Abstract
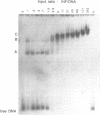
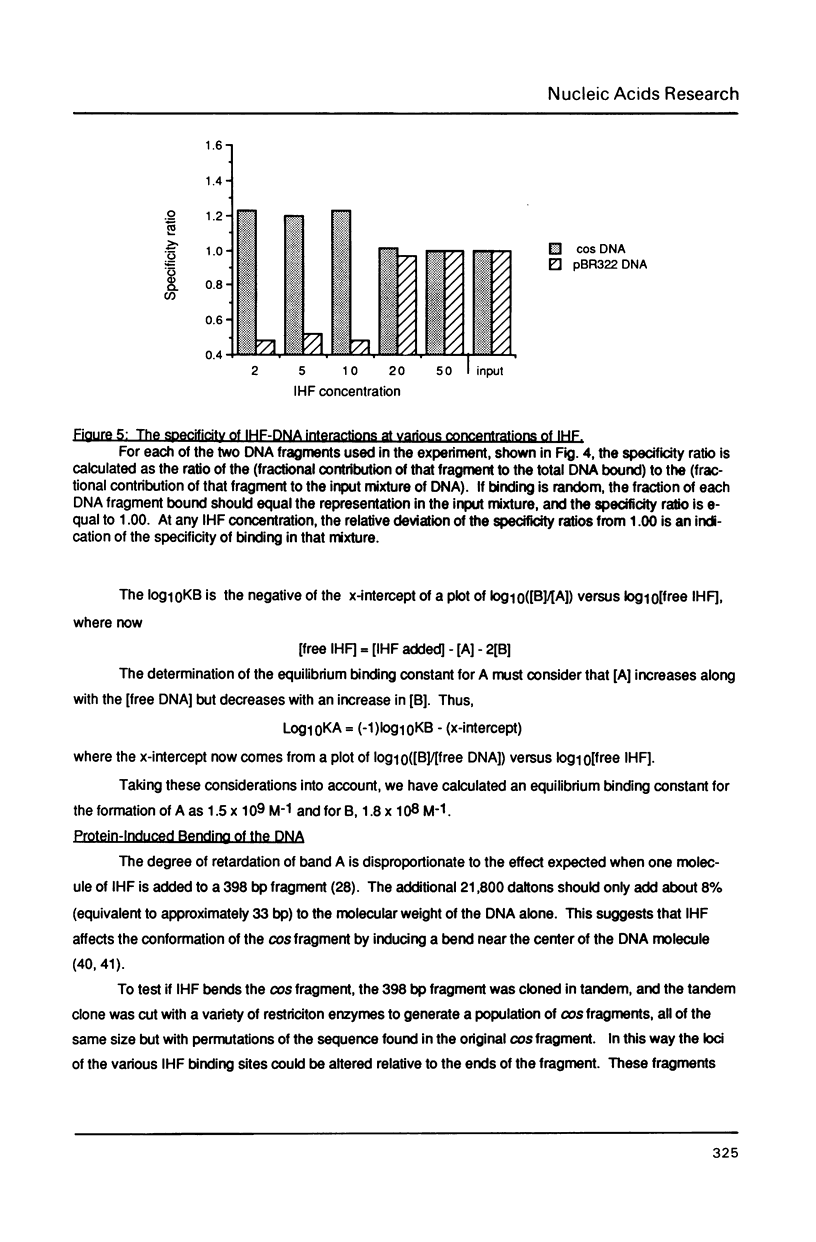
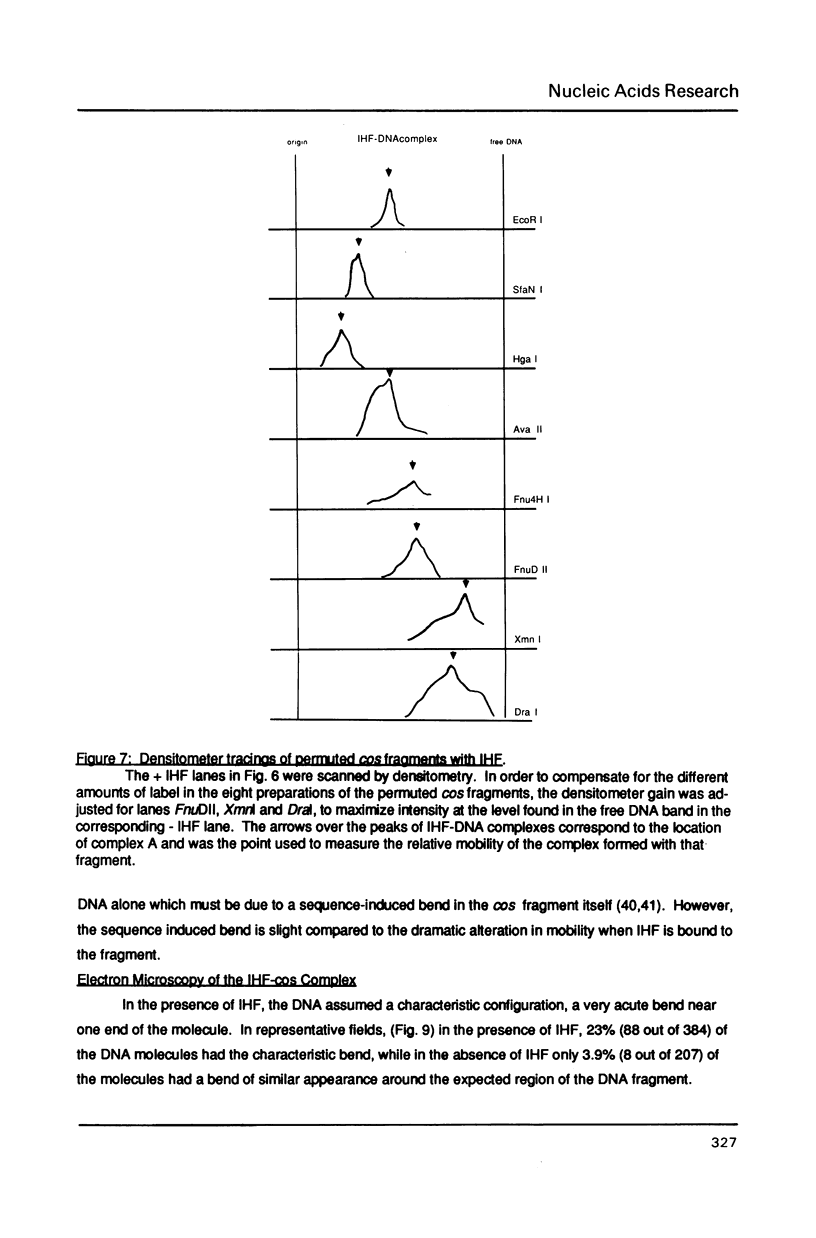
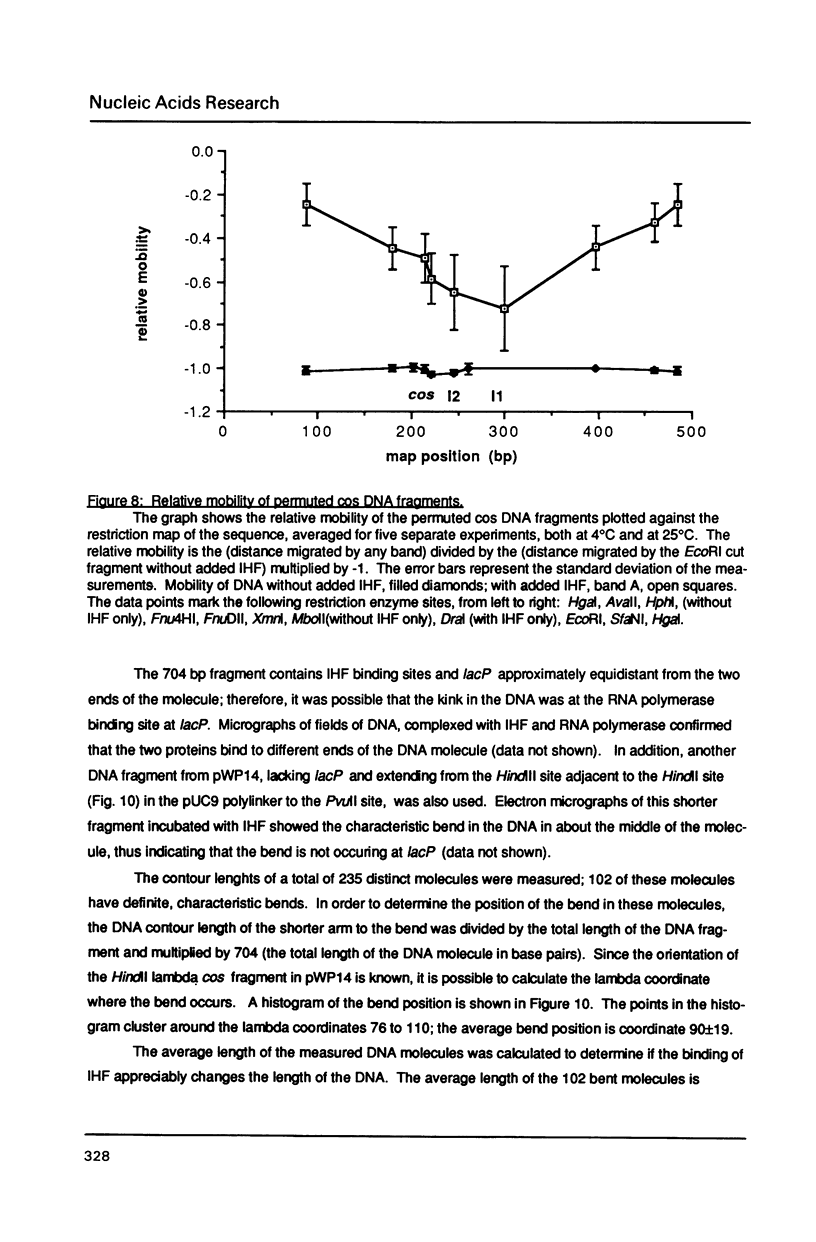
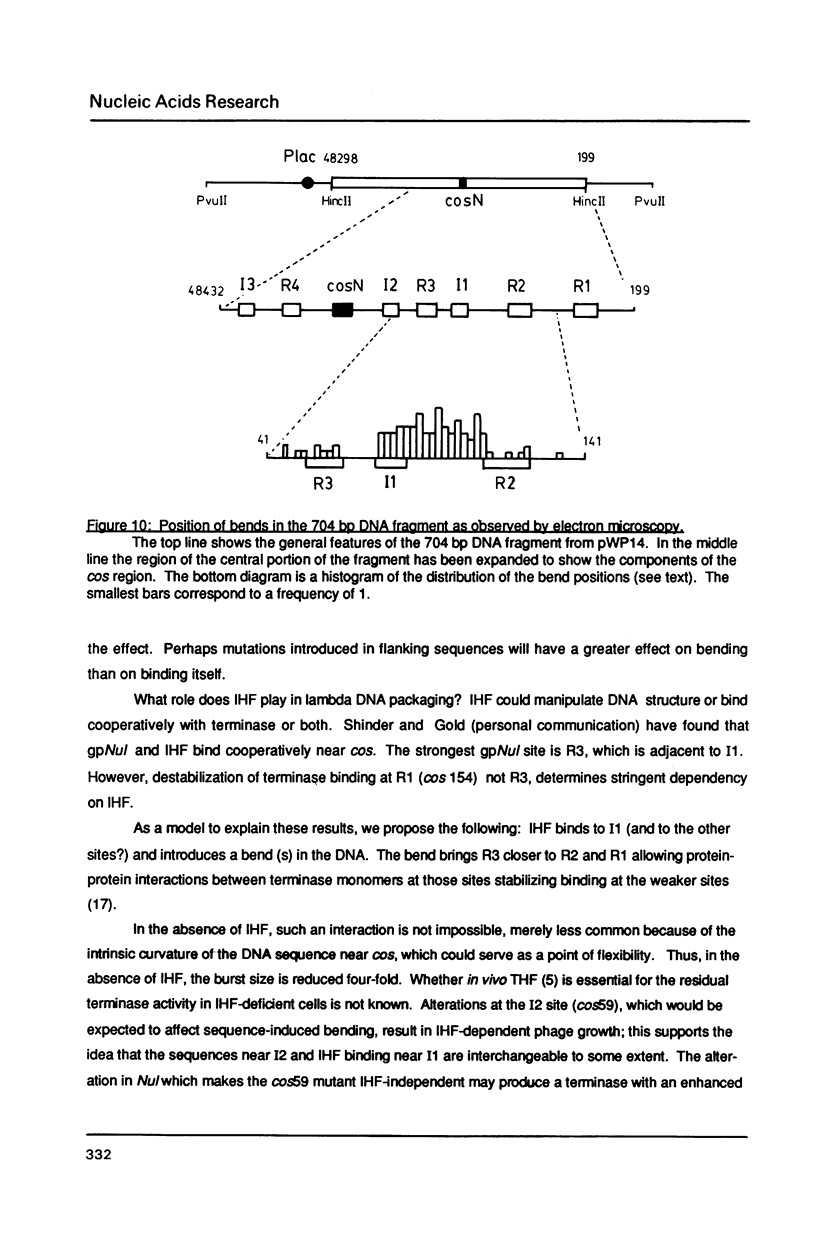
The interaction of E. coli's integration Host Factor (IHF) with fragments of lambda DNA containing the cos site has been studied by gel-mobility retardation and electron microscopy. The cos fragment used in the mobility assays is 398 bp and spans a region from 48,298 to 194 on the lambda chromosome. Several different complexes of IHF with this fragment can be distinguished by their differential mobility on polyacrylamide gels. Relative band intensities indicate that the formation of a complex between IHF and this DNA fragment has an equilibrium binding constant of the same magnitude as DNA fragments containing lambda's attP site. Gel-mobility retardation and electron microscopy have been employed to show that IHF sharply bends DNA near cos and to map the bending site. The protein-induced bend is near an intrinsic bend due to DNA sequence. The position of the bend suggests that IHF's role in lambda DNA packaging may be the enhancement of terminase binding/cos cutting by manipulating DNA structure.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bear S. E., Court D. L., Friedman D. I. An accessory role for Escherichia coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J Virol. 1984 Dec;52(3):966–972. doi: 10.1128/jvi.52.3.966-972.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Gold M. Enzymatic breakage of the cohesive end site of phage lambda DNA: terminase (ter) reaction. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4199–4203. doi: 10.1073/pnas.75.9.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Gold M. Isolation of the bacteriophage lambda A-gene protein. Proc Natl Acad Sci U S A. 1975 Feb;72(2):581–585. doi: 10.1073/pnas.72.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R. Principles of sequence-dependent flexure of DNA. J Mol Biol. 1986 Dec 20;192(4):907–918. doi: 10.1016/0022-2836(86)90036-7. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986 Sep 5;233(4768):1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Feiss M., Fogarty S., Christiansen S. Bacteriophage lambda DNA packaging: a mutant terminase that is independent of integration host factor. Mol Gen Genet. 1988 Apr;212(1):142–148. doi: 10.1007/BF00322457. [DOI] [PubMed] [Google Scholar]
- Feiss M., Widner W., Miller G., Johnson G., Christiansen S. Structure of the bacteriophage lambda cohesive end site: location of the sites of terminase binding (cosB) and nicking (cosN). Gene. 1983 Oct;24(2-3):207–218. doi: 10.1016/0378-1119(83)90081-1. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Equilibrium studies of the cyclic AMP receptor protein-DNA interaction. J Mol Biol. 1984 Jan 25;172(3):241–262. doi: 10.1016/s0022-2836(84)80025-x. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. F., Nash H. A. Role of Escherichia coli IHF protein in lambda site-specific recombination. A mutational analysis of binding sites. J Mol Biol. 1986 Sep 20;191(2):181–189. doi: 10.1016/0022-2836(86)90255-x. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Gold M., Parris W. A bacterial protein requirement for the bacteriophage lambda terminase reaction. Nucleic Acids Res. 1986 Dec 22;14(24):9797–9809. doi: 10.1093/nar/14.24.9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N., van de Putte P. Regulation of Mu transposition. I. Localization of the presumed recognition sites for HimD and Ner functions controlling bacteriophage Mu transcription. Gene. 1984 Oct;30(1-3):41–46. doi: 10.1016/0378-1119(84)90103-3. [DOI] [PubMed] [Google Scholar]
- Granston A. E., Alessi D. M., Eades L. J., Friedman D. I. A point mutation in the Nul gene of bacteriophage lambda facilitates phage growth in Escherichia coli with himA and gyrB mutations. Mol Gen Genet. 1988 Apr;212(1):149–156. doi: 10.1007/BF00322458. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986 Mar 15;261(8):3744–3752. [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S., Lesser C. F., Youderian P., Susskind M. M., Landy A. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem. 1985 Apr 10;260(7):4468–4477. [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller G., Feiss M. The bacteriophage lambda cohesive end site: isolation of spacing/substitution mutations that result in dependence on Escherichia coli integration host factor. Mol Gen Genet. 1988 Apr;212(1):157–165. doi: 10.1007/BF00322459. [DOI] [PubMed] [Google Scholar]
- Miller H. I. Multilevel regulation of bacteriophage lambda lysogeny by the E. coli himA gene. Cell. 1981 Jul;25(1):269–276. doi: 10.1016/0092-8674(81)90252-x. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Nash H. A. Direct role of the himA gene product in phage lambda integration. Nature. 1981 Apr 9;290(5806):523–526. doi: 10.1038/290523a0. [DOI] [PubMed] [Google Scholar]
- Morisato D., Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987 Oct 9;51(1):101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A., Flamm E., Weisberg R. A., Miller H. I. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J Bacteriol. 1987 Sep;169(9):4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J Biol Chem. 1981 Sep 10;256(17):9246–9253. [PubMed] [Google Scholar]
- Parris W., Davidson A., Keeler C. L., Jr, Gold M. The Nu1 subunit of bacteriophage lambda terminase. J Biol Chem. 1988 Jun 15;263(17):8413–8419. [PubMed] [Google Scholar]
- Prentki P., Chandler M., Galas D. J. Escherichia coli integration host factor bends the DNA at the ends of IS1 and in an insertion hotspot with multiple IHF binding sites. EMBO J. 1987 Aug;6(8):2479–2487. doi: 10.1002/j.1460-2075.1987.tb02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. The interaction of recombination proteins with supercoiled DNA: defining the role of supercoiling in lambda integrative recombination. Cell. 1986 Sep 26;46(7):1011–1021. doi: 10.1016/0092-8674(86)90700-2. [DOI] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Shinder G., Gold M. The Nul subunit of bacteriophage lambda terminase binds to specific sites in cos DNA. J Virol. 1988 Feb;62(2):387–392. doi: 10.1128/jvi.62.2.387-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinder G., Parris W., Gold M. Terminase host factor: a histone-like E. coli protein which can bind to the cos region of bacteriophage lambda DNA. Nucleic Acids Res. 1988 Apr 11;16(7):2765–2785. doi: 10.1093/nar/16.7.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Waechter-Brulla D., Gumport R. I., Gardner J. F., Moitoso de Vargas L., Landy A. Mutations in an integration host factor-binding site: effect on lambda site-specific recombination and regulatory implications. J Bacteriol. 1986 Dec;168(3):1343–1351. doi: 10.1128/jb.168.3.1343-1351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I. T., Steitz T. A. Model of specific complex between catabolite gene activator protein and B-DNA suggested by electrostatic complementarity. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3973–3977. doi: 10.1073/pnas.81.13.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. C. Use of polylysine for adsorption of nuclei acids and enzymes to electron microscope specimen films. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2311–2315. doi: 10.1073/pnas.74.6.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Xin W. N., Feiss M. The interaction of Escherichia coli integration host factor with the cohesive end sites of phages lambda and 21. Nucleic Acids Res. 1988 Mar 25;16(5):2015–2030. doi: 10.1093/nar/16.5.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]