Abstract
The remarkable discovery of small noncoding microRNAs (miRNAs) and their role in posttranscriptional gene regulation have revealed another fine-tuning step in the expression of genetic information. A large number of cellular pathways, which act in organismal development and are important in health and disease, appear to be modulated by miRNAs. At the molecular level, miRNAs restrain the production of proteins by affecting the stability of their target mRNA and/or by down-regulating their translation. This review attempts to offer a snapshot of aspects of miRNA coding, processing, target recognition and function in animals. Our goal here is to provide the readers with a thought-provoking and mechanistic introduction to the miRNA world rather than with a detailed encyclopedia.
Keywords: MicroRNA, Noncoding RNA, Splicing, Intron, RNA interference, RISC, Knock down, Gene silencing, Argonaute, Dicer, Drosha
Introduction
The discovery of miRNA is one of the most significant landmarks in modern molecular biology. MicroRNAs (miRNAs) are small, regulatory, noncoding RNA molecules that control the expression of their target mRNAs predominantly by binding to the 3' untranslated region (UTR). A single UTR may have binding sites for many miRNAs or multiple sites for a single miRNA, suggesting a complex post-transcriptional control of gene expression exerted by these regulatory RNAs. The founding members of the miRNA family were identified in the Ambrose and Ruvkun laboratories and originally termed "small temporal RNAs" (1, 2). The genome-wide identification and expression of miRNA in mammals, fly, and worm has catapulted the field of small RNAs to the forefront of modern biology (3–5). Furthermore, differential miRNA expression in diverse cell and tissue types makes these molecules ideal biomarkers of disease detection and progression as well as targets for therapeutic intervention. Considerable amount of data generated in a short period show that miRNAs mainly exert their function post-transcriptionally by modulating mRNA stability and/or protein translation.
Conservation, coding and transcription of miRNA genes
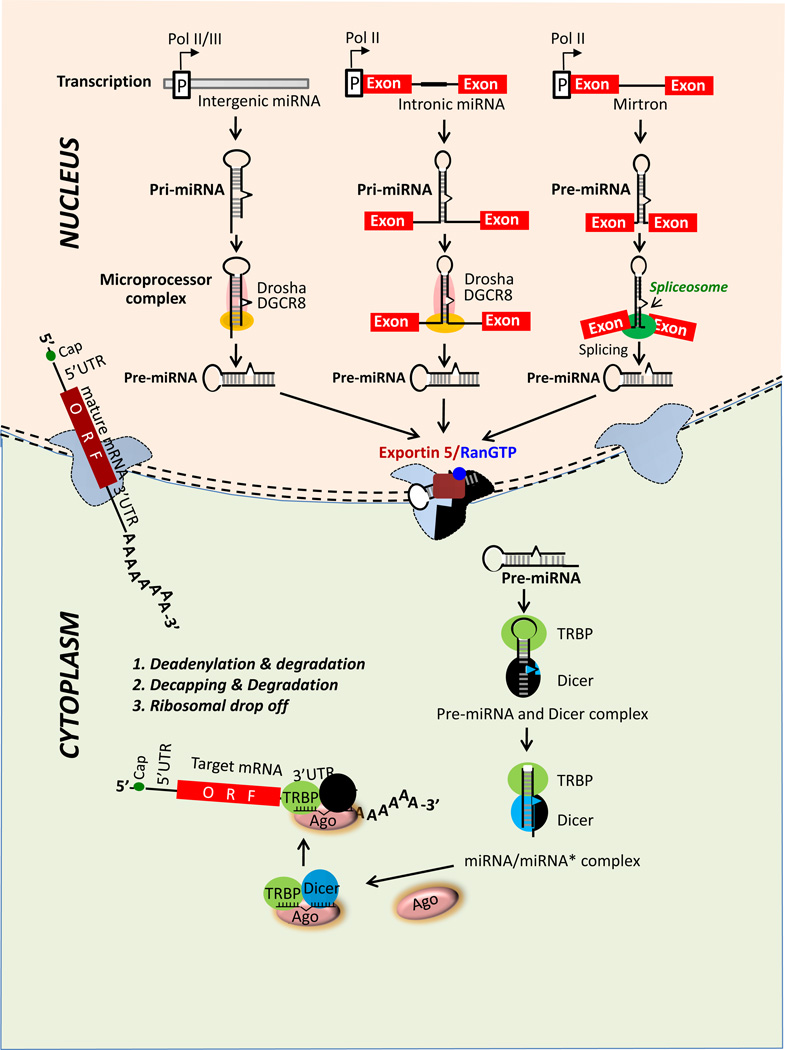
miRNAs form a class of small RNAs, which appear to play a central role in silencing the gene expression connected with complex gene regulatory networks. miRNA are conserved from amoeba to humans. Thus far, use of large-scale small RNA cloning strategies have led to the discovery of miRNAs in a wide variety of organisms including brown alga, nematode, mollusk, tunicates (Ciona intestinalis), sea lamprey, vertebrates, insects, amoeba, monocots, dicots, large DNA viruses (Epstein-Barr and herpes viruses), and recently in T. gondii, a unicellular protozoan parasite of the Apicomplexan family (6–10). More than two-thirds of all human miRNAs are encoded in the intervening regions (introns) of protein-coding genes as well as in long noncoding transcripts (Fig. 1). miRNAs can also be encoded in exons or introns, depending upon the alternative splicing status of the pre-mRNA (11). In addition, miRNAs can be of intergenic origin, i.e. found in the chromosomal regions between two genes (Fig. 1) (12, 13).
Figure 1. General molecular mechanisms of miRNA biogenesis.
miRNAs are transcribed by RNA polymerases II or III as pri-mRNA that are modified with cap structure and polyadenylation. Initial processing of pri-miRNA occurs in nucleus by the Drosha complex which crops the miRNA into a hairpin-shaped pre-miRNA. Next, pre-miRNA is exported to cytoplasm with Exportin-5/Ran-GTP complex for Dicer processing. Following this, one of the strands of the miRNA duplex is incorporated with Ago to form miRNA-RISC that engages on the target mRNA to mediate gene silencing either by translational repression or by mRNA degradation/deadenylation (detailed in Figs. 3, 4). Noncanonical intronic miRNAs, "mirtrons", are processed by the spliceosome, which is independent of the Drosha microprocessor complex.
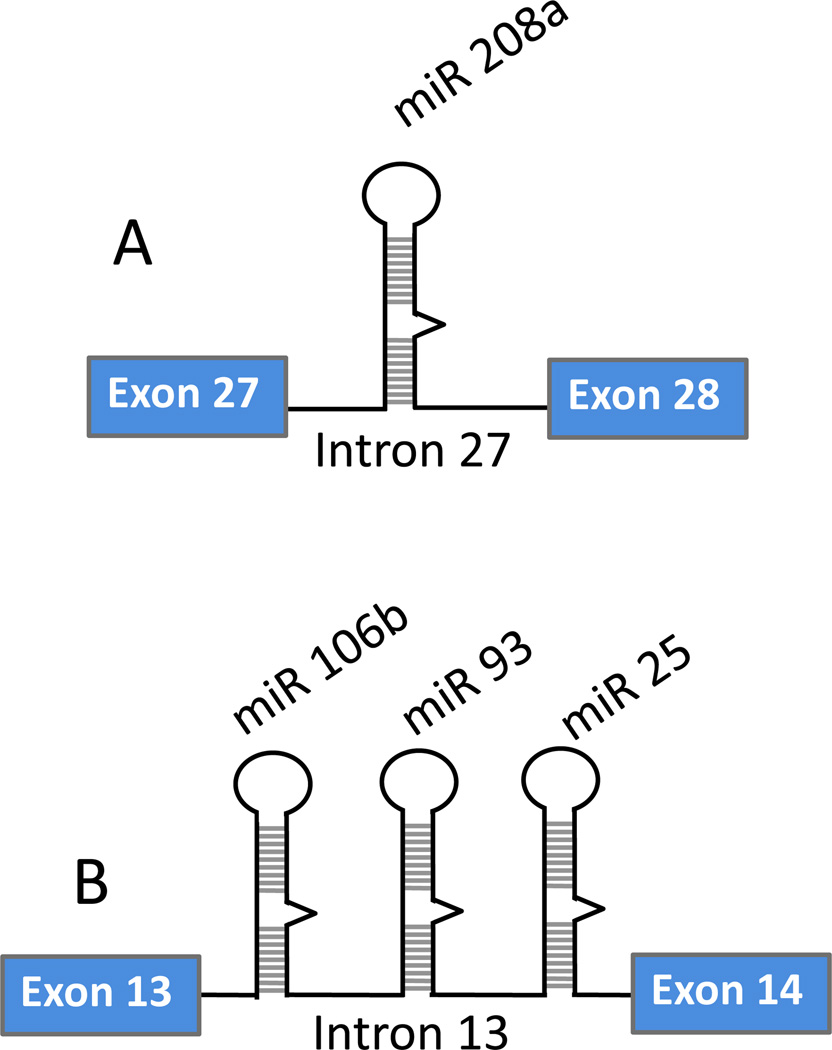
As mentioned above, a large number of miRNAs are encoded in the introns of host genes, suggesting that the RNA polymerase II-mediated transcription of these miRNAs and the host genes is coupled (Fig. 1) (14–15). There are also examples of miRNA genes with RNA polymerase III promoter. C19MC, the largest known miRNA cluster containing 46 tandemly repeated miRNAs, appears to be transcribed by RNA polymerase III (16). These miRNAs are presumably formed by repetitive sequences and interspersed by positive- and negative-strand Alu repeats (17, 18). Intergenic miRNA clusters are likely to be associated with complex transcriptional loci; however, their transcriptional activation and processing remain undefined. Interestingly, another report suggested that C19MC cluster is derived from an intronic non-protein-coding RNA Polymerase II transcript (19). Many miRNAs are coded from single standalone genes however, and clusters containing multiple miRNAs (e.g., miR-106b-25, miR 17–92; miR 302–367) appear to be common (Fig. 2). The question remains unanswered as to why a significant number of miRNAs are expressed from clusters, although there is little evidence supporting a common pathway or regulatory function of miRNAs originating from the same cluster. It has been suggested that the expression of the intronic miRNAs may be coupled with that of the host mRNA and the two may be functionally related (20–25). In studies of other genes, expression of the intronic miRNAs differed from their host gene expression, especially in the pathological states (26). A significant intraclustral differential expression pattern of miRNA has also been observed (27). Overall, these observations point to diverse mechanisms and the existence of additional steps in the posttranscriptional processing of miRNA involving proteins in the maturation process. Many of the RNA-binding proteins, including Lin 28, hnRNP L, KSRP, and hnRNP A1, bind to conserved loop sequences in the miRNA, and therefore, may form part of the gene regulatory network that controls the level of miRNA expression in response to intra- or extra-cellular stimuli (28–35).
Figure 2. Coding structure of intronic miRNAs.
The Figure illustrates the coding patterns of intronic miRNAs. Intronic or intragenic miRNAs can be coded as: (A) a single standalone gene, or (B) in a cluster containing multiple miRNA genes.
A set of miRNAs is subjected to posttranscriptional regulation by cell signaling pathways including those involving Transforming Growth Factor-beta (TGF-β) and Ras/MAP Kinase (MAPK) (36–38). Overall, the signaling pathways related to TGF-β, the bone morphogenetic protein (BMP) and SMAD modulate the expression of as many as 20 miRNAs post-transcriptionally via Drosha and p68 microprocessor complex (36). SMADs are directly recruited to the stem region of this subset of miRNAs, and the recruitment of Drosha and DGCR8 to the pri-miRNA facilitates further maturation of this subset (36, 39–40). miRNA regulation mediated by TGF-β, BMP and SMAD proteins appear to orchestrate the contractile phenotype in human vascular smooth muscle cells.
Interestingly, the tumor suppressor protein p53 modulates p68- and Drosha-mediated miRNA processing. Both Drosha and p68 associate with a subset of miRNAs and their processing is enhanced in the presence of the p53 transcription factor (41). This explains why TDNA damage leads to posttranscriptional induction of several miRNAs as p53 is induced in DNA damage.
The p68 protein mentioned above is a DEAD-box RNA helicase and found to be associated with the Drosha complex in prostate cancer cells (42). It is in fact highly expressed in prostate cancer cells and is also associated with human androgen receptor (AR) coactivator complexes in the nucleus. These findings give rise to the notion that AR may be involved in the post-transcriptional modification of a set of miRNAs that are likely associated with castration-resistant prostate cancer. However, this hypothesis is yet to be tested experimentally.
Maturation of miRNAs–Post-transcriptional processing of the precursor
In the metazoa (such as human), miRNAs are processed from the primary transcript using a two-step sequential mechanism involving two RNase III nucleases (Fig. 1). As indicated before, miRNAs are generated either from the processing of a host intron or by transcription from their own dedicated promoters. The primary precursor (pri-miRNA) is processed into an approximately 70 nucleotide long stem-loop structure by nuclear RNase III Drosha present in the microprocessor complex, which in mammals also contains the double-stranded RNA-binding protein, DGCR8. In Drosophila and C. elegans, the DGCR8 homolog is known as Pasha (43, 44). The two RNase domains of Drosha help cleave the 5' and 3' ends of the pri-miRNA, which determines the length of pre-miRNA (44). The resultant pre-miRNA is exported to the cytoplasm by a complex of Exportin-5 and Ran-GTP (45). The final maturation of miRNA occurs with the help of Dicer, another RNase III nuclease that processes the pre-miRNA into a 22 bp double-stranded RNA. The processing is often coupled with the formation of the ribonucleoprotein complex known as RISC (miRNA-Induced Silencing Complex) (Figs. 1, 3, 4). The RISC minimally consists of one strand of the miRNA (called "guide strand") in addition to Dicer, TRBP, PACT and Argonaute (Ago) proteins. The complex engages with the target to execute silencing while the other strand of the miRNA (called "passenger strand") is generally, but not always, destroyed. It is unclear how the asymmetric strand selection and passenger strand destruction occurs (extensively reviewed in 46–48 and references therein).
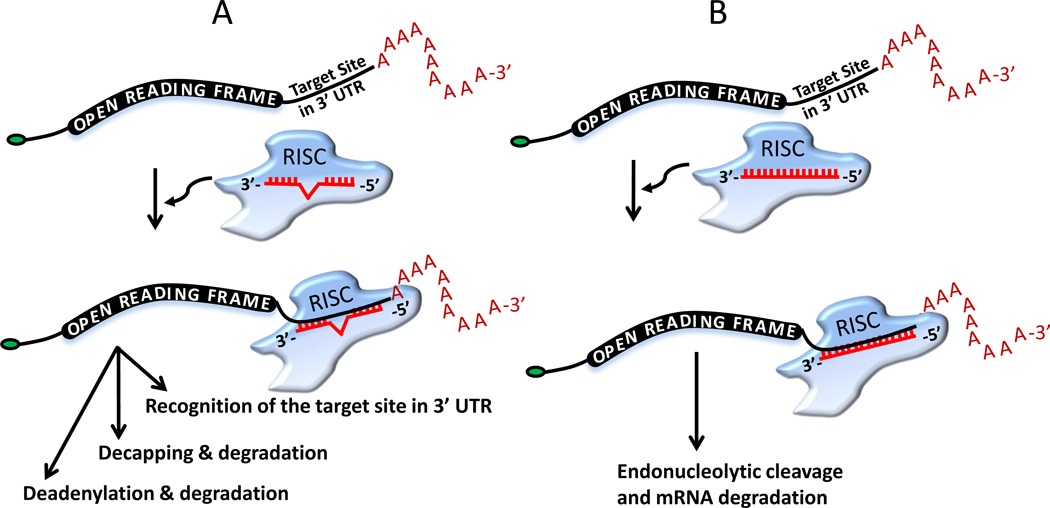
Figure 3. Two modes of action of miRNAs. miRNAs can establish (A) imperfect or (B) perfect complementarity within the 3’ UTR of its target mRNAs.
Depending upon the complementarity, miRNA can then modulate the translation and mRNA destabilization (mostly in animals) or degrade the target message (common in plants). Also see Fig. 4 for detailed mechanisms.
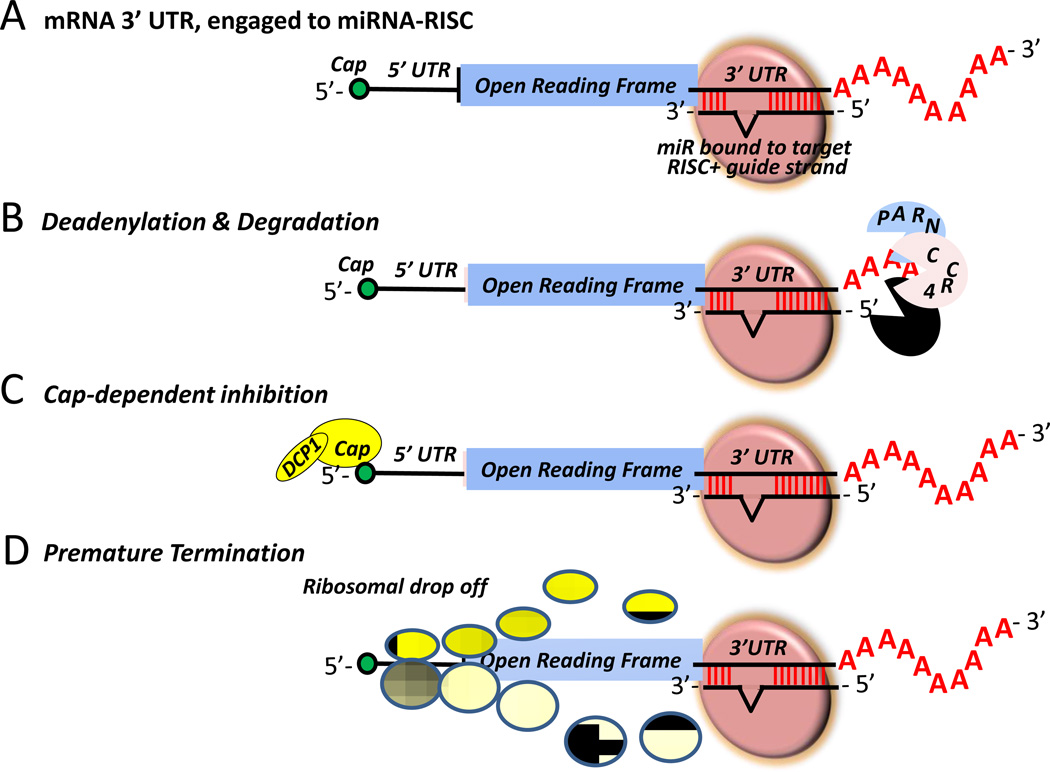
Figure 4. Suggested mechanisms of miRNA-mediated translational repression.
(A) 3’ UTR of a mRNA engaged to RISC and miRNA; (B) CAF1/CCR4/NOT1 complex-mediated deadneylation and degradation of the target mRNA; (C) Dcp1/2 complex-mediated decapping; and (D) translational repression in actively translating polysomes at initiation and elongation stages by ribosome drop off.
Interestingly, there exists a subset of intronic miRNA, known as "mirtron" (49–52), whose processing appears to be independent of Drosha (Fig. 1). Mirtrons were initially identified in Drosophila and C. elegans (49, 50), but more recently identified in human, macaque, chimpanzee and mouse as well (52). Unlike the pri-miRNAs, the mirtrons lack the lower hairpin and the flanking single-stranded regions that recruit the microprocessor complex. Instead, they contain 5' and 3' splice sites that are presumably recognized by small nuclear RNAs (snRNA). After the debranching of the lariate intron, the spliced introns fold back into a stem-loop structure that obviates the need for the Drosha-mediated microprocessor function. It will not be surprising if refined methods of high-resolution deep-sequencing reveal many more mirtrons in mammalian genomes, including human.
Apparently, Drosha-dependent pre-miRNA processing may be independent of pre-mRNA processing as splicing-inactive substrates are active for pre-miRNA processing in assays in vitro (53). Another study showed a functional link between nuclear pre-mRNA splicing and processing. Although microprocessor-dependent pre-miRNA cropping can occur faster than pre-mRNA splicing, Drosha-mediated miRNA processing and pre-mRNA splicing may occur concurrently involving functionally linked RNA:protein complexes (53). It is likely that this situation may exist in minority cases where the miRNA is encoded in a small intron, causing interference with splice site recognition by snRNAs. In the majority of cases, in contrast, the miRNA constitute only a tiny fraction of information content within the intron that could be several hundred kilobases long and hence, additional internal intronic promoter-mediated transcription, independent of the host gene, cannot be ruled out. Finally, these in vitro studies are unable to answer if mutations of splice sites of miRNA hosting pre-mRNA introns may abolish the splicing, hence creating a stable transcript that might "over-produce” the resident miRNAs, resulting in increased translational repression of its target mRNA. Only in vivo experiments will be able to shed light on the so-called “fine-tuned” gene expression by intronic miRNAs in response to splicing defects of host pre-mRNAs.
Lastly, additional mechanisms of miRNA biogenesis continue to emerge. For example, a few miRNAs are produced from a functionally distinct RNA family, namely the small nucleolar RNAs or snoRNAs, an ancient class of noncoding RNAs present in all eukaryotes (55–57). The snoRNAs bind specific conserved proteins, forming snoRNP complexes that carry out methylation and pseudouridylation of ribosomal RNA. In recent years, a number of snoRNAs have been found to be processed into smaller non-coding RNAs, especially microRNAs. It is currently not clear how the same noncoding RNA precursor is funneled either into the snoRNP assembly pathway or to the miRNA processing pathway. In another example, an atypical mechanism has recently been reported for murine gamma-herpesvirus 68 (MHV68) miRNA (58). Specifically, these miRNAs are transcribed from RNA polymerase III promoters located within adjacent viral tRNA-like sequences. The resultant pri-miRNAs bear a 5' tRNA moiety and are not processed by Drosha but instead by cellular tRNase Z, which cleaves 3' to the tRNA to liberate pre-miRNA hairpins that are then processed by Dicer to yield the mature viral miRNAs (58).
Target recognition by miRNA
miRNAs bind to their cognate target RNA through RNA-RNA base pairing that involves not only the Watson-Crick A:U and G:C pairs but also the G:U pair. The miRNA-binding sequence in the target is generally referred to as the miRNA Recognition Element or MRE. In animals, however, recognition of an MRE by miRNA is not as straightforward as one might anticipate. This is mainly because the full length of the miRNA is almost never perfectly complementary to the MRE. As a first rule, pairing of roughly the 6–8 nucleotide stretch of the 5'-end of the miRNA, known as the "seed" or "core" sequence, is generally considered necessary and sufficient for functional RISC formation. However, many exceptions to this rule have been experimentally unraveled, whereby a variety of imperfect base-pairing patterns between a miRNA and its MRE have emerged. A classic example is the base-pairing between let-7, one of the first miRNAs discovered, and the two highly confirmed MREs in its target, lin-41, neither of which has perfect complementarity with the 5′-end of let-7 (60). Thus, in spite of multiple Bioinformatic suites for miRNA-target prediction, the ultimate proof of a miRNA-MRE pair must await experimental verification.
As the coding sequence of an actively translated mRNA is covered by ribosomes, one would expect that the MREs would be located in the untranslated regions. Indeed, a comprehensive study (60) showed that >45,000 miRNA target seed sites are particularly conserved in the 3' UTR of target mRNAs, suggesting that the predominant regulatory functions of miRNAs are exerted from the 3' UTR. An interesting study (61) found that after cell proliferation is activated, mRNAs tend to terminate upstream to normal polyadenylation sites. Not unexpectedly, the resultant shorter 3' UTRs lost some MRE sequences, in agreement with a generally negative role of miRNA acting through the 3' UTR and a need for elevated gene expression in proliferating cells. However, subsequent studies revealed that functional MREs may also occur in the 5' UTR (62–64) as well as in the coding sequence (65). In either case, it is possible that the efficiency of the MRE to engage the miRNA will be modulated by the kinetics of ribosomal movement across the site and/or the local secondary structure of the mRNA, which has not been investigated thoroughly.
miRNA function: molecular mechanisms
The mechanism of how miRNAs silence their target mRNAs remains fuzzy (46–48; 66–70). In vitro and in vivo methods in various experimental models including Drosophila, C. elegans, zebrafish and numerous types of mammalian cells suggested apparently multiple mechanisms (Figs. 3, 4). The pioneering initial studies of miRNAs (tmRNA) showed that the lin-4 noncoding RNA of C. elegans actively repressed lin-14 protein synthesis (1, 71–73). As mentioned, the let-7 miRNA of C. elegans also regulated the expression of target gene lin-41, which regulated the developmental timing in the worm (2, 74, 75). These and other studies suggested that lin-4 and let-7 only act by translation repression of the target mRNA without promoting its degradation. Furthermore, studies unraveled the association of miRNAs at post-initiation as well as in the polyribosomal steps, confirming that miRNA acts to repress the translation of its target gene. It was also determined that miRNA-mediated translation repression requires the 5' cap and 3' poly(A) tail (66, 69, 76–82).
In contrast, recent studies provide overwhelming evidence that the miRNA-bound mRNA is also subject to degradation, mainly through deadenylation (Figs. 3, 4) (1, 2, 83, 84). These studies suggested that in addition to translation repression, miRNA-mediated mRNA degradation exerts an enhanced and robust block in the production of the target gene’s protein product. Many studies also suggested that miRNA-supported mRNA deadenylation, degradation and translation repression may occur independently of each other (85, 86). Interestingly, miRNA has also been implicated in the removal of the 5'-cap structure (86, 88). In two recent genome-wide studies, both target protein and mRNA levels of miRNAs were studied (89, 90). By overexpression and knockdown of various miRNAs, these studies together demonstrated the downregulation of both target protein and mRNA in HeLa cells and mouse neutrophils. A more recent study showed that mammalian miRNA predominantly act to decrease target mRNA levels (91). This study used a novel method of ribosome profiling which essentially determines the positions of ribosomes at specific codon nucleotide resolution coupled with deep-sequencing (92). The results suggested that the miRNA-mediated destabilization of target mRNAs is likely the predominant reason for reduced protein production in mammalian cells.
Multiplicity is a common feature in miRNA-mediated regulation such that: (a) one 3' UTR often contains multiple MREs, and (b) the same miRNA affects dozens, if not hundreds, of targets. Multiple MREs on the same 3' UTR generally promote stronger silencing when situated at an optimal distance from each other (93, 94); although the mechanism for this is unknown, it has been speculated that multiple RISC complexes may stabilize each other through cooperative contacts (93).
Intriguingly, recent evidence showed that miRNAs may target the promoter regions as well (48) and have the potential to activate gene expression (95). Finally, the gene silencing properties of miRNAs appear to be cell cycle-dependent such that they may globally up-regulate translation in times of cell growth (96). The molecular mechanisms of these apparently novel functions of miRNA remain to be elucidated.
In summing up, miRNAs appear to use multiple mechanisms for silencing of gene expression. Nevertheless, the majority of studies overwhelmingly support two consensus models: (i) translation repression at the initiation or elongation stages (Fig. 3), and (ii) deadenylation and subsequent degradation of the message (Figs. 3, 4). It may be envisioned that these mechanisms function together and are also regulated in a gene- or cell type-specific manner.
Conclusion
Naturally occurring miRNAs have emerged as important regulators of cell differentiation, proliferation and survival. Reciprocally, aberrant and altered miRNA expression has been linked to many pathological conditions including cardiovascular disorders, diabetes and cancer. The biogenesis of miRNAs may be regulated not only by transcriptional regulation of their promoters that generate the precursor but also by proteins representing important cellular pathways that participate in the processing of the precursor to generate the mature miRNAs. Although remarkable progress has been made on miRNA biogenesis and mode of action, it is still unclear how miRNAs find and bind to their cognate mRNAs and how other cis-acting elements within the close proximity of an MRE may affect miRNA function. It is also unclear how miRNAs in some context actually stimulate gene expression. A comprehensive pathway-focused analysis of miRNA expression and function should aid in our understanding of the cellular and molecular processes that are modified during the course of pathogenesis.
Acknowledgements
Authors like to thank Dr. Ajay Singh for offering the opportunity to contribute to this special issue of Molecular and Cellular Pharmacology, and to Dr. Kavleen Sikan for critical reading of the manuscript. Research in the authors' laboratory was supported by grants from Department of Defense prostate and breast cancer research programs as well as from National Science Foundation (to GCS), and by National Institute of Health (AI059267 to SB).
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Lee RC, Feinbaum RL, Ambros V. TheC elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Missal K, Rose D, Stadler PF. Non-coding RNAs in Ciona intestinalis. Bioinformatics. 2005;21 Suppl 2:ii77– ii78. doi: 10.1093/bioinformatics/bti1113. [DOI] [PubMed] [Google Scholar]
- 9.Shi W, Hendrix D, Levine M, Haley B. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nat Struct Mol Biol. 2009;16:183–189. doi: 10.1038/nsmb.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun L, Cannella D, Ortet P, et al. A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like Argonaute in the single-cell human parasite Toxoplasma gondii. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000920. e1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saini HK, Enright AJ, Griffiths-Jones S. Annotation of mammalian primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 17.Gu TJ, Yi X, Zhao XW, Zhao Y, Yin JQ. Alu-directed transcriptional regulation of some novel miRNAs. BMC Genomics. 2009;10:563. doi: 10.1186/1471-2164-10-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehnert S, Van Loo P, Thilakarathne PJ, Marynen P, Verbeke G, Schuit FC. Evidence for co-evolution between human microRNAs and Alu-repeats. PLoS One. 2009;4:e4456. doi: 10.1371/journal.pone.0004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bortolin-Cavaillé ML, Dance M, Weber M, Cavaillé J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–3473. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36:5232–5241. doi: 10.1093/nar/gkn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhnert F, Mancuso MR, Hampton J, et al. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 23.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med. 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 25.Rayner KJ, Suárez Y, Dávalos A, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell ML, Buvoli M, Leinwand LA. Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol Cell Biol. 2010;30:1937–1945. doi: 10.1128/MCB.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sikand K, Slane SD, Shukla GC. Intrinsic expression of host genes and intronic miRNAs in prostate carcinoma cells. Cancer Cell Int. 2009;9:21. doi: 10.1186/1475-2867-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.TGuil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 29.Michlewski G, Guil S, Semple CA, Caceres JF. TTPosttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trabucchi M, Briata P, Garcia-Mayoral M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruggiero T, Trabucchi M, De Santa F, et al. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 32.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processingTT. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heo I, Joo C, Kim YK, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Michlewski G, Cáceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piskounova E, Viswanathan SR, Janas M, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 36.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;TT139:TT112–TT122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hata A, Davis BN. Control of microRNA biogenesis by TGFbeta signaling pathway-A novel role of Smads in the nucleus. Cytokine Growth Factor Rev. 2009;20:517–521. doi: 10.1016/j.cytogfr.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;T39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 42.Clark EL, Coulson A, Dalgliesh C, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68:7938–7946. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han J, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 45.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 46.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 47.Fabian MR, Sundermeier TR, Sonenberg N. Understanding how miRNAs post-transcriptionally regulate gene expression. Prog Mol Subcell Biol. 2010;50:1–20. doi: 10.1007/978-3-642-03103-8_1. [DOI] [PubMed] [Google Scholar]
- 48.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kataoka N, Fujita M, Ohno M. Functional association of the Microprocessor complex with the spliceosome. Mol Cell Biol. 2009;29:3243–3254. doi: 10.1128/MCB.00360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott MS, Ono M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie. 2011 Jun 2; doi: 10.1016/j.biochi.2011.05.026. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holley CL, Topkara VK. An introduction to small non-coding RNAs: miRNA and snoRNA. Cardiovasc Drugs Ther. 2011;25:151–159. doi: 10.1007/s10557-011-6290-z. [DOI] [PubMed] [Google Scholar]
- 56.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5'UTR as in the 3'UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 64.Ajay SS, Athey BD, Lee I. Unified translation repression mechanism for microRNAs and upstream AUGs. BMC Genomics. 2010;11:155. doi: 10.1186/1471-2164-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 66.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13(12):1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 67.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 68.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 69.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 70.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 71.Wightman B, Bürglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3'-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991;5:1813–1824. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- 72.Arasu P, Wightman B, Ruvkun G. Temporal regulation of lin-14 by the antagonistic action of two other heterochronic genes, lin-4 and lin-28. Genes Dev. 1991;5:1825–1833. doi: 10.1101/gad.5.10.1825. [DOI] [PubMed] [Google Scholar]
- 73.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 74.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 75.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 76.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 77.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 78.Kim J, Krichevsky A, Grad Y, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10:387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pillai RS, Bhattacharyya SN, Artus CG, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 81.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 82.Mathonnet G, Fabian MR, Svitkin YV, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 83.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. Erratum in: Genes Dev 2007;21 2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wakiyama M, Yokoyama S. MicroRNA-mediated mRNA deadenylation and repression of protein synthesis in a mammalian cell-free system. Prog Mol Subcell Biol. 2010;50:85–97. doi: 10.1007/978-3-642-03103-8_6. [DOI] [PubMed] [Google Scholar]
- 87.Eulalio A, Rehwinkel J, Stricker M, et al. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 90.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]