | |
| KEN | Box | 1,045 | 362 | 387 | 1,966 | 2,683 | 6,443 |
| Pro. | 891 | 331 | 340 | 1,668 | 2,206 | 5,436 | |
| Motif-KEN | Box | 143 | 42 | 35 | 159 | 222 | 601 |
| Pro. | 138 | 42 | 34 | 157 | 220 | 591 | |
| GPS 2.2 | Box | 641 | 227 | 241 | 1,191 | 1,632 | 3,932 |
| Pro. | 571 | 216 | 217 | 1,052 | 1,403 | 3,459 |
Box, the number of the predicted boxes;
Pro., the number of the predicted D-box or KEN-box proteins.
Functional abundance and diversity of the D-box and KEN-box proteins
It is generally believed that APC/C-mediated degradation plays a predominant role in the cell cycle, especially mitosis [3]–[7], [9]. However, this long-standing view has been challenged by recent discoveries that APC/C is involved in other biological processes [10]–[15]. With GPS-ARM, we predicted thousands of potential D-boxes and KEN-boxes in eukaryotes (Table 2). Although a substantial proportion of the results might not be bona fide hits, they still afford a great opportunity to systematically evaluate the functional abundance and distribution of D-box and KEN-box proteins, from the point of view that such prediction results will advance the effort to determine the real box proteins.
From H. sapiens, we predicted a total of 4,841 D-boxes in 3,832 proteins and 1,632 KEN-boxes in 1,403 proteins (Table 2). With a hypergeometric distribution [32], we statistically analyzed the enriched biological processes, molecular functions and cellular components with GO annotations for the predicted D-box (Table S2) and KEN-box (Table S3) proteins. Interestingly, for the D-box proteins, the top five most over-represented biological processes are regulation of small GTPase mediated signal transduction (GO:0051056), protein phosphorylation (GO:0006468), regulation of Rho protein signal transduction (GO:0035023), microtubule-based movement (GO:0007018) and axon guidance (GO:0007411) (Table 3). These results suggest that D-box proteins are preferentially implicated in Rho GTPase regulation and axon growth, which is consistent with recently reported experimental observations [10]–[12]. Furthermore, we observed that the GO terms of cell adhesion (GO:0007155) and regulation of glucose transport (GO:0010827) are significantly present (Table 3), and these analyses are also supported by recent studies [13]–[15]. In contrast with the D-box, the functions of the KEN-box proteins are more closely related to the cell cycle and mitosis, although the GO terms for cell adhesion and axon guidance are also enriched (Table 3). Excluding proteins in the training data set did not influence the significance of statistical results for D-boxes (Table S4) and KEN-boxes (Table S5).
Table 3. Statistical analysis of the functional abundance and diversity of the D-box and the KEN-box proteins in H. sapiens.
| Description of GO term | D- or KEN-box | Proteome | E-ratioc | p-value | ||
| Num.a | Per.b | Num. | Per. | |||
| The top 15 most enriched biological processes in D-box proteins | ||||||
| Regulation of small GTPase mediated signal transduction (GO:0051056) | 70 | 2.02% | 169 | 0.92% | 2.18 | 1.19E-11 |
| Protein phosphorylation (GO:0006468) | 116 | 3.34% | 371 | 2.03% | 1.65 | 7.27E-09 |
| Regulation of Rho protein signal transduction (GO:0035023) | 35 | 1.01% | 72 | 0.39% | 2.56 | 1.20E-08 |
| Microtubule-based movement (GO:0007018) | 39 | 1.12% | 93 | 0.51% | 2.21 | 2.74E-07 |
| Axon guidance (GO:0007411) | 93 | 2.68% | 303 | 1.66% | 1.62 | 5.54E-07 |
| Intracellular signal transduction (GO:0035556) | 82 | 2.36% | 269 | 1.47% | 1.61 | 3.40E-06 |
| G2/M transition of mitotic cell cycle (GO:0000086) | 41 | 1.18% | 110 | 0.60% | 1.96 | 5.28E-06 |
| Mitotic cell cycle (GO:0000278) | 90 | 2.59% | 306 | 1.67% | 1.55 | 5.95E-06 |
| Intracellular protein kinase cascade (GO:0007243) | 35 | 1.01% | 89 | 0.49% | 2.07 | 6.38E-06 |
| Cell adhesion (GO:0007155) | 141 | 4.06% | 546 | 2.99% | 1.36 | 4.11E-05 |
| Positive regulation of Rho GTPase activity (GO:0032321) | 13 | 0.37% | 23 | 0.13% | 2.98 | 6.84E-05 |
| Peptidyl-serine phosphorylation (GO:0018105) | 20 | 0.58% | 45 | 0.25% | 2.34 | 8.14E-05 |
| Mitotic metaphase/anaphase transition (GO:0007091) | 9 | 0.26% | 13 | 0.07% | 3.65 | 1.08E-04 |
| Nerve growth factor receptor signaling pathway (GO:0048011) | 63 | 1.82% | 215 | 1.18% | 1.54 | 1.56E-04 |
| Regulation of glucose transport (GO:0010827) | 15 | 0.43% | 31 | 0.17% | 2.55 | 1.98E-04 |
| The top 15 most enriched biological processes in KEN-box proteins | ||||||
| Cell cycle (GO:0007049) | 71 | 5.56% | 416 | 2.28% | 2.44 | 1.64E-12 |
| Cell division (GO:0051301) | 50 | 3.92% | 267 | 1.46% | 2.68 | 1.20E-10 |
| Mitotic cell cycle (GO:0000278) | 54 | 4.23% | 306 | 1.67% | 2.53 | 2.21E-10 |
| M phase of mitotic cell cycle (GO:0000087) | 26 | 2.04% | 93 | 0.51% | 4.00 | 5.69E-10 |
| Microtubule-based movement (GO:0007018) | 24 | 1.88% | 93 | 0.51% | 3.70 | 1.48E-08 |
| Flavonoid metabolic process (GO:0009812) | 8 | 0.63% | 11 | 0.06% | 10.41 | 7.55E-08 |
| Mitosis (GO:0007067) | 33 | 2.59% | 181 | 0.99% | 2.61 | 3.22E-07 |
| Golgi to plasma membrane protein transport (GO:0043001) | 6 | 0.47% | 7 | 0.04% | 12.27 | 7.55E-07 |
| Flavone metabolic process (GO:0051552) | 5 | 0.39% | 5 | 0.03% | 14.32 | 1.65E-06 |
| Sulfation (GO:0051923) | 6 | 0.47% | 8 | 0.04% | 10.74 | 2.84E-06 |
| Mitotic prometaphase (GO:0000236) | 19 | 1.49% | 85 | 0.47% | 3.20 | 4.81E-06 |
| Cell adhesion (GO:0007155) | 64 | 5.02% | 546 | 2.99% | 1.68 | 3.08E-05 |
| G2/M transition of mitotic cell cycle (GO:0000086) | 20 | 1.57% | 110 | 0.60% | 2.60 | 6.78E-05 |
| Axon guidance (GO:0007411) | 40 | 3.13% | 303 | 1.66% | 1.89 | 7.58E-05 |
| Protein targeting to lysosome (GO:0006622) | 5 | 0.39% | 8 | 0.04% | 8.95 | 7.72E-05 |
The top15 most over-represented biological processes are shown.
the number of proteins annotated;
the proportion of proteins annotated;
E-ratio, enrichment ratio, the D-box or KEN-box proportion divided by the human proteome proportion.
To confirm this analysis, we compared the functional diversity of the D-box and KEN-box proteins using the Yates' Chi-square (χ2) test [32] (Table S6). Indeed, KEN-box proteins were found to be preferentially involved in mitosis-related processes (Table S6). Taken together, although most of the experimental efforts to date have been performed in an effort to elucidate the regulatory roles of APC/C substrates in the cell cycle and mitosis, our results suggest that the D-box and KEN-box proteins in fact modulate a broad spectrum of biological processes. Again, excluding proteins in the training data set did not influence the significance of final results (Table S7). In this regard, the functional abundance and diversity of newly predicted D-box and KEN-box proteins is similar with total predictions.
Systematic prediction of mitosis-specific APC/C substrates
The ab initio prediction of D-boxes and KEN-boxes inevitably generates a substantial number of false positive hits, because most of these potential boxes may only bind to or be recognized by APC/C in vitro and not in vivo. It is believed that Cdh1, Cdc20 and APC/C have to co-localize and “kiss” their substrates for interaction to take place in a cell. In this regard, the accurate prediction of in vivo APC/C substrates is still a great challenge.
During mitosis, the accumulated evidence suggests that Cdh1, Cdc20 and the core subunits of APC/C complex (e.g., Apc10) localize in various distinct regions, such as the midbody, centrosome, and kinetochore [33]–[36]. Previously, we reported the MiCroKit 3.0 database that contains proteins that localize in the midbody, centrosome and/or kinetochore (microkit proteins) [36]. All of the microkit proteins were experimentally identified with directly corroborating evidence for subcellular localization under fluorescent microscopy [36]. Given the functional importance of the midbody, centrosome and kinetochore in mitosis and co-localization, we hypothesized that the D-boxes and KEN-boxes would likely be enriched in the microkit proteins.
Using the MiCroKit database as a reference and the default thresholds, we predicted a total of 608 potential D-boxes (File S1) and 298 KEN-boxes (File S2) in 421 and 234 proteins, respectively (Table 4). With the hypergeometric test, the statistical results clearly indicated that the D-box and KEN-box proteins are significantly over-represented in the microkit proteins (p≪0.01) (Table 4). In this regard, it is proposed that the midbody, centrosome and kinetochore are potential hotspots of APC/C substrates. The detailed prediction results can also be downloaded at: http://arm.biocuckoo.org/faq.php.
Table 4. Statistical results of the potential D-box and KEN-box substrates predicted from the microkit proteins.
| Organism | MiCroKit proteins | D-box | E-ratioc | p-value | KEN-box | E-ratio | p-value | ||
| Pro.a | Boxb | Pro. | Box | ||||||
| S. cerevisiae | 266 | 74 | 93 | 1.92 | 5.93E-09 | 58 | 70 | 2.53 | 1.31E-11 |
| C. elegans | 95 | 30 | 37 | 2.04 | 5.24E-05 | 21 | 23 | 3.41 | 3.55E-07 |
| D. melanogaster | 112 | 50 | 80 | 2.19 | 3.19E-09 | 30 | 35 | 3.86 | 2.67E-11 |
| M. musculus | 132 | 52 | 83 | 2.00 | 1.22E-07 | 24 | 32 | 2.83 | 3.17E-06 |
| H. sapiens | 677 | 215 | 315 | 1.68 | 3.25E-16 | 101 | 138 | 2.15 | 1.51E-13 |
| Total | 1,282 | 421 | 608 | 1.78 | 5.50E-36 | 234 | 298 | 2.62 | 1.18E-42 |
Pro., the number of predicted D-box or KEN-box proteins;
Box, the number of predicted D-boxes or KEN-boxes;
E-ratio, enrichment ratio, the MiCroKit D-box or KEN-box proportion in comparison with the proteomic proportion.
Discussion
As one of the most complicated cascades in eukaryotes, the cell cycle is precisely orchestrated by protein biosynthesis, phosphorylation and ubiquitin-dependent degradation in both a temporal and spatial manner [1]–[8]. Identification of APC/C-mediated degradation substrates is crucial for clearly elucidating the molecular mechanisms of the cell cycle. Previous studies suggested that the two APC/C co-activators Cdh1 and Cdc20 are responsible for the recognition of specific targets [16]–[18], [21], [22]. However, a recent analysis using single-particle electron microscopy and NMR spectroscopy reported that a core APC/C subunit of Apc10 also contributes to substrate recognition as a co-receptor of Cdh1 [20]. The efficient identification of APC/C is largely dependent on the discovery of specific boxes or motifs in its substrates. Although a variety of non-consensus motifs have been identified [23]–[25], the D-box and the KEN-box are the two major APC/C recognition motifs [3]–[6], [26]. However, either a too relaxed [5], [17], [18], [21] or too stringent a set of [3]–[5], [16], [22] simple motifs are only of limited use because of their weak predictive power (Table 1).
Previously, the GPS algorithm we developed was mainly used for the prediction of post-translational modification sites in proteins [29]. For the proper usage of the GPS algorithm, the prerequisites are that both the positions of the potentially modified residues and the motif length should be determined and fixed. For example, the lysine residues were regarded as potential pupylation sites in GPS-PUP, while the pupylation site peptide was determined as PSP(8, 18) [29]. In this work, we used the core motifs of RXXL and KEN in a different manner to determine the APC/C recognition motifs for the D-boxes and KEN-boxes, respectively. The default thresholds stand for the highest MCC values of the LOO validations, with an Ac of 87.29%, a Pr of 82.46%, a Sn of 63.51%, a Sp of 95.39% and an MCC of 0.6463 under the high threshold condition for the D-box, and an Ac of 95.00%, a Pr of 100.00%, a Sn of 93.18%, a Sp of 100% and an MCC of 0.8858 under the low threshold condition for the KEN-box (Table 1). Since the false positive rates (Type I error in statistics, equal to 1-Sp) are quite low due to high Sp values, the prediction performance of GPS-ARM is satisfactory. In addition, we collected nine D-boxes and KEN-boxes from recently published articles, while GPS-ARM can predict six of them as positive hits (Table S8).
With the GPS-ARM version 1.0, we directly predicted 11,417 potential D-boxes in 9,166 proteins and 3,932 potential KEN-boxes in 3,459 proteins from five eukaryotic organisms (Table 2). It is proposed that a considerable proportion of the RXXL (∼8%) and KEN (∼61%) motifs are real and functional boxes. Since a single predicted protein only contains 1.25 D-box and 1.14 KEN-box, it is concluded that one or two boxes per protein are sufficient for recognition and degradation by APC/C. Beyond the functions of the cell cycle and mitosis, our statistical results indicated that the D-box and KEN-box are involved in additional biological processes (Table 3), and these results are consistent with recently reported experimental observations [10]–[15]. Furthermore, we systematically predicted mitosis-specific APC/C substrates with the localization information from the MiCroKit 3.0 database [36]. Statistical analysis suggested that the D-box and KEN-box proteins are significantly enriched in the midbody, centrosome and kinetochore (Table 4). Taken together, although further improvement should be carried out as new experimental data are available, the GPS-ARM and subsequent analyses provide useful information for further experimental manipulation.
Supporting Information
Prediction results of D-boxes in proteins which were localized at MiCroKit (Centrosome, Midbody, and Kinetochore).
(TXT)
Prediction results of KEN-boxes in proteins which were localized at MiCroKit (Centrosome, Midbody, and Kinetochore).
(TXT)
We manually collected 74 experimentally identified D-boxes in 68 unique proteins and 44 experimentally identified KEN-boxes in 42 unique proteins from the scientific literature (PubMed). a. UniProt, the UniProt accession numbers of the D-box and KEN-box proteins; b. Position, the position of the D-box or KEN-box; c. Motif type, the type of known motif that the box follows in. d. PMID, the primary references for the known D-boxes or KEN-boxes.
(XLS)
The top 15 most enriched biological processes, molecular functions and cellular components of the D-box proteins in H. sapiens.
(XLS)
The top 15 most enriched biological processes, molecular functions and cellular components of the KEN-box proteins in H. sapiens.
(XLS)
The top 15 most enriched biological processes, molecular functions and cellular components of newly predicted D-box proteins in H. sapiens. The proteins in the training data set were excluded.
(XLS)
The top 15 most enriched biological processes, molecular functions and cellular components of newly predicted KEN-box proteins in H. sapiens. The proteins in the training data set were excluded.
(XLS)
Statistical comparison of the GO terms for the substrates between the predicted KEN-box and D-box proteins in the human proteome. Yates' Chi-square (χ2) test was performed (p-value<0.05) [32]. The entries with the grey background indicate the Enrichment_ratio ≤1.
(XLS)
Statistical comparison of the GO terms for newly predicted KEN-box and D-box proteins in the human proteome. The proteins in the training data set were excluded.
(XLS)
From recently published papers, we collected nine D-boxes and KEN-boxes in six proteins. The data set was not used for training. The default parameters were used for the GPS-ARM.
(XLS)
Acknowledgments
The authors are thankful for two anonymous reviewers, whose suggestions have greatly improved the presentation of this manuscript. Pacific Edit reviewed the manuscript prior to submission.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported, in whole or in part, by the National Basic Research Program (973 project) (2010CB945400, 2012CB911200, 2012CB910101), Natural Science Foundation of China (90919001, 31071154, 30900835, 30830036, 91019020, 31171263), and Fundamental Research Funds for the Central Universities (HUST: 2010JC049, 2010ZD018, 2011TS085; SYSU: 11lgzd11). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Milestones in cell division. Nat Cell Biol. 2001;3:E265. doi: 10.1038/ncb1201-e265. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K. A prize for proliferation. Cell. 2001;107:689–701. doi: 10.1016/s0092-8674(01)00604-3. [DOI] [PubMed] [Google Scholar]
- 3.Barford D. Structure, function and mechanism of the anaphase promoting complex (APC/C). Q Rev Biophys. 2011;44:153–190. doi: 10.1017/S0033583510000259. [DOI] [PubMed] [Google Scholar]
- 4.Thornton BR, Toczyski DP. Precise destruction: an emerging picture of the APC. Genes Dev. 2006;20:3069–3078. doi: 10.1101/gad.1478306. [DOI] [PubMed] [Google Scholar]
- 5.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 6.Fang G, Yu H, Kirschner MW. Control of mitotic transitions by the anaphase-promoting complex. Philos Trans R Soc Lond B Biol Sci. 1999;354:1583–1590. doi: 10.1098/rstb.1999.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pagano M. Cell cycle regulation by the ubiquitin pathway. FASEB J. 1997;11:1067–1075. doi: 10.1096/fasebj.11.13.9367342. [DOI] [PubMed] [Google Scholar]
- 8.Peters JM. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 9.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, et al. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liot C, Seguin L, Siret A, Crouin C, Schmidt S, et al. APC Mediates Degradation of the Oncogenic Rho-GEF Ect2 after Mitosis. PLoS One. 2011;6:e23676. doi: 10.1371/journal.pone.0023676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naoe H, Araki K, Nagano O, Kobayashi Y, Ishizawa J, et al. The anaphase-promoting complex/cyclosome activator Cdh1 modulates Rho GTPase by targeting p190 RhoGAP for degradation. Mol Cell Biol. 2010;30:3994–4005. doi: 10.1128/MCB.01358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim AH, Bonni A. Thinking within the D box: initial identification of Cdh1-APC substrates in the nervous system. Mol Cell Neurosci. 2007;34:281–287. doi: 10.1016/j.mcn.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Silies M, Klambt C. APC/C(Fzr/Cdh1)-dependent regulation of cell adhesion controls glial migration in the Drosophila PNS. Nat Neurosci. 2010;13:1357–1364. doi: 10.1038/nn.2656. [DOI] [PubMed] [Google Scholar]
- 14.Tudzarova S, Colombo SL, Stoeber K, Carcamo S, Williams GH, et al. Two ubiquitin ligases, APC/C-Cdh1 and SKP1-CUL1-F (SCF)-beta-TrCP, sequentially regulate glycolysis during the cell cycle. Proc Natl Acad Sci U S A. 2011;108:5278–5283. doi: 10.1073/pnas.1102247108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colombo SL, Palacios-Callender M, Frakich N, De Leon J, Schmitt CA, et al. Anaphase-promoting complex/cyclosome-Cdh1 coordinates glycolysis and glutaminolysis with transition to S phase in human T lymphocytes. Proc Natl Acad Sci U S A. 2010;107:18868–18873. doi: 10.1073/pnas.1012362107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 17.Owens TJ, Hoyt MA. The D box asserts itself. Mol Cell. 2005;18:611–612. doi: 10.1016/j.molcel.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 18.King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 20.da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurden MD, Holland AJ, van Zon W, Tighe A, Vergnolle MA, et al. Cdc20 is required for the post-anaphase, KEN-dependent degradation of centromere protein F. J Cell Sci. 2010;123:321–330. doi: 10.1242/jcs.062075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen HG, Chinnappan D, Urano T, Ravid K. Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: identification of an aneuploidy-promoting property. Mol Cell Biol. 2005;25:4977–4992. doi: 10.1128/MCB.25.12.4977-4992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro A, Vigneron S, Bernis C, Labbe JC, Lorca T. Xkid is degraded in a D-box, KEN-box, and A-box-independent pathway. Mol Cell Biol. 2003;23:4126–4138. doi: 10.1128/MCB.23.12.4126-4138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reis A, Levasseur M, Chang HY, Elliott DJ, Jones KT. The CRY box: a second APCcdh1-dependent degron in mammalian cdc20. EMBO Rep. 2006;7:1040–1045. doi: 10.1038/sj.embor.7400772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davey NE, Haslam NJ, Shields DC, Edwards RJ. SLiMSearch 2.0: biological context for short linear motifs in proteins. Nucleic Acids Res. 2011;39:W56–60. doi: 10.1093/nar/gkr402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael S, Trave G, Ramu C, Chica C, Gibson TJ. Discovery of candidate KEN-box motifs using cell cycle keyword enrichment combined with native disorder prediction and motif conservation. Bioinformatics. 2008;24:453–457. doi: 10.1093/bioinformatics/btm624. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Ma Q, Cao J, Gao X, Ren J, et al. GPS-PUP: computational prediction of pupylation sites in prokaryotic proteins. Mol Biosyst. 2011;7:2737–2740. doi: 10.1039/c1mb05217a. [DOI] [PubMed] [Google Scholar]
- 30.Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, et al. ELM–the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40:D242–251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuster-Bockler B, Schultz J, Rahmann S. HMM Logos for visualization of protein families. BMC Bioinformatics. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, Cao J, Ma Q, Gao X, Ren J, et al. GPS-YNO2: computational prediction of tyrosine nitration sites in proteins. Mol Biosyst. 2011;7:1197–1204. doi: 10.1039/c0mb00279h. [DOI] [PubMed] [Google Scholar]
- 33.Kurasawa Y, Todokoro K. Identification of human APC10/Doc1 as a subunit of anaphase promoting complex. Oncogene. 1999;18:5131–5137. doi: 10.1038/sj.onc.1203133. [DOI] [PubMed] [Google Scholar]
- 34.Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, Ching YP, Chun AC, Jin DY. Nuclear localization of the cell cycle regulator CDH1 and its regulation by phosphorylation. J Biol Chem. 2003;278:12530–12536. doi: 10.1074/jbc.M212853200. [DOI] [PubMed] [Google Scholar]
- 36.Ren J, Liu Z, Gao X, Jin C, Ye M, et al. MiCroKit 3.0: an integrated database of midbody, centrosome and kinetochore. Nucleic Acids Res. 2010;38:D155–160. doi: 10.1093/nar/gkp784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prediction results of D-boxes in proteins which were localized at MiCroKit (Centrosome, Midbody, and Kinetochore).
(TXT)
Prediction results of KEN-boxes in proteins which were localized at MiCroKit (Centrosome, Midbody, and Kinetochore).
(TXT)
We manually collected 74 experimentally identified D-boxes in 68 unique proteins and 44 experimentally identified KEN-boxes in 42 unique proteins from the scientific literature (PubMed). a. UniProt, the UniProt accession numbers of the D-box and KEN-box proteins; b. Position, the position of the D-box or KEN-box; c. Motif type, the type of known motif that the box follows in. d. PMID, the primary references for the known D-boxes or KEN-boxes.
(XLS)
The top 15 most enriched biological processes, molecular functions and cellular components of the D-box proteins in H. sapiens.
(XLS)
The top 15 most enriched biological processes, molecular functions and cellular components of the KEN-box proteins in H. sapiens.
(XLS)
The top 15 most enriched biological processes, molecular functions and cellular components of newly predicted D-box proteins in H. sapiens. The proteins in the training data set were excluded.
(XLS)
The top 15 most enriched biological processes, molecular functions and cellular components of newly predicted KEN-box proteins in H. sapiens. The proteins in the training data set were excluded.
(XLS)
Statistical comparison of the GO terms for the substrates between the predicted KEN-box and D-box proteins in the human proteome. Yates' Chi-square (χ2) test was performed (p-value<0.05) [32]. The entries with the grey background indicate the Enrichment_ratio ≤1.
(XLS)
Statistical comparison of the GO terms for newly predicted KEN-box and D-box proteins in the human proteome. The proteins in the training data set were excluded.
(XLS)
From recently published papers, we collected nine D-boxes and KEN-boxes in six proteins. The data set was not used for training. The default parameters were used for the GPS-ARM.
(XLS)