Abstract
Uracil methylation is essential for survival of organisms and passage of information from generation to generation with high fidelity. Two alternative uridyl methylation enzymes, flavin-dependent thymidylate synthase and folate/FAD-dependent RNA methyltransferase, have joined the long-known classical enzymes, thymidylate synthase and SAM-dependent RNA methyltransferase. These alternative enzymes differ significantly from their classical counterparts in structure, cofactor requirements and chemical mechanism. This review covers the available structural and mechanistic knowledge of the classical and alternative enzymes in biological uracil methylation, and offers a possibility of using inhibitors specifically aiming at microbial thymidylate production as antimicrobial drugs.
Keywords: Uracil methylation, thymidylate synthase, flavin, enzyme mechanism
1. Introduction
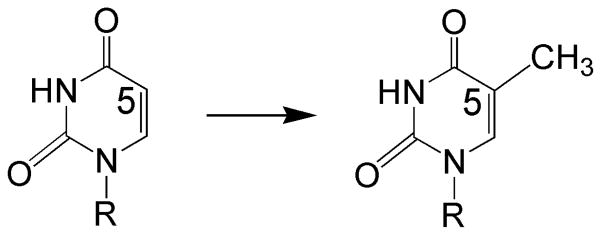
Methylation of uracil at C5 to form thymine moieties is an important transformation in both DNA biosynthesis and posttranslational modification of RNA (Scheme 1). The DNA building block 2′-deoxythymidine-5′-monophosphate (dTMP, or thymidylate) is formed by methylation of 2′-deoxyuridine-5′-monophosphate (dUMP). Thymidylate is essential for survival of all organisms, since without a pool of thymidylate for DNA biosynthesis cellular reproduction ceases.1 In RNA, methylated uracils are found at a single site in tRNAs and two sites in the ribosome.2 The 5-methyluridyl at position 54 in the T-loop of tRNAs in almost all living cells (designated as T54) plays a role in maintaining the tertiary structure of tRNA.3 One of the methylated uracils in rRNA, U1939 located at the interface of ribosomal subunits, has been proposed to be involved in interaction between the subunits and in sensing uncharged tRNAs.4, 5 The second methylated uracil in rRNA (U747) is situated in the ribosomal tunnel through which the emerging peptide chain passes during protein synthesis, and hence might be involved in regulatory interactions with the synthesized peptide.6
Scheme 1.
Uracil methylation reaction catalyzed by the enzymes reviewed here. R = 2′-deoxyribose or RNA.
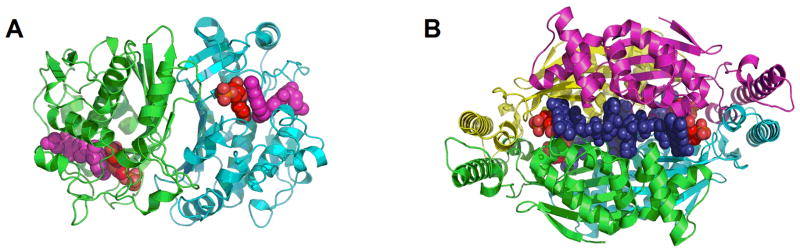
Different enzymes are responsible for uracil methylation in DNA biosynthesis and RNA modification. Conversion of dUMP to dTMP is catalyzed by thymidylate synthase (TSase) enzymes. Two different classes of TSases are known which are dissimilar in their sequence, structure, cofactor requirements and chemical mechanism. The extensively studied ThyA TSase is a homodimeric enzyme (Fig. 1A) that utilizes (R)-N5,N10-methylene-5,6,7,8-tetrahydrofolate (CH2H4folate) as both a methylene and a hydride source for the C7 methyl of dTMP (Scheme 2A). On the other hand, a recently discovered ThyX TSase is a homotetrameric catalyst (Fig. 1B) that uses CH2H4folate only for its methylene, and acquires the reducing hydride from nicotinamide adenine dinucleotide phosphate (NADPH) or other reducing agents (e.g. dithionite, ferredoxin, etc.). The redox chemistry in ThyX TSase is mediated by flavin adenine dinucleotide (FAD) cofactor (Scheme 2B). Many human pathogens, including biological warfare agents, lack the genes for ThyA TSase and rely on this alternative flavin-dependent TSase instead.7–10 Since ThyX differs from ThyA TSase in structure and chemistry and is absent in humans, it can potentially serve as a target of antibiotics with low toxicities.
Fig. 1. Structures of thymidylate synthases.
(A) Classical thymidylate synthase dimer from E. coli (PDB ID 2KCE). The dUMP substrate is red and the CH2H4folate analog (Zd 1694, Raltitrexed) is magenta, shown as space filling shapes. (B) Flavin-dependent thymidylate synthase tetramer from T. maritima (PDB ID 1O26). FAD prosthetic group is blue and dUMP is red.
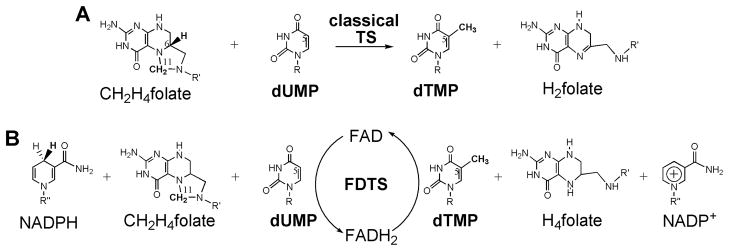
Scheme 2. Reactions catalyzed by thymidylate synthases.
(A) The overall reaction catalyzed by classical TSase. The atoms of importance are numbered. (B) The overall reaction catalyzed by FDTS. R=2′-deoxyribose-5′-phosphate; R′=(p-aminobenzoyl)-glutamate; R″=adenosine-5′-pyrophosphate-ribityl. Reproduced with permission from ref. 28.
The methylation of U1939 and U747 in rRNA of the ribosome is carried out by RumA and RumB methyltransferases, respectively. In tRNA, the enzyme TrmA catalyzes the production of T54. Although RumA, RumB and TrmA share little sequence and structure homology, all three enzymes use S-adenosylmethionine (SAM) as a methyl donor and follow the same chemical mechanism, described in detail later.
In late 1970s, an alternative class of enzymes has been discovered for T54 production in tRNA. Unlike TrmA, the enzyme responsible for catalysis in this alternative pathway does not depend on SAM but rather uses reduced FAD and CH2H4folate, and is therefore designated as TrmFO methyltransferase. Even though TrmA and TrmFO phylogenic distribution is mutually exclusive, with TrmFO present in several pathogens, early studies11 showed little effect of TrmFO on bacterial growth. If further investigation demonstrates some essential activity of TrmFO in the cell, this enzyme could become of interest as an antibiotic target.
This review highlights the available structural and mechanistic knowledge of the classical and alternative enzymes in biological uracil methylation, and offers a possibility of using inhibitors specifically aiming at microbial thymidylate production as antimicrobial drugs.
2. Classical thymidylate synthase (TSase, EC 2.1.1.45) and flavin-dependent thymidylate synthase (FDTS, EC 2.1.1.148)
ThyA thymidylate synthase has been intensively studied for decades, with over a hundred crystal structures available to date, and consequently is referred to as classical TSase. ThyA is a homodimer with one active site per subunit, as shown in Fig. 1A. The enzyme uses CH2H4folate as both the source of a one-carbon unit and the reducing hydride to form the C7 methyl of dTMP, and produces 7,8-dihydrofolate (H2folate, Scheme 2A). CH2H4folate is regenerated for subsequent TSase turnovers by reduction of H2folate by dihydrofolate reductase (DHFR, encoded by folA gene) to form H4folate and then conversion to CH2H4folate catalyzed by serine hydroxymethyl transferase (SHMT). The TSase/DHFR cycle is central to thymidylate biosynthesis in the organisms relying on ThyA.
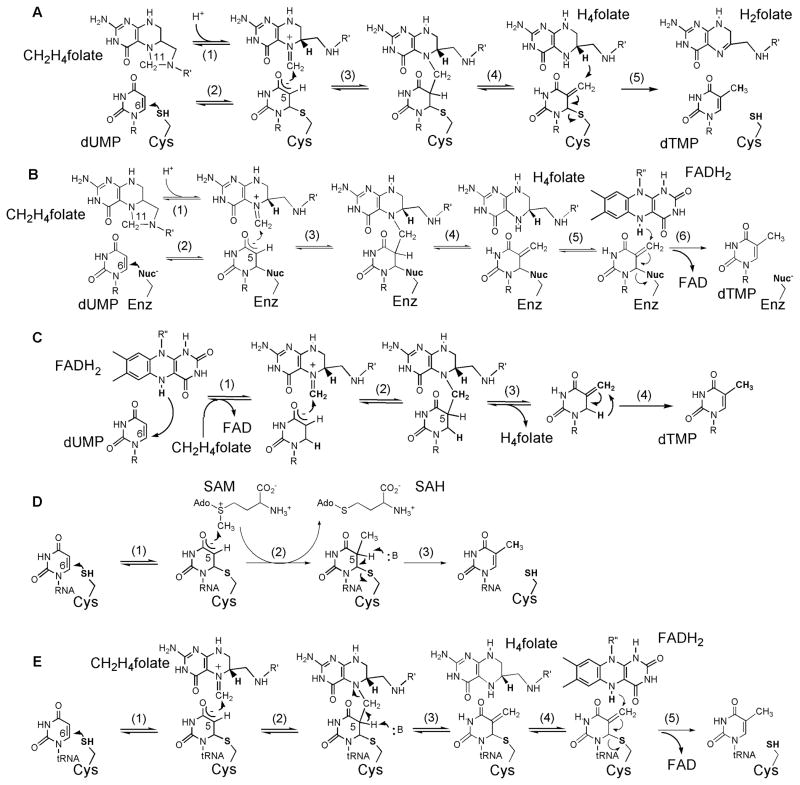
The currently proposed chemical mechanism of classical TSase is presented in Scheme 3A.1, 12 Upon binding, N10 protonation of CH2H4folate results in a reactive iminium cation (step 1). A conserved active site cysteine covalently activates dUMP via Michael addition (step 2), and the C5 of the resulting enolate reacts in a Mannich-type condensation with the N5 imine of CH2H4folate (step 3). The enzyme-bound bridged intermediate undergoes Hofmann elimination of H4folate (step 4) to form an exocyclic methylene intermediate. Finally, the C7 of this intermediate is reduced by the 6S hydride from H4folate (step 5) producing H2folate and dTMP.
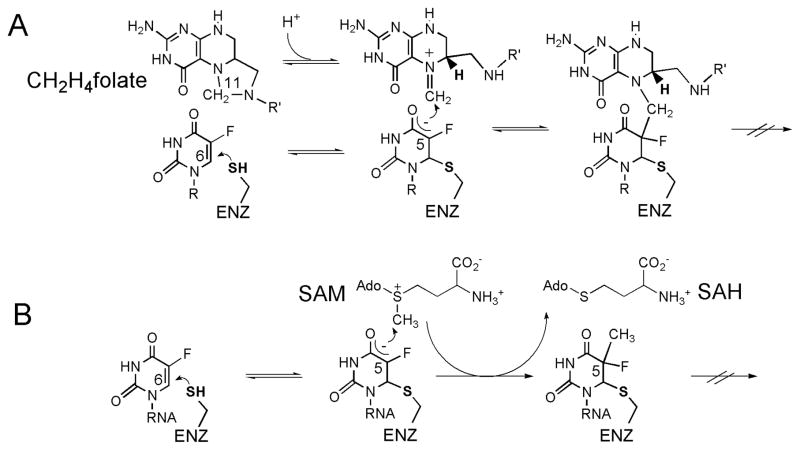
Scheme 3. Proposed chemical mechanisms of various uracil methylation enzymes.
(A) The mechanism of classical TSase-catalyzed reaction. (B) The mechanism of FDTS-catalyzed reaction involving an enzymatic nucleophile. (C) The mechanism of FDTS-catalyzed reaction where reduced flavin acts as the nucleophile. R=2′-deoxyribose-5′-phosphate; R′=(p-aminobenzoyl)-glutamate; R″=adenosine-5′-pyrophosphate-ribityl. (D) The mechanism of S-adenosylmethionine-dependent RNA methyltransferases (Rum A and TrmA). SAM = S-adenosylmethionine; SAH = S-adenosylhomocysteine. (E) The mechanism of folate/FAD-dependent tRNA methyltransferase (TrmFO). Note that in cases of the flavin-dependent enzymes, the FAD cofactor remains non-covalently bound to the enzyme throughout the catalytic turnover.
Establishment of the chemical mechanism for classical TSase relied on key kinetic, chemical and structural studies. Michael-addition (Scheme 3A, step 2) and subsequent enolate condensation (Scheme 3A, step 3) of the mechanism are supported by the crystal structure of the wild-type E. coli enzyme in a covalent complex with 5-fluorouridylate and CH2H4folate (Protein Data Bank ID 1TLS).13 The mechanism for the formation of this complex is outlined in Scheme 4A. This covalent ternary intermediate (Scheme 3A, between steps 3 and 4) has also been detected in quenching experiments with wild-type TSase14 and by isolation on SDS-PAGE in reactions of E60A and E60L mutants of L. casei TSase with radiolabeled substrates.15 The formation of the exocyclic methylene intermediate (Scheme 3A, between steps 4 and 5) was confirmed in experiments with a W82Y mutant of L. casei TSase,16 which allowed premature release of H4folate from the active-site and subsequent chemical trapping of the intermediate with β-mercaptoethanol under steady-state conditions.
Scheme 4. Mechanism of inhibition of SAM- and folate-dependent uracil methyltransferases by 5-fluorouridylate.
(A) Mechanism of formation of an inhibitory covalent complex in CH2H4folate-dependent enzymes (classical TSase and TrmFO). (B) Mechanism of formation of an inhibitory covalent complex in SAM-dependent enzymes (RumA and TrmA). SAM = S-adenosylmethionine; SAH = S-adenosylhomocysteine.
The kinetic mechanism of classical TSase is generally sequential with dUMP binding first. This is supported by structural studies and monitoring the release of 5F-dUMP as a function of CH2H4folate concentration. The ordered mechanism is also supported by the complete suppression of dUMP kinetic isotope effects (KIEs) at high CH2H4folate concentrations.17 Under certain conditions, however, such as when using polyglutamyl CH2H4folates and with some mutants, the order of binding can become random. For this reason, it is accepted that the binding order of substrates to TSase is mostly preferential but subject to change based on reaction conditions.12
Several clinical drugs (e.g. 5-fluorouracil, 5-trifluoromethyl 2′-deoxyuridine, etc.), used for chemotherapy and treatment of other conditions, have been shown to act through inhibition of classical TSase.18 Competitive inhibitors of TSase have been developed based both on the pyrimidine (5-substituted uridylates) and folate (raltitrexed, 10-methylfolate, 10-propargyl-5,8-dideazafolate and others) moieties. Many of the 5-substituted substrate, intermediate, and product nucleotide analogs function by reaction with the enzyme, thus causing mechanism-based inactivation. Covalent anchoring of these inhibitors by the active-site cysteine of classical TSase is a key feature of their mechanism of inactivation. A well-studied example, 5F-dUMP (Kd ≈ 10−12 M), reacts with the active site cysteine and undergoes condensation with CH2H4folate but prevents proton abstraction and elimination of the H4folate, as a consequence stalling the reaction at the covalent ternary complex (Scheme 4A). Other dUMP analogs carry C5 substituents that upon cysteine attack at C6 become reactive and covalently bind to other proximal residues of the enzyme, or react with solvent water. The enzyme inactivation by this type of inhibitors is independent of CH2H4folate. The same is true for substrate analogs with electrophilic C5 substituents that serve as a sink for the negative charge generated in cysteine attack at C6 (e.g. 5-NO2-dUMP, dAzMP and others).12, 19
The extensive study of classical TSase resulted in its well-characterized structure and mechanism, and identification of key compounds that inhibit its activity. This knowledge has been useful not only clinically in light of the drugs that target TSase, but also in establishing a model of catalysis for many other enzymes. For instance, such enzymes as dUMP and dCMP hydroxymethyltransferases, DNA and RNA cytosine methyltransferases, and RNA uridyl methyltransferases (considered later) share similar catalytic features.12, 20 The use of classical TSase as a model system has recently extended even further with detailed mechanistic study using KIEs and other methods to probe the enzyme’s role in activating and catalyzing C-H bond cleavage.17, 21 This underscores the importance of classical TSase and continued studies of this enzyme to understand general features of biological catalysts.
In 2002, several organisms were identified that lacked the genes coding for classical TSase, DHFR, or thymidine kinase (a thymidine-scavenging enzyme), yet produced thymidine as indicated by their growth in thymidine-depleted media.7, 22, 23 This finding led to the discovery of an alternative thymidylate synthase enzyme, ThyX. ThyX thymidylate synthase catalyzes the same net conversion of dUMP to dTMP, as classical TSase (Scheme 2B). However, unlike classical TSase, ThyX uses CH2H4folate only as a methylene donor and employs a flavin adenine dinucleotide (FAD) prosthetic group to catalyze the redox chemistry, and is therefore referred to as flavin-dependent thymidylate synthase (FDTS).
FDTS accomplishes the combined activities of classical TSase and DHFR, converting dUMP to dTMP and producing H4folate instead of H2folate (Scheme 2B). Since some organisms that depend on FDTS lack the gene for DHFR, it was suggested that FDTS might be a bifunctional enzyme, with classical TSase and flavin-dependent DHFR activities. Such bifunctional classical TSase-DHFR enzymes are common in protozoa24 and plants25. However, several observations have ruled out the possibility of FDTS being a bifunctional catalyst: (i) in studies with (R)-[6-3H]-CH2H4folate, the tritium was retained by H4folate and not transferred to dTMP,26 as with the classical TSase;1, 27 (ii) in contrast to classical TSase, reactions carried out in D2O produced deuterated dTMP, which points to proton exchange between the reduced flavin and the solvent prior to a hydride transfer from the flavin directly to the nucleotide; and (iii) when using tritiated NADPH to reduce the flavin, the tritium was found in water and not H4folate,26 ruling out a flavin-dependent DHFR functionality of FDTS. Overall these findings support a mechanism in which NADPH reduces FAD to FADH2 and the uracil moiety accepts a hydride from the FADH2 to form dTMP.28
Very few crystal structures of FDTS enzymes are available today. The first crystal structure of FDTS, obtained from the organism Thermotoga maritima, showed no structural similarity with classical TSase.29 Unlike dimeric classical TSase, FDTS is a homotetramer with four active sites, each at the interface of three of the subunits (Fig. 1B). Since then, crystal structures for FDTSs from Mycobacterium tuberculosis,30 Paramecium bursaria chlorella virus-1,31 and more recently Helicobacter pylori32 have been solved, with FAD only and FAD in combination with dUMP, 5F-dUMP or 5Br-dUMP. Additionally, a structure with NADP+ has been solved for M. tuberculosis FDTS where nicotinamide replaced the flavin cofactor in the enzyme during crystallization. However, the mechanistic importance of this finding has yet to be realized.33 Structures with CH2H4folate or any other folate moieties, on the other hand, have not been obtained. Although FDTSs from different organisms differ in sequence and size, tetrameric structure and key residues involved in substrate binding and catalysis are conserved.
Structural comparison of classical TSase and FDTS provides insight into the differences in catalysis of these two enzymes. The binding sites for dUMP and FAD within the FDTS active-site have been characterized. In the structures, the N5 of isoalloxazine ring of FAD is located sufficiently close to the uracil moiety to donate its hydride to the nucleotide (<4 Å). The conserved cysteine crucial to classical TSase activity is absent from the FDTS active site, and some FDTSs lack cysteines altogether. The only likely candidate for cysteine’s role as an enzymatic nucleophile in FDTS is a conserved serine residue located 4 Å away from the electrophilic C6 of dUMP. The hypothesis of serine acting as a nucleophile was supported by studies of FDTS from M. tuberculosis and H. pylori (MtbFDTS34 and HpFDTS35, respectively). A chemical mechanism where serine acts as the catalytic nucleophile similar to the cysteine of classical TSase is presented in Scheme 3B.26 In this mechanism, serine activates dUMP (step 2) for subsequent reaction with CH2H4folate (steps 3–5). In the final step, the enzyme-bound exocyclic methylene intermediate is reduced by a hydride from FADH2 (step 6), releasing the product dTMP.
The mechanism with serine as an active-site nucleophile was further investigated by conducting mutation studies with T. maritima FDTS (TmFDTS)36, similar to those with MtbFDTS34 and HpFDTS35. Surprisingly, mutation of the only conserved serine in the TmFDTS active site (S88) to alanine resulted in an active enzyme. Moreover, an S88C mutant was 400 times less active than the wild-type enzyme and did not display any classical TSase activity (i.e. catalysis without the reducing equivalents from NADPH or dithionite). The search for other potential nucleophiles in active site of TmFDTS revealed the conserved residues S83 and Y91 as possible candidates. Tyrosine mutation to phenylalanine in HpFDTS, however, yields an enzyme with 50% more, not less activity than the wild type.35 The S83, on the other hand, is too far away (17 Å) from the electrophilic C6 of dUMP to activate it and is hydrogen-bonded to the adenosine moiety of FAD at the core of the FDTS tetramer, and is thus not a capable nucleophile. These observations suggested that FDTS catalysis does not rely on an enzymatic nucleophile.36
Experiments with halogenated dUMP analogs further strengthen the lack of an enzymatic anchor in FDTS-catalyzed reaction. With classical TSase, isolation of a covalent complex of the enzyme with 5F-dUMP and CH2H4folate (PDB ID 1TLS) served as an evidence for Michael addition. No such complex has been identified for FDTS either in MALDI-TOF mass spectrometry36 or X-ray crystallography37 analyses. Furthermore, 5F-dUMP was shown to inhibit FDTS only at micromolar concentrations – orders of magnitude higher than observed for classical TSase – and inhibition was completely reversible. Another test for a Michael nucleophile in enzyme’s active site is the dehalogenation of 5Br-dUMP. While classical TSase catalyzes this dehalogenation,38 FDTS fails to do so.
Other possible non-enzymatic nucleophiles include a hydroxide from the water and the N5 of the flavin, proposed to be the nucleophile in UDP-galactopyranose mutase catalysis.39 However, the basic environment in the FDTS active site necessary for deprotonation of water to form hydroxide is lacking. Studies with 5-carba-5-deaza-FAD, i.e. FAD with a carbon in place of the N5, resulted in an active enzyme, thus eliminating the N5 nucleophilic involvement.36
With the data supporting the absence of an enzymatic nucleophile, a revised version of mechanism in Scheme 3B is necessary for FDTS-catalyzed reaction. To follow the flow of hydrogens from the reduced flavin, isotopic labeling experiments were conducted with TmFDTS.36 In reactions in D2O at 65°C (close to the physiological temperature of T. maritima), 7-D-dTMP was identified as the sole product. However, similar reactions done at 37°C produced up to 60% of 6-D-dTMP, which suggested that the hydrogen could be directly transferred from the reduced flavin to C6 of the uracil.36
In light of the studies described above, a mechanism consistent with all current mechanistic and structural data was proposed where a hydride from FADH2 nucleophilically attacks the C6 of dUMP (Scheme 3C, step 1). The subsequent steps lead to the formation of a putative exocyclic methylene intermediate (Scheme 3C, between steps 3 and 4), analogous to the classical TSase intermediate (Scheme 3A, between steps 4 and 5) except for the lack of the covalent bond to the enzyme. This intermediate is an isomer of dTMP and needs to undergo rearrangement to form that product (Scheme 3C, step 4). Interestingly, the chemistry proposed in Scheme 3C is quite different from that of classical TSase and uridyl methyltransferases in general.
In addition to efforts to establish the chemical cascade for FDTS-catalyzed reaction, its kinetic mechanism has been explored over the past several years. Altogether, FDTS seems to follow a sequential order of substrate binding, with little data available on the order of product release. The multisubstrate nature of the FDTS reaction makes its comprehensive kinetic analysis challenging. Furthermore, the activation kinetics observed for dUMP, substrate inhibition and negative cooperativity observed for CH2H4folate, and the ability for FDTS to function as an oxidase (reducing molecular oxygen to hydrogen peroxide) have also added to the complications involved in deconvolution of the entire kinetic mechanism.28 While the oxidase activity of FDTS can compete with the synthase activity and potentially interfere with dTMP production, it has proved instrumental in probing the nature of substrate binding. In studies of FDTS oxidase activity, it was demonstrated that the rate of H2O2 formation increases as a function of dUMP concentration when CH2H4folate is not present suggesting that dUMP may activate the redox chemistry of the flavin. The observation that CH2H4folate and O2 compete for the same activated enzyme complex allowed an approximation of binding affinities for these species. Importantly, this has led to better approximations for functional binding constants for both dUMP and CH2H4folate, giving a more realistic picture of binding than offered by standard Michaelis analysis.40
Potent classical TSase inhibitors (as mentioned above) have been tested in the past against FDTS activity but showed no or little effect.41 This enhances the promise for the development of inhibitors specific to FDTS with low effects on the classical enzyme, and thus lower toxicity to the host. One avenue for such selective inhibition is the development of analogs that mimic the non-covalent intermediates in FDTS mechanism (Scheme 3C) or the transition states for their formation. These molecules might bind tightly to FDTS, but not to classical TSase. Another possibility is to exploit the structural differences in the active sites of classical TSase and FDTS. Namely, classical TSase has a deep and well organized active site that closely interacts with the functional groups of the bound folate and dUMP moieties.26, 42 FDTS, on the other hand, seems to have much larger, flexible and solvent-exposed active site. As a result, bulkier analogs of the substrates may selectively bind to the accessible FDTS active site, but not to the sheltered classical TSase pocket. To date there are very few compounds that are known to inhibit FDTS and none have proven to be highly specific or mechanism-based. Recently, a series of derivatives based on a thiazolidine core were synthesized and shown to inhibit FDTS activity.43 This resulted in the identification of two classes of submicromolar inhibitors, which either competed with dUMP or inactivated FDTS independently of dUMP concentration. To date, none of these compounds have been demonstrated to be selective for FDTS lowering their potential to serve as drug leads. The only report of a selective FDTS inhibitor is that of a 5-propynyl-dUMP derivative, with additional eight-carbon amide attached to the side chain (N-(3-(5-(2′-deoxyuridine-5′-monophosphate))prop-2-ynyl)-octanamide),44 which inhibited MtbFDTS almost 100 times more than classical MtbTSase enzyme. The mechanism for the selective inhibition for this compound is not known but could be due to the ability of the accessible FDTS active site, but not the sheltered TSase pocket, to accommodate the bulky inhibitor.
3. SAM-dependent RNA methyltransferases (EC 2.1.1.35) and folate/FAD-dependent tRNA methyltransferase (EC 2.1.1.74)
Methylation of uracil moeties also occurs in posttranscriptional modification of RNA. These important biological modifications are carried out by either the S-adenosylmethionine (SAM)-dependent enzymes
RumA, RumB and TrmA, or folate-dependent TrmFO enzymes. RumA and RumB methyltransferases methylate U1939 and U747, respectively, of 23S rRNA. The first reported RNA uridine methyltransferase crystal structure was that of E. coli RumA, in complex with S-adenosylhomocysteine (i.e. SAM without its S-methyl) and a 37-nucleotide fragment of 23S rRNA (PDB ID 1UWV and 2BH2).45 Interestingly, the structure showed RumA to be a monomeric metalloprotein containing a [4Fe4S] cluster, which was proposed to play a role in correct protein folding and/or rRNA binding. In the structure, the C5 of U1939 to be methylated within the nucleotide fragment was replaced with fluorine. This substitution stalled the methylation and led to formation of a stable covalent enzyme-RNA complex (Scheme 4B), which indicated involvement of an enzymatic nucleophile analogous to cysteine in classical thymidylate synthase mechanism (Scheme 3A). The proposed chemical mechanism for RumA with Michael addition of a catalytic cysteine (C389 in E. coli) to C6 of the uridine is shown in Scheme 3D.46 Based on the structure, the conserved glutamate (E424) was suggested to be the general base in step 3 of the mechanism, and this hypothesis was confirmed by mutagenesis.45
In most Gram negative bacteria, some archae and all eukaryotes, TrmA enzyme is responsible for catalyzing SAM-dependent U54 methylation within tRNA. Although TrmA shares little sequence homology with RumA, a recently obtained crystal structure of E. coli TrmA E358Q mutant complexed with a 19-nucleotide tRNA fragment (PDB ID 3BT7) showed RNA binding in a manner similar to RumA,47 and suggested TrmA chemical mechanism to be analogous to that of RumA (Scheme 3D). Mutation of the glutamate 358 to glutamine arrested catalysis and allowed crystallization of the covalent complex (Scheme 3D, between steps 2 and 3). Isolation of this complex supported the role of E358 as a general base in TrmA, similarly to E424 in RumA.
Over three decades ago, it was reported that tRNA U54 methylation in the Gram-positive bacteria Bacillus subtilis and pathogenic Enterococcus faecalis does not depend on SAM. Instead the purified methytransferases from these organisms use CH2H4folate as a methylene donor and FADH2 as a reductant, as supported by incorporation of tritium from [5-3H]-5-deaza-FMNH2 into the methyl of thymidine product.48 More recently, a gene coding for the folate/FAD-dependent tRNA methyltransferase, named TrmFO, has been identified in most Gram-positive and some Gram-negative bacteria, including T. maritima and Thermus thermophilus.49, 50 Notably, the phylogenic distribution of TrmFO and TrmA enzymes is mutually exclusive.
The chemical mechanism of TrmFO-catalyzed reaction has remained elusive due to lack of structural information. The recent crystal structure of T. thermophilus TrmFO-H4folate complex (PDB ID 3G5R) provided some insight into the methylene transfer step in this enzyme’s catalysis.51 In the complex, the pterin ring of the folate is sandwiched between the isoalloxazine moiety of FAD and the imidazole of a histidine residue. Modeling of CH2H4folate and manual docking of a tRNA into TrmFO-H4folate structure places the target U54 in close enough proximity for a direct methylene transfer from CH2H4folate to the uracil. Additionally, a conserved cysteine (C51 in T. thermophilus TrmFO) located in the vicinity of flavin ring was speculated to nucleophilically activate U54, analogously to the catalytic mechanisms of ThyA and TrmA uridyl methylases (Schemes 3A and 3D, respectively). Indeed, mutation of C51 to alanine abolished TrmFO activity.
The possibility of covalent catalysis in TrmFO reaction, and the role of the active-site cysteine in particular, were further explored in studies with B. subtilis enzyme,52 whose structure has not yet been solved. Using a gel mobility-shift assay, it was shown that the wild-type enzyme and several of its mutants formed a covalent adduct with a substrate analog (5F-U54-miniRNA). Contrary to expectations, the alanine mutant of cysteine-53 (equivalent to C51 in T. thermophilus TrmFO) was still capable of forming the covalent complex, despite the inability of alanine to act as a nucleophile. Instead, a conserved cysteine-226 far away from the active site (>20 Å in T. thermophilus enzyme) appears to fulfill the nucleophilic function, since C226A mutant failed to produce the covalent adduct and to methylate tRNA. On the basis of these observations, a catalytic cascade for TrmFO shown in Scheme 3E was proposed. Because C53A mutation abolishes the methylation activity of TrmFO, this residue was suggested to be the general base in step 3 of the mechanism. The C226A/C51A double mutant, just like C226A, formed neither the protein-RNA adduct nor the methylated tRNA product.
Although C226 proposed to activate the uracil is a large distance away from the active site, one possible scenario considered52 was that a conformational change in protein occurs upon tRNA binding to bring this cysteine closer to the site of chemistry. Partial proteolysis experiments with B. subtilis TrmFO showed that enzyme becomes more susceptible to proteolysis in the presence of tRNA, indeed pointing to the changes in protein conformation. In another scenario, TrmFO could dimerize upon binding of tRNA, and C226 of one monomer could activate the C6 of U54-tRNA bound in the active site of the other monomer. Modeling of T. thermophilus TrmFO dimer with the target uridine of tRNA in one of the active sites put the uridine C6 atom ~ 6 Å away from the C223 nucleophile, suggesting that a structural change would still be needed to allow C223 to participate in the catalysis. Certainly, a crystal structure of TrmFO enzyme in complex with tRNA substrate could clarify the aspects of its catalysis in the future.
Conclusion
Over the years, structural and mechanistic studies of uracil methylating enzymes provided an insight into how these enzymes work. Majority of these methyltransferases appear to share a catalytic theme (e.g., RumA, TrmA, TrmFO and classical TSase), while others (e.g., FDTS) seem to perform their function via unique chemistry. Such studies are of broad potential utility, ranging from using uracil methyltransferases as potential antibiotic targets to conducting basic research on how various enzymes catalyze the same transformation using different chemical mechanisms. Studies of FDTS enzymes in particular are still in their infancy, and many aspects of their catalysis need further elucidation, which is a major reason for the current scarcity of potent and selective FDTS inhibitors.43, 44 Considering that FDTS represents an important but under-characterized antibiotic target, future investigation of its chemical mechanism and interactions with the substrates will likely provide the basis for rational design of mechanism-based inhibitors. To this date, the relevance of the folate/FAD-dependent tRNA methyltransferase (TrmFO) to the survival of pathogens has not been elucidated. If future studies reveal its specific activity in the cell, TrmFO might join FDTS on the list of potential enzymatic targets for future antibiotics.
Acknowledgments
This work was supported by NIH R01 GM065368 and NSF CHE 0715448 to AK, the Iowa CBB to TVM and EMK, and NSF GRFP to EMK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finer-Moore JS, Santi DV, Stroud RM. Biochemistry. 2003;42:248–256. doi: 10.1021/bi020599a. [DOI] [PubMed] [Google Scholar]
- 2.Myllykallio H, Skouloubris S, Grosjean H, Liebl U. DNA and RNA modification enzymes: structure, mechanism, function and evolution. Landes Bioscience; 2009. [Google Scholar]
- 3.Bjork GR. Prog Nucleic Acid Res Mol Biol. 1995;50:263–338. doi: 10.1016/s0079-6603(08)60817-x. [DOI] [PubMed] [Google Scholar]
- 4.Agarwalla S, Kealey JT, Santi DV, Stroud RM. J Biol Chem. 2002;277:8835–8840. doi: 10.1074/jbc.M111825200. [DOI] [PubMed] [Google Scholar]
- 5.Persaud C, Lu Y, Vila-Sanjurjo A, Campbell JL, Finley J, O’Connor M. Biochem Biophys Res Comm. 2010;392:223–227. doi: 10.1016/j.bbrc.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 6.Madsen CT, Mengel-Jorgensen J, Kirpekar F, Douthwaite S. Nucleic Acids Res. 2003;31:4738–4746. doi: 10.1093/nar/gkg657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. http://www.cdc.gov.
- 9.Leduc D, Graziani S, Meslet-Cladiere L, Sodolescu A, Liebl U, Myllykallio H. Biochem Soc Trans. 2004;32:231–235. doi: 10.1042/bst0320231. [DOI] [PubMed] [Google Scholar]
- 10.Myllykallio H, Leduc D, Filee J, Liebl U. Trends Microbiol. 2003;11:220–223. doi: 10.1016/s0966-842x(03)00101-x. [DOI] [PubMed] [Google Scholar]
- 11.Persson BC, Grustafsson C, Berg DE, Bjork GR. Proc Natl Acad Sci USA. 1992;89:3995–3998. doi: 10.1073/pnas.89.9.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carreras CW, Santi DV. Annu Rev Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 13.Matthews DA, Villafranca JE, Janson CA, Smith WW, Welsh K, Freer S. J Mol Biol. 1990;214:937–948. doi: 10.1016/0022-2836(90)90347-O. [DOI] [PubMed] [Google Scholar]
- 14.Moore MA, Ahmed F, Dunlap RB. Biochemistry. 1986;25:3311–3317. doi: 10.1021/bi00359a034. [DOI] [PubMed] [Google Scholar]
- 15.Huang W, Santi DV. J Biol Chem. 1994;269:31327–31329. [PubMed] [Google Scholar]
- 16.Barrett JE, Maltby DA, Santi DV, Schultz PG. JACS. 1998;120:449. [Google Scholar]
- 17.Wang Z, Kohen A. J Am Chem Soc. 2010;132:9820–9825. doi: 10.1021/ja103010b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Touroutoglou N, Pazdur R. Clin Cancer Res. 1996;2:227–243. [PubMed] [Google Scholar]
- 19.Kalman TI, Goldman D. Biochem Biophys Res Comm. 1981;102:682–689. doi: 10.1016/s0006-291x(81)80186-6. [DOI] [PubMed] [Google Scholar]
- 20.Gabbara S, Sheluho D, Bhagwat AS. Biochemistry. 1995;34:8914–8923. doi: 10.1021/bi00027a044. [DOI] [PubMed] [Google Scholar]
- 21.Kanaan N, Ferrer S, Marti S, Garcia-Viloca M, Kohen A, Moliner V. J Am Chem Soc. 2011;133:6692–6702. doi: 10.1021/ja1114369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews II, Kreusch AT, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elslinger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, Stevens RC. Proc Natl Acad Sci U S A. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murzin AG. Science. 2002;297:61–62. doi: 10.1126/science.1073910. [DOI] [PubMed] [Google Scholar]
- 24.Ivanetich KM, Santi DV. FASEB. 1990;4:1591–1597. doi: 10.1096/fasebj.4.6.2180768. [DOI] [PubMed] [Google Scholar]
- 25.Cella R, Parisi B. Physiol Plant. 1993;88:509–521. [Google Scholar]
- 26.Agrawal N, Lesley SA, Kuhn P, Kohen A. Biochemistry. 2004;43:10295–10301. doi: 10.1021/bi0490439. [DOI] [PubMed] [Google Scholar]
- 27.Hong B, Maley F, Kohen A. Biochemistry. 2007;46:14188–14197. doi: 10.1021/bi701363s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koehn EM, Kohen A. Arch Biochem Biophys. 2010;493:96–102. doi: 10.1016/j.abb.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn P, Lesley SA, Mathews II, Canaves JM, Brinen LS, Dai X, Deacon AM, Elslinger MA, Eshaghi S, Floyd R, Godzik A, Grittini C, Grzechnik SK, Guda C, Hodgson KO, Jaroszewski L, Karlak C, Klock HE, Koesema E, Kovarik JM, Kreusch AT, McMullan D, McPhillips TM, Miller MA, Miller M, Morse A, Moy K, Ouyang J, Robb A, Rodrigues K, Selby TL, Spraggon G, Stevens RC, Taylor SS, van den Bedem H, Velasquez J, Vincent J, Wang X, West B, Wolf G, Wooley J, Wilson IA. Proteins: Struct Funct Genet. 2002;49:142–145. doi: 10.1002/prot.10202. [DOI] [PubMed] [Google Scholar]
- 30.Sampathkumar P, Turley S, Ulmer JE, Rhie HG, Sibley CH, Hol WGJ. J Mol Biol. 2005;352:1091–1104. doi: 10.1016/j.jmb.2005.07.071. [DOI] [PubMed] [Google Scholar]
- 31.Graziani SB, Bernauer J, Skouloubris S, Graille M, Zhou CZ, Marchand C, Decottignies P, Tilbeurgh H, Myllykallio H, Liebl U. J Biol Chem. 2006;281:24048–24057. doi: 10.1074/jbc.M600745200. [DOI] [PubMed] [Google Scholar]
- 32.Wang K, Wang Q, Chen J, Chen L, Jiang H, Shen X. Prot Sci. 2011 doi: 10.1002/pro.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampathkumar P, Turley S, Sibley C, Hol WGJ. J Mol Biol. 2006;360:1–6. doi: 10.1016/j.jmb.2006.04.061. [DOI] [PubMed] [Google Scholar]
- 34.Ulmer JE, Boum Y, Thouvenel CD, Myllykallio H, Sibley CH. J Bacteriol. 2008;190:2056–2064. doi: 10.1128/JB.01094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leduc D, Graziani S, Lipowski G, Marchand C, Le Marechal P, Liebl U, Myllykallio H. Proc Natl Acad Sci U S A. 2004;101:7252–7257. doi: 10.1073/pnas.0401365101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koehn EM, Fleischmann T, Conrad JA, Palfey BA, Lesley SA, Mathews II, Kohen A. Nature. 2009;458:919–924. doi: 10.1038/nature07973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathews II, Deacon AM, Canaves JM, McMullan D, Lesley SA, Agarwalla S, Kuhn P. Structure (Camb) 2003;11:677–690. doi: 10.1016/s0969-2126(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 38.Wataya Y, Santi DV. Biochem Biophys Res Comm. 1975;67:818–823. doi: 10.1016/0006-291x(75)90886-4. [DOI] [PubMed] [Google Scholar]
- 39.Soltero-Higgin M, Carlson EE, Gruber TD, Kiessling LL. Nat Struc Mol Biol. 2004;11:539–543. doi: 10.1038/nsmb772. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Chernyshev A, Koehn EM, Manuel TD, Lesley SA, Kohen A. FEBS Journal. 2009;276:2801–2810. doi: 10.1111/j.1742-4658.2009.07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graziani SB, Xia Y, Gurnon JR, Van Etten JL, Leduc D, Skouloubris SP, Myllykallio H, Liebl U. J Biol Chem. 2004;279:54340–54347. doi: 10.1074/jbc.M409121200. [DOI] [PubMed] [Google Scholar]
- 42.Chernyshev A, Fleischmann T, Kohen A. Appl Microbiol Biotechnol. 2007;74:282–289. doi: 10.1007/s00253-006-0763-1. [DOI] [PubMed] [Google Scholar]
- 43.Esra Önen F, Boum Y, Jacquement C, Spanedda MV, Jaber N, Scherman D, Myllykallio H, Herscovici J. Bioorg Med Chem Lett. 2008;18:3628–3631. doi: 10.1016/j.bmcl.2008.04.080. [DOI] [PubMed] [Google Scholar]
- 44.Kogler M, Vanderhoydonck B, De Jonghe S, Rozenski J, Van Belle K, Herman J, Louat T, Parchina A, Sibley C, Lescrinier E, Herdewijn P. J Med Chem. 2011;54:4847–4862. doi: 10.1021/jm2004688. [DOI] [PubMed] [Google Scholar]
- 45.Griffin J, Roshick C, Iliffe-Lee E, McClarty G. J Biol Chem. 2005;280:5456–5467. doi: 10.1074/jbc.M412415200. [DOI] [PubMed] [Google Scholar]
- 46.Kealey JT, Gu X, Santi DV. Biochimie. 1994;76:1133–1142. doi: 10.1016/0300-9084(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 47.Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. Proc Natl Acad Sci USA. 2008;105:6876–6881. doi: 10.1073/pnas.0802247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delk AS, Nagle DP, Jr, Rabinowitz JC. J Biol Chem. 1980;255:4387–4390. [PubMed] [Google Scholar]
- 49.Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Nucleic Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urbonavicius J, Brochier-Armanet C, Skouloubris S, Myllykallio H, Grosjean H. Methods Enzymol. 2007;425:103–119. doi: 10.1016/S0076-6879(07)25004-9. [DOI] [PubMed] [Google Scholar]
- 51.Nishimasu H, Ishitani R, Yamashita K, Iwashita C, Hirata A, Hori H, Nureki O. Proc Natl Acad Sci USA. 2009;106:8180–8185. doi: 10.1073/pnas.0901330106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamdane D, Argentini M, Cornu D, Myllykallio H, Skouloubris S, Hui-Bon-Hoa G, Golinelli-Pipaneau B. J Biol Chem. 2011 doi: 10.1074/jbc.M111.256966. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]