Abstract
Background.
Polymorphisms in the non-muscle myosin IIA gene (MYH9) are associated with focal segmental glomerulosclerosis (FSGS) and non-diabetic end-stage renal disease (ESRD) in African Americans and FSGS in European Americans. We tested for association of single nucleotide polymorphisms (SNPs) in MYH9 with T2DM–ESRD in European Americans; additionally, three APOL1 gene variants were evaluated.
Methods.
Fifteen MYH9 SNPs and two APOL1 SNPs plus a 6-bp deletion were genotyped in 1963 European Americans, 536 cases with T2DM–ESRD and 1427 non-nephropathy controls (467 with T2DM and 960 without diabetes).
Results.
Comparing T2DM–ESRD cases with the 467 T2DM non-nephropathy controls, single variant associations trending toward significance were detected with SNPs rs4821480, rs2032487 and rs4281481 comprising part of the major MYH9 E1 risk haplotype [P-values 0.053–0.055 recessive, odds ratio (OR) 6.08–6.14]. Comparing T2DM–ESRD cases to all 1427 non-nephropathy controls, we confirmed evidence of association in these three SNPs as well as in the fourth E1 SNP (rs3752462) (P-values 0.017–0.035, OR 1.41–3.72). APOL1 G1/G2 nephropathy risk variants were rare in individuals of European American heritage, present in 0.28% of chromosomes in T2DM–ESRD cases and 0.32% of controls.
Conclusions.
MYH9 SNPs rs4821480, rs2032487, rs4281481 and rs3752462 are associated with T2DM–ESRD susceptibility in European Americans. The APOL1 risk variants are not present at appreciable frequency in this cohort with T2DM–ESRD. Therefore, polymorphisms in MYH9 appear to influence nephropathy risk in this sample.
Keywords: APOL1, diabetic nephropathy, end-stage renal disease, MYH9, type 2 diabetes mellitus
Introduction
Type 2 diabetes mellitus (T2DM) remains the most common cause of end-stage renal disease (ESRD) in the USA with an annual incidence rate of 273 per million population in European Americans [1]. Marked familial clustering of renal disease has repeatedly been documented in T2DM-affected European Americans [2–4] as well as in other ethnic groups [5].
The MYH9 gene encodes non-muscle myosin IIA and is expressed in glomerular podocytes and mesangial cells [6, 7]. Polymorphisms in MYH9 are strongly associated with idiopathic and HIV-associated forms of focal segmental glomerulosclerosis (FSGS), clinically diagnosed ‘hypertensive nephropathy’ (focal global glomerulosclerosis) and non-diabetes-associated ESRD in African Americans [8–11], idiopathic FSGS and non-diabetic kidney disease in European Americans [11, 12] and non-diabetic ESRD in Hispanic Americans [13]. The MYH9 haplotypes designated E1 (GCCT) and E2 (TTTC) showed replicated association with risk and protection, respectively, for kidney disease in African Americans and European Americans [8–11]. MYH9 also influences kidney function in Europeans [14]. Variations in APOL1, the gene encoding apolipoprotein L-1, have recently been shown to be associated with kidney disease in African Americans [15–18]; however, it was reported that these variants are not present on European chromosomes [16]. Although not fully elucidated, a variation in the Chromosome 22 gene region encompassing MYH9 and APOL1 contributes to nephropathy risk.
Gene polymorphisms in APOL1 and MYH9 have not been evaluated for association with T2DM–ESRD in European Americans. We evaluated 15 MYH9 single nucleotide polymorphisms (SNPs) and 3 APOL1 variants for association with T2DM–ESRD in European Americans to determine whether genetic factors contributing to the development and progression of non-diabetic kidney disease in African Americans also mediate T2DM–ESRD susceptibility in European Americans.
Materials and methods
The cases included 536 unrelated and self-reported European Americans with clinically diagnosed T2DM-associated ESRD residing in the Southeastern USA (T2DM–ESRD cases). All T2DM–ESRD cases were receiving renal replacement therapy. Type 2 diabetes was considered to be the primary cause of nephropathy in subjects developing diabetes after 30 years of age, in the presence of diabetic retinopathy and/or proteinuria exceeding 500 mg/24 h, or if diabetes was present >5 years before initiation of renal replacement therapy, in the absence of quantitated urine protein or retinopathy data. The mean ± SD diabetes duration in the T2DM–ESRD cases was 20.07 ± 10.12 years.
Four hundred and sixty-seven unrelated self-reported European Americans with T2DM lacking renal disease were also recruited (T2DM non-nephropathy controls). These individuals were actively receiving oral hypoglycemic agents and/or insulin, had a serum creatinine concentration ≤1.5 mg/dL and urine albumin:creatinine ratio <30 mg/g. The mean ± SD diabetes duration in the T2DM non-nephropathy controls was 11.98 ± 7.59 years. An additional 960 control subjects lacking diabetes and nephropathy were recruited from medical clinics and health fairs in NC (non-diabetic non-nephropathy controls). These were self-reported European Americans who were born in the Southeastern USA, ≥18 years of age, denied a personal history of diabetes or renal disease and had a serum creatinine concentration <1.5 mg/dL. Albuminuria was not measured in this low-risk control group.
DNA was isolated from blood using an AutoPure LS automated DNA extraction robot (Gentra Systems, Minneapolis, MN). Recruitment and sample collection procedures were approved by the Institutional Review Board at the Wake Forest School of Medicine and all subjects provided written informed consent.
SNP genotyping
Sixteen SNPs spanning ∼40 kb of the MYH9 gene were chosen based on prior analyses in African Americans [9–11]. In addition, two SNPs (G1, rs73885319 and rs60910145) and one deletion (G2, rs71785323) in APOL1 were selected based on significant association with nephropathy in African Americans [15–18]. SNPs were genotyped using the MassARRAY genotyping system (Sequenom, San Diego, CA) and polymerase chain reaction (PCR) primers were designed using the MassARRAY Design 3.4 Software (Sequenom) (provided upon request). One SNP (rs11549907) was not successfully genotyped due to failure of PCR design. Seventeen SNPs (15 MYH9; 2 APOL1 G1) and the APOL1 deletion (G2) were successfully genotyped in 1003 T2DM-affected European Americans, 536 with T2DM–ESRD and 467 with T2DM lacking nephropathy and 960 control subjects lacking T2DM. The genotyping efficiency of each of the variants exceeded 95%.
DNA sequencing
Twenty-seven individual DNA samples were not successfully genotyped on the Sequenom MassARRAY. Therefore, SNPs rs2032487 and rs4821480 were genotyped by DNA sequence analysis. Nineteen individuals for whom genotypes were successfully obtained at these SNPs were also sequenced to verify the accuracy of genotype calls. The 365 nucleotide region surrounding the two SNPs was PCR amplified and products directly sequenced using Big Dye Ready Reaction Mix on an ABI3730xl sequencer (Applied Biosystems, Foster City, CA). Sequence data were visualized using Sequencher Software version 4.9 (GeneCodes corporation, Ann Arbor, MI). Genotype calls were verified by sequencing for individuals whose genotypes were successfully identified from Sequenom with 100% concordance. Association analyses were performed using genotypes obtained from sequencing results, in addition to those calls obtained from Sequenom.
Ancestry informative markers
To assess whether our sample was enriched for individuals with significant proportions of African ancestry, 70 ancestry informative markers (AIMs) (described in [19]) were genotyped in all reportedly European American cases and controls with APOL1 G1 or G2 nephropathy risk variants. Ancestral allele frequencies were estimated from the results of genotyping the 70 AIMs in Yoruba Nigerians and European Americans. Individual ancestral proportions were generated for each subject and evaluated under a two-population model using FRAPPE [20], an EM algorithm-based approach. This led to the removal of 12 individuals, all with >9% African ancestry. Statistical analyses were performed on remaining individuals for association between T2DM-associated ESRD and MYH9 variants.
Statistical analysis
Age and body mass index (BMI) were compared using a Kruskal–Wallis one-way analysis of variance on ranks (SigmaStat; Systam Software, San Jose, CA) with a Dunn’s method multiple comparison test. Age at T2DM diagnosis in the T2DM–ESRD cases and T2DM non-nephropathy subjects was compared using a Mann–Whitney rank-sum test (SigmaStat). Each SNP was tested for departure from Hardy–Weinberg equilibrium using the χ2 goodness-of-fit test in the statistical analysis program SNPGWA (www.phs.wfubmc.edu). Tests for genotypic association were performed on each SNP individually, using the large sample test. The primary inference is based on the 2 degrees of freedom (2df) global test of genotypic association (not shown). If significant, the individual genetic models (dominant, additive, recessive and lack-of-fit to additive) were examined. This is consistent with the Fisher’s protected least significant difference multiple comparisons procedure. The influence of possible covariates (age and sex) on evidence of association was evaluated using SNPGWA (www.phs.wfubmc.edu). Four SNP haplotype analyses were computed using the program Dandelion, using 10 000 permutations (www.phs.wfubmc.edu). The test of statistical significance for the individual haplotypes includes the continuity correction factor of c = 0.5 (i.e. a value of 0.5 was added to each cell). Thus, for low-frequency haplotypes, the confidence interval (CI) for the odds ratio (OR) may not overlap with 1 and the P-value may still not be significant.
Results
Table 1 summarizes the demographic characteristics of the study sample. T2DM–ESRD cases and T2DM non-nephropathy controls were older than non-diabetic non-nephropathy controls. T2DM non-nephropathy controls and T2DM–ESRD cases also had higher mean BMI compared to non-diabetic non-nephropathy controls. The mean age at T2DM diagnosis was lower in subjects with T2DM–ESRD compared to T2DM non-nephropathy controls. Non-diabetic non-nephropathy controls were more often female, relative to the T2DM–ESRD cases and T2DM non-nephropathy controls.
Table 1.
Demographic characteristics of European cohorta
| n | Age of examination (years) | BMI (kg/m2) | Age at T2DM diagnosis (years) | % Female | Age at ESRD diagnosis (years) | Urine albumin:creatinine ratio (mg/g) | Serum Creatinine (mg/dL) | |
| Non-diabetic non-nephropathy controls | 960 | 53.0 ± 14.73 | 28.41 ± 5.73 | 64.5 | 0.90 ± 0.19 | |||
| T2DM–ESRD cases | 536 | 65.57 ± 10.45bc | 29.24 ± 6.73c | 45.52 ± 14.00c | 50.4 | 63.28 ± 10.67 | ||
| T2DM, non-nephropathy controls | 467 | 62.95 ± 9.23b | 32.59 ± 7.10b | 51.00 ± 11.01 | 56.3 | 9.06 ± 7.89 | 0.99 ± 0.22 |
Data are n, % or means ± SD.
Mean is significantly different from non-diabetic non-nephropathy control (P < 0.001).
Mean is significantly different (P < 0.001) from T2DM non-nephropathy control.
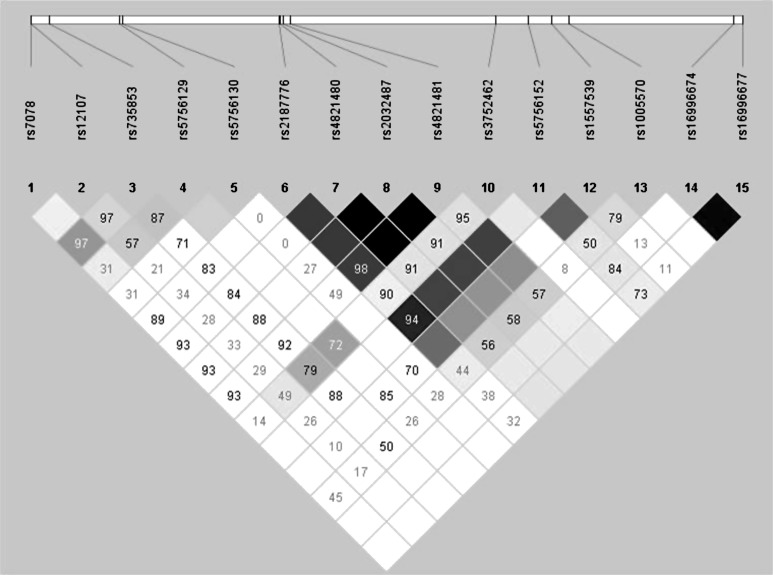
The 15 genotyped SNPs spanned 49.3 kb of MYH9 (Figure 1). Of these SNPs, rs4821480, rs2032487, rs4821481 and rs3752462 were used to determine the E1 risk and E2 protective haplotypes [8, 9, 11]. No SNPs in the non-diabetic non-nephropathy or the T2DM non-nephropathy control samples deviated from Hardy–Weinberg equilibrium after correction for multiple comparisons (P = 0.05/15 = 0.003). Cases with T2DM–ESRD were modestly deviated from Hardy–Weinberg equilibrium at SNPs rs2187776 (P = 8.960 × 10−4), rs4821480 (P = 1.597 × 10−3), rs2032487 (P = 2.732 × 10−3) and rs4821481 (P = 2.262 × 10−3); however, these SNPs were associated in the T2DM–ESRD cohort when compared to the combined non-nephropathy control groups.
Fig. 1.
Gene structure and linkage disequilibrium plot generated by 15 SNPs genotyped in European Americans. Inter-SNP D’-values are displayed on the plot.
In the association analysis comparing 536 T2DM–ESRD cases and 960 non-diabetic non-nephropathy controls, rs4821480, rs2032487 and rs4821481 and rs3752462 comprising the E1 and E2 haplotypes, trended toward association in the 2df test (0.016 ≤ P ≤ 0.200) and also trended toward association in the recessive model (0.045 ≤ P ≤ 0.107) (Table 2). The minor allele of each SNP was associated with increased risk for T2DM–ESRD; OR 1.42–3.13.
Table 2.
MYH9 SNP associations in 536 European American T2DM–ESRD cases compared to 960 non-diabetic non-nephropathy controls
| Risk allele frequencies |
Dominant model |
Additive model |
Recessive model |
Lack-of-fit to additive model | ||||||
| SNP | Risk allele | T2DM–ESRD | Controls | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
| rs7078 | A | 0.706 | 0.721 | 0.538 | 0.88 (0.60–1.31) | 0.731 | 0.97 (0.82–1.15) | 0.917 | 0.99 (0.80–1.22) | 0.606 |
| rs12107 | G | 0.874 | 0.867 | 0.047 | 2.85 (0.97–8.39) | 0.573 | 1.07 (0.85–1.34) | 0.972 | 1.00 (0.78–1.29) | 0.050 |
| rs735853 | C | 0.535 | 0.522 | 0.452 | 0.91 (0.70–1.17) | 0.477 | 1.06 (0.91–1.23) | 0.071 | 1.25 (0.98–1.58) | 0.030 |
| rs5756129 | T | 0.789 | 0.778 | 0.855 | 1.05 (0.64–1.73) | 0.508 | 1.06 (0.89–1.28) | 0.474 | 1.08 (0.87–1.35) | 0.786 |
| rs5756130 | C | 0.949 | 0.958 | 0.116 | 0.24 (0.04–1.65) | 0.263 | 0.82 (0.58–1.16) | 0.388 | 0.85 (0.59–1.23) | 0.212 |
| rs2187776 | C | 0.041 | 0.034 | 0.612 | 1.11 (0.73–1.69) | 0.340 | 1.20 (0.83–1.73) | 0.055 | 3.89 (0.87–17.44) | 0.097 |
| rs4821480 | G | 0.061 | 0.047 | 0.228 | 1.24 (0.87–1.77) | 0.131 | 1.27 (0.93–1.74) | 0.107 | 2.50 (0.79–7.92) | 0.333 |
| rs2032487 | C | 0.063 | 0.048 | 0.185 | 1.27 (0.89–1.79) | 0.091 | 1.31 (0.96–1.79) | 0.056 | 3.13 (0.91–10.73) | 0.217 |
| rs4821481 | C | 0.062 | 0.046 | 0.145 | 1.30 (0.91–1.85) | 0.071 | 1.33 (0.97–1.83) | 0.058 | 3.11 (0.91–10.66) | 0.252 |
| rs3752462 | T | 0.321 | 0.323 | 0.181 | 0.86 (0.70–1.07) | 0.929 | 0.99 (0.84–1.17) | 0.045 | 1.42 (1.01–2.00) | 0.004 |
| rs5756152 | A | 0.033 | 0.032 | 0.908 | 1.03 (0.67–1.58) | 0.844 | 1.04 (0.69–1.57) | 0.621 | 1.76 (0.18–16.94) | 0.649 |
| rs1557539 | G | 0.979 | 0.981 | 0.774 | 0.57 (0.01–28.62) | 0.631 | 0.88 (0.52–1.48) | 0.649 | 0.88 (0.52–1.51) | 0.864 |
| rs1005570 | A | 0.110 | 0.094 | 0.156 | 1.22 (0.93–1.59) | 0.179 | 1.18 (0.93–1.50) | 0.762 | 1.14 (0.49–2.65) | 0.645 |
| rs16996674 | T | 0.005 | 0.003 | 0.309 | 1.79 (0.57–5.57) | 0.325 | 1.66 (0.6–4.61) | 0.770 | 1.78 (0.04–89.86) | 0.801 |
| rs16996677 | A | 0.005 | 0.003 | 0.454 | 1.52 (0.51–4.53) | 0.455 | 1.45 (0.54–3.9) | 0.774 | 1.76 (0.03–88.92) | 0.921 |
Additional association analysis comparing 536 T2DM–ESRD subjects and 467 T2DM non-nephropathy subjects revealed similar results (Table 3). SNPs rs4821480, rs2032487 and rs4821481 trended toward association under the 2df test (0.086 ≤ P ≤ 0.099) and in the recessive model (0.053 ≤ P ≤ 0.055), with evidence that the minor alleles contributed to risk, indicated by the OR (6.08–6.14). Since it is possible that some T2DM non-nephropathy controls with short diabetes durations will develop future nephropathy, an analysis restricting this group to the 419 participants with diabetes durations >5 years was performed; results were comparable (data not shown). In addition, inclusion of controls destined to develop diabetic nephropathy would reduce our power to detect association.
Table 3.
MYH9 SNP associations in 536 European American T2DM–ESRD cases compared to 467 T2DM non-nephropathy controls
| Risk allele frequencies |
Dominant model |
Additive model |
Recessive model |
Lack-of-fit to additive model | ||||||
| SNP | Risk allele | T2DM–ESRD | T2DM | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
| rs7078 | A | 0.706 | 0.711 | 0.206 | 0.73 (0.45–1.19) | 0.805 | 0.98 (0.8–1.19) | 0.720 | 1.05 (0.81–1.35) | 0.148 |
| rs12107 | G | 0.874 | 0.876 | 0.021 | 3.53 (1.13–11.03) | 0.933 | 0.99 (0.76–1.29) | 0.434 | 0.89 (0.66–1.19) | 0.007 |
| rs735853 | C | 0.535 | 0.507 | 0.401 | 1.13 (0.85–1.52) | 0.215 | 1.12 (0.94–1.33) | 0.236 | 1.18 (0.90–1.57) | 0.781 |
| rs5756129 | T | 0.789 | 0.779 | 0.798 | 0.92 (0.51–1.69) | 0.601 | 1.06 (0.85–1.31) | 0.464 | 1.10 (0.85–1.42) | 0.484 |
| rs5756130 | C | 0.949 | 0.954 | 0.175 | 0.16 (0.01–3.2) | 0.637 | 0.91 (0.61–1.36) | 0.834 | 0.96 (0.62–1.46) | 0.203 |
| rs2187776 | C | 0.041 | 0.027 | 0.225 | 1.38 (0.82–2.31) | 0.149 | 1.4 (0.88–2.2) | 0.184 | 3.19 (0.52–19.51) | 0.540 |
| rs4821480 | G | 0.061 | 0.042 | 0.147 | 1.38 (0.89–2.13) | 0.070 | 1.44 (0.97–2.13) | 0.053 | 6.14 (0.75–50.1) | 0.246 |
| rs2032487 | C | 0.063 | 0.044 | 0.149 | 1.37 (0.89–2.09) | 0.071 | 1.42 (0.97–2.09) | 0.054 | 6.09 (0.75–49.71) | 0.243 |
| rs4821481 | C | 0.062 | 0.042 | 0.113 | 1.42 (0.92–2.18) | 0.053 | 1.47 (0.99–2.17) | 0.055 | 6.08 (0.75–49.6) | 0.281 |
| rs3752462 | T | 0.321 | 0.302 | 0.804 | 1.03 (0.80–1.33) | 0.371 | 1.09 (0.90–1.31) | 0.125 | 1.38 (0.91–2.08) | 0.201 |
| rs5756152 | A | 0.033 | 0.026 | 0.365 | 1.28 (0.75–2.19) | 0.415 | 1.23 (0.75–2.02) | 0.900 | 0.86 (0.09–8.34) | 0.606 |
| rs1557539 | G | 0.979 | 0.988 | 0.938 | 1.17 (0.02–59.03) | 0.175 | 0.63 (0.32–1.24) | 0.153 | 0.60 (0.29–1.22) | 0.584 |
| rs1005570 | A | 0.110 | 0.099 | 0.669 | 1.07 (0.78–1.47) | 0.445 | 1.12 (0.84–1.49) | 0.137 | 2.61 (0.70–9.70) | 0.201 |
| rs16996674 | T | 0.005 | 0.001 | 0.221 | 2.63 (0.53–13.08) | 0.306 | 1.99 (0.51–7.78) | 0.944 | 0.87 (0.02–43.9) | 0.389 |
| rs16996677 | A | 0.005 | 0.001 | 0.223 | 2.16 (0.52–13.01) | 0.310 | 1.98 (0.51–7.74) | 0.942 | 0.86 (0.02–43.68) | 0.391 |
To ascertain whether these markers exhibited stronger evidence of association when compared to larger numbers of non-nephropathy controls, genotypes from the 1427 non-nephropathy controls (with and without T2DM) were combined and contrasted to those in the 536 T2DM–ESRD cases (Table 4). In this analysis, rs4821480 trended toward association under the 2df test (P = 0.064) and recessive model (P = 0.033) with OR 3.11 (95% CI 1.04–9.29); rs2032487 was associated under the 2df test (P =0.034) and recessive model (P = 0.017) with OR 3.72 (95% CI 1.18–11.77); rs4821481 was associated under the 2df test (P = 0.029) and recessive model (P = 0.017) with OR 3.70 (95% CI 1.17–11.71) and rs3752462 was associated under the 2df test (P = 0.031) and recessive model (P = 0.035) with OR 1.41 (95% CI 1.02–1.93). Analyses performed after adjustment for age and gender yielded similar results (P-values ranged from 0.037 to 0.194 under the recessive model for these four SNPs; data not shown).
Table 4.
MYH9 SNP associations in 536 European American T2DM–ESRD cases compared to 1427 non-diabetic non-nephropathy controls
| Risk allele frequencies |
Dominant model |
Additive model |
Recessive model |
Lack-of-fit to additive model | ||||||
| SNP | Risk allele | T2DM–ESRD | Controls | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value |
| rs7078 | A | 0.706 | 0.712 | 0.333 | 0.83 (0.58–1.20) | 0.727 | 0.97 (0.83–1.14) | 0.942 | 1.01 (0.82–1.23) | 0.331 |
| rs12107 | G | 0.874 | 0.870 | 0.027 | 3.07 (1.08–8.74) | 0.714 | 1.04 (0.84–1.28) | 0.770 | 0.97 (0.76–1.22) | 0.019 |
| rs735853 | C | 0.535 | 0.517 | 0.860 | 0.98 (0.77–1.24) | 0.303 | 1.08 (0.93–1.25) | 0.073 | 1.23 (0.98–1.53) | 0.096 |
| rs5756129 | T | 0.789 | 0.778 | 0.977 | 1.01 (0.63–1.61) | 0.491 | 1.06 (0.86–1.26) | 0.417 | 1.09 (0.89–1.34) | 0.634 |
| rs5756130 | C | 0.949 | 0.957 | 0.034 | 0.16 (0.02–1.11) | 0.282 | 0.84 (0.61–1.16) | 0.456 | 0.88 (0.62–1.24) | 0.061 |
| rs2187776 | C | 0.041 | 0.032 | 0.356 | 1.20 (0.81–1.78) | 0.162 | 1.28 (0.90–1.82) | 0.024 | 4.15 (1.08–15.9) | 0.072 |
| rs4821480 | G | 0.061 | 0.045 | 0.139 | 1.28 (0.92–1.79) | 0.060 | 1.33 (0.99–1.78) | 0.033 | 3.11 (1.04–9.29) | 0.163 |
| rs2032487 | C | 0.063 | 0.046 | 0.117 | 1.30 (0.94–1.80) | 0.044 | 1.35 (1.01–1.81) | 0.017 | 3.72 (1.18–11.77) | 0.099 |
| rs4821481 | C | 0.062 | 0.045 | 0.084 | 1.34 (0.96–1.86) | 0.031 | 1.38 (1.03–1.86) | 0.017 | 3.70 (1.17–11.71) | 0.122 |
| rs3752462 | T | 0.321 | 0.316 | 0.393 | 0.92 (0.75–1.12) | 0.755 | 1.02 (0.88–1.19) | 0.035 | 1.41 (1.02–1.93) | 0.009 |
| rs5756152 | A | 0.033 | 0.030 | 0.613 | 1.11 (0.74–1.67) | 0.575 | 1.12 (0.76–1.65) | 0.659 | 1.57 (0.21–11.93) | 0.794 |
| rs1557539 | G | 0.979 | 0.984 | 0.617 | 0.38 (0.01–19.28) | 0.303 | 0.77 (0.47–1.27) | 0.322 | 0.77 (0.46–1.29) | 0.790 |
| rs1005570 | A | 0.110 | 0.096 | 0.234 | 1.17 (0.91–1.50) | 0.197 | 1.16 (0.93–1.45) | 0.417 | 1.40 (0.62–3.16) | 0.833 |
| rs16996674 | T | 0.005 | 0.002 | 0.129 | 2.28 (0.76–6.82) | 0.135 | 2.08 (0.77–5.63) | 0.613 | 2.65 (0.05–133.62) | 0.780 |
| rs16996677 | A | 0.005 | 0.003 | 0.200 | 1.98 (0.68–5.73) | 0.199 | 1.85 (0.70–4.88) | 0.616 | 2.62 (0.05–132.46) | 0.889 |
A haplotype analysis was performed to evaluate whether the major MYH9 E1 risk haplotype was associated with T2DM–ESRD in European Americans. The risk alleles of the four SNPs comprising this haplotype (G at rs4821480, C at rs2032487, C at rs4821481 and T at rs3752462) were evaluated; results indicated that the E1 GCCT risk haplotype was present at a frequency of 6.04% in T2DM–ESRD cases and 4.43% in non-nephropathy controls, with P = 0.140 and OR = 1.39 (95% CI 1.02–1.90). Three percent of the total cohort was missing SNP data for at least one E1 SNP, which could have affected our ability to detect significance, given that P-values for the four individual SNPs were nominal. The seven potential haplotypes arising from allele combinations at the four SNPs were evaluated. Of these, the most common was the E2 protective haplotype (TTTC), with 68.00% frequency in the overall European American cohort, supporting that the protective MYH9 E2 haplotype is much more frequent in European Americans than the E1 risk haplotype [11].
APOL1 G1 and G2 variants were also genotyped [15–17]. Among cases and controls with APOL1 nephropathy risk variants (with ‘European American’ defined as possessing <9% global African ancestry based upon 70 AIMs), 11 APOL1 G1 and/or G2 risk variants were detected; four in 960 non-T2DM non-nephropathy controls (0.21%), four in 467 non-T2DM non-nephropathy controls (0.43%) and three in 536 T2DM–ESRD cases (0.28%). One European American T2DM non-nephropathy control had two APOL1 G2 variants.
Discussion
This report is the first to identify MYH9 SNP associations with presumed diabetic ESRD in European Americans. We evaluated the presence of APOL1 risk variants (0.28% of chromosomes in T2DM–ESRD cases and 0.32% of the chromosomes in controls) confirming the relative rarity of APOL1 nephropathy risk variants in European Americans. APOL1 G1 and G2 variants and MYH9 SNPs rs4821480, rs2032487, rs4821481 and rs3752462 are known to be strongly associated with a spectrum of related, predominantly non-diabetic kidney diseases in African Americans as well as idiopathic FSGS and non-diabetic kidney disease in European Americans and non-diabetic ESRD in Hispanic Americans [8–12, 18]. Additionally, there is evidence that MYH9 accounts for residual risk beyond that attributable to these APOL1 variants in African Americans [C. Langefeld (personal communication)]. Although a recessive model has been emphasized in the literature for these MYH9 associations [8–11], in the current analyses we detected an association between MYH9 SNPs and T2DM–ESRD in European Americans in a recessive as well as an additive model. Due to the low frequency of MYH9 risk alleles in European Americans, few individuals homozygous for risk alleles will be detected. Only 10.26% (55/536) of T2DM–ESRD cases had at least one copy of the MYH9 risk alleles at the four E1 haplotype SNPs and 1.31% (7/536) had two copies of risk alleles. Of all 1963 European Americans in the analyses, 8.66% (170) had at least one copy of the MYH9 risk alleles at the four SNPs and 0.61% (12) had two copies of risk alleles at the four SNPs. It was apparent that individuals homozygous for risk alleles were over-represented in T2DM–ESRD cases, consistent with prior reports. Additional analyses limited to individuals not homozygous for the risk allele revealed a lack of statistical evidence for association (rs4821480 P = 0.941, OR = 0.89; rs2032487 P = 0.612, OR = 2.65; rs4821481 P = 0.614, OR = 2.64; rs3752462 P = 0.113, OR = 1.31), indicating that the recessive model drove the association observed in the additive model.
To account for the possibility that some T2DM non-nephropathy controls will develop future kidney disease, only those with serum creatinine concentrations <1.5 mg/dL and urine albumin:creatinine ratio <30 mg/g were evaluated. Since a small percentage of these controls will likely develop progressive nephropathy, a subsequent analysis excluded those with <5 year diabetes duration; results were similar to those in the initial analysis. Furthermore, comparing T2DM–ESRD to T2DM non-nephropathy controls, the recessive model P-value for the strongest signal (rs4821480) did not change when excluding individuals with diabetes duration <5 years. Combined non-diabetic non-nephropathy controls and T2DM non-nephropathy controls with diabetes duration ≥5 years were compared to T2DM–ESRD cases, the P-value for the strongest signal (rs2032487) increased only slightly, from 0.017 to 0.020. We attribute this slight increase to the decreased sample size associated with removing 48 individuals with <5 years diabetes duration.
Although previous studies indicated that MYH9 polymorphisms were not associated with diabetic nephropathy [10, 11], our findings indicate that these variants contribute to risk for ‘clinically diagnosed’ type 2 diabetes-associated ESRD in European Americans, although clearly with smaller effect size as compared to that in non-diabetic kidney disease in African Americans. Our samples were recruited in the southern USA and likely represent a homogeneous sample consistent with Northern European heritage [21]. Genome-wide ancestry analyses would be required to comprehensively rule out the unlikely possibility that the observed signal is actually marking cryptic population substructure. In our report evaluating MYH9 in African Americans with T2DM–ESRD, we proposed that association could have resulted from inclusion of MYH9-associated FSGS in the sample of cases with coincident T2DM and ESRD [8]; an observation that was subsequently proven [22]. Concurrent FSGS and diabetic nephropathy would appear less likely in European Americans, as the prevalence of both T2DM and FSGS are markedly lower than in African Americans (as is the frequency of MYH9 E1 risk haplotypes); nonetheless, it remains possible that some of these European Americans with clinically diagnosed T2DM–ESRD had MYH9-related FSGS. Renal biopsies are required—to determine the true cause of nephropathy in these clinically diagnosed cases, a procedure that is infrequently performed.
Prior reports have detected associations of these SNPs and haplotypes primarily with non-diabetes-associated kidney diseases [9–12]. A genetic predisposition to T2DM–ESRD is clearly established in African Americans and European Americans. Polymorphisms in the non-muscle myosin heavy chain IIA gene, expressed in glomerular podocytes and mesangial cells [6, 7], may lead to alterations in the cytoskeleton, impair the glomerular filtration barrier and result in proteinuria with progressive renal failure in subjects with T2DM [7]. Genetic variants which have the potential to alter risk for diabetic nephropathy, such as TCF7L2, ACACB, ELMO1 and FRMD3, in addition to others [23], have previously been described [23–29]. It would not be unreasonable to postulate that these and other variants may be interacting with one another and/or with MYH9 risk variants to modify risk for diabetic and non-diabetic ESRD. The existence of ‘second hits’ genes that may be interacting with MYH9 to mediate either genetic predisposition to, or protection from kidney disease of various causes needs to be explored in African- and European-derived populations.
This manuscript reports the results of a focused a priori hypothesis that E1 haplotype SNPs were associated with diabetic nephropathy in European Americans. Although the statistical power of the tests in this paper are low for a recessive model for the E1 haplotype (∼10%), significant association was observed for the individual SNPs comprising the E1 haplotype. The strength of the association between MYH9 polymorphisms and T2DM-associated ESRD in this European American cohort is weaker than that detected in African Americans with diabetic and non-diabetic forms of kidney disease [8–11]. In conclusion, this is the first report investigating and identifying MYH9 SNP association with presumed diabetic nephropathy in European Americans. Variation at the MYH9–APOL1 gene region on Chromosome 22q is associated with nephropathy susceptibility, including in European American populations.
Acknowledgments
We wish to thank the patients, their relatives and staff of the Southeastern Kidney Council, Inc./ESRD Network 6 for their participation. This work was supported by National Institutes of Health grants RO1 DK066358 (D.W.B.), R01 DK053591 (D.W.B.), R01 HL56266 (B.I.F.), RO1 DK070941 (B.I.F.), R01 DK084149 (B.I.F.) and in part by the General Clinical Research Center of the Wake Forest School of Medicine grant M01 RR07122.
Conflict of interest statement. None declared.
References
- 1.U.S. Renal Data System. USRDS 2009 Annual Data Report. Atlas of chronic kidney Disease and End-Stage Renal Disease in the United States. Bethesda, MD: National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; 2009. [Google Scholar]
- 2.Bowden DW. Genetics of kidney disease. Kidney Int Suppl. 2003;83:S8–S12. doi: 10.1046/j.1523-1755.63.s83.3.x. [DOI] [PubMed] [Google Scholar]
- 3.O'Dea DF, Murphy SW, Hefferton D, et al. Higher risk for renal failure in first-degree relatives of white patients with end-stage renal disease: a population-based study. Am J Kidney Dis. 1998;32:794–801. doi: 10.1016/s0272-6386(98)70135-0. [DOI] [PubMed] [Google Scholar]
- 4.Spray BJ, Atassi NG, Tuttle AB, et al. Familial risk, age at onset, and cause of end-stage renal disease in white Americans. J Am Soc Nephrol. 1995;5:1806–1810. doi: 10.1681/ASN.V5101806. [DOI] [PubMed] [Google Scholar]
- 5.Satko SG, Sedor JR, Iyengar SK, et al. Familial clustering of chronic kidney disease. Semin Dial. 2007;20:229–236. doi: 10.1111/j.1525-139X.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- 6.Arrondel C, Vodovar N, Knebelmann B, et al. Expression of the nonmuscle myosin heavy chain IIA in the human kidney and screening for MYH9 mutations in Epstein and Fechtner syndromes. J Am Soc Nephrol. 2002;13:65–74. doi: 10.1681/ASN.V13165. [DOI] [PubMed] [Google Scholar]
- 7.Singh N, Nainani N, Arora P, et al. CKD in MYH9-related disorders. Am J Kidney Dis. 2009;54:732–740. doi: 10.1053/j.ajkd.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Freedman BI, Hicks PJ, Bostrom MA, et al. Non-muscle myosin heavy chain 9 gene MYH9 associations in African Americans with clinically diagnosed type 2 diabetes mellitus-associated ESRD. Nephrol Dial Transplant. 2009;24:3366–3371. doi: 10.1093/ndt/gfp316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman BI, Kopp JB, Winkler CA, et al. Polymorphisms in the nonmuscle myosin heavy chain 9 gene (MYH9) are associated with albuminuria in hypertensive African Americans: the HyperGEN study. Am J Nephrol. 2009;29:626–632. doi: 10.1159/000194791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kao WH, Klag MJ, Meoni LA, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopp JB, Smith MW, Nelson GW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Seaghdha CM, Parekh RS, Hwang SJ, et al. The MYH9/APOL1 region and chronic kidney disease in European-Americans. Hum Mol Genet. 2011;20:2450–2456. doi: 10.1093/hmg/ddr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behar DM, Rosset S, Tzur S, et al. African ancestry allelic variation at the MYH9 gene contributes to increased susceptibility to non-diabetic end-stage kidney disease in Hispanic Americans. Hum Mol Genet. 19:1816–1827. doi: 10.1093/hmg/ddq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattaro C, Aulchenko YS, Isaacs A, et al. Genome-wide linkage analysis of serum creatinine in three isolated European populations. Kidney Int. 2009;76:297–306. doi: 10.1038/ki.2009.135. [DOI] [PubMed] [Google Scholar]
- 15.Freedman BI, Kopp JB, Langefeld CD, et al. The apolipoprotein L1 (APOL1) gene and nondiabetic nephropathy in African Americans. J Am Soc Nephrol. 2010;21:1422–1426. doi: 10.1681/ASN.2010070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genovese G, Tonna SJ, Knob AU, et al. A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kidney Int. 2010;78:698–704. doi: 10.1038/ki.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzur S, Rosset S, Shemer R, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keene KL, Mychaleckyj JC, Leak TS, et al. Exploration of the utility of ancestry informative markers for genetic association studies of African Americans with type 2 diabetes and end stage renal disease. Hum Genet. 2008;124:147–154. doi: 10.1007/s00439-008-0532-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang H, Peng J, Wang P, et al. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 21.Bento JL, Palmer ND, Zhong M, et al. Heterogeneity in gene loci associated with type 2 diabetes on human chromosome 20q13.1. Genomics. 2008;92:226–234. doi: 10.1016/j.ygeno.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gopalakrishnan I, Iskandar SS, Daeihagh P, et al. Coincident idiopathic focal segmental glomerulosclerosis collapsing variant and diabetic nephropathy in an African American homozygous for MYH9 risk variants. Hum Pathol. 2011;42:291–294. doi: 10.1016/j.humpath.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pezzolesi MG, Poznik GD, Mychaleckyj JC, et al. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009;58:1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kottgen A, Hwang SJ, Rampersaud E, et al. TCF7L2 variants associate with CKD progression and renal function in population-based cohorts. J Am Soc Nephrol. 2008;19:1989–1999. doi: 10.1681/ASN.2007121291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leak TS, Perlegas PS, Smith SG, et al. Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Ann Hum Genet. 2009;73:152–159. doi: 10.1111/j.1469-1809.2008.00498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda S, Kobayashi MA, Araki S, et al. A single nucleotide polymorphism within the acetyl-coenzyme A carboxylase beta gene is associated with proteinuria in patients with type 2 diabetes. PLoS Genet. 2010;6:e1000842. doi: 10.1371/journal.pgen.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pezzolesi MG, Katavetin P, Kure M, et al. Confirmation of genetic associations at ELMO1 in the GoKinD collection supports its role as a susceptibility gene in diabetic nephropathy. Diabetes. 2009;58:2698–2702. doi: 10.2337/db09-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimazaki A, Kawamura Y, Kanazawa A, et al. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes. 2005;54:1171–1178. doi: 10.2337/diabetes.54.4.1171. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida T, Kato K, Yokoi K, et al. Association of genetic variants with myocardial infarction in individuals with or without hypertension or diabetes mellitus. Int J Mol Med. 2009;24:701–709. doi: 10.3892/ijmm_00000282. [DOI] [PubMed] [Google Scholar]