Abstract
Three different parasites of the phylum Parabasala (Tritrichomonas foetus, Trichomitus rotunda and Tetratrichomonas buttreyi) have been described in pigs. In a previous study (Mostegl et al., 2011) approximately 47% of 91 paraffin wax-embedded intestinal samples of pigs which were Trichomonas-positive by in situ hybridization using a probe with a broad reactivity spectrum contained other species than T. foetus. Out of these, intestinal trichomonads from three pigs (pigs 1–3) were further analyzed by gene sequencing of a part of the 18S ribosomal RNA (rRNA) gene using primer walking. Subsequently, the partial sequences achieved by the different primer pairs were combined to a sequence of about 1000 bp for each trichomonad. In all three pigs unique sequences were acquired which showed only moderate similarities to sequences available in the GenBank. Alignments and the BLAST analysis showed a high degree of homology between sequences of trichomonads from pig 1 and pig 3 with only 1% difference. These sequences were found to be 92% similar to Hypotrichomonas acosta, a trichomonad isolated from squamate reptiles. The trichomonad sequence detected in the intestine of pig 2 showed about 10% nucleotide differences compared to pigs 1 and 3. This sequence was 97% similar to two Trichomitus batrachorum (a frog symbiont) sequences. A phylogenetic analysis using the neighbor-joining and maximum likelihood methods supported the data of the BLAST analysis. These results suggest the presence of at least two as yet undescribed trichomonad species in the intestinal contents of pigs.
Keywords: Trichomonads, Pig, Intestine, Tritrichomonas sp., Tetratrichomonas sp., PCR, 18S ribosomal RNA
1. Introduction
In veterinary medicine infections with trichomonads of the phylum Parabasala are commonly found. Trichomonads are anaerobic protozoa, possess up to six flagella and harbor hydrogenosomes instead of mitochondria.
In pigs several trichomonads are described inhabiting the digestive tract. Tetratrichomonas buttreyi (Rivera et al., 2008) is not only found in pigs but also in cattle, where it is considered to be non-pathogenic (Castella et al., 1997). Trichomitus rotunda was first described in Hibler et al. (1960). It is not distinguishable from Tritrichomonas foetus by light microscopy and there are no gene sequences of this species available. Therefore, the definite existence of this species is considered doubtful (Rivera et al., 2008). Most information is available for T. foetus, which was shown to be the same species as Tritrichomonas suis (Parsonson et al., 1976; Lun et al., 2005). T. foetus can be found in the colon and occasionally in the small intestine of pigs (Lun et al., 2005). It has been commonly regarded as commensalic and non-pathogenic (Tachezy et al., 2002), however, a recent study showed that it may be considered a facultative pathogen (Mostegl et al., 2011). In that study, a chromogenic in situ hybridization (ISH) using a probe able to detect the parabasalid classes Cristamonadea, Tritrichomonadea, Hypotrichomonadea and Trichomonadea (except for most species from the families Hexamastigidae and Tricercomitidae) (Cepicka et al., 2010) was applied to screen 192 pigs for the presence of trichomonads. In a second ISH all positive samples were further examined with another probe specific for the family of Tritrichomonadidae (except for Tritrichomonas muris). In several pigs it could be demonstrated that other trichomonads than T. foetus were present in the intestinal contents. Based on the fact that trichomonads are not only classified using morphology but also by gene sequences of the 18S ribosomal RNA (rRNA) gene (Cepicka et al., 2010; Hampl et al., 2006), a further analysis of these trichomonads present in pigs was intended.
In this study, a part of the 18S rRNA gene of trichomonads found in the intestinal contents of three different pigs and shown not to be T. foetus, is analyzed by gene sequencing.
2. Materials and methods
2.1. Sampling
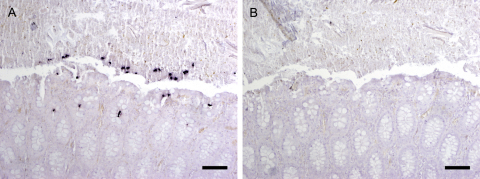
In a previous retrospective study (Mostegl et al., 2011) paraffin-wax embedded intestinal samples of 192 pigs which were affected with chronic diseases, the major symptoms of which were wasting, diarrhea, dyspnea and skin lesions, were screened for the presence of intestinal trichomonads using two different ISH probes either detecting the parabasalid classes Cristamonadea, Tritrichomonadea, Hypotrichomonadea and Trichomonadea (except for most species from the families Hexamastigidae and Tricercomitidae) (OT probe) (Mostegl et al., 2010) or the family of Tritrichomonadidae (except for Tritrichomonas muris) (Tritri probe) (Mostegl et al., 2011). In 91 cases trichomonads were found. In 43 pigs it could clearly be shown that other trichomonads than T. foetus were present. Three pigs (pigs 1–3) out of these 43 pigs were chosen for further analyses, based on the significant amount of trichomonads identified in the intestinal contents. In pigs 1 and 3 ISH using the OT probe showed the presence of low to moderate amounts of specifically stained trichomonads only within the intestinal lumen of the colon section. In pig 2 a moderate number of trichomonads, not only in the intestinal lumen but also in the crypts, were present (Fig. 1). In the second ISH with the Tritri probe none of the three pigs displayed any positively stained parasites (Fig. 2, pig 2), suggesting the presence of other trichomonads than T. foetus. All three pigs came from different farms.
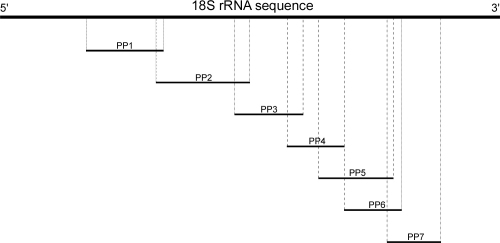
Fig. 1.
Scheme of the 18S rRNA gene in correlation with the seven primer pairs (PP) used for primer walking.
Fig. 2.
ISH of the colon of pig 2. (A) ISH using the OT probe reveals trichomonads within the crypts and the gut lumen easily discernible by their purple to black staining. Bar = 150 μm. (B) ISH of the corresponding region in a serial section with the Tritri probe shows no positively stained trichomonads. Bar = 150 μm.
2.2. Primer design
PCR followed by gene sequencing analysis was carried out on DNA extracted from formalin-fixed and paraffin-embedded intestinal tissue from all three pigs. In this context it was necessary to design primers which generate several overlapping short PCR products (<300 bp), because the DNA degradation caused by the formalin fixation does not allow reliable amplification of longer fragments (Greer et al., 1991; Farrugia et al., 2010). In a first PCR step an already published primer pair able to amplify a region in the 18S rRNA gene of many members of the six classes of parabasalia was used (Mostegl et al., 2010), referred to as primer pair (PP) 1 (Table 1), and the acquired sequence was subjected to the Basic Local Alignment Search Tool (BLAST). Based on the BLAST analysis data 18S rRNA gene sequences from different trichomonads, such as T. foetus (accession nos. M81842 and U17509), T. buttreyi (accession nos. AY886865 and AY886869) and Trichomitus batrachorum (accession nos. AF076958 and AF124610) showed the highest similarities and thus were chosen to be used for all further alignments. Based on this alignment a primer walking was conducted (Fig. 2), which included using five newly designed primers (PP2, PP3, PP4, PP6 and PP7) and another previously published primer pair (PP5) (Table 1). All newly designed primer pairs were submitted to BLAST to rule out cross reactivity.
Table 1.
List of all used primer pairs (PP), their forward and reverse sequences, previous publications if available, and GenBank accession numbers of the sequences used for the primer design.
| Primer pair | Designed on | Forward sequence | Reverse sequence | Publication |
|---|---|---|---|---|
| PP1 | M81842 | 331F: 5′-GGT AGG CTA TCA CGG GTA AC-3′ | 578R: 5′-ACT YGC AGA GCT GGA ATT AC-3′ | (Mostegl et al., 2010) |
| PP2 | M81842 | 553F: 5′-GCT GCG GTA ATT CCA GCT CT-3′ | 848R: 5′-GCC CTT GAT CGA CAG AAA CC-3′ | |
| PP3 | AF076958 | 726F: 5′-AGA AAC GAA AGC GAA GGC AT-3′ | 941R: 5′-TTC CGT CAA TTC CTT CAA GT-3′ | |
| PP4 | M81842 | 962F: 5′-GGG CTC TGG GGG AAC TAC GA-3′ | 1141R: 5′-GGC CAT GCA CCA CCA AAA GT-3′ | |
| PP5 | M81842 | 1064F: 5′-AAC TTA CCA GGA CCA GAT GT-3′ | 1297R: 5′-CAC GGA CCT GTT ATT GCT AC-3′ | (Mostegl et al., 2011) |
| PP6 | M81842 | 1142F: 5′-GTT GGT GGT GCG TGG GTT GA-3′ | 1325R: 5′-CGT GCA GCC CAG AGC ATC TA-3′ | |
| PP7 | M81842 | 1279F: 5′-AGC AAT AAC AGG TCC GTG AT-3′ | 1445R: 5′-ACA AGG GAT TCC TGG TTC AT-3′ |
2.3. PCR and gene sequencing analysis
Prior to PCR DNA was extracted from three 10 μm thick tissue sections of formalin-fixed and paraffin-embedded intestinal tissue. First, the sections were dewaxed using xylene, washed with ethanol and air-dried. Subsequently, a DNA extraction step using the Nexttec Clean Column kit (Nexttec, Leverkusen, Germany) according to the manufacturer's instructions was performed. The PCR reaction master mixture of all PCR reactions consisted of 10 μl HotMasterMix (5Prime, Eppendorf, Hamburg, Germany), 0.4 μM of each primer, 2 μl template DNA and distilled water to a total volume of 25 μl. The PCR reaction was started with a first heat denaturation step at 94 °C for 2 min, followed by 40 cycles of heat denaturation at 94 °C for 30 s, primer annealing at 60 °C (except for PP1 where an annealing temperature of 58 °C was used) and DNA elongation at 72 °C for 1 min. Finally, a last DNA elongation step was carried out at 72 °C for 10 min. No positive control was used. As negative control 2 μl of distilled water instead of template DNA was added to the PCR reaction. An aliquot of 10 μl of each PCR product was analyzed by gel electrophoresis using a 2% Tris acetate–EDTA–agarose gel. Subsequently, the agarose gel was stained with ethidium bromide and bands were visualized using the BioSens gel imaging system software (GenXpress, Wiener Neudorf, Austria). PCR products showing the expected size were sequenced in both directions according to Bakonyi et al. (2004), except for DNA purification which was carried out using the DyeEx 2.0 Spin Kit (QIAGEN, Hilden, Germany) instead of precipitation with ethanol. The acquired forward and reverse sequences were combined excluding the outer primer regions and subjected to BLAST to search against GenBank sequences.
2.4. Phylogenetic analysis
In the phylogenetic analysis the three new sequences (JF742056, JF742057 and JF742058), as well as known sequences of the classes Trichomitea, Trichomonadea, Tritrichomonadea, Tetratrichomonadea, Spirotrichonymphea and Trichonymphea were included. Staurojoenina assimilis was used as outgroup. The sequences were aligned using Align plus for Windows 95 (Scientific and Educational Software, Cary, NC). The resulting alignments were manually edited using Align plus.
The phylogenetic investigation was performed with the MEGA4 program (Tamura et al., 2007) using the neighbor-joining (NJ) method (Saitou and Nei, 1987). The stability of the tree was tested by bootstrap analysis of 1000 replicates and the nucleotide substitution model of Kimura-2-parameter was used to calculate the genetic distance between each pair of sequences. In addition, the same taxa were subjected to a phylogenetic analysis with the Phylip-3.69 program using the maximum likelihood (ML) method (Felsenstein and Churchill, 1996). The stability of the tree was tested by bootstrap analysis of 1000 replicates and the F84 model was used to calculate the genetic distance between each pair of sequences (Thorne et al., 1992; Felsenstein, 1993).
3. Results
3.1. Gene sequencing analysis
For the genetic analysis of trichomonads from pig 1 a nucleotide sequence of 1082 bp (accession no. JF742056), for those from pig 2 of 1074 bp (accession no. JF742057), and for those from pig 3 of 1083 bp (accession no. JF742058) could be achieved, excluding the outermost primers. An alignment of all three acquired sequences revealed a high similarity between the nucleotide sequences of pig 1 and pig 3, showing only 1% difference. The trichomonad sequences found in pig 2 showed about 10% nucleotide differences compared to the sequences of pigs 1 and 3. The BLAST analysis (Table 2) showed a 92% similarity for the sequences of pig 1 and pig 3 with H. acosta and a 97% similarity for the sequence of pig 2 with T. batrachorum.
Table 2.
Result of the BLAST analysis of the partial 18S rRNA gene of the trichomonads from the three pigs; included are all sequences showing a query coverage of 100% and a similarity of at least 90%. Shown are the respective similarity to the new sequences, the trichomonad species and the GenBank accession number.
| Pig | Similarity | Species | Accession no. |
|---|---|---|---|
| 1 | 92% | Hypotrichomonas acosta | AF076959 |
| 91% | Hypotrichomonadidae sp. | GQ254642 | |
| 91% | Trichomitus batrachorum | AF124610 | |
| 90% | Trichomitus batrachorum | AF076958 | |
| 2 | 97% | T. batrachorum | AF124610 |
| 96% | T. batrachorum | AF076958 | |
| 92% | Hypotrichomonadidae sp. | GQ254642 | |
| 3 | 92% | H. acosta | AF076959 |
| 91% | T. batrachorum | AF124610, AF076958 | |
| 91% | Hypotrichomonadidae sp. | GQ254642 | |
3.2. Phylogenetic analysis
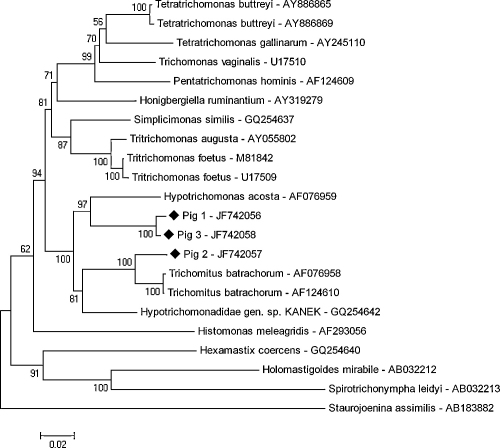
In the NJ-phylogenetic tree a strong similarity between the trichomonad sequence from pig 1 and pig 3 could be clearly shown, supported by a bootstrap value of 100. The cluster of these sequences seems to be most closely related to H. acosta showing a bootstrap value of 97. In the phylogenetic analysis the sequence of pig 2 was placed closely to T. batrachorum sequences, supported by a bootstrap value of 100 (Fig. 3). These assignments were clearly confirmed by the phylogenetic analysis based on the ML-algorithm (not shown). The sequence of Hypotrichomonadidae gen. sp. KANEK was only distantly related, showing a bootstrap value of 81 (NJ) and 57 (ML), respectively. Sequences of other classes than Hypotrichomonadea could be clearly allocated to other clusters.
Fig. 3.
Phylogenetic tree of trichomonads based on the partial nucleotide sequence of the 18S rRNA gene. The tree was conducted using the neighbor-joining method. The three novel pig trichomonad sequences are marked with diamonds.
4. Discussion
In this study, the examination of intestinal tissue from three different pigs with ISH using two different probes clearly showed that the trichomonads found in the intestinal contents were not homologous to trichomonads from the class Tritrichomonadea. According to the literature it could be expected that protozoa from the porcine intestine belong to T. foetus, which is considered to be the most prevalent species in this environment (Lun et al., 2005). However, alignments and a BLAST analysis also proved that the recovered sequences did not match sequences from T. foetus. The sequences found in pig 1 and pig 3 were almost identical but significantly differed from the trichomonad sequences of pig 2. This suggests that the protozoa found in pig 1 and pig 3 belong to the same species. The phylogenetic analysis of these sequences groups them together with a bootstrap value of 100, strengthening this hypothesis. The BLAST analysis and the phylogenetic analysis showed that H. acosta (Edgcomb et al., 1998), a member of the class Hypotrichomonadea isolated from squamate reptiles, has the highest similarity with these sequences. No such species has ever been described in pigs.
The trichomonad sequence of pig 2 was found to be at least 10% different to the sequences of pig 1 and pig 3. The BLAST analysis revealed a sequence similarity of 97% with T. batrachorum (Edgcomb et al., 1998), a frog symbiont also from the class Hypotrichomonadea. In the phylogenetic tree the trichomonad found in pig 2 was grouped together with the two T. batrachorum (accession no. AF076958 strain BUB, AF124610 strain R105) strains available in the GenBank. It seems that the protozoal sequence found in pig 2 is Trichomitus-like, reminding of T. rotunda, the only Trichomitus species ever described in pigs but with no sequence data available. All three achieved sequences showed no significant similarity with T. foetus or with T. buttreyi, the third trichomonad described in pigs.
The fact that the closest relatives of these trichomonads inhabit reptilian and amphibian hosts raises the question whether pigs are only aberrant hosts of these organisms. The affected pigs, however, were conventionally raised and did not have any outdoor access. So contacts with these classes of vertebrates are very unlikely. Whether contacts with other mammals, such as rodents can be entirely excluded, has to remain open.
The detected trichomonads seem to be apathogenic and thus of no clinical relevance due to the facts that only low numbers of parasites in pig 2 were found in the crypt lumina and no trichomonads were found to emigrate into the lamina propria mucosae. In both other pigs trichomonads were only present in the intestinal lumen.
The three generated sequences are unique and have to be considered to belong to not yet properly described species. Due to the fact that all three trichomonads were found in formalin-fixed and paraffin-embedded tissue sections, unfortunately, no complete species description could be carried out, which would require morphological description and cultivation of the found protozoa. Taken together, this study makes clear that as yet undescribed trichomonad sequences are present in the intestine of pigs. It is very likely that that other presumably apathogenic trichomonads exist in pigs and that further examinations are rewarding.
All sequences generated have been submitted to the GenBank with the following preliminary GenBank accession numbers JF742056 (pig 1), JF742057 (pig 2) and JF742058 (pig 3).
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
The authors wish to thank Jolanta Kolodziejek for helping with the phylogenetic analyses and Karin Fragner and Klaus Bittermann for their excellent technical support.
This work was funded by the Austrian Science Fund (FWF) grant P20926.
References
- Bakonyi T., Gould E.A., Kolodziejek J., Weissenböck H., Nowotny N. Complete genome analysis and molecular characterization of Usutu virus that emerged in Austria in 2001: comparison with the South African strain SAAR-1776 and other flaviviruses. Virology. 2004;328:301–310. doi: 10.1016/j.virol.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Castella J., Muńoz E., Ferrer D., Gutiérrez J.F. Isolation of the trichomonad Tetratrichomonas buttreyi (Hibler et al., 1960) Honigberg, 1963 in bovine diarrhoeic faeces. Vet. Parasitol. 1997;70:41–45. doi: 10.1016/s0304-4017(96)01140-5. [DOI] [PubMed] [Google Scholar]
- Cepicka I., Hampl V., Kulda J. Critical taxonomic revision of parabasalids with description of one new genus and three new species. Protist. 2010;161:400–433. doi: 10.1016/j.protis.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Edgcomb V., Viscogliosi E., Simpson A.G.B., Delgado-Viscogliosi P., Roger A.J., Sogin M.L. New insights into the phylogeny of trichomonads inferred from small subunit rRNA sequences. Protist. 1998;149:359–366. doi: 10.1016/S1434-4610(98)70042-2. [DOI] [PubMed] [Google Scholar]
- Farrugia A., Keyser C., Ludes B. Efficiency evaluation of a DNA extraction and purification protocol on archival formalin-fixed and paraffin-embedded tissue. Forensic Sci. Int. 2010;194:25–28. doi: 10.1016/j.forsciint.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Department of Genetics, University of Washington; Seattle: 1993. PHYLIP. Phylogenetic Inference Package, Version 3.5c. [Google Scholar]
- Felsenstein F., Churchill G.A. A hidden Markov model approach to variation among sites in rate of evolution. Mol. Biol. Evol. 1996;13:93–104. doi: 10.1093/oxfordjournals.molbev.a025575. [DOI] [PubMed] [Google Scholar]
- Greer C.E., Lund J.K., Manos M.M. PCR amplification from paraffin-embedded tissues: recommendations on fixatives for long-term storage and prospective studies. Genome Res. 1991;1:46–50. doi: 10.1101/gr.1.1.46. [DOI] [PubMed] [Google Scholar]
- Hampl V., Vrlík M., Cepicka I., Pecka Z., Kulda J., Tachezy J. Affiliation of Cochlosoma to trichomonads confirmed by phylogenetic analysis of the small-subunit rRNA gene and a new family concept of the order Trichomonadida. Int. J. Syst. Evol. Microbiol. 2006;56:305–312. doi: 10.1099/ijs.0.63754-0. [DOI] [PubMed] [Google Scholar]
- Hibler C.P., Hammmond D.M., Caskey F.H., Johnson A.E., Fitzgerald P.R. The morphology and incidence of the trichomonads in swine, Tritrichomonas suis (Gruby & Delafond), Tritrichomonas rotunda, n. sp. and Trichomonas buttreyi, n. sp. J. Protozool. 1960;7:159–171. [Google Scholar]
- Lun Z.-R., Chen X.-G., Zhu X.-Q., Li X.-R., Xie M.-Q. Are Tritrichomonas foetus and Tritrichomonas suis synonyms? Trends Parasitol. 2005;21:122–125. doi: 10.1016/j.pt.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Mostegl M.M., Richter B., Nedorost N., Maderner A., Dinhopl N., Kulda J., Liebhart D., Hess M., Weissenböck H. Design and validation of an oligonucleotide probe for the detection of protozoa from the order Trichomonadida using chromogenic in situ hybridization. Vet. Parasitol. 2010;171:1–6. doi: 10.1016/j.vetpar.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostegl M.M., Richter B., Nedorost N., Maderner A., Dinhopl N., Weissenböck H. Investigations on the prevalence and potential pathogenicity of intestinal trichomonads in pigs using in situ hybridization. Vet. Parasitol. 2011 doi: 10.1016/j.vetpar.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonson I.M., Clark B.L., Dufty J.H. Early pathogenesis and pathology of Tritrichomonas foetus infection in virgin heifers. J. Comp. Pathol. 1976;86:59–66. doi: 10.1016/0021-9975(76)90028-1. [DOI] [PubMed] [Google Scholar]
- Rivera W.L., Lupisan A.J.B., Baking J.M.P. Ultrastructural study of a tetratrichomonad isolated from pig fecal samples. Parasitol. Res. 2008;103:1311–1316. doi: 10.1007/s00436-008-1134-x. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Tachezy J., Tachezy R., Hampl V., Sedinová M., Vanácová S., Vrlík M., Van Ranst M., Flegr J., Kulda J. Cattle pathogen Tritrichomonas foetus (Riedmüller, 1928) and pig commensal Tritrichomonas suis (Gruby & Delafond, 1843) belong to the same species. J. Eukaryot. Microbiol. 2002;49:154–163. doi: 10.1111/j.1550-7408.2002.tb00360.x. [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thorne J.L., Kishino H., Felsenstein J. Inching toward reality: an improved likelihood model of sequence evolution. J. Mol. Evol. 1992;34:3–16. doi: 10.1007/BF00163848. [DOI] [PubMed] [Google Scholar]